Summary: The abscisic acid receptor PYL8 plays an important role for regulation of root abscisic acid sensitivity, and abscisic acid-dependent inhibition of PP2Cs by PYR/PYLs is required for root hydrotropism.
Abstract
Abscisic acid (ABA) signaling plays a critical role in regulating root growth and root system architecture. ABA-mediated growth promotion and root tropic response under water stress are key responses for plant survival under limiting water conditions. In this work, we have explored the role of Arabidopsis (Arabidopsis thaliana) PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS for root ABA signaling. As a result, we discovered that PYL8 plays a nonredundant role for the regulation of root ABA sensitivity. Unexpectedly, given the multigenic nature and partial functional redundancy observed in the PYR/PYL family, the single pyl8 mutant showed reduced sensitivity to ABA-mediated root growth inhibition. This effect was due to the lack of PYL8-mediated inhibition of several clade A phosphatases type 2C (PP2Cs), since PYL8 interacted in vivo with at least five PP2Cs, namely HYPERSENSITIVE TO ABA1 (HAB1), HAB2, ABA-INSENSITIVE1 (ABI1), ABI2, and PP2CA/ABA-HYPERSENSITIVE GERMINATION3 as revealed by tandem affinity purification and mass spectrometry proteomic approaches. We also discovered that PYR/PYL receptors and clade A PP2Cs are crucial for the hydrotropic response that takes place to guide root growth far from regions with low water potential. Thus, an ABA-hypersensitive pp2c quadruple mutant showed enhanced hydrotropism, whereas an ABA-insensitive sextuple pyr/pyl mutant showed reduced hydrotropic response, indicating that ABA-dependent inhibition of PP2Cs by PYR/PYLs is required for the proper perception of a moisture gradient.
Control of abscisic acid (ABA) signaling by PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS (RCAR) ABA receptors involves direct ABA-dependent inhibition of clade A phosphatases type 2C (PP2Cs), such as ABA-INSENSITIVE1 (ABI1), HYPERSENSITIVE TO ABA1 (HAB1), and PP2CA, which are key negative regulators of the pathway (Saez et al., 2004, 2006; Rubio et al., 2009). Under resting conditions, clade A PP2Cs can interact with and dephosphorylate SnRK2.2, SnRK2.3, and SnRK2.6/OST1, reducing their catalytic activity (Fujii et al., 2009; Umezawa et al., 2009; Vlad et al., 2009). The increase of ABA levels in the plant cell leads to PYR/PYL receptor-mediated inhibition of PP2C activity, which results in the activation of the three SnRK2s and ultimately of the ABA signaling pathway (Ma et al., 2009; Park et al., 2009; Umezawa et al., 2009; Vlad et al., 2009; Gonzalez-Guzman et al., 2012). Biochemically, SnRK2s are activated through the phosphorylation of certain Ser residues of their activation loop, including Ser-175, either by autophosphorylation or by yet unidentified upstream activating kinases that are staurosporine resistant (Boudsocq et al., 2007; Fujii et al., 2009; Umezawa et al., 2009; Vlad et al., 2009, 2010). Next, the SnRK2s directly phosphorylate transcription factors that bind to ABA-responsive promoter elements and components of the machinery regulating stomatal aperture, like the slow anion channel SLAC1, K+ inward channel KAT1, or reactive oxygen species biosynthetic enzymes such as the NADPH oxidase AtrbohF (Kobayashi et al., 2005; Fujii et al., 2009; Geiger et al., 2009; Lee et al., 2009; Sato et al., 2009; Sirichandra et al., 2009).
PYR/PYL ABA receptors constitute a 14-member family, and all of them (except PYL13) are able to activate ABA-responsive gene expression using protoplast transfection assays (Fujii et al., 2009). However, gene expression patterns obtained from public databases and GUS reporter gene analyses have revealed substantial differences among them (Gonzalez-Guzman et al., 2012). Thus, the expression of PYL3 and PYL10 to PYL13 is very low to undetectable in different whole-genome microarrays, whereas the expression of PYR1 and the rest of PYL1 to PYL9 could be detected in both vegetative and reproductive tissues, although at different levels (Gonzalez-Guzman et al., 2012). From a biochemical point of view, recent studies reveal at least two families of PYR/PYL receptors characterized by a different oligomeric state, some being dimeric (PYR1, PYL1, and PYL2), whereas others are monomeric (PYL5, PYL6, and PYL8; Dupeux et al., 2011). The dimeric receptors show a higher dissociation constant for ABA (greater than 50 μm; lower affinity) than monomeric ones (approximately 1 μm); however, in the presence of the PP2C, both groups of receptors form ternary complexes with high affinity for ABA (dissociation constant of 30–60 nm; Ma et al., 2009; Santiago et al., 2009), and physiological characterization of some ABA responses in different multiple pyr/pyl mutants did not reveal a clear difference between dimeric and monomeric receptors (Gonzalez-Guzman et al., 2012). Finally, both the biochemical properties of the PYR/PYL receptors and in silico modeling of the ABA activation pathway reveal adequate coverage of the full spectrum of physiological ABA concentrations, ranging from basal ABA levels (nanomolar range) to high levels induced by water stress (micromolar range; Priest et al., 2006).
Gene expression patterns, biochemical analysis of different PP2C-PYL receptor complexes, and genetic analysis of different pyr/pyl mutants suggest that the function of PYR/PYL proteins is not completely redundant (Santiago et al., 2009; Szostkiewicz et al., 2010; Antoni et al., 2012; Gonzalez-Guzman et al., 2012). However, some functional redundancy exists, since the generation of a pyr1pyl1pyl2pyl4 quadruple mutant, 1124, was required to obtain robust ABA-insensitive phenotypes (Park et al., 2009), and a pyr1pyl1pyl2pyl4pyl5pyl8 sextuple mutant, 112458, is at least 1 order of magnitude more ABA insensitive than 1124 (Gonzalez-Guzman et al., 2012). Recently, analysis of mutants lacking three, four, five, or six PYR/PYLs has revealed quantitative regulation of ABA signaling by this family of receptors (Gonzalez-Guzman et al., 2012). Finally, GUS reporter analyses of PYR1, PYL1, PYL2, PYL4, PYL5, and PYL8 promoters has shown both overlapping and differential expression in different tissues (Gonzalez-Guzman et al., 2012). For instance, in 5-d-old seedlings, only PYR1 and PYL5 were expressed in the cortex of the upper part of the root, whereas PYL1, PYL4, and PYL8 were expressed in the columella cells. On the other side, overlapping expression of PYR1, PYL1, PYL2, PYL4, and PYL8 in root vascular tissue was observed (Gonzalez-Guzman et al., 2012).
ABA regulates root growth and root architecture, likely interacting with other hormones in these processes, such as auxins, gibberellins, or brassinosteroids (Deak and Malamy, 2005; Swarup et al., 2005; Péret et al., 2009; Rodrigues et al., 2009; Ubeda-Tomás et al., 2009, 2012; Hacham et al., 2011). ABA signaling in the root is required for different processes, such as the maintenance of primary root elongation and the repression of lateral root formation when water availability is reduced (Sharp et al., 2004; Deak and Malamy, 2005). Recent results in 17 natural accessions of Arabidopsis (Arabidopsis thaliana) revealed increased root-versus-shoot biomass partitioning as a crucial plant response to cope with water stress (Des Marais et al., 2012). Several mechanisms dependent on ABA signaling have been proposed to maintain root elongation at low water potentials, such as osmotic adjustment in the root tip, restriction of ethylene production, and control of K+ translocation from root to shoot (Gaymard et al., 1998; Sharp et al., 2004). Enhanced cell wall loosening is required to maintain root elongation at low water potential, and indeed, ABA induces xyloglucan endotransglycosylase, which is a cell wall-degrading enzyme (Wu et al., 1994). Thus, the role of ABA in maintaining root growth under water deficits has been well established (Sharp et al., 2004); however, high concentrations of ABA inhibit root growth. Another important function of ABA is the regulation of the hydrotropic response (i.e. a genuine response of roots to a moisture gradient). Results from Takahashi et al. (2002) indicate that ABA constitutes an intrinsic signal in hydrotropism, since both aba1-1 and abi2-1 mutants were less sensitive to hydrotropic stimulation, whereas the addition of ABA to aba1-1 restored its capacity to perceive the moisture gradient. Additionally, the no hydrotropic response1 mutant of Arabidopsis showed reduced ABA sensitivity in root (Eapen et al., 2003), and ABA induces the expression of MIZ1, a gene essential for hydrotropism (Kobayashi et al., 2007). The regulation of root growth by ABA must be closely connected with hydrotropism, since the hydrotropic response likely involves asymmetric transmission of ABA signaling to the root sides that are in contact with different water potentials.
To further understand ABA perception and signaling in the root, we have analyzed the root ABA sensitivity of nine pyr/pyl mutants (i.e. pyr1, pyl1, pyl2, pyl4, pyl5, pyl6, pyl7, pyl8, and pyl9) as well as different combinations among them. Unexpectedly, given the partial functional redundancy among PYR/PYL genes, we found that pyl8 single knockout showed reduced sensitivity to ABA-mediated inhibition of root growth. The combination of different pyr/pyl mutations enhanced this phenotype. This genetic evidence together with root expression analyses served to pinpoint relevant ABA receptors in the root, particularly PYL8. Moreover, using tandem affinity purification (TAP) technology and mass spectrometry (MS) analyses, we were able to identify clade A PP2Cs that interacted in vivo with PYL8, providing new in vivo evidence for the mechanism of action of PYL8 to regulate ABA signaling. Finally, we found that PYR/PYLs and clade A PP2Cs play an important role for the ABA-mediated root hydrotropic response.
RESULTS
Regulation of ABA Signaling in Root by PYR/PYL Receptors
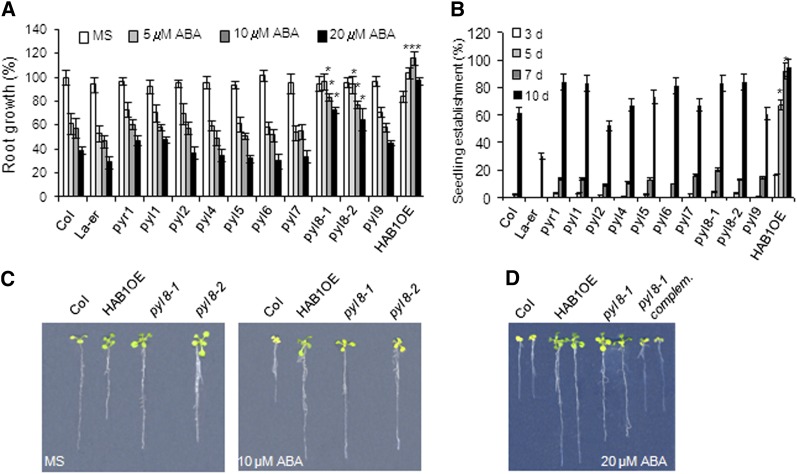
To further understand ABA perception in the root, we have analyzed the root ABA sensitivity of different pyr/pyl single mutants, namely pyr1-1, pyl1, pyl2, pyl4, pyl5, pyl6, pyl7, pyl8-1, and pyl9 (Fig. 1A; Supplemental Fig. S1). All the mutants were generated in the Columbia (Col) background, with the exception of pyl2, which is in the Landsberg erecta background (Park et al., 2009). We found that pyl8-1 showed reduced inhibition of root growth compared with the wild type, whereas the rest of the single mutants did not show significant differences from the wild type in that response (Fig. 1A). ABA-mediated inhibition of seedling establishment in pyr/pyl single mutants was quite similar to the wild type, although slight differences could be noticed in some mutants (Fig. 1B). Root growth of pyl8-1 was even resistant to 20 μm ABA compared with the wild type, whereas the other single mutants did not show significant differences (P < 0.01) compared with the wild type at 10 and 20 μm ABA (Fig. 1A). To further verify that the phenotype observed in pyl8-1 was a consequence of impaired PYL8 expression, we analyzed the phenotype of a second pyl8 allele, pyl8-2, and found that it also showed a reduced sensitivity to ABA-mediated inhibition of root growth (Fig. 1, A–C). Finally, the phenotype of pyl8-1 was complemented by the introduction of a 35S:PYL8 transgene (Fig. 1D).
Figure 1.
PYL8 plays a nonredundant role in root sensitivity to ABA. A, Quantification of ABA-mediated root growth inhibition of pyr/pyl mutants compared with the wild type. Data are averages ± se from three independent experiments (n = 15 each). *P < 0.01 (Student’s t test) with respect to the wild type in the same experimental condition. B, Seedling establishment of pyr/pyl mutants compared with Col and Landsberg erecta (La-er) wild types in medium supplemented with 0.5 μm ABA at 3, 5, 7, and 10 d after sowing. Data show the percentage of seeds that germinated and developed green cotyledons. Values are averages ± se for three independent experiments (100 seeds each). *P < 0.01 (Student’s t test) with respect to the wild type in the same experimental condition. C, ABA-insensitive phenotypes of pyl8-1 and pyl8-2 alleles compared with the Col wild type. Photographs show representative seedlings 10 d after the transfer of 4-d-old seedlings from Murashige and Skoog medium (MS) to plates lacking or supplemented with 10 μm ABA. D, Complementation of the pyl8-1 allele by introduction of a 35S:3HA-PYL8 transgene (pyl8-1 complemented). The photograph shows representative seedlings 12 d after the transfer of 4-d-old seedlings from Murashige and Skoog medium to plates supplemented with 20 μm ABA. [See online article for color version of this figure.]
During the course of these experiments, we analyzed HAB1-overexpressing lines (HAB1 OE), where additional copies of HAB1 have been introduced under the control of its own promoter, ProHAB1:HAB1-dHA, and found that they showed reduced sensitivity to ABA-mediated inhibition of root growth, which mimicked the pyl8 phenotype (Saez et al., 2004; Fig. 1, A and C). Interestingly, these lines showed lower root growth than the wild type in medium lacking exogenous ABA, and ABA supplementation improved the root growth of HAB1 OE plants between 20% and 30% compared with growth in medium lacking ABA (Fig. 1, A and C). Recently, improved root growth by ABA supplementation has also been reported for strong ABA-insensitive mutants lacking five or six PYR/PYL receptors (Gonzalez-Guzman et al., 2012).
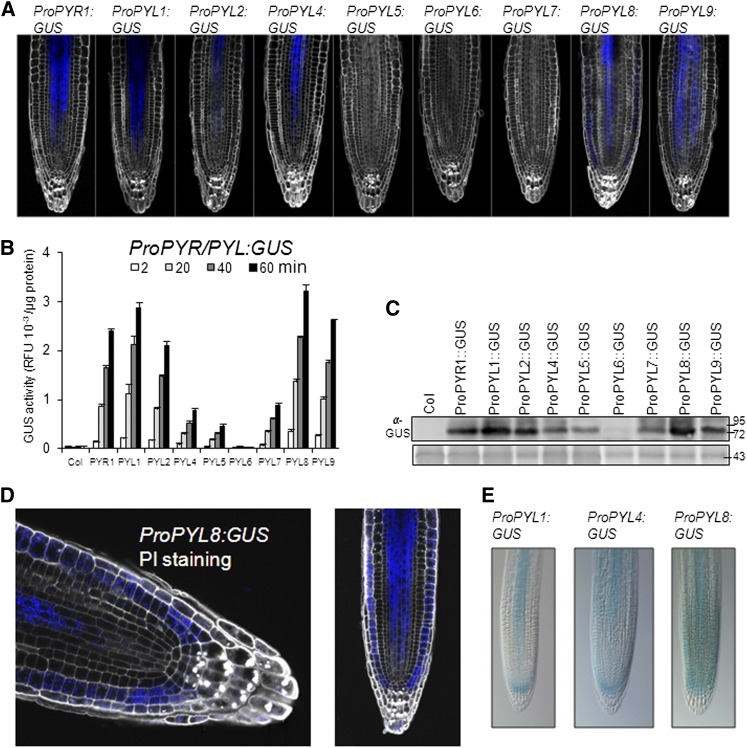
GUS reporter analysis of PYR1, PYL1, PYL2, PYL4, PYL5, and PYL8 promoters has been recently reported (Gonzalez-Guzman et al., 2012). We have completed this analysis by generating additional GUS reporter lines for PYL6, PYL7, and PYL9 promoters (Supplemental Fig. S2). Root expression of GUS driven by the PYL6 promoter was almost undetectable, expression driven by the PYL7 promoter was weak and could only be detected after 6 h of incubation with the GUS substrate, whereas ProPYL9:GUS lines showed GUS staining after 3 h (Supplemental Fig. S2). Additionally, we used a modified pseudo-Schiff propidium iodide (PS-PI) staining method to get a detailed GUS staining of the apical root (Fig. 2A). After 3 h of incubation with the GUS substrate, we could detect GUS staining in stele cells of the ProPYR1:GUS, ProPYL1:GUS, ProPYL2:GUS, ProPYL4:GUS, ProPYL8:GUS, and ProPYL9:GUS lines as well as root epidermis and lateral root cap for PYL8 (Fig. 2, A and D). PS-PI staining combined with confocal laser scanning microscopy produced high-resolution images; however, it eliminated GUS staining of columella cells in ProPYL1:GUS, ProPYL4:GUS, and ProPYL8:GUS lines, which was detected previously (Gonzalez-Guzman et al., 2012; Fig. 2E). In order to get an estimation of GUS expression in the whole root, we performed a quantitative GUS activity assay in extracts of root tissue prepared from 15-d-old seedlings by using 4-methylumbelliferyl β-d-glucuronide as a substrate (Fig. 2B). GUS activity was particularly high for ProPYL8:GUS, ProPYL1:GUS, ProPYR1:GUS, ProPYL9:GUS, and ProPYL2:GUS genes, whereas the expression of ProPYL4:GUS, ProPYL5:GUS, and ProPYL7:GUS genes was lower and ProPYL6:GUS was almost undetectable (Fig. 2B). These results were in good agreement with immunoblot analysis of the corresponding protein extracts using anti-GUS antibody (Fig. 2C), and they provide a quantitative estimation on the expression of the different PYR/PYL receptors in root.
Figure 2.
GUS expression driven by ProPYR1:GUS, ProPYL1:GUS, ProPYL2:GUS, ProPYL4:GUS, ProPYL5:GUS, ProPYL6:GUS, ProPYL7:GUS, ProPYL8:GUS, and ProPYL9:GUS genes in the apical root. A, GUS expression visualized using modified PS-PI staining and confocal laser scanning microscopy. B, Quantification of GUS activity in 15-d-old roots using 4-methylumbelliferyl β-d-glucuronide as a substrate. RFU, Relative fluorescence units. C, Immunoblot analysis of protein extracts from 15-d-old roots using anti-GUS antibody. Ponceau staining from a 43-kD protein is shown as a loading control. D, Magnification of the apical root from ProPYL8:GUS lines that were stained as described in A. E, GUS expression driven by ProPYL1:GUS, ProPYL4:GUS, and ProPYL8:GUS genes in columella cells. GUS staining was observed in the absence of subsequent PS-PI staining.
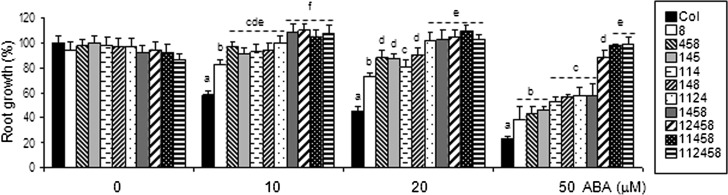
Since different PYR/PYL receptors are expressed at high levels in the root, we performed root growth assays of different triple, quadruple, pentuple, and sextuple mutants (Fig. 3). To this end, we transferred 4-d-old seedlings to Murashige and Skoog medium plates lacking or supplemented with 10, 20, or 50 μm ABA. Some mutants were more resistant to ABA-mediated root growth inhibition than pyl8, and the combination of pyl8 with other mutant loci enhanced the pyl8 phenotype, particularly evident at 50 μm ABA, when compared with pentuple and sextuple mutants (Fig. 3). Triple mutants lacking the pyl8 locus, such as 114 or 145, also showed an ABA-insensitive phenotype, which indicates that, in addition to PYL8, other loci contribute to root ABA sensitivity, and additional pyr/pyl combinations will be required for a more comprehensive analysis. In spite of this limitation, in different combined mutants, we observed additive effects by progressive inactivation of PYR/PYL genes (Fig. 3). For instance, the ABA-insensitive phenotype was clearly increased when pentuple and sextuple mutants were compared with pyl8 in 50 μm ABA, indicating a quantitative regulation of root ABA sensitivity by PYR/PYL genes (Fig. 3). Finally, as described previously for 12458 and 112458 mutants (Gonzalez-Guzman et al., 2012), ABA supplementation (10–20 μm) slightly improved the root growth of 1458 and 11458 mutants compared with growth in the absence of supplemented ABA (Fig. 3).
Figure 3.
Quantification of ABA-mediated inhibition of the root growth of pyr/pyl mutants compared with the wild type. Data are averages ± se from three independent experiments (n = 15 each). The letters denote significant differences among the different genetic backgrounds (P < 0.05, Fisher’s lsd tests). Primary root lengths of 15 plants per genotype (three independent experiments) were measured after 8 d in medium lacking or supplemented with 10, 20, or 50 μm ABA. The 145, 148, 1458, and 12458 mutants contain the pyr1-1 allele; the 114, 1124, 11458, and 112458 mutants contain both pyr1-1 and pyl1 alleles. The rest of the abbreviations reflect the corresponding pyl number.
PYL8 Regulates Root ABA Signaling through Interaction with Clade A PP2Cs
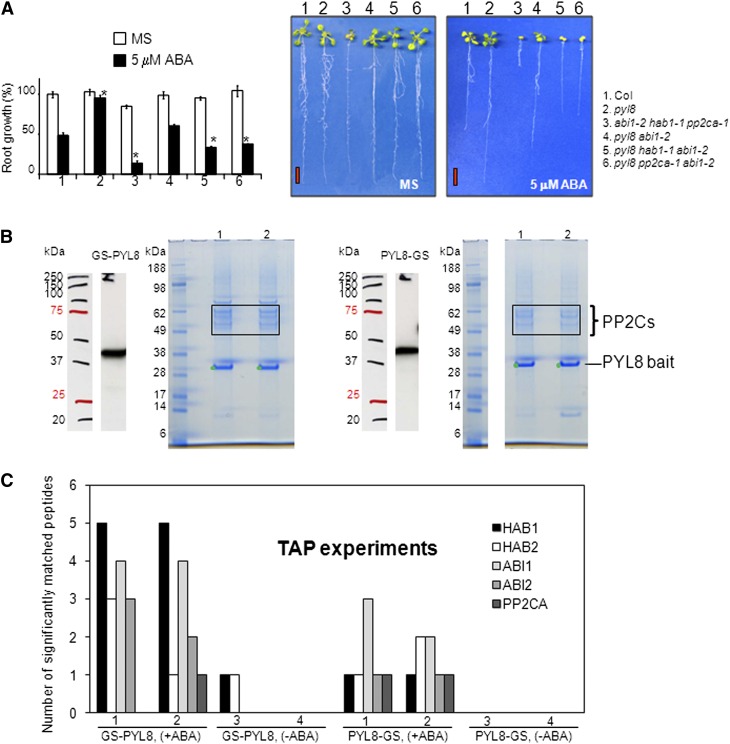
Given the important role of PYL8 to control root sensitivity to ABA, we decided to further pursue the study of its mechanism of action. To this end, we followed both a genetic and a biochemical approach. First, since PYR/PYL ABA receptors inhibit PP2Cs in an ABA-dependent manner and PYL8 inhibited several PP2Cs in vitro (Santiago et al., 2009; Antoni et al., 2012), we reasoned that the pyl8 phenotype might be due to an enhanced in vivo activity of PP2Cs. To test this hypothesis, we crossed pyl8-1 with the hab1-1abi1-2pp2ca-1 triple mutant to generate different combinations of pyl8-1 with pp2c mutants. The reduced sensitivity of pyl8 to ABA-mediated inhibition of root growth was notably diminished when pyl8 was combined with abi1-2, hab1-1, or pp2ca-1 (Fig. 4A). Therefore, these results are in agreement with the in vitro inhibition of clade A PP2Cs by PYL8 and reveal a genetic interaction of PYL8 with different clade A PP2Cs.
Figure 4.
Genetic and biochemical interaction of PYL8 with clade A PP2Cs. A, The reduced sensitivity of pyl8 to ABA-mediated inhibition of root growth is abrogated by knocking out clade A PP2Cs. The quantification of ABA-mediated root growth inhibition of the indicated genotypes compared with the wild type is shown. Data are averages ± se from three independent experiments (n = 15 each). *P < 0.01 (Student’s t test) with respect to the wild type in the same growth conditions. Photographs show representative seedlings 10 d after the transfer of 4-d-old seedlings to Murashige and Skoog plates (MS) lacking or supplemented with 5 µm ABA. Bars = 1 cm. B, Clade A PP2Cs interact in vivo with PYL8 in the presence of ABA. GS-PYL8 and PYL8-GS interact with clade A PP2Cs expressed in Arabidopsis cell suspension cultures. The SDS-PAGE analysis shows the zone where HAB1/HAB2 (top two bands) and ABI1/ABI2/PP2CA (other bands) were recovered as interacting partners of PYL8 when extracts and TAP purification buffers were supplemented with 50 μm ABA. C, Quantification of significantly (P > 95%) matched peptides of clade A PP2Cs recovered in independent TAP experiments using either GS-PYL8 or PYL8-GS as bait. ABA supplementation (50 μm; +ABA) dramatically increased the recovery of clade A PP2Cs compared with samples lacking ABA supplementation (−ABA). Detailed results of the peptides identified by MS analyses are provided as Supplemental Table S1. [See online article for color version of this figure.]
Second, we followed a biochemical approach to identify PYL8-interacting proteins in vivo using Arabidopsis suspension cells that express a protein G-streptavidin (GS)-tagged PYL8 as a bait for TAP (Van Leene et al., 2008). The GS tag combines two IgG-binding domains of protein G with a streptavidin-binding peptide, separated by two tobacco etch virus cleavage sites, and it has proved to be a good tag for TAP approaches (Van Leene et al., 2008). Previously, PYL8 and other PYR/PYL ABA receptors have been recovered in vivo as ABI1-interacting proteins using affinity protein complex purifications and subsequent identification by MS analyses (Nishimura et al., 2010). Therefore, it has been demonstrated that a single PP2C interacts in vivo with different ABA receptors. We wondered whether a single ABA receptor might be able to interact in vivo with different PP2Cs. To this end, we performed TAP of protein complexes in Arabidopsis suspension cells that express PYL8 fused to the N- or C-terminal GS tag (GS-PYL8 or PYL8-GS, respectively) as bait, and subsequently, we proceeded to the identification of interacting partners by MS analyses (Supplemental Table S1). Recombinant GS-PYL8 or PYL8-GS protein production was confirmed by immunoblot analysis (Fig. 4B). PYL8-bound complexes were recovered from both cultures (two technical repeats per culture) treated with 50 μm ABA, and finally they were eluted from streptavidin beads and analyzed by SDS-PAGE (Fig. 4B). Matrix-assisted laser-desorption ionization time of flight/time of flight (MALDI-TOF/TOF)-MS analysis of these complexes revealed that five clade A PP2Cs, HAB1, HAB2, ABI1, ABI2, and PP2CA/ABA-HYPERSENSITIVE GERMINATION3, were able to interact with PYL8 in samples supplemented with 50 μm ABA (Fig. 4, B and C; Supplemental Table S1). Therefore, these results provide evidence that a PYR/PYL ABA receptor can form complexes with several clade A PP2Cs in vivo in Arabidopsis.
We did not recover any ABA-activated SnRK2 in these complexes, which suggests that the ABA-dependent interaction of PYL8 with the PP2Cs effectively competed in planta with the interaction of PP2Cs and SnRK2s or, alternatively, that the residual interaction of the SnRK2s (via the ABA box) and PP2Cs was not enough to withstand the double purification of the TAP procedure. When samples were not supplemented with exogenous ABA, only one peptide that significantly matched both to HAB1 and HAB2 (VIQWQGAR; identical sequence in both PP2Cs) could be recovered from PYL8-interacting proteins in one experiment (Fig. 4C; Supplemental Table S1). Thus, exogenous ABA supplementation dramatically increased the recovery of PP2Cs as PYL8-interacting proteins in TAP complexes.
PYR/PYL Receptors and Clade A PP2Cs Mediate the Root Hydrotropic Response
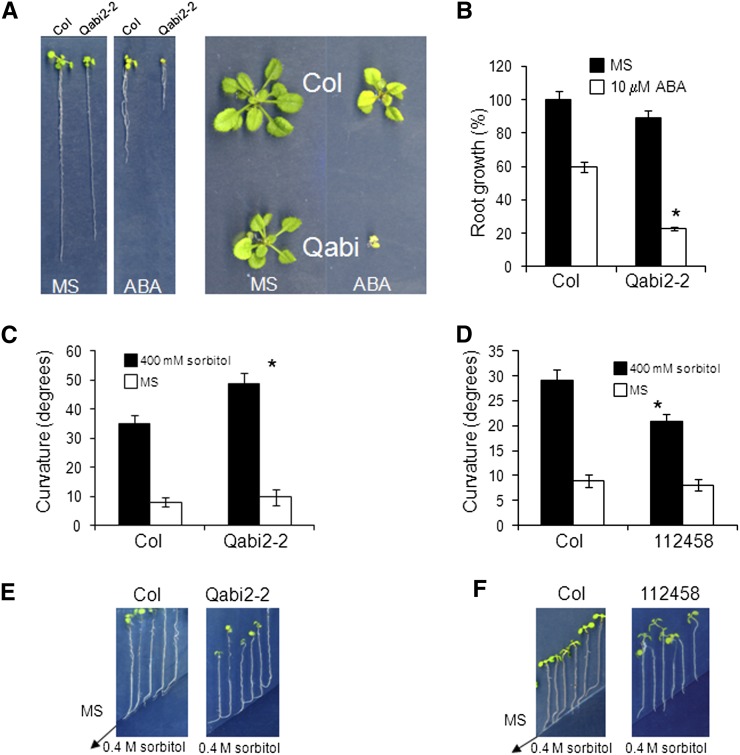
PYR/PYL receptors and clade A PP2Cs are key players for ABA signaling in root, and taking into account the important role of ABA for hydrotropism, we decided to investigate their role in the root hydrotropic response. Since PYL8 plays an important role for ABA signaling in root and interacts at least with five clade A PP2Cs, we generated an abi1-2abi2-2hab1-1pp2ca-1 quadruple mutant, abbreviated as Qabi2-2 (Fig. 5, A and B). The Qabi2-2 mutant is impaired in four PYL8-interacting PP2Cs, and it turned out to be very hypersensitive to ABA-mediated inhibition of root and shoot growth (Fig. 5, A and B). Using the experimental system developed by Takahashi et al. (2002), which uses split agar plates containing sorbitol in the region with low water potential, we measured the hydrotropic response of mutants showing enhanced or impaired ABA signaling (Fig. 5, C and D). In this assay, Murashige and Skoog medium containing 1% agar and agar containing 400 mm sorbitol are placed side by side, which generates a water potential gradient at the border between the two media (Fig. 5, E and F). Thus, we analyzed the hydrotropic responses of the strongly ABA-hypersensitive Qabi2-2 mutant and the ABA-insensitive 112458 sextuple mutant, which is strongly impaired in ABA perception through PYR/PYL receptors (Gonzalez-Guzman et al., 2012). As a result, we found that the Qabi2-2 mutant showed enhanced root curvature compared with the wild type when faced with a medium containing −1 MPa sorbitol (Fig. 5C). Conversely, the 112458 mutant showed reduced root curvature compared with the wild type (Fig. 5D). This response had important consequences, since seedlings of the Qabi2-2 mutant avoided better than the wild type the entrance in medium with low water potential, whereas seedlings of the 112458 mutant were impaired in that response (Fig. 5, E and F).
Figure 5.
Enhanced hydrotropic response of the pp2c quadruple mutant and reduced hydrotropic response of the pyr/pyl sextuple mutant. A, ABA-hypersensitive phenotype of the hab1-1abi1-2pp2ca-1abi2-2 quadruple mutant, abbreviated as Qabi2-2, compared with the Col wild type. Photographs show representative seedlings 10 d (left) or 20 d (right) after the transfer of 4-d-old seedlings to Murashige and Skoog plates lacking or supplemented with 10 µm ABA. B, ABA-hypersensitive root growth inhibition of the Qabi2-2 mutant compared with the Col wild type. C, Enhanced hydrotropic response of the Qabi2-2 mutant compared with the wild type. D, Reduced hydrotropic response of the pyr/pyl sextuple mutant compared with the wild type. C and D show hydrotropism assays with 7-d-old Arabidopsis seedlings. Data represent measures of the root curvature angle taken 14 h after the transfer of 7-d-old seedlings to split agar plates containing 0.4 m sorbitol in the region with low water potential. Values are averages from three independent experiments ± se (n = 42 each). *P < 0.05 (Student’s t test) when comparing data from each genotype and the wild type in the same assay conditions. E and F, Photographs show the experiments described in C and D, respectively, at 3 d after the transfer of 7-d-old seedlings to split agar plates containing 0.4 m sorbitol. The arrows mark the limit between Murashige and Skoog medium and medium supplemented with 0.4 m sorbitol. [See online article for color version of this figure.]
DISCUSSION
Previous analyses of loss-of-function mutants indicated that the combination of several pyr/pyl loci was required to impair ABA signaling (Park et al., 2009; Gonzalez-Guzman et al., 2012). Thus, the generation of pyr1pyl1pyl4 triple or pyr1pyl1pyl2pyl4 quadruple mutants (Park et al., 2009) or different combinations of triple mutants (Gonzalez-Guzman et al., 2012) was at least required to obtain a robust ABA-insensitive phenotype, which suggested certain functional redundancy among PYR/PYL genes. Only the pyr1 mutant was reported to show a phenotype by inactivation of a single PYR/PYL gene (i.e. pyrabactin resistance; Park et al., 2009). Given these precedents, we were surprised to find that the single pyl8 mutant showed reduced ABA-mediated inhibition of root growth compared with the wild type. Several reasons might explain the nonredundant role of PYL8 to regulate root sensitivity to ABA. First, the root expression pattern of PYL8 shows some specificity with respect to other PYR/PYL receptors (discussed below). Second, biochemically, PYL8 is a monomeric receptor (higher affinity for ABA than dimeric receptors) that shows higher capacity (lower 50% inhibitory concentration) to inhibit in vitro certain PP2Cs (ABI1, PP2CA, and HAI1) than other PYR/PYLs (Santiago et al., 2009; Antoni et al., 2012). For instance, the ABA-dependent inhibition of PP2CA and HAI1 by PYL8 was almost 1 order of magnitude higher than by other PYR/PYLs (Antoni et al., 2012). Third, we have shown that at least five clade A PP2Cs interacted in vivo with PYL8 in an ABA-dependent manner. We cannot exclude that other clade A PP2Cs that are strongly induced by ABA (HAI1, HAI2, and HAI3) but otherwise expressed at low levels could also interact with PYL8. The Arabidopsis suspension cells used in TAP experiments were supplemented with ABA to prepare protein extracts, but they were not grown in the presence of ABA to avoid growth impairment and overexpression of the other PP2Cs.
Therefore, lack of PYL8 function likely leads to a globally enhanced activity of PP2Cs or diminished capacity to inhibit them. Indeed, analyses of the root ABA sensitivity of both pyl8pp2c mutants (Fig. 4A) and HAB1 OE plants (Fig. 1, A and C) support this idea. Thus, the root ABA sensitivity of HAB1 OE plants mimicked pyl8 or pyr/pyl combined mutants, which suggests that these phenotypes are due to enhanced clade A PP2C activity. Interestingly, HAB1 OE plants as well as severe ABA-insensitive pyr/pyl mutants (1458, 11458, 12458, and 112458) showed improved root growth upon supplementation of the growth medium with ABA (Figs. 1 and 3; Gonzalez-Guzman et al., 2012). Therefore, although ABA has an inhibitory role in root growth depending on the concentration and plant growth conditions, it is clear that ABA contributes to root growth stimulation under certain conditions. Low concentrations of ABA (less than 1 μm) are known to stimulate root growth in wild-type Arabidopsis (Ephritikhine et al., 1999; Fujii et al., 2007). ABA is also required to maintain primary root growth at low water potentials in maize (Zea mays) seedlings, and one of the factors that explain it is the restriction of ethylene production by ABA (for review, see Sharp et al., 2004). Finally, mutants whose threshold for ABA perception and signaling has been dramatically altered can reveal the ABA stimulatory effect on root growth at concentrations that would be inhibitory for the wild type. Different physiological mechanisms have been offered to explain this effect at low water potentials (Sharp et al., 2004). The interaction of ABA with other hormones that regulate root growth, although not well known yet, will be another possible explanation.
Our TAP experiments, together with those of Nishimura et al. (2010) using yellow fluorescent protein-ABI1 as a bait, provide evidence that multiple interactions among PP2Cs and PYR/PYLs occur in vivo, generating a regulatory network that offers a wide sensitivity and combinatorial possibilities to modulate ABA signaling. Mathematical modeling will be required to provide quantitative insights on the complex combinatory PP2C-ABA-PYL interactions. However, our results differed from those of Nishimura et al. (2010) because we did not recover any ABA-activated SnRK2 in our complexes. Although in vitro experiments have suggested the possible existence of quaternary PYL-ABA-PP2C-SnRK2 complexes (Soon et al., 2012), the fact is that such complexes have not yet been recovered from plant extracts. The ABI1-interacting proteins identified in MS analyses by Nishimura et al. (2010) might simply reflect the recovery of independent PYL-ABA-ABI1 and ABI1-SnRK2 complexes. Alternatively, TAP is a more stringent technique than single GFP affinity purification, which might result in the loss of the SnRK2 ABA box-PP2C interaction or weaker interactions than ternary ABA complexes. This could explain another divergence with respect to Nishimura et al. (2010): the dramatic increase in the recovery of PP2C peptides by exogenous addition of ABA. Thus, without ABA supplementation, GS-PYL8 only recovered one PP2C peptide in one experiment (none with PYL8 tagged at the C terminus), compared with 28 peptides when 50 μm ABA was supplemented (Fig. 4C; Supplemental Table S1). These results suggest that, in vivo, PYL8 shows only a weak/transient interaction with clade A PP2Cs when ABA levels are low, further supporting the current model for PYR/PYL-mediated signaling able to perceive changes in ABA levels (Cutler et al., 2010).
The expression in the root of PYR1, PYL1, PYL2, PYL4, PYL8, and PYL9 was predominant in the vascular tissue (Fig. 2; Gonzalez-Guzman et al., 2012), where ABA biosynthetic enzymes are localized as well (Cheng et al., 2002; Tan et al., 2003). Active ABA signaling in the root vascular tissue that carries out ABA biosynthesis might act as a positive feedback for ABA production or play a regulatory role for different transport processes (Gaymard et al., 1998; Barrero et al., 2006). Additionally, expression in columella cells could also be detected for PYL1, PYL4, and PYL8. Active pools of ABA have been detected in the columella cells by a ProRD29B:GUS reporter system, which suggests that even in the absence of stress, ABA signaling occurs in these cells (Christmann et al., 2005). Root columella cells play a key role for sensing gravity in a process governed by auxins, and the presence of ABA receptors in this region suggests that ABA signaling might somehow affect auxin signaling in this area. For instance, it has been proposed that the degradation of starch grains in amyloplasts in columella cells is required to have a hydrotropic response, since gravitropism would be inhibitory to hydrotropism (Takahashi et al., 2003). It has also been suggested that starch degradation in the columella cells of roots subjected to osmotic stress might be an osmoregulatory mechanism to increase osmolite concentration and to sustain Glc supply under water stress (Ponce et al., 2008). Since ABA signaling is required both for hydrotropism and osmoregulation of water-stressed roots (Takahashi et al., 2002; Sharp et al., 2004; this work), the presence of PYR/PYL receptors in columella cells might contribute to regulate both processes.
Finally, the expression of PYL8 was also documented in the root epidermis and lateral root cap. The localization of the moisture-gradient-sensing apparatus has not been precisely defined, likely because different root tissues might be required for proper hydrotropic perception and response. However, the root cap has been suggested to play a role for moisture-gradient perception (Eapen et al., 2003; Kobayashi et al., 2007). The localization of at least three ABA receptors in this area and the presence of PYL8 in the root epidermis and the lateral root cap fit well with the requirement of ABA signaling for root hydrotropism. Moreover, we provide evidence that pyr/pyl mutants are impaired in hydrotropism, indicating that ABA perception by these receptors is required for a proper response. Interestingly, this response can be enhanced by multiple knocking out of the PP2Cs that represses ABA signaling under basal conditions. Thus, the enhanced hydrotropic response of pp2c knockouts together with their reduced water loss and enhanced transcriptional response to ABA constitute a powerful mechanism to cope with water stress (Saez et al., 2006; Rubio et al., 2009). Future studies on the role played by ABA signaling for hydrotropism should answer important questions, such as how ABA generates the asymmetric growth required to escape from low-water-potential regions of the soil or whether ABA gradients are generated in the root in an analogous manner to auxins.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were routinely grown under greenhouse conditions in pots containing a 1:3 vermiculite:soil mixture. For plants grown under growth chamber conditions, seeds were surface sterilized by treatment with 70% ethanol for 20 min, followed by commercial bleach (2.5% sodium hypochlorite) containing 0.05% Triton X-100 for 10 min, and finally, four washes with sterile distilled water. Stratification of the seeds was conducted in the dark at 4°C for 3 d. Then, seeds were sown on Murashige and Skoog (1962) plates composed of Murashige and Skoog basal salts, 0.1% MES, and 1% agar. The pH was adjusted to 5.7 with KOH before autoclaving. Plates were sealed and incubated in a controlled-environment growth chamber at 22°C under a 16-h-light/8-h-dark photoperiod at 80 to 100 μE m−2 s−1. The pyr1-1 allele and the T-DNA insertion lines for pyl1, pyl2, pyl4, pyl5, and pyl8-1 have been described previously (Park et al., 2009; Lackman et al., 2011; Gonzalez-Guzman et al., 2012). Seeds of pyl6, pyl7, and pyl8-2 insertion lines, SAIL_1179_D01, SALK_012096, and SALK_033867, respectively, were obtained from the Nottingham Arabidopsis Stock Centre. The abi1-2abi2-2hab1-1pp2ca-1 quadruple mutant was generated by crossing two triple pp2c mutants described by Rubio et al. (2009). The pyl8-1 allele was crossed with the abi1-2hab1-1pp2ca-1 triple mutant to generate different combinations of pyl8 and pp2c knockout alleles.
Generation of Transgenic Lines and GUS Analyses
To construct the ProPYL6:GUS and ProPYL8:GUS genes, a fragment comprising 1.5 kb 5′ upstream of the ATG start codon plus the first 30 bp of the PYL6 or PYL8 coding sequence, respectively, was amplified by PCR and cloned into pCR8/GW/TOPO T/A. Next, it was recombined by Gateway LR reaction into pMDC163 destination vector (Curtis and Grossniklaus, 2003). To generate the ProPYL7:GUS gene, the upstream sequence amplified was of 0.5 kb to avoid overlapping with regulatory sequences of the At4g01023 neighboring gene. The different pMDC163-based constructs carrying ProPYR/PYL:GUS genes were transferred to Agrobacterium tumefaciens pGV2260 (Deblaere et al., 1985) by electroporation and used to transform Col wild-type plants by the floral dipping method. Seeds of transformed plants were harvested and plated on hygromycin (20 μg mL−1) selection medium to identify T1 transgenic plants, and T3 progeny homozygous for the selection marker were used for further studies. Imaging of GUS and GUS quantitative assays were performed as described by Jefferson et al. (1987). Root GUS staining was also visualized using modified PS-PI staining and confocal laser scanning microscopy as described previously (Truernit et al., 2008).
The coding sequence of PYL8 was amplified by PCR and cloned into pCR8/GW/TOPO (Santiago et al., 2009). Next, it was recombined by LR reaction into the ALLIGATOR2 destination vector (Bensmihen et al., 2004). The ALLIGATOR2-35S:3HA-PYL8 construct was transferred to A. tumefaciens C58C1 (pGV2260; Deblaere et al., 1985) by electroporation and used to transform pyl8-1 plants by the floral dip method. T1 transgenic seeds were selected based on GFP visualization, and T3 progeny homozygous for the selection marker were used for further studies. The generation of ProHAB1:HAB1-dHA lines was described previously (Saez et al., 2004).
Seed Germination and Seedling Establishment Assays
After surface sterilization of the seeds, stratification was conducted in the dark at 4°C for 3 d. Next, approximately 100 seeds of each genotype were sown on Murashige and Skoog plates lacking or supplemented with 0.5 μm ABA. To score seed germination, radicle emergence was analyzed at 72 h after sowing. Seedling establishment was scored as the percentage of seeds that developed green expanded cotyledons and the first pair of true leaves at 7 d.
Root Growth Assays
Seedlings were grown on vertically oriented Murashige and Skoog plates for 4 to 5 d. Afterward, 20 plants were transferred to new Murashige and Skoog plates lacking or supplemented with the indicated concentrations of ABA. The plates were scanned on a flatbed scanner after 10 d to produce image files suitable for quantitative analysis of root growth using the NIH Image software ImageJ version 1.37.
TAP
Cloning of transgenes encoding GS-tagged fusions under the control of the constitutive cauliflower mosaic virus 35S promoter and transformation of Arabidopsis cell suspension cultures were carried out as described previously (Van Leene et al., 2007). Briefly, the GS tag combines two IgG-binding domains of protein G with a streptavidin-binding peptide, separated by two tobacco etch virus cleavage sites. In GS-PYL8, the two IgG-binding domains of protein G are placed in front of the streptavidin-binding peptide; in PYL8-GS, the order of these domains is opposite. TAP of protein complexes was done using the GS tag (Bürckstümmer et al., 2006) followed by protein precipitation and separation, according to Van Leene et al. (2008). For the protocols of proteolysis and peptide isolation, acquisition of mass spectra by a 4800 MALDI-TOF/TOF Proteomics Analyzer (AB SCIEX), and MS-based protein homology identification based on The Arabidopsis Information Resource genomic database, we refer to Van Leene et al. (2010). Experimental background proteins were subtracted based on approximately 40 TAP experiments on wild-type cultures and cultures expressing the TAP-tagged mock proteins GUS, red fluorescent protein, and GFP (Van Leene et al., 2010).
Hydrotropism Assay
The hydrotropic response was analyzed in 7-d-old Arabidopsis seedlings as described by Takahashi et al. (2002). Briefly, plastic square plates were filled with 1% agar containing Murashige and Skoog medium. After solidification of the agar, one-half of the medium was removed by cutting with a scalpel at an angle of 36° and replaced with 1% agar containing Murashige and Skoog medium supplemented with 400 mm sorbitol. Root tips were placed on the border between these two media, where a water potential gradient was generated, and plates were positioned vertically. After 14 h, the hydrotropic response was calculated by measuring the root curvature angle.
The Arabidopsis Genome Initiative locus identifiers for PYR1, PYL1, PYL2, PYL3, PYL4, PYL5, PYL6, PYL7, PYL8, PYL9, PYL10, PYL11, PYL12, and PYL13 are At4g17870, At5g46790, At2g26040, At1g73000, At2g38310, At5g05440, At2g40330, At4g01026, At5g53160, At1g01360, At4g27920, At5g45860, At5g45870, and At4g18620, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Schematic diagram of the PYR1, PYL1, PYL2, PYL4, PYL5, PYL6, PYL7, PYL8, and PYL9 genes showing the position of the T-DNA insertion or the nonsense mutation in the pyr1-1 allele.
Supplemental Figure S2. GUS expression driven by ProPYL6:GUS, ProPYL7:GUS, and ProPYL9:GUS genes in roots of 5-d-old seedlings.
Supplemental Table S1. Summary of results obtained in TAP experiments and peptide identification using the 4800 MALDI-TOF/TOF Proteomics Analyzer (AB SCIEX) and the GPS Explorer version 6 (AB SCIEX) software package.
Supplemental Table S2. List of oligonucleotides used in this work.
Acknowledgments
We thank J. Ecker and the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants as well as the Syngenta and SAIL Arabidopsis Biological Resource Center/Nottingham Arabidopsis Stock Centre for distributing these seeds. We thank Ebe Merilo for help with statistical analysis.
Glossary
- ABA
abscisic acid
- PP2C
phosphatase type 2C
- TAP
tandem affinity purification
- MS
mass spectrometry
- Col
Columbia
- PS-PI
pseudo-Schiff propidium iodide
- GS
protein G-streptavidin
- MALDI-TOF/TOF
matrix-assisted laser-desorption ionization time of flight/time of flight
References
- Antoni R, Gonzalez-Guzman M, Rodriguez L, Rodrigues A, Pizzio GA, Rodriguez PL. (2012) Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol 158: 970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero JM, Rodriguez PL, Quesada V, Piqueras P, Ponce MR, Micol JL. (2006) Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ 29: 2000–2008 [DOI] [PubMed] [Google Scholar]
- Bensmihen S, To A, Lambert G, Kroj T, Giraudat J, Parcy F. (2004) Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett 561: 127–131 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Droillard MJ, Barbier-Brygoo H, Laurière C. (2007) Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol Biol 63: 491–503 [DOI] [PubMed] [Google Scholar]
- Bürckstümmer T, Bennett KL, Preradovic A, Schütze G, Hantschel O, Superti-Furga G, Bauch A. (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods 3: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, et al. (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann A, Hoffmann T, Teplova I, Grill E, Muller A. (2005) Generation of active pools of abscisic acid revealed by in vivo imaging of water-stressed Arabidopsis. Plant Physiol 137: 209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Deak KI, Malamy J. (2005) Osmotic regulation of root system architecture. Plant J 43: 17–28 [DOI] [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, De Greve H, Deboeck F, Schell J, Van Montagu M, Leemans J. (1985) Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res 13: 4777–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais DL, McKay JK, Richards JH, Sen S, Wayne T, Juenger TE. (2012) Physiological genomics of response to soil drying in diverse Arabidopsis accessions. Plant Cell 24: 893–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupeux F, Santiago J, Betz K, Twycross J, Park SY, Rodriguez L, Gonzalez-Guzman M, Jensen MR, Krasnogor N, Blackledge M, et al. (2011) A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO J 30: 4171–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eapen D, Barroso ML, Campos ME, Ponce G, Corkidi G, Dubrovsky JG, Cassab GI. (2003) A no hydrotropic response root mutant that responds positively to gravitropism in Arabidopsis. Plant Physiol 131: 536–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephritikhine G, Fellner M, Vannini C, Lapous D, Barbier-Brygoo H. (1999) The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant J 18: 303–314 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK. (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaymard F, Pilot G, Lacombe B, Bouchez D, Bruneau D, Boucherez J, Michaux-Ferrière N, Thibaud JB, Sentenac H. (1998) Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 94: 647–655 [DOI] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al. (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Pizzio GA, Antoni R, Vera-Sirera F, Merilo E, Bassel GW, Fernández MA, Holdsworth MJ, Perez-Amador MA, Kollist H, et al. (2012) Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24: 2483–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S. (2011) Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Takahashi A, Kakimoto Y, Miyazawa Y, Fujii N, Higashitani A, Takahashi H. (2007) A gene essential for hydrotropism in roots. Proc Natl Acad Sci USA 104: 4724–4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T. (2005) Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J 44: 939–949 [DOI] [PubMed] [Google Scholar]
- Lackman P, Gonzalez-Guzman M, Tilleman S, Carqueijeiro I, Perez AC, Moses T, Seo M, Kanno Y, Hakkinen ST, Van Montagu MC, et al. (2011) Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proc Natl Acad Sci USA 108: 5891–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S. (2009) A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA 106: 21419–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Plant Physiol 15: 473–497 [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, et al. (2010) PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 61: 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TFF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, De Rybel B, Casimiro I, Benková E, Swarup R, Laplaze L, Beeckman T, Bennett MJ. (2009) Arabidopsis lateral root development: an emerging story. Trends Plant Sci 14: 399–408 [DOI] [PubMed] [Google Scholar]
- Ponce G, Rasgado FA, Cassab GI. (2008) Roles of amyloplasts and water deficit in root tropisms. Plant Cell Environ 31: 205–217 [DOI] [PubMed] [Google Scholar]
- Priest DM, Ambrose SJ, Vaistij FE, Elias L, Higgins GS, Ross AR, Abrams SR, Bowles DJ. (2006) Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. Plant J 46: 492–502 [DOI] [PubMed] [Google Scholar]
- Rodrigues A, Santiago J, Rubio S, Saez A, Osmont KS, Gadea J, Hardtke CS, Rodriguez PL. (2009) The short-rooted phenotype of the brevis radix mutant partly reflects root abscisic acid hypersensitivity. Plant Physiol 149: 1917–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, Santiago J, Flexas J, Schroeder JI, Rodriguez PL. (2009) Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol 150: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL. (2004) Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J 37: 354–369 [DOI] [PubMed] [Google Scholar]
- Saez A, Robert N, Maktabi MH, Schroeder JI, Serrano R, Rodriguez PL. (2006) Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol 141: 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Márquez JA, Cutler SR, Rodriguez PL. (2009) Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 60: 575–588 [DOI] [PubMed] [Google Scholar]
- Sato A, Sato Y, Fukao Y, Fujiwara M, Umezawa T, Shinozaki K, Hibi T, Taniguchi M, Miyake H, Goto DB, et al. (2009) Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem J 424: 439–448 [DOI] [PubMed] [Google Scholar]
- Sharp RE, Poroyko V, Hejlek LG, Spollen WG, Springer GK, Bohnert HJ, Nguyen HT. (2004) Root growth maintenance during water deficits: physiology to functional genomics. J Exp Bot 55: 2343–2351 [DOI] [PubMed] [Google Scholar]
- Sirichandra C, Gu D, Hu HC, Davanture M, Lee S, Djaoui M, Valot B, Zivy M, Leung J, Merlot S, et al. (2009) Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett 583: 2982–2986 [DOI] [PubMed] [Google Scholar]
- Soon FF, Ng LM, Zhou XE, West GM, Kovach A, Tan MH, Suino-Powell KM, He Y, Xu Y, Chalmers MJ, et al. (2012) Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335: 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kramer EM, Perry P, Knox K, Leyser HM, Haseloff J, Beemster GT, Bhalerao R, Bennett MJ. (2005) Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol 7: 1057–1065 [DOI] [PubMed] [Google Scholar]
- Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, Assaad FF, Christmann A, Grill E. (2010) Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J 61: 25–35 [DOI] [PubMed] [Google Scholar]
- Takahashi N, Goto N, Okada K, Takahashi H. (2002) Hydrotropism in abscisic acid, wavy, and gravitropic mutants of Arabidopsis thaliana. Planta 216: 203–211 [DOI] [PubMed] [Google Scholar]
- Takahashi N, Yamazaki Y, Kobayashi A, Higashitani A, Takahashi H. (2003) Hydrotropism interacts with gravitropism by degrading amyloplasts in seedling roots of Arabidopsis and radish. Plant Physiol 132: 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR. (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthélémy J, Palauqui JC. (2008) High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S, Beemster GT, Bennett MJ. (2012) Hormonal regulation of root growth: integrating local activities into global behaviour. Trends Plant Sci 17: 326–331 [DOI] [PubMed] [Google Scholar]
- Ubeda-Tomás S, Federici F, Casimiro I, Beemster GT, Bhalerao R, Swarup R, Doerner P, Haseloff J, Bennett MJ. (2009) Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr Biol 19: 1194–1199 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van De Slijke E, Stals H, Van Isterdael G, Verkest A, Neirynck S, Buffel Y, et al. (2010) Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol 6: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Stals H, Eeckhout D, Persiau G, Van De Slijke E, Van Isterdael G, De Clercq A, Bonnet E, Laukens K, Remmerie N, et al. (2007) A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol Cell Proteomics 6: 1226–1238 [DOI] [PubMed] [Google Scholar]
- Van Leene J, Witters E, Inzé D, De Jaeger G. (2008) Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci 13: 517–520 [DOI] [PubMed] [Google Scholar]
- Vlad F, Droillard MJ, Valot B, Khafif M, Rodrigues A, Brault M, Zivy M, Rodriguez PL, Merlot S, Laurière C. (2010) Phospho-site mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. Plant J 63: 778–790 [DOI] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S. (2009) Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Spollen WG, Sharp RE, Hetherington PR, Fry SC. (1994) Root growth maintenance at low water potentials (increased activity of xyloglucan endotransglycosylase and its possible regulation by abscisic acid). Plant Physiol 106: 607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]