Summary: The expression domains and phenotypes associated with distinct members of the GLV/RFG/CLEL secreted peptide family indicate that they function in multiple developmental programs, including meristem maintenance, gravitropism, lateral root emergence, and the formation of root hairs.
Abstract
The GOLVEN (GLV)/ROOT GROWTH FACTORS/CLE-Like small signaling peptide family is encoded by 11 genes in Arabidopsis (Arabidopsis thaliana). Some of them have already been shown to control root meristem maintenance, auxin fluxes, and gravitropic responses. As a basis for the detailed analysis of their function, we determined the expression domains for each of the 11 GLV genes with promoter-reporter lines. Although they are collectively active in all examined plant parts, GLV genes have highly specific transcription patterns, generally restricted to very few cells or cell types in the root and shoot and in vegetative and reproductive tissues. GLV functions were further investigated with the comparative analysis of root phenotypes induced by gain- and loss-of-function mutants or in treatments with GLV-derived synthetic peptides. We identified functional classes that relate to the gene expression domains in the primary root and suggest that different GLV signals trigger distinct downstream pathways. Interestingly, GLV genes transcribed at the early stages of lateral root development strongly inhibited root branching when overexpressed. Furthermore, transcription patterns together with mutant phenotypes pointed to the involvement of GLV4 and GLV8 in root hair formation. Overall, our data suggest that nine GLV genes form three subgroups according to their expression and function within the root and offer a comprehensive framework to study the role of the GLV signaling peptides in plant development.
In plants, intercellular signals are in part relayed by small-molecule phytohormones, such as auxin, cytokinin, ethylene, gibberellin, abscisic acid, and brassinosteroids. Recent biochemical and genetic studies have shown that plant cell-to-cell communication also involves peptide signaling pathways, similar to those previously identified in animals, regulating processes as diverse as plant-pathogen interaction, cell division and expansion, meristematic stem cell homeostasis, and pollen self-incompatibility (Butenko et al., 2009; Murphy et al., 2012). The recent analysis of the Arabidopsis (Arabidopsis thaliana) genome showed that plants code for hundreds of secreted small peptides, among which are numerous potential peptide hormones (Lease and Walker, 2006). Nevertheless, their characterization is not trivial, since most are encoded in multigene families, and the study of the corresponding loss-of-function (lof) mutants may not provide information on the function due to genetic redundancy. The expression patterns of all members of a given family, therefore, can serve as a guide for functional studies and suggest which genes are involved in identical or distinct developmental processes.
Alternatively, phenotypic analysis based on ectopic expression is another useful approach to gain insight into the molecular mechanisms controlled by peptide hormones, but it needs to be interpreted with caution as in any gain-of-function (gof) approach. In addition, synthetic peptides derived from the short conserved C-terminal domain of secreted peptide precursors (Matsubayashi, 2011) often mimic overexpression phenotypes, as is the case for the members of the large CLV3/ESR-RELATED (for CLAVATA3/EMBRYO SURROUNDING REGION [CLE]) family (Fiers et al., 2005, 2006; Strabala et al., 2006; Whitford et al., 2008; Jun et al., 2010).
In Arabidopsis, the GOLVEN (GLV)/ROOT GROWTH FACTORS (RGF) family coding for secretory peptides includes 11 genes that share the same structure (Matsuzaki et al., 2010; Whitford et al., 2012). They code for small precursor proteins of 79 to 163 amino acids containing a variable middle region that links two conserved domains: the N terminus that bears the signature of a signal peptide of 21 to 33 amino acids, with a cleavage site presumably processed when the GLV preproproteins enter the secretory pathway (Bendtsen et al., 2004), and a conserved motif of 13 to 16 amino acids at or near the C terminus that defines the GLV family, found in all higher plant genomes analyzed so far. The mature GLV1, GLV2, GLV3, and RGF1/GLV11 peptides were shown to be posttranslationally modified by sulfation of a conserved Tyr residue and hydroxylation of a Pro residue in the second half of the GLV motif (Matsuzaki et al., 2010; Whitford et al., 2012). The GLV/RGF peptides have also been associated with CLE18, a peculiar member of the CLV3/ESR family, and therefore named CLE-Like (CLEL; Meng et al., 2012). For clarity, related genes, proteins, and peptides are referred to hereafter following the GLV nomenclature. Table I lists the correspondence between Arabidopsis names reported in the three initial studies about the GLV/RGF/CLEL family.
Table I. Summary of GLV expression patterns and mutant phenotypes.
Web sites for software programs are as follows: AtGenExpress Visualization Tool, http://www.weigelworld.org/resources/microarray/AtGenExpress; eFP browser, http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Genevestigator, https://www.genevestigator.com/gv/plant.jsp. fl, Flowers; le, leaves; lr, lateral roots; n.a., not assayed; n.d., not detected; n.p., no probe found in the ATH1 GeneChip; ro, root; sh, shoot; st, stem.
| Expression Pattern |
gof Phenotypes |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Names | Promoter Reporter | qRT-PCR | AtGen Express | eFP Browser | Genevestigator | Matsuzaki et al. (2010) | Wavy/Curly | Increased RAM Size | Lower LR Density | Branched Root Hairs | lof Phenotypes | Alteration of PIN2 Traffic |
| At4g16515 | sh, le, st, fl | sh, le, fl | sh, le, st, fl | fl | sh, le, st, fl | n.d. | +++abc | +++abcd | ++ad | n.a. | Reduced hypocotyl bendingb | ++b |
| GLV1 | ||||||||||||
| RGF6 | ||||||||||||
| CLEL6 | ||||||||||||
| At5g64770 | sh, le, st, fl, lr | sh, le, fl | sh, le, st, fl | sh, le, st | sh, le, st, fl | n.d. | +++ab | +++acd | +++a | n.a. | Reduced hypocotyl bendingb | n.a. |
| GLV2 | ||||||||||||
| RGF9 | ||||||||||||
| CLEL9 | ||||||||||||
| At3g30350 | ro, lr | ro | ro, fl | ro | ro | n.d. | ++ab | +ad | +a | +a | Altered root gravitropismab | ++ab |
| GLV3 | ||||||||||||
| RGF4 | ||||||||||||
| At3g02240 | ro, lr | ro | ro, sh, fl | ro | ro, lr | n.d. | +/−a | ++ad | +a | ++a | Shorter root hairsa | − |
| GLV4 | ||||||||||||
| RGF7 | ||||||||||||
| CLEL4 | ||||||||||||
| At1g13620 | ro, lr | ro | n.p. | n.p. | n.p. | ro | ++a | +++ad | +++a | +a | Short RAMde | +++a |
| GLV5 | ||||||||||||
| RFG2 | ||||||||||||
| CLEL1 | ||||||||||||
| At2g03830 | ro, sh, le, fl, lr | ro, sh, le, fl | ro, fl | ro, sh, le, fl | ro | n.d. | +++a | +++a | +++a | n.a. | n.a. | +a |
| GLV6 | ||||||||||||
| RGF8 | ||||||||||||
| CLEL2 | ||||||||||||
| At2g04025 | ro, lr | ro | ro, fl | ro | ro, lr | ro | ++a | +++ad | +++a | n.a. | Short RAMde | n.a. |
| GLV7 | ||||||||||||
| RGF3 | ||||||||||||
| CLEL3 | ||||||||||||
| At3g02242 | ro, sh, le, fl, lr | ro | n.p. | n.p. | n.p. | n.a. | −a | +/−a | +/−a | +++a | Shorter and fewer root hairsa | n.a. |
| GLV8 | ||||||||||||
| CLEL5 | ||||||||||||
| At5g15725 | ro, lr | ro | ro | ro, sh | ro, lr | n.a. | ++a | +a | +a | n.a. | n.a. | n.a. |
| GLV9 | ||||||||||||
| At5g51451 | ro, lr | ro, sh, le, fl | n.p. | n.p. | n.p. | n.d. | ++ac | ++acd | +++ac | n.a. | n.a. | n.a. |
| GLV10 | ||||||||||||
| RGF5 | ||||||||||||
| CLEL7 | ||||||||||||
| At5g60810 | ro, lr | ro | n.p. | n.p. | n.p. | ro | ++ac | +++ad | +++a | n.a. | Short RAMde | +++a |
| GLV11 | ||||||||||||
| RGF1 | ||||||||||||
| CLEL8 | ||||||||||||
This report.
Observed for the glv5 glv7 glv11 triple mutant (Matsuzaki et al., 2010).
Promoter-reporter line analysis showed that GLV1 and GLV2 are transcribed in the epidermis and cortex of the hypocotyl and are induced at its lower side upon reorientation, probably as part of the auxin transcriptional response induced by gravistimulation (Whitford et al., 2012). The promoters of GLV1 and GLV2 are also active in cotyledon, leaf, and flower tissues but not in the root. Instead, GLV3 expression was only detected in the root apical meristem (RAM), more precisely in precursor cells of the cortex, endodermis, and stele, close to the quiescent center (QC). In situ hybridization in RAM tissues showed that GLV11 is expressed in the QC and columella stem cells, while high levels of the GLV5 and GLV7 transcripts were detected mainly in the innermost layer of the central columella (Matsuzaki et al., 2010). Interestingly, the expression domains described so far provided clues about the GLV gene functions, because glv1, glv2, and glv3 lof mutants show impaired gravitropic response in the hypocotyl and the root, while the rgf1 rgf2 rgf3 triple mutant is defective in root stem cell maintenance (Matsuzaki et al., 2010; Whitford et al., 2012). Proteins identified as targets of the GLV/RGF signaling pathway(s) include the PLT1 and PLT2 transcription factors that are stabilized by the addition of the GLV11/RGF1 peptide (Matsuzaki et al., 2010) and the PIN2 auxin efflux carrier whose intracellular trafficking is regulated by the GLV3 peptide (Whitford et al., 2012).
As a first step toward a systematic analysis of GLV function, we mapped the transcription of the 11 Arabidopsis GLV genes in primary roots, young shoots, and inflorescences. In addition, we analyzed phenotypes in overexpression lines, focusing our analysis on organs and tissues highlighted by the GLV transcriptional patterns. Following this approach, we found that GLV genes are involved in multiple root development programs. This study offers a useful scaffold to guide the future genetic and biochemical studies necessary for the dissection of the signaling pathways triggered by the GLV peptides.
RESULTS
GLV Expression in the Primary Root
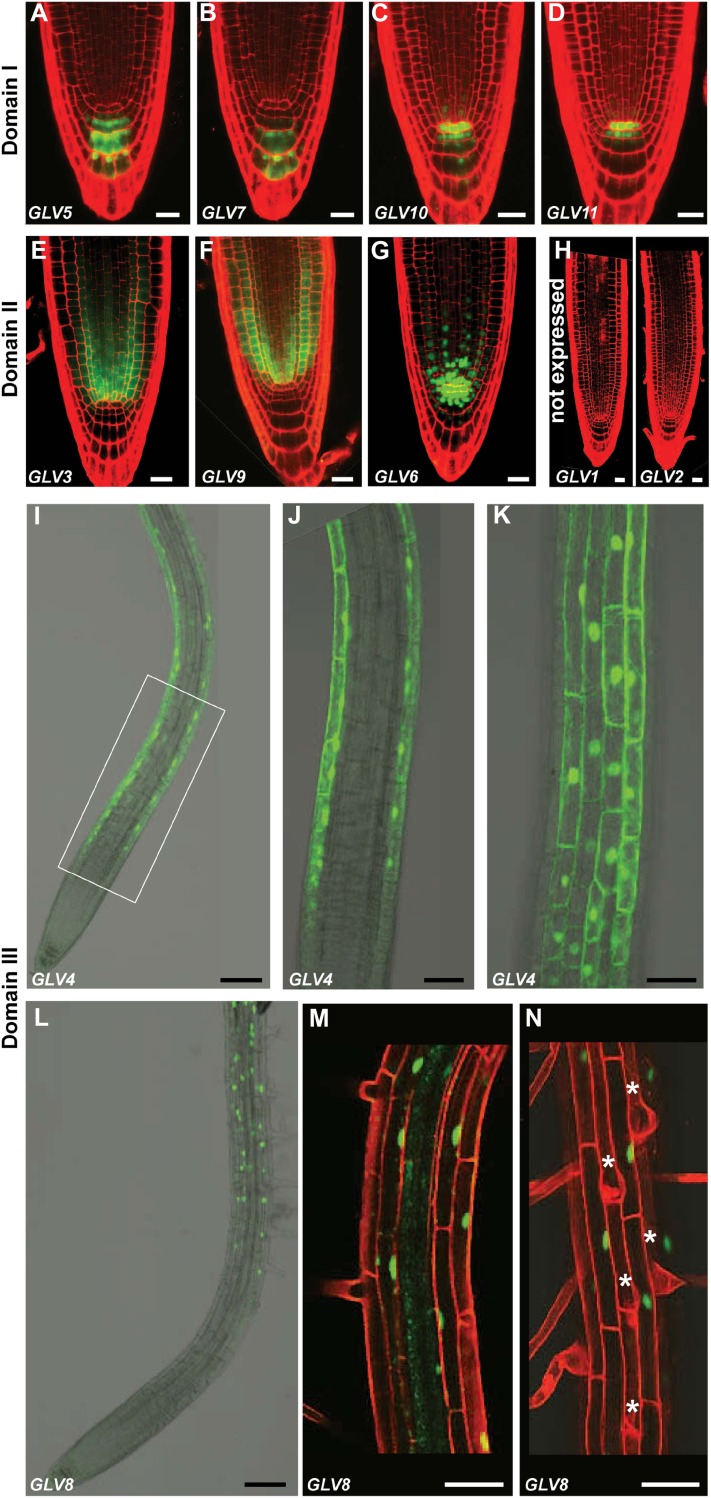
GLV expression in the primary root was characterized with promoter-reporter lines. GLVpro::NLS-2xGFP or GLVpro::GUS-GFP Arabidopsis seedlings were grown for 4 to 5 d after germination (dag), and the GFP signal was analyzed with confocal microscopy (Fig. 1). The GLV root transcription profiles were surprisingly diverse yet always restricted to very few cell types, except for GLV1 and GLV2, which were not found to be expressed in any part of the primary root (Whitford et al., 2012; Fig. 1H). Three main GLV expression domains were defined as follows. I, GLV5, GLV7, GLV10, and GLV11 were transcribed in the QC and/or columella cells (CCs; Fig. 1, A–D). II, GLV3 and GLV9 patterns were positioned just above the QC, detectable in most cell layers, and restricted to the root meristem (Fig. 1, E and F). GLV6 was present in both domains I and II (Fig. 1G). III, GLV4 and GLV8 were transcribed only in the root portion above the meristem (Fig. 1, I–N).
Figure 1.
GLV transcription profiles in the primary root. A to D, GLV genes expressed in the QC and/or CCs (domain I): GLV5pro::NLS-2xGFP (A), GLV7pro::NLS-2xGFP (B), GLV10pro::NLS-2xGFP (C), and GLV11pro::NLS-2xGFP (D). E and F, GLV genes active in the RAM (domain II): GLV3pro::GUS-GFP (E) and GLV9pro::GUS-GFP (F). G, GLV6pro::NLS-GFP-GUS, GFP signal in the meristem, QC, and CCs (domains I and II). H, GLV1 and GLV2 not expressed in the primary root, GLVpro::NLS-2xGFP. I to N, GLV genes expressed above the meristem (domain III): GLV4pro::NLS-2xGFP (I–K) and GLV8pro::NLS-GFP-GUS (L–N). J, K, M, and N are higher magnification images, and the rectangle in I indicates the area enlarged in J and K. J and M represent median sections. K and N show epidermal cell files. Stars mark hair cells. Roots in A to H, M, and N were counterstained with PI (red). Bars = 20 μm (A–H), 100 μm (I and L), and 50 μm (J, K, M, and N).
GLV promoters within the same domain had somewhat distinct profiles. GLV5 expression was detected in the second, third, and fourth CC layers, at a lower level in undifferentiated CCs, but not, or only marginally, in the QC (Fig. 1A). GLV7 was also excluded from the QC but present only in the third and fourth CC layers (Fig. 1B). GLV10 and GLV11 patterns were largely overlapping: both genes were expressed in the QC and the CC initials (Fig. 1, C and D). Weak GLV10 transcriptional activity was also detected in additional underlying CCs and in the vascular initials above the QC (Fig. 1C).
GLV3 transcription was detected mainly in the endodermis, the cortex, and the vascular tissues (Fig. 1E). Expression was strongest within two to three cells in the QC vicinity. Limited expression was also observed in the epidermis. The GLV9 signal was strongest in the meristematic cortical cells but also detected in the epidermis and within the vasculature (Fig. 1F). Neither GLV3 nor GLV9 was transcriptionally active in the QC. GLV6 was transcribed within the meristem, mainly in the cortex but also in the epidermal and procambial cells closest to the QC (Fig. 1G). It was also present in the QC and the first and second CC layers. GLV6 expression was very strong in the initials surrounding the QC.
GLV4 transcription was only detected in the root elongation zone and the proximal region of the differentiation zone (Fig. 1I) and was restricted to all cells of the epidermis (Fig. 1, J and K; Supplemental Movie S1). The lower limit of the GLV8 expression domain coincided with the border of the maturation zone and extended shootward throughout the entire root, all the way up to the colet (Fig. 1L). The reporter GFP signal was observed in the cortex and the epidermis (Fig. 1, M and N; Supplemental Movie S2). Unlike GLV4, GLV8 was restricted to the nonhair cell files in the epidermis, although it was present in all cortical cells regardless of their position.
To summarize, nine of the 11 GLV genes are transcribed with high tissue specificity in different domains of the Arabidopsis primary root.
GLV Expression during Lateral Root Formation
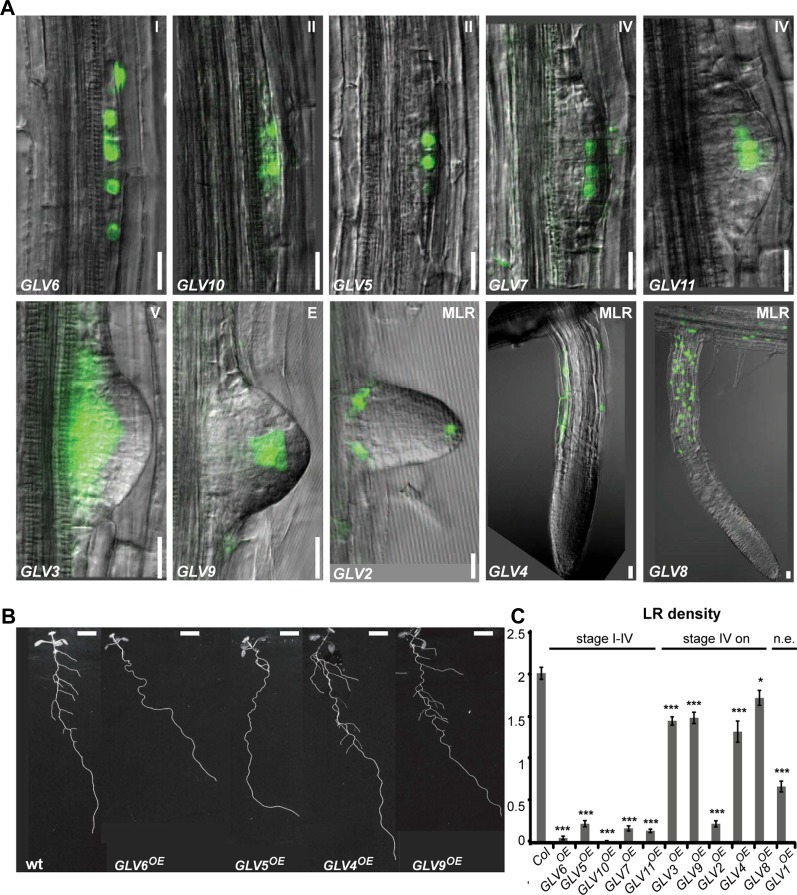
GLV transcriptional activity was also analyzed in lateral roots (LRs). We found that all GLV genes, except GLV1, were expressed during LR formation. However, their respective spatial and temporal patterns were very different (Fig. 2A). According to the developmental stages defined by Malamy and Benfey (1997), GLV6 was the earliest gene and the only one active from stage I on, followed by GLV5 and GLV10 (stage II), GLV7 and GLV11 (stage IV), GLV3 (stage V), and finally GLV9 and GLV2 after emergence of the primordium. GLV4 and GLV8 expression was only detected in mature LRs in the same tissue as described for the primary root (Fig. 1). Surprisingly, GLV2 expression was also detected in LRs but was not found in the primary root.
Figure 2.
Lateral root GLV expression and gof phenotype. A, First stage at which transcriptional activity is detected for each promoter. Corresponding developmental stages are indicated in the top right corner either as a number or a letter. E, Emerged LR; MLR, LR with established apical meristem. GLV1 expression was not detected in LRs (data not shown). B, LRs in wild-type (wt) and selected GLVOE plants at 12 dag. Bars = 20 μm (A) and 0.5 cm (B). C, Number of emerged LRs per 1 cm in GLVOE lines. GLV genes were organized according to their expression onset, from earliest to latest. Two categories are defined on top of the graph corresponding to GLV genes active before and after stage IV, respectively. Col, Columbia-0; n.e., not expressed in LRs. The chart shows data from two independent experiments ± se (n = 22–64). Stars indicate significant differences compared with the wild type (*P < 0.05, ***P < 0.001).
GLV Expression in Shoot Tissues of Seedlings
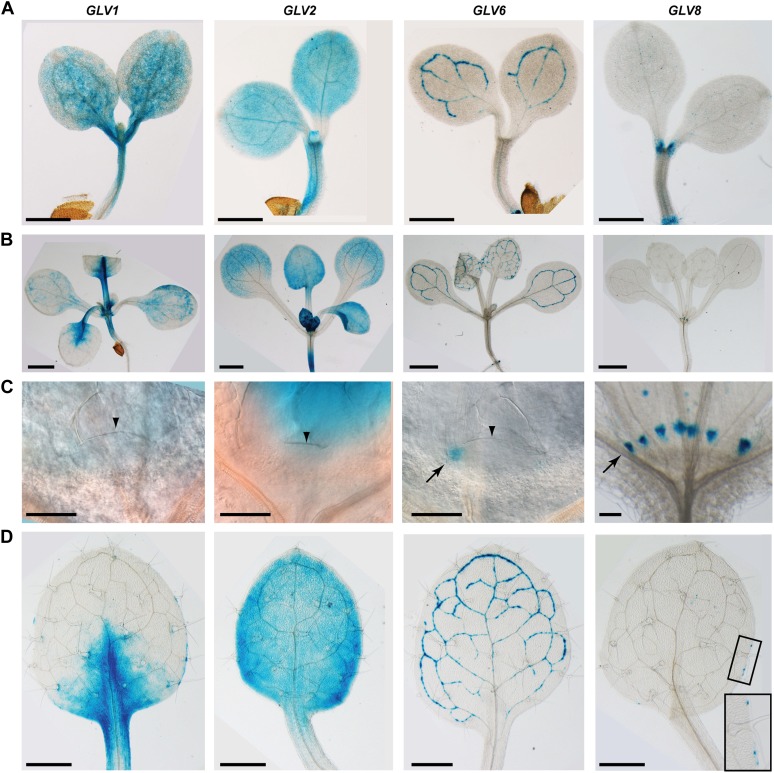
GLV transcription in shoot tissues was analyzed in young seedlings transformed with a GLVpro::GUS-GFP or GLVpro::NLS-GFP-GUS construct and stained for GUS enzymatic activity at 5, 10, and 15 dag to image the cotyledons and the first two true leaves, corresponding to developmental stages 0.7, 1.02, and 1.04, respectively (Boyes et al., 2001; Fig. 3). We previously reported that GLV1 and GLV2 were transcribed in cotyledons, leaves, and hypocotyls of 5-dag seedlings (Whitford et al., 2012). The two genes could already be distinguished at that stage based on the GUS staining of cotyledons: GLV1 was transcribed irregularly throughout the cotyledon and at a higher level at the base of the organ; GLV2 was transcribed homogeneously across the cotyledon (Fig. 3A). In the true leaves of older plants, GLV1 and GLV2 transcription patterns were partly complementary: GLV1 was expressed at a high level in the basal part of the leaf, close to the main veins, irregularly in the lamina joining these veins, and in separate segments of the leaf margin; GLV2 was expressed in the whole leaf lamina, with a stronger signal in the outer part of the leaf (Fig. 3, B and D; Whitford et al., 2012). GLV2 transcriptional activity was higher in younger leaves and decreased as leaves expanded (Fig. 3B). GLV1 transcription level was similar in all leaves regardless of age. Neither of the two genes was found to be expressed in the shoot apical meristem (SAM; Fig. 3C).
Figure 3.
GLV expression in shoot tissues. Images show GLV1pro::GUS-GFP, GLV2pro::GUS-GFP, GLV6pro::NLS-GFP-GUS, and GLV8pro::NLS-GFP-GUS. The expression of other GLV genes was not detected in these tissues (data not shown). A, Cotyledons of 5-dag seedlings. B, Shoots of 10-dag seedlings. C, Differential interference contrast images showing GLV expression in the SAM (GLV1, GLV2, GLV6) and the stipules (GLV8). Arrowheads point to the SAM, and arrows point to GLV6 expression at the border between the meristem and the leaves and to GLV8 expression in stipules. D, GLV expression in first and second leaves (10 dag). Bars = 1 mm (A and B), 50 μm (C), and 0.5 mm (D).
Two additional GLV genes were transcribed in the aerial part of the Arabidopsis plants. GLV6 promoter activity was detected in the vasculature of cotyledons and leaves (Fig. 3, A, B, and D) but also in the SAM, more precisely at the border between the meristem and the leaf primordia (Fig. 3C). The GLV8 pattern was irregular. The restricted GUS staining of cotyledons and leaves marked patches that could not be associated with lamina substructures (Fig. 3, A and D). Expression was also observed at the base of the cotyledon petioles and in stipules, but not in the SAM (Fig. 3, A–C). Furthermore, no promoter activity was detected in the rosette for GLV3 (Whitford et al., 2012), GLV4, GLV5, GLV7, GLV9, GLV10, and GLV11 (data not shown).
GLV Expression in the Inflorescence
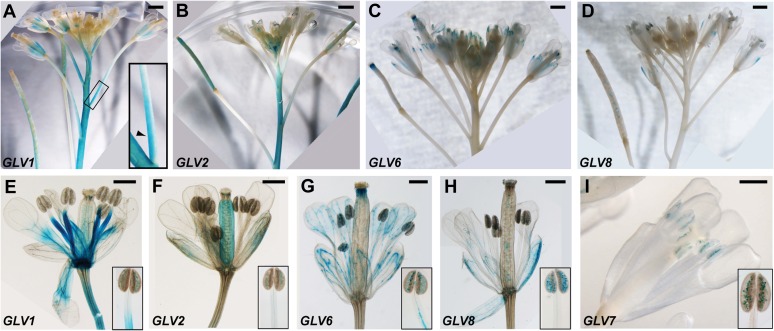
GLV1 promoter activity was detected in the stem, the rachis of the inflorescence, and the lower portion of the flower pedicel. Noticeably, GUS staining was asymmetric in the pedicel and only detected at its upper side (Fig. 4A), a pattern reminiscent of the asymmetric expression of the gene in gravistimulated hypocotyls (Whitford et al., 2012). Within the flower, GLV1 transcription was detected throughout flower development at the base of the sepals, in the petals, the filament of the stamens, and the gynoecium (Fig. 4E). GLV2 transcription was observed in the stem and sepals and in the gynoecium of the developing flowers (Fig. 4, B and F). Both genes were also expressed later in the silique (Fig. 4, A and B). GLV6 transcription was only detected in the flower, the vasculature of sepals, stamens, and petals (Fig. 4, C and G) as well as at the tip of the gynoecium and later in the siliques. GLV8 was expressed irregularly in the sepals, similar to its leaf pattern, as well as in pollen grains (Fig. 4, D and H). Although GLV7 activity was not detected in other shoot tissues, it was transcribed in the pollen (Fig. 4I). None of the other GLV promoters was found to be active in the inflorescence (data not shown).
Figure 4.
GLV transcription profiles in GUS-stained inflorescences. A to D, Whole inflorescences. E to I, Flowers. Plants shown are as follows: GLV1pro::GUS-GFP (A and E), GLV2pro::GUS-GFP (B and F), GLV6pro::NLS-GFP-GUS (C and G), GLV8pro::NLS-GFP-GUS (D and H), and GLV7pro::GUS-GFP (I). Insets in E to I show a separate stamen. Bars = 1 mm (top panels) and 0.5 mm (bottom panels).
GLV Promoter-Reporter Profiles Agree with Quantitative Reverse Transcription-PCR and Other Transcriptome Data
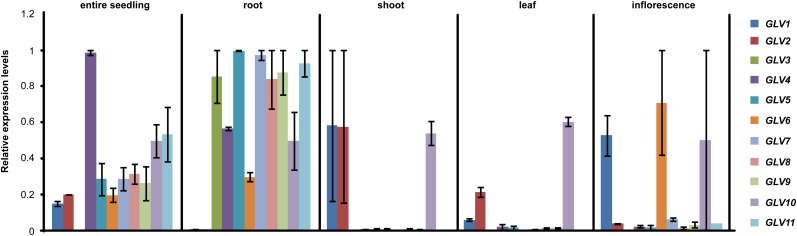
To determine whether the results obtained with GLV promoter-reporter lines correspond to endogenous gene transcription, we extracted RNA from wild-type plants and measured GLV transcript levels by quantitative reverse transcription (qRT)-PCR in entire seedlings (5 dag), shoots (5 dag), roots (5 dag), true leaves (14 dag), and inflorescences (30 dag; Fig. 5). All GLV transcripts were detected in young seedlings, except GLV3, whose level was close to the detection limit. In agreement with reporter line analysis, the GLV3 to GLV11 transcripts were all detected in the main root, while GLV1 and GLV2 were absent from this organ. Only the GLV1, GLV2, and GLV10 transcripts were detected in aerial tissues (5-dag seedlings and true leaves), reflecting reporter data for GLV1 and GLV2 but not GLV10. GLV1, GLV6, and GLV10 were highly expressed in the inflorescence, while the other genes had low or undetectable transcript levels. Reporter lines and qRT-PCR data matched in most cases. However, GLV10 transcripts were detected in the root, as determined with reporter lines, but also in other shoot tissues, suggesting that the 5′ promoter region analyzed in this study may not contain all the cis-regulatory sequences controlling GLV10 tissue specificity (Lee et al., 2006).
Figure 5.
Relative GLV transcript levels in different parts of the plant. GLV transcript levels were measured by qRT-PCR. The data are shown for two independent biological replicates ± se.
In a previous report, in situ hybridization analysis showed that the RGF1/GLV11 transcript is present in the QC and CCs, while RGF2/GLV5 and RGF3/GLV7 transcripts were mainly detected in the innermost layer of central CCs (Matsuzaki et al., 2010). These results are in accordance with the data obtained with the corresponding GLVpro::NLS-2xGFP lines (Fig. 1, A, B, and D). Finally, transcript-level data are available in public ATH1 microarray compendia for seven GLV genes: GLV1, GLV2, GLV3, GLV4, GLV6, GLV7, and GLV9 (Schmid et al., 2005; Winter et al., 2007; Hruz et al., 2008). As summarized in Table I and Figure 6, the expression profiles deduced from microarray data agreed in almost all cases with the patterns described above (Figs. 1–4). To conclude, taking the available data into consideration, our promoter-reporter lines reliably defined the expression domain of all but one GLV gene.
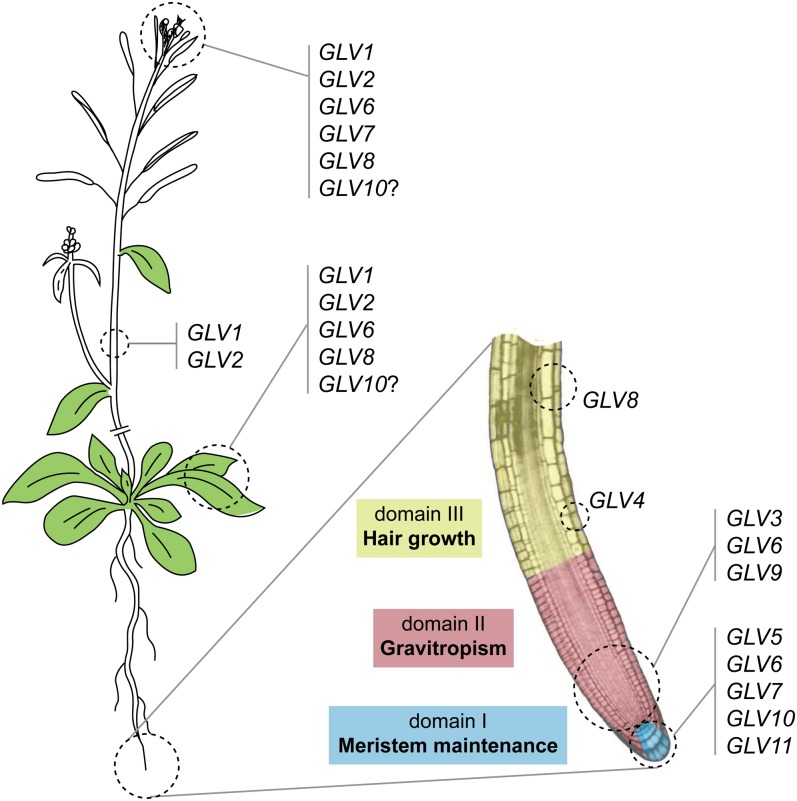
Figure 6.
Schematic representation of the GLV gene expression domains and functions.
gof Root Phenotypes Point to the Involvement of GLV Genes in Diverse Developmental Processes
Because genes coding for plant signaling peptides are in most cases part of multigene families, single lof mutants do not often display developmental defects due to redundancy. In such cases, gof mutants are useful to initiate the functional classification of these genes, for example in developmental processes. For this purpose, transgenic lines were created in which each of the GLV genes was overexpressed under the control of the Cauliflower mosaic virus 35S promoter. At least five independent single-locus lines were obtained per gene (referred to as GLVOE). We confirmed that the corresponding GLV transcript was up-regulated in most lines (Supplemental Table S2). To score defects in GLVOE mutants, seedlings were grown on inclined plates (1% agarose). We have previously reported gof phenotypes for the GLV1, GLV2, and GLV3 genes (Whitford et al., 2012). Transgenic Arabidopsis seedlings in which one of these genes was overexpressed showed a wavy-root phenotype when grown on inclined plates, which can be explained by a defective gravitropic response (Oliva and Dunand, 2007; Whitford et al., 2012). With this study, we continued the detailed analysis of root growth and development in Arabidopsis lines overexpressing one of the GLV1 to GLV11 genes (Table I).
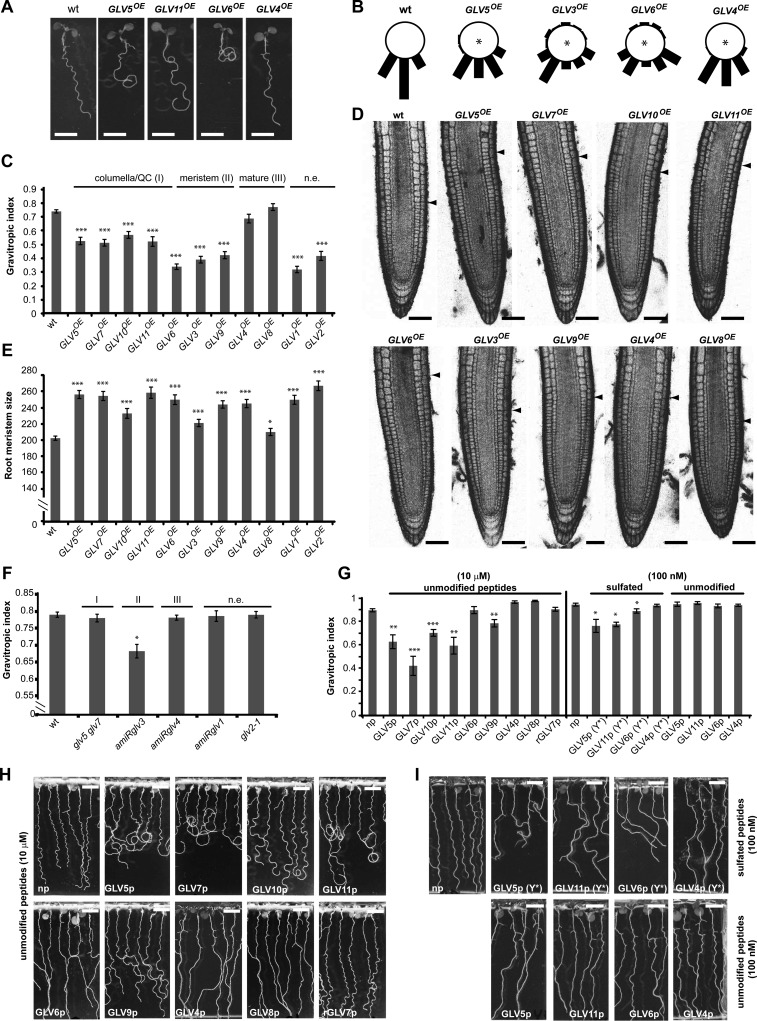
Overexpression of all GLV genes, except GLV4 and GLV8, markedly affected the growth of the primary root on the agar surface of inclined plates, resulting in dramatically larger and more irregular waves than in the wild type, and leading in many cases to root curling (Fig. 7A; Table I; data not shown). Root growth was quantified with the gravitropic index (GI), defined as the ratio of the linear distance connecting the colet and the root tip over the root length (Grabov et al., 2005). Two independent T3 homozygous lines were measured for each GLV gene (Fig. 7C). GI analysis showed that all GLV overexpression lines, except GLV4OE and GLV8OE (corresponding to domain III genes), have significant gravitropic growth defects and can be ranked according to phenotype severity. Overexpression of genes normally active in the meristematic zone (domain II) induced the strongest alteration: first GLV6, followed by GLV3 and GLV9. The phenotype of GLV5OE, GLV7OE, GLV10OE, and GLV11OE lines (domain I) was moderate. GLV1 and GLV2 overexpression also resulted in lower GI, although neither gene is expressed in the main root.
Figure 7.
GLV gof and lof root phenotypes. A, Representative GLVOE lines grown on inclined plates showing increasing defects in root gravitropic growth. B, Gravitropic response of GLVOE lines following gravistimulation. Stars indicate significant differences compared with the wild type (wt; for odds ratio estimates of root tip angle distribution, see Supplemental Table S5). C, Quantification of the GLVOE root growth phenotype according to the GI ± se (n = 17–45). For clarity, the results for only one of two analyzed independent lines are shown (for details, see “Materials and Methods”). D, Representative GLVOE root tips. Arrowheads show the boundary between the root meristem and the elongation zone. E, Quantification of root meristem size (μm) in GLVOE lines ± se (n = 14–84). For clarity, only one of two analyzed independent lines is shown. F, GI quantification of GLV lof mutants roots ± se (n = 25–30). G, GI quantification of wild-type roots treated with GLV-derived synthetic peptides ± se (n = 6–8), as shown in H and I. H and I, Root growth defects induced by GLV synthetic peptides. Wild-type roots were treated with either unmodified peptides at 10 μm (H) or 100 nm (I, bottom panel) or Tyr-sulfated peptides (Y*) at 100 nm (I, top panel). As indicated at the top of C, E, and F, GLV genes or peptides were grouped according to their expression domains within the root: I, columella/QC; II, meristem; III, maturation zone. n.e., Not expressed in the main root; np, no-peptide treatment; rGLVp, random GLVp. Stars indicate significant differences compared with the wild type (*P < 0.05, **P < 0.01, ***P < 0.001). Bars = 0.5 cm (A, H, and I) and 50 μm (D).
In addition, we studied the gravitropic response of selected GLVOE lines in experiments where vertical plates were reoriented by a 90° angle. The root curvature was measured 6 h after gravistimulation. The chosen GLVOE lines represented the different phenotypic classes as quantified with the GI (Fig. 7B) and included genes expressed in different parts of the root (Fig. 1). The results confirmed that some GLVOE lines did not respond properly to gravity, as reported previously for GLV3OE (Fig. 7B). In agreement with GI measurements, GLV3OE and GLV6OE lines had the strongest agravitropic phenotype, GLV5OE showed an intermediate defect, whereas GLV4 overexpression had little effect.
In summary, the distribution of the GLV genes in three gof phenotypic classes matched their corresponding domains of expression (compare Figs. 1 and 7C) and suggests that GLV genes active in domain II (GLV3, GLV6, and GLV9) are involved in gravitropic responses. To further test this hypothesis, we measured the GI of available GLV lof mutants grown under the same conditions and representing each defined expression domain: a double glv5 glv7 transfer DNA (T-DNA) insertion mutant (domain I); lines silenced for GLV3 (amiRglv3; domain II), GLV4 (amiRglv4; domain III), and GLV1 (amiRglv1; not expressed in the root); and the glv2-1 T-DNA insertion mutant (not expressed in the main root). Only amiRglv3 plants had a reduced GI (Fig. 7F), confirming the role of domain II GLV genes in gravitropism.
Previous reports showed that the overexpression of certain GLV genes also resulted in a larger RAM (Table I and refs. therein). We expanded this analysis to all the lines overexpressing GLV genes. The size of the RAM in the GLVOE lines was larger than that in the wild type, although GLV8OE displayed only mild defects (Fig. 7, D and E). The ranking and classification based on either GI or RAM size were not strictly equivalent (e.g. see GLV4OE phenotypes), suggesting that the GLV peptides may be recognized by distinct receptors and trigger different cellular responses.
Finally, we investigated root-branching defects in GLVOE lines, since 10 of the GLV genes were found to be expressed during LR development (Fig. 2A). In wild-type plants, several emerged LRs are already visible at 12 dag. In some GLVOE lines, however, only a few or no LRs were observed at that time, while root length was not significantly different between lines (Fig. 2, B and C; data not shown). Interestingly, the timing of GLV expression onset during LR initiation and emergence correlated in most cases with the defects observed in gof lines (i.e. overexpression of GLV genes transcribed at earlier stages resulted in a smaller number of LRs [Fig. 2, A and C], except for GLV2).
Treatments with GLV Peptides Mimic Overexpression Phenotypes
Treatment of roots with synthetic peptides encompassing the GLV conserved motif at the C terminus of GLV1, GLV2, and GLV3 preproproteins caused gravitropic defects (Meng et al., 2012; Whitford et al., 2012). On the other hand, roots grown on all GLV/RGF-derived peptides, except GLV6/RGF8p, resulted in the enlargement of the RAM (Matsuzaki et al., 2010), reminiscent of the corresponding overexpression phenotypes. Building on these previous studies, we wished to confirm that the functional domain in the other GLV precursors was also encoded in their C-terminal region by comparing peptide-induced and gof phenotypes. We analyzed the root growth pattern of wild-type roots transferred onto solid medium containing synthetic peptides (GLVp) corresponding to all members of the GLV family expressed in roots, except for GLV3p, whose effect on root growth was previously reported (Whitford et al., 2012). As a control, roots were also treated with randomized peptides (rGLVp), with identical amino acid content but a scrambled sequence.
For this assay, 3-dag wild-type seedlings were transferred on the surface of inclined plates in which the solid medium contained peptides at a concentration of 10 μm (for amino acid sequences, see Supplemental Table S4). Five days after transfer, GLVp-treated roots had wavy or agravitropic phenotypes, whereas roots grown without peptide or with randomized peptides did not (Fig. 7H). However, the severity of the peptide-induced growth pattern varied. From weakest to strongest: GLV8p, GLV6p, and GLV4p treatments resulted in a partial loss of wave formation; GLV9 and GLV10p induced exaggerated waving; and GLV11p, GLV5p, and GLV7p induced the formation of loops, indicative of a poor gravitropic response (Fig. 7, G and H). Most peptides induced the formation of waves and loops similar to those observed in GLVOE roots. Addition of GLV4p or GLV8p altered only slightly the root growth direction, in accordance with the phenotype of the corresponding overexpression lines. However, the addition of GLV6p had a similar effect, contradicting in this case the phenotype of the GLV6OE lines that showed the highest GI alteration. Also, opposite to the overexpression lines, peptides corresponding to genes in the meristematic region (GLV6 and GLV9) did not induce the strongest phenotype (Fig. 7, G and H).
Four endogenous mature GLV peptides have been shown to carry posttranslational modifications (GLV1, GLV2, GLV3, and GLV11/RGF1), and synthetic Tyr-sulfated versions of these peptides have a higher bioactivity than their unmodified counterparts (Matsuzaki et al., 2010; Whitford et al., 2012). Therefore, we tested whether additional GLV peptides carrying a sulfated Tyr, noted GLVp(Y*), may induce root growth defects at lower concentrations (100 nm) than the unmodified ones. The growth of roots treated with GLV4p(Y*) was not significantly altered. However, GLV5p(Y*) and GLV11p(Y*) induced a wavy phenotype similar to the unmodified peptides at higher concentrations (Fig. 7, G–I). Interestingly, GLV6p(Y*) treatment resulted in markedly slanted roots, in contrast to its unmodified counterpart, which did not significantly alter root growth at any of the assayed concentrations (Fig. 7, G and I).
We have noticed that peptide addition may affect root growth somewhat differently than overexpression of the corresponding gene (i.e. gene overexpression sometimes resulted in altered root waving, while peptide treatment instead induced root slanting). Such a discrepancy might be explained in several ways. The level of signaling peptides accumulating in the cellular microenvironment under the transcriptional control of the 35S cauliflower mosaic virus promoter can be higher than the local concentration of the synthetic peptides added exogenously. Alternatively, the maturation process might differ between GLV peptides, and some of them may require unknown posttranslational modifications to be active, for example, to bind to their cognate receptor protein with high affinity.
In summary, root growth phenotypes induced by Tyr-sulfated and unmodified GLV-derived peptides resemble in most cases those resulting from overexpression of the corresponding gene, indicating that the functional domain of the GLV protein is contained within the C-terminal portion of the precursor proteins. Furthermore, as reported in previous studies, synthetic peptides are more active when carrying a sulfated Tyr, confirming the importance of such posttranslational modifications for GLV functions (Fig. 7, G and I).
Effect of GLV Peptides on PIN2 Distribution in RAM Epidermal Cells
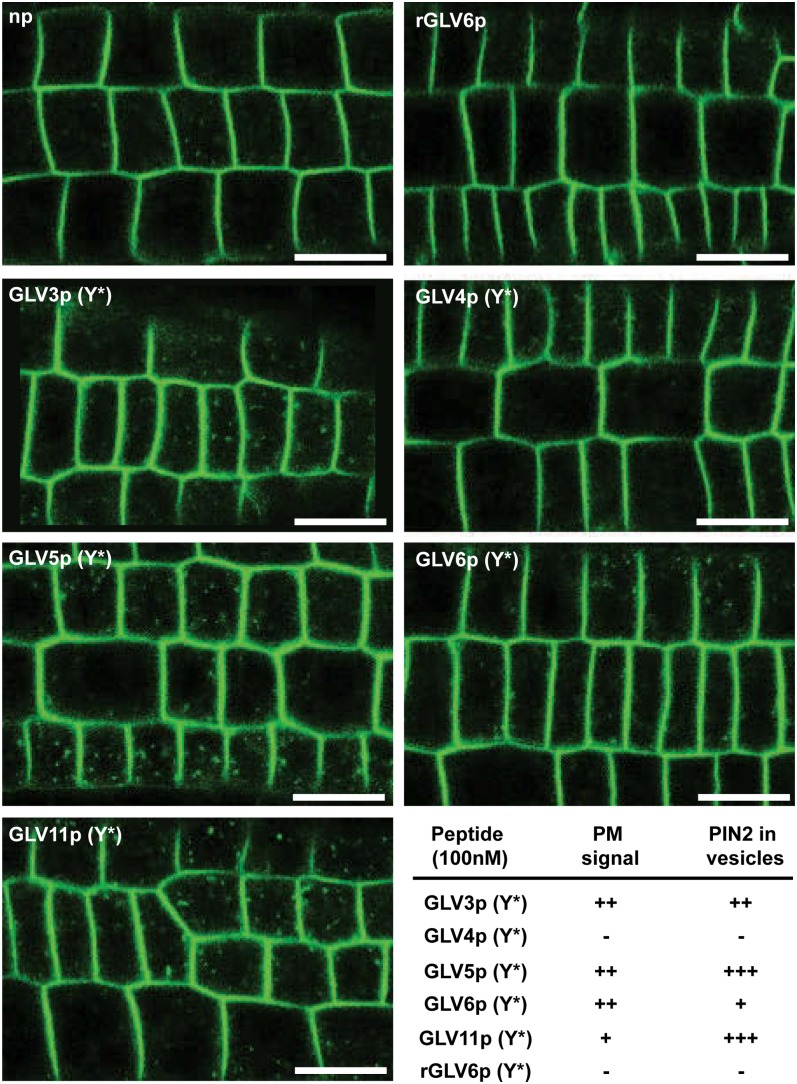
We previously reported that the GLV1 and GLV3 peptides induced a rapid increase in the level of PIN2 auxin efflux carrier in intracellular vesicles as well as in the plasma membrane (PM) of epidermal cells in the RAM (Whitford et al., 2012), thereby explaining how GLV gain of function prevents the formation of an asymmetric auxin gradient required for normal gravitropic responses. To study whether other GLV peptides have similar effects on PIN2 distribution, we analyzed the dynamics of the PIN2-GFP fusion protein immediately after peptide addition. PIN2pro:PIN2-GFP roots were treated with GLV peptides carrying a sulfated Tyr and derived from the GLV4, GLV5, and GLV6 precursor proteins (Supplemental Table S4). These were selected as representatives of each of the three main root expression domains (Table I). We also included RGF1/GLV11p as a reference to the work of Matsuzaki et al. (2010) and a random GLV6 peptide (rGLV6p) as a control.
The GLV3p-induced changes in PIN2-GFP signals associated with the PM and vesicles already reached a plateau 10 min after peptide treatment (Whitford et al., 2012). Therefore, the effect of the other GLV peptides was analyzed at the same time following peptide addition. Similar to the effect of GLV3p(Y*), GLV6p(Y*), GLV5p(Y*), and GLV11p(Y*), treatments resulted in the accumulation of PIN2-GFP in the PM (Fig. 8). Like GLV3p(Y*), GLV5p(Y*) and GLV11p(Y*) strongly induced PIN2 accumulation in intracellular vesicles, while GLV6p resulted in more subtle differences. GLV4p(Y*) and the rGLV6p control did not show any change in the PIN2-GFP signal. As summarized in Table I, the effect of synthetic GLV peptides on PIN2 distribution is consistent with their respective effect on root growth direction.
Figure 8.
GLV peptides alter PIN2 distribution. Roots expressing the PIN2-GFP fusion protein were treated with 100 nm of the indicated peptides. The image shows the GFP signal in RAM epidermal cells 10 min after peptide addition. Bars = 20 μm.
Root Hair Development Is Impaired in GLV gof and lof Plants
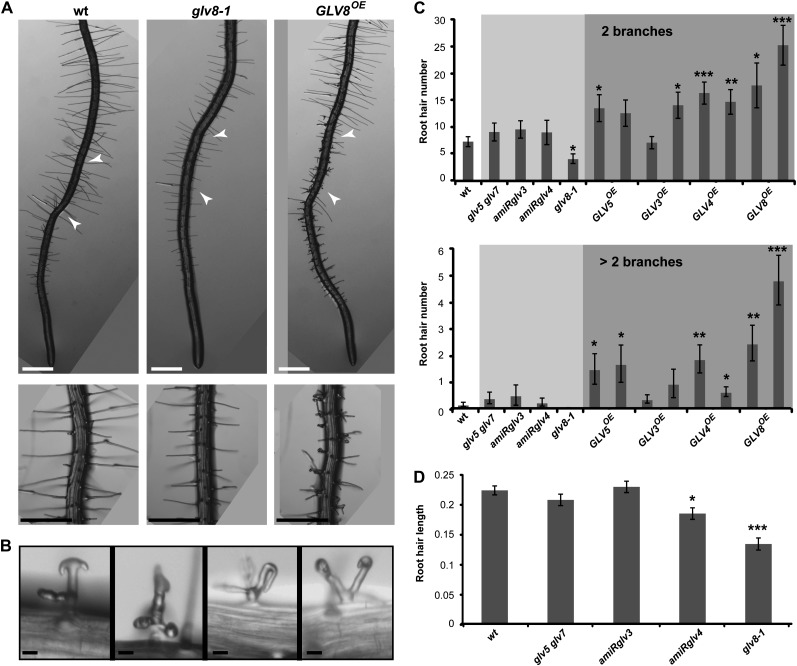
GLV8OE lines had a weak root meristem phenotype, and GLV8 transcription was restricted to the cortical and nonhair epidermal cells of the root maturation zone. Therefore, we examined whether GLV8 is involved in the developmental programs specific to these tissues, including the formation and growth of the root hairs (Tominaga-Wada et al., 2011). While some wild-type hairs branch during cell differentiation, they rarely form more than two prongs. In contrast, GLV8OE roots carried a large number of bifurcated and higher order branched root hairs (Fig. 9A). The GLV3OE, GLV4OE, and GLV5OE lines had also more branched hairs than the wild type, but always fewer than GLV8OE. The shape of GLV8OE root hairs was also sometimes aberrant, including bent tips (Fig. 9B).
Figure 9.
Root hair defects in GLV8 gof and lof mutants. A, Primary 4-dag roots of the indicated genotypes grown on agar medium. The bottom panels show root portions of the same genotypes at higher magnification. Arrowheads indicate the region with fully elongated root hairs, magnified in the bottom panels, where the root hair length was quantified. B, Abnormal root hair structures in GLV8OE roots. Bars = 0.5 mm (A) and 20 μm (B). C, Quantification of branched root hairs in the wild type (wt) and GLV mutants. The top panel refers to root hairs with two branches, and the bottom panel refers to root hairs with more than two branches. Light and dark shading indicate lof and gof lines, respectively. The chart shows data from two independent replicates ± se (n = 25–30). D, Quantification of root hair length (mm) in GLV lof mutants compared with the wild type. The chart shows data from two independent replicates ± se (n = 24–70). Stars indicate significant differences compared with the wild type (*P < 0.05, **P < 0.01, ***P < 0.001).
To confirm the role of GLV8 in root hair development, we analyzed hair morphology in lof mutant plants. The SALK_054452 insertional line carries a T-DNA in the GLV8 promoter region approximately 80 bp before the start codon. The GLV8 transcript could not be detected by qRT-PCR in this line, hereafter renamed glv8-1 (Supplemental Table S2). In glv8-1 mutant roots, the hairs were shorter and the number of branched hairs was reduced compared with the wild type (Fig. 9, C and D). The other tested lof mutants, comprising lines silenced for GLV3 (amiRglv3) or GLV4 (amiRglv4), and a double glv5 glv7 T-DNA insertion mutant had a normal number of branched hairs (Fig. 9C). While root hair length was not affected in the amiRglv3 and glv5 glv7 mutants, amiRglv4 plants had shorter root hairs than the wild type, but this phenotype was weaker than in glv8-1 (Fig. 9D). Because GLV4 is expressed in the epidermal cells of the elongation and proximal maturation zones, it may act redundantly with GLV8 in that region of the root.
In summary, both the peculiar expression patterns of GLV4 and GLV8 and the morphological analysis of related mutants indicate that GLV signaling is also involved in root hair development.
DISCUSSION
Definition of the GLV Expression Domains
Transcription pattern analysis with promoter-reporter lines provides high-resolution expression maps, including for genes not represented in the ATH1 GeneChip. In the case of the GLV/RGF/CLEL family, ATH1 transcript profile data are available for only seven of the 11 genes (GLV1, GLV2, GLV3, GLV4, GLV6, GLV7, and GLV9). This study showed that nine of them are transcribed in the Arabidopsis primary root with specific patterns, grouped in three distinct domains: I, the QC and CCs; II, the root meristem; and III, the maturation zone (Fig. 6). Furthermore, transcription was also detected in the course of LR development for 10 GLV genes, again each with specific patterns. Finally, localized GLV expression was detected in shoot tissues and in the inflorescence for five GLV members (Fig. 6).
Based on in situ hybridization, Matsuzaki et al. (2010) reported that GLV11/RGF1, GLV5/RGF2, and GLV7/RGF3 are transcribed within the QC and CCs. The data we obtained with the corresponding GLV promoter-reporter fusions corroborate these results. Taking into consideration our qRT-PCR results and expression data available in public microarray compendia, it is likely that transcripts from additional GLV genes expressed at low levels failed to be detected by in situ hybridization. In comparison, the promoter fusions used in this study driving the accumulation of double GFP proteins in the nucleus may provide better sensitivity. Alternatively, the GLV transcripts may be unusually unstable.
GLV Functions Are Difficult to Dissect Genetically But May Be Classified According to Expression Domains
The diversity of GLV expression patterns suggests that GLV peptides relay cell-to-cell signals in diverse developmental processes. However, several members of the family are transcribed in close or overlapping domains, implying that single lof mutants may not yield altered phenotypes because of genetic redundancy. The combination of multiple GLV lof mutations is not trivial, because signaling peptides are encoded by small-size genes and T-DNA insertion lines are lacking for some of them. Furthermore, silencing multiple GLV genes with a single artificial mRNA (Schwab et al., 2006) is not possible, because they share little DNA sequence homology and only in short stretches. Therefore, the comparative analysis of gof mutant lines is a useful approach to gain the first insights into GLV gene function.
Our phenotypic classification focused on root gof defects detected in young seedlings. Although older soil-grown GLVOE plants were not exhaustively characterized, we did not observe any obvious phenotypes in their aerial parts. However, expression data suggest that a closer inspection is needed to investigate the involvement of GLV signaling in developmental processes that take place in the shoot.
The overexpression of most GLV genes resulted in root gravitropic defects as well as increased meristem size, probably because the corresponding mature peptides bind identical receptor(s), as reported for other peptide families (Ogawa et al., 2008). Thus, multiple GLV genes are likely to carry partly redundant signals during gravitropic responses (Whitford et al., 2012; this report) and in meristem maintenance mechanisms (Matsuzaki et al., 2010; this report), according to their expression pattern in the RAM. The severity of the phenotypes varied, possibly reflecting ligand-receptor affinity and sequence similarity between mature GLV peptides. Based on GI measurements, GLV genes could be classified into functional groups corresponding to their expression domains, but this was not strictly the case when RAM sizes were compared. These results suggest that two different receptors might relay GLV signals involved in gravitropic responses and RAM maintenance and that one may be more specific or saturable than the other. Furthermore, because they do not act cell autonomously, secreted peptide signals may consist of integrated gradients resulting from the addition of redundant molecules encoded by genes with overlapping, but distinct, expression profiles.
Although gof studies, via overexpression or peptide application, cannot determine the precise contribution of each GLV family member to specific processes, our results indicate that a larger number than initially reported may trigger the same pathway. For example, RGF1/GLV11, RGF2/GLV5, and RGF3/GLV7 were shown to control meristem maintenance (Matsuzaki et al., 2010). But GLV6 and GLV10 are also expressed in the QC and CCs, and the corresponding gof lines also displayed increased RAM size, suggesting redundancy.
Similarly, GLV3, GLV6, and GLV9 may all be involved in root gravitropism because they have similar expression domains and gof phenotype as indicated by their GI. An important site for the response to a gravistimulus is the RAM epidermal cell layer, in which the GLV signal controls the level of PIN2 in the PM (Abas et al., 2006; Whitford et al., 2012). In that framework, the closest cells expressing GLV genes are those found in the inner root tissues defining domain II within the RAM, where GLV3, GLV6, and GLV9 are transcribed. Interestingly, the overexpression of these genes led to the strongest defects in GI, possibly reflecting a high affinity of the encoded signaling peptides for their cognate receptor(s). Furthermore, only the down-regulation of GLV3 resulted in a lower GI than the wild type, while the loss of function of GLV genes expressed in other domains of the root did not. These results support a model in which the signal encoded by the GLV3, GLV6, and GLV9 peptides, secreted from inner cell layers, is perceived in the epidermis, where it regulates the differential root tip growth resulting in root gravibending and waving. Interestingly, GLV1 and GLV2 are involved in the regulation of the gravitropic curvature in the hypocotyl (Whitford et al., 2012), and the molecular pathways downstream of the GLV signals in the two organs may be related.
GLV Signals Are Involved in LR Formation
As recently reported for two members of the gene family (GLV1/CLEL6 and GLV10/CLEL7; Meng et al., 2012), overexpression resulted in impaired LR development, suggesting that GLV signals take part in this process. Our transcriptional and mutant analysis confirms that this indeed is the case. Interestingly, with the exception of GLV2, genes transcribed at early stages of LR initiation and yielding the strongest gof LR inhibition phenotype are also active at the core of the RAM, including the CCs, the QC, and the surrounding initials (domain I). GLV genes with lower or no inhibitory activity are either transcribed in tissues farther up in the primary root or absent from it. Remarkably, GLV5, GLV6, GLV7, GLV10, and GLV11 expressed in domain I in the primary root also showed increased RAM size when overexpressed (Matsuzaki et al., 2010; Meng et al., 2012; this report), suggesting that they may control meristematic activity in both the pericycle and the RAM. Because the expression of GLV genes is turned on at successive stages of LR formation, the corresponding peptides may be involved in different steps of the developmental program or may have an additive effect. By analogy with other signaling ligands, GLV peptides may be perceived by membrane receptor kinases involved in LR initiation. In that context, ARABIDOPSIS CRINKLY4 is an interesting candidate because it is expressed at the core of the LR primordium and controls cell division in the pericycle cell layer at the early stages of LR development (De Smet et al., 2008).
The GLV8 Signal Regulates Root Hair Development
GLV8 gain of function resulted in no or minor primary root growth defects. Furthermore, GLV8 is transcribed in the maturation zone, specifically in cortical and nonhair cells, outside the root meristem. Thus, this gene may have evolved to fulfill specific functions different from its paralogous counterparts active in the RAM and in the course of LR initiation. In Arabidopsis, the differentiation of root epidermal cells into root hair or nonhair cells depends on their position relative to the underlying cortical cells. Root hair cells are in contact with two cortical cells, whereas epidermal cells in contact with only one cortical cell remain hairless (Dolan et al., 1994). The peculiar transcription pattern of GLV8 prompted us to search for root hair phenotypes in related mutant lines. The GLV8 gof and lof phenotypes are partly opposite: GLV8OE lines produced hairs with more complex and irregular shapes, whereas glv8-1 root hairs were simpler and shorter than those in the wild type. Like GLV8, GLV4 is absent from the meristem, and the expression of both genes overlaps in the proximal maturation zone. In addition, GLV4 silencing also resulted in shorter root hairs.
These observations indicate that GLV signaling is involved in the formation of the root hairs, possibly through the perception of the GLV4 and GLV8 mature peptides secreted by epidermal and cortical cells. Such ligands may bind receptors present in the PM of the hair cells and regulate hair development (Duan et al., 2010). For example, the SCRAMBLE (SCM) Leu-rich repeat receptor-like kinase has been shown to be required for epidermal cell specification (Kwak et al., 2005). SCM is supposed to perceive a signal secreted from the cortex that would relay positional cues to epidermal cells, determining their hair or nonhair fate. Accordingly, the SCM gene is expressed very early during development. However, GLV4 and GLV8 are only transcribed in the elongation and maturation zones of the root, where epidermal cells are already differentiated. In addition, the organization of the hair and nonhair cell files is not affected in GLV8OE and glv8-1 roots (data not shown), and it is thus unlikely that the encoded peptides act as a paracrine signal binding SCM to specify the hair cell fate. Instead, they may be involved in root hair growth. By analogy with the defects observed in PIN2 trafficking in the RAM upon GLV peptide addition, GLV4 and GLV8 may control the vesicular trafficking necessary to sustain the hair tip growth.
GLV Functions and Auxin Signaling
The molecular mechanisms downstream of GLV perception in the LR meristem and the epidermal cells of the root maturation zone remain to be analyzed. We speculate that similar processes may be at play as described in the root meristem, where GLV signaling has been shown to regulate the intracellular vesicle trafficking and PM concentration of the PIN2 auxin efflux carrier, thereby affecting the formation of auxin gradients in the root tip (Whitford et al., 2012). Likewise, changes of auxin concentration at the core of the LR primordium (De Rybel et al., 2010) or in root hair cells (Pitts et al., 1998; Rahman et al., 2002; Jones et al., 2009) depending on GLV activity may control their respective development.
CONCLUSION
We have conducted an exhaustive analysis of the domains of expression for each member of the GLV family coding for signaling peptides in Arabidopsis. We have also studied related gof and lof phenotypes focusing on root development. The combined data sets suggest that nine GLV genes form three subgroups according to their expression and function within the root (Fig. 6). Our study provides useful tools to predict the functions of specific GLV genes in particular developmental processes, as demonstrated in the case of LR and root hair formation.
MATERIALS AND METHODS
Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were sown on one-half-strength Murashige and Skoog (MS) medium (Duchefa Biochemie) supplemented with 1.5% (w/v) Suc and 1% (w/v) agarose, pH 5.8, and stratified for 2 d at 4°C before germination. Seedlings were germinated in climate-controlled growth chambers at 22°C under long-day conditions (100 μmol m−2 s−1). To score root growth defects and GI measurements, seedlings were grown on plates inclined at an angle of approximately 45°. For gravistimulation experiments, 4-dag light-grown seedlings germinated on vertical plates were placed in the dark and reoriented by a 90° angle.
Recombinant DNA Constructs and Arabidopsis Lines
In most cases, the reporter protein driven by a GLV promoter was either a double GFP fused to a nuclear localization signal (NLS-2xGFP) or a translational fusion between GUS and GFP (GUS-GFP). In GLVpro::NLS-2xGFP lines, transcriptionally active cells were marked with a bright fluorescent nucleus, yielding high spatial resolution and high sensitivity in confocal microscopy analysis. However, GLVpro::GUS-GFP lines were preferred to study expression patterns in shoot tissues to avoid chlorophyll autofluorescence. In some cases, the reporter gene consisted of an open reading frame (ORF) coding for a nuclear GFP-GUS fusion protein (GLVpro::NLS-GFP-GUS lines; for additional information, see Supplemental Table S1). Absence of a reporter line in Figures 2 to 4, documenting GLV expression, indicates that transcriptional activity was not detected for the corresponding gene in the studied plant organ.
The GLV1pro::GUS-GFP, GLV1pro::NLS-GFP-GFP, GLV2pro::GUS-GFP, GLV2pro::NLS-GFP-GFP, and GLV3pro::GUS-GFP lines were described previously (Whitford et al., 2012). The GLV4 to GLV11 promoter sequences (700–1,700 bp upstream of the ORF initiation codon) were PCR amplified with Arabidopsis genomic DNA as template. Primers were designed so that the promoter amplicons carried a BamHI restriction site at the 5′ end and a SalI (GLV5, GLV6, GLV7, GLV9, GLV10) or XhoI (GLV4, GLV8, GLV11) site at the 3′ end (Supplemental Table S1). Each promoter amplicon was digested with BamHI and SalI or XhoI enzymes and ligated into pEN-L4-R1 digested with the same restriction enzymes, resulting in eight GLV promoter entry clones (pEn-L4-GLVpro-R1; Karimi et al., 2007; Fernandez et al., 2009). The pEN-L4-R1 vector carries a multiple cloning site flanked by the attL4 and attR1 Gateway recombination sites (attL4-XmnI-SalI-BamHI-KpnI-ccdB-XhoI-attR1; O. Nyabi, M. Karimi, P. Hilson, and J. Haigh, unpublished data). The GLV promoters were subcloned upstream of reporter genes via MultiSite LR clonase reaction (Invitrogen; http://www.invitrogen.com/) in the pK7m34GW destination vector (Karimi et al., 2005). GLVpro::GFP-GUS expression clones were created by combining pEn-L4-GLVpro-R1 with pEn-L1-F-L2 (GFP) and pEn-R2-SI-L3 (GUS with intron). GLVpro::NLS-GFP-GUS expression clones were created by combining pEn-L4-GLVpro-R1 with pEnL1-NF-L2 (nuclear GFP) and pEn-R2-SI-L3. GLVpro::NLS-GFP-GFP expression clones were created by combining pEn-L4-GLVpro-R1 with pEnL1-NF-L2 and pEn-R2-F-L3.
GLV1OE, GLV2OE, GLV3OE, amiRglv1, glv2-1, and amiRglv3 lines were described previously (Whitford et al., 2012). The GLV4 to GLV11 ORF sequences were PCR amplified with primers carrying attB1 (5′) and attB2 (3′) sites (Supplemental Table S3) and Arabidopsis complementary DNA (cDNA) as template and captured by BP clonase reaction in an entry clone (pEN-L1-GLV-L2) derived from pDONR221. Overexpression constructs were obtained by LR recombination between pEN-L1-GLV-L2 and the destination vector pK7GW2 (Karimi et al., 2002). GLV4 was silenced by expression of an artificial microRNA constructed with the online tool WMD3 (http://wmd3.weigelworld.org/; Ossowski et al., 2008) and the primers Glv4I-miR-s (5′-GATTTAACCATAATTTGCCCGGCTCTCTCTTTTGTATTCC-3′), glv4II-miR-a (5′-GAGCCGGGCAAATTATGGTTAAATCAAAGAGAATCAATGA-3′), glv4III-miRs (5′-GAGCAGGGCAAATTAAGGTTAATTCACAGGTCGTGATATG-3′), and glv4IV-miRa (5′-GAATTAACCTTAATTTGCCCTGCTCTACATATATATTCCT-3′). All constructs were transformed into the Columbia-0 accession by Agrobacterium tumefaciens (strain C58C1) flower dip (Clough and Bent, 1998). In all experiments, the wild-type accession was Columbia-0.
The mutant lines glv8-1 (SALK_054452), glv5-1 (same as rgf2-1; Matsuzaki et al., 2010; SALK_145834), and glv7-1 (same as rgf3-1; Matsuzaki et al., 2010; SALK_053439) were obtained from the European Arabidopsis Stock Center. The presence of the relevant T-DNA was confirmed by PCR with a primer corresponding to the T-DNA left border, GLV gene-specific primers, and genomic DNA as template. The relevant T-DNA is in the promoter of glv8-1 and in an intron in glv5-1 and glv7-1. The glv5 and glv7 lines were crossed to obtain the glv5 glv7 double mutant.
qRT-PCR Analysis
To determine GLV expression levels in different plant organs, 5-dag seedlings were split in two: the shoot part (above the colet) and the whole main root (below the colet). Whole 5-dag seedlings were also included in the analysis. The first/second leaves were harvested in 14-dag seedlings, and inflorescences (approximately 30 dag) were cut approximately 3 cm below the apex. Total RNA was isolated with Trizol (Invitrogen). Residual DNA contaminants were removed by treating the RNA samples with RNase-free DNase (Roche). One microgram of RNA from each sample was used as a template to synthesize the first cDNA strand with the iScript cDNA Synthesis Kit (Bio-Rad). Expression levels were analyzed by qRT-PCR. Reactions were performed on 384-well plates with a LightCycler real-time thermocycler (Roche) and SYBR Green to monitor double-stranded DNA synthesis. Each quantitative PCR was performed in three technical replicates. Data were analyzed with the “delta-delta method” (Pfaffl, 2001), taking primer efficiency into consideration, and normalized with UBIQUITIN, CKIIa2, and CDKA as reference transcripts. The sample with the maximum value for each gene was chosen as the calibrator (set to 1), and the results of two biological replicates were averaged.
Expression levels of GLV genes in gof and lof Arabidopsis lines were quantified in a similar way. Transcript fold change was calculated with respect to the wild type. At least five independent T3 homozygous lines were obtained for each construct. Supplemental Table S2 indicates the level of GLV transcripts in the glv T-DNA insertional mutants and independent gof and lof T3 homozygous transgenic lines that were characterized phenotypically.
Peptide Treatment
GLV peptides derived from the C-terminal conserved region of the GLV preproprotein were either obtained from a commercial provider (GenScript; http://www.genscript.com/) or synthesized in house as described previously (Whitford et al., 2012). To study root phenotypes induced by peptide treatment, seedlings were germinated for 3 d on vertical plates without peptide and then transferred for 4 to 5 d onto fresh plates with medium containing a peptide at the indicated concentration.
Microscopic Analysis
PIN2pro:PIN2-GFP seedlings (Xu and Scheres, 2005) were germinated on solid MS medium and treated for 10 min in liquid MS medium containing no peptide or supplemented with 100 nm Tyr-sulfated GLV peptides. Immediately after treatment, seedlings were mounted in the same liquid medium on a microscopic glass slide and imaged on a Zeiss Axiovert 100M confocal laser scanning microscope with the software package LSM 510 (version 3.2; Zeiss) equipped with 25-mW argon and 2-mW helium-neon lasers. Other images were taken with an Olympus FluoView FV1000 microscope. GFP signals were detected with a 488-nm filter for excitation and 520 nm for detection. Cell membranes were counterstained with propidium iodide (PI) and imaged with a 543-nm filter for excitation and 590 to 620 nm for detection. For the three-dimensional reconstruction of root tissues, longitudinal sections of root segments (approximately 300 μm long) were imaged with a 40× objective across one-half of the root cylinder (from epidermis to vasculature). The Z-stack step size was 1 to 1.5 μm. Movies were generated with the Volocity software (version 6.2; Perkin-Elmer) to show successive transverse planes and to visualize the nuclear GFP marker in the different root cell files.
Tissues were stained for GUS activity as described previously (Beeckman and Engler, 1994), and stained plants were analyzed and imaged with either a binocular Leica microscope or an Olympus microscope (DIC-BX51). Photographs were taken with a CAMEDIA C-3040 zoom digital camera (Olympus).
Morphological Analysis
Imaged root features were measured with the ImageJ software (http://rsbweb.nih.gov/ij/). Two independent T3 homozygous lines were analyzed for each GLV transgenic line in all quantification experiments. The phenotype was very similar in the two lines analyzed, indicating that the GLV transcript levels are saturating in both of them. Therefore, for clarity in Figure 7, only one is shown. Meristem size was measured in GLVOE seedlings germinated on solid medium. For this purpose, 5-dag roots were stained with PI for 2 min, mounted in water, and directly imaged with the confocal microscope. For GI calculation, the root length and the distance between the colet and the root tip were measured in 7-dag seedlings grown on inclined plates. Root hairs of 4- to 5-dag seedlings were imaged with a binocular Leica microscope. Root hair length was quantified in a 1-mm segment, approximately at the same distance from the root tip, where hairs are fully elongated.
Statistical Analysis
Statistical differences between mutant lines and the wild type were assessed with Student’s t test. Results were obtained by pooling data from two independent experiments. Error bars show se. In gravistimulation experiments, statistical differences were analyzed using a regression model as described previously (Whitford et al., 2012; Supplemental Table S5): *P < 0.05, **P < 0.01, ***P < 0.001.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. GLV primers for amplification of promoter sequences.
Supplemental Table S2. Transcript levels in GLV mutant lines.
Supplemental Table S3. GLV primers for amplification of ORF sequences.
Supplemental Table S4. Synthetic peptides.
Supplemental Table S5. Odds ratio estimates of root tip angle distribution.
Supplemental Movie S1. Expression of GLV4 in the root epidermal cells.
Supplemental Movie S2. Expression of GLV8 in the root cortical and nonhair epidermal cells.
Acknowledgments
We thank Annick Bleys for help in preparing the manuscript, Rebecca De Clercq and Wilson Ardiles-Diaz for assistance with gene constructs and transgenic lines, Nicholas Provart and Jamie Waese for the SVG image of an Arabidopsis plant, and Eef Parthoens for help with confocal microscopy.
Glossary
- lof
loss-of-function
- gof
gain-of-function
- RAM
root apical meristem
- QC
quiescent center
- CC
columella cell
- dag
d after germination
- LR
lateral root
- SAM
shoot apical meristem
- cDNA
complementary DNA
- qRT
quantitative reverse transcription
- GI
gravitropic index
- PM
plasma membrane
- MS
Murashige and Skoog
- ORF
open reading frame
- PI
propidium iodide
- T-DNA
transfer DNA
References
- Abas L, Benjamins R, Malenica N, Paciorek T, Wiśniewska J, Moulinier-Anzola JC, Sieberer T, Friml J, Luschnig C. (2006) Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat Cell Biol 8: 249–256 [DOI] [PubMed] [Google Scholar]
- Beeckman T, Engler G. (1994) An easy technique for the clearing of histochemically stained plant tissue. Plant Mol Biol Rep 12: 37–42 [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko MA, Vie AK, Brembu T, Aalen RB, Bones AM. (2009) Plant peptides in signalling: looking for new partners. Trends Plant Sci 14: 255–263 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S, et al. (2010) A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol 20: 1697–1706 [DOI] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, Levesque MP, Grunewald W, Van Damme D, Van Noorden G, Naudts M, Van Isterdael G, De Clercq R, et al. (2008) Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322: 594–597 [DOI] [PubMed] [Google Scholar]
- Dolan L, Duckett CM, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig S, Roberts K. (1994) Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120: 2465–2474 [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu H-M. (2010) FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA 107: 17821–17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AI, Viron N, Alhagdow M, Karimi M, Jones M, Amsellem Z, Sicard A, Czerednik A, Angenent G, Grierson D, et al. (2009) Flexible tools for gene expression and silencing in tomato. Plant Physiol 151: 1729–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, van der Schors R, van der Geest L, Li KW, Stiekema WJ, Liu C-M. (2006) The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking sequences. Plant Physiol 141: 1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu C-M. (2005) The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17: 2542–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabov A, Ashley MK, Rigas S, Hatzopoulos P, Dolan L, Vicente-Agullo F. (2005) Morphometric analysis of root shape. New Phytol 165: 641–651 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Kramer EM, Knox K, Swarup R, Bennett MJ, Lazarus CM, Leyser HMO, Grierson CS. (2009) Auxin transport through non-hair cells sustains root-hair development. Nat Cell Biol 11: 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J, Fiume E, Roeder AHK, Meng L, Sharma VK, Osmont KS, Baker C, Ha CM, Meyerowitz EM, Feldman LJ, et al. (2010) Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol 154: 1721–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Bleys A, Vanderhaeghen R, Hilson P. (2007) Building blocks for plant gene assembly. Plant Physiol 145: 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P. (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kwak S-H, Shen R, Schiefelbein J. (2005) Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307: 1111–1113 [DOI] [PubMed] [Google Scholar]
- Lease KA, Walker JC. (2006) The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol 142: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-Y, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN. (2006) Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA 103: 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y. (2011) Small post-translationally modified peptide signals in Arabidopsis. The Arabidopsis Book 9: e0150, doi/10.1199/tab.0150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. (2010) Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329: 1065–1067 [DOI] [PubMed] [Google Scholar]
- Meng L, Buchanan BB, Feldman LJ, Luan S. (2012) CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. Proc Natl Acad Sci USA 109: 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Smith S, De Smet I. (2012) Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. Plant Cell 24: 3198–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. (2008) Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294. [DOI] [PubMed] [Google Scholar]
- Oliva M, Dunand C. (2007) Waving and skewing: how gravity and the surface of growth media affect root development in Arabidopsis. New Phytol 176: 37–43 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D. (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts RJ, Cernac A, Estelle M. (1998) Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J 16: 553–560 [DOI] [PubMed] [Google Scholar]
- Rahman A, Hosokawa S, Oono Y, Amakawa T, Goto N, Tsurumi S. (2002) Auxin and ethylene response interactions during Arabidopsis root hair development dissected by auxin influx modulators. Plant Physiol 130: 1908–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strabala TJ, O’Donnell PJ, Smit A-M, Ampomah-Dwamena C, Martin EJ, Netzler N, Nieuwenhuizen NJ, Quinn BD, Foote HCC, Hudson KR. (2006) Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol 140: 1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga-Wada R, Ishida T, Wada T. (2011) New insights into the mechanism of development of Arabidopsis root hairs and trichomes. In WJ Kwang, ed, International Review of Cell and Molecular Biology, Vol 286. Academic Press, San Diego, pp 67–106 [DOI] [PubMed]
- Whitford R, Fernandez A, De Groodt R, Ortega E, Hilson P. (2008) Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc Natl Acad Sci USA 105: 18625–18630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford R, Fernandez A, Tejos R, Pérez AC, Kleine-Vehn J, Vanneste S, Drozdzecki A, Leitner J, Abas L, Aerts M, et al. (2012) GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Dev Cell 22: 678–685 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Scheres B. (2005) Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17: 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]