Abstract
Background
Fermentative bacteria offer the potential to convert lignocellulosic waste-streams into biofuels such as hydrogen (H2) and ethanol. Current fermentative H2 and ethanol yields, however, are below theoretical maxima, vary greatly among organisms, and depend on the extent of metabolic pathways utilized. For fermentative H2 and/or ethanol production to become practical, biofuel yields must be increased. We performed a comparative meta-analysis of (i) reported end-product yields, and (ii) genes encoding pyruvate metabolism and end-product synthesis pathways to identify suitable biomarkers for screening a microorganism’s potential of H2 and/or ethanol production, and to identify targets for metabolic engineering to improve biofuel yields. Our interest in H2 and/or ethanol optimization restricted our meta-analysis to organisms with sequenced genomes and limited branched end-product pathways. These included members of the Firmicutes, Euryarchaeota, and Thermotogae.
Results
Bioinformatic analysis revealed that the absence of genes encoding acetaldehyde dehydrogenase and bifunctional acetaldehyde/alcohol dehydrogenase (AdhE) in Caldicellulosiruptor, Thermococcus, Pyrococcus, and Thermotoga species coincide with high H2 yields and low ethanol production. Organisms containing genes (or activities) for both ethanol and H2 synthesis pathways (i.e. Caldanaerobacter subterraneus subsp. tengcongensis, Ethanoligenens harbinense, and Clostridium species) had relatively uniform mixed product patterns. The absence of hydrogenases in Geobacillus and Bacillus species did not confer high ethanol production, but rather high lactate production. Only Thermoanaerobacter pseudethanolicus produced relatively high ethanol and low H2 yields. This may be attributed to the presence of genes encoding proteins that promote NADH production. Lactate dehydrogenase and pyruvate:formate lyase are not conducive for ethanol and/or H2 production. While the type(s) of encoded hydrogenases appear to have little impact on H2 production in organisms that do not encode ethanol producing pathways, they do influence reduced end-product yields in those that do.
Conclusions
Here we show that composition of genes encoding pathways involved in pyruvate catabolism and end-product synthesis pathways can be used to approximate potential end-product distribution patterns. We have identified a number of genetic biomarkers for streamlining ethanol and H2 producing capabilities. By linking genome content, reaction thermodynamics, and end-product yields, we offer potential targets for optimization of either ethanol or H2 yields through metabolic engineering.
Background
Fuel derived from waste-stream lignocellulosic biomass via consolidated bioprocessing is a renewable and carbon-neutral alternative to current petroleum-based fuels [1-3]. Consequently, considerable effort is being made to characterize species capable of efficiently converting lignocellulosic substrates into biofuels. An ideal biofuel producing microorganism should posses several key features, including: (i) high yields of the desired product, (ii) simultaneous utilization of sugars (cellulose, hemicellulose, pectin), and (iii) growth at elevated temperatures, and (iv) low product inhibition. Recent studies have focused on the characterization of numerous cellulose and hemicellulose degrading species of bacteria [4-6]. To fully exploit the biofuel producing potential of these organisms, several genomes have been sequenced and are now available for analysis (http://genome.jgi-psf.org/). While some hemicellulolytic or cellulolytic microorganisms are capable of hydrogen (H2) or ethanol production via fermentation, end-product yields typically are far lower than their maximum theoretical values (4 mol H2 or 2 mol ethanol per mol glucose) when cells are grown in pure culture. This is due to the presence of branched catabolic pathways that divert carbon and/or electrons away from a particular desired end-product [7]. Strategies that optimize yields for a single biofuel (H2 or ethanol) can only be developed through a detailed knowledge of the relationships between genome content, gene and gene product expression, pathway utilization, and end-product synthesis patterns.
Given that our primary focus is to optimize H2 and/or ethanol yields, we restricted our meta-analysis to sequenced organisms with limited branched end-product pathways (i.e. organisms that do not produce butyrate, butanol, propionate, propanol, and acetoin) for which end-product data was available. These included members of the Firmicutes (Clostridium, Caldicellulosiruptor, Thermoanaerobacter, Caldanaerobacter, Ethanoligenens, Geobacillus, and Bacillus species), Euryarchaeota (Thermococcus and Pyrococcus species), and Thermotogae (Thermotoga species). A list of species analyzed and corresponding GenBank accession numbers are summarized in Table 1. With the exception of Caldanaerobacter subterraneus subsp. tengcongensis, Thermoanaerobacter pseudethanolicus, Pyrococcus furiosus, Geobacillus thermoglucosidasius, and Bacillus cereus, all organisms were capable of cellulose and/or xylan saccharification.
Table 1.
H2and ethanol producing organisms included in meta-analysis of end-product yields and genome content
| Organism | Synonyms | Taxon ID | GenBank # | Sequencing Center | Phyla | C sources |
|---|---|---|---|---|---|---|
|
Caldicellulosiruptor saccharolyticus DSM 8903 |
|
351627 |
NC_009437 |
DOE Joint Genome Institute |
F |
S,C,X |
|
Caldicellulosiruptor besci DSM 6725 |
Anaerocellum thermophilum; Z-1320 |
521460 |
NC_012036 |
DOE Joint Genome Institute |
F |
S,C,X |
|
Pyrococcus furiosus DSM 3638 |
|
186497 |
AE009950 |
Univ of Maryland, Univ of Utah |
E |
S,C,X |
|
Thermococcus kodakaraensis KOD1 |
|
69014 |
NC_006624 |
Kwansei Gakuin Univ, Kyoto University |
E |
S |
|
Thermotoga neapolitana DSM 4359 |
ATCC 49049; JCM 10099; NS-E |
309803 |
NC_011978 |
Genotech corp. |
T |
S,C |
|
Thermotoga petrophila RKU-1 |
|
390874 |
NC_009486 |
DOE Joint Genome Institute |
T |
S,C,X |
|
Thermotoga maritima MSB8 |
DSM 3109 |
243274 |
NC_000853 |
J. Craig Venter Institute |
T |
S,C,X |
|
Caldanaerobacter subterraneus subsp. tengcongensis MB4 |
Thermoanaerobacter tencongensis |
273068 |
NC_003869 |
Beijing Genomics Institute, The Institute of Microbiology, China |
F |
S |
|
Ethanoligenens harbinense YUAN-3 T |
DSM 18485 |
663278 |
NC_014828 |
DOE Joint Genome Institute |
F |
S,C |
|
Clostridium cellulolyticum H10 |
|
394503 |
NC_011898 |
DOE Joint Genome Institute |
F |
S,C,X |
|
Clostridium phytofermentans ISDg |
ATCC 700394 |
357809 |
NC_010001 |
DOE Joint Genome Institute |
F |
S,C,X |
|
Clostridium thermocellum ATCC 27405 |
DSM 1237 |
203119 |
NC_009012 |
DOE Joint Genome Institute, University of Rochester |
F |
S,C,X |
|
Clostridium thermocellum DSM 4150 |
JW20 |
492476 |
ABVG00000000 |
DOE Joint Genome Institute |
F |
S,C,X |
|
Thermoanaerobacter pseudethanolicus 39E |
ATCC 33223 |
340099 |
NC_010321 |
DOE Joint Genome Institute |
F |
S,X |
|
Geobacillus thermoglucosidasius C56-YS93 |
|
634956 |
NC_015660 |
DOE Joint Genome Institute |
F |
S |
| Bacillus cereus ATCC 14579 | DSM 31 | 226900 | NC_004721 | Integrated Genomics Inc. | F | S |
National Center for Biotechnology Information taxon IDs, GenBank accession numbers, corresponding sequencing centers responsible for the generation of the genome sequences data analyzed in this study are provided. Phyla (F; Firmicutes: E;Euryarchaeota: T; Thermotogae), and polymeric carbon sources degraded (S; starch: C; cellulose: X; xylose) by each organism are indicated).
We focused on the various metabolic branches involved in pyruvate formation from phosphoenolpyruvate (PEP) and subsequent catabolism of pyruvate into end-products. Although studies comparing the H2 and ethanol-producing potential of several cellulose degrading bacteria have been previously published [8-10], a comprehensive comparison of the major biofuel producing pathways at the genome level has not yet been reported. Here we present a comparison of the genes encoding proteins involved in (i) pyruvate metabolism, (ii) ethanol synthesis, and (iii) H2 metabolism, in order to rationalize reported end-product yields. Results indicate that the presence or absence of specific genes dictating carbon and electron flow towards end-products may be used to infer end-product synthesis patterns and help develop informed metabolic engineering strategies for optimization of H2 and ethanol yields. Furthermore, certain genes may be suitable biomarkers for screening novel microorganisms’ capability of producing optimal H2 or ethanol yields, and may be suitable targets for metabolic engineering strategies for optimization of either ethanol or H2 yields
Methods
Comparative analysis of genome annotations
All sequence data and gene annotations were accessed using the Joint Genome Institute’s Integrated Microbial Genomes (IMG) database [11]. Gene annotations presented in this paper reflect the numbering of the final assembly or most recent drafts available (July, 2012). Comparative analyses were performed using the IMG database. In brief, analyses of all genomes (Table 1) were conducted using three annotation databases independently: i) Clusters of Orthologs Groups (COGs) [12], ii) KEGG Orthology assignments (KO) [13], and (iii) TIGRFAMs [14]. Genes identified using a single database were cross-referenced against the others to identify genes of interest. Functional annotations of the identified genes were evaluated on a case-by-case basis and decisions regarding the annotation accuracy were made using a combination of manual analysis of genomic context, literature searches, and functional prediction through RPS-BLAST using the Conserved Domain Database website [15].
Hydrogenases were classified based on phylogenetic relationships of hydrogenase large subunits according to Calusinska et al. [16]. The evolutionary history was inferred using the Neighbor-Joining method [17]. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed [18]. The evolutionary distances were computed using the Poisson correction method [19] and are in the units of the number of amino acid substitutions per site. The analysis involved 50 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 863 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 [20]. Thermodynamic calculations were performed using values provided by Thauer et al.[21] and the CRC Handbook of Chemistry and Physics [21,22]. BioEdit v.7.0.9.0 [23] was used to perform sequence alignments.
Results and discussion
Survey of End-product yields
A literature survey of end-product yields (normalized to mol end-product per mol hexose equivalent) of the species surveyed in this study is summarized in Table 2. While it is difficult to perform a direct comparison of end-product yields from available literature due to different growth conditions employed (ex. growth substrate, carbon loading, reactor conditions, etc.), and further difficult to validate these data due to incomplete end-product quantifications and lack of corresponding carbon balances and oxidation/reduction (O/R) ratios, it still provides a good approximation of molar end-product yields based on substrate utilization. Calculated end-product yields reveal that the Caldicellulosiruptor, Pyrococcus, Thermococcus, and Thermotoga species surveyed, produced, in most cases, near-maximal H2 yields with concomitant CO2 and acetate production, and little or no ethanol, formate, and lactate [24-40]. It is important to note that while some studies [29-31,34,35,39] report lower overall end-product yields, likely due to a large amount of carbon flux being directed towards biomass production under a given growth condition, H2:ethanol ratios remain high. Cal. subterraneus subsp. tengcongensis, E. harbinense, and Clostridium species displayed mixed end-product fermentation patterns, with comparatively lower H2, CO2, and acetate yields, higher ethanol yields, and generally low formate and lactate yields [10,41-47]. Ta. pseudethanolicus produced the highest ethanol yields of the organisms surveyed with little concomitant H2, acetate, and lactate production, and no formate synthesis [48-50]. G. thermoglucosidasius and B. cereus produced the highest lactate and formate yields, moderate ethanol and acetate yields, and low H2 and CO2 yields [51,52].
Table 2.
Summary of end-product yields, optimal growth temperatures, total molar reduction values of H2 + ethanol (RVEP), and growth conditions employed
|
Organism |
Growth temp (°C) |
End products (mol/mol hexose equivalent) |
|
Growth condition |
Ref |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| H2 | CO2 | Acetate | Ethanol | Formate | Lactate | RVEP | ||||
|
Ca. saccharolyticus DSM 8903 |
70 |
4.0 |
1.8 |
NR |
ND |
ND |
ND |
4.0 |
Cont., 1.1 g l-1 glucose (D = 0.09 h-1) |
[24] |
| |
|
3.6 |
1.5 |
1.6 |
ND |
ND |
ND |
3.6 |
Cont., 4.1 g l-1 glucose (D = 0.1 h-1) |
[24] |
| |
|
3.5 |
NR |
2.1 |
NR |
NR |
NR |
3.5 |
Batch, 10 g l-1 sucrose |
[25] |
| |
|
2.5 |
1.4 |
1.4 |
ND |
ND |
0.1 |
2.5 |
Batch, 10 g l-1 glucose |
[26] |
|
Ca. bescii DSM 6725 |
75 |
✓ |
✓ |
✓ |
NR |
NR |
✓ |
NA |
|
[27,28] |
|
P. furiosus DSM 3638 |
90 |
3.8 |
1.9 |
1.5 |
0.1 |
NR |
NR |
4.0 |
Cont, cellobiose (D = 0.45 h-1) |
[29]A |
| |
|
3.5 |
1.0 |
1.4 |
ND |
NR |
ND |
3.5 |
Batch, 1.9 g l-1, maltose |
[30]A |
| |
|
2.9 |
1.9 |
0.8 |
0.1 |
NR |
ND |
3.1 |
Batch, 2 g l-1 maltose |
[31]B |
| |
|
2.8 |
0.9 |
1.2 |
ND |
NR |
ND |
2.8 |
Batch, 3.5 g l-1, cellobiose |
[30]A |
| |
|
2.6 |
1.4 |
1.0 |
ND |
NR |
NR |
2.6 |
Cont, maltose (D = 0.45 h-1) |
[29]A |
|
Th. kodakaraensis KOD1 |
85 |
3.3 |
1.8 |
1.1 |
NR |
NR |
NR |
3.3 |
Cont, starch (D = 0.2 h-1) |
[32]C |
|
T. neapolitana DSM 4359 |
80-85 |
3.8 |
2.0 |
1.8 |
ND |
NR |
0.1 |
3.8 |
Batch, 2.5 g l-1 glucose |
[33] |
| |
|
3.2 |
NR |
1.9 |
NR |
NR |
NR |
3.2 |
Batch (N2 sparged), 7.0 g l-1 glucose |
[34] |
| |
|
2.4 |
NR |
1.1 |
NR |
NR |
0.7 |
2.4 |
Batch, 1.1 g l-1 glucose |
[35] |
| |
|
1.8 |
NR |
1.0 |
NR |
NR |
NR |
1.8 |
Batch, 7.5 g l-1 glucose |
[40] |
| |
|
1.8 |
NR |
1.5 |
NR |
NR |
NR |
1.8 |
Batch, 7.0 g l-1 glucose |
[34] |
|
T. petrophila RKU-1 |
80 |
3.7 |
0.4 |
1.8 |
NR |
NR |
0.3 |
3.7 |
Batch, 1 g l-1 glucose |
[36] |
|
T. maritima MSB8 |
80 |
4.0 |
2.0 |
2.0 |
NR |
ND |
NR |
4.0 |
Batch, 2 g l-1 glucose |
[38] |
| |
|
2.2 |
1.1 |
1.0 |
ND |
NR |
0.3 |
2.2 |
Batch, 3 g l-1 glucose |
[39] |
| |
|
1.7 |
NR |
1.0 |
NR |
NR |
NR |
1.7 |
Batch, 7.5 g l-1 glucose |
[40] |
|
Cal. subterraneus subsp. tengcongensis MB4 |
75 |
2.8 |
NR |
1.4 |
0.6 |
NR |
ND |
4.0 |
Cont, starch (D = 0.27 h-1) |
[42] |
| |
|
NR |
NR |
2.0 |
ND |
NR |
ND |
NA |
Cont (N2 sparged), glucose (D = 0.24 h-1) |
[42] |
| |
|
0.3 |
1.5 |
1.0 |
0.7 |
NR |
ND |
1.7 |
Batch, 4 g l-1 glucose |
[41] |
|
E. harbinense YUAN-3 T |
35 |
2.8 |
✓ |
0.7 |
1.1 |
ND |
ND |
5.0 |
Batch, 20 g l-1 glucose |
[43] |
|
C. cellulolyticum H10 |
37 |
1.6 |
1.0 |
0.8 |
0.3 |
ND |
NR |
2.2 |
Batch, 5 g l-1 cellulose |
[44] |
| |
|
1.8 |
1.1 |
0.8 |
0.4 |
ND |
NR |
2,6 |
Batch, 5 g l-1 cellobiose |
[44] |
|
C. phytofermentans ISDg |
35-37 |
Major |
Major |
0.6 |
1.4 |
0.1 |
0.3 |
NA |
Batch, 34 g l-1 cellobiose |
[45] |
| |
|
1.0 |
0.9 |
0.6 |
0.5 |
0.1 |
NR |
2.0 |
Batch, 5 g l-1 cellulose |
[44] |
| |
|
1.6 |
1.2 |
0.6 |
0.6 |
ND |
NR |
2.8 |
Batch, 5 g l-1 cellobiose |
[44] |
|
C. thermocellum ATCC 27405 |
60 |
0.8 |
1.1 |
0.7 |
0.8 |
0.3 |
ND |
2.4 |
Batch, 1.1 g l-1 cellobiose |
[10] |
| |
|
1.0 |
0.8 |
0.8 |
0.6 |
0.4 |
0.4 |
2.2 |
Batch, 4.5 g l-1 cellobiose |
[46] |
|
C. thermocellum DSM 4150 |
60 |
1.8 |
1.7 |
0.9 |
0.8 |
ND |
0.1 |
3.4 |
Batch, 2 g l-1 glucose |
[47] |
| |
|
0.6 |
1.8 |
0.3 |
1.4 |
ND |
0.2 |
3.4 |
Batch, 27 g l-1 cellobiose |
[47] |
|
Ta. pseudethanolicus 39E |
65 |
0.1 |
2.0 |
0.1 |
1.8 |
NR |
0.1 |
3.7 |
Batch, 8 g l-1 glucose |
[50] |
| |
|
NR |
NR |
NR |
1.6 |
NR |
<0.1 |
3.2 |
1 g l-1 xylose |
[48] |
| |
|
NR |
NR |
0.4 |
1.0 |
NR |
<0.1 |
2.0 |
Batch, 20 g l-1 xylose |
[49] |
| |
|
NR |
NR |
0.2 |
0.4 |
NR |
1.1 |
0.8 |
Batch, 20 g l-1 glucose |
[49] |
|
G. thermoglucosidasius M10EXGD |
60 |
NR |
NR |
0.6 |
0.4 |
1.0 |
0.9 |
0.8 |
Batch, 10 g l-1 glucose |
[52] |
| B cereus ATCC 14579 | 35 | NR | 0.1 | 0.2 | 0.2 | 0.3 | 1.1 | 0.4 | Batch, 3.6 g l-1 glucose | [51] |
A ~ 0.5 mol alanine per mol-hexose produced on cellobiose and maltose.
BProduces H2, CO2, volatile fatty acids, and NH3 on peptides in the absence of carbon source.
C ~ 0.5 mol alanine per mol-hexose produced on starch.
DOnly G. thermoglucosidasuis strain C56-TS93 has been sequenced but no end-product data is available. Strain M10EXG was used for end-product yield comparisons instead.
Abbreviations: NR, not reported; ND, not detected; NA, not applicable; Major, reported as major product without absolute values; ✓, reported as present with no values indicated; Cont, continuous culture; D, dilution rate.
While reported yields vary considerably for each organisms, it is important to note that different growth conditions may influence end-product yields through regulation of gene and gene product expression [42,53], and modulation of metabolic flux and intracellular metabolite levels [54,55] that may act as allosteric regulators [56,57]. Variations in fermentation conditions including substrate availability/dilution rates [46,53-55,58-61], substrate composition [54,62-67], media composition [55], pH [68], gas partial pressures [34,42,69,70], growth phase [57], and accumulation of end-products [47,62,69,71,72] have been shown to influence end-product yields. Hence, while genome content alone cannot be used to predict end-product yields with accuracy, it can reflect end-product distribution profiles.
Genome comparison of pyruvate metabolism and end-product synthesis pathways
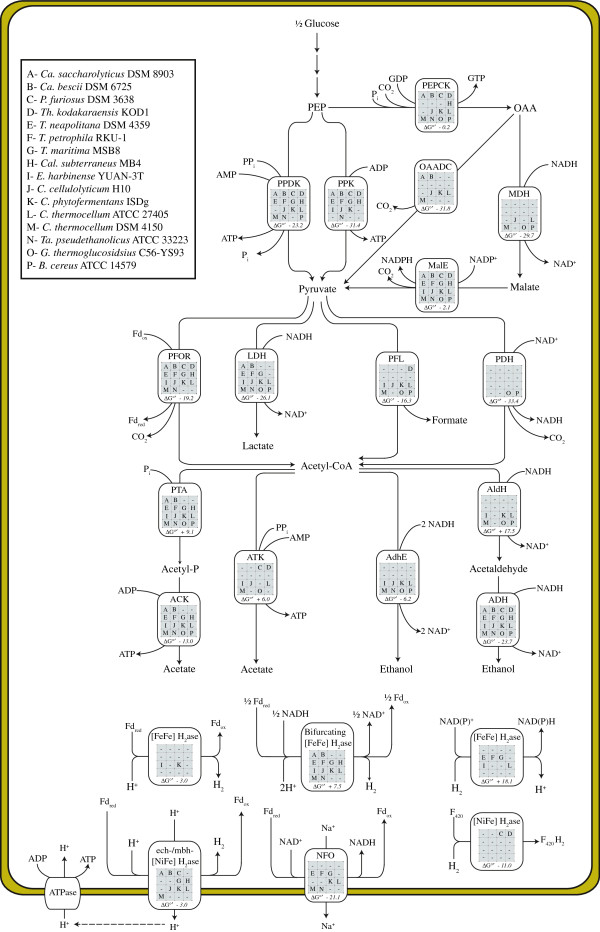
The assemblage of genes encoding proteins involved in pyruvate metabolism and end-product synthesis dictate, in part, how carbon and electron flux is distributed between the catabolic, anabolic, and energy producing pathways of the cell. The flow of carbon and electrons from PEP towards end-products may be separated into branch-points or nodes which include (i) the PEP/oxaloacetate/pyruvate node, (ii) the pyruvate/lactate/acetyl-CoA node, (iii) the acetyl-CoA/acetate/ethanol node, and the (iv) ferredoxin/NAD(P)H/H2 node [73]. Several different enzymes may be involved in the conversion of intermediate metabolites within these nodes. These enzymes, and the presence of corresponding genes encoding these proteins in each of the organisms surveyed, are summarized in Figure 1. The oxidation of electron carriers (NADH and/or reduced ferredoxin) is required for maintaining glycolytic flux and leads to the ultimate production of reduced products (ethanol, lactate, and H2). Thus, distribution of carbon and electron flux among different pathways can influence levels of reduced electron carrier pools, which in turn can dictate end-product distribution patterns. Genome content can be used to resolve the relationship between carbon and electron flux with end-product distribution.
Figure 1.
Comparison of putative gene products involved in pyruvate metabolism and end-product synthesis among select hydrogen and ethanol-producing species. Presence of putative gene products are indicated in matrix with respective letters corresponding to selected organism (see legend). Numbers indicate standard free energies of reaction (△G°’) corresponding to a particular enzyme. Abbreviations: PEPCK, phosphoenolpyruvate carboxykinase; OAADC, oxaloacetate decarboxylase; MDH, malate dehydrogenase; MalE, malic enzyme; PPK, pyruvate kinase; PPDK, pyruvate phosphate dikinase; LDH, lactate dehydrogenase; PFL, pyruvate formate lyase; PFOR, pyruvate:ferredoxin oxidoreductase; PDH, pyruvate dehydrogenase; ADH, alcohol dehydrogenase; ALDH, acetaldehyde dehydrogenase; AdhE, bifinctional acetaldehyde/alcohol dehydrogenase; ACK, acetate kinase; PTA, phosphotransacetylase; NFO, NADH:Fd oxidoreductase.
Genes involved in pyruvate synthesis
All organisms considered in this study utilize the Embden-Meyerhof-Parnas pathway for conversion of glucose to PEP with the following notable variations. Alignments of key residues of phosphofructokinase (PFK) according to Bapteste et al.[74,75], suggest that P. furiosus, Th. kodakaraensis, Cal. subterraneus subsp. tengcongensis, E. harbinense, G. thermoglucosidasius, and B. cereus encode an ATP-dependent PFK, while Thermotoga, Caldicellulosiruptor, Clostridium, and Thermoanaerobacter species encode both an ATP-dependent PFK, as well as a pyrophosphate (PPi)-dependent PFK [74,75] (Additional file 1). Furthermore, while bacteria catalyze the oxidation of glyceraldehyde-3-P to 3-phosphoglycerate (yielding NADH and ATP) with glyceraldehydes-3-phosphate dehydrogenase (GAPDH) and phosphoglycerate kinase (PGK), archea (P. furiosus and Th. kodakaraensis) preferentially catalyze the same reaction via glyceraldehyde-3-phosphate ferredoxin oxidoreductase (GAPFOR). This enzyme reduces ferredoxin (Fd) rather than NAD+ and does not produce ATP [76].
In contrast to the generally conserved gene content required for the production of PEP, a number of enzymes may catalyze the conversion of PEP to pyruvate [73] (Figure 1; Table 3). PEP can be directly converted into pyruvate via an ATP-dependent pyruvate kinase (PPK), or via an AMP-dependent pyruvate phosphate dikinase (PPDK). All strains considered in this review encode both ppk and ppdk, with the exception of C. thermocellum strains, which do not encode a ppk, and E. harbinense, G. thermoglucosidasius, and B. cereus, which do not encode ppdk. Given that the formation of ATP from ADP and Pi is more thermodynamically favorable than from AMP and PPi (△G°’ = 31.7 vs. 41.7 kJ mol-1), production of pyruvate via PPK is more favorable than via PPDK [21].
Table 3.
Genes encoding proteins involved in interconversion of phosphenolpyruvate and pyruvate
|
Organism |
Gene |
||||||
|---|---|---|---|---|---|---|---|
| eno | ppk | ppdk | pepck | oaadc | mdh | malE | |
| Standard free energy (ΔG°’) |
ND |
−31.4 |
−23.2 |
−0.2 |
−31.8 |
−29.7 |
−2.1 |
|
Ca. saccharolyticus DSM 8903 |
Athe_1403 |
Athe_1266 |
Athe_1409 |
Athe_0393 |
Athe_1316-1319 |
|
Athe_1062 |
|
Ca. bescii DSM 6725 |
Csac_1950 |
Csac_1831 |
Csac_1955 |
Csac_0274 |
Csac_2482-2485 |
|
Csac_2059 |
|
P. furiosus DSM 3638 |
PF0215 |
PF1188 |
PF0043 |
PF0289 |
|
|
PF1026 |
| |
PF1641 |
|
|
|
|
|
|
|
Th. kodakaraensis KOD1 |
TK1497 |
TK0511 |
TK0200 |
TK1405 |
|
|
TK1963 |
| |
TK2106 |
|
TK1292 |
|
|
|
|
|
T. neapolitana DSM 4359 |
CTN_1698 |
CTN_0477 |
CTN_0413 |
|
|
|
CTN_0126 |
|
T. petrophila RKU-1 |
Tpet_0050 |
Tpet_0716 |
Tpet_0652 |
|
|
|
Tpet_0379 |
|
T. maritima MSB8 |
TM0877 |
TM0208 |
TM0272 |
|
|
|
TM0542 |
|
Cal. subterraneus subsp. tengcongensis MB4A |
TTE1759 |
TTE1815 |
TTE0164 |
TTE1783 |
|
|
TTE2332 |
| |
|
|
TTE0981 |
|
|
|
|
|
E. harbinense YUAN-3 T |
Ethha_2662 |
Ethha_0305 |
|
|
|
|
Ethha_0739 |
|
C. cellulolyticum H10 |
Ccel_2254 |
Ccel_2569 |
Ccel_2388 |
Ccel_0212 |
Ccel_1736-1738 |
Ccel_0137 |
Ccel_0138 |
|
C. phytofermentans ISDg |
Cphy_3001 |
Cphy_0741 |
Cphy_0651 |
Cphy_3853 |
Cphy_2433-2434 |
|
Cphy_0409 |
| |
|
Cphy_2900 |
|
|
|
|
|
|
C. thermocellum ATCC 27405 |
Cthe_0143 |
|
Cthe_1253 |
Cthe_2874 |
Cthe_0699-0701 |
Cthe_0345 |
Cthe_0344 |
| |
|
|
Cthe_1308 |
|
|
|
|
|
C. thermocellum DSM 4150 |
|
|
CtherDRAFT_1661 |
CtherDRAFT_1742 |
CtherDRAFT_0819-0822 |
YesA |
YesA |
| |
|
|
CtherDRAFT_1896 |
|
|
|
|
|
Ta. pseudethanolicus 39E |
Teth39_0735 |
Teth39_0684 |
Teth39_1358 |
Teth39_0711 |
|
|
Teth39_0337 |
| |
|
|
Teth39_2098 |
|
|
|
|
|
G. thermoglucosidasius C56-YS93 |
Geoth_0446 |
Geoth_0898 |
|
Geoth_0811 |
|
Geoth_0904 |
Geoth_1713 |
| |
|
|
|
|
|
Geoth_3508 |
Geoth_2444 |
|
B.cereus ATCC 14579 |
BC5135 |
BC3323 |
BC3087 |
BC4762 |
|
BC4592 |
BC0580 NAD) |
| |
|
BC4599 |
|
|
|
BC2959 |
BC1741 (NAD) |
| BC4604 (NADP) | |||||||
AGenes have been verified by PCR amplification (unpublished).
Abbreviations: eno, enolase; ppk, pyruvate kinase; ppdk, pyruvate phosphate dikinase; pepck, phosphoenolpyruvate carboxykinase; oaadc, oxaloacetate decarboxylase; mdh, malate dehydrogenase; malE, malic enzyme.
Flux balance analysis integrated with RNAseq data suggests higher carbon and electron flux in C. thermocellum ATCC 27405 is directed through enzymes capable of direct, rather than indirect, conversion of PEP to pyruvate [77]. However, C. cellulolyticum mutation studies suggests that a portion of PEP can also be converted to pyruvate via the “malate shunt” [78]. This PPK/PPDK bypass system utilizes either (i) phosphoenolpyruvate carboxykinase (PEPCK), malate dehydrogenase (MDH), and malic enzyme (MalE), or (ii) PEPCK and oxaloacetate decarboxylase (OAADC), for the interconversion of PEP and pyruvate (Figure 1). While PEPCK provides a pathway for energy conservation via ATP (or GTP) production, MDH and MalE permit transhydrogenation from NADH to NADP+[71], generating additional reducing equivalents required for biosynthesis. G. thermoglucosidasius, B. cereus, C. thermocellum (ATCC 27405), and C. cellulolyticum contain pepck, mdh and malE suggesting that they are capable of transhydrogenation using these proteins. Although the draft genome of C. thermocellum DSM 4150 does not include genes encoding MDH and MalE, we have verified their presence via PCR amplification (unpublished results). Deletion of mdh in C. cellulolyticum resulted in significant increases in lactate, and to a lesser extent ethanol yields, and reduced acetate production when grown on cellulose demonstrating carbon and electron flux through MDH in wild type strains [78]. It seems evident that in the absence of MDH, transhydrogenation was reduced, and thus the resulting increase in NADH:NADPH ratios promote lactate and ethanol production, while decreasing NADPH levels for biosynthesis.
A number of organisms analyzed encode pepck and oaadc (Ca. bescii, Ca. saccharolyticus, C. cellulolyticum, C. phytofermentans, and C. thermocellum), also allowing for indirect conversion of PEP to pyruvate via an oxaloacetate intermediate. While the redirection of carbon and electron flux through this pathway likely has little effect on product yields, synthesis of GTP, versus ATP, may promote transcription and protein synthesis. Finally, Cal. subterraneus, E. harbinense, P. furiosus, Th. kodakaraensis, Ta. pseudethanolicus, and Thermotoga species do not encode all of the proteins required for a “malate shunt” and consequentially the catalysis of PEP to pyruvate must be achieved via PPK and/or PPDK.
Genes involved in pyruvate catabolism
The pyruvate/lactate/acetyl-CoA node plays an important role in regulating carbon flux and electron distribution and dramatically affects end-product distribution. The NADH-dependent reduction of pyruvate to lactate via fructose-1,6-bisphosphate activated lactate dehydrogenase (LDH) [56] diverts reducing equivalents away from biofuels such as H2 and ethanol. Alternatively, the oxidative decarboxylation of pyruvate to acetyl-CoA via pyruvate dehydrogenase (pdh) or pyruvate:ferreodoxin oxidoreductase (pfor) generate NADH and reduced Fd, respectively. These reducing equivalents may then be oxidized during the production of H2 or ethanol (Figure 1). Pyruvate may also be catabolised to acetyl-CoA via pyruvate:formate lyase (pfl) yielding formate in the process. In some enterobacteria, formate is further oxidized to CO2, releasing H2, through the action of a multisubunit formate hydrogen lyase (FHL) complex [79]. However, pfl was not encoded in any of the organisms analysed.
With the exception of Cal. subterraneus subsp. tengcongensis, P. furiosus, and Th. kodakaraensis, ldh genes were identified in all organisms studied (Table 4). Surprisingly, while the production of lactate from pyruvate is highly favorable thermodynamically (△G°’ = − 26.1 kJ mol-1-), only B. cereus, G. thermoglucosidasius, and, under some conditions, Ta. pseudethanolicus and T. neapolitana produce high yields of lactate (> 0.5 mol mol-glucose-1). In all other organisms surveyed lactate production was either a minor end-product, not detected, or not reported under the reported growth conditions (Table 2). This suggests that the presence of ldh cannot be used to predict lactate production.
Table 4.
Genes encoding proteins directly involved in pyruvate catabolism
|
Organism |
Gene |
|||
|---|---|---|---|---|
| ldh | pdh | pfor | pfl | |
| Standard free energy (G°’) |
−26.1 |
−33.4 |
−19.2 |
−16.3 |
|
Ca. saccharolyticus DSM 8903 |
Csac_1027 |
|
Csac_1458-1461 |
|
| |
|
|
Csac_2248-2249 |
|
|
Ca. bescii DSM 6725 |
Athe_1918 |
|
Athe_0874-0877 |
|
| |
|
|
Athe_1708-1709 |
|
|
P. furiosus DSM 3638 |
|
|
PF0965-PF0967, PF0971 |
|
|
Th. kodakaraensis KOD1 |
|
|
TK1978, TK1982-1984 |
TK0289 |
|
T. neapolitana DSM 4359 |
CTN_0802 |
|
CTN_0680-CTN_0683 |
|
|
T. petrophila RKU-1 |
Tpet_0930 |
|
Tpet_0905-Tpet_0908 |
|
|
T. maritima MSB8 |
TM1867 |
|
TM0015-TM0018 |
|
|
Cal. subterraneus subsp. tengcongensis MB4 |
|
|
TTE0445 |
|
| |
|
TTE0960 |
|
|
|
E. harbinense YUAN-3 T |
Ethha_1350 |
|
Ethha_0231-0234 |
Ethha_1657 |
| |
Ethha_2705 |
|
|
|
|
C. cellulolyticum H10 |
Ccel_2485 |
|
Ccel_0016 |
Ccel_2224 |
| |
|
|
Ccel_1164 |
Ccel_2582 |
|
C. phytofermentans ISDg |
Cphy_1117 Cphy_1232 |
|
Cphy_0603 Cphy_3558 |
Cphy_1174 |
| |
|
|
|
Cphy_1417 |
| |
|
|
|
Cphy_2823 |
|
C. thermocellum ATCC 27405 |
Cthe_1053 |
|
Cthe_2390-2393 |
Cthe_0505 |
| |
|
|
Cthe_2794-2797 |
|
| |
|
|
Cthe_3120 |
|
|
C. thermocellum DSM 4150 |
CtherDRAFT_2943 |
|
CtherDRAFT_0414-0417 |
CtherDRAFT_2234 |
| |
|
|
CtherDRAFT_1182-1185 |
|
| |
|
|
CtherDRAFT_1311 |
|
|
Ta. pseudethanolicus 39E |
Teth39_1997 |
|
Teth39_0289 |
|
| |
|
|
Teth39_1842 |
|
| |
|
|
|
|
|
G. thermoglucosidasius C56-YS93 |
Geoth_3351 |
Geoth_0237-0239 |
|
Geoth_3895 |
| |
|
Geoth_1595-1597 |
|
|
| |
|
Geoth_2366-2368 |
|
|
| |
|
Geoth_2479-2480 |
|
|
| |
|
Geoth_2860-2863 |
|
|
|
B.cereus ATCC 14579 |
BC1924 |
BC3970-3973 |
|
BC0491 |
| |
BC4870 |
|
|
|
| BC4996 | ||||
Abbreviations: ldh, lactate dehydrogenase; pdh, pyruvate dehydrogenase; pfor, pyruvate:ferredoxin oxidoreductase; pfl, pyruvate formate lyase.
LDH is, in fact, allosterically activated by fructose-1,6-bisphosphate in C. thermocellum ATCC 27405, Ca. saccharolyticus, and Thermoanaerobacter brockii[56,57,62,80]. While enzyme assays reveal high LDH activity in C. thermocellum[10,72], most studies report only trace amounts of lactate. Islam et al. [46], however, demonstrated that lactate production was triggered in stationary-phase batch cultures only under excess cellobiose conditions. In Thermoanaerobacter brockii, Ben-Bassat et al. reported elevated lactate production as a consequence of accumulated intracellular fructose-1,6-bisphosphate (FDP) when cultures were grown on glucose compared to starch [62]. Finally, Willquist and van Niel [57] reported that LDH in Ca. saccharolyticus was activated by FDP and ATP, and inhibited by NAD+ and PPi. An increase in fructose-1,6-bisphosphate, NADH:NAD+ ratios, and ATP:PPi ratios was observed during the transition from exponential to stationary phase in Ca. saccharolyticus cultures, and was accordingly accompanied by lactate production [57].
All organisms analyzed encode either pdh or pfor, but not both (Table 4). While G. thermoglucosidasius and B. cereus encode pdh, all other organisms analyzed encode pfor. Although Caldicellulosiruptor, Clostridia, and Thermoanaerobacter species studied appear to encode a putative pdh, there has been no enzymatic evidence to support the presence of PDH in these species. Thus far, only PFOR activity has been verified in C. cellulolyticum[58,60] and C. thermocellum[10,72]. The putative E1, E2, and E3 subunits of the pdh complex (Csac_0874-0872) in Ca. saccharolyticus were designated simply as a keto-acid dehydrogenase by van de Werken et al. [81]. Similarly, while genes encoding a putative pdh (Teth_0790-0793) are present in Ta. pseudethanolicus, genomic context strongly supports that this putative pdh is part of an acetoin dehydrogenase complex, despite the absence of reported acetoin production. In Clostridia species, putative pdh’s (Cthe_3449-3450, Cthe_1543) may actually encode 2-oxo acid dehydrogenase complexes, which share a common structure and homology to pyruvate dehydrogenase. These include 2-oxoglutarate dehydrogenase, branched-chain alpha-keto acid dehydrogenase, acetoin dehydrogenase complex, and the glycine cleavage complex. All organisms that encode a pfor also encode a Fd-dependent hydrogenase (H2ase), bifurcating H2ase, and/or a NADH:Fd oxidoreductase (NFO), and are thus capable of reoxidizing reduced Fd produced by PFOR. Conversely, G. thermoglucosidasius and B. cereus, which encode pdh but not pfor, do not encode enzymes capable of reoxidizing reduced Fd, and thus do not produce H2. While the presence of PDH allows for additional NADH production that could be used for ethanol production, G. thermoglucosidasius and B. cereus end-product profiles suggest that this NADH is preferentially rexodized through lactate production rather than ethanol production. Pyruvate decarboxylase, a homotetrameric enzyme that catalyzes the decarboxylation of pyruvate to acetaldehyde was not encoded by any of the species considered in this study.
Given the requirement of reduced electron carriers for the production of ethanol/H2, the oxidative decarboxylation of pyruvate via PDH/PFOR is favorable over PFL for the production of these biofuels. Genome analyses revealed that a number of organisms, including P. furiosus, Ta. pseudethanolicus, Cal. subterraneus subsp. tencongensis, and all Caldicellulosiruptor and Thermotoga species considered, did not encode PFL. In each of these species, the production of formate has neither been detected nor reported. Unfortunately, many studies do not report formate production, despite the presence of PFL. This may be a consequence of the quantification methods used for volatile fatty acid detection. When formate is not produced, the total oxidation value of 2 CO2 per mole glucose (+4), must be balanced with the production of H2 and/or ethanol. Thus, the “total molar reduction values of reduced end-products (H2 + ethanol)”, termed RVEP, should be −4, providing that all carbon and electron flux is directed towards end-product formation and not biosynthesis. Indeed, RVEP’s were usually greater than 3.5 in organisms that do not encode pfl (T. maritima, Ca. saccharolyticus), and below 3.5 in those that do encode pfl (C. phytofermentans, C. thermocellum, G. thermoglucosidasius, and B. cereus; Table 2). In some studies, RVEP’s were low due to a large amount of carbon and electron flux directed towards biosynthesis. In G. thermoglucosidasius and B. cereus RVEP’s of H2 plus ethanol ranged from 0.4 to 0.8 due to higher reported formate yields. The large differences in formate yields between organisms that encode pfl may be due to regulation of pfl. In Escherichia coli[82,83] and Streptococcus bovis[84,85], pfl expression has been shown to be negatively regulated by AdhE. Thus presence of pfl alone is not a good indicator of formate yields.
Genes involved in acetyl-CoA catabolism, acetate production, and ethanol production
The acetyl-CoA/acetate/ethanol node represents the third major branch-point that dictates how carbon and electrons flow towards end-products (Figure 1). Acetyl-CoA may be converted to acetate, with the concomitant production of ATP, either indirectly through an acetyl phosphate intermediate using phosphotransacetylase (pta) and acetate kinase (ack), or directly via acetate thiokinase (atk). Although both reactions produce ATP, the former uses ADP and Pi whereas the latter uses AMP and inorganic PPi as substrates for ATP synthesis. As a result, acetate production via pta and ack is more thermodynamically favorable than via atk (△G°’ = −3.9 vs. +6.0 kJ/mol, respectively) which is typically used for acetate assimilation. Of the organisms surveyed, E. harbinense, G. thermodenitrificans, C. cellulolyticum, both C. thermocellum strains, and G. thermoglucosidasius contain all three genes capable of converting pyruvate to acetate (Table 5). Conversely, Cal. subterraneus subsp. tengcongensis, Thermotoga and Caldicellulosiruptor species, C. phytofermentans, Ta. pseudethanolicus, and B. cereus encode only pta and ack, whereas P. furiosus and Th. kodakaraensis encode only atk.
Table 5.
Genes encoding proteins involved in end-product synthesis from acetyl-CoA
|
Organism |
gene |
|||||
|---|---|---|---|---|---|---|
| pta | ack | atk | aldH | adh | adhE | |
| Standard free energy (G°’) |
9.1 |
−13.0 |
6.0 |
17.5 |
−23.7 |
−6.2 |
|
Ca. saccharolyticus DSM 8903 |
Csac_2041 |
Csac_2040 |
|
|
Csac_0407 |
|
| |
|
|
|
|
Csac_0554 |
|
| |
|
|
|
|
Csac_0622 |
|
| |
|
|
|
|
Csac_0711 |
|
| |
|
|
|
|
Csac_1500 |
|
|
Ca. bescii DSM 6725 |
Athe_1494 |
Athe_1493 |
|
|
Athe_0928 |
|
| |
|
|
|
|
Athe_0224 |
|
|
P. furiosus DSM 3638 |
|
|
PF1540 |
|
PF0075 |
|
| |
|
|
PF1787 |
|
PF0608 |
|
|
Th. kodakaraensis KOD1 |
|
|
TK0465 |
|
TK1008 |
|
| |
|
|
TK0665 |
|
TK1569 |
|
|
T. neapolitana DSM 4359 |
CTN_0945 CTN_1440 |
CTN_0411 |
|
|
CTN_0257 |
|
| |
|
|
|
|
CTN_0369 |
|
| |
|
|
|
|
CTN_0385 |
|
| |
|
|
|
|
CTN_0580 |
|
| |
|
|
|
|
CTN_1655 |
|
| |
|
|
|
|
CTN_1756 |
|
|
T. petrophila RKU-1 |
Tpet_1042 Tpet_1615 |
Tpet_0650 |
|
|
Tpet_0007 |
|
| |
|
|
|
|
Tpet_0107 |
|
| |
|
|
|
|
Tpet_0484 |
|
| |
|
|
|
|
Tpet_0508 |
|
| |
|
|
|
|
Tpet_0563 |
|
| |
|
|
|
|
Tpet_0614 |
|
| |
|
|
|
|
Tpet_0813 |
|
|
T. maritima MSB8 |
TM1130 TM1755 |
TM0274 |
|
|
TM0111 |
|
| |
|
|
|
|
TM0298 |
|
| |
|
|
|
|
TM0412 |
|
| |
|
|
|
|
TM0436 |
|
| |
|
|
|
|
TM0820 |
|
| |
|
|
|
|
TM0920 |
|
|
Cal. subterraneus subsp. tengcongensis MB4 |
TTE1482 |
TTE1481 |
|
|
TTE0313 |
|
| |
|
|
|
|
TTE0695 |
|
| |
|
|
|
|
TTE0696 |
|
| |
|
|
|
|
TTE1591 |
|
|
E. harbinense YUAN-3 T |
Ethha_2711 |
Ethha_2004 |
Ethha_1333 |
Ethha_0578 |
Ethha_0051 |
Ethha_1385 |
| |
|
|
|
Ettha_0635 |
Ethha_0580 |
|
| |
|
|
|
|
Ethha_1164 |
|
| |
|
|
|
|
Ethha_2217 |
|
| |
|
|
|
|
Ethha_2239 |
|
|
C. cellulolyticum H10 |
Ccel_2137 |
Ccel_2136 |
Ccel_0494 Ccel_1469 |
|
Ccel_0894 |
Ccel_3198 |
| |
|
|
|
|
Ccel_1083 |
|
| |
|
|
|
|
Ccel_3337 |
|
|
C. phytofermentans ISDg |
Cphy_1326 |
Cphy_132 |
|
Cphy_0958 |
Cphy_1029 |
Cphy_3925 |
| |
|
|
|
Cphy_1178 |
Cphy_1421 |
|
| |
|
|
|
Cphy_1416 |
Cphy_2463 |
|
| |
|
|
|
Cphy_1428 |
Cphy_2463 |
|
| |
|
|
|
Cphy_2418 |
|
|
| |
|
|
|
Cphy_2642 |
|
|
| |
|
|
|
Cphy_3041 |
|
|
|
C. thermocellum ATCC 27405 |
Cthe_1029 |
Cthe_1028 |
Cthe_0551 |
Cthe_2238 |
Cthe_0101 |
Cthe_0423 |
| |
|
|
|
|
Cthe_0394 |
|
| |
|
|
|
|
Cthe_2579 |
|
|
C. thermocellum DSM 4150 |
CtherDRAFT_2741 |
CtherDRAFT_2742 |
CtherDRAFT_2349 |
CtherDRAFT_1042 |
CtherDRAFT_0189 |
CtherDRAFT_1096 |
| |
|
|
|
|
CtherDRAFT_0616 |
|
| |
|
|
|
|
CtherDRAFT_2833 |
|
|
Ta. pseudethanolicus 39E |
Teth39_1296 |
Teth39_1295 |
|
|
Teth39_0220 |
Teth39_0206 |
| |
|
|
|
|
Teth39_1597 |
|
| |
|
|
|
|
Teth39_1979 |
|
|
G. thermoglucosidasius C56-YS93 |
Cthe_3862 |
Geoth_0875 |
Geoth_0855 |
Geoth_0268 |
Geoth_1572 |
Geoth_3879 |
| |
|
|
Geoth_0879 |
Geoth_0652 |
Geoth_1941 |
|
| |
|
|
Geoth_2349 |
Geoth_3494 |
Geoth_0631 |
|
|
B. cereus ATCC 14579 |
BC5387 |
BC4637 |
|
BC2832 |
BC0802 |
BC4365 |
| |
|
|
|
BC3555 |
BC2529 |
|
| BC1285 | BC2220 | |||||
Abbreviations: pta, phosphotransacetylase; ack, acetate kinase; atk, acetate thiokinase; aldH, acetaldehyde dehydrogenase; adh, alcohol dehydrogenase; adhE; bifunctional acetylaldehyde/alcohol dehydrogenase.
Alternatively, acetyl-CoA may be converted into ethanol, during which 2 NADH (or NADPH) are oxidized, either directly via a fused acetaldehyde/alcohol dehydrogenase encoded by adhE, which has been proposed to be the key enzyme responsible for ethanol production [86,87], or indirectly through an acetaldehyde intermediate via acetaldehyde dehydrogenase (aldH) and alcohol dehydrogenase (adh). While all organisms surveyed encoded multiple class IV Fe-containing ADHs (Table 5), the functions of these ADHs may vary with respect to substrate specificity (aldehyde length and substitution), coenzyme specificity (NADH vs. NADPH), and the catalytic directionality favored (ethanol formation vs. consumption) [10,57-59,72,88-91]. Although there are reports of in silico determinations of substrate and cofactor specificity amongst ADHs, in our experience such resolutions are problematic [92,93]. Often times, the gene neighborhoods of identified ADHs were suggestive that the physiological role of many enzymes was not ethanol production. This is evident in Ca. saccharolyticus, which does not produce ethanol despite reported NADPH-dependent ADH activity [57].
P. furiosus, Th. kodakaraensis, and all Thermotoga and Caldicellulosiruptor species do not encode adhE or aldH, and therefore produce negligible or no ethanol. Given the absence of ethanol producing pathways in these species, reducing equivalents are disposed of through H2 production via H2ases and/or lactate production via LDH. Surprisingly, while Cal. subterraneus subsp. tengcongensis also does not appear to encode aldH or adhE, NADPH-dependent AldH and both NADH and NADPH-dependent ADH activities, as well as ethanol production, have been reported by Soboh et al. [42]. Similarly, Caldicellulosiruptor obsidiansis, which does not encode aldH or adhE, does produce trace levels of ethanol, suggesting that the various encoded ADHs may have broad substrate specificities [94]. Although C. cellulolyticum and Ta. pseudethanolicus do not encode aldH, they do encode adhE, and thus are capable of ethanol production. Of the organisms surveyed, only G. thermoglucosidasius and C. cellulolyticum encoded aldH and adh but no adhE, and produced moderate amounts of ethanol (~0.4 mol per mol hexose). Conversely, a number of organisms (E. harbinense, C. phytofermentans, both C. thermocellum strains, G. thermoglucosidasius, and B. cereus) encoded aldH, adh, and adhE, all of which produce varying ethanol yields.
Hydrogenases
In addition to disposal of reducing equivalents via alcohol and organic acid production, electrons generated during conversion of glucose to acetyl-CoA can be used to produce molecular hydrogen via a suite of [FeFe] and/or [NiFe] H2ases. The incredible diversity of H2ases has been extensively reviewed by Vignais et al. and Calusinska et al. [16,95,96]. H2ases may be (i) monomeric or multimeric, (ii) can catalyze the reversible production of H2 using various electron donors, including reduced Fd and NAD(P)H, or (iii) can act as sensory H2ases capable of regulating gene expression [97]. While most H2ases can reversibly shuttle electrons between electron carriers and H2, they are typically committed to either H2-uptake or evolution, depending on reaction thermodynamics and the requirements of the cell in vivo[95]. While Fd-dependent H2 production remains thermodynamically favorable at physiological concentrations (△G°’ ~ −3.0 kJ mol-1), potential production of H2 from NAD(P)H (△G°’ = +18.1 kJ mol-1) becomes increasingly unfavorable with increasing hydrogen partial pressure [98]. Hence, Fd-dependent H2ases are associated with H2 evolution, whereas NAD(P)H-dependent H2ases are more likely to catalyze H2 uptake. Recent characterization of a heterotrimeric “bifurcating” H2ase from Thermotoga maritma demonstrated that it can simultaneously oxidize reduced Fd and NADH to H2 (△G°’ ~ +7.5 kJ mol-1), which drives the endergonic production of H2 from NADH by coupling it to the exergonic oxidation of reduced Fd [99].
With the exception of G. thermoglucosidasius and B. cereus, which did not contain putative H2ase genes, the genomes of all of the organisms surveyed encode multiple H2ases. These H2ases were classified based on i) the phylogenetic relationship of H2ase large subunits (Additional file 2 and Additional file 3), according to Calusinska et al. [16], ii) H2ase modular structure, and iii) subunit composition, based on gene neighbourhoods. Encoded [NiFe] H2ases fell into 3 major subgroups including: (i) Fd-dependent, H2-evolving, membrane-bound H2ases (Mbh) and/or energy conserving [NiFe] H2ases (Ech) capable of generating sodium/proton motive force (Group 4) [42], (ii) Soluble cofactor-dependent (F420 or NAD(P)H), bidirectional, cytoplasmic, heteromultimeric H2ases (Group 3), and (iii) H2-uptake, membrane bound H2ases (Group 1) [96] (Additional file 2). Similarly, encoded [FeFe] H2ases fell into 5 major subgroups including: (i) heterotrimeric bifurcating H2ases, (ii) dimeric, NAD(P)H-dependent uptake H2ases, (iii) monomeric, putatively Fd-dependent H2ases, (iv) dimeric sensory H2ases containing PAS/PAC sensory domains which may be involved in redox sensing, and (v) monomeric sensory H2ases (Additional file 3). These sensory H2ases are usually encoded upstream of trimeric bifurcating H2ases (Table 6) and are often separated by a histidine/serine kinase suggesting a regulatory relationship between these two enzymes [16].
Table 6.
Genes encoding putative hydrogenases, sensory hydrogenases, and NADH:Fd oxidoreductases using ferredoxin, coenzyme F420, and NAD(P)H as electron carriers
|
Organism |
Hydrogenase and NADH:Fd oxidoreductase classification and corresponding genes |
||||||
|---|---|---|---|---|---|---|---|
| |
[NiFe] H2ase |
[FeFe] H2ase |
NFO |
||||
| Fd-dependent ech and mbhG4 | F420-dependentG3and otherG1 | Bifurcating | SensoryA | NAD(P)H-dependent | Fd-dependent | rnf-type | |
| Standard free energy (ΔG°’)* |
−3.0 |
11 |
+7.5** |
NA |
18.1 |
18.1 |
−21.1*** |
|
Ca. bescii DSM 6725 |
Athe_1082-Athe_1087 |
|
Athe_1297- Athe_1299 A1 TR(M3) |
Athe_1292 D M2e |
|
|
|
|
Ca. saccharolyticus DSM 8903 |
Csac_1534-Csac_1539 |
|
Csac_1862- Csac_1864 A1 TR(M3) |
Csac_1857 D M2e |
|
|
|
|
P. furiosus DSM 3638 |
PF1423- PF1436 |
PF0891- PF0894 G3 |
|
|
|
|
|
| |
|
PF1329- PF1332 G3 |
|
|
|
|
|
|
Th. kodakaraensis KOD1 |
TK2080- TK2093 |
TK2069-TK2072 G3 |
|
|
|
|
|
|
T. neapolitana DSM 4359 |
|
|
CTN_1067- CTN1069 TTH |
CTN_1071- CTN_1072 CD(M2f) |
CTN_0485 TTH |
|
CTN_0437-CTN_0442 |
|
T. petrophila RKU-1 |
|
|
Tpet_1367- Tpet_1369 TTH |
Tpet_1371- Tpet_1372 CD(M2f) |
Tpet_0723 TTH |
|
Tpet_0675-Tpet_0680 |
|
T. maritima MSB8 |
|
|
TM1424- TM1426 TTH |
TM1420- TM1422 CD(M2f) |
TM0201 TTH |
|
TM0244- TM0249 |
|
Cal.subterraneus subsp. tengcongensis MB4 |
TTE0123- TTE0134 |
|
TTE0892- TTE0894 A1 TR(M3) |
TTE0887 D M2e |
|
|
|
| |
|
|
|
TTE0697 CD(M2f) |
|
|
|
|
E. harbinense YUAN-3 T |
|
|
Ethha_2614- Ethha_2616 A8 TR(M3) |
Ethha_0052 CD(M2f) |
Ethha_2293 A7 D(M3) |
Ethha_0031 B2 M2a |
|
|
C. cellulolyticum H10 |
Ccel_1686- Ccel_1691 |
Ccel_1070-Ccel_1071 G1 |
Ccel_2303- Ccel_2305 A8 TR(M3) |
Ccel_2300- Ccel_2301 CD(M2f) |
|
Ethha_2695 B3 M3a |
|
| |
Ccel_3363- Ccel_3371 |
|
Ccel_2232- Ccel_2234 A1 TR(M3) |
|
|
|
|
| |
|
|
Ccel_2467- Ccel_2468 A1 TR(M3) |
|
|
|
|
|
C. phytofermentans ISDg |
Cphy_1730-Cphy_1735 |
|
Cphy_0087- Cphy_0089 A8 TR(M3) |
Cphy_0092- Cphy_0093 CD(M2f) |
|
Cphy_2056 A5 M2c |
Cphy_0211-Cphy_0216 |
| |
|
|
Cphy_3803- Cphy_3805 A1 TR(M3) |
Cphy_3798 D M2e |
Cthe_3003-Cthe_3004 |
Cphy_0090 B1 M3a |
|
|
C. thermocellum ATCC 27405 |
Cthe_3013-Cthe_3024 |
|
Cthe_0428- Cthe_0430 A8 TR(M3) |
Cthe_0425- Cthe_0426 CD(M2f) |
|
|
Cthe_2430-Cthe_2435 |
| |
|
|
Cthe_0340- Cthe_0342 A1 TR(M3) |
Cthe_0335 D M2e |
|
|
|
|
C. thermocellum DSM 4150 |
CtherDRAFT_2162-CtherDRAFT_2173 |
|
CtherDRAFT_1101-CtherDRAFT_1103 A8 TR(M3) |
CtherDRAFT_1098-CtherDRAFT_1099 CD(M2f) |
YesB |
|
CtherDRAFT_0369-CtherDRAFT_0375 |
| |
|
|
CtherDRAFT_2978 A1 TR(M3) |
|
|
|
|
|
Ta. pseudethanolicus 39E |
|
|
|
Teth39_0221 CD(M2f) |
|
|
Teth39_2119-Teth39_2124 |
| |
|
|
Teth39_1456- Teth39_1458 A1 TR(M3) |
Teth39_1463 D M2e |
|
|
|
|
G. thermoglucosidasius C56-YS93 | |||||||
| B. cereus ATCC 14579 | |||||||
AGroup D M2e hydrogenases are poorly characterized and do not contain a PAS/PAC-sensory domain. However, given their proximity to protein kinases and bifurcating hydrogenases, and their phylogenetic proximity to group C D(M2f) sensory hydrogenases (Additional file 3) we have classified them as sensory hydrogenases.
BVerified by microarray and proteomic analysis (unpublished).
Characterization of hydrogenase specificity was based metallocenter composition ([NiFe] or [FeFe]), modular structure, subunit composition, and large (catalytic) subunit phylogeny according to Vignais et al. and Calusinska et al. [16,95,96]. Phylogenetic cluster groupings are indicated in superscript, and corresponding phylogenetic trees are provided in Additional file 1 and Additional file 2. Abbreviations: H2ase, hydrogenase; NFO, NADH:ferredoxin oxidoreductase; ech, energy conserving hydrogenase; mbh, membrane bound hydrogenase; rnf, Rhodobacter nitrogen fixation.
With the exception of P. furiosus and Th. kodakaranesis, which encode only Fd-dependent and putative F420-dependent [NiFe] H2ases, all other H2ase encoding organisms surveyed are capable of H2ase-mediated oxidation/reduction of both Fd and NAD(P)H. This seems fitting given that P. furiosus and Th. kodakaraensis preferentially catalyze the oxidation of glyceraldedhyde-3-P via GAPFOR rather than GAPDH and PGK, and thus must reoxidize reduced Fd, rather than NADH, during fermentative product synthesis. All other H2ase encoding organisms produce NADH during glycolysis and reduced Fd via PFOR. In these organisms, the oxidation of these electron carriers may be carried out using various different types of H2ases. All of these species encoded at least a single putative bifurcating H2ase (Table 6). The majority of these bifurcating H2ases were found downstream dimeric or monomeric sensory [FeFe] H2ases that may be involved in their regulation (Table 6). Soboh et al. have demonstrated that NADH-dependent H2ase activities in Cal. subterraneus subsp. tengcongensis are affected by H2 partial pressures [42] suggesting possible regulation of these H2ases via a two-component signal transduction mechanism in response changes in redox levels [16,97]. It is important to note that these NADH-dependent H2ase activities may reflect bifurcating H2ase activities given that Cal. subterraneus subsp. tengcongensis encodes only a Fd-dependent and a putative bifurcating H2ase, and no NAD(P)H-dependent H2ases.
While Ta. pseudethanolicus only encodes a bifurcating H2ase, all other organisms that encode a bifurcating H2ase also encode Fd-dependent H2ases. Putative Fd-dependent, [NiFe] Ech/Mbh-type H2ases were identified in the genomes of Cal. subterraneus subsp. tengcongensis, P. furiosus, Th. kodakaraensis, and all Caldicellulosiruptor and Clostridium species (Table 6). A pair of putative Fd-dependent [FeFe] H2ases were identified in both E. harbinense and C. phytofermentans. With the exception of Ta. pseudethanolicus, Cal. subterraneus subsp. tengcongensis, and Caldicellulosiruptor species, all organisms surveyed containing a bifurcating H2ase also appear to be capable of NADH and/or NADPH oxidation using NADH/NADPH-dependent H2ases. As with ADHs, however, we could not determine H2ase cofactor specificity exclusively using in silico sequence analysis, stressing the importance of activity characterization of enzyme substrate specificity. While C. cellulolyticum achieves NAD(P)H oxidation using a putative H2-uptake [NiFe] H2ases, E. harbinense, Thermotoga species, and C. thermocellum ATCC 27405 achieve this using [FeFe] H2ases. Although the draft genome of C. thermocellum DSM 4150 does not encode an NAD(P)H-dependent H2ase, our proteomic and microarray data reveal the presence of Cthe_3003/Cthe_3004 homologues (Rydzak, unpublished results).
In addition to H2ase-mediated electron transfer between Fd and/or NADH and H2, electrons may be transferred directly between Fd and NAD(P)H via an Rnf-like (Rhodobacter nitrogen fixation) NADH:ferredoxin oxidoreductase (NFO), a membrane-bound enzyme complex capable of generating a sodium motive force derived from the energy difference between reduced Fd and NADH. Only Thermotoga species, C. phytofermentans, C. thermocellum, and Ta. pseudethanolicus encode putatively identified NFO. Proteomic analysis of C. thermocellum, however, revealed low, or no, expression of NFO subunits, suggesting it does not play a major factor in electron exchange between Fd and NADH [100].
While the presence/absence of genes encoding pathways that lead to reduced fermentation products (i.e. formate, lactate, and particularly ethanol) is a major determinant of H2 yields, we can make some inferences with respect to H2 yields based on the types of H2ases encoded. Given the thermodynamic efficiencies of H2 production using different cofactors, we can say that Fd-dependent H2ases are conducive for H2 production while NAD(P)H-dependent H2ases are not. However, organisms that do not encode ethanol-producing pathways (i.e. Caldicellulosiruptor and Thermotoga species) may generate high intracellular NADH:NAD+ ratios, making NADH-dependent H2 production thermodynamically feasible under physiological conditions. Conversely, in organisms capable of producing both H2 and ethanol (Ethanoligenens, Clostridium, and Thermoanaerobacter species), the presence of Fd-dependent H2ases appears to be beneficial for H2 production. For example, E. harbinense and Clostridium species, which encode Fd-dependent, as well as bifurcating and NAD(P)H-dependent H2ases, produce much higher H2 yields when compared to those of Ta. pseudethanolicus, which encodes only one bifurcating H2ase and no Fd or NAD(P)H-dependent H2ases. Interestingly, organisms that do not encode H2ases (G. thermoglucosidasius and B. cereus) produce low ethanol and high lactate (and/or formate yields), suggesting that H2 production can help lower NADH:NAD+ ratios, and thus reduce flux through LDH.
Influence of overall genome content on end-product profiles
The presence and absence of genes encoding proteins involved in pyruvate metabolism and end-product synthesis may be used as an indicator of end-product distribution. By comparing genome content to end-product yields, we identified key markers that influence ethanol and H2 yields. These include (i) MDH (ii) LDH, (iii) PFL vs. PFOR and/or PDH (iv) Aldh and AdhE, and (v) bifurcating, Fd-dependent, and NAD(P)H dependent H2ase.
While it is difficult to elucidate how differences in “malate shunt” genes affect end-product synthesis patterns by comparing reported yields, eliminating MDH has been shown to increase lactate and ethanol production, and decrease acetate production in C. cellulolyticum[78]. The elimination of this transhydrogenation pathway may increase NADH:NAD+ ratios for reduced end-product synthesis and reduce NADPH:NADP+ ratios for biosynthesis. While presence of LDH is not a good predictor of lactate yields, LDH, when activated, diverts reducing equivalents away from H2 and ethanol. In contrast to PFL, PFOR and PDH produce additional reducing equivalents (reduced Fd and NADH, respectively), and thus promote reduced end-product synthesis. Organisms that do not encode pfl generally produce more ethanol and H2 (based on sum redox value) compared to those that do encode pfl. Of the organisms surveyed, those that did not encode (or express) both adhE and aldH produced near-maximal H2 yields and little to no ethanol. While the type(s) of encoded H2ases appear to have little impact in organisms that do not encode ethanol producing pathways, they do seem to influence reduced end-product yields in those that do. For example, Ta. pseudethanolicus, which encodes an adhE, NFO, and a single bifurcating H2ase, but no discernable Fd or NAD(P)H-dependent H2ases, generates low H2 and near-optimal ethanol yields. The inability to oxidize reduced Fd via Fd-dependent H2ases may elevate reduced Fd levels, which in turn can be used by NFO to produce additional NADH for ethanol synthesis. Interestingly, in the absence of H2ases, lactate production was favoured over ethanol production, suggesting that H2 production can help lower NADH:NAD+ ratios, and thus reduce flux through LDH.
Given the impact that MDH, PFL, Aldh, AdhE, and the different H2ases have on end-product yields, screening for these biomarkers can streamline ethanol and H2 producing potential of sequenced and novel organisms through in silico gene mining and the use of universal primers, respectively. Furthermore, understanding how end-product yields are affected by (i) the framework of genes encoding pathways catalyzing pyruvate into end-products, and (ii) thermodynamic efficiencies of these reactions, we can begin to develop informed metabolic engineering strategies for optimization of either ethanol or H2 (Figure 2). For example, in order to optimize either ethanol or H2, we would recommend elimination of ldh and pfl in order to allow accumulation of additional reducing equivalents. Given that ethanol and H2 compete for reducing equivalents, elimination of one product should direct carbon/and or electron flux towards the other.
Figure 2.
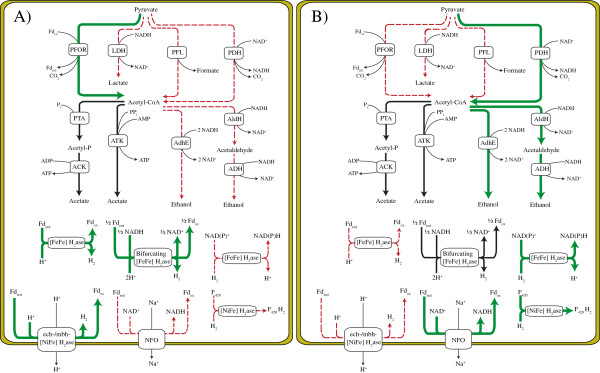
Differentiation between fermentation pathways that favor (A) hydrogen and (B) ethanol production based on comparative genomics and end-product profiles. Pathways that favor (green lines), disfavor (broken red lines), and appear to have little impact (black lines) on production of H2 or ethanol are indicated. Correlation of reaction thermodynamics and genome content with reported end-product yields suggest that reduction, and subsequent reoxidation, of ferredoxin via PFOR and Fd-dependent (and/or bifurcating) H2ases, respectively, support H2 production. Alternatively, reduction, of NAD+ via PDH (and/or NADH generating uptake H2ases) generate NADH conducive for ethanol production. Abbreviations (see figure 1 legend).
For optimization of H2 yields (Figure 2A), deletion of aldH and adhE is likely most effective. Although conversion of pyruvate to acetyl-CoA is more thermodynamically favorable using PDH versus PFOR (△G°’ = −33.4 vs. -19.2 kJ mol-1), production of H2 from NADH is highly unfavorable compared to the use of reduced Fd (△G°’ = +18.1 vs. -3.0 kJ mol-1). This in turn demonstrates that reduction of Fd via PFOR and subsequent H2 production via a Fd-dependent H2ase (△G°’ = −21.2 kJ mol-1) is more favorable than NADH production via PDH and subsequent H2 production via NAD(P)H-dependent H2ases (△G°’ = −15.3 kJ mol-1). Therefore, we propose that conversion of pyruvate to acetyl-CoA via PFOR is favorable for H2 production, and pdh (and pfl) should be deleted. Given that 2 NADH (per glucose) are produced during glycolysis in most anaerobic microorganisms, the presence of a bifurcating H2ase, which would simultaneously oxidize the 2 NADH generated during and 2 reduced Fd produced by PFOR, would be required to achieve theoretically maximal H2 yields of 4 mol per mol glucose. A Fd-dependent H2ase would also be conducive for H2 production during times when reducing equivalents generated during glycolysis are redirected towards biosynthetic pathways, resulting in a disproportionate ratio of reduced ferredoxin to NAD(P)H. Alternatively, in organisms such as P. furiosus and Th. kodakaraensis, which generate high levels of reduced Fd and low levels of NADH, the presence of Fd-dependent H2ases, rather than bifurcating H2ases, would be more conducive for H2 production. In all cases, NFO and NAD(P)H-dependent H2ases should be deleted to prevent oxidation of reduced Fd and uptake of H2, respectively, which would generate NAD(P)H.
The metabolic engineering strategies employed for optimization of ethanol (Figure 2B) are much different than those used for the production of H2. First, adhE and/or aldH and adh genes that encode enzymes with high catalytic efficiencies in the direction of ethanol formation should be heterologously expressed. Given that ethanol production is NAD(P)H dependent, increasing NADH production should be optimized, while Fd reduction should be eliminated. Through deletion of pfl and pfor, and expression of pdh, up to 4 NADH can be generated per glucose, allowing for the theoretical maximum of 2 mol ethanol per mol glucose to be produced. To prevent NADH reoxidation, lactate and H2 production should be eliminated by deleting ldh and NAD(P)H-dependent H2ases. While this strategy is theoretically sound, low AldH/Adh catalytic efficiencies may cause NADH/NAD+ ratios to rise so high that they may impede glycolysis. In these situations, the presence of a NFO or NAD(P)H-dependent H2ase may intermittently alleviate these high NADH/NAD+ ratios through generation of reduced Fd pools or H2 production, respectively, albeit it would decrease reducing equivalents for ethanol production.
While some attempts to increase H2 and/or ethanol yields through genetic engineering have been successful in a number of lignocellulolytic organisms (reviewed elsewhere; [101]) engineering of strains discussed here has only been marginally successful. Heterologous expression of Zymomonas mobilis pyruvate decarboxylase and Adh in C. cellulolyticum increased cellulose consumption and biomass production, and decreased lactate production and pyruvate overflow due to a more efficient regulation of carbon and electron flow at the pyruvate branchpoint [102]. However, despite higher levels of total ethanol produced, ethanol yields (per mol hexose consumed) actually decreased when compared to the wild-type strain. Similarly, deletion of PTA in C. thermocellum drastically reduced acetate production, but had minimal impact on lactate or ethanol production [103]. This suggests that genome content alone cannot exclusively dictate the extent of end-product yields observed in literature, and thus growth conditions must be optimized in order to moderate regulatory mechanisms that direct carbon and electron flux. This could only be attained through a thorough understanding of regulatory mechanisms that mediate gene and gene-product expression and activity levels under various growth conditions through a combination of genomics, transcriptomics, proteomics, metabolomics, and enzyme characterization.
Conclusions
Fermentative bacteria offer the potential to convert biomass into renewable biofuels such as H2 and ethanol through consolidated bioprocessing. However, these bacteria display highly variable, branched catabolic pathways that divert carbon and electrons towards unwanted end products (i.e. lactate, formate). In order to make fermentative H2 and/or ethanol production more economically feasible, biofuel production yields must be increased in lignocellulolytic bacteria capable of consolidated bioprocessing. While the cellulolytic and, to a lesser extent, H2 and ethanol producing capabilities of cellulolytic bacteria have been reviewed [8,9,44], a comprehensive comparison between genome content and corresponding end-product distribution patterns has not been reported. While reported end-product yields vary considerably in response to growth conditions, which may influence gene and gene product expression and metabolic flux, we demonstrate that composition of genes encoding pyruvate catabolism and end-product synthesis pathways alone can be used to approximate potential end-product distribution patterns. We have identified a number of genetic biomarkers, including (i) MDH (ii) LDH, (iii) PFL vs. PFOR and/or PDH (iv) Aldh and AdhE, and (V) bifurcating, Fd-dependent, and NAD(P)H dependent H2ases, that can be used for streamlining H2 and/or ethanol producing capabilities in sequenced and novel isolates. By linking genome content, reaction thermodynamics, and end-product yields, we offer potential targets for optimization of either ethanol or H2 yields via metabolic engineering. Deletion of LDH and PFL could potentially increase both H2 and ethanol yields. While deletion of ethanol producing pathways (aldH, adh, adhE), increasing flux through PFOR, overexpression of Fd -dependent H2ases, and elimination of potential H2-uptake (NAD(P)H-dependent) H2ases could lead to increased H2 production, eliminating H2 production and redirecting flux through PDH would be beneficial for ethanol production. Although gene and gene-product expression, functional characterization, and metabolomic flux analysis remains critical in determining pathway utilization, insights regarding how genome content affects end-product yields can be used to direct metabolic engineering strategies and streamline the characterization of novel species with potential industrial applications.
Abbreviations
ACK: Acetate kinase; ADH: Alcohol dehydrogenase; AdhE: Acetaldehyde/alcohol dehydrogenase (bifunctional); AldH: Aldehyde dehydrogenase; ATK: Acetate thiokinase; Ech: Energy conserving hydrogenase; Fd: Ferredoxin; FDP: Fructose-1,6-bisphosphate; FHL: Formate hydrogen lyase; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; GAPFOR: Glyceraldehydes-3-phosphate ferredoxin oxidoreductase; H2ase: Hydrogenase; IMG: Integrated Microbial Genomes; KO: KEGG Orthology; LDH: Lactate dehydrogenase; MalE: Malic enzyme; Mbh: Membrane-bound hydrogenase; MDH: Malate dehydrogenase; NFO: NADH:ferredoxin oxidoreductase; O/R: (Oxidation/reduction); OAADC: Oxaloacetate decarboxylase; PDH: Pyruvate dehydrogenase; PEP: Phosphoenolpyruvate; PEPCK: Phosphoenolpyruvate carboxykinase; PFK: Phosphofructokinase; PFL: Pyruvate:formate lyase; PFOR: Pyruvate:ferredoxin oxidoreductase; PGK: Phosphoglycerate kinase; PPDK: Pyruvate phosphate dikinase; PPK: Pyruvate kinase; PTA: Phosphotransacetylase; Rnf: Rhodobacter nitrogen fixation; RVEP: Total molar reduction values of reduced end-products (H2 + ethanol).
Authors’ contributions
TR and CRC co-authored the manuscript. TV, CRC and TR performed genomic meta-analysis. TR performed end-product comparisons and thermodynamic calculations. CRC performed phylogenetic analysis. RS, NC, and DBL conceived of the study, participated in its design, and helped draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Cofactor specificity (ATP or PPi) of phosphofructokinases based on sequence alignments. Alignments of key residues determining ATP or PPi specificity, as determined by Bapteste et al. [74] and Bielen et al. [75], were performed using BioEdit v.7.0.9.0. The P. furiosus and Th. kodakarensis genes are very distinct (different COG and different KO) and are annotated as Archaeal phosphofructokinases.
Phylogenetic clustering of [NiFe] hydrogenases large (catalytic) subunits. Catalytic (large) subunits of [NiFe] H2ases were identified based upon the modular signatures as described by Calusinska et al. [16], Species considered in this manuscript are highlighted and corresponding H2ase gene loci are provided.
Phylogenetic clustering of [FeFe] hydrogenases large (catalytic) subunits. Catalytic (large) subunits of [FeFe] H2ases were identified based upon the modular signatures as described by Calusinska et al. [16]. Species considered in this manuscript are highlighted and corresponding H2ase gene loci are provided.
Contributor Information
Carlo R Carere, Email: umcarerc@cc.umanitoba.ca.
Thomas Rydzak, Email: rydzak.t@gmail.com.
Tobin J Verbeke, Email: umverbe0@cc.umanitoba.ca.
Nazim Cicek, Email: nazim_cicek@cc.umanitoba.ca.
David B Levin, Email: levindb@cc.umanitoba.ca.
Richard Sparling, Email: Richard_sparling@umanitoba.ca.
Acknowledgements
This work was supported by funds provided by the Natural Sciences and Engineering Research Council of Canada (NSERC), through a Strategic Programs grant (STPGP 306944–04), by Genome Canada, through the Applied Genomics Research in Bioproducts or Crops (ABC) program for the grant titled, “Microbial Genomics for Biofuels and CoProducts from Biorefining Processes”, and by the Province of Manitoba, Agricultural and Rural Development Initiative (ARDI), grant 09–986.
References
- Carere C, Kalia V, Sparling R, Cicek N, Levin D. Pyruvate catabolism and hydrogen synthesis pathway genes of Clostridium thermocellum ATCC 27405. Indian J Microbiol. 2008;48(2):252–266. doi: 10.1007/s12088-008-0036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DB, Pitt L, Love M. Biohydrogen production: prospects and limitations to practical application. Int J Hydrogen Energy. 2004;29(2):173–185. doi: 10.1016/S0360-3199(03)00094-6. [DOI] [Google Scholar]
- Lynd LR, van Zyl WH, McBride JE, Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol. 2005;16(5):577–583. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Desvaux M. Clostridium cellulolyticum: model organism of mesophillic cellulolytic clostridia. FEMS Microbiol Rev. 2005;29:741–764. doi: 10.1016/j.femsre.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Islam R, Cicek N, Sparling R, Levin D. Influence of initial cellulose concentration on the carbon flow distribution during batch fermentation by Clostridium thermocellum ATCC 27405. Appl Microbiol Biotechnol. 2009;82(1):141–148. doi: 10.1007/s00253-008-1763-0. [DOI] [PubMed] [Google Scholar]
- Yang SJ, Kataeva I, Hamilton-Brehm SD, Engle NL, Tschaplinski TJ, Doeppke C, Davis M, Westpheling J, Adams MWW. Efficient degradation of lignocellulosic plant biomass, without pretreatment, by the thermophilic anaerobe "anaerocellum thermophilum" DSM 6725. Appl Environ Microbiol. 2009;75(14):4762–4769. doi: 10.1128/AEM.00236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallenbeck PC, Benemann JR. Biological hydrogen production; fundamentals and limiting processes. Int J Hydrogen Energy. 2002;27:1123–1505. doi: 10.1016/S0360-3199(02)00123-4. [DOI] [Google Scholar]
- Bruggemann H, Gottschalk G. Comparative genomics of clostridia: link between the ecological niche and cell surface properties. Ann N Y Acad Sci. 2008;1125:73–81. doi: 10.1196/annals.1419.021. [DOI] [PubMed] [Google Scholar]
- Desvaux M. Unravelling carbon metabolism in anaerobic cellulolytic bacteria. Biotechnol Prog. 2006;22(5):1229–1238. doi: 10.1002/bp060016e. [DOI] [PubMed] [Google Scholar]
- Rydzak T, Levin DB, Cicek N, Sparling R. Growth phase-dependant enzyme profile of pyruvate catabolism and end-product formation in Clostridium thermocellum ATCC 27405. J Biotechnol. 2009;140(3–4):169–175. doi: 10.1016/j.jbiotec.2009.01.022. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Korzeniewski F, Palaniappan K, Szeto E, Werner G, Padki A, Zhao X, Dubchak I, Hugenholtz P, Anderson I. et al. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 2006;34(Database issue):D344-D348. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN. et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T. et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480-D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT, White O. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res. 2001;29(1):41–43. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Calusinska M, Happe T, Joris B, Wilmotte A. The surprising diversity of clostridial hydrogenases: a comparative genomic perspective. Microbiology. 2010;156(Pt 6):1575–1588. doi: 10.1099/mic.0.032771-0. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. In: Evolving Genes and Proteins. Bryson V, Vogel H, editor. Academic Press, New York; 1965. Evolutionary divergence and convergence in proteins; pp. 97–166. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41(1):100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemical Rubber Company. CRC handbook of chemistry and physics. CRC Press, Cleveland, OH; 1977. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;1999:95–98. [Google Scholar]
- de Vrije T, Mars AE, Budde MAW, Lai MH, Dijkema C, de Waard P, Claassen PAM. Glycolytic pathway and hydrogen yield studies of the extreme thermophile Caldicellulosiruptor saccharolyticus. Appl Microbiol Biotechnol. 2007;74(6):1358–1367. doi: 10.1007/s00253-006-0783-x. [DOI] [PubMed] [Google Scholar]
- Bredholt S, Sonne-Hansen J, Nielsen P, Mathrani IM, Ahring BK. Caldicellulosiruptor kristjanssonii sp nov., a cellulolytic extremely thermophilic, anaerobic bacterium. Int J Syst Bacteriol. 1999;49:991–996. doi: 10.1099/00207713-49-3-991. [DOI] [PubMed] [Google Scholar]
- Kadar Z, De Vrijek T, van Noorden GE, Budde MAW, Szengyel Z, Reczey K, Claassen PAM. Yields from glucose, xylose, and paper sludge hydrolysate during hydrogen production by the extreme thermophile Caldicellulosiruptor saccharolyticus. Appl Biochem Biotechnol. 2004;113-16:497–508. doi: 10.1385/abab:114:1-3:497. [DOI] [PubMed] [Google Scholar]
- Kataeva IA, Yang SJ, Dam P, Poole FL, Yin Y, Zhou FF, Chou WC, Xu Y, Goodwin L, Sims DR. et al. Genome sequence of the anaerobic, thermophilic, and cellulolytic bacterium "anaerocellum thermophilum" DSM 6725. J Bacteriol. 2009;191(11):3760–3761. doi: 10.1128/JB.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetlichnyi VA, Svetlichnaya TP, Chernykh NA, Zavarzin GA. Anaerocellum-thermophilum Gen-Nov Sp-Nov - an extremely thermophilic cellulolytic eubacterium isolated from hot-springs in the Valley of Geysers. Microbiology. 1990;59(5):598–604. [Google Scholar]
- Chou CJ, Shockley KR, Conners SB, Lewis DL, Comfort DA, Adams MW, Kelly RM. Impact of substrate glycoside linkage and elemental sulfur on bioenergetics of and hydrogen production by the hyperthermophilic archaeon Pyrococcus furiosus. Appl Environ Microbiol. 2007;73(21):6842–6853. doi: 10.1128/AEM.00597-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kengen SW, de Bok FA, van Loo ND, Dijkema C, Stams AJ, de Vos WM. Evidence for the operation of a novel Embden-Meyerhof pathway that involves ADP-dependent kinases during sugar fermentation by Pyrococcus furiosus. J Biol Chem. 1994;269(26):17537–17541. [PubMed] [Google Scholar]
- Schicho RN, Ma K, Adams MW, Kelly RM. Bioenergetics of sulfur reduction in the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1993;175(6):1823–1830. doi: 10.1128/jb.175.6.1823-1830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai T, Imanaka H, Nakajima A, Uwamori K, Omori Y, Fukui T, Atomi H, Imanaka T. Continuous hydrogen production by the hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1. J Biotechnol. 2005;116(3):271–282. doi: 10.1016/j.jbiotec.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Munro SA, Zinder SH, Walker LP. The fermentation stoichiometry of Thermotoga neapolitana and influence of temperature, oxygen, and pH on hydrogen production. Biotechnol Prog. 2009;25(4):1035–1042. doi: 10.1002/btpr.201. [DOI] [PubMed] [Google Scholar]
- Nguyen TA, Han SJ, Kim JP, Kim MS, Sim SJ. Hydrogen production of the hyperthermophilic eubacterium, Thermotoga neapolitana under N2 sparging condition. Bioresour Technol. 2010;101(Suppl 1):S38–S41. doi: 10.1016/j.biortech.2009.03.041. [DOI] [PubMed] [Google Scholar]
- Eriksen NT, Nielsen TM, Iversen N. Hydrogen production in anaerobic and microaerobic Thermotoga neapolitana. Biotechnol Lett. 2008;30(1):103–109. doi: 10.1007/s10529-007-9520-5. [DOI] [PubMed] [Google Scholar]
- Takahata Y, Nishijima M, Hoaki T, Maruyama T. Thermotoga petrophila sp. nov. and Thermotoga naphthophila sp. nov., two hyperthermophilic bacteria from the Kubiki oil reservoir in Niigata, Japan. Int J Syst Evol Microbiol. 2001;51(Pt 5):1901–1909. doi: 10.1099/00207713-51-5-1901. [DOI] [PubMed] [Google Scholar]
- Nguyen TN, Borges KM, Romano AH, Noll KM. Differential gene expression in Thermotoga neapolitana in response to growth substrate. FEMS Microbiol Lett. 2001;195(1):79–83. doi: 10.1111/j.1574-6968.2001.tb10501.x. [DOI] [PubMed] [Google Scholar]
- Schröder C, Selig M, Schönheit P. Glucose fermentation to acetate, CO2, and H2 in the anaerobic hyperthermophilic eubacterium thermotoga maritima: involvement of the embden-meyerhof pathway. Arch Microbiol. 1994;161(6):460–470. [Google Scholar]
- Lakhal R, Auria R, Davidson S, Ollivier B, Dolla A, Hamdi M, Combet-Blanc Y. Effect of oxygen and redox potential on glucose fermentation in thermotoga maritima under controlled physicochemical conditions. Int J Microbiol. 2010;2010:896510. doi: 10.1155/2010/896510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TAD, Pyo Kim J, Sun Kim M, Kwan Oh Y, Sim SJ. Optimization of hydrogen production by hyperthermophilic eubacteria, thermotoga maritima and thermotoga neapolitana in batch fermentation. Int J Hydrogen Energy. 2008;33(5):1483–1488. doi: 10.1016/j.ijhydene.2007.09.033. [DOI] [Google Scholar]
- Xue Y, Xu Y, Liu Y, Ma Y, Zhou P. Thermoanaerobacter tengcongensis sp. nov., a novel anaerobic, saccharolytic, thermophilic bacterium isolated from a hot spring in Tengcong, China. Int J Syst Evol Microbiol. 2001;51(Pt 4):1335–1341. doi: 10.1099/00207713-51-4-1335. [DOI] [PubMed] [Google Scholar]
- Soboh B, Linder D, Hedderich R. A multisubunit membrane-bound [NiFe] hydrogenase and an NADH-dependent Fe-only hydrogenase in the fermenting bacterium Thermoanaerobacter tengcongensis. Microbiology. 2004;150(7):2451–2463. doi: 10.1099/mic.0.27159-0. [DOI] [PubMed] [Google Scholar]
- Xing D, Ren N, Li Q, Lin M, Wang A, Zhao L. Ethanoligenens harbinense gen. nov., sp. nov., isolated from molasses wastewater. Int J Syst Evol Microbiol. 2006;56(Pt 4):755–760. doi: 10.1099/ijs.0.63926-0. [DOI] [PubMed] [Google Scholar]
- Ren Z, Ward TE, Logan BE, Regan JM. Characterization of the cellulolytic and hydrogen-producing activities of six mesophilic Clostridium species. J Appl Microbiol. 2007;103(6):2258–2266. doi: 10.1111/j.1365-2672.2007.03477.x. [DOI] [PubMed] [Google Scholar]
- Warnick TA, Methe BA, Leschine SB. Clostridium phytofermentans sp. nov., a cellulolytic mesophile from forest soil. Int J Syst Evol Microbiol. 2002;52(Pt 4):1155–1160. doi: 10.1099/00207713-52-4-1155. [DOI] [PubMed] [Google Scholar]
- Islam R, Cicek N, Sparling R, Levin D. Effect of substrate loading on hydrogen production during anaerobic fermentation by Clostridium thermocellum 27405. Appl Microbiol Biotechnol. 2006;72(3):576–583. doi: 10.1007/s00253-006-0316-7. [DOI] [PubMed] [Google Scholar]
- Freier D, Mothershed CP, Wiegel J. Characterization of Clostridium thermocellum JW20. Appl Environ Microbiol. 1988;54(1):204–211. doi: 10.1128/aem.54.1.204-211.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacis LS, Lawford HG. Ethanol-production from xylose by thermoanaerobacter-ethanolicus in batch and continuous culture. Arch Microbiol. 1988;150(1):48–55. doi: 10.1007/BF00409717. [DOI] [Google Scholar]
- Lacis LS, Lawford HG. Thermoanaerobacter ethanolicus growth and product yield from elevated levels of xylose or glucose in continuous cultures. Appl Environ Microbiol. 1991;57(2):579–585. doi: 10.1128/aem.57.2.579-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegel J, Ljungdahl LG. Thermoanaerobacter ethanolicus gen. nov., spec. nov., a new, extreme thermophilic, anaerobic bacterium. Arch Microbiol. 1981;128(4):343–348. doi: 10.1007/BF00405910. [DOI] [Google Scholar]
- Ouhib-Jacobs O, Lindley ND, Schmitt P, Clavel T. Fructose and glucose mediates enterotoxin production and anaerobic metabolism of Bacillus cereus ATCC14579(T) J Appl Microbiol. 2009;107(3):821–829. doi: 10.1111/j.1365-2672.2009.04254.x. [DOI] [PubMed] [Google Scholar]
- Tang YJ, Sapra R, Joyner D, Hazen TC, Myers S, Reichmuth D, Blanch H, Keasling JD. Analysis of metabolic pathways and fluxes in a newly discovered thermophilic and ethanol-tolerant Geobacillus strain. Biotechnol Bioeng. 2009;102(5):1377–1386. doi: 10.1002/bit.22181. [DOI] [PubMed] [Google Scholar]
- Stevenson DM, Weimer PJ. Expression of 17 genes in Clostridium thermocellum ATCC 27405 during fermentation of cellulose or cellobiose in continuous culture. Appl Environ Microbiol. 2005;71(8):4672–4678. doi: 10.1128/AEM.71.8.4672-4678.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel HJ. Growth of the thermophilic bacterium Clostridium thermocellum in continuous culture. Curr Microbiol. 1995;31(4):210–214. doi: 10.1007/BF00298375. [DOI] [Google Scholar]
- Guedon E, Payot S, Desvaux M, Petitdemange H. Carbon and electron flow in Clostridium cellulolyticum grown in chemostat culture on synthetic medium. J Bacteriol. 1999;181(10):3262–3269. doi: 10.1128/jb.181.10.3262-3269.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özkan M, Ylmaz E, Lynd LR, Özcengiz G. Cloning and expression of the Clostridium thermocellum L-lactate dehydrogenase in Escherichia coli and enzyme characterization. Can J Microbiol. 2004;50:845–851. doi: 10.1139/w04-071. [DOI] [PubMed] [Google Scholar]
- Willquist K, Zeidan AA, van Niel EW. Physiological characteristics of the extreme thermophile Caldicellulosiruptor saccharolyticus: an efficient hydrogen cell factory. Microb Cell Fact. 2010;9:89. doi: 10.1186/1475-2859-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvaux M, Guedon E, Petitdemange H. Metabolic flux in cellulose batch and cellulose-fed continuous cultures of Clostridium cellulolyticum in response to acidic environment. Microbiology. 2001;147(Pt 6):1461–1471. doi: 10.1099/00221287-147-6-1461. [DOI] [PubMed] [Google Scholar]
- Desvaux M, Petitdemange H. Flux analysis of the metabolism of Clostridium cellulolyticum grown in cellulose-fed continuous culture on a chemically defined medium under ammonium-limited conditions. Appl Environ Microbiol. 2001;67(9):3846–3851. doi: 10.1128/AEM.67.9.3846-3851.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvaux M, Guedon E, Petitdemange H. Kinetics and metabolism of cellulose degradation at high substrate concentrations in steady-state continuous cultures of Clostridium cellulolyticum on a chemically defined medium. Appl Environ Microbiol. 2001;67(9):3837–3845. doi: 10.1128/AEM.67.9.3837-3845.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedon E, Payot S, Desvaux M, Petitdemange H. Relationships between cellobiose catabolism, enzyme levels, and metabolic intermediates in Clostridium cellulolyticum grown in a synthetic medium. Biotechnol Bioeng. 2000;67(3):327–335. doi: 10.1002/(SICI)1097-0290(20000205)67:3<327::AID-BIT9>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Ben-Bassat A, Lamed R, Zeikus JG. Ethanol production by thermophilic bacteria: metabolic control of end product formation in Thermoanaerobium brockii. J Bacteriol. 1981;146(1):192–199. doi: 10.1128/jb.146.1.192-199.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DB, Islam R, Cicek N, Sparling R. Hydrogen production by Clostridium thermocellum 27405 from cellulosic biomass substrates. Int J Hydrogen Energy. 2006;31(11):1496–1503. doi: 10.1016/j.ijhydene.2006.06.015. [DOI] [Google Scholar]
- Strobel HJ, Caldwell FC, Dawson KA. Carbohydrate transport by the anaerobic thermophile Clostridium thermocellum LQRI. Appl Environ Microbiol. 1995;61(11):4012–4015. doi: 10.1128/aem.61.11.4012-4015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YH, Lynd LR. Regulation of cellulase synthesis in batch and continuous cultures of Clostridium thermocellum. J Bacteriol. 2005;187(1):99–106. doi: 10.1128/JB.187.1.99-106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girbal L, Soucaille P. Regulation of Clostridium acetobutylicum metabolism as revealed by mixed-substrate steady-state continuous cultures: role of NADH/NAD ratio and ATP pool. J Bacteriol. 1994;176(21):6433–6438. doi: 10.1128/jb.176.21.6433-6438.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos I, Girbal L, Soucaille P. Regulation of carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH on mixtures of glucose and glycerol. J Bacteriol. 1994;176(5):1443–1450. doi: 10.1128/jb.176.5.1443-1450.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ml D, Guedon E, Petitdemange H. Metabolic flux in cellulose batch and cellulose-fed continuous cultures of Clostridium cellulolyticum in response to acidic environment. Microbiology. 2001;147(6):1461–1471. doi: 10.1099/00221287-147-6-1461. [DOI] [PubMed] [Google Scholar]
- Lamed RJ, Lobos JH, Su TM. Effects of stirring and hydrogen on fermentation products of Clostridium thermocellum. Appl Environ Microbiol. 1988;54(5):1216–1221. doi: 10.1128/aem.54.5.1216-1221.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothun GD, Knutson BL, Berberich JA, Strobel HJ, Nokes SE. Metabolic selectivity and growth of Clostridium thermocellum in continuous culture under elevated hydrostatic pressure. Appl Microbiol Biotechnol. 2004;65(2):149–157. doi: 10.1007/s00253-004-1554-1. [DOI] [PubMed] [Google Scholar]
- Lamed R, Zeikus JG. Ethanol production by thermophilic bacteria: relationship between fermentation product yields of and catabolic enzyme activities in Clostridium thermocellum and Thermoanaerobium brockii. J Bacteriol. 1980;144(2):569–578. doi: 10.1128/jb.144.2.569-578.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydzak T, Levin DB, Cicek N, Sparling R. End-product induced metabolic shifts in Clostridium thermocellum ATCC 27405. Appl Microbiol Biotechnol. 2011;92(1):199–209. doi: 10.1007/s00253-011-3511-0. [DOI] [PubMed] [Google Scholar]
- Sauer U, Eikmanns BJ. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol Rev. 2005;29(4):765–794. doi: 10.1016/j.femsre.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Bapteste E, Moreira D, Philippe H. Rampant horizontal gene transfer and phospho-donor change in the evolution of the phosphofructokinase. Gene. 2003;318:185–191. doi: 10.1016/s0378-1119(03)00797-2. [DOI] [PubMed] [Google Scholar]
- Bielen AAM, Willquist K, Engman J, Van Der Oost J, Van Niel EWJ, Kengen SWM. Pyrophosphate as a central energy carrier in the hydrogen-producing extremely thermophilic Caldicellulosiruptor saccharolyticus. FEMS Microbiol Lett. 2010;307(1):48–54. doi: 10.1111/j.1574-6968.2010.01957.x. [DOI] [PubMed] [Google Scholar]
- Mukund S, Adams MW. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase, a novel tungsten-containing enzyme with a potential glycolytic role in the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1995;270(15):8389–8392. doi: 10.1074/jbc.270.15.8389. [DOI] [PubMed] [Google Scholar]
- Gowen CM, Fong SS. Genome-scale metabolic model integrated with RNAseq data to identify metabolic states of Clostridium thermocellum. Biotechnol J. 2010;5(7):759–767. doi: 10.1002/biot.201000084. [DOI] [PubMed] [Google Scholar]
- Li Y, Tschaplinski TJ, Engle NL, Hamilton CY, Rodriguez M Jr, Liao JC, Schadt CW, Guss AM, Yang Y, Graham DE. Combined inactivation of the Clostridium cellulolyticum lactate and malate dehydrogenase genes substantially increases ethanol yield from cellulose and switchgrass fermentations. Biotechnol Biofuels. 2012;5(1):2. doi: 10.1186/1754-6834-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axley MJ, Grahame DA, Stadtman TC. Escherichia coli formate-hydrogen lyase. Purification and properties of the selenium-dependent formate dehydrogenase component. J Biol Chem. 1990;265(30):18213–18218. [PubMed] [Google Scholar]
- Garvie EI. Bacterial lactate dehydrogenases. Microbiol Rev. 1980;44(1):106–139. doi: 10.1128/mr.44.1.106-139.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Werken HJ, Verhaart MR, VanFossen AL, Willquist K, Lewis DL, Nichols JD, Goorissen HP, Mongodin EF, Nelson KE, van Niel EW. et al. Hydrogenomics of the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl Environ Microbiol. 2008;74(21):6720–6729. doi: 10.1128/AEM.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Membrillo-Hernandez J, Echave P, Cabiscol E, Tamarit J, Ros J, Lin EC. Evolution of the adhE gene product of Escherichia coli from a functional reductase to a dehydrogenase. Genetic and biochemical studies of the mutant proteins. J Biol Chem. 2000;275(43):33869–33875. doi: 10.1074/jbc.M005464200. [DOI] [PubMed] [Google Scholar]
- Zhu J, Shimizu K. Effect of a single-gene knockout on the metabolic regulation in Escherichia coli for D-lactate production under microaerobic condition. Metab Eng. 2005;7(2):104–115. doi: 10.1016/j.ymben.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Asanuma N, Hino T. Effects of pH and energy supply on activity and amount of pyruvate formate-lyase in Streptococcus bovis. Appl Environ Microbiol. 2000;66(9):3773–3777. doi: 10.1128/AEM.66.9.3773-3777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asanuma N, Yoshii T, Hino T. Molecular characteristics and transcription of the gene encoding a multifunctional alcohol dehydrogenase in relation to the deactivation of pyruvate formate-lyase in the ruminal bacterium Streptococcus bovis. Arch Microbiol. 2004;181(2):122–128. doi: 10.1007/s00203-003-0638-0. [DOI] [PubMed] [Google Scholar]
- Brown SD, Guss AM, Karpinets TV, Parks JM, Smolin N, Yang S, Land ML, Klingeman DM, Bhandiwad A, Rodriguez M Jr. et al. Mutant alcohol dehydrogenase leads to improved ethanol tolerance in Clostridium thermocellum. Proc Natl Acad Sci USA. 2011;108(33):13752–13757. doi: 10.1073/pnas.1102444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh CT, Li J, Blanch HW, Clark DS. Redesigning Escherichia coli metabolism for anaerobic production of isobutanol. Appl Environ Microbiol. 2011;77(14):4894–4904. doi: 10.1128/AEM.00382-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Dong Y, Zhang J, Zhang A, Wang L, Feng L. Two novel metal-independent long-chain alkyl alcohol dehydrogenases from Geobacillus thermodenitrificans NG80-2. Microbiology. 2009;155(Pt 6):2078–2085. doi: 10.1099/mic.0.027201-0. [DOI] [PubMed] [Google Scholar]
- Pei J, Zhou Q, Jiang Y, Le Y, Li H, Shao W, Wiegel J. Thermoanaerobacter spp. control ethanol pathway via transcriptional regulation and versatility of key enzymes. Metab Eng. 2010;12(5):420–428. doi: 10.1016/j.ymben.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Burdette D, Zeikus JG. Purification of acetaldehyde dehydrogenase and alcohol dehydrogenases from Thermoanaerobacter ethanolicus 39E and characterization of the secondary-alcohol dehydrogenase (2 degrees Adh) as a bifunctional alcohol dehydrogenase--acetyl-CoA reductive thioesteras. Biochem J. 1994;302(Pt 1):163–170. doi: 10.1042/bj3020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovitt RW, Shen GJ, Zeikus JG. Ethanol production by thermophilic bacteria: biochemical basis for ethanol and hydrogen tolerance in Clostridium thermohydrosulfuricum. J Bacteriol. 1988;170(6):2809–2815. doi: 10.1128/jb.170.6.2809-2815.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard N, Johnsen K, Holbrook JJ, Delcour J. D175 Discriminates between NADH and NADPH in the coenzyme binding site of Lactobacillus delbrueckii subsp. bulgaricus D-lactate dehydrogenase. Biochem Biophys Res Commun. 1995;208(3):895–900. doi: 10.1006/bbrc.1995.1419. [DOI] [PubMed] [Google Scholar]
- Nair RV, Bennett GN, Papoutsakis ET. Molecular characterization of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. J Bacteriol. 1994;176(3):871–885. doi: 10.1128/jb.176.3.871-885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton-Brehm SD, Mosher JJ, Vishnivetskaya T, Podar M, Carroll S, Allman S, Phelps TJ, Keller M, Elkins JG. Caldicellulosiruptor obsidiansis sp. nov., an anaerobic, extremely thermophilic, cellulolytic bacterium isolated from Obsidian Pool, Yellowstone National Park. Appl Environ Microbiol. 2009;76(4):1014–1020. doi: 10.1128/AEM.01903-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais PM, Billoud B, Meyer J. Classification and phylogeny of hydrogenases. FEMS Microbiol Rev. 2001;25:455–501. doi: 10.1111/j.1574-6976.2001.tb00587.x. [DOI] [PubMed] [Google Scholar]
- Vignais PM. Hydrogenases and H(+)-reduction in primary energy conservation. Results Probl Cell Differ. 2008;45:223–252. doi: 10.1007/400_2006_027. [DOI] [PubMed] [Google Scholar]
- Buhrke T, Lenz O, Porthun A, Friedrich B. The H2-sensing complex of Ralstonia eutropha: interaction between a regulatory [NiFe] hydrogenase and a histidine protein kinase. Mol Microbiol. 2004;51(6):1677–1689. doi: 10.1111/j.1365-2958.2003.03933.x. [DOI] [PubMed] [Google Scholar]
- Angenent LT, Karim K, Al-Dahhan MH, Wrenn BA, Domiguez-Espinosa R. Production of bioenergy and biochemicals from industrial and agricultural wastewater. Trends Biotechnol. 2004;22(9):477–485. doi: 10.1016/j.tibtech.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Schut GJ, Adams MW. The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J Bacteriol. 2009;191(13):4451–4457. doi: 10.1128/JB.01582-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydzak T, McQueen PD, Krokhin OV, Spicer V, Ezzati P, Dwivedi RC, Shamshurin D, Levin DB, Wilkins JA, Sparling R. Proteomic analysis of Clostridium thermocellum core metabolism: Relative protein expression profiles and growth phase-dependent changes in protein expression. BMC Microbiol. 2012;12(1):214. doi: 10.1186/1471-2180-12-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T, Yao S. Thermophilic, lignocellulolytic bacteria for ethanol production: current state and perspectives. Appl Microbiol Biotechnol. 2011;92(1):13–27. doi: 10.1007/s00253-011-3456-3. [DOI] [PubMed] [Google Scholar]
- Guedon E, Desvaux M, Petitdemange H. Improvement of cellulolytic properties of Clostridium cellulolyticum by metabolic engineering. Appl Environ Microbiol. 2002;68(1):53–58. doi: 10.1128/AEM.68.1.53-58.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi SA, Olson DG, Argyros DA, Miller BB, Barrett TF, Murphy DM, McCool JD, Warner AK, Rajgarhia VB, Lynd LR. et al. Development of pyrF-based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant. Appl Environ Microbiol. 2010;76(19):6591–6599. doi: 10.1128/AEM.01484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cofactor specificity (ATP or PPi) of phosphofructokinases based on sequence alignments. Alignments of key residues determining ATP or PPi specificity, as determined by Bapteste et al. [74] and Bielen et al. [75], were performed using BioEdit v.7.0.9.0. The P. furiosus and Th. kodakarensis genes are very distinct (different COG and different KO) and are annotated as Archaeal phosphofructokinases.
Phylogenetic clustering of [NiFe] hydrogenases large (catalytic) subunits. Catalytic (large) subunits of [NiFe] H2ases were identified based upon the modular signatures as described by Calusinska et al. [16], Species considered in this manuscript are highlighted and corresponding H2ase gene loci are provided.
Phylogenetic clustering of [FeFe] hydrogenases large (catalytic) subunits. Catalytic (large) subunits of [FeFe] H2ases were identified based upon the modular signatures as described by Calusinska et al. [16]. Species considered in this manuscript are highlighted and corresponding H2ase gene loci are provided.