Abstract
Dark-grown seedlings exhibit skotomorphogenic development. Genetic and molecular evidence indicates that a quartet of Arabidopsis Phytochrome (phy)-Interacting bHLH Factors (PIF1, 3, 4, and 5) are critically necessary to maintaining this developmental state and that light activation of phy induces a switch to photomorphogenic development by inducing rapid degradation of the PIFs. Here, using integrated ChIP–seq and RNA–seq analyses, we have identified genes that are direct targets of PIF3 transcriptional regulation, exerted by sequence-specific binding to G-box (CACGTG) or PBE-box (CACATG) motifs in the target promoters genome-wide. In addition, expression analysis of selected genes in this set, in all triple pif-mutant combinations, provides evidence that the PIF quartet members collaborate to generate an expression pattern that is the product of a mosaic of differential transcriptional responsiveness of individual genes to the different PIFs and of differential regulatory activity of individual PIFs toward the different genes. Together with prior evidence that all four PIFs can bind to G-boxes, the data suggest that this collective activity may be exerted via shared occupancy of binding sites in target promoters.
Author Summary
An important issue in understanding mechanisms of eukaryotic transcriptional regulation is how members of large transcription-factor families, with conserved DNA–binding domains (such as the 162-member Arabidopsis bHLH family), discriminate between target genes. However, the specific question of whether, and to what extent, closely related sub-family members, with potential overlapping functional redundancy (like the quartet of Phytochrome (phy)-Interacting bHLH transcription Factors (PIF1, 3, 4, and 5) studied here), share regulation of target genes through shared binding to promoter-localized consensus motifs does not appear to have been widely investigated. Here, using ChIP–seq analysis, we have identified genes that bind PIF3 to conserved, sequence-specific sites in their promoters; and, using RNA–seq, we have identified those genes displaying altered expression in various pif mutants. Integration of these data identifies those genes that are likely direct targets of transcriptional regulation by PIF3. Our data suggest that the PIF quartet members share directly in transcriptional activation of numerous target genes, potentially via redundant promoter occupancy, in a manner that varies quantitatively from gene to gene. This finding suggests that these PIFs function collectively as a signaling hub, selectively partitioning common upstream signals from light-activated phys at the transcriptional-network interface.
Introduction
A key component of the successful colonization of land by terrestrial flowering plants was the evolution of a developmental strategy termed skotomorphogenesis (etiolated growth). This strategy enabled post-germinative seedlings emerging from buried seed to grow heterotrophically, on seed reserves, rapidly upwards through the subterranean darkness to the soil surface. Coupled with this was the evolution of a photosensory mechanism to trigger a switch to autotrophic, photomorphogenic (deetiolated) development upon emergence into sunlight.
Genetic evidence indicates that a small subfamily of basic helix-loop-helix (bHLH) transcription factors, termed PIFs (for Phytochrome (phy)-Interacting Factors) are centrally critical to the promotion of such skotomorphogenic development in dark-grown seedlings [1]. A quadruple pif mutant (pifq), lacking PIF-family members PIF1, PIF3, PIF4 and PIF5 (termed the PIF quartet), displays morphogenic development in total darkness that strongly phenocopies that of normal light-grown seedlings [2], [3]. This observation establishes that these factors act constitutively to promote skotomorphogenic development and that their absence induces the switch to photomorphogenic development. All four quartet members have been shown individually to bind preferentially to a core G-box DNA-sequence motif (CACGTG) (a variant of the canonical E-box motif (CANNTG)) [4]–[9], and to function as transcriptional activators in transfection or heterologous systems [4]–[7], [10]. Because monogenic mutants at each of these loci have no, or minimal, visible effects on skotomorphogenesis, and the various double and triple pif-mutant combinations progressively exhibit increasingly photomorphogenic phenotypes in darkness, it appears that the PIF quartet members act with partially additive or overlapping redundancy to drive the skotomorphogenic pathway [2], [3], [11]–[13].
The phy family of sensory photoreceptors (especially phyA and phyB) has a central role in inducing the switch from skotomorphogenic to photomorphogenic development (deetiolation) in response to initial exposure of dark-grown seedlings to light [1], [14], [15]. The existing evidence indicates that this is achieved in large part by rapid phy-triggered degradation of the PIF proteins. The mechanism underlying this process involves the rapid, light-induced translocation of the activated (Pfr) conformer of the phy molecule from the cytoplasm into the nucleus, where it physically interacts with PIF-quartet members. This interaction induces phosphorylation of the PIF proteins which in turn triggers ubiquitylation and proteolytic degradation of the transcription factors (half-lives of 5–20 min) via the proteasome system. The altered transcriptional landscape resulting from the consequent robust reduction in steady-state abundance of these factors is the major driving force in the switch from heterotrophic to autotrophic development inherent in the deetiolation process.
A limited number of transcriptome analyses, using Affymetrix ATH1 microarrays, aimed at identifying genes regulated by the phy-PIF signaling pathway during deetiolation have been reported [3], [12], [16]–[19]. The data show that 80% of the genes that display altered expression in the pifq mutant in the dark are normally altered by prolonged light in fully deetiolated wild-type (WT) seedlings [17], but that only a relatively small fraction of these are misexpressed in dark-grown pif1 [19], pif3 [16]–[18] and pif4pif5 mutants [12]. These results affirm the central collective regulatory function of these four PIFs in regulating the overall transcriptional network that drives the developmental switch from skotomorphogenesis to photomorphogenesis, and provide initial indications of functional redundancy at the gene expression level. These genes could be either direct or indirect targets of PIF transcriptional regulatory activity [20]. Identification of those genes that respond rapidly (within 1 h) to initial light exposure has defined a subset of PIF-regulated genes that are likely to be enriched for loci that are directly transcriptionally regulated by the PIF-quartet proteins [17]. PIF-regulated genes that conversely respond rapidly to vegetative shade in fully-green, light-grown plants have also been identified by microarray-based expression profiling [11], [21]. It is notable that these early-response genes are enriched for transcription-factor-encoding loci, suggesting a potential hierarchal network that drives a transcriptional cascade. However, rapid responsiveness alone obviously does not establish that transcriptional regulation is direct.
The advent of ChIP-chip and ChIP-seq technology has provided the opportunity to identify genes that contain binding sites for transcription factors of interest, on a genome-wide scale [20], [22], [23]. When combined with full transcriptome analysis, the data provide identification of genes that are direct targets of transcriptional regulation by the factor(s) under study. A number of such studies have recently been reported for a diversity of factors in Arabidopsis, using either ChIP-chip or ChIP-seq analysis of factor binding sites, coupled predominantly with Affymetrix ATH1 microarrays (representing about 80% of the protein-coding genes in the genome) for expression analysis [21], [23]–[29]. These data have begun to provide insight into the complexity of the transcriptional networks that coordinate a variety of the fundamental processes underlying plant growth and development.
Despite these advances, the use of the ATH1 microarray for expression analysis in many of these studies means that important expression changes in genes not present on this array might have been missed. In addition, the question of whether, and to what extent, closely related transcription-factor family members, such as the PIF quartet, with apparently shared DNA-target-sequence specificity, contribute toward the transcriptional regulation of common target genes does not appear to have been addressed in many existing studies of eukaryotic systems [27], [30]–[34], although a recent report by Hornitschek et al shows differential binding of recombinant PIF4 and PIF5 to various E-box variants in vitro using protein-binding microarrays, as well as shared binding in vivo to four selected promoters using ChIP-PCR analysis [21]. Here, using ChIP-seq analysis, we have identified PIF3-binding sites, genome wide, and, in parallel, using RNA-seq analysis of selected pif-mutant lines, we have defined the genes regulated by PIF3, genome-wide, in dark-grown seedlings. By merging these datasets, we have identified those genes whose expression is, at least partially, directly regulated by promoter-bound PIF3. In addition, by profiling the expression of a selected subset of these direct PIF3-targets in multiple additional pif-mutant combinations, we have addressed the question of whether PIF1, 3, 4 and 5 display qualitative and/or quantitative functional divergence in regulating shared target genes.
Results
ChIP–seq–based identification of PIF3-binding sites
Two-day-old dark-grown wild-type (WT) and MYC-epitope-tagged-PIF3 (P3M)-expressing, pif3-3 null-mutant seedlings were used for ChIP-seq analysis. DNA prepared from MYC-antibody-generated immunoprecipitates from four independent biological replicates of each genotype was subjected to high-throughput sequencing. Statistically-significant binding peaks were defined by comparing the parallel P3M and WT ChIP samples within each replicate using the MACS algorithm [35]. Replicate-specific peaks (Table S1) were defined as reproducible if they were identified at the same genomic location in two or more biological replicates (overlapping Venn sectors in Figure 1A; also Table S2). For each reproducible peak, we assigned a common summit as the mean of the individual replicate-specific summits.
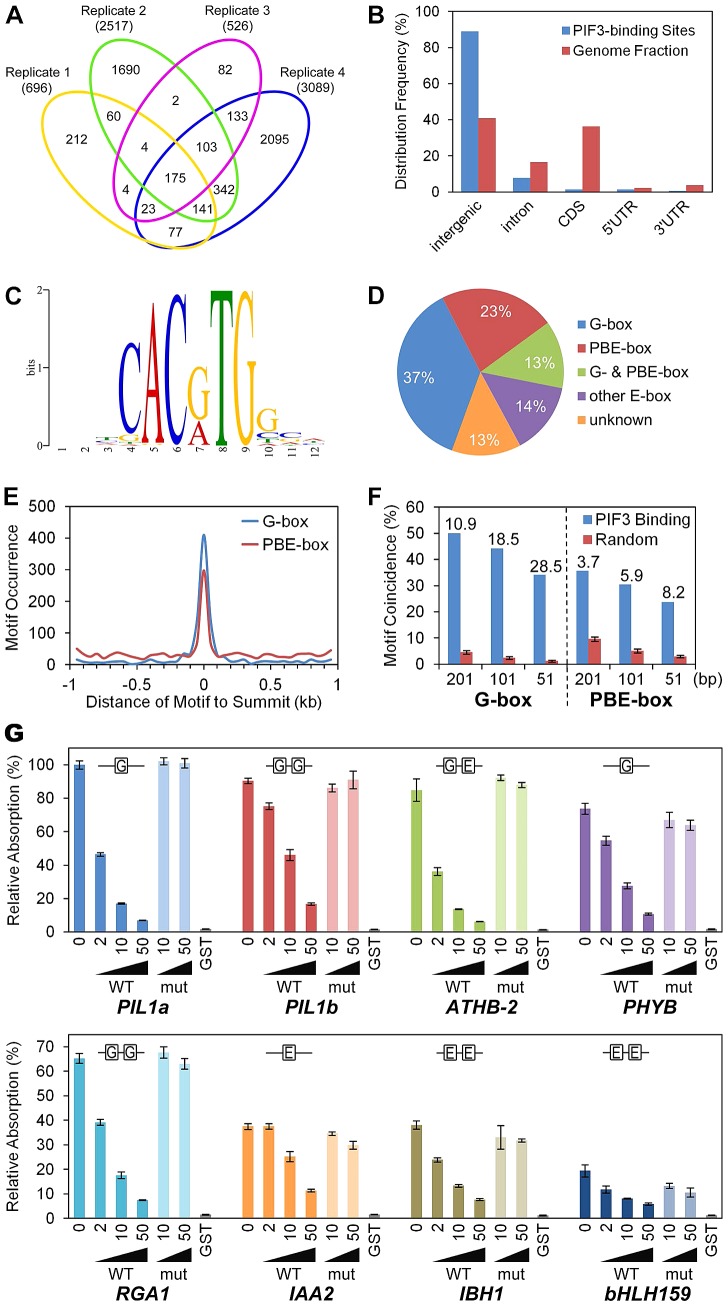
Figure 1. Genome-wide identification of PIF3-binding sites and motifs.
(A) Venn diagram depicting total numbers (parentheses) and reproducible presence (overlapping sectors) of statistically significant PIF3-binding peaks in ChIP-seq analysis of four biological replicates (Venn ovals) of dark-grown seedlings. (B) Relative binding-peak distribution across genomic regions. (C) MEME motif search identifies two dominant PIF3-binding motifs, defined as G-box (CACGTG) and PBE-box (CACATG) motifs. (D) Percentage of PIF3 binding sites containing designated motifs. Other E-box: Variants of E-box (CANNTG) motif other than G- or PBE-box. Unknown: Unknown and/or non-statistically-overrepresented motif. (E) Distribution of the G- and PBE-box motifs in the 1 kb regions surrounding the PIF3-binding peak-summits. (F) G- and PBE-box-motif coincidence with PIF3-binding peaks (% within 201, 101, and 51 bp centered at the peak-summits) is significantly higher than in other random genomic regions of the same size. Internal numbers indicate the relative fold motif-enrichment at PIF3 binding-sites. Error bars represent the standard deviation of 100 random simulations. (G) DPI-ELISA assays of in vitro binding of recombinant GST-PIF3 to the G- and PBE-box motifs. Binding activity (Relative Absorbance) for each DNA probe is expressed as a percentage of each reaction relative to GST-PIF3 binding to the PIL1a WT probe. Data represent the mean of independent duplicates +/− SEM. WT: wild-type competitor probes. mut: competitor probes mutated at the G-box and PBE-box motifs. GST: GST negative-control binding to the biotinylated WT probes.
This analysis identified 1064 reproducible peaks which form our “high-confidence” set of PIF3-binding sites (Table S2). These sites are evenly distributed on the five chromosomes and 89% are located in intergenic regions (Figure 1B). In ChIP-qPCR validation assays, all but 1 of the 38 tested regions exhibited strong binding enrichment in the P3M samples compared to the WT controls (Figure S1), indicating a low false positive rate for our ChIP-seq procedure.
G-box and PBE-box motifs dominate PIF3-binding DNA sequences
Using the MEME program [36], we performed de novo motif discovery on the +/−100 bp regions surrounding the 1064 “high-confidence” PIF3 binding-peak summits described above. Two E-box (CANNTG) variants were identified as statistically overrepresented motifs within these PIF3-binding regions (Figure 1C). The CACGTG (‘G-box’) variant is well-established as a preferred PIF-binding motif [4]–[9]. By contrast, the CACATG variant is previously undescribed as a PIF3-binding motif, although PIF1 [37] and, recently, PIF4 [21] have been reported to bind. We conclude that this variant is a strong candidate for being a general alternative binding motif for PIF3 across the genome, and define it, therefore, as the PIF-binding E-box (PBE-box). The relative distribution of these two motifs across the 1064 PIF3 binding-sites is summarized in Figure 1D. A majority (73%) of the sites contain one or both motifs (G-box 50% and PBE-box 36%, with 13% overlap) within the 200-bp window.
A broader analysis shows that 64% of the G-box and 30% of the PBE-box motifs present in the 2 kb windows surrounding the PIF3-binding summits cluster within the designated 200-bp binding sites (Figure 1E). Similarly, both motifs are strongly enriched in these 200-bp windows compared to random 200-bp genome segments, and this enrichment increases toward the PIF3-binding summit (Figure 1F). These data establish the highly significant coincidence between PIF3 and these two specific cis-elements.
To examine the potential direct interaction of PIF3 with the newly-identified PBE-box compared to that of the G-box, we performed DNA-Protein-Interaction (DPI)-ELISA [38]. We tested the binding of PIF3 to several G-box- (PIL1, PHYB, and RGA1) or PBE-box- (IAA2, IBH1, and AT4G30410) containing probes generated from various genomic PIF3-binding sites identified in the ChIP-seq analysis. Figure 1G shows that recombinant PIF3 binds sequence-specifically to all G-box- and PBE-box-containing probes, although the apparent affinity for the G-box seems overall to be higher than for the E-box. An EMSA analysis showed similar results (Figure S2). These in vitro binding-assay data indicate that the coincidence of PIF3-binding sites with the G-box or PBE-box motif in the ChIP-seq assay likely results from their direct interaction in vivo, and that the PBE-box is indeed another sequence-specific PIF-binding, E-box variant genome wide. Because all of the PIF3-binding sites tested by ChIP-qPCR in Figure S1 contain coincident G- or PBE-box motifs, these data validate the in vivo-binding of PIF3 to these motifs.
The binding of PIF3 to the ATHB-2 probe, which contains one G-box and one PBE-box, provides an interesting insight. Neither the competitor mutated in both motifs (Figure 1G; also Figure S2), nor the competitor mutated only in the G-box motif (Figure S3) displayed competitive activity, whereas the probe mutated only in the PBE-box motif showed competitive efficiency similar to that of the WT sequence (Figure S2). These findings suggest that PIF3 may have differential binding affinity toward these two motifs in specific genomic contexts.
Identification of genes associated with PIF3-binding sites
Although all ChIP-defined transcription factor binding sites may prove to be functionally significant, we have chosen here to focus on identifying those genes displaying motif-coincident PIF3-binding sites located in conventional promoter regions (defined here as “PIF3-bound genes”). Initially, from the 1064 binding sites defined above, we identified 709 sites that are both intergenic and G- and/or PBE-box-coincident (Table S2). For these 709 sites, we defined PIF3-bound genes as having a binding site in the 5′ flanking DNA, within 5 kb of the transcription start site (TSS), in the absence of intervening genes. This analysis identified 596 PIF3-binding sites, with 828 associated genes, where some sites are associated with two genes on opposite strands. Of these genes, 88% have PIF3-binding sites within 3 kb of TSS, whereas the remaining 12% have sites between 3 and 5 kb upstream (Table S3). These 828 genes thus constitute a set of PIF3-bound genes whose transcription is potentially directly regulated by PIF3.
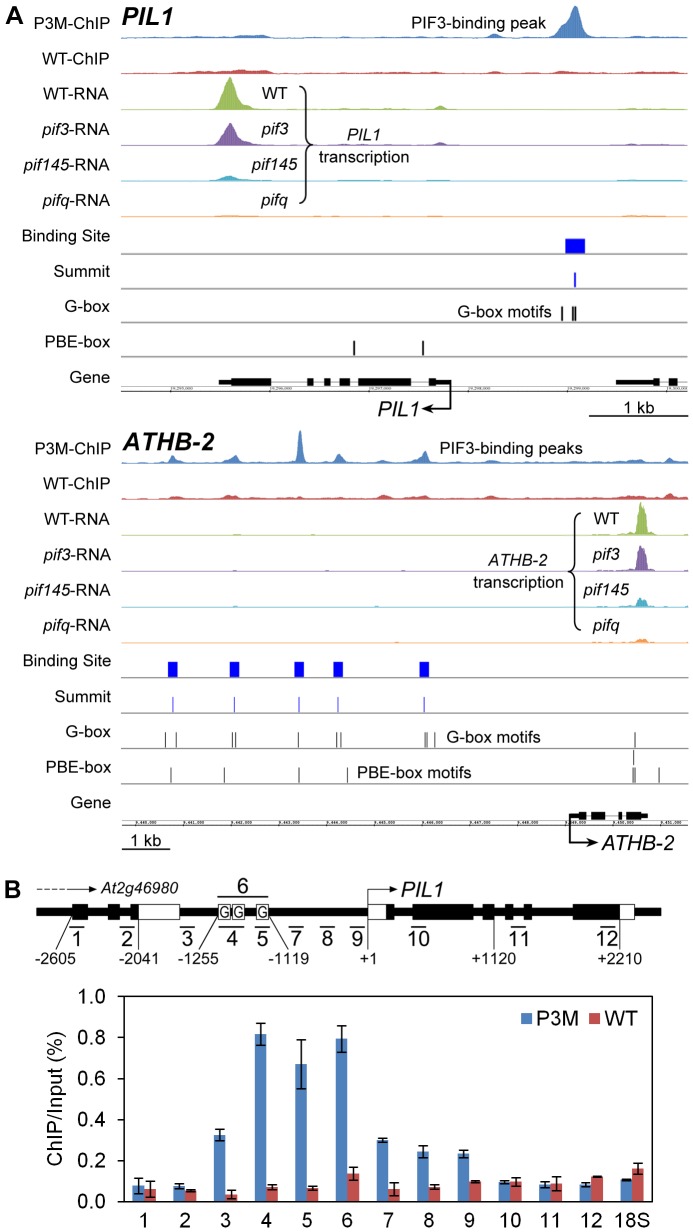
To provide genome-wide visualization of the ChIP-seq analysis, we developed a platform using the Integrated Genome Browser [39]. Figure 2A shows the chromosomal regions around PIL1 and ATHB-2, as examples. The chromosomal region surrounding the PIL1 gene shows a single PIF3-binding peak that is coincident with three G-box motifs located in the PIL1 promoter region. ATHB-2 is somewhat unusual in that it displays five specific PIF3-binding peaks in its extensive 5′-upstream region, each coincident with 1 to 3 G-box motifs (Figure 2A). ChIP-qPCR analysis scanning across the PIL1 genomic region provides robust validation of the ChIP-seq data for this gene (Figure 2B).
Figure 2. Compiled ChIP–seq and RNA–seq data identify PIL1 and ATHB2 as direct targets of PIF3 transcriptional regulation.
(A) Visualization of ChIP-seq and RNA-seq data in the genomic regions encompassing two representative genes, PIL1 and ATHB-2. The ChIP and RNA tracks show the pile-up distribution of the combined raw reads from four biological replicates of ChIP-seq data and three replicates of RNA-seq data, respectively. P3M- and WT-ChIP: DNA immunoprecipitated from PIF3-Myc-expressing and from wild-type seedlings, respectively. WT-, pif3-, pif145- and pifq-RNA: RNA from genotypes used for expression analysis. Binding Site: 201 bp defined as the PIF3-binding site. Summit: predicted PIF3-binding center defined from the binding-peak maximum. G- and PBE-box: Vertical lines indicate motif positions. (B) ChIP-qPCR verification of specific PIF3 binding to the G-box-located promoter region of PIL1. The schematic diagram illustrates the genomic region around the PIL1 locus. The short bars with numbers show 12 specific qPCR products. The black and white rectangular boxes represent CDS and UTR, respectively. Boxes labeled ‘G’ indicate the approximate locations of three G-box motifs in the PIL1 promoter. The relative enrichment level is represented by the percentage of co-immunoprecipitated DNA to the input control in the P3M and WT samples. Data are represented as the mean of biological triplicates +/− SEM. 18S: 18S rRNA as internal control.
Definition of PIF3 contribution to the transcriptional pattern collectively regulated by the PIF-quartet during skotomorphogenic development
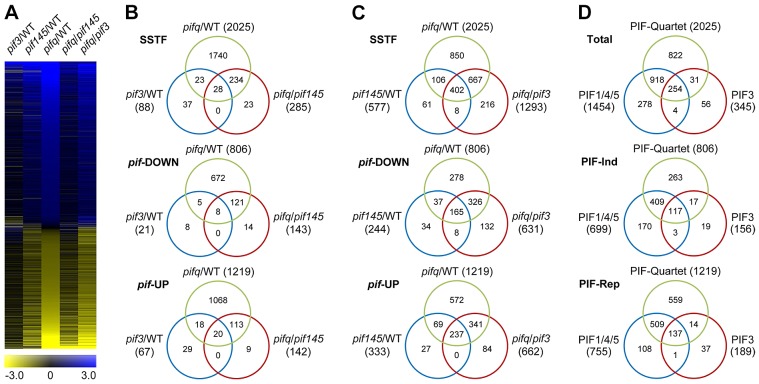
To identify the genes regulated by PIF3, genome-wide, in the promotion of skotomorphogenic development, we performed 3′-end-capture directional RNA-seq analysis, comparing the expression profiles of 2-d dark-grown WT, pif3, pif1pif4pif5 (pif145) and pif1pif3pif4pif5 (pifq) Arabidopsis seedlings. Genes displaying Statistically-Significant Two-Fold (SSTF) expression changes in the three mutant genotypes compared to the WT and each other were identified as being regulated by the relevant mutated PIF(s) (Figure 3; listed in Tables S4, S5, S6, S7, S8). The degree of overlap between SSTF genes identified in these comparisons is depicted in the Venn diagrams in Figure 3B, 3C and 3D. We defined a combined total of 345 genes in the pif3/WT and pifq/pif145 gene-sets as the composite PIF3-regulated gene-set (Table S9). Similarly, a combined total of 1454 genes in the pif145/WT and pifq/pif3 gene-sets were defined as the composite PIF1/4/5-trio regulated gene-set (Table S10). Comparison of these composite gene-sets is displayed in Figure 3D.
Figure 3. RNA–seq analysis of selected pif-mutants identifies PIF-regulated genes genome wide.
(A) Hierarchical clustering of differentially expressed genes by fold-change in expression (log2 scale), in the five pairwise genotypic comparisons indicated (pif3/WT, pif145/WT. pifq/WT, pifq/pif145 and pifq/pif3). Data shown are for the genes identified here as displaying Statistically-Significant, Two-Fold (SSTF) differences in any pairwise comparison. (B and C) Venn diagrams depicting total gene numbers (parentheses) and genes in common (overlapping sectors) that display SSTF differences in expression in the pairwise genotypic comparisons shown (Top Venn). Separation of genes into down-regulated (pif-DOWN; Middle Venn) and up-regulated (pif-UP; Bottom Venn) is based on the direction of the change in expression displayed in each pairwise comparison. (D) Venn diagrams summarizing the overlap between all identified PIF-quartet-, PIF1/4/5-trio- and PIF3-regulated genes. PIF3-regulated genes are defined as the combined total of the non-overlapping genes in the pif3/WT and pifq/pif145 sets in (B). PIF1/4/5-trio-regulated genes are defined as the combined total of the non-overlapping genes in pif145/WT and pifq/pif3 sets in (C). Separation of genes into PIF-induced (PIF-Ind; Middle Venn) and PIF-repressed (PIF-Rep; Bottom Venn) is based on the deduced action of the PIF proteins when present in WT seedlings.
The data indicate that 254 (74%) PIF3-regulated genes are also redundantly transcriptionally regulated by one or more of the other three PIF-quartet proteins. Conversely, 1740 (86%) PIF-quartet-regulated genes show no significant PIF3 dependence (Figure 3B), whereas 918 (45%) do display PIF1/4/5 regulation (Figure 3D), indicating that one or more of the other PIF-quartet members function non-redundantly with PIF3 in regulating the expression of many target genes. The general robustness of our genome-wide RNA-seq expression profiling is demonstrated by the extensive RT-qPCR validation data presented in Figure S3.
Definition of genes that are likely direct targets of transcriptional regulation by PIF3
To identify the genes that both physically bind PIF3 in their promoters, in a G-box- or PBE-box-coincident manner, and display PIF-regulated expression (defined here as “direct-target genes”), we merged our ChIP-seq and RNA-seq data. This permitted gene-by-gene visualization of the PIF3-binding peaks and PIF-dependent transcription, genome-wide, as shown for PIL1 and ATHB2 in Figure 2A. The expression data for these two genes show a clear difference in transcript levels between the WT and pifq mutant, demonstrating the robust dependence of full expression on the presence of the PIF-quartet. Comparison of the expression peaks for the pif145 and pifq mutants also suggests that PIF3 acts in the absence of the other three quartet members to promote a moderate increase in transcript levels. Overall, the combined PIF-regulated expression-patterns and promoter-located PIF3-binding sites displayed by these two genes render them likely direct-targets of transcriptional regulation by PIF3 and one or more other quartet members in promoting skotomorphogenic development.
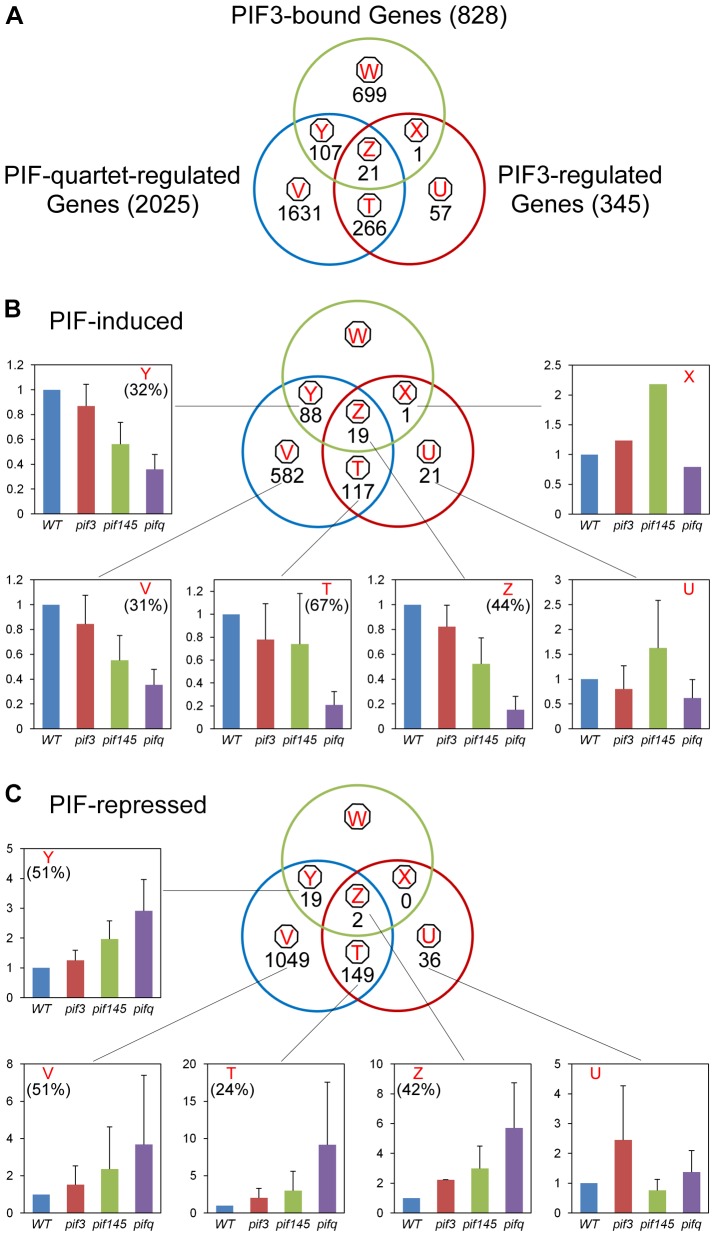
The Venn diagrams in Figure 4 show the genome-wide overlap of the genes identified independently as displaying PIF-quartet- and/or PIF3-dependent expression in a SSTF manner, with those exhibiting promoter-located, motif-coincident PIF3-binding sites (Figure 4A, Classes X, Y and Z; listed in Table S11). By these criteria, a total of 22 genes (Classes X and Z) were identified as robustly-likely, direct-target genes of autonomous-PIF3 transcriptional regulation. Of these, 21 genes (19 PIF3-induced; 2 PIF3-repressed) also display collective PIF-quartet regulation (Class Z). The 19 PIF3-induced Z-Class genes are listed in Table 1. The bar graphs in Figure 4B and 4C portray the mean expression level (relative to WT) of all the genes in each class, for each pif genotype. The quantitatively robust responsiveness of the PIF3-bound genes to the presence of PIF3 in the pif145 mutant background is evident from these data (Class Z-associated bar graphs). This robust PIF3-responsiveness was validated using RT-qPCR for selected members of the 19 PIF3-induced, Z-Class gene-set, having a range of quantitative dependence on this bHLH factor (Figure S3A).
Figure 4. Merging of ChIP–seq and RNA–seq data identifies apparent direct targets of PIF3 transcriptional regulation.
(A) Venn diagrams showing the overlap of genes displaying promoter-localized, G- or PBE-box-coincident, PIF3-binding peaks (PIF3-bound genes), with those displaying PIF-quartet- and/or PIF3-regulated expression (as defined in Figure 3D). This comparison defines seven classes of genes corresponding to the diagram sectors (circled red letters). The number of genes in each class is indicated. (B and C) Genes indicated in (A) divided into PIF-induced (B) and PIF-repressed (C) sets based on the direction of the transcriptional response elicited by the designated PIFs in dark-grown WT seedlings. The average fold-change in expression of all genes in each class relative to WT (set at unity) is shown in the bar graphs. Error bars represent the standard error for the genes averaged in each class. The percentage indicates the relative contribution of PIF3 to the total PIF-regulated expression in each class, defined by comparing pif145 with pifq.
Table 1. Direct-target genes of PIF3-induced transcription (Class Z and YZ1.5 genes).
| Functional Category/Locus | Annotation | Fold Induction (FI) | Transcriptional-response Category | |||||
| PIF-quartet Induction | PIF3 Induction | PIF145 Induction | Direct PIF3 Inductiond | Rapid Light Repressione | Reciprocal Light/Shade Regulationf | |||
| PIF3 L.O.F.a | PIF3 G.O.F.b | PIF145 G.O.F.c | ||||||
| (WT/pifq) | (WT/pif3) | (pif145/pifq) | (pif3/pifq) | |||||
| Transcription | ||||||||
| AT2G46970 | PIL1/bHLH124 | 97.58 | 1.52 | 17.71 | 65.55 | Class Z | Not ATH1g | Class M |
| AT5G54470 | BBX29 | 21.04 | 1.01 | 7.20 | 21.19 | Class Z | ||
| AT1G02340 | HFR1/bHLH26 | 24.56 | 1.39 | 4.41 | 18.00 | Class Z | ||
| AT3G21330 | bHLH87 | 3.09 | 1.17 | 2.07 | 2.69 | Class Z | Not ATH1 | |
| AT4G16780 | ATHB-2 | 7.36 | 1.25 | 2.06 | 5.98 | Class YZ1.5 | Class 7 | Class M |
| AT3G61830 | ARF18 | 3.59 | 1.33 | 2.03 | 2.75 | Class YZ1.5 | Class 7 | |
| AT4G27310 | BBX28 | 2.93 | 1.28 | 2.00 | 2.33 | Class YZ1.5 | Not ATH1 | |
| AT5G53980 | ATHB52 | 3.72 | 1.20 | 1.92 | 3.15 | Class YZ1.5 | Class 7 | Class M |
| AT4G32280 | IAA29 | 6.43 | 1.03 | 1.91 | 6.35 | Class YZ1.5 | Class 7 | Class M |
| AT4G01250 | WRKY22 | 2.62 | 1.18 | 1.80 | 2.25 | Class YZ1.5 | ||
| AT2G42870 | PAR1/HLH1 | 2.01 | 0.96 | 1.69 | 2.13 | Class YZ1.5 | ||
| AT1G69690 | TCP15 | 2.53 | 1.06 | 1.66 | 2.43 | Class YZ1.5 | Class 7 | Class M |
| AT3G25730 | EDF3 | 4.01 | 1.61 | 1.66 | 2.53 | Class YZ1.5 | Class 7 | |
| AT3G15540 | IAA19/MSG2 | 3.21 | 1.15 | 1.66 | 2.84 | Class YZ1.5 | Class 7 | Class M |
| AT1G77200 | AP2-EREBP family | 2.96 | 1.28 | 1.66 | 2.36 | Class YZ1.5 | Class 7 | |
| AT3G62090 | PIF6/PIL2/bHLH132 | 13.38 | 1.15 | 1.51 | 11.79 | Class YZ1.5 | Class 7 | |
| Stress/Defense | ||||||||
| AT2G33380 | RD20/CLO3 | 25.22 | 1.96 | 15.47 | 13.07 | Class Z | ||
| AT5G63650 | SNRK2.5 | 6.72 | 1.23 | 3.62 | 5.53 | Class Z | Class 7 | Class M |
| AT4G36010 | Thaumatin family | 3.74 | 1.30 | 1.83 | 2.91 | Class YZ1.5 | Class 7 | Class M |
| Signaling | ||||||||
| AT3G48260 | WNK3 | 8.54 | 1.28 | 3.33 | 6.78 | Class Z | Not ATH1 | |
| Photosynthesis | ||||||||
| AT1G34630 | Tim17/22/23 family | 3.31 | 1.52 | 1.87 | 2.22 | Class YZ1.5 | Class 7 | |
| Growth/Development | ||||||||
| AT4G14130 | XTR7/XTH15 | 19.37 | 1.00 | 6.07 | 19.66 | Class Z | Class 7 | Class M |
| AT1G67265 | RTFL21/DVL3 | 9.92 | 1.21 | 2.71 | 8.31 | Class Z | Not ATH1 | |
| AT1G10550 | XTH33 | 3.11 | 1.46 | 2.21 | 2.16 | Class Z | Class 7 | |
| Cellular Metabolism | ||||||||
| AT5G04120 | dPGM-like | 9.35 | 1.53 | 8.04 | 6.22 | Class Z | ||
| AT4G11050 | GH9C3 | 11.38 | 1.40 | 6.90 | 8.25 | Class Z | Class 7 | |
| AT5G51210 | OLEO3 | 6.54 | 1.54 | 4.42 | 4.32 | Class Z | ||
| AT2G31980 | CYS2 | 7.85 | 0.80 | 3.28 | 9.92 | Class Z | Class 7 | |
| AT5G02540 | SDR | 12.09 | 1.40 | 2.18 | 8.76 | Class YZ1.5 | Class 7 | Class M |
| AT2G34020 | Ca-binding EF-hand | 2.14 | 1.04 | 1.96 | 2.10 | Class YZ1.5 | Class 7 | |
| AT5G07010 | ST2A | 4.80 | 0.92 | 1.95 | 5.32 | Class YZ1.5 | Class 7 | |
| Unknown | ||||||||
| AT4G35720 | Unknown protein | 8.30 | 1.04 | 3.98 | 8.10 | Class Z | Class 7 | Class M |
| AT5G02580 | Unknown protein | 4.68 | 1.01 | 2.38 | 4.68 | Class Z | Class 7 | Class M |
| AT3G17580 | Unknown protein | 3.94 | 1.05 | 2.17 | 3.82 | Class Z | Not ATH1 | |
| Non-coding | ||||||||
| AT5G26146 | Natural antisense | 3.08 | 1.35 | 2.63 | 2.33 | Class Z | Not ATH1 | |
| AT3G17110 | Pseudogene | 2.97 | 1.10 | 2.35 | 2.74 | Class Z | ||
A striking feature of our data is the relatively large number of PIF-quartet-regulated genes (107 total genes; 88 PIF-induced and 19 PIF-repressed) that display promoter-located, PIF3-binding sites, but lack evidence of SSTF-level PIF3 regulation in our RNA-seq analysis (Figure 4, Class Y; also Table S11). Nevertheless, the bar graphs of the mean expression of these genes suggest a tendency toward a consistent difference in expression between the pif145 and pifq mutants, across the gene-set. In addition, combined analysis of the full set of PIF-induced, PIF3-bound genes (Classes Y and Z together) shows that there is a reciprocal continuum in the magnitude of the relative contributions of PIF3 and the PIF1/4/5-trio to the collective activity of the PIF quartet in transcriptionally activating these genes (Figure S4). To more closely examine the PIF3 contribution to the total PIF-quartet activity in the Y-Class genes, we therefore arrayed these genes by the pif145/pifq fold-change value and assayed the relative expression levels in 20 selected PIF-induced loci by RT-qPCR. Figure S3B shows that the 17 genes in this group with fold-changes >1.5 by the RNA-seq analysis, all exhibit statistically-significant (Student's t-test, P<0.05), PIF3-promoted expression increases in the pif145 mutant compared to the pifq mutant by RT-qPCR. This suggests that a subset of Y-Class genes may represent additional bona fide autonomously-PIF3-regulated genes that are below the resolution of the SSTF criteria we imposed on our RNA-seq analysis. We have therefore designated these 17 as Class YZ1.5 genes, having moderate (>1.5-fold), but statistically significant, regulation by PIF3 (Table 1). The evidence indicates, therefore, that a combined total of at least 38 YZ1.5- and Z-Class genes are direct targets of moderate to robust transcriptional regulation by promoter-bound PIF3. Because an additional 34 of the 88 Y-Class genes also display >1.5-fold PIF3-induced expression (Figure S3B and Table S11), it is possible that the number of direct targets of partial PIF3 transcriptional regulation is yet larger.
The W-class genes are those that display promoter-localized, G- or PBE-box-coincident PIF3-binding peaks, but no differential expression between the pifq mutant and wild type (Figure 4A). This observation is consistent with data from a variety of organisms that have shown that transcription factors vary greatly in their number of genomic binding sites, and that binding events can vastly exceed the number of known or possible direct gene targets [40]. The reasons for this phenomenon here are unclear but could include functional redundancy with other factors, including other PIF proteins. Consistent with this possibility, a subset of 41 of the total 699 W-class genes exhibit rapid light responsiveness [17] upon initial exposure (Table S12).
Definition of genes that are potential direct targets of transcriptional regulation by PIF1, 4, and/or 5, as well as PIF3
The reciprocal continuum in relative PIF3 and PIF1/4/5-trio contributions to the collective PIF-quartet transcriptional activation of PIF-induced, Y- and Z-class genes referred to above (Figure S4), indicates that PIF1, 4 and/or 5 contribute substantially to the regulation of these PIF3-bound genes. To identify the individual genes in this set displaying a significant PIF1/4/5 contribution, we compared the Y- and Z-class genes (Table S11) with those defined above as PIF1/4/5-regulated (Figure 3D; also Table S10). Overall, 92 (72%) of the 128 combined Y- and Z-class genes exhibit regulation by PIF1, 4 and/or 5 (Table S11), as shown by significant differences in the pif145/WT and/or pifq/pif3 comparisons. More notably, all 38 PIF3-induced direct-target genes (Class YZ1.5 and Z) are also PIF1/4/5-induced (Table 1). Because all four PIFs have been shown to bind to the G-box motif in sequence-specific fashion [4]–[9], it appears probable that these PIF-quartet-regulated genes, displaying promoter-located PIF3-binding sites (Figure 4; also Table S11), may be directly regulated by one or more of the other quartet members, in addition to, or instead of, PIF3.
Transcription-factor genes are the dominant targets of direct transcriptional regulation by the PIFs
Categorization of the YZ-class genes by the known or predicted functions of their encoded products reveals substantial enrichment in a diversity of transcription-factor-encoding genes (Table 1; also Figure S5 and Table S11), consistent with the concept that these multiple direct targets of the PIF quartet function at the apex of a primary transcriptional-cascade to regulate the downstream transcriptional network. It is also notable, however, that a considerable number of the YZ-class genes that have other cellular functions are also apparent direct targets of transcriptional regulation by the PIFs, including two non-protein-encoding genes of unknown function (Table 1).
Definition of light-regulated direct-target genes of PIF3
Previously, by microarray profiling, we identified a subset of genes (designated Class 7) that, in dark-grown seedlings, exhibit a PIF-quartet-dependent expression pattern, that is rapidly reversed (within 1 h) upon initial exposure to phy-activating R light [17]. Of the 24 rapidly light-repressed Class 7 genes displaying promoter-localized, G- or PBE-box-coincident PIF3-binding peaks, 21 (88%) are either Class YZ1.5 or Z genes here (Table 1). These genes are thus identified as a subset whose expression is directly promoted, at least partially, by PIF3 transcriptional activation in the dark, and is rapidly reduced in the light, at least in part, by photoactivated-phyB-induced PIF3 degradation. It is notable that 9 of these genes (43%) encode transcription factors (Table 1), indicative of being master regulators at the apex of the downstream transcriptional cascade controlled by the phy signaling pathway.
In striking contrast to the light-repressed Class 7 genes, only 7 of the 115 rapidly light-induced Class 7 genes (6%) [17] display PIF3-binding peaks that are coincident with a G-box or PBE-box, and of these only 2 genes (<2%) (PSY and KAI2) exhibit derepression here in the dark-grown pifq mutant. No individual PIF3 contribution to this repression was detectable here. Collectively, these data indicate that PIF3 acts predominantly, if not exclusively, to activate the expression of direct-target genes in dark-grown seedlings. Conversely, the 94% (108/115) of light-induced Class 7 genes that do not display G- or PBE-box coincident PIF3-binding peaks, might suggest that one or more of the proposed direct targets of the PIF quartet (Table 1) can act as key repressor(s) that regulate a diverse set of light-induced genes.
Recently, we defined a small core set of 14 Class 7 PIF-quartet-regulated genes (called M-Class genes) that display rapid, reciprocal, transcriptional responsiveness to light and vegetative shade in dark-grown and light-grown seedlings, respectively [11]. Our present analysis shows that 11 (79%) of these M-Class genes are identical to those identified here as dual Class 7 and YZ1.5-/Z-Class genes (Table 1), indicating that they are likely direct targets of PIF3 regulation, not only during skotomorphogenesis and deetiolation, but also subsequently, on a continuing basis, through juvenile vegetative development. The correlated PIF3-binding and PIF-regulated transcriptional behavior of several of these M-Class genes, determined by merging the ChIP-seq and RNA-seq data, is depicted in Figure S6.
Individual PIF-quartet members display quantitatively differential regulation of different direct-target genes
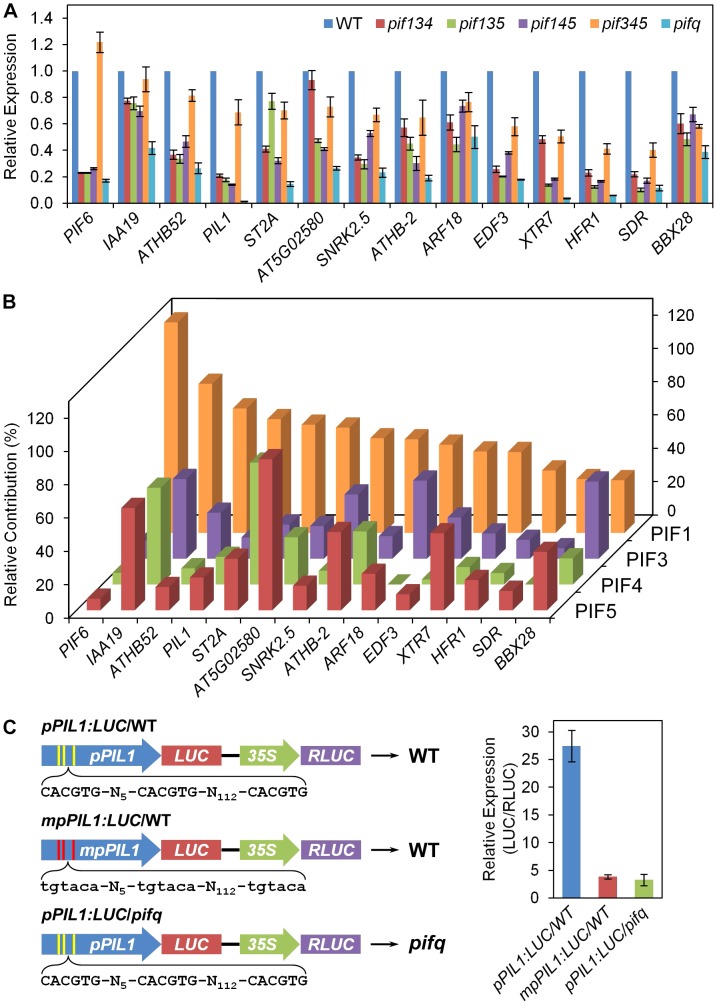
Because our data indicate that the contribution of PIF3 to the total level of expression collectively regulated by the PIF-quartet is quantitatively variable between genes (Figure 2A; Figure 4B, 4C; Figures S3 and S4), we wished to determine whether the other members of the PIF-quartet display a similarly variable pattern of regulation. For this purpose, we assayed the expression by RT-qPCR of a selected set of apparently PIF3 direct-target genes (Classes Z and YZ1.5), in the four different pif triple mutants compared to the pifq mutant and WT (Figure 5A). The relative autonomous contribution of each individual PIF (in the absence of the other three quartet members) to the total, collective PIF-quartet-supported expression was calculated as a percentage of the total difference in expression between the WT and pifq mutant, for each separate gene. The data reveal a striking diversity of relative contributions, both between the individual PIFs, and between genes for any individual PIF, in two-dimensional-matrix fashion (Figure 5B). Particularly notable is the dominant role played by PIF1 in promoting the expression of the majority of these genes. On the other hand, PIF3 contributes strongly to ARF18, SNRK2.5 and BBX28 expression, PIF4 strongly activates ST2A and ATHB-2, PIF5 contributes actively to AT5G02580, ATHB-2 and XTR7 expression, while all four PIFs contribute substantially to IAA19 transcription. Because all these tested genes are prospective direct-target genes of multiple PIF-quartet members, our findings suggest that there is an intricate combinatorial network, in which the individual PIF-quartet factors collaborate to transcriptionally regulate an array of direct-target genes, through potentially common DNA binding sites, with quantitatively differential regulatory activity.
Figure 5. Differential regulation of PIF3 direct-target genes by individual PIF-quartet proteins.
(A) Individual PIF-quartet members display diverse patterns of shared regulatory activity toward genes defined as direct targets of PIF3 transcriptional activation. Expression levels in the pifq and pif-triple mutants indicated, were determined by RT-qPCR, normalized to an internal PP2AA3 control, and presented relative to WT levels set at unity. Data are represented as the mean of biological triplicates +/− SEM. (B) Matrix of relative contributions from individual PIF proteins toward the shared transcriptional activation of individual, potentially-shared direct-target genes. Percent contribution is calculated as the proportion of the total differential expression between pifq and WT, that is contributed by the differential expression between pifq and each pif-triple mutant. (C) In planta PIL1 promoter activity requires both G-box motifs and PIF-quartet members. Left: Schematic of pPIL1:LUC constructs expressed transgenically in either WT or pifq plants, as indicated. Yellow and red stripes represent the locations of three native (pPIL1) and mutated (mpPIL1) G-box motifs, respectively, in variants of the PIL1 promoter, as shown by the DNA sequences displayed below each construct. A contiguous 35S-promoter driven RLUC reporter was included as an internal control in each construct. Right: Mean expression of the LUC reporter gene is shown as LUC enzyme activity normalized to the RLUC control in the same transgenic plant. Data represent the means of 6 or 7 independent transgenic lines +/− SEM.
Comparison of our data with recently published ChIP-seq-identified PIF4- and PIF5-binding sites [21], [24] supports this conclusion (Figure S7). Although the three studies were performed under contrasting experimental conditions, our analysis shows that 82% of genes with promoter-located, G- or PBE-box-associated PIF3-binding peaks identified here, also display PIF4- and/or PIF5-binding peaks (Figure S7A). Perhaps more striking, 89% of the 128 PIF3-binding, PIF-quartet-regulated genes identified here (Y- and Z-class genes in Figure 4A), are also bound by PIF4 and or PIF5, with 52% being bound by all three PIFs (Figure S7B; Table S11).
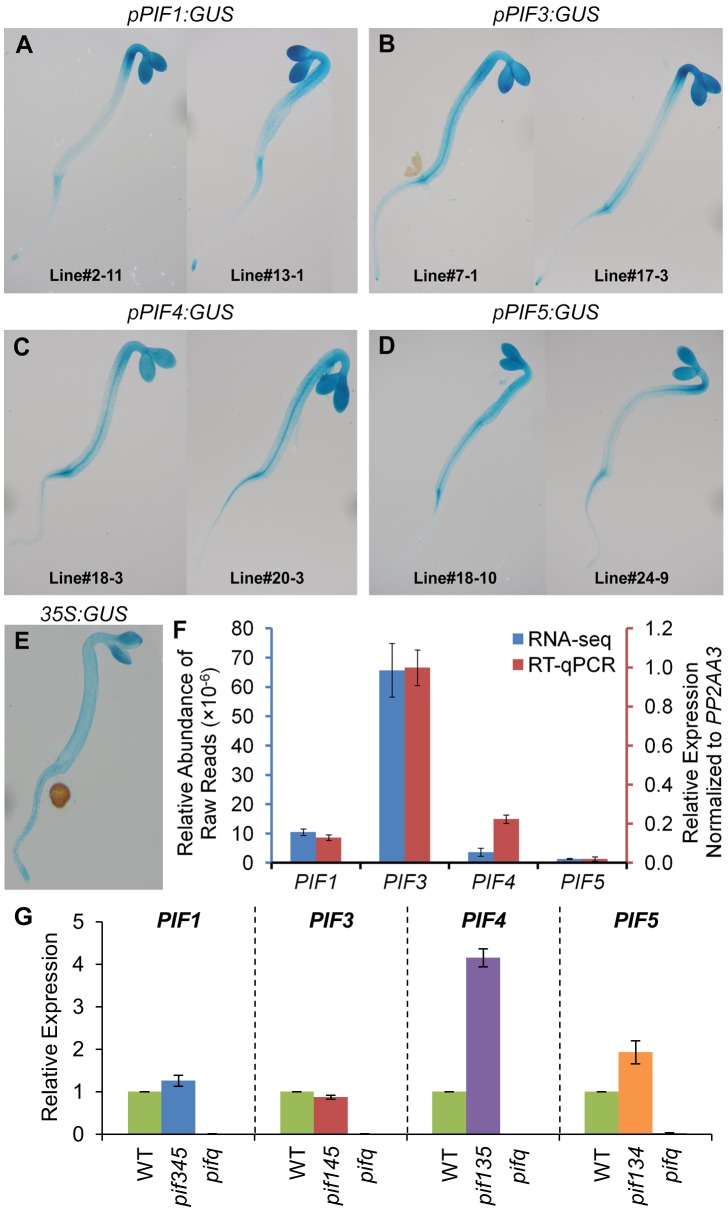
One possible mechanistic basis for the differential control of shared target genes by the individual PIF-quartet members described above (Figure 5) is that each PIF transcription factor has a different spatial expression pattern across the plant. To examine this possibility, we expressed pPIF:GUS fusions for each of the PIF genes transgenically in Arabidopsis seedlings, and assayed the distribution of GUS expression histochemically. The data show that all four PIF promoters support expression broadly throughout the seedling shoot tissue, with largely similar distribution patterns between the quartet members, within the resolution of this procedure (Figure 6A–6D).
Figure 6. Expression patterns of PIF-encoding genes.
(A–E) Representative images of histochemical staining of GUS activity in 2-d-old dark-grown transgenic seedlings. The GUS reporter gene is driven by PIF1 (A), PIF3 (B), PIF4 (C), PIF5 (D) and CaMV 35S (E) promoters, respectively. Data of biological triplicates were collected from two independent transgenic lines, and representative images are shown for each transgene. (F) Steady-state transcript levels of PIF-encoding genes defined by RNA-seq and RT-qPCR analyses in 2-d-old dark-grown WT seedlings. Data are presented as the mean of biological triplicates +/− SEM. (G) Relative expression of PIF-encoding genes in 2-d-old dark-grown pifq and pif-triple mutants. Expression was determined by RT-qPCR, normalized to an internal PP2AA3 control, and presented relative to WT levels set at unity. Data are represented as the mean of biological triplicates +/− SEM.
In principle, differences in absolute expression levels among the PIF-quartet members could also be a fundamental determinant of differences in PIF-promoted expression of target genes (Figure 5). However, this does not appear to be the case here. Examination of the RNA-seq profiles, and independent RT-qPCR analyses, of PIF1, PIF3, PIF4 and PIF5 expression, shows that, while there are marked differences in expression between these genes in wild-type seedlings (Figure 6F), these are not strongly correlated with the respective patterns of target-gene expression (Figure 5). In particular, PIF1 and PIF3 display expression levels that are robustly converse to their respective general levels of transcriptional activation. Similarly, and more importantly, although the expression levels of PIF4 and PIF5 are significantly elevated in the relevant triple mutant compared to wild-type (Figure 6G), these differences also do not correlate with the overall differential expression patterns of the target genes. While these elevated levels could indicate that the computation in Figure 5B overestimates the normal, relative contributions of these two PIFs to the collective PIF-quartet activity, displayed when all four PIFs are present, they do not account for the apparent dominance of PIF1 or the diversity of response-patterns between the genes.
Taken together, these results suggest that the sometimes strikingly different quantitative contributions of the individual PIFs to the expression of a given target gene appears unlikely to be primarily due to either differences in transcriptionally-driven PIF abundance or differences in spatially-determined abundance of the PIF-quartet-members. It appears more likely that these differences are due to intrinsic differential activities of the individual PIFs in the context of the individual target-gene promoters. In addition, because the GUS expression pattern driven by the CaMV 35S promoter (Figure 6E) overlaps substantially with that driven by the PIF promoters (Figure 6A–6D), it seems reasonable to expect that the majority of PIF3-binding sites detected by ChIP-seq analysis here, using 35S-driven PIF3-Myc expression, will reflect sites that are normally available to PIF3 generated by endogenous PIF3 promoter activity.
The G-box motifs in the PIL1 promoter are functionally necessary for PIF-quartet promoted expression
The robust binding of PIF3 to the G-box-containing region of the PIL1 promoter detected by ChIP-seq analysis (Figure 2) and in vitro assay (Figure 1G and Figure S2), and the partial autonomous promotion of PIL1 expression by PIF3 observed by RNA-seq analysis (Figure 2A, Figure 5, and Figure S3A), provides strong evidence that PIL1 is a direct target of PIF3 transcriptional regulation via physical interaction of the bHLH factor with these cis-elements. Conversely, because the non-PIF3 quartet members contribute robustly to the collective PIF-quartet-dependent expression of PIL1 (Figure 2A and Figure 5), and given that these non-PIF3 members also bind selectively to G-box motifs [4]–[9], it might be predicted that PIF1, 4 and/or 5 transcriptional activation of PIL1 will, like PIF3, be exerted through interaction with the G-boxes in the PIL1 promoter [5], [6]. To examine this prediction, we tested the functional necessity of these G-box motifs to PIL1 expression using reporter constructs in transgenic seedlings. The data show that activation of the PIL1 promoter requires both the presence of one or more of the PIF quartet and one or more of the G-boxes (Figure 5C), indicating that the G-box elements are the major, if not sole, targets of PIF-quartet transcriptional activation activity. By contrast, it is notable that, although a recent report shows that PIF7 also binds to the G-box region of the PIL1 promoter in a manner that is functionally important for shade-induced expression of this gene in light-grown seedlings [41], the extremely low residual levels of PIL1 expression in dark-grown pifq seedlings compared to wild-type (Figure 2A and Figure 5A) indicate that PIF7 has minimal, if any, contribution under these conditions. Together, the evidence suggests that, to the extent that the PIF-quartet members share transcriptional activation of PIL1 (Figure 5A and 5B), they do so by sharing the G-box motifs as interaction sites. This conclusion is consistent with the demonstration that PIF3 binds to all three G-box motifs in the PIL1-promoter cluster, both in vivo (Figure 2B) and in vitro (Figure 1G and Figure S2). By extrapolation, the other Y- and Z-Class, PIF-quartet-regulated genes, established here as being direct-targets of PIF3 transcriptional regulation through G- or PBE-box binding motifs, are strong candidates for likewise being targets of functionally active, direct binding-site sharing among the four PIF factors.
Discussion
Previous genetic studies have established that the overarching biological function of the PIF quartet is to promote skotomorphogenic growth and development in post-germinative seedlings in darkness, and to promote shade-avoidance behavior in deetiolated seedlings in response to exposure to neighboring vegetation [2], [3], [11], [42]. The evidence shows that the quadruple pifq mutant is strongly impaired in skotomorphogenic growth and development in dark-grown seedlings and has reduced shade-avoidance responsiveness to signals from neighboring vegetation in green seedlings [2], [3], [11]. In addition, there are indications that the contributions of individual PIF members to the collective activities of the quartet vary quantitatively, both between the PIFs for a given morphogenic-response feature, and between morphogenic-response features for a given PIF. For example, experiments comparing single, double, triple and quadruple pif-mutant combinations indicate that the individual PIFs appear to contribute additively or synergistically, in more or less equivalent fashion, to the promotion of hypocotyl-cell elongation growth in dark-grown seedlings [2], [3], [11]. By contrast, PIF1 appears to dominate the concomitant suppression of cotyledon separation that occurs in these same seedlings during dark-growth [2], [11]. In green seedlings, on the other hand, PIF4 and/or PIF5 appear to have a major role in promoting the stem and petiole elongation intrinsic to shade-avoidance in response to vegetative shade [21], [42], whereas PIF3 [43], together with PIFs 4 and 5 [44], contribute strongly to growth during the night period under diurnal light/dark cycles. Consistent with this general pattern, another related bHLH factor, PIF7, displays only moderate involvement in seedling deetiolation [45], but has a prominent role in shade avoidance [41]. Although a limited number of previous studies have examined the transcriptome regulated by PIF-quartet members in seedlings in darkness [3], [12], [16]–[19] and vegetative shade [11], [21] using the Affymetrix ATH1 array, these studies did not provide full genome coverage and did not permit dissection of potential quantitative differences in transcriptome profiles controlled by the individual PIFs.
The RNA-seq analysis performed here defines, with full genome coverage, the transcriptome collectively regulated by the PIF quartet in promoting skotomorphogenesis, and provides initial definition of the extent, and quantitative partitioning, of shared transcriptional control of the genes within of this network between PIF3 and the PIF1/4/5-trio. Superimposed on these data, our ChIP-seq analysis has identified a subset of these genes that are likely direct targets of PIF3 transcriptional regulation, exerted by physical binding of this factor to promoter-localized G- or PBE-box recognition-motifs (Class X, Y and Z genes, combined; Figure 4). The predominant pattern of PIF-regulated expression of these PIF3-bound genes (108 (84%) of 129 total) is one of high levels in the presence of the wild-type PIF factors, and reduced levels in the genetically-imposed absence of these factors in dark-grown seedlings, indicative of transcriptional activation by PIF3 and/or one or more of the other three PIF-quartet members. This pattern is consistent with the existing reports that all four factors function intrinsically as transcriptional activators, at least in transfection or heterologous expression systems [4]–[9], and with the demonstration here and elsewhere [21] that these PIFs function to activate PIL1-promoter-driven expression in transgenic seedlings (Figure 5C). We have therefore focused here primarily on this predominant class of PIF-transcriptionally-activated genes.
Our data indicate that there is a continuum, from robust to marginal, in the extent of the contribution of PIF3 to the combined transcriptional regulatory activity of the PIF quartet toward the PIF-induced, Class Y and Z genes (Figure S4). Conversely, by definition, there is a complimentary continuum in the share of this combined activity provided by the collective actions of PIFs 1, 4 and 5. These data imply at least some degree of shared, but quantitatively differential, transcriptional-regulatory activity among the PIF-quartet members toward individual genes that are apparent direct targets of PIF3-induced expression. Our RT-qPCR analysis of the expression patterns of selected genes from this subset, in all pif triple-mutant combinations, confirms that all four quartet members display such intra-subfamily differential activity toward individual genes in this set. Moreover, this analysis shows, conversely, that the individual PIF proteins induce differential levels of transcription in each different gene (Figure 5A). The three-dimensional response surface generated by this comparison (Figure 5B) suggests that this pattern may be iterated across all PIF-regulated genes genome wide, and points to the potential for considerable signaling and regulatory complexity at the PIF-target-gene interface.
Because it has been shown that all four PIF-quartet members bind robustly to the G-box motif [4]–[9], it appears likely that many of the direct targets of PIF3 transcriptional regulation are also direct targets of these other PIFs [5], [6], and that the shared activation of genes by the individual quartet members observed here will involve some degree of shared occupancy of these binding sites by the different PIFs. This may also apply to the newly discovered PBE-box motif, as there is recent evidence that PIF4 also recognizes this motif [21]. However, there is also evidence of potential divergence in motif recognition, as PIF5 was shown in the same report not to bind to the PBE-box motif [21]. The prominent contribution of PIF1 to the transcriptional activation of many of the genes examined here (Figure 5), despite its apparent considerably lower expression level than PIF3 (Figure 6F), is particularly intriguing in this respect, as this may imply that PIF1 may dominate promotion of target gene expression in dark-grown seedlings.
Comparison of the genes identified here as direct targets of PIF3 transcriptional activation (Class Z and YZ1.5 genes), with those previously identified as being rapidly (within 1 h) repressed by initial exposure of dark-grown seedlings to red light [17], has defined an overlapping subset of 21 genes (22 including PIL1) (Table 1). The evidence is strong, therefore, that these 22 genes form a core set that are directly transcriptionally activated by PIF3 in darkness and repressed in light, at least in part, by direct, photoactivated-phy-induced PIF3 degradation. Moreover, because all of these genes are also transcriptionally activated, either collectively (Table 1 and Table S11), or individually (Figure 5A and 5B) by PIF1, 4 and/or 5 in darkness, it appears likely that these PIFs share similarly directly in the light-reversible trans-activation of this core gene-set via photoactivated-phyB-induced degradation of the PIF-trio members.
The predicted or established functional diversity of the PIF direct-target genes identified here (Figure S5) suggests that PIF3 and/or one or more other PIF-quartet members act pleiotropically to directly regulate the transcription of a diversity of genes involved in a spectrum of cellular processes that sustain the skotomorphogenic developmental pathway. Consistent with previous analyses [3], [11], [17], [21], the PIF-induced genes are strikingly enriched for transcription-factor-encoding loci (40% of the annotated genes in this set). These data support the proposition, therefore, that the PIFs regulate an extensive transcriptional network via direct activation of a battery of primary target-genes in a hierarchal transcriptional cascade [20]. Because the encoded target-proteins represent multiple major classes of transcription factors (including bHLH, homeobox, bZIP, ARF, AUX/IAA, AP2-EREBP, BBX and TCP), it appears likely that they act concomitantly to activate multiple, diverse downstream pathways in parallel. Interestingly, however, many apparent PIF direct-target genes are involved in other cellular processes (including cytokinin metabolism, auxin-responsiveness, protein phosphorylation and cell-wall metabolism), suggesting a more immediate mode of PIF regulation of these processes.
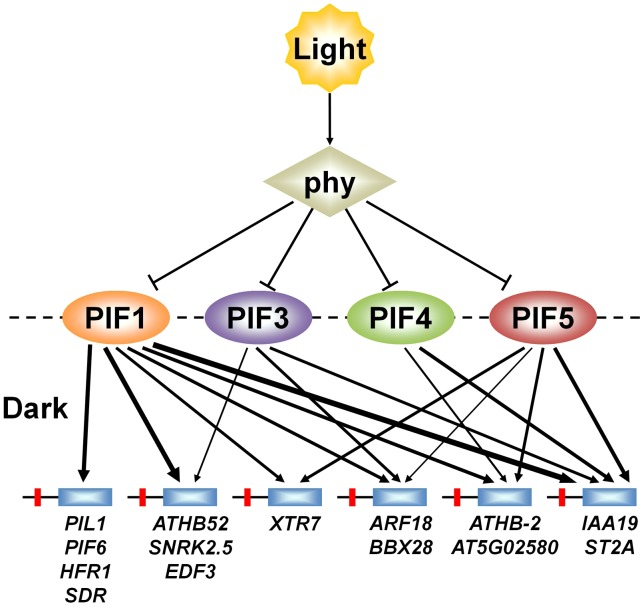
A central issue in understanding mechanisms of eukaryotic transcriptional regulation is how members of large transcription-factor families, with conserved DNA-binding domains (such as the 162-member Arabidopsis bHLH family [46]), discriminate between target genes [22], [30], [47]. However, the specific question of whether, and to what extent, closely-related sub-family members, with potential overlapping functional redundancy (like the PIF quartet), share regulation of target genes through shared binding to promoter-localized consensus motifs, does not appear to have been widely investigated [31]–[34]. Our data, together with those of others [21], [24], provide evidence suggesting that the PIF quartet members share directly in transcriptional activation of numerous target genes, potentially via redundant promoter occupancy, in a manner that varies quantitatively from gene to gene (Figure 7). This finding suggests that these PIFs function collectively as a signaling hub, selectively partitioning common upstream signals from light-activated phys at the transcriptional-network interface. Definition of the mechanistic basis and functional consequences of this apparent complexity will require further investigation.
Figure 7. PIFs direct differential light-signal channeling to the phy-regulated transcriptional network.
Model depicting proposed quantitatively differential partitioning of transcriptional activation activity to shared direct-target genes, both by and between individual PIF-quartet members. Arrows represent the presence or absence and relative level (line thickness) of shared transcriptional activation of different direct-target genes by the individual PIFs in the dark. This representation is based on the data in Figure 5A and 5B. Light-activation of phy photoreceptors induces rapid proteolysis of the PIFs, reversing this transcriptional activity.
Materials and Methods
Plant materials and growth conditions
The Colombia-0 ecotype of Arabidopsis thaliana was used for all experiments. The 35S:6×His-PIF3-5×MYC (P3M) transgenic line [48], pif3 [18], pif1pif4pif5 (pif145) [11], pif1pif3pif4 (pif134), pif1pif3pif5 (pif135), pif3pif4pif5 (pif345), and pif1pif3pif4pif5 (pifq) [2] were as described.
Stratified seeds were irradiated with WL at 21°C for 3 h to induce germination, followed by a FR pulse for 15 min to suppress pseudo dark effects [2], and grown in darkness at 21°C for 2 d before harvest.
ChIP–seq
The ChIP assay was performed using about 2 g of Arabidopsis 2-d-old dark-grown whole seedlings as described [49] under green safelight. Polyclonal anti-MYC antibodies (Abcam, ab9132) were used with BSA-blocked Protein G Agarose beads (Millipore) to immunoprecipitate the P3M-DNA complex. Wild-type Arabidopsis seedlings grown under the same conditions were used as the negative control following the same assay procedure.
The ChIP-seq library was constructed according to Illumina's instructions (www.illumina.com) with some modifications. Four ChIP samples from technical replicates of each biological replicate were pooled together and concentrated to increase the starting amount of DNA. The end repair of DNA fragments was performed using End-It DNA End-Repair Kit (Epicentre). The A-tailing was added to the end-repaired DNA fragments using Klenow Fragment (NEB), and then Illumina's PE adapters were ligated by T4 DNA Ligase (Promega) at 16°C overnight. The adapter-ligated DNA fragments in the 200–300 bp size-range were selected by the gel purification, and then were amplified using Phusion High-Fidelity DNA Polymerase (NEB) with the Illumina PE PCR primer set. The library was purified using an Agencourt AMPure XP system (Beckman Coulter Genomics), and then validated by Bioanalyzer 2000 (Agilent).
The parallel libraries from P3M and WT ChIP samples were assayed by single-end sequencing on an Illumina GAIIx platform. The 36-nt reads were aligned to the TAIR9 assembly of the Arabidopsis genome using Bowtie [50] with up to 2 mismatches allowed. Only reads mapped uniquely to the nuclear genome with the lowest number of mismatches were retained for binding-peak identification. To increase the uniformity of read-counts across biological replicates, two technical-replicate sequencing runs were performed on the 4 libraries from the 1st and 2nd ChIP experiments (two of the four biological replicates). The aligned reads from the two technical sequencing replicates of each library were combined and processed as single biological replicate data.
The statistical identification of PIF3-binding peaks was performed separately for each biological replicate using MACS [35] with the default 10−5 P-value cutoff. MACS analysis was customized to ensure a more uniform analysis across biological replicates, and to decrease the size of the window for detecting background enrichment (due to the small size of the Arabidopsis genome) by employing modified parameters (gsize = 1.1e8, bw = 100, nomodel, shiftsize = 50, slocal = 1000, and llocal = 2000).
Four independent biological replicates of ChIP-seq data were collected, and replicate-specific binding peaks, identified in at least one other replicate, were defined as reproducible, if the distance between the summits of each replicate were less than 100 bp. For each reproducible peak, a mean summit position was assigned as the average position of the individual replicate-specific summits, and the PIF3-binding sites were defined as the 201 bp windows centered at each reproducible mean-summit position.
De novo PIF3-binding motif discovery was performed on the 201-bp defined binding sites using MEME [36], and the enrichment significance of identified G-box and PBE-box motifs beyond the genome background was quantified by 100 random simulations, where in each simulation 1064 randomly selected genomic regions of the same size were searched for the occurrence of each motif. The tight association of PIF3 binding with a specific motif was defined as the distance between the peak summit and the closest motif less than 100 bp.
Definition of the closest neighboring genes to each binding peak was approached by scanning the regions within +/−5 kb centered at each peak summit, using CisGenome [51], and the potential target genes downstream of each summit with no intervening genes were selected manually.
RNA–seq
Total RNA was extracted from 2-d-old dark-grown seedlings using QIAshredder column and RNeasy Plus Mini Kit (Qiagen) according to the manufacturer's instructions. The sequencing library construction was adapted from 3′-end RNA-seq protocol [52]. The mRNA was fractionated from 20 µg of total RNA using Dynabeads Oligo (dT)25 (Invitrogen), and fragmented using Fragmentation Reagents (Ambion) at 70°C for 2.5 min in 20 µl of reaction. The polyA-tailed 3′-end fragments were captured by another run of mRNA purification as described above, and then treated by Antarctic Phosphatase (NEB) and T4 Polynucleotide Kinase (NEB) at 37°C for 1 h and 2 h, respectively. The sample was purified using RNeasy MinElute Cleanup Kit (Qiagen) according to Illumina's protocol. The eluted mRNA fragments were ligated with 2.5 µM of Illumina's SRA 5′ adaptor by T4 RNA Ligase 1 (NEB) at 20°C for 4 h. The 3′ cDNA adapter derived from Illumina's v1.5 sRNA 3′ adapter was conjugated with the anchored oligo (dT)20 primer, and introduced through reverse transcription using the SuperScript III First-Strand Synthesis System (Invitrogen). The first-strand cDNA was purified using the Agencourt AMPure XP system, and then amplified by PCR reaction using Phusion High-Fidelity DNA Polymerase with Illumina's sRNA PCR primer set. The size of purified library DNA was validated by Bioanalyzer 2000.
Libraries from the 1st biological replicate were assayed by 36-cycle single-end sequencing on the Illumina GAIIx platform, while libraries from the 2nd and 3rd biological replicates were assayed by 50-cycle single-end sequencing on the HiSeq2000 platform. For consistency, only the 5′-end 36-nt trimmed reads from the 2nd and 3rd replicates, as well as the full-length 36-nt reads from the 1st replicate, were aligned to the TAIR9 representative transcriptome using Bowtie with zero mismatches allowed. Only reads mapping uniquely to the 3′-end 500-bp region of the coding strand were counted for gene expression. Differentially expressed genes were identified using the edgeR package [53], and SSTF genes were defined as those that differ by ≥2-fold with an adjusted P value ≤0.05 as described [17].
In vitro protein–DNA binding assay
The recombinant protein GST-PIF3-Flag and the GST control were purified from E. coli as described previously [9]. DNA probes were generated by annealing a 5′ biotinylated oligonucleotide (IDT) to a complementary unmodified oligonucleotide (IDT). The complementary oligonucleotides were diluted in annealing buffer (10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA) to a final concentration of 40 µM, heated to 95°C for 5 min, and cooled down slowly (0.1°C/second) to 12°C. The same procedure was followed to generate unmodified dsDNA fragments for competition assays. Probes are listed in Table S13.
The DPI-ELISA assays were performed as described [38]. Biotinylated probes were bound to Reacti-Bind Streptavidin High Binding Capacity Coated 96-Well Plates (Thermo Scientific) by applying 2 pmol/well of the probes in TBS-T buffer (20 mM Tris-HCl (pH 7.5), 180 mM NaCl, 0.1% (v/v) Tween 20) for 1 h at 37°C. The wells were blocked with 5% (w/v) non-fat dry milk in TBS-T buffer for 30 min, and then incubated with 100 ng of GST-PIF3-Flag or GST for 1 h. For competition assays, 2, 10 or 50 pmol/well of the unlabeled probes were added at the same time with the proteins. After incubation, the wells were washed 3 times with TBS-T/PBS-T buffer, and then were incubated with 1∶2000 diluted THE GST Antibody [HRP] (GenScript, A00866) in PBS-T buffer for 1 h. The wells were then washed twice with PBS-T and PBS buffers, respectively, after incubation. The protein binding was detected by adding the OPD solution (Thermo Scientific), and the reaction was stopped by 2.5 M sulfuric acid. The color extinction was measured at 490 nm, using 650 nm as a reference wavelength in the ELISA reader.
The EMSA assays were performed as described [9]. 100 ng of recombinant proteins and the biotinylated DNA probes were used in each assay. Gel electrophoresis using native 5% PAGE gel in ice cold 0.5× TBE buffer (280 V, 15 min) was followed by wet-transfer electro blotting to Biodyne B Nylon membrane (Pierce) in 0.5× TBE buffer (80 V, 1 h). The Lightshift Chemiluminescent DNA EMSA kit (Pierce) was used for detection of the biotinylated probes according to the manufacturer's instructions.
Quantitative PCR (qPCR)
RT-qPCR was performed as described [17]. Each PCR reaction was repeated at least twice, and the mean value of technical replicates was recorded for each biological replicate. Data from biological triplicates were collected, and the mean value with standard error is represented in the bar graphs. Primers and gene accession numbers are listed in Table S13.
Construction of pPIL1:LUC and pPIF:GUS transgenic plants
The 1.8 kb PIL1 promoter region (pPIL1) upstream of the ATG was amplified by PCR using the pPIL1-F1/R1 primer set, and then the XhoI/EcoRI fragment was cloned into the pBluescript II SK (pBSK) vector (Stratagene) to produce pBSK-pPIL1. The G-box mutations were introduced by two-step PCR amplification (using pPIL1-F7/R7, pPIL1-F8/R8, pPIL1-F9/R9, and pPIL1-F10/R10 primer sets), and the XhoI/MfeI fragment from the pPIL1-F1/R5 primer set was cloned to replace the unmutated fragment of pBSK-pPIL1 to produce pBSK-mpPIL1. The HindIII/BamHI fragment containing the omega-LUC+-rbcS terminator from the pENTR/D-TOPO\arGIp::LUC+ construct was cloned into pBSK-pPIL1 and pBSK-mpPIL1, respectively, to produce pBSK-pPIL1:LUC and pBSK-mpPIL1:LUC. The CDS of Renilla Luciferase (RLUC) was amplified by PCR using the Rluc-F1/R1 primer set, and then the NcoI/PmlI fragment was cloned into the pCAMBIA1302 binary vector to produce pC1302-35S:RLUC. The PstI/SacI fragments from pBSK-pPIL1:LUC and pBSK-mpPIL1:LUC were then sub-cloned into pC1302-35S:RLUC to produce pC1302-pPIL1:LUC-35S:RLUC (pPIL1:LUC) and pC1302-mpPIL1:LUC-35S:RLUC (mpPIL1:LUC), respectively.
The 2.5–3.0 kb promoter regions upstream of the ATG of PIF3, PIF4 and PIF5 were amplified from Arabidopsis (Col-0 ecotype) genomic DNA by PCR using the TOPO-PIF3p-LP1/RP1, TOPO-PIF4p-LP1/RP1 and TOPO-PIF5p-LP1/RP1 primer sets, respectively. The PCR products were cloned into the pENTR/D-TOPO vector (Invitrogen) to produce the pPIF3, pPIF4 and pPIF5 entry clones. For the PIF1 promoter, the first 2 kb fragment upstream of the ATG was amplified by PCR using the TOPO-PIF1p-LP3/RP1 primer set, and then was cloned into the pENTR/D-TOPO vector to produce the intermediate entry clone. The second fragment of 2–4 kb upstream of ATG was amplified using the NotI-PIF1p-LP/XcmI-PIF1p-RP primer set, and then the NotI/XcmI fragment of the PCR product was subcloned into the intermediate entry clone to produce the pPIF1 entry clone. All four entry clones were subcloned into the gateway compatible pGWB3 binary vector [54] using Gateway LR Clonase II Enzyme Mix (Invitrogen) to produce pPIF:GUS constructs.
The constructs were transformed into Arabidopsis plants as described [55], and the individual transgenic lines were selected on MS medium containing 25 mg/L of Hygromycin B (Invitrogen).
Luciferase assay
The 2-d dark-grown seedlings of independent transgenic lines were ground in liquid nitrogen, and total protein was extracted in LUC extraction buffer (1× PBS, 4 mM EDTA, 2 mM DTT, 5% glycerol, 1 mg/ml BSA, 2 mM PMSF and 1× complete protease inhibitor cocktail (Roche) at 3× w/v) as described [7]. 20 µl of the supernatant were used to measure the LUC and RLUC activity using a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instruction. The relative expression of LUC was represented by its enzyme activity compared to the RLUC internal control.
Histochemical GUS staining
Histochemical GUS staining assays were performed on 2-d-old dark-grown seedlings as described [56] using a modified substrate buffer (1× PBS (pH 7.0), 1 mM K3Fe(III)(CN)6, 0.5 mM K4Fe(II)(CN)6, 1 mM EDTA, 1% Triton X-100, 1 mg/ml X-gluc). Data of biological triplicates were collected from two independent transgenic lines, and representative images are shown for each transgene.
Accession number
ChIP-seq and RNA-seq data reported in this study have been deposited in the Gene Expression Omnibus database under the accession number GSE39217.
Supporting Information
ChIP-qPCR validation of PIF3-binding sites. (A–C) PIF3-binding sites containing the G-box (A), PBE-box (B), or both motifs (C) were validated using independent ChIP-qPCR tests. Sites are named according to adjacent genes. The relative enrichment level is presented as the percentage of input-control DNA that is immunoprecipitated for the P3M and WT samples. Data are presented as the mean of biological triplicates +/− SEM.
(TIF)
EMSA assays of PIF3 in vitro binding to the G-box and PBE-box motifs. EMSA assays show that the recombinant GST-PIF3 protein (Lane 3), but not GST by itself (Lane 2), specifically binds to the wild-type (WT) biotinylated G-box and PBE-box probes. For probes containing single G-box or PBE-box motif (PIL1a, PHYB, IAA2, IBH1 and AT4G30410), the cold competitor probes mutated at the motif (mut; Lane 5) did not exhibit competitive effects similar to the WT competitor (Lane 4). For probes containing two G-box motifs (PIL1b and RGA1), the mut probes at either one motif (Land 6 and 7) showed competitive effects similar to the WT probes (Lane 4), whereas the mut probe at both motifs (Lane 5) did not show efficient competition. The ATHB-2 probe contains one G-box and one PBE-box motif separated by 5 bp. The mut probe at only the G-box (Lane 6) displayed no competition, as did the probe mutated at both G-box and PBE-box motifs (Lane 5), while the mut probe at only PBE-box motif (Lane 7) showed a competitive effect similar to the WT probe (Lane 4). 1 both G-boxes mutated, 2 only 1st G-Box mutated, 3 only 2nd G-Box mutated, 4 G-box and PBE-box mutated, 5 only 1st PBE-box mutated.
(TIF)
RT-qPCR validation of PIF3- and PIF-quartet-regulated genes. (A and B) The orange horizontal bar graphs show the relative PIF3-induced expression of 19 Z-Class (A) and 88 Y-Class (B) PIF-induced genes as determined by RNA-seq analysis. The relative PIF3-induced expression value was calculated as the ratio of the level in the pif145 mutant compared to that in the pifq mutant (pif145/pifq), and the genes are arrayed according to this ratio. We assayed the expression patterns of 12 (A) and 20 (B) genes in each class using RT-qPCR, and the expression relative to WT was normalized to the PP2AA3 internal control, for the pif3, pif145 and pifq mutants. The data are presented as the mean of biological triplicates +/− SEM in the vertical bar-graph boxes, compared to the RNA-seq data for these genes. Genes highlighted-in-red exhibited statistically-significant differences between pif145 and pifq in the RT-qPCR tests, by Student's t-test (P<0.05) (indicated by red star above the pif145 bar), while genes highlighted-in-blue did not. Data from RNA-seq are shown as the fold-change determined by the edgeR package in each pif mutant compared to WT. The internal percentage values in each box represent the relative contribution of PIF3 to the total PIF-quartet-induced transcription for each tested gene, as shown in Figure 5.
(TIF)
Relative contribution of PIF3 and the PIF1/4/5-trio to the collective regulation of the PIF quartet. The relative contribution of PIF3 and the PIF1/4/5-trio to the collective regulation of the PIF quartet is plotted for the 107 PIF3-bound, PIF-induced genes (Classes Y and Z). The degree of collective regulation by the PIF quartet is defined as the difference in expression between pifq and WT. The separate activities of PIF3 and the combined PIF1/4/5-trio, are defined as the differences in expression between pif145 and pifq, and between pif145 and WT, respectively. The relative contribution is then defined as the percentage of the collective PIF-quartet regulation contributed by the separate activities of PIF3 and the combined PIF1/4/5-trio, respectively.
(TIF)
Functional categorization of PIF3-bound, PIF-regulated genes. (A) The total PIF3-bound, PIF-regulated genes identified by integrating our ChIP-seq and RNA-seq analyses (Classes X, Y and Z) were assigned to functional categories, color-coded as shown. This assignment was based on the Gene Ontology annotations for biological and/or molecular function in the TAIR database. The percentage of the total annotated genes within each category was calculated after excluding the genes annotated as having unknown function. (B) PIF-induced and PIF-repressed genes in Classes X, Y and Z were assigned separately to the functional categories as described in (A). The first numbers in parentheses represent the genes with functional annotation, and the second numbers represent the total genes in each class.
(TIF)
Correlated PIF3-binding and PIF-regulation of eight M-class genes. (A–H) Visualization of ChIP-seq and RNA-seq data in the genomic regions encompassing TCP15 (A), IAA29 (B), ATHB52 (C), CKX5 (D), XTR7 (E), SNRK2.5 (F), AT5G02580 (G), and SDR (H), respectively. The ChIP and RNA tracks show the pile-up distribution of the combined raw reads from four biological replicates of ChIP-seq data and three replicates of RNA-seq data, respectively. P3M- and WT-ChIP: DNA immunoprecipitated from PIF3-Myc-expressing and from wild-type seedlings, respectively. WT-, pif3-, pif145- and pifq-RNA: RNA from genotypes used for expression analysis. Binding Site: 201 bp defined as the PIF3-binding site. Summit, predicted PIF3-binding center defined from the binding-peak maximum. G- and PBE-box: Vertical lines indicate motif positions.
(TIF)
Comparison of potential direct targets of PIF3, PIF4 and/or PIF5. (A) Venn diagram shows overlap of genes associated with PIF3, PIF4 and/or PIF5 binding sites. (B) Venn diagram shows PIF-quartet-regulated genes bound by PIF3, PIF4 and/or PIF5.
(TIF)
List of PIF3-binding peaks from four ChIP-seq replicates. (A) List of PIF3-binding peaks from the 1st replicate. (B) List of PIF3-binding peaks from the 2nd replicate. (C) List of PIF3-binding peaks from the 3rd replicate. (D) List of PIF3-binding peaks from the 4th replicate.
(XLS)
List of reproducible PIF3-binding sites.
(XLS)
List of PIF3-bound genes.
(XLS)
List of SSTF genes from pifq/WT comparison (PIF-quartet-regulated genes).
(XLS)
List of SSTF genes from pif3/WT comparison (PIF3-regulated genes (loss-of-function)).
(XLS)
List of SSTF genes from pif145/WT comparison (PIF1/4/5-trio-regulated genes (loss-of-function)).
(XLS)
List of SSTF genes from pifq/pif145 comparison (PIF3-regulated genes (gain-of-function)).
(XLS)
List of SSTF genes from pifq/pif3 comparison (PIF1/4/5-trio-regulated genes (gain-of-function)).
(XLS)
Composite list of PIF3-regulated genes.
(XLS)
Composite list of PIF1/4/5-trio-regulated genes.
(XLS)
List of PIF3-, PIF4 and/or PIF5-bound, PIF-regulated (Class X, Y and Z) genes.
(XLS)
List of rapidly-light-regulated W-class genes.
(XLS)
List of oligonucleotides used in this study. (A) List of ChIP-qPCR primers. (B) List of RT-qPCR primers. (C) List of cloning primers. (D) List of in vitro binding probes.
(XLS)
Acknowledgments
We thank the Vincent J. Coates Genomics Sequencing Laboratory and the Functional Genomics Laboratory at UC Berkeley for high-throughput sequencing service, Dr. Feng Li for bioinformatics assistance, and all lab members for technical support and critical discussion.
Funding Statement
This work was supported by a Feodor Lynen Research Fellowship from the Alexander von Humboldt Foundation to AP; by China Scholarship Council to HS; by NIH Grant 5R01 GM083084-03 to TPS; and by NIH Grant 2R01 GM-047475, DOE Grant DEFG03-87ER13742, and USDA ARS Current Research Information System Grant 5335-21000-027-00D to PHQ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leivar P, Monte E, Oka Y, Liu T, Carle C, et al. (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, et al. (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci U S A 106: 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Al-Sady B, Kikis EA, Monte E, Quail PH (2008) Mechanistic duality of transcription factor function in phytochrome signaling. Proc Natl Acad Sci U S A 105: 2232–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, et al. (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- 6. Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C (2009) Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J 28: 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huq E, Al-Sady B, Hudson M, Kim C, Apel K, et al. (2004) Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941. [DOI] [PubMed] [Google Scholar]
- 8. Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Martinez-Garcia JF, Huq E, Quail PH (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863. [DOI] [PubMed] [Google Scholar]
- 10. Shen H, Zhu L, Castillon A, Majee M, Downie B, et al. (2008) Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leivar P, Tepperman JM, Cohn MM, Monte E, Al-Sady B, et al. (2012) Dynamic Antagonism between Phytochromes and PIF Family Basic Helix-Loop-Helix Factors Induces Selective Reciprocal Responses to Light and Shade in a Rapidly Responsive Transcriptional Network in Arabidopsis. Plant Cell 24: 1398–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lorrain S, Trevisan M, Pradervand S, Fankhauser C (2009) Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J 60: 449–461. [DOI] [PubMed] [Google Scholar]
- 13. Stephenson PG, Fankhauser C, Terry MJ (2009) PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci U S A 106: 7654–7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Quail PH (2010) Phytochromes. Curr Biol 20: R504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li J, Li G, Wang H, Deng XW (2011) Phytochrome Signaling Mechanisms. The Arabidopsis Book e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sentandreu M, Martin G, Gonzalez-Schain N, Leivar P, Soy J, et al. (2011) Functional profiling identifies genes involved in organ-specific branches of the PIF3 regulatory network in Arabidopsis. Plant Cell 23: 3974–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, et al. (2009) Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, et al. (2004) The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci U S A 101: 16091–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moon J, Zhu L, Shen H, Huq E (2008) PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci U S A 105: 9433–9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaufmann K, Pajoro A, Angenent GC (2010) Regulation of transcription in plants: mechanisms controlling developmental switches. Nat Rev Genet 11: 830–842. [DOI] [PubMed] [Google Scholar]
- 21. Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, et al. (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711. [DOI] [PubMed] [Google Scholar]
- 22. Farnham PJ (2009) Insights from genomic profiling of transcription factors. Nat Rev Genet 10: 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ferrier T, Matus JT, Jin J, Riechmann JL (2011) Arabidopsis paves the way: genomic and network analyses in crops. Curr Opin Biotechnol 22: 260–270. [DOI] [PubMed] [Google Scholar]
- 24. Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ouyang X, Li J, Li G, Li B, Chen B, et al. (2011) Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23: 2514–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun Y, Fan XY, Cao DM, Tang W, He K, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tao Z, Shen L, Liu C, Liu L, Yan Y, et al. (2012) Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J 70: 549–561. [DOI] [PubMed] [Google Scholar]
- 28. Yu X, Li L, Zola J, Aluru M, Ye H, et al. (2011) A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65: 634–646. [DOI] [PubMed] [Google Scholar]
- 29. Zhang H, He H, Wang X, Yang X, Li L, et al. (2011) Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J 65: 346–358. [DOI] [PubMed] [Google Scholar]
- 30. Hollenhorst PC, McIntosh LP, Graves BJ (2011) Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem 80: 437–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hollenhorst PC, Shah AA, Hopkins C, Graves BJ (2007) Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev 21: 1882–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weinmann AS, Bartley SM, Zhang T, Zhang MQ, Farnham PJ (2001) Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol Cell Biol 21: 6820–6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boros J, O'Donnell A, Donaldson IJ, Kasza A, Zeef L, et al. (2009) Overlapping promoter targeting by Elk-1 and other divergent ETS-domain transcription factor family members. Nucleic Acids Res 37: 7368–7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, et al. (2007) A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res 17: 1550–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34: W369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim DH, Yamaguchi S, Lim S, Oh E, Park J, et al. (2008) SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20: 1260–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brand LH, Kirchler T, Hummel S, Chaban C, Wanke D (2010) DPI-ELISA: a fast and versatile method to specify the binding of plant transcription factors to DNA in vitro. Plant Methods 6: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nicol JW, Helt GA, Blanchard SG Jr, Raja A, Loraine AE (2009) The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. MacQuarrie KL, Fong AP, Morse RH, Tapscott SJ (2011) Genome-wide transcription factor binding: beyond direct target regulation. Trends Genet 27: 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, et al. (2012) Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26: 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323. [DOI] [PubMed] [Google Scholar]
- 43. Soy J, Leivar P, Gonzalez-Schain N, Sentandreu M, Prat S, et al. (2012) Phytochrome-imposed oscillations in PIF3-protein abundance regulate hypocotyl growth under diurnal light-dark conditions in Arabidopsis. Plant J 71: 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, et al. (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361. [DOI] [PubMed] [Google Scholar]
- 45. Leivar P, Monte E, Al-Sady B, Carle C, Storer A, et al. (2008) The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bailey PC, Martin C, Toledo-Ortiz G, Quail PH, Huq E, et al. (2003) Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15: 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boyd KE, Wells J, Gutman J, Bartley SM, Farnham PJ (1998) c-Myc target gene specificity is determined by a post-DNAbinding mechanism. Proc Natl Acad Sci U S A 95: 13887–13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446. [DOI] [PubMed] [Google Scholar]
- 49. Gendrel AV, Lippman Z, Martienssen R, Colot V (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- 50. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ji H, Jiang H, Ma W, Johnson DS, Myers RM, et al. (2008) An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol 26: 1293–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yoon OK, Brem RB (2010) Noncanonical transcript forms in yeast and their regulation during environmental stress. RNA 16: 1256–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, et al. (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41. [DOI] [PubMed] [Google Scholar]
- 55. Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646. [DOI] [PubMed] [Google Scholar]
- 56. Somers DE, Quail PH (1995) Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis. Plant J 7: 413–427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ChIP-qPCR validation of PIF3-binding sites. (A–C) PIF3-binding sites containing the G-box (A), PBE-box (B), or both motifs (C) were validated using independent ChIP-qPCR tests. Sites are named according to adjacent genes. The relative enrichment level is presented as the percentage of input-control DNA that is immunoprecipitated for the P3M and WT samples. Data are presented as the mean of biological triplicates +/− SEM.
(TIF)
EMSA assays of PIF3 in vitro binding to the G-box and PBE-box motifs. EMSA assays show that the recombinant GST-PIF3 protein (Lane 3), but not GST by itself (Lane 2), specifically binds to the wild-type (WT) biotinylated G-box and PBE-box probes. For probes containing single G-box or PBE-box motif (PIL1a, PHYB, IAA2, IBH1 and AT4G30410), the cold competitor probes mutated at the motif (mut; Lane 5) did not exhibit competitive effects similar to the WT competitor (Lane 4). For probes containing two G-box motifs (PIL1b and RGA1), the mut probes at either one motif (Land 6 and 7) showed competitive effects similar to the WT probes (Lane 4), whereas the mut probe at both motifs (Lane 5) did not show efficient competition. The ATHB-2 probe contains one G-box and one PBE-box motif separated by 5 bp. The mut probe at only the G-box (Lane 6) displayed no competition, as did the probe mutated at both G-box and PBE-box motifs (Lane 5), while the mut probe at only PBE-box motif (Lane 7) showed a competitive effect similar to the WT probe (Lane 4). 1 both G-boxes mutated, 2 only 1st G-Box mutated, 3 only 2nd G-Box mutated, 4 G-box and PBE-box mutated, 5 only 1st PBE-box mutated.
(TIF)
RT-qPCR validation of PIF3- and PIF-quartet-regulated genes. (A and B) The orange horizontal bar graphs show the relative PIF3-induced expression of 19 Z-Class (A) and 88 Y-Class (B) PIF-induced genes as determined by RNA-seq analysis. The relative PIF3-induced expression value was calculated as the ratio of the level in the pif145 mutant compared to that in the pifq mutant (pif145/pifq), and the genes are arrayed according to this ratio. We assayed the expression patterns of 12 (A) and 20 (B) genes in each class using RT-qPCR, and the expression relative to WT was normalized to the PP2AA3 internal control, for the pif3, pif145 and pifq mutants. The data are presented as the mean of biological triplicates +/− SEM in the vertical bar-graph boxes, compared to the RNA-seq data for these genes. Genes highlighted-in-red exhibited statistically-significant differences between pif145 and pifq in the RT-qPCR tests, by Student's t-test (P<0.05) (indicated by red star above the pif145 bar), while genes highlighted-in-blue did not. Data from RNA-seq are shown as the fold-change determined by the edgeR package in each pif mutant compared to WT. The internal percentage values in each box represent the relative contribution of PIF3 to the total PIF-quartet-induced transcription for each tested gene, as shown in Figure 5.
(TIF)
Relative contribution of PIF3 and the PIF1/4/5-trio to the collective regulation of the PIF quartet. The relative contribution of PIF3 and the PIF1/4/5-trio to the collective regulation of the PIF quartet is plotted for the 107 PIF3-bound, PIF-induced genes (Classes Y and Z). The degree of collective regulation by the PIF quartet is defined as the difference in expression between pifq and WT. The separate activities of PIF3 and the combined PIF1/4/5-trio, are defined as the differences in expression between pif145 and pifq, and between pif145 and WT, respectively. The relative contribution is then defined as the percentage of the collective PIF-quartet regulation contributed by the separate activities of PIF3 and the combined PIF1/4/5-trio, respectively.
(TIF)
Functional categorization of PIF3-bound, PIF-regulated genes. (A) The total PIF3-bound, PIF-regulated genes identified by integrating our ChIP-seq and RNA-seq analyses (Classes X, Y and Z) were assigned to functional categories, color-coded as shown. This assignment was based on the Gene Ontology annotations for biological and/or molecular function in the TAIR database. The percentage of the total annotated genes within each category was calculated after excluding the genes annotated as having unknown function. (B) PIF-induced and PIF-repressed genes in Classes X, Y and Z were assigned separately to the functional categories as described in (A). The first numbers in parentheses represent the genes with functional annotation, and the second numbers represent the total genes in each class.
(TIF)
Correlated PIF3-binding and PIF-regulation of eight M-class genes. (A–H) Visualization of ChIP-seq and RNA-seq data in the genomic regions encompassing TCP15 (A), IAA29 (B), ATHB52 (C), CKX5 (D), XTR7 (E), SNRK2.5 (F), AT5G02580 (G), and SDR (H), respectively. The ChIP and RNA tracks show the pile-up distribution of the combined raw reads from four biological replicates of ChIP-seq data and three replicates of RNA-seq data, respectively. P3M- and WT-ChIP: DNA immunoprecipitated from PIF3-Myc-expressing and from wild-type seedlings, respectively. WT-, pif3-, pif145- and pifq-RNA: RNA from genotypes used for expression analysis. Binding Site: 201 bp defined as the PIF3-binding site. Summit, predicted PIF3-binding center defined from the binding-peak maximum. G- and PBE-box: Vertical lines indicate motif positions.
(TIF)
Comparison of potential direct targets of PIF3, PIF4 and/or PIF5. (A) Venn diagram shows overlap of genes associated with PIF3, PIF4 and/or PIF5 binding sites. (B) Venn diagram shows PIF-quartet-regulated genes bound by PIF3, PIF4 and/or PIF5.
(TIF)
List of PIF3-binding peaks from four ChIP-seq replicates. (A) List of PIF3-binding peaks from the 1st replicate. (B) List of PIF3-binding peaks from the 2nd replicate. (C) List of PIF3-binding peaks from the 3rd replicate. (D) List of PIF3-binding peaks from the 4th replicate.
(XLS)
List of reproducible PIF3-binding sites.
(XLS)
List of PIF3-bound genes.
(XLS)
List of SSTF genes from pifq/WT comparison (PIF-quartet-regulated genes).
(XLS)
List of SSTF genes from pif3/WT comparison (PIF3-regulated genes (loss-of-function)).
(XLS)
List of SSTF genes from pif145/WT comparison (PIF1/4/5-trio-regulated genes (loss-of-function)).
(XLS)
List of SSTF genes from pifq/pif145 comparison (PIF3-regulated genes (gain-of-function)).
(XLS)
List of SSTF genes from pifq/pif3 comparison (PIF1/4/5-trio-regulated genes (gain-of-function)).
(XLS)
Composite list of PIF3-regulated genes.
(XLS)
Composite list of PIF1/4/5-trio-regulated genes.
(XLS)
List of PIF3-, PIF4 and/or PIF5-bound, PIF-regulated (Class X, Y and Z) genes.
(XLS)
List of rapidly-light-regulated W-class genes.
(XLS)
List of oligonucleotides used in this study. (A) List of ChIP-qPCR primers. (B) List of RT-qPCR primers. (C) List of cloning primers. (D) List of in vitro binding probes.
(XLS)