Summary: The mitochondrial folylpolyglutamate synthetase DFC is involved in nitrogen utilization through its participation in photorespiration during early seedling development in Arabidopsis.
Abstract
Investigations into the biochemical processes and regulatory mechanisms of nitrogen (N) utilization can aid in understanding how N is used efficiently in plants. This report describes a deficiency in N utilization in an Arabidopsis (Arabidopsis thaliana) transfer DNA insertion mutant of the mitochondrial folylpolyglutamate synthetase gene DFC, which catalyzes the conjugation of glutamate residues to the tetrahydrofolate during folate synthesis. The mutant seedlings displayed several metabolic changes that are typical of plant responses to low-N stress, including increased levels of starch and anthocyanin synthesis as well as decreased levels of soluble protein and free amino acid, as compared with those in wild-type seedlings when external N was sufficient. More striking changes were observed when dfc seedlings were grown under N-limited conditions, including shorter primary roots, fewer lateral roots, higher levels of glycine and carbon-N ratios, and lower N content than those in wild-type seedlings. Gene expression studies in mutant seedlings revealed altered transcript levels of several genes involved in folate biosynthesis and N metabolism. The biochemical and metabolic changes also suggested that N assimilation is drastically perturbed due to a loss of DFC function. The observation that elevated CO2 partly rescued the dfc phenotypes suggests that the alterations in N metabolism in dfc may be mainly due to a defect in photorespiration. These results indicate that DFC is required for N utilization in Arabidopsis and provide new insight into a potential interaction between folate and N metabolism.
Nitrogen (N) is an essential macronutrient for plants and a major limiting factor for crop growth (Diaz et al., 2006). Investigations into the biochemical processes and regulatory mechanisms of N utilization can aid in understanding how N is used efficiently in plants. Low inorganic N results in numerous perturbations in plant metabolism, such as decreases in nitrate (NO3−), Gln, malate, and fumarate content, a decrease in the Gln-Glu ratio, reduced nitrate reductase and phosphoenolpyruvate carboxylase activities, higher Gln synthetase and Glu dehydrogenase activities, and a greater concentration of starch in leaf rosettes (Diaz et al., 2008; Lemaître et al., 2008). An imbalance between carbon (C) fixation and N assimilation caused by N limitation leads to exogenous senescence-triggering factors that facilitate systematic N remobilization (Lemaître et al., 2008; Tschoep et al., 2009). Seedling-stage responses to low-N stress are particularly striking (Martin et al., 2002).
In recent years, considerable effort has been exerted toward understanding the molecular mechanisms underlying N uptake and utilization as well as adaptations to low-N conditions in plants. For example, nitrate transporters and genes downstream of N nutrient receptors are affected by low N and have been identified in analyses of root system architecture (Zhang and Forde, 1998; Gan et al., 2005; Krouk et al., 2006; Remans et al., 2006; Vidal and Gutiérrez, 2008; Ho et al., 2009; Hu et al., 2009; Vert and Chory, 2009; Vidal et al., 2010). Additionally, the expression of many other genes involved in various biological processes, including glutamate receptor1.1, E3 ubiquitin ligase, nodule inception-like protein, and even microRNAs, are altered during adaptation to N-limited conditions (Kang and Turano, 2003; Peng et al., 2007b; Castaings et al., 2009; Pant et al., 2009; Zheng, 2009). Quantitative trait locus mapping has identified several genetic loci involved in response to low-N stress (Loudet et al., 2003; Diaz et al., 2006). Additionally, genomics-based approaches have been used to characterize global plant responses to N limitation. The data thus generated suggest that many genes involved in photosynthesis, chlorophyll synthesis, plastidic protein synthesis, secondary metabolism, RNA synthesis and processing, amino acid activation, and protein synthesis are either repressed or induced under N deprivation (Scheible et al., 2004; Bi et al., 2007; Peng et al., 2007a; Gutiérrez et al., 2008).
N metabolism during early seedling establishment is critical for plants to become N- and C-autotrophic seedlings (Gaufichon et al., 2010). These processes are accompanied by many distinct metabolic, cellular, and physiological changes (Boyes et al., 2001; Andre and Benning, 2007). Folate synthesis also plays an important role during the transition from heterotrophic to photoautotrophic growth during seedling development (Jabrin et al., 2003). Folate is synthesized preferentially in highly dividing tissues and in photosynthetic leaves during seedling organ differentiation. For example, folate accumulates in pea (Pisum sativum) cotyledons and embryos during seed germination, and the initial folate content even supports root elongation during the first 1 to 2 d in the presence of sulfanilamide, an inhibitor of folate biosynthesis (Gambonnet et al., 2001).
Much progress has been made toward understanding the biochemistry of plant enzymes that are involved in folate biosynthesis and turnover, and consequently, the biosynthetic pathway of folates in plants has been constructed (Blancquaert et al., 2010; Hanson and Gregory, 2011). But few reports provided genetic evidence of the in planta functions of these folate biosynthesis enzymes during seed and seedling development (Ravanel et al., 2001; Ishikawa et al., 2003; Mehrshahi et al., 2010; Srivastava et al., 2011). For example, Arabidopsis (Arabidopsis thaliana) DIHYDROFOLATE SYNTHETASE FOLYLPOLYGLUTAMATE SYNTHETASE (DHFS-FPGS) HOMOLOG A (DFA) is a single-copy gene that encodes a functional mitochondrial matrix-localized dihydrofolate synthetase, which catalyzes the addition of the first glutamyl side chain to dihydropteroate to form dihydrofolate, and the globular arrest1 mutant, which carries a DFA mutation, exhibits defective embryo development (Ravanel et al., 2001; Ishikawa et al., 2003). Glu residues are attached to folate molecules via the catalytic activity of FPGS, which is specified by three FPGS genes in Arabidopsis, DHFS-FPGS HOMOLOG B (DFB), DHFS-FPGS HOMOLOG C (DFC), and DHFS-FPGS HOMOLOG D (DFD), which encode the plastidic, mitochondrial, and cytosolic isoforms, respectively (Ravanel et al., 2001; Mehrshahi et al., 2010). Different double-mutant combinations of the Arabidopsis FPGS isoforms are known to result in dramatic developmental abnormalities, including embryo lethality, seedling lethality, late flowering, and dwarf plants with various reproductive defects (Mehrshahi et al., 2010). In addition, the dfb mutant has a short primary root with a disorganized quiescent center, suggesting DFB as the predominant FPGS isoform that generates the physiologically active folate cofactors required to sustain postembryonic root growth in developing Arabidopsis seedlings (Srivastava et al., 2011). Taken together, the above findings suggest that variations in folylpolyglutamate derivatives are vital for Arabidopsis seedling development.
Folate derivatives are tripartite molecules containing pteridine, ρ-aminobenzoate, and Glu moieties with a short γ-linked chain of glutamyl residues attached to the first Glu (Cossins, 2000). In plants, folylpolyglutamate derivatives are central cofactors in folate-dependent enzymes, including those essential for photorespiration (glycine decarboxylase [GDC] and serine hydroxymethyltransferase [SHMT]), Met synthetase (the rate-limiting step of methyl donor synthesis), and magnesium-protoporphyrin IX methyltransferase, which is required for chlorophyll synthesis (Besson et al., 1993; Rebeille et al., 1994; Ravanel et al., 2004; Van Wilder et al., 2009). Among these folate-dependent reactions, the photorespiratory GDC/SHMT reaction leads to an important production of ammonium (NH4+) that must be either reassimilated or lost to the atmosphere, and photorespiratory NH4+ production by Gly oxidative decarboxylation exceeds the primary NO3− reduction in vegetative leaves in C3 plants by about 10-fold (Masclaux-Daubresse et al., 2006). These findings indicate that N utilization and photorespiration are closely associated. Additionally, photorespiration is inhibited in Arabidopsis under N-limited conditions (Bi et al., 2007; Tschoep et al., 2009); elevated CO2 also inhibits photorespiration in Arabidopsis and decreases NO3− absorption and assimilation (Rachmilevitch et al., 2004; Bloom et al., 2010). Thus, whether and how folylpolyglutamates are involved in these processes should be explored.
In this report, an Arabidopsis mutant carrying a T-DNA insertion in the gene (DFC) encoding mitochondrial FPGS is characterized for its altered N metabolism and enhanced phenotype under low-N stress. Also, elevated CO2 levels partly rescued the dfc mutant phenotypes. These results suggest that the alteration in N metabolism in the dfc mutant is likely mainly due to a defect in photorespiration and that an intact folate biosynthesis pathway is required for N utilization during early seedling development.
RESULTS
Isolation and Characterization of the dfc Mutant
To understand the role of FPGS in plant growth and development, a transfer DNA (T-DNA) line with an insertion in exon 15 of the mitochondrial FPGS gene DFC (At3g10160) was obtained from the Arabidopsis Biological Resource Center (Ohio State University) and named dfc (Supplemental Fig. S1A). DFC transcripts could not be detected in dfc plants with primers either ahead of or spanning the T-DNA insertion site (Supplemental Fig. S1B). No significant differences in growth, development, or fertility were observed between wild-type and dfc plants in soil (Supplemental Fig. S1, C–E), which was consistent with a previous report that vegetative phenotypes of the fpgs2 (i.e. dfc) mutant do not differ visually from those of wild-type plants (Mehrshahi et al., 2010).
Developmental Responses to N Supply in dfc Seedlings
Because dfc showed no striking morphological differences from wild-type plants under normal growth conditions, we grew the mutant under various nutrient-deficient conditions to examine its response to nutrient stress. The concentrations of supplementary C and N are expressed in millimolar; 1 mm Suc and 1 mm NO3− are designated 1 C and 1 N, respectively.
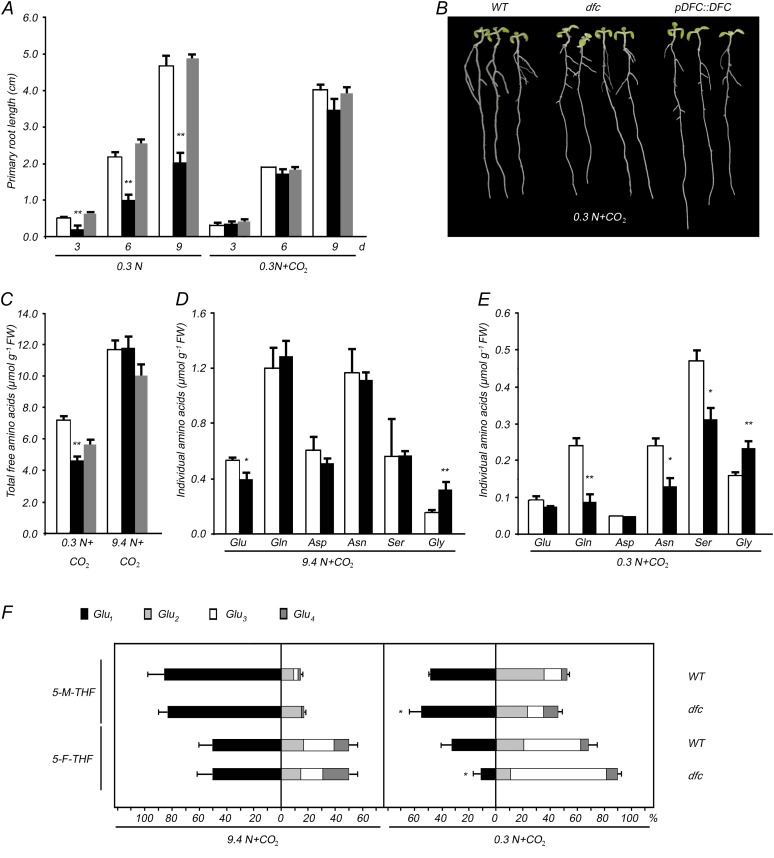
First, we examined if sulfur or phosphorus deficiency affected dfc development. When seedlings were grown on one-half-strength Murashige and Skoog (MS) medium in the absence of sulfur or phosphorus, the primary root length of dfc and wild-type seedlings was similar (Supplemental Fig. S2A). Similarly, no differences under various C conditions were observed (Supplemental Fig. S2B). However, primary root elongation in dfc and the wild type responded differently to the various external N concentrations when 30 C was chosen for all subsequent experiments. For example, the difference in primary root length between dfc and wild-type seedlings became evident (0.24 ± 0.05 cm for mutant versus 0.58 ± 0.08 cm for the wild type) on day 3 when 0.3 n was supplemented in the medium (Fig. 1, A and B) and remained at 3 n, albeit somewhat attenuated (Fig. 1A). However, the difference was no longer observed when NO3− was increased to 9.4 n (Fig. 1, A and B). Therefore, in subsequent experiments, 0.3 n was regarded as an N-limited condition and 9.4 n was considered an N-sufficient condition. To determine whether the difference in primary root length was caused by a delay in primary root emergence in the mutant, seed germination was investigated in dfc and the wild type under both N conditions (Supplemental Fig. S2C). Only a subtle difference was observed between dfc and the wild type under both N conditions. For example, 26.7% to 30.2% of the dfc seeds had germinated after 24 h, which was one-half the number of germinated wild-type seeds, but all of the germination percentages exceeded 90% after 36 h (Supplemental Fig. S2C).
Figure 1.
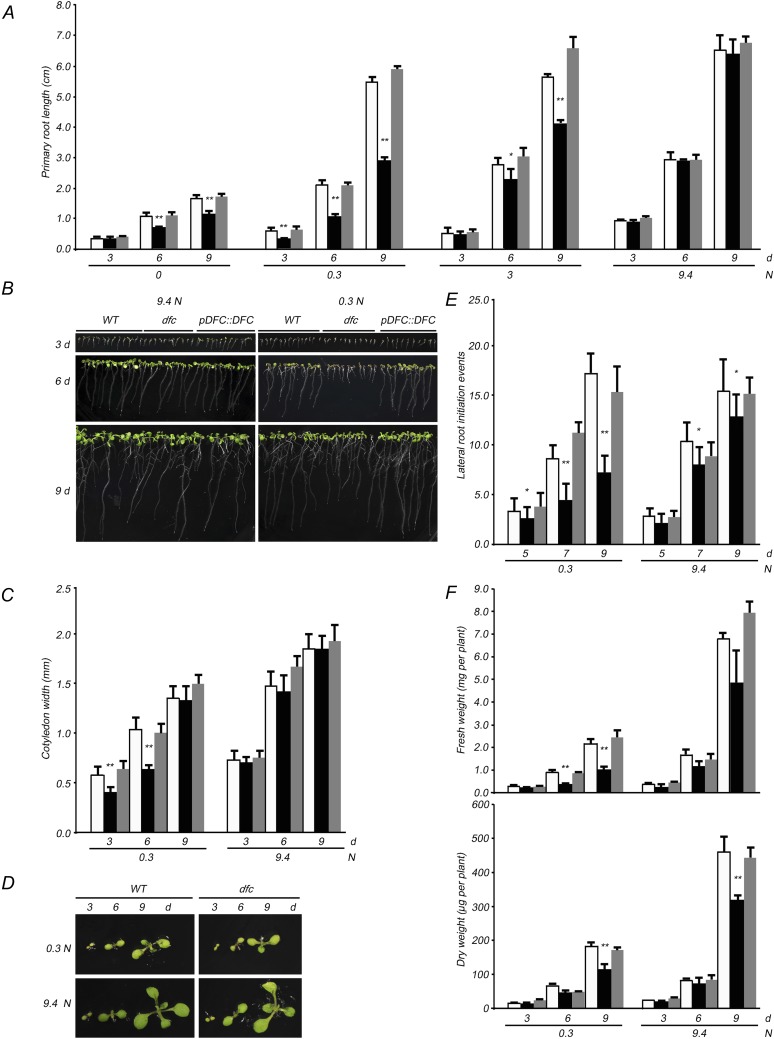
Effects of NO3− on the growth of wild-type (white bars), dfc (black bars), and DFC complementation (gray bars) seedlings. A, Effects of different NO3− levels (0, 0.3, 3, and 9.4 n) on the primary root length with 30 mm Suc in the medium and NO3− as the sole N source. B, Wild-type (WT), dfc, and DFC complementation (pDFC::DFC) seedlings grown on 0.3 or 9.4 n medium for 3, 6, and 9 d. C, Cotyledon width of wild-type, dfc, and DFC complementation seedlings grown on 0.3 or 9.4 n medium. D, Cotyledon and leaves of wild-type and dfc seedlings grown on 0.3 or 9.4 n medium for 3, 6, and 9 d. E, Lateral root initiation events of wild-type, dfc, and DFC complementation seedlings grown on 0.3 or 9.4 n medium for 5, 7, and 9 d. F, Fresh weight and dry weight per plant of wild-type, dfc, and DFC complementation seedlings grown on 0.3 or 9.4 n medium for 3, 6, and 9 d. Data are means ± sd (n = 4), and each replicate consisted of a pool of 10 to 15 plants. Black bars with single asterisks indicate significant differences at P < 0.05, and those with double asterisks indicate highly significant differences at P < 0.01 (Student’s t test). [See online article for color version of this figure.]
Subsequently, we investigated some other developmental responses of dfc seedlings under both N conditions. Cotyledon growth is closely related to the N response in Arabidopsis (Martin et al., 2002). Wild-type and dfc seedlings had a similar cotyledon size under N-sufficient conditions, but the mutant cotyledon width was approximately 60% of that of the wild type before day 6 under N-limited conditions (Fig. 1C). Even though cotyledon width was similar between dfc and the wild type on day 9, dfc cotyledons were paler than those of the wild type (Fig. 1D). Lateral root initiation is a determinant of plant root architecture and a secondary effect of overall N starvation (Malamy and Ryan, 2001). Similar to the case in cotyledons, a drastic reduction in lateral root initiation in the dfc mutant was observed under N-limited conditions as compared with the wild type, whereas only a slight difference was observed under N-sufficient conditions (Fig. 1E). Differences in biomass were also observed in wild-type and dfc seedlings. Low-N-induced reductions in fresh weight and dry weight were observed in both wild-type and dfc seedlings (Fig. 1F). On the 9.4 n medium, the fresh weight of the dfc mutant was approximately 70% that of the wild type, and it was less than one-half that of the wild type on the 0.3 n medium on days 6 and 9 (Fig. 1F, top panel). A similar pattern was observed in dry weight (Fig. 1F, bottom panel).
A complementation experiment was performed to confirm that the observed dfc phenotypes were due to the disruption of the DFC gene. A plasmid containing the DFC genomic fragment was introduced into dfc plants (Supplemental Fig. S2D). DFC transcripts were observed in the transformed plants and were of the same size and abundance as those in wild-type plants (Supplemental Fig. S2E). These results indicate that DFC transcription was restored in the complemented plants. Additionally, the DFC complementation transformants were always similar to the wild type in terms of primary root length, cotyledon width, lateral root initiation, fresh weight, and dry weight under both conditions (Fig. 1). These observations indicate that the dfc mutation could have a detrimental effect on seedling growth under N-sufficient conditions and that N limitation exaggerated these differences.
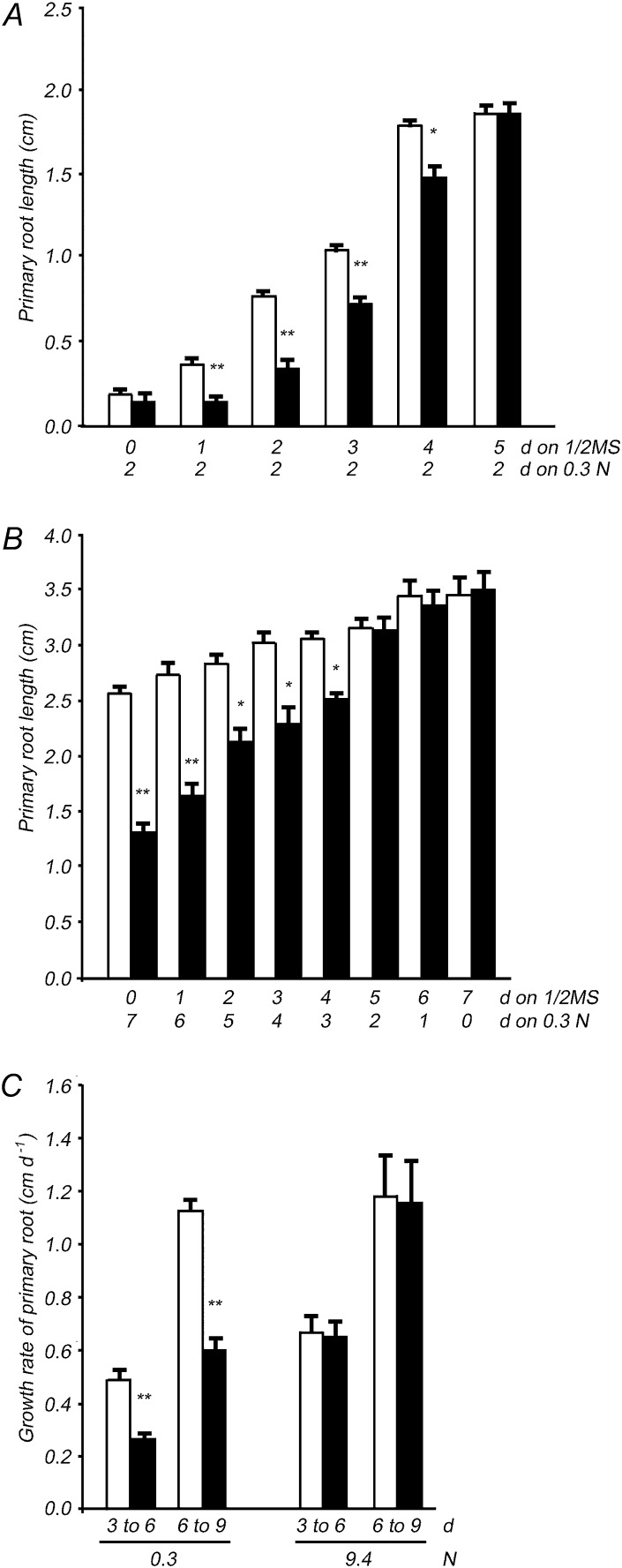
To determine the stages at which low-N stress has the greatest effects on dfc seedling development, the maladjustment time window in the mutant was investigated by removing N from the medium at various time points after sowing. The difference in primary root length between dfc and the wild type was highly significant (P < 0.01) only in seedlings grown on one-half-strength MS for 0 to 4 d. When N deprivation was applied on day 5, no significant difference was observed (Fig. 2A). In further time-course experiments, up to 4-d-old dfc seedlings grown on one-half-strength MS arrested root elongation under the N deprivation treatment compared with that in the wild type, whereas this phenomenon was not observed in the dfc mutant if N deprivation began on day 5 (Fig. 2B). The responses of seedlings to N limitation seemed to fall within a narrow window of developmental sensitivity. Primary root growth rates from days 3 to 6 and from days 6 to 9 were calculated to further characterize the development of mutant seedlings under N-limited conditions. There was no difference in the growth rates of the wild type and dfc when grown under N-sufficient conditions, whereas the growth rate in dfc was only one-half the rate of the wild type under N-limited conditions (Fig. 2C). These results indicate that intact folate synthesis is required at early seedling stages, especially under N limitation.
Figure 2.
The maladjustment time window in the dfc mutant (black bars) compared with that in wild-type (white bars) seedlings, and the growth rates of those seedlings on 0.3 or 9.4 n medium. A, Primary root length of wild-type and dfc seedlings grown on the 0.3 n medium for 2 d after transfer from one-half-strength (1/2) MS medium on different days (0–5). B, Primary root length of wild-type and dfc seedlings on day 7 from the start of incubation. Both types of seedlings were grown on one-half-strength MS medium for 0 to 7 d and then transferred onto the 0.3 n medium for the remaining time (7–0 d). C, Primary root growth rates of wild-type and dfc seedlings from days 3 to 6 and from days 6 to 9 on 0.3 or 9.4 n medium. Data are means ± sd (n = 4), and each replicate consisted of a pool of 10 to15 plants. Black bars with single asterisks indicate significant differences at P < 0.05, and those with double asterisks indicate highly significant differences at P < 0.01 (Student’s t test).
N Metabolism Was Perturbed in the dfc Mutant
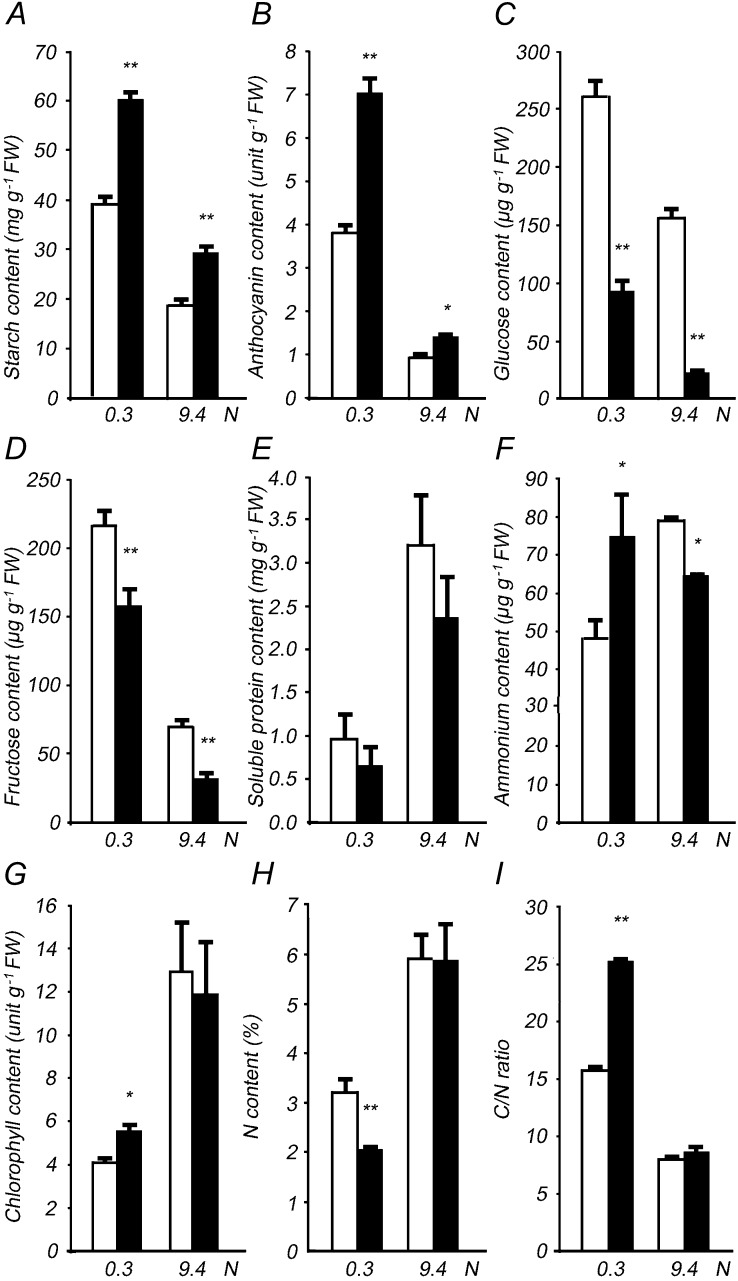
Given that dfc seedlings showed enhanced morphological changes under low-N stress, C and N metabolite levels were determined in the mutant to evaluate the perturbation of N metabolism. N limitation led to extra accumulation of starch, anthocyanin, Glc, Fru, and a higher C-N ratio as well as a decrease in soluble protein, NH4+, NO3−, chlorophyll, and N content in wild-type seedlings (Fig. 3). Under N-sufficient conditions, dfc seedlings had higher levels of starch and anthocyanin but lower levels of Glc, Fru, soluble protein, and NH4+ than did the wild type (Fig. 3, A–F). All these differences between dfc and the wild type remained under N-limited conditions, although these parameters/metabolites were either up-regulated or down-regulated, and additional NH4+ accumulation was detected in the dfc (Fig. 3F). dfc and the wild type had similar chlorophyll levels, N contents, and C-N ratios under N-sufficient conditions but not under low-N stress (Fig. 3, G–I). The NO3− concentrations in both wild-type and dfc seedlings under N-limited conditions were approximately 1.7% of those under N-sufficient conditions, without obvious differences between the two genotypes (data not shown). Additionally, the soluble protein in dfc seeds was only 32% of that in the wild type (Supplemental Fig. S2F). These results suggest that the mutant seedlings exhibited altered N metabolite profiles even under N-sufficient conditions, most of which were further enhanced under N-limited conditions.
Figure 3.
Altered C and N metabolite levels in 12-d-old wild-type (white bars) and dfc (black bars) seedlings on 0.3 or 9.4 n medium. The contents of starch (A), anthocyanin (B), Glc (C), Fru (D), soluble protein (E), NH4+ (F), chlorophyll (G), and N (H) as well as C-N ratio (I) in seedlings grown on 0.3 or 9.4 n medium are shown. Data are means ± sd (n = 4), and each replicate consisted of a pool of 30 to 50 plants. Black bars with single asterisks indicate significant differences at P < 0.05, and those with double asterisks indicate highly significant differences at P < 0.01 (Student’s t test). FW, Fresh weight.
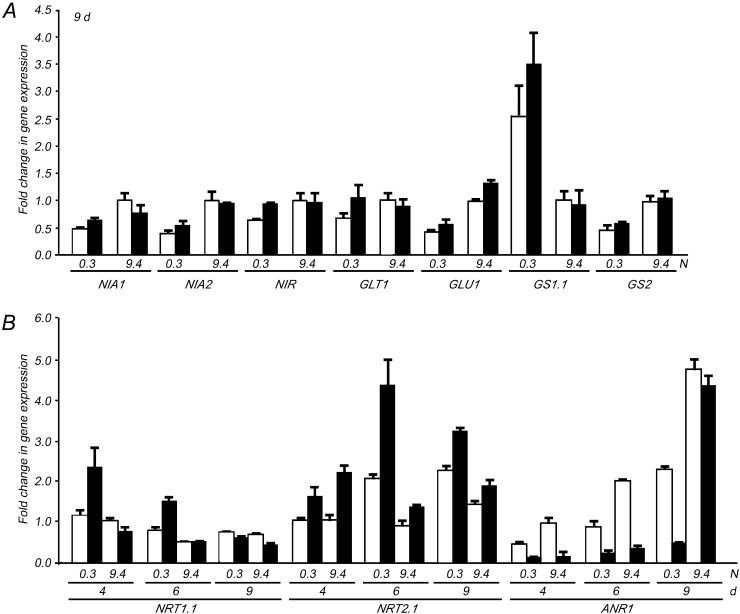
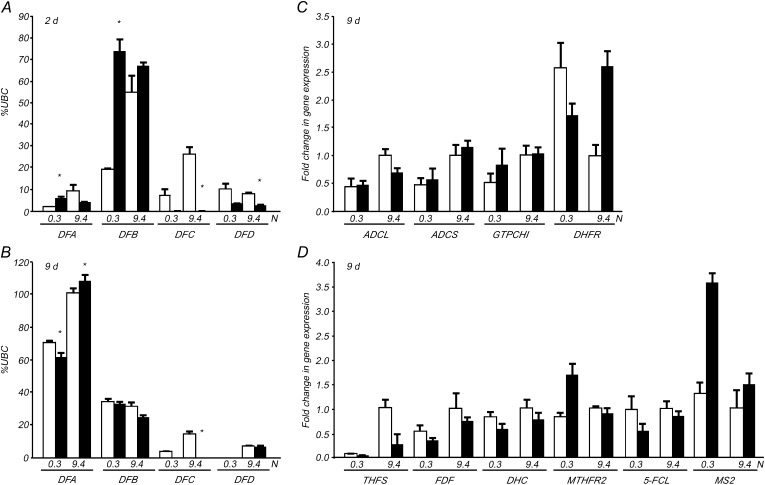
Next, we determined whether the expression of N metabolism-related genes was also affected in dfc. The relative comparison revealed that transcript levels of the genes involved in nitrate reduction (NITRATE REDUCTASE1 [NIA1], NIA2, and NITRITE REDUCTASE [NIR]) and N assimilation (NADH-DEPENDENT GLUTAMATE SYNTHASE1 [GLT1], FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE1 [Fd-GOGAT GLU1], GLUTAMINE SYNTHETASE1.1 [GS1.1], and GS2) were similar between dfc and the wild type under both N conditions and that low N caused slight decreases in transcript levels of most of these genes in both genotypes, with the exception of GS1.1 (Fig. 4A). However, genes involved in nitrate uptake and root system architecture were differentially expressed in dfc and the wild type during the early stage of seedling development, with the control treatment (9.4 n in wild-type seedlings) on day 4. The mutation in DFC consistently led to an increase in NITRATE TRANSPORTER2.1 (NRT2.1) expression and a decrease in ARABIDOPSIS NITRATE REGULATED1 (ANR1) expression under both N conditions, whereas an increase in NRT1.1 expression was only observed under N-limited conditions on days 4 and 6 (Fig. 4B).
Figure 4.
Altered transcript levels of the genes involved in N metabolism in wild-type (white bars) and dfc (black bars) seedlings on 0.3 or 9.4 n medium. A, Fold change in expression of the genes involved in NO3− reduction and N assimilation in 9-d-old seedlings. The transcript level of each gene is normalized to that in the wild type on the 9.4 n medium. B, Fold change in expression of NRT1.1, NRT2.1, and ANR1 on days 4, 6, and 9. The transcript level of each gene is normalized to that of the wild type grown on the 9.4 n medium on day 4.
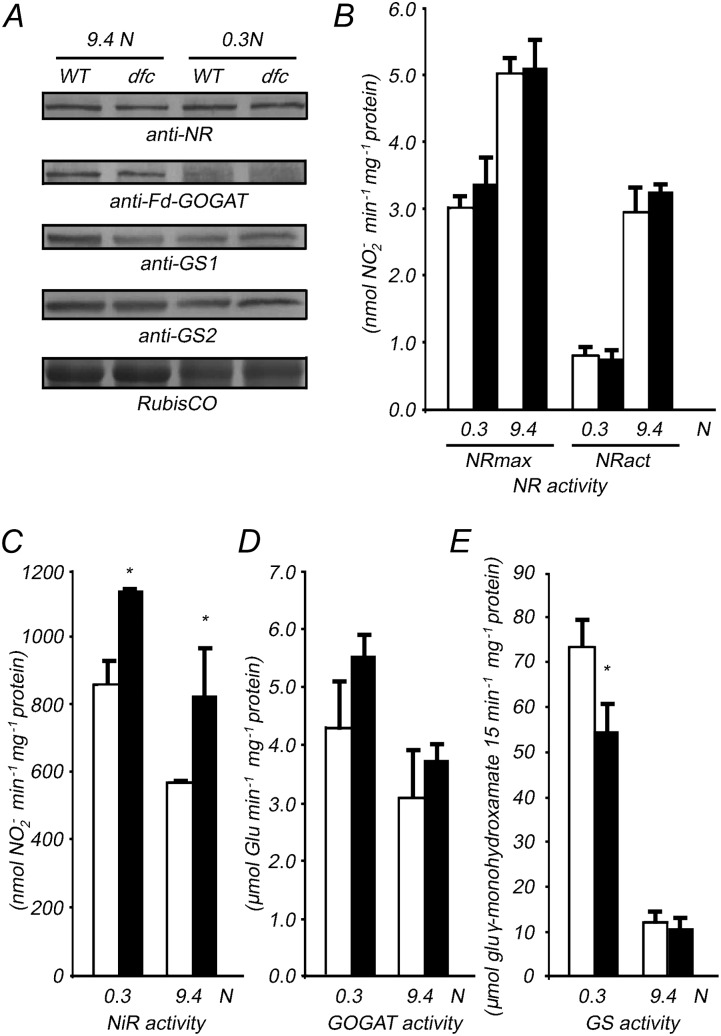
Levels of proteins involved in nitrate reduction and assimilation in dfc and the wild type were evaluated using the corresponding antibodies. The analyzed proteins included nitrate reductase (NR), GOGAT, a cytosolic form of glutamine synthetase (GS1), and a chloroplastic form of glutamine synthetase (GS2). NR and GS2 protein contents were similar under both N conditions in wild-type seedlings; ferredoxin-dependent GOGAT, GS1, and GS2 levels were lower under N-limited compared with N-sufficient conditions. However, no obvious difference in protein distribution was observed between dfc and the wild type, with the exception of GS1, which was lower in the mutant on the 9.4 n medium (Fig. 5A).
Figure 5.
Altered biochemical characteristics of N assimilation in 12-d-old wild-type (white bars) and dfc (black bars) seedlings on 0.3 or 9.4 n medium. A, Relative protein content involved in NO3− reduction and N assimilation (NR, GOGAT, GS1, and GS2) in seedlings detected with the corresponding antibodies. Coomassie blue staining of a large subunit of the Rubisco subunit was used as the loading control. WT, Wild type. B to E, NR (including NRmax and NRact; B), NiR (C), GOGAT (D), and GS (E) activities in the seedlings. Data are means ± sd (n = 4), and each replicate consisted of a pool of 30 to 50 plants. Black bars with single asterisks indicate significant differences at P < 0.05, and those with double asterisks indicate highly significant differences at P < 0.01 (Student’s t test).
Nitrate assimilation in dfc and the wild type grown under both N conditions was evaluated by analyzing NR, nitrite reductase (NiR), GOGAT, and glutamine synthetase (GS) activities. Maximum NR activity (NRmax) determines the total amount of functional NR, and NR activation activity (NRact) is assumed to reflect the actual nitrate reduction enzyme activity (Kaiser et al., 2000). Both NRmax and NRact decreased significantly under N-limited conditions but were similar between the wild type and dfc (Fig. 5B). NiR and GOGAT activities were slightly higher in the mutant than in the wild type under both N conditions (Fig. 5, C and D). No obvious difference in GS activity was observed between dfc and the wild type under N-sufficient conditions; however, significantly lower GS activity (P < 0.05) was detected in the mutant compared with the wild type under N-limited conditions (Fig. 5E). Taken together, these results indicate that N metabolism was perturbed in the dfc mutant.
Altered Amino Acid Profiling in the dfc Mutant
Free amino acids are the key point between N assimilation and protein synthesis (Kant et al., 2011). Therefore, free amino acid content was determined in the seeds and seedlings of dfc and the wild type. Overall, a similar amount of total free amino acids was detected in both seeds, which were collected from plants in N-sufficient soil. High levels of Gln and Leu and low levels of Glu, Asp, and Arg were detected in the mutant seeds compared with those of the wild type (Supplemental Table S1), implying altered N utilization during the formation of dfc seeds.
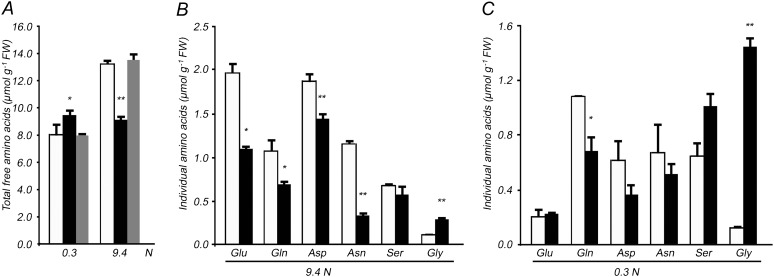
In N-sufficient seedlings, the total of all free amino acids in the dfc was only 69% of that in the wild type (Fig 6A; Supplemental Table S2). Over one-half of the detected individual amino acids, such as Glu, Gln, Asp, Asn, Trp, and Met, were accumulated less in dfc than in the wild type; for Gly, the level was 2.6-fold that in the wild type (Fig 6B; Supplemental Table S2). Most strikingly, in dfc, Asn and Met were around one-fourth the levels in the wild type (Fig 6B; Supplemental Table S2). Even though the Gln-Glu ratio in dfc (0.63) was slightly higher than that in the wild type (0.55), the presence of fewer major N-storage amino acids indicated that N utilization was impaired in dfc. Low N stress led to a decrease in total free amino acids in wild-type seedlings but not in dfc (Fig 6A; Supplemental Table S2). Most of the individual amino acid contents decreased in the wild type, whereas some showed different changes in dfc (Supplemental Table S2). First, there were similar amounts of Gln under both N conditions, and the level in the mutant was always 63% of that in the wild type. Second, Glu, Asp, and Ala in the mutant decreased to less than one-half the level under N-sufficient conditions, whereas more did in the wild type. Thus, there were similar amounts of Glu and Ala in dfc and the wild type under N limitation. Third, most of the individual amino acids increased by 1.4- to 2.5-fold in the mutant compared with N-sufficient conditions, including Ser, Asn, Trp, Leu, and Met, whereas levels in the wild type did not change very much. As a result, amounts of Trp, Leu, Arg, and His in dfc and the wild type were similar. Ser and Met increased 1.5-fold, and Asn was 77% of that in the wild type under N-limited conditions. Fourth, Gly content in the mutant under N-limited conditions increased 5-fold compared with N-sufficient conditions and was 11-fold greater than that observed in the wild type under N-limited conditions (Fig. 6, B and C; Supplemental Table S2). Therefore, the indicator of photorespiration (Gly-Ser ratio) in the mutant differed significantly from that in the wild type under both N conditions: the Gly-Ser ratio increased from 0.50 (N-sufficient conditions) to 1.42 (N-limited conditions) in dfc, whereas it increased only slightly, from 0.16 to 0.19, in the wild type. These results imply that Gly oxidation may be impaired, in addition to the altered N utilization, in dfc under N-limited conditions.
Figure 6.
Altered amino acid profiles in 12-d-old wild-type (white bars), dfc (black bars), and DFC complementation (gray bars) seedlings on 0.3 or 9.4 n medium. Total free amino acid (A) and some individual amino acid (B and C) contents in seedlings are shown. Data are represented as means ± sd (n = 4), and each replicate consisted of 200 mg of pooled plant material. Black bars with single asterisks indicate significant differences at P < 0.05, and those with double asterisks indicate highly significant differences at P < 0.01 (Student’s t test). FW, Fresh weight.
Folate Metabolism Was Disturbed in the dfc Mutant
We investigated how folate metabolism was disturbed in the dfc mutant at both the transcriptional and metabolic levels.
We were interested in determining whether the dfc mutation had altered the expression of genes involved in folate biosynthesis and how these genes responded to low-N stress. First, transcript levels of the genes encoding DHFS (DFA) and FPGS (DFB, DFC, and DFD) were analyzed at different time points and normalized to the UBC gene. The DFB transcript level was the highest in 2-d-old germinating seeds, and the DFA transcript level was the highest in 9-d-old seedlings under both N conditions. The expression levels of DFA, DFB, and DFC were lower under N-limited conditions than under N-sufficient conditions (Fig. 7, A and B). Interestingly, a significant increase in DFB expression (P < 0.05) was observed in 2-d-old mutant seeds germinating under N limitation as compared with the wild type, but this increase was not observed in 9-d-old mutant seedlings (Fig. 7, A and B). These results indicate that DFC and DFB play a role during the early seedling morphogenesis stage and that the latter could somehow compensate for a loss of DFC function when the dfc mutant was under low-N stress. Second, the other genes involved in folate synthesis were analyzed relative to those in the wild type under sufficient N supply. Most of the genes, such as genes encoding aminodeoxychorismate lyase (ADCL), aminodeoxychorismate synthase (ADCS), and GTP cyclohydrolase I (GTPCHI), showed similar expression patterns in dfc and the wild type under both N conditions, and low N levels reduced the transcript levels in both dfc and wild-type seedlings (Fig. 7C). DIHYDROFOLATE REDUCTASE (DHFR) was an exception, as it was expressed at a higher level in dfc than in the wild type under N-sufficient conditions and at a lower level than the wild type under N-limited conditions. DHFR expression increased in wild-type seedlings under low-N stress, which was also in contrast to the other genes (Fig. 7C).
Figure 7.
Altered transcript levels of the genes involved in folate metabolism and the synthesis of one-carbon units in wild-type (white bars) and dfc (black bars) seedlings on 0.3 or 9.4 n medium. A and B, The transcript levels of DFA, DFB, DFC, and DFD in seedlings on day 2 (A) and day 9 (B) are shown as percentages of that of the UBC gene. Black bars with single asterisks indicate significant differences at P < 0.05 (Student’s t test). C and D, Transcript levels of other genes involved in folate metabolism (C) and genes involved in the synthesis of one-carbon units (D) in 9-d-old seedlings are shown, normalized to those of the wild type on the 9.4 n medium. FDF, FORMYTETRAHYDROFOLATE DEFORMYLASE; DHC, METHYLENETETRAHYDROFOLATE DEHYDROGENASE; 5-FCL, 5-FORMYLTETRAHYDROFOLATE CYCLOLIGASE.
Because one-carbon (C1) unit transfer reactions are mediated by folates, we examined whether the expression of genes responsible for the synthesis of C1 units was altered in the dfc mutant. No significant changes in expression were observed in most of the studied genes in low-nitrate-treated wild-type seedlings, except for 10-formyl-tetrahydrofolate synthetase (THFS), which was down-regulated (Fig. 7D). These results indicate that N limitation has little impact on the expression of most genes that are essential for C1 unit synthesis at the transcriptional level. The expression of some genes was affected by the dfc mutation. For example, THFS expression was down-regulated in dfc under both N conditions, and METHYLENETETRAHYDROFOLATE REDUCTASE2 (MTHFR2) and cytosolic METHIONINE SYNTHASE2 (MS2) were up-regulated in dfc under N limitation (Fig. 7D).
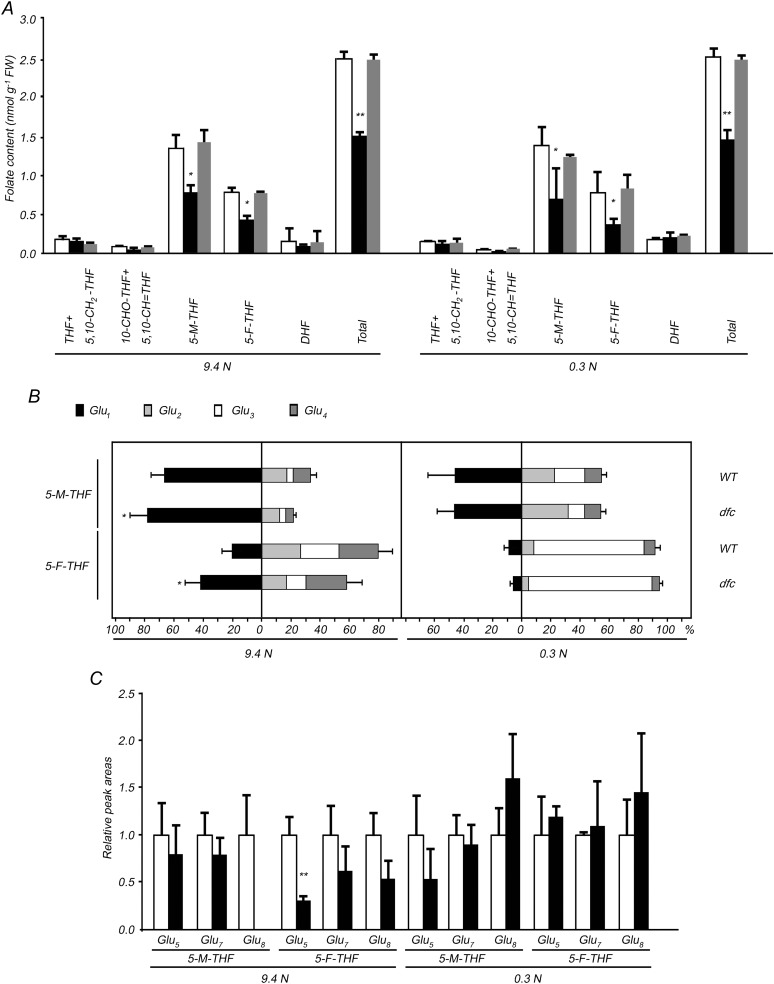
Liquid chromatography-tandem mass spectroscopy (LC-MS) was used to profile total folate and various folate types. As expected, no differences in amounts and types of folate were observed between wild-type and DFC complementation seedlings under both N conditions, and there were significant reductions (P < 0.05) in 5-methyl-tetrahydrofolate (5-M-THF) and 5-fromyl-tetrahydrofolate (5-F-THF) in dfc, to levels that were about one-half those in the wild type (Fig. 8A; Supplemental Table S3). The percentage of each folate species was similar between dfc and the wild type under both N conditions (Supplemental Table S3), which implies that the ratio of different folate species was not affected by the external N supply or the dfc mutation.
Figure 8.
Altered folate profiles in 12-d-old wild-type (WT; white bars), dfc (black bars), and DFC complementation (gray bars) seedlings on 0.3 or 9.4 n medium. A, Various folate types and total folate levels of seedlings. The folate species detected were as follows: THF, 5,10-methenyltetrahydrofolate (5,10-CH=THF), 5-M-THF, 5-F-THF, and dihydrofolate (DHF). Note that THF and 5,10-CH2-THF are grouped, and 10-formyl-THF (10-CHO-THF) and 5,10-CH=THF are grouped because the procedure used for folate analysis results in interconversions in these two pairs of folate species. B, Folate glutamylation profiles are shown as percentages of each class in the sum of the amount of folylpolyglutamates (5-M-THF-Glun and 5-F-THF-Glun; n = 1–4). C, LC-MS relative peak areas of folylpolyglutamates (5-M-THF-Glun and 5-F-THF-Glun; n = 5, 7, or 8) normalized to the levels of the wild type. Data are means ± sd (n = 4), and each replicate consisted of 100 mg of pooled plant material. Black bars with single asterisks indicate significant differences at P < 0.05, and those with double asterisks indicate highly significant differences at P < 0.01 (Student’s t test). FW, Fresh weight.
Next, we determined whether the folate poly-Glu profile changed because of either low-N stress or the dfc mutation using LC-MS with different extraction methods and liquid chromatography conditions. Due to technical limitations, only compounds with standards can be quantitatively determined by LC-MS, and we obtained 5-M-THF-Glun and 5-F-THF-Glun (n = 1–4) from Schircks Laboratories. Thus, poly-Glu profiles of 5-M-THF and 5-F-THF with 1-, 2-, 3-, and 4-Glu tails were measured, and the results are shown as percentages of the sum of Glu1 + Glu2 + Glu3 + Glu4 (Fig. 8B; Supplemental Table S4). Because we did not obtain standards for 5-M-THF and 5-F-THF with 5-, 7-, or 8-Glu tails, these were analyzed using the predicted mass-to-charge ratio (m/z), and their peak areas were obtained and normalized to the wild type under the same conditions (Fig. 8C). The percentages of folylpolyglutamates with 1-, 2-, 3-, and 4-Glu tails in dfc responded to low-N stress in a manner similar to that of the wild type, with marked decreases in 5-M-THF-Glu1 and 5-F-THF-Glu1 and dramatic increases in 5-M-THF-Glu3 and 5-F-THF-Glu3 being detected in both genotypes as compared with N-sufficient conditions (Fig. 8B, right panel). When N was sufficient, the percentages of 5-M-THF-Glu1 and 5-F-THF-Glu1 in dfc were higher than those in the wild type, and the percentages of those molecules with 2-, 3-, and 4-Glu tails decreased in the mutant, except for 5-M-THF-Glu3 and 5-F-THF-Glu4 (Fig. 8B, left panel). The relative peak areas of folylpolyglutamates with 5-, 7-, or 8-Glu tails were consistently lower in dfc than in the wild type; 5-F-THF-Glu5 was significantly lower and 5-M-THF-Glu8 was hardly detected in the dfc mutant when grown with a sufficient N supply (Fig. 8C). These results indicate that polyglutamylation levels of these two types of folates were significantly reduced due to the dfc mutation when N was sufficient. But under N-limited conditions, the differences in 5-M-THF-Glu1 and 5-F-THF-Glu1 between dfc and the wild type were no longer observed, and the relative 5-M-THF-Glu8 and 5-F-THF-Glu8 peak areas in dfc were somewhat higher than those in the wild type (Fig. 8, B and C). These results suggest that polyglutamylation levels of 5-M-THF and 5-F-THF may be unrelated to the phenotypes observed in dfc. However, it remains to be determined whether the polyglutamylation patterns of as yet uncharacterized species are associated with these phenotypes.
Elevated CO2 Partly Rescued the dfc Phenotype
A CO2 concentration lower than the CO2 compensation point or strong light stimulates photorespiration (hence the Gly-to-Ser flux rate in mitochondria), whereas a high CO2 concentration (4,000 μmol mol−1), which is a nonphotorespiration condition, suppresses photorespiration (Goyer et al., 2005). Photorespiration mutants can often be rescued under elevated CO2 concentrations (Ogren, 1984). Given the obvious link between folate and Gly metabolism and the observation that Gly and Ser were the most affected amino acids in the dfc under N-limited conditions (Fig. 8, B and C), the potential role of photorespiration in the mutant was evaluated, especially under the low-N condition.
First, we investigated the morphological changes in the mutant under N-limited conditions when elevated CO2 was applied. dfc primary root length was about 45% of that in the wild type under N-limited conditions with ambient CO2 (Figs. 1A and 9A). When grown on the 0.3 n medium supplied with a high CO2 concentration (4,000 μmol mol−1), three genotypes of seedlings (i.e. the wild type, dfc, and DFC complementation) developed similar morphologies with respect to primary root length, cotyledon size, and cotyledon greening (Fig. 9, A and B). We next examined free amino acids in the elevated-CO2-treated dfc and wild-type seedlings. Under N-sufficient conditions, the total free amino acid content was significantly lower in the mutant than in the wild type with ambient CO2, and elevated CO2 treatment conferred a similar level of total amino acids (and similar levels for most individual amino acids) between dfc and the wild type (Figs. 6A and 9C; Supplemental Table S5). However, differing from the elevated CO2-treated root phenotype observed in dfc under N-limited conditions, a greater degree of change in amino acids was observed between dfc and the wild type; levels not only of the major N-storage amino acids such as Glu, Gln, Asp, and Asn but also of Gly and Ser decreased dramatically, and total free amino acid levels in dfc were lower in the mutant than in the wild type (Figs. 6C and 9, C and E; Supplemental Table S5). It was also evident that elevated CO2 resulted in a significant reduction in total free amino acids regardless of external N availability (Figs. 6A and 9C; Supplemental Table S5). Additionally, there were no obvious differences in two indicators in the high-CO2-treated wild-type seedlings between N-sufficient and N-limited conditions (0.30 at 9.4 n and 0.34 at 0.3 n for Gly-Ser ratio; 2.22 under sufficient N and 2.57 under N limitation for Gln-Glu ratio), whereas in the mutant, the Gly-Ser ratio had increased from 0.57 to 0.75 and the Gln-Glu ratio had decreased from 3.17 to 1.20 due to the low-N stress. These results suggest that intact folate synthesis is required for N utilization under nonphotorespiration conditions.
Figure 9.
Effects of elevated CO2 on wild-type (WT; white bars), dfc (black bars), and DFC complementation (gray bars) seedlings on 0.3 or 9.4 n medium. A, The primary root length of seedlings under N-limited conditions with ambient air or elevated CO2. B, Wild-type, dfc, and DFC complementation seedlings grown on the 0.3 n medium for 9 d with elevated CO2. C, Total free amino acid contents of 12-d-old seedlings on 0.3 or 9.4 n medium with elevated CO2. D, Contents of some individual amino acids in 12-d-old seedlings on the 9.4 n medium with elevated CO2. E, Contents of some individual amino acids in 12-d-old seedlings on the 0.3 n medium with elevated CO2. F, The percentage of each folate in the sum of the amount of folylpolyglutamates (5-M-THF-Glun and 5-F-THF-Glun; n = 1–4) in 12-d-old seedlings on 0.3 or 9.4 n medium with elevated CO2. Data are means ± sd (n = 4), and each replicate consisted of a pool of 30 to 50 plants. Black bars with single asterisks indicate significant differences at P < 0.05, and those with double asterisks indicate highly significant differences at P < 0.01 (Student’s t test). FW, Fresh weight. [See online article for color version of this figure.]
The polyglutamylation profiles of 5-M-THF and 5-F-THF in elevated-CO2-treated seedlings were also analyzed. Compared with responses to low-N stress with ambient air, dramatic increases in the percentages of 5-M-THF-Glu3 and 5-F-THF-Glu3 and marked decreases in the percentages of 5-M-THF-Glu1 and 5-F-THF-Glu1 were also observed in both dfc and the wild type with elevated CO2. These results indicate that the responses of these two types of folate to low-N stress in dfc were not affected by the CO2 concentration (Fig. 9F). However, the polyglutamylation levels in elevated-CO2-treated dfc and wild-type seedlings differed from seedlings with ambient air. Profile differences in the percentages of 5-M-THF-Glu1 or 5-F-THF-Glu1 were no longer observed between dfc and the wild type when N was sufficient (Fig. 9F, left panel; Supplemental Table S4). Under N-limited conditions with elevated CO2, the percentage of dfc 5-M-THF-Glu1 was significantly higher (P < 0.05) and that of dfc 5-F-THF-Glu1 was significantly lower (P < 0.05) than in the wild type (Fig. 9F, right panel; Supplemental Table S4). These results suggest that polyglutamylation levels of 5-M-THF and 5-F-THF responded in similar ways to low-N stress but that the levels changed to a different extent in dfc and the wild type with elevated-CO2 treatment (Fig. 9F). However, the polyglutamylation levels of these two types of folate may not be related to the recovery of the dfc phenotype under N limitation with elevated CO2.
DISCUSSION
We investigated novel links between folate and N metabolism using the T-DNA insertion mutant of the mitochondrial DFC gene. We showed that the dfc mutant had defects in N utilization and that low-N stress enhanced the alteration causing the obvious phenotypes during early seedling development. In addition, the observation that elevated CO2 could partly rescue the dfc phenotype under N-limited conditions implied a defect in the photorespiratory system in the mutant.
Low NO3− Enhanced the Alteration in N Utilization in the dfc Mutant
N availability affects root growth, and 0.1 to 0.6 mm N conditions limit growth (Martin et al., 2002). Low inorganic N typically results in the accumulation of starch and anthocyanin, decreased levels of soluble protein and chlorophyll, and large decreases in Gln and Asn (Fig. 3; Stitt and Krapp, 1999; Peng et al., 2007b; Tschoep et al., 2009). A similar metabolite profile was observed in dfc even under N-sufficient conditions (Figs. 3 and 6; Supplemental Table S2). This indicates that the dfc mutant likely already suffered from internal N starvation despite the sufficient external N supply. Normally, the internal N pools of amino acids such as Gln in plants may indicate N status by providing a signal that regulates nitrate uptake by plants (Miller et al., 2008). The NRT2.1 transcript level is decreased by the direct application of amino acids to the roots of barley (Hordeum vulgare) and Arabidopsis and is inversely correlated with Glu and Gln concentrations in plants (Zhuo et al., 1999; Vidmar et al., 2000). The higher level of NRT2.1 expression in dfc under both N conditions implied that the internal Gln concentration was lower than that in the wild type, which was confirmed by the free amino acid analysis (Figs. 4B and 6; Supplemental Table S2). Additionally, a lower level of soluble protein and some variations in individual amino acids were observed in mutant seeds obtained from the soil (Supplemental Fig. S2F). Together with the slight delay in germination, the metabolic data suggest an altered N utilization during the formation of dfc seeds and that the impaired N utilization, at least in part, results in the phenotype that is observed under N limitation (Fig. 1; Supplemental Fig. S2C).
Under N-limited conditions, the accumulated NH4+, lower N content and GS activity, higher C-N ratio, fewer major N storage amino acids, and accumulated Gly in dfc as compared with the wild type were indicative of an enhanced alteration in N utilization (Figs. 3, 5E, and 6C; Supplemental Table S2). The responses of free amino acids to low-N stress reveal an alteration in N utilization in the dfc, because most of the individual amino acids responded differently from those in the wild type (Fig. 6, B and C; Supplemental Table S2). However, it is unlikely that N sensing or signaling was blocked in dfc, as it responded to low-N stress to the same extent as the wild type, although there were some differences between the mutant and the wild type in terms of absolute C-N metabolite contents and the transcript abundance of most of the genes (Figs. 3, 4, and 6). Two genes are involved in the transcriptional regulation of lateral root growth under different N supply: NRT2.1 and ANR1 (Zhang and Forde, 1998; Little et al., 2005). The NRT2.1 transcript accumulates in nitrate-deprived plants, and ANR1 plays a role as a feedback regulator of lateral root growth rates based on the N status of the plant (Vidmar et al., 2000; Orsel et al., 2002; Gan et al., 2005). In our study, higher NRT2.1 expression levels and lower ANR1 expression levels that were not N dependent were observed in dfc, but dfc lateral root growth decreased significantly under N-limited conditions, indicating that the root phenotype is not due to the altered expression of ANR1 and NRT2.1. This also implies that intact folate synthesis is required for NRT2.1- and ANR1-regulated signaling mechanisms that govern lateral root formation.
The dfc responses to low-N stress were also different from those of N-sensing or N-signaling mutants, such as chl1-5, lin1, nla, and nlp7 (Tsay et al., 1993; Little et al., 2005; Peng et al., 2007b; Castaings et al., 2009). First, dfc seedling development was more retarded than that of the wild type, particularly at the early stage, and the difference between dfc and the wild type only occurred during a limited developmental window (Figs. 1 and 2). Second, the root growth rate of the dfc mutant was only one-half that of the wild type under N-limited conditions (Fig. 2C). Third, NRT1.1 is normally involved in local signaling of the external NO3− concentration (Gojon et al., 2011), and NO3− stimulates primary root growth mediated by NRT1.1 (Krouk et al., 2010). A significantly higher NRT1.1 transcript level was observed in dfc on the 0.3 n medium during early seedling development, but this was not observed when N was sufficient (Fig. 4B), indicating that the primary root growth regulated by NRT1.1 under N-limited conditions was also disturbed by the dfc mutation.
Consequently, the differences in N utilization between dfc and the wild type could result in altered internal metabolite concentrations, gene expression, and responses to low-N stress; these results also indicated that the interference in N utilization became more serious under N-limited conditions in the dfc mutant.
Folate Metabolism Was Impaired Due to the dfc Mutation, and the Polyglutamylation Status of 5-M-THF and 5-F-THF Was Affected by Low-N Stress
Differing from the electrochemical method for folate assays used in the studies by Mehrshahi et al. (2010) and Srivastava et al. (2011), LC-MS was used to detect profiles of both various folate types and polyglutamylation status in dfc and the wild type under different N supply. When we compared the data for the folate species profiles and those for poly-Glu profiles, an interesting phenomenon was observed: in some cases, sums of Glu1 through Glu4 for 5-M-THF and 5-F-THF (extracted in methanol) were greater than the data for folate species under the same conditions. Given that we used methotrexate (MTX) as an external standard and that these two methods had different loss rates during sample preparation, it is hard to directly compare the data. For this reason, we focused on the proportions of these folylpolyglutamates and compared the data for dfc with those for the wild type.
When the function of the mitochondria-targeted FPGS isoform was disrupted, the profiles of both the folate types and polyglutamylation status differed from those in the wild type. The contents of 5-M-THF, 5-F-THF, and total folates in the dfc mutant were approximately one-half those in the wild type under both N conditions (Fig. 8A; Supplemental Table S3), indicating that the changes in folate levels are due to the mutation. Further analysis of 5-M-THF and 5-F-THF polyglutamylation revealed an increase in monopoly-Glu and a decrease in polyglutamylation in dfc when N was sufficient (Fig. 8, B and C; Supplemental Table S4). These results indicate that folate metabolism was impaired and polyglutamylation status was perturbed due to the dfc mutation.
Differing from the enhanced dfc phenotypes under N limitation, the amounts of tetrahydrofolic acid (THF) and methylenetetrahydrofolate (5,10-CH2-THF) and the similar folate composition ratio showed no response to low-N stress in either genotype (Fig. 8A; Supplemental Table S3). This suggests that the alterations in N utilization and morphological phenotypes in dfc relied not on lower amounts of these cofactors but on their polyglutamylation status. However, we could not obtain standards for THF or 5,10-CH2-THF-Glun (n = 2–8). Also, the THF and 5,10-CH2-THF pools were too small to investigate. It is difficult to evaluate interrelations between polyglutamylation levels and dfc phenotypes under N limitation by only analyzing Gly oxidation status in the mutant.
Interestingly, low-N stress did influence folate metabolism, and the responses in dfc and the wild type were different. First, low N decreased the expression of most of the genes involved in folate synthesis and C1 units in both dfc and the wild type, but there were some exceptions in dfc. Specifically, the transcript levels of MTHFR2 and MS2 at day 9 and DFB at day 2 were much higher (Fig. 7), implying that reactions that use folates as cofactors may have been disturbed due to the dfc mutation under N-limited conditions. A higher expression of DFB, which encodes the plastically targeted FPGS isoform, was observed in 2-d-old dfc compared with the wild type under N-limited conditions, but it later disappeared. This result indicates that the plastidial FPGS isoform (DFB) could have a compensating effect on folate polyglutamylation in the dfc mutant (Fig. 7, A and B). Second, higher proportions of 5-M-THF-Glu3 and 5-F-THF-Glu3 were induced under N-limited conditions in both dfc and the wild type, and the percentage of 5-M-THF-Glu in dfc was always one-half that in the wild type, either with or without elevated CO2 (Figs. 8B and 9F). These observations suggest a special demand for folates with Glu tails in plants adapting to low-N stress (Fig. 8B). Third, the varied proportions of 5-M-THF-Glu1 and 5-F-THF-Glu1 under both N conditions, either with or without elevated CO2, were not consistent with the dfc phenotype under N limitation or the recovery with elevated CO2, indicating that the altered polyglutamylation pattern of those as yet uncharacterized species may be the cause of dfc phenotypes in response to low-N stress.
Photorespiration Is Probably a Link between Folate Metabolism and N Metabolism
Several plant metabolic processes have a requirement for long-chain-length folate poly-Glu residues (Mehrshahi et al., 2010). Arabidopsis Met synthase uses poly-Glu but not mono-Glu 5-M-THF to catalyze the methylation of homo-Cys to produce Met (Ravanel et al., 2004). Photorespiration also involves folate-dependent fluxes in C3 plants (Hanson and Roje, 2001). During photorespiration, the GDC complex catalyzes the conversion of Gly to 5,10-CH2-THF, which then acts as a C1 donor for Ser synthesis, a reaction catalyzed by SHMT. GDC releases ammonia, CO2, and 5,10-CH2-THF during photorespiration by catalyzing the oxidative decarboxylation of Gly to form Ser. To avoid toxic NH4+ accumulation and to prevent N loss through volatilization, photosynthetic plant cells must reassimilate the ammonia through the GS/GOGAT cycle in chloroplasts (Siedow and Day, 2000). Photorespiration, the tricarboxylic acid cycle, and C/N metabolism are tightly coordinated through N assimilation in plants to sustain optimal growth and development (Zheng, 2009). As folate poly-Glu residues are the preferred substrates for both GDC and SHMT, FPGS activity could indirectly play an important role in photorespiration (Besson et al., 1993).
Folates are known to be involved in the Gly-to-Ser transition during photorespiration. For example, the level of Gly was decreased slightly in the wild type as plant photorespiration was inhibited, but the Gly-to-Ser transition in folate synthesis mutants was accompanied by the accumulation of Gly in ambient air, such as the 5-fcl mutant and the 10-fromyl-tetrahydrofolate deformylases double-knockout (dKO) mutant, and a nonphotorespiration condition (elevated CO2) decreased their Gly levels and Gly-Ser ratios (Goyer et al., 2005; Collakova et al., 2008). Similar Gly level and Gly-Ser ratio phenotypes were observed in dfc in this study. Elevated CO2 suppressed the accumulated Gly and lowered the Gly-Ser ratio in dfc under both N conditions (Figs. 6, B and C, and 9, D and E). This was accompanied by the rescue of dfc phenotypes under N-limited conditions, such as short primary roots and anthocyanin accumulation in seedlings (Fig. 9, A and B), indicating that impaired photorespiration (Gly-to-Ser transition) could be related to growth defects in the mutant under N-limited conditions.
The dfc mutant had some features in common with photorespiratory mutants, such as shm1 and dKO, which are mutants with respect to their profiles of soluble sugars and free amino acids, including decreased Glc and Fru, increased Gly, and decreased Glu and Asp, when N was sufficient (Figs. 3, C and D, and 6B; Collakova et al., 2008), although dfc did not display phenotypes typical of photorespiratory mutants. When N was sufficient, the elevated CO2 recovered the polyglutamylation status of 5-M-THF and 5-F-THF and most of the individual amino acid contents in dfc (Fig. 9, D and F; Supplemental Table S3). However, levels of N-storage amino acids, such as Gln and Asn, remained less than one-half of those in the wild type under N-limited conditions, although the Gly-to-Ser transition was greatly improved (Fig. 9E). This implies that DFC may have a role in other biochemical processes besides photorespiration and may participate in N metabolism. Interestingly, when the differences in the percentages of 5-M-THF-Glu1 and 5-F-THF-Glu1 between dfc and the wild type were enlarged (Figs. 8B, left panel, and 9F, right panel), the levels of total and most individual amino acids in the dfc were much lower than in the wild type (Supplemental Tables S2 and S5), indicating that there may be a negative correlation between the polyglutamylation levels of 5-M-THF and 5-F-THF and the differences between dfc and the wild type in most of the amino acid profiles (Figs. 6, B and C, 8B, and 9, D–F).
Levels of most individual amino acids were lower in the dfc mutant than in the wild type under sufficient N supply, and the total free amino acid level was about 20% higher than that in the wild type under N-limited conditions, with Gly being the most affected (Fig. 6; Supplemental Table S2). This was different from findings with respiration mutants such as cytoplasmic male sterile tobacco (Nicotiana sylvestris) and onset of leaf death5, which contain elevated total free amino acid levels and little accumulation of Gly and Ser (Dutilleul et al., 2005; Schippers et al., 2008). Such a difference suggests that the mechanism underlying the altered N utilization in the dfc mutant is probably different from that in respiration mutants.
Taken together, these genetic data suggest that an intact folate synthesis pathway is required for correct N utilization in plants. The dfc mutation leads to an alteration in N utilization and variation in folate profiles, which are enhanced under low-N stress, and the variation in folylpolyglutamates in the dfc could impact N metabolism through defective photorespiration. Although the functional characterization of the dfc mutant indicates that an intact folate synthesis pathway is required for correct N utilization in plants, the underlying mechanism through which DFC affects N metabolism awaits further detailed investigations.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana ecotype Columbia) seeds were grown on one-half-strength MS medium under a 16-h photoperiod (photosynthetic photon flux density of 60 μE m−2 s−1) and day/night temperatures of 22°C/16°C. For N limitation experiments, NH4+ was removed from the one-half-strength MS medium and NO3− was used as the sole N source. The medium contained various concentrations of NO3− and Suc, pH 5.8, 0.7% phytagel, and basal nutrients (1.5 mm CaCl2, 0.75 mm MgSO4, 0.63 mm KH2PO4, 0.05 mm FeSO4-EDTA, 0.05 mm Na2EDTA, 0.066 mm MnSO4, 0.050 mm H3BO3, 0.0025 mm potassium iodide, 0.015 mm ZnSO4, 0.005 mm Na2MoO4, 0.00005 mm CuSO4, and 0.00005 mm CoCl2; Murashige and Skoog, 1962). All wild-type, dfc mutant, and DFC complementation seedlings were grown on the same plate, and whole seedlings were harvested, frozen in liquid N2, and stored at −80°C until use. We first conducted experiments to determine the optimal nutrient-limiting conditions for Arabidopsis. Plants were grown on the plates without one of the following nutrients: sulfur, phosphorus, C, and N. Then, plants were also grown at different levels of C or N. The concentrations of supplementary C and N are expressed as millimolar; 1 mm Suc and 1 mm NO3− are designated as 1 C and 1 N, respectively. 30 C was chosen for all subsequent experiments, 0.3 n was chosen for an N-limited condition, and 9.4 n was chosen as an N-sufficient condition. Potassium level was balanced with potassium chloride to maintain 9.4 mm potassium. In the experiments at various CO2 levels, seedlings were grown under both N conditions for 2 weeks in either ambient air (400 μmol CO2 mol−1) or under an elevated CO2 condition (4,000 μmol CO2 mol−1). CO2 levels were monitored with a Senseair CO2 sensor (www.senseair.se).
Identification of the dfc Mutant and DFC Complementation Transformation
The SALK_008883 T-DNA insertion line was obtained from the Arabidopsis Biological Resource Center. T-DNA genotypes were confirmed with PCR using the gene-specific primers LP and RP or a T-DNA-specific primer (LBa1) and RP. After confirmation, the SALK_008883 T-DNA line was designated as the dfc mutant. For reverse transcription (RT)-PCR analysis, the DFC primers ahead of the T-DNA insertion were A and B, those spanning the T-DNA insertion site were C and D, and the ACTIN2 (ACT2) primers were ACTIN2-F and ACTIN2-R.
A 6,147-bp genomic DNA fragment containing the 1,448-bp promoter fragment ahead of the ATG and DFC (AT3G10160) genomic DNA sequences was amplified and cloned into the binary vector pHWG for molecular complementation (Karimi et al., 2002; Curtis and Grossniklaus, 2003). The construct was introduced into dfc plants using the floral dipping method (Clough and Bent, 1998). DFC complementation-transformed plants were screened on one-half-strength MS plates containing 25 mg L−1 hygromycin B and identified with PCR-based genotyping using the primer pairs HYG-LP and HYG-RP for the transgene, LP and RP for the wild-type genomic locus, and LBa1 and RP for the T-DNA insert locus in separate PCR procedures. All primers used in the study are listed in Supplemental Table S6.
Real-Time RT-PCR Expression Analysis
Total RNA was isolated from whole seedlings using Trizol reagent (Invitrogen; www.invitrogen.com). To eliminate any residual genomic DNA, total RNA was treated with RNase-free DNase I (New England Biolabs; www.neb.com) and used to synthesize first-strand complementary DNA (cDNA) using the RevertAid First Strand cDNA Synthesis kit (Fermentas; www.thermoscientificbio.com/fermentas/?redirect=true). ACT2 primers were used to detect genomic DNA contamination. Primer premier 5.0 (www.premierbiosoft.com) was used to design the primers.
Relative quantification values for each target gene were calculated by the 2-ΔΔCT method (Livak and Schmittgen, 2001) using ACT2 as an internal reference gene to compare data from different PCR runs or cDNA samples. To ensure the validity of the 2-ΔΔCT method, 3-fold serial dilutions of cDNA from control Arabidopsis plants were used to create standard curves, and the amplification efficiencies of the target and reference genes were approximately equal (Livak and Schmittgen, 2001). Expression of most of the genes was also confirmed by real-time relative quantitative RT-PCR using ACT2 as an internal control. Real-time relative quantitative RT-PCR analysis provided relative changes in gene expression, with the control treatment (9.4 n in wild-type seedlings) normalized to a value of 1. To compare the DFA, DFB, DFC, and DFD transcripts, the results given are percentages of the level of mRNA for the UBC gene (At5g25760). Data were statistically analyzed using Student’s t test. The results shown are representative of three independent experiments, and within each experiment, treatments were replicated three times, unless otherwise stated.
Measurement of Folate Profiles in Arabidopsis Seedlings
The following folates were purchased from Schircks Laboratories (www.schircks.com): 5-methyltetrahydrofolate, THF, 5-F-THF, 5,10-methenyltetrahydrofolate, dihydrofolate, 5-M-THF-Glun, and 5-F-THF-Glun (n = 1–4). Folic acid and MTX were purchased from Sigma (www.sigmaaldrich.com).
The preparation of samples for folate composition was carried out according to Zhang et al. (2005) with minor modifications. About 0.1 g of plant material (fresh weight) after 12 d of growth on the medium was ground to a fine powder in liquid N. The powder was transferred to a 1.5-mL tube, and 0.5 mL of extraction buffer (0.05 m phosphate buffer containing 1.0% ascorbic acid and 0.1% 2-mercaptoethanol, pH 6.5; freshly prepared) and 0.1 mL of the 10 μg mL−1 external standard (MTX) solution was added. The capped tube was placed at 100°C for 10 min (inhibition of enzymatic interconversion) and flash cooled on ice for another 10 min. Primary centrifugation was at 13,000 rpm for 10 min. The supernatant was transferred to a new tube, and then another 0.5 mL of extraction buffer was added to the pellet. The capped tube was placed at 100°C for 10 min (inhibition of enzymatic interconversion) and again flash cooled on ice for another 10 min. After centrifugation, the supernatants were combined. For deconjugation of polyglutamylated folates, 12 μL of rat serum was added to the extraction solution, which was then incubated at 37°C for 4 h. An additional treatment of 10 min at 100°C was carried out, followed by cooling on ice and centrifugation. The supernatant at the bottom of the centrifuge tube was then ready for LC-MS analysis. All sample preparation manipulations were carried out under subdued lighting.
The HPLC system was a TSQ Quantum LC-MS apparatus (Thermo Scientific; www.thermo.com) including a quaternary pump (flow rate of 0.2 mL min−1), an autosampler with column oven, and a degasser and electrospray ionization apparatus. The needle wash solvent was a mixture of methanol and water (50:50, v/v). A Zorbax RP-C18 column (2.1 mm × 100 mm i.d.; octadecylsilyl, 3.5-μm particle size [Agilent; www.home.agilent.com]) was used for all analyses. The mobile phase consisted of eluent A (0.1% formic acid in water) and eluent B (0.1% formic acid in acetonitrile). The starting eluent was 95% A and 5% B. The proportion of B was increased linearly to 10% over 3 min and then to 80% over 4 min. The proportion of B was then increased immediately to 100% and held for 3 min. The mobile phase was then immediately adjusted to its initial composition and held for 12 min in order to reequilibrate the column. The injection volume was 12 μL. The column was kept at 25°C in a column oven. The autosampler (kept at 4°C) was equipped with a black door to avoid exposing the sample to light. Under these conditions, the retention times for 5,10-methenyltetrahydrofolate, THF, 5-M-THF, MTX, folic acid, 5-F-THF, and dihydrofolate were 0.86, 2.56, 3.13, 10.61, 11.29, 11.36, and 11.49 min, respectively.
Sample preparation for the folylpolyglutamates of 5-M-THF-Glun and 5-F-THF-Glun (n = 1–4, 5, 7, and 8) was carried out as follows (Garratt et al., 2005; Lu et al., 2007). About 0.1 g of plant material (fresh weight) after 12 d of growth on the medium was ground to a fine powder in liquid N. The powder was transferred to an ice-cold 1.5-mL tube containing 0.25 mL of extraction buffer (50:50 methanol:water, 0.1% ascorbic acid, and 0.8% 2-mercaptoethanol, pH 6.0; freshly prepared, ice cold). The capped tube was centrifuged at 4°C and 13,000 rpm for 10 min. The supernatant was transferred to a new amber tube and centrifuged again. After centrifugation, 10 μL of MTX (500 ng mL−1) was added to 100 μL of supernatant, and the supernatant was ready for LC-MS analysis. All sample preparation manipulations were carried out under subdued lighting.
The needle wash solvent was a mixture of methanol and water (50:50, v/v). A Zorbax RP-C18 column (2.1 mm × 50 mm i.d.; octadecylsilyl, 3.5-μm particle size) from Agilent was used for all analyses. The mobile phase consisted of eluent A (0.1% formic acid in water) and eluent C (0.1% formic acid in acetonitrile). The starting eluent was 96% A and 4% C. The proportion of C was increased linearly to 19% over 1.5 min and then to 90% over 3.2 min. Eluent C was restored to 4% rapidly from 3.2 to 3.3 min. Then, the mobile phase was immediately adjusted to its initial composition and held for 12 min to reequilibrate the column. Injection volume was 6 μL. The column was maintained at 25°C in a column oven. The autosampler (kept at 4°C) was equipped with a black door to avoid exposing the samples to light.
The electrospray ionization source was operated in positive mode (4,800 V), and capillary temperature was maintained at 300°C. High-purity N served both as sheath and auxiliary gas and was set to 37 and 5 (arbitrary units), respectively. Argon (1.5 mTorr) was used as the collision-induced dissociation gas. Detection was carried out in the selected reaction monitoring mode. The mass spectroscopy transitions and fragmentation conditions selected for individual analytes are as follows: m/z 460.2→313 at 30 eV for 5-M-THF-Glu1, m/z 589.1→313 at 32 eV for 5-M-THF-Glu2, m/z 718.1→312.9 at 41 eV for 5-M-THF-Glu3, m/z 847.3→313.1 at 45 eV for 5-M-THF-Glu4, m/z 474.2→327.1 at 27 eV for 5-F-THF-Glu1, m/z 603.1→327 at 36 eV for 5-F-THF-Glu2, m/z 732→326.8 at 36 eV for 5-F-THF-Glu3, m/z 861.3→327 at 48 eV for 5-F-THF-Glu4, m/z 455→308 at 30 eV for MTX (external standard). The peak full width at half-maximum was set to 0.5 for both Q1 and Q3. The scan time for each analyte was 0.5 s. Under these conditions, the retention times of 5-M-THF-Glu1, 5-M-THF-Glu2, 5-M-THF-Glu3, 5-M-THF-Glu4, 5-F-THF-Glu1, 5-F-THF-Glu2, 5-F-THF-Glu3, 5-F-THF-Glu4, and MTX, were 1.74, 2.83, 7.14, 7.67, 7.90, 7.96, 8.00, 8.19, and 8.37 min, respectively. 5-M-THF-Glun and 5-F-THF-Glun (n = 5, 7, and 8) were analyzed using the predicted m/z, and the peak area was calculated and normalized to that of the wild type due to the lack of the standards. The predicted m/z values of 5-M-THF-Glu5, 5-M-THF-Glu7, 5-M-THF-Glu8, 5-F-THF-Glu5, 5-F-THF-Glu7, and 5-F-THF-Glu8 used in this study were 976.2, 617.7, 682.2, 990.0, 624.7, and 689.2, respectively.
Biochemical Analysis
Whole seedlings from dfc and the wild type grown on 0.3 or 9.4 n medium for 12 d were harvested for the following biochemical analyses. Soluble proteins were extracted from the frozen seedling powder with 100 mm HEPES-KOH (pH 7.5) and 0.1% Triton X-100 and assayed using a commercial protein assay kit (Bio-Rad; www.bio-rad.com). Soluble sugar (Glc and Fru) and starch were extracted and assayed using a commercially available kit (Megazyme; www.megazyme.com). Chlorophyll and anthocyanin in the frozen leaf powder were extracted and assayed according to Peng et al. (2007b). Free and total amino acids were analyzed with an ASI.KAUNER amino acid analyzer A200 (www.knauer.net). C and N contents were analyzed with a Perkin-Elmer 2400 Series II CHNS/O Elemental Analyzer (www.perkinelmer.com).
Measurement of NO3− and NH4+ Contents
Twelve-day-old seedlings were collected and immediately frozen in liquid N. Samples were boiled in water (20 mL g−1 fresh weight) and then frozen and thawed once to extract NO3−. After filtering through a 0.2-µm polyvinylidene fluoride membrane (Millipore; www.millipore.com), the extracted NO3− was reduced inline to NO2− in a copperized cadmium column and determined as NO2−. According to the Griess-Ilosvay reaction, NO2− is diazotized with sulfanilamide and coupled with N-(1-naphthyl)-ethylenediamine dihydrochloride to form a purple-red azo dye monitored at 538 nm (Oliveira et al., 2007). NH4+ was measured according to Andrew et al. (1995).
Antibody and Immunoblot Analysis
Anti-rabbit polyclonal antibodies were obtained from Agrisera (www.agrisera.com). For protein immunoblot analysis, 0.2 g of tissue was ground with liquid N and then homogenized in 200 μL of ice-cold 2× Laemmli buffer with minor changes (Laemmli, 1970). The buffer consisted of 125 mm Tris-HCl, pH 8.0, 20% glycerol, 2% SDS, 2% 2-mercaptoethanol, and 1% protease inhibitor cocktail (Invitrogen; www.invitrogen.com). The homogenate was heated at 65°C for 20 min, centrifuged at 10,000g for 10 min, and the supernatant was collected. Ten micrograms of protein was analyzed by SDS-PAGE. Anti-NR, anti-GOGAT, anti-GS1, anti-GS2 and alkaline phosphatase-labeled anti-rabbit IgG antibodies were obtained from Agrisera and used at dilutions of 1:1,000, 1:1,000, 1:10,000, 1:5,000, and 1:5,000, respectively. Bands were detected using 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium.
Extraction and NR and NiR Enzyme Activity Analysis
Whole 12-d-old seedlings (approximately 100 mg) were frozen in liquid N and ground with a mortar and pestle. The powdered tissues for NR analysis were added to 0.3 mL of extraction buffer (per 100 mg of tissue) containing 50 mm HEPES, pH 7.5, 1 mm EDTA, 10 mm MgCl2, 5 mm dithiothreitol, and 1% proteinase inhibitor (www.sigmaaldrich.com). The mixture was vortexed and then centrifuged at 14,000 rpm for 10 min at 4°C. The supernatant was used directly for NR assays with modifications to measure the increase in NO2− in the assay mixture (Lea et al., 2006; Wang et al., 2010). The assay mixture contained 50 mm HEPES, pH 7.5, 100 μm NADH, 5 mm KNO3, with 2 mm EDTA for NRmax or 5 mm MgCl2 for NRact. The assay volume was 1 mL, assays were run at 30°C for 30 min, and activity was measured in crude extracts by determining NO2− formation by adding 1% sulfanilamide and 0.02% N-(1-naphthyl)-ethylenediamine dihydrochloride. Absorbance was measured at 540 nm. NR enzyme activity was expressed as nmol of NO2− formed per min per mg of protein. The powdered tissues for NiR analysis were added to 0.3 mL of extraction buffer (per 100 mg of tissue) containing 50 mm potassium phosphate buffer (pH 7.5), 1 mm EDTA, 10 mm 2-mercaptoethanol, 100 mm phenylmethanesulfonyl fluoride, and 5 mg of polyvinylpyrrolidone and then homogenized. The homogenate was centrifuged, and the supernatant (crude enzyme solution) was used for the NiR activity analysis. A blank sample, in which sulfanilamide was added prior to the extract, was used for background reading. NiR activity was assayed following Takahashi et al. (2001), with modifications, to measure the decrease of NO2− in the assay mixture. A 45-μL sample of the crude enzyme solution was transferred to a 1.5-mL centrifuge tube, and 195 μL of the assay solution containing 50 mm potassium phosphate buffer (pH 7.5), 1 mm NaNO2, and 1 mm methyl viologen was added. The reaction was started by adding 60 μL of 57.4 mm Na2S2O4 in 290 mm NaHCO3 (final Na2S2O4 concentration in the assay solution, 11.5 mm), and the reaction was run for 5 min at 30°C. A 0.3-mL aliquot was transferred to a new tube containing 0.7 mL of water and mixed vigorously to stop the reaction, after which 1 mL of 1% (w/v) sulfanilamide in 3 n HCl and 1 mL of 0.02% (w/v) N-1-naphthylethylenediamine dihydrochloride were added. The absorbance of this mixture at 540 nm was measured. NiR enzyme activity was expressed as nmol of NO2− used per min per mg of protein.
GS and GOGAT Enzyme Activity Analysis
For the assessment of total GS activities, freshly harvested samples (500 mg) were ground on ice with extraction buffer consisting of 100 mm Tris-HCl (pH 7.6), 1 mm MgCl2, 1 mm EDTA, and 10 mm 2-mercaptoethanol. Semisynthetase GS activity was assayed, with NH2OH used as an artificial substrate, by quantifying the formation of Glu γ-monohydroxamate. The homogenates were centrifuged at 12,000g for 30 min at 4°C, and the supernatant was analyzed for total GS activities. Total leaf GS activity was measured in a preincubation assay buffer (30°C) consisting of 37.5 mm imidazole buffer (pH 7.0), 30 mm sodium Glu, 25 mm MgSO4, 10 mm NH2OH, and 30 mm ATP. The reaction was terminated after 15 min at 30°C by the addition of acidic FeCl3 solution (88 mm FeCl3, 670 mm HCl, and 200 mm TCA). After allowing 10 min for the color development, the reaction mixture was centrifuged at 4,000g at room temperature for 10 min, and 2 mL of supernatant was then transferred from each well into a new tube. The A540 was measured in a spectrophotometer quantification reader (O’Neal and Joy, 1973). GS enzyme activity was expressed as μmol of Glu γ-monohydroxamate formed per 15 min per mg of protein.
For the assessment of GOGAT activities, freshly harvested samples (100 mg) were ground on ice with extraction buffer consisting of 100 mm potassium phosphate buffer (pH 7.4), 1.28 mm EDTA, and 10 mm 2-mercaptoethanol. GOGAT activity was assayed by quantifying the formation of Glu and using NADH as the substrate. The reaction mixture consisted of 100 mm potassium phosphate buffer (pH 7.4), 10 mm Gln, 10 mm 2-oxoglutarate, 0.05 mm NADH, and extract. After a 5-min preincubation at 30°C, the reaction was started by adding the reductant solution (1.68 mg Na2S2O4 and 3.48 mg of NaHCO3 in 1 mL of reaction solution). After 15 min of incubation at 30°C, the reaction was terminated by heating to 98°C for 5 min. The Glu concentration was then determined using the ninhydrin reaction (Lancien et al., 2002). GOGAT activity was expressed as μmol of Glu formed per min per mg of protein.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At3g10160 (DFC), At3g18780 (ACT2), At1g77760 (NIA1), At1g37130 (NIA2), At2g15620 (NIR), At5g53460 (GLT1), At5g04140 (GLU1), At5g37600 (GS1.1), At5g35630 (GS2), At1g12110 (NRT1.1), At1g08090 (NRT2.1), At2g14210 (ANR1), At5g41480 (DFA), At5g05980 (DFB), At3g55630 (DFD), At5g57850 (ADCL), At2g28880 (ADCS), At3g07270 (GTPCHI), At2g21550 (DHFR), At1g50480 (THFS), At4g17360 (FDF), At2g38660 (DHC), At2g44160 (MTHFR2), At5g13050 (5-FCL), At3g03780 (MS2), and At5g25760 (UBC).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Arabidopsis DFC locus and characterization of the dfc mutant.
Supplemental Figure S2. Effects of the absence of sulfur, phosphorus, or C on the growth of wild-type, dfc, and DFC complementation seedlings, the germination percentage, the analysis of DFC complementation, and soluble protein concentrations of wild-type and dfc seeds.
Supplemental Table S1. Free amino acid profiles of wild-type, dfc, and DFC complementation seeds.
Supplemental Table S2. Free amino acid profiles of wild-type, dfc, and DFC complementation seedlings on 0.3 or 9.4 n medium in ambient air.
Supplemental Table S3. Profiles of total folate and various folate types of wild-type, dfc, and DFC complementation seedlings on 0.3 or 9.4 n medium in ambient air.
Supplemental Table S4. Profiles of 5-M-THF-Glun and 5-F-THF-Glun (n = 1–4) of wild-type and dfc seedlings on 0.3 or 9.4 n medium in ambient air or elevated CO2.
Supplemental Table S5. Free amino acid profiles of wild-type, dfc, and DFC complementation seedlings on 0.3 or 9.4 n medium under elevated CO2.
Supplemental Table S6. Primer sequences used for genotyping and gene expression analysis.
Acknowledgments
We are grateful to Flanders Interuniversity Institute for Biotechnology for providing the pHWG plasmid. We thank Dr. Jin Si (Animal Nutrition and Feed Research Institute, Chinese Academy of Agricultural Sciences) for help in the analyses of free amino acids, NO3−, nitrite, and NH4+. We thank Dr. Chengjun Ji (Department of Ecology, Peking University) for help with the C and N content analyses. We thank Profs. Dominique Van Der Straeten (Department of Physiology, Ghent University), Christopher A. Makaroff (Department of Chemistry and Biochemistry, Miami University), and Guofang Zhang (Department of Nutrition, Case Western Reserve University) for stimulating discussions and critical reading of the manuscript. We are grateful to two anonymous reviewers for their critical comments to improve the manuscript.
Glossary
- N
nitrogen
- C
carbon
- FPGS
folylpolyglutamate synthetase
- GDC
glycine decarboxylase
- SHMT
serine hydroxymethyltransferase
- MS
Murashige and Skoog
- NRmax
maximum nitrate reductase activity
- NRact
nitrate reductase activation activity
- NR
nitrate reductase
- NiR
nitrite reductase
- GOGAT
glutamine oxoglutarate aminotransferase
- GS
glutamine synthetase
- GS1
cytosolic form of glutamine synthetase
- GS2
chloroplastic form of glutamine synthetase
- THF
tetrahydrofolic acid
- LC-MS
liquid chromatography-tandem mass spectroscopy
- 5-M-THF
5-methyl-tetrahydrofolate
- 5-F-THF
5-fromyl-tetrahydrofolate
- m/z
mass-to-charge ratio
- MTX
methotrexate
- 5,10-CH2-THF
methylenetetrahydrofolate
- RT
reverse transcription
- T-DNA
transfer DNA
- cDNA
complementary DNA
References
- Andre C, Benning C. (2007) Arabidopsis seedlings deficient in a plastidic pyruvate kinase are unable to utilize seed storage compounds for germination and establishment. Plant Physiol 145: 1670–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew KN, Worsfold PJ, Comber M. (1995) On-line flow injection monitoring of ammonia in industrial liquid effluents. Anal Chim Acta 314: 33–43 [Google Scholar]
- Besson V, Rébeillé F, Neuburger M, Douce R, Cossins EA. (1993) Effects of tetrahydrofolate polyglutamates on the kinetic parameters of serine hydroxymethyltransferase and glycine decarboxylase from pea leaf mitochondria. Biochem J 292: 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi YM, Wang RL, Zhu T, Rothstein SJ. (2007) Global transcription profiling reveals differential responses to chronic nitrogen stress and putative nitrogen regulatory components in Arabidopsis. BMC Genomics 8: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancquaert D, Storozhenko S, Loizeau K, De Steur H, De Brouwer V, Viaene J, Ravanel S, Rébeillé F, Lambert W, Van Der Straeten D. (2010) Folates and folic acid: from fundamental research toward sustainable health. Crit Rev Plant Sci 29: 14–35 [Google Scholar]
- Bloom AJ, Burger M, Rubio Asensio JS, Cousins AB. (2010) Carbon dioxide enrichment inhibits nitrate assimilation in wheat and Arabidopsis. Science 328: 899–903 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet-Mercey S, Taconnat L, Renou JP, Daniel-Vedele F, Fernandez E, et al. (2009) The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J 57: 426–435 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collakova E, Goyer A, Naponelli V, Krassovskaya I, Gregory JF, III, Hanson AD, Shachar-Hill Y. (2008) Arabidopsis 10-formyl tetrahydrofolate deformylases are essential for photorespiration. Plant Cell 20: 1818–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossins EA. (2000) The fascinating world of folate and one-carbon metabolism. Can J Bot 78: 691–708 [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C, Lemaître T, Christ A, Azzopardi M, Kato Y, Sato F, Morot-Gaudry JF, Le Dily F, Masclaux-Daubresse C. (2008) Nitrogen recycling and remobilization are differentially controlled by leaf senescence and development stage in Arabidopsis under low nitrogen nutrition. Plant Physiol 147: 1437–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C, Saliba-Colombani V, Loudet O, Belluomo P, Moreau L, Daniel-Vedele F, Morot-Gaudry JF, Masclaux-Daubresse C. (2006) Leaf yellowing and anthocyanin accumulation are two genetically independent strategies in response to nitrogen limitation in Arabidopsis thaliana. Plant Cell Physiol 47: 74–83 [DOI] [PubMed] [Google Scholar]
- Dutilleul C, Lelarge C, Prioul JL, De Paepe R, Foyer CH, Noctor G. (2005) Mitochondria-driven changes in leaf NAD status exert a crucial influence on the control of nitrate assimilation and the integration of carbon and nitrogen metabolism. Plant Physiol 139: 64–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambonnet B, Jabrin S, Ravanel S, Karan M, Douce R, Rébeillé F. (2001) Folate distribution during higher plant development. J Sci Food Agric 81: 835–841 [Google Scholar]
- Gan Y, Filleur S, Rahman A, Gotensparre S, Forde BG. (2005) Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 222: 730–742 [DOI] [PubMed] [Google Scholar]
- Garratt LC, Ortori CA, Tucker GA, Sablitzky F, Bennett MJ, Barrett DA. (2005) Comprehensive metabolic profiling of mono- and polyglutamated folates and their precursors in plant and animal tissue using liquid chromatography/negative ion electrospray ionisation tandem mass spectrometry. Rapid Commun Mass Spectrom 19: 2390–2398 [DOI] [PubMed] [Google Scholar]
- Gaufichon L, Reisdorf-Cren M, Rothstein SJ, Chardon F, Suzuki A. (2010) Biological functions of asparagine synthetase in plants. Plant Sci 179: 141–153 [Google Scholar]
- Gojon A, Krouk G, Perrine-Walker F, Laugier E. (2011) Nitrate transceptor(s) in plants. J Exp Bot 62: 2299–2308 [DOI] [PubMed] [Google Scholar]
- Goyer A, Collakova E, Díaz de la Garza R, Quinlivan EP, Williamson J, Gregory JF, III, Shachar-Hill Y, Hanson AD. (2005) 5-Formyltetrahydrofolate is an inhibitory but well tolerated metabolite in Arabidopsis leaves. J Biol Chem 280: 26137–26142 [DOI] [PubMed] [Google Scholar]
- Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Gregory JF., III (2011) Folate biosynthesis, turnover, and transport in plants. Annu Rev Plant Biol 62: 105–125 [DOI] [PubMed] [Google Scholar]
- Hanson AD, Roje S. (2001) One-carbon metabolism in higher plants. Annu Rev Plant Physiol Plant Mol Biol 52: 119–137 [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Hu HC, Wang YY, Tsay YF. (2009) AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J 57: 264–278 [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Machida C, Yoshioka Y, Kitano H, Machida Y. (2003) The GLOBULAR ARREST1 gene, which is involved in the biosynthesis of folates, is essential for embryogenesis in Arabidopsis thaliana. Plant J 33: 235–244 [DOI] [PubMed] [Google Scholar]
- Jabrin S, Ravanel S, Gambonnet B, Douce R, Rébeillé F. (2003) One-carbon metabolism in plants: regulation of tetrahydrofolate synthesis during germination and seedling development. Plant Physiol 131: 1431–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WM, Kandlbinder A, Stoimenova M, Glaab J. (2000) Discrepancy between nitrate reduction rates in intact leaves and nitrate reductase activity in leaf extracts: what limits nitrate reduction in situ? Planta 210: 801–807 [DOI] [PubMed] [Google Scholar]
- Kang J, Turano FJ. (2003) The putative glutamate receptor 1.1 (AtGLR1.1) functions as a regulator of carbon and nitrogen metabolism in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 6872–6877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Bi YM, Rothstein SJ. (2011) Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J Exp Bot 62: 1499–1509 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. (2010) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Krouk G, Tillard P, Gojon A. (2006) Regulation of the high-affinity NO3− uptake system by NRT1.1-mediated NO3− demand signaling in Arabidopsis. Plant Physiol 142: 1075–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lancien M, Martin M, Hsieh MH, Leustek T, Goodman H, Coruzzi GM. (2002) Arabidopsis glt1-T mutant defines a role for NADH-GOGAT in the non-photorespiratory ammonium assimilatory pathway. Plant J 29: 347–358 [DOI] [PubMed] [Google Scholar]
- Lea US, Leydecker MT, Quilleré I, Meyer C, Lillo C. (2006) Posttranslational regulation of nitrate reductase strongly affects the levels of free amino acids and nitrate, whereas transcriptional regulation has only minor influence. Plant Physiol 140: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaître T, Gaufichon L, Boutet-Mercey S, Christ A, Masclaux-Daubresse C. (2008) Enzymatic and metabolic diagnostic of nitrogen deficiency in Arabidopsis thaliana Wassilewskija accession. Plant Cell Physiol 49: 1056–1065 [DOI] [PubMed] [Google Scholar]
- Little DY, Rao H, Oliva S, Daniel-Vedele F, Krapp A, Malamy JE. (2005) The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci USA 102: 13693–13698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Merigout P, Talbotec J, Daniel-Vedele F. (2003) Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis. Plant Physiol 131: 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Kwon YK, Rabinowitz JD. (2007) Isotope ratio-based profiling of microbial folates. J Am Soc Mass Spectrom 18: 898–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Ryan KS. (2001) Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol 127: 899–909 [PMC free article] [PubMed] [Google Scholar]
- Martin T, Oswald O, Graham IA. (2002) Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol 128: 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Reisdorf-Cren M, Pageau K, Lelandais M, Grandjean O, Kronenberger J, Valadier MH, Feraud M, Jouglet T, Suzuki A. (2006) Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol 140: 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrshahi P, Gonzalez-Jorge S, Akhtar TA, Ward JL, Santoyo-Castelazo A, Marcus SE, Lara-Núñez A, Ravanel S, Hawkins ND, Beale MH, et al. (2010) Functional analysis of folate polyglutamylation and its essential role in plant metabolism and development. Plant J 64: 267–279 [DOI] [PubMed] [Google Scholar]
- Miller AJ, Fan X, Shen Q, Smith SJ. (2008) Amino acids and nitrate as signals for the regulation of nitrogen acquisition. J Exp Bot 59: 111–119 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Ogren WL. (1984) Photorespiration: pathways, regulation, and modification. Annu Rev Plant Physiol Plant Mol Biol 35: 415–442 [Google Scholar]
- Oliveira SM, Lopes TIMS, Rangel AOSS. (2007) Sequential injection determination of nitrate in vegetables by spectrophotometry with inline cadmium reduction. Commun Soil Sci PLAN 38: 533–544 [Google Scholar]
- O’Neal D, Joy KW. (1973) Glutamine synthetase of pea leaves. I. Purification, stabilization, and pH optima. Arch Biochem Biophys 159: 113–122 [DOI] [PubMed] [Google Scholar]
- Orsel M, Krapp A, Daniel-Vedele F. (2002) Analysis of the NRT2 nitrate transporter family in Arabidopsis: structure and gene expression. Plant Physiol 129: 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible WR. (2009) Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol 150: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Bi YM, Zhu T, Rothstein SJ. (2007a) Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol Biol 65: 775–797 [DOI] [PubMed] [Google Scholar]
- Peng M, Hannam C, Gu H, Bi YM, Rothstein SJ. (2007b) A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J 50: 320–337 [DOI] [PubMed] [Google Scholar]
- Rachmilevitch S, Cousins AB, Bloom AJ. (2004) Nitrate assimilation in plant shoots depends on photorespiration. Proc Natl Acad Sci USA 101: 11506–11510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel S, Cherest H, Jabrin S, Grunwald D, Surdin-Kerjan Y, Douce R, Rébeillé F. (2001) Tetrahydrofolate biosynthesis in plants: molecular and functional characterization of dihydrofolate synthetase and three isoforms of folylpolyglutamate synthetase in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 15360–15365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel SP, Block MA, Rippert P, Jabrin S, Curien G, Rébeillé F, Douce R. (2004) Methionine metabolism in plants: chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J Biol Chem 279: 22548–22557 [DOI] [PubMed] [Google Scholar]
- Rebeille F, Neuburger M, Douce R. (1994) Interaction between glycine decarboxylase, serine hydroxymethyltransferase and tetrahydrofolate polyglutamates in pea leaf mitochondria. Biochem J 302: 223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, Gojon A. (2006) A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol 140: 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M. (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schippers JHM, Nunes-Nesi A, Apetrei R, Hille J, Fernie AR, Dijkwel PP. (2008) The Arabidopsis onset of leaf death5 mutation of quinolinate synthase affects nicotinamide adenine dinucleotide biosynthesis and causes early ageing. Plant Cell 20: 2909–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedow JN, Day DA. (2000) Respiration and photorespiration. In B Buchanan, W Gruissem, R Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Biologists, Rockville, MD, pp 676–729
- Srivastava AC, Ramos-Parra PA, Bedair M, Robledo-Hernández AL, Tang Y, Sumner LW, Díaz de la Garza RI, Blancaflor EB. (2011) The folylpolyglutamate synthetase plastidial isoform is required for postembryonic root development in Arabidopsis. Plant Physiol 155: 1237–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Krapp A. (1999) The interaction between elevated carbon dioxide and nitrogen nutrition: the physiological and molecular background. Plant Cell Environ 22: 583–621 [Google Scholar]
- Takahashi M, Sasaki Y, Ida S, Morikawa H. (2001) Nitrite reductase gene enrichment improves assimilation of NO2 in Arabidopsis. Plant Physiol 126: 731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM. (1993) The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713 [DOI] [PubMed] [Google Scholar]
- Tschoep H, Gibon Y, Carillo P, Armengaud P, Szecowka M, Nunes-Nesi A, Fernie AR, Koehl K, Stitt M. (2009) Adjustment of growth and central metabolism to a mild but sustained nitrogen-limitation in Arabidopsis. Plant Cell Environ 32: 300–318 [DOI] [PubMed] [Google Scholar]
- Vert G, Chory J. (2009) A toggle switch in plant nitrate uptake. Cell 138: 1064–1066 [DOI] [PubMed] [Google Scholar]
- Vidal EA, Gutiérrez RA. (2008) A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr Opin Plant Biol 11: 521–529 [DOI] [PubMed] [Google Scholar]
- Vidal EA, Tamayo KP, Gutierrez RA. (2010) Gene networks for nitrogen sensing, signaling, and response in Arabidopsis thaliana. Wiley Interdiscip Rev Syst Biol Med 2: 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidmar JJ, Zhuo D, Siddiqi MY, Schjoerring JK, Touraine B, Glass ADM. (2000) Regulation of high-affinity nitrate transporter genes and high-affinity nitrate influx by nitrogen pools in roots of barley. Plant Physiol 123: 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Du Y, Li Y, Ren D, Song CP. (2010) Hydrogen peroxide-mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. Plant Cell 22: 2981–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wilder V, De Brouwer V, Loizeau K, Gambonnet B, Albrieux C, Van Der Straeten D, Lambert WE, Douce R, Block MA, Rebeille F, et al. (2009) C1 metabolism and chlorophyll synthesis: the Mg-protoporphyrin IX methyltransferase activity is dependent on the folate status. New Phytol 182: 137–145 [DOI] [PubMed] [Google Scholar]
- Zhang GF, Storozhenko S, Van Der Straeten D, Lambert WE. (2005) Investigation of the extraction behavior of the main monoglutamate folates from spinach by liquid chromatography-electrospray ionization tandem mass spectrometry. J Chromatogr A 1078: 59–66 [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG. (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
- Zheng ZL. (2009) Carbon and nitrogen nutrient balance signaling in plants. Plant Signal Behav 4: 584–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo D, Okamoto M, Vidmar JJ, Glass AD. (1999) Regulation of a putative high-affinity nitrate transporter (Nrt2;1At) in roots of Arabidopsis thaliana. Plant J 17: 563–568 [DOI] [PubMed] [Google Scholar]