Abstract
Moloney murine leukemia virus (MLV)-derived gamma-retroviral vectors integrate preferentially near transcriptional regulatory regions in the human genome, and are associated with a significant risk of insertional gene deregulation. Self-inactivating (SIN) vectors carry a deletion of the U3 enhancer and promoter in the long terminal repeat (LTR), and show reduced genotoxicity in pre-clinical assays. We report a high-definition analysis of the integration preferences of a SIN MLV vector compared to a wild-type-LTR MLV vector in the genome of CD34+ human hematopoietic stem/progenitor cells (HSPCs). We sequenced 13,011 unique SIN-MLV integration sites and compared them to 32,574 previously generated MLV sites in human HSPCs. The SIN-MLV vector recapitulates the integration pattern observed for MLV, with the characteristic clustering of integrations around enhancer and promoter regions associated to H3K4me3 and H3K4me1 histone modifications, specialized chromatin configurations (presence of the H2A.Z histone variant) and binding of RNA Pol II. SIN-MLV and MLV integration clusters and hot spots overlap in most cases and are generated at a comparable frequency, indicating that the reduced genotoxicity of SIN-MLV vectors in hematopoietic cells is not due to a modified integration profile.
Introduction
Retroviral integration is a non-random process, whereby the viral RNA genome, reverse transcribed into double-stranded DNA and assembled in pre-integration complexes (PICs), associates with the host cell chromatin and integrates through the activity of the viral integrase [1]. Large-scale surveys of retroviral integration in murine and human cells uncovered some genomic features systematically and specifically associated with retroviral insertions, and revealed that each retrovirus type has a unique, characteristic pattern of integration within mammalian genomes [2]. Target site selection depends on both viral and cellular determinants, poorly defined for most retroviruses. The Moloney murine leukemia virus (MLV) and its derived vectors integrate preferentially in transcriptionally active promoters and regulatory regions [2], [3], [4], while the simian (SIV) and human immunodeficiency virus (HIV) and their derived lentiviral vectors target gene-dense regions and the transcribed portion of expressed genes, away from regulatory elements [2], [4], [5].
Recent clinical studies have shown that transplantation of stem cells genetically modified by retroviral vectors may cure severe genetic diseases such as immunodeficiencies [6], [7], [8], skin adhesion defects [9] and lysosomal storage disorders [10]. Some of these studies showed also the genotoxic consequences of retroviral gene transfer technology. Insertional activation of proto-oncogenes by MLV-derived vectors caused T-cell lymphoproliferative disorders in patients undergoing gene therapy for X-linked severe combined immunodeficiency [11], [12] and Wiskott-Aldrich syndrome [13], and pre-malignant expansion of myeloid progenitors in patients treated for chronic granulomatous disease (CGD) [14], [15]. The strong transcriptional enhancers present in the MLV LTR played a major role in deregulating gene expression. Pre-clinical studies showed that enhancer-less, self-inactivating (SIN) MLV-derived vectors are less prone to insertional oncogenesis and cell immortalization than their full-LTR counterparts, with a genotoxic profile closer to that of SIN-HIV vectors [16], [17], [18], [19]. The MLV U3 enhancer contains repeated binding sites for cellular transcription factors (TF), which may play a role in tethering retroviral pre-integration complexes to transcriptionally active regulatory regions and contribute to the MLV genotoxic characteristics [20].
In this study, we have analyzed the integration profiles of a MLV and SIN MLV vectors in the genome of a clinically relevant target cell population, cord blood-derived CD34+ hematopoietic stem/progenitor cells (HSPCs), by ligation-mediated PCR (LM-PCR) and high-throughput sequencing. We show that SIN-MLV and MLV vectors have very similar integration preferences, with the typical clustering around enhancer and promoter regions associated to specific histone modifications, specialized chromatin configurations and binding of RNA Pol II. Strikingly, SIN-MLV and MLV integration clusters and hot spots overlap in most cases and are generated at a similar frequency, indicating that the U3 enhancer has no role in targeting MLV PICs to the genome, at least in hematopoietic cells.
Results
MLV and SIN-MLV vectors share the same integration profile in the genome of human HSPCs
To generate a high-definition integration profile of SIN MLV integrations in human HSPCs, we transduced umbilical cord blood-derived CD34+ cells with a previously described SIN-MLV vector carrying a GFP expression cassette under the control of the human elongation factor 1α (EFS) promoter [16], [21]. Cells were transduced at 40 to 60% efficiency, and were selected for GFP expression by cell sorting 10 days after infection, to dilute unintegrated vectors. Vector-genome junctions were amplified from genomic DNA by ligation-mediated (LM)-PCR and pyrosequenced as previously described [4]. Raw sequences (available at the NCBI Sequence Read Archive with the accession number SRA061405) were processed by a previously described bioinformatic pipeline [4] and mapped on the University of California at Santa Cruz (UCSC) hg19 release of the human genome (http://genome.ucsc.edu), to obtain 13,011 unique insertion sites. Two previously generated datasets of full-LTR MLV vector integrations (32,574) and random sites (40,000) normalized for a number of parameters [4] were re-annotated on the hg19 release of the human genome and used for comparison.
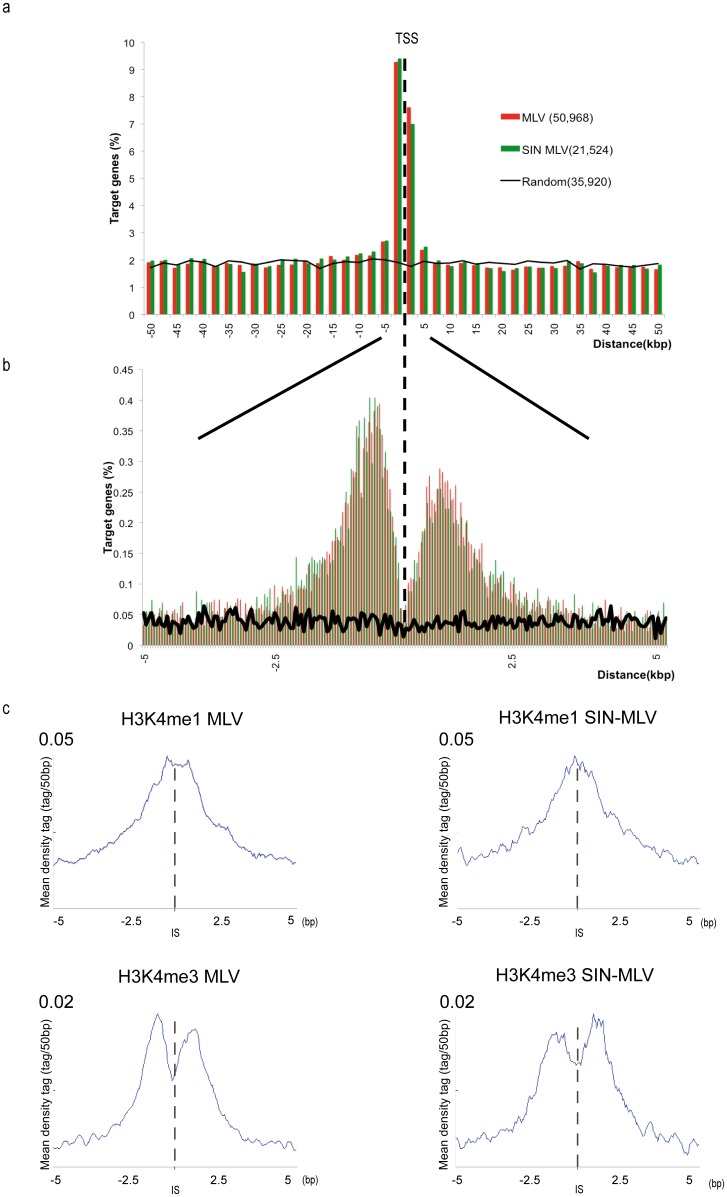
To identify differences in the integration preferences of MLV and SIN-MLV in HSPCs, we first analyzed the relationship between integration sites and Known Genes (UCSC definition) in the human genome: integration were annotated as TSS-proximal when occurring in an interval of ±2.5 kb from the TSS of any Known Gene, intragenic when occurring inside a Known Gene >2.5 kb from the TSS, and intergenic in all other cases. Intergenic and intragenic integrations were <40% for both MLV and SIN-MLV, while TSS-proximal integrations were 22.9% and 23.8% respectively (P>0.1 for all comparisons) ( Table 1 ). Plotting the relative distance of all integration sites in an interval of ±50 kb from any TSS showed two virtually overlapping distributions with the characteristic enrichment in the ±2.5 kb interval and, at 50-bp resolution, the characteristic drop in frequency in close proximity (±0.2 kb) of a TSS ( Figure 1 ). Similarly, integrations were enriched around annotated CpG islands (UCSC track) for both MLV and SIN-MLV vectors (12.8 and 12.4% respectively, P>0.1), and showed again overlapping, bimodal distributions in the ±2.5 kb interval around the island midpoint (Figure S1A). Finally, the moderate enrichment in the frequency of integration around mammalian, evolutionarily conserved non-coding sequences (CNC) [22] was the same for both vectors (8.0 and 7.7% respectively, P>0.1), and showed a virtually identical distribution at ±2.5 kb around the feature midpoint (Figure S1B). In all cases, there were significant differences between the distributions of the two MLV vectors and that of the random controls (P<10−15).
Table 1. Distribution of SIN-MLV and MLV integrations in the genome of human HSPCs.
| Intergenic (%) | TSS-proximal (%) | Intragenic (%) | CpG islands (%) | CNCs (%) | Total hits | |
| SIN-MLV | 38.0 | 23.8 | 38.1 | 12.4 | 7.7 | 13,011 |
| MLV | 38.2 | 22.9 | 38.8 | 12.8 | 8.0 | 32,574 |
| Random | 59.1 | 3.0 | 37.8 | 1.2 | 5.4 | 40,000 |
Percentage of intergenic, TSS-proximal or intragenic integration sites in the SIN-MLV, MLV and random control site datasets, together with the percentage of sites at a distance of ±1,000 bp from at least one CpG island or conserved non coding region (CNC).
Figure 1. Genomic distribution and association with histone modifications of MLV and SIN-MLV integrations in human HSPCs.
Distribution of the distance of SIN-MLV (green bars) and MLV (red bars) integration sites from the TSS of targeted genes at 2,500 bp (a) or 50 bp (b) resolution. The percentage of genes targeted at each position is plotted on the y axis. The number of plotted sites is higher than the actual number of mapped integrations sites, since each site may relate to more than one gene. The black line indicates the distribution of random control sites. (c) The distribution of H3K4me1 (top panels) and H3K4me3 (lower panels) epigenetic marks in a 10-kb window around vector integration sites (IS) is shown for MLV integrations (left panels) or SIN-MLV integrations (right panels). The mean density of tags (tag/50 bp) of the reference dataset (i.e. each chromatin feature) is plotted on the y axis. The scale of the graph is shown at the top left of each panel.
We previously reported that MLV integrations are strongly associated with histone modifications marking transcriptionally active promoters and enhancers, with the specialized H2A.Z histone variant, and with binding sites for RNA Pol II and transcription factors in both HSCs and T cells [4], [20], [23]. Taking advantage of published ChIP-Seq datasets on epigenetic features in the chromatin of human CD34+ HSPCs [24], we analyzed the association of MLV and SIN-MLV integrations with histone modifications marking active enhancers and promoters (H3K4me1, H3K4me3) and heterochromatin (H3K27me3), H2A.Z and binding of Pol II. In all cases, there was no obvious difference between the two vectors: we observed a strong association with H3K4me1, H3K4me3, Pol II and H2A.Z, and no correlation with H3K27me3 ( Figure 1C and Figure S1C).
All together, these data indicate that the absence of the U3 region of the LTR of the SIN-MLV vector causes no significant change in the integration preferences of MLV in the genome of human HSPCs.
MLV and SIN-MLV vectors integrate in the same hot spots in the HSPC genome
MLV and SIN-MLV integrations showed the same non-random, highly clustered distribution in the human genome, with integration hot and cold spots. Integration clusters were statistically defined as described [4], obtaining a numerosity-adjusted threshold for cluster definition of 3 integrations within 31,525 bp for SIN-MLV and 12,587 bp for MLV at a P-value<0.01. We identified 1,415 clusters containing 56.0% (7,318) of the total SIN-MLV integration sites, an overall clustering highly comparable (P>0.1) to that showed by the MLV vector (3,497 clusters containing 65.3% of the integrations). Most of the SIN-MLV clusters (75.9%) overlapped for at least 1 bp with MLV clusters, and up to 69% overlapped for at least 1,000 bp. The non-overlapping clusters contained only 3 integrations and mapped less than 200-bp apart in the MLV and SIN-MLV datasets.
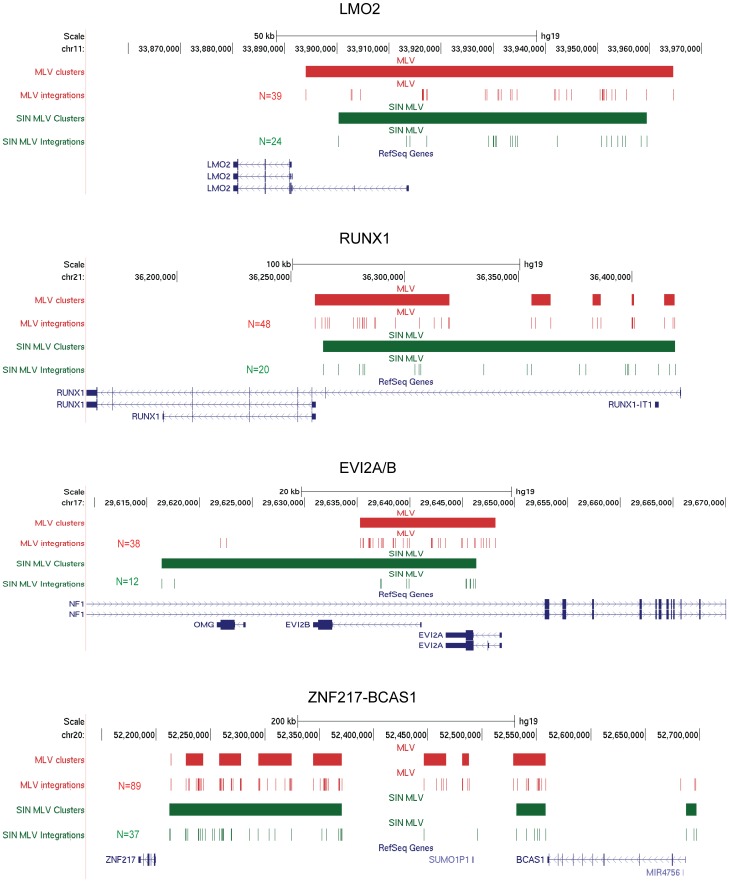
We then looked at the integration clusters of both vectors in a number of individual loci characteristically hit at high frequency by MLV integration. Most of the loci highly targeted by MLV were targeted also by SIN-MLV. Whenever the numerosity was sufficient, we observed a striking overlap between the integration hot spots of the two vectors within the same loci. As examples, SIN-MLV integrations faithfully reproduced the MLV integration patterns in the LMO2, EVI2A/B, RUNX1, RUNX2, ZNF217-BCAS1, CD34, ELF1 and NFE2 loci, which were targeted at the same overall frequency and at the same hot spots within each locus ( Figure 2 and Figure S2). We also found SIN-MLV integrations mapping very closely to the MLV integrations in two hot spots (MECOM and PRDM16) observed in the CGD and WAS clinical trials [13], [14], [15] (Figure S2).
Figure 2. MLV and SIN-MLV integration sites and clusters in CD34+ HSPC-specific loci.
Distribution of MLV (red) and SIN-MLV (green) integration clusters (horizontal solid bars) and integrations (vertical marks) in the LMO2, RUNX1, EVI2A/B, and ZNF217-BCAS1 loci as displayed by the UCSC Genome Browser. The base position feature at the top (scale bar and chromosome number) identifies the genomic coordinates of the displayed region.
Discussion
Retroviruses select their target integration sites by tethering their PICs to the host cell chromatin through protein-protein interactions that appear to be specific for each retrovirus type [2]. The chromatin component LEDGF/p75 has a major role in tethering HIV PICs to the body of active transcription units, associated with H4K12ac, H2BK5me1, H3K27me1, H3K36me3, and H4K20me1 histone modifications [5], [25]. LEDGF/p75 is directly bound by the HIV integrase [26], a major viral determinant of target site selection [27]. Much less is known for the MLV PICs: regions preferred by MLV are marked by acetylations of histones H2A, H2B, H3 and H4, methylations of H3 (H3K4me1, H3K4me2, H3K4me3), and binding of Pol II, CTCF, histone acetyl transferases (p300 and CBP) and H2A.Z [4], [23], a histone variant enriched at targets of the Polycomb complex and marking elements involved in the regulation of cell commitment and differentiation [28]. Most of these associations are statistically significant for all MLV integrations, independently from their location with respect to promoters and in all analyzed cell types, possibly reflecting the engagements of PICs by basal components of the enhancer-binding and/or RNA Pol II transcriptional machinery. A yeast two-hybrid analysis of proteins potentially interacting with the MLV integrase provided biochemical evidence in this direction [29].
The peculiar characteristics of MLV integration, coupled with the strong transcriptional enhancer activity of the LTR U3 region, explain the high risk of insertional gene activation and genotoxicity observed in pre-clinical [19], [30], [31], [32] as well as clinical [11], [12], [13], [14], [15] studies. SIN vectors have been developed to reduce the genotoxic potential of MLV vectors, and have indeed shown a reduced cell immortalization and tumorigenic activity by sensitive pre-clinical assays [18], [19], [33]. In this study, we compared the integration preferences of a SIN and a traditional MLV vector in human CD34+ HSPCs, the most used target cell in clinical applications of retroviral transgenesis. By an LM-PCR coupled to pyrosequencing approach we show that the lack of the U3 enhancer/promoter in the LTR has no impact on the integration pattern of MLV in human HSPCs. Indeed, the SIN-MLV integration map reproduced with remarkable precision that of the unmodified MLV, including the association with TSSs, CpG islands, CNCs and representative epigenetic marks of active and highly regulated enhancers and promoters. SIN-MLV and MLV integrations cluster into hot spots at approximately the same frequency, and generate almost overlapping integration maps at the level of highly targeted loci, including the gene involved in most of the severe adverse events observed in clinical trials, i.e., LMO2 [11], [12]. On the basis of the integration pattern, a SIN-MLV vector therefore maintains the same genotoxic potential of a traditional MLV vector. Insertion of either provirus has a high chance of altering gene regulation by disrupting the physical continuity of enhancers and promoters, and by altering the chromatin configuration induced by the binding of transcription factors and the basal transcriptional machinery (the enhanceosome). This type of effect is not expected to differ between SIN-MLV and MLV vectors. On the contrary, the lack of the two copies of the U3 enhancer may significantly reduce the dominant activity in overcoming gene regulation typical of oncogenic retroviruses in vivo [19], although studies based on in vitro immortalization of bone marrow-derived cells provided conflicting evidence on this point [18], [34]. The choice of a cellular, possibly restricted enhancer to drive the internal transgene cassette may therefore overcome the LTR-specific component of the MLV genotoxicity. Indeed, most of the gene deregulation and genotoxic activity of SIN vectors appears to be due to the characteristics of the enhancer driving transgene expression more than by the SIN design per se [16], [17], [18], [19].
Genotoxicity of retroviral vectors has many components, including the vector design, the nature of the target cell and the genetic background of the patient, all ultimately affecting the risk of a specific gene therapy approach [35]. Target site selection is just one of these components. Based on current knowledge, SIN lentiviral vectors appear to combine an integration profile that does not target regulatory elements with the lack of strong viral enhancers. SIN-MLV vectors share with SIN lentiviral vectors only the latter component. On the other hand, the lower propensity to integrate within transcribed regions may reduce the recently emerged post-transcriptional component of insertional gene deregulation [23], [36], [37], [38], [39], [40]. For SIN-MLV vectors, the designs of the transgene expression cassette, and particularly the choice of its transcriptional regulatory elements, appear to be the most relevant determinants of their biosafety characteristics.
Materials and Methods
Vectors and cells
Human CD34+ HSPCs were purified form umbilical cord blood, pre-stimulated for 48 hours in serum-free Iscove modified Dulbecco medium supplemented with 20% FCS, 20 ng/ml human thrombopoietin, 100 ng/ml Flt-3 ligand, 20 ng/ml interleukin-6, and 100 ng/ml stem cell factor, as previously described [3]. HSPCs were transduced with the SIN-MLV vector pSRS11.EFS.GFP.pre, expressing GFP under the control of the elongation factor 1α promoter, pseudotyped in an amphotropic envelope by three-plasmid transfection in 293 cells, as previously described [16], [21]. Cells were infected by 3 rounds of spinoculation (1,500 rpm for 45 min) in the presence of 4 µg/ml polybrene. Transduction efficiency was evaluated by cytofluorimetric analysis of GFP expression 48 hrs after infection. All human studies were approved by the San Raffaele Scientific Institute Ethical Committee. Written informed consent was received from participants prior to inclusion in the study.
Amplification, sequencing, and analysis of retroviral integration sites
Genomic DNA was extracted from a pool of 2×106 CD34+/GFP+ cells enriched by fluorescence-activated cell sorting, after a brief period in culture to dilute unintegrated vectors. 3′-LTR vector-genome junctions were amplified by LM-PCR adapted to the GS-FLX Genome Sequencer (Roche/454 Life Sciences) pyrosequencing platform, as previously described [3], [4]. Raw sequence reads were processed by an automated bioinformatic pipeline that eliminated small and redundant sequences [4] and mapped on the UCSC hg19 release of the human genome. All UCSC Known Genes having their TSS at ±50 kb from an integration site were annotated as targets. Genomic features were annotated when their genomic coordinates overlapped for ≥1 nucleotide with a ±50-kb interval around each integration site. We used UCSC tracks for both cytosine-phosphate-guanosine (CpG) islands and conserved TFBSs. The genomic coordinates of 82,335 mammalian conserved non-coding sequences (CNCs) were previously described [22]. For the association of the integrations with epigenetic marks, we re-annotated published ChIP-Seq data [24] in the UCSC hg19 release of the human genome, and analyzed the distribution of histone modifications (H3K4me1, H3K4me3, H3K27me3) H2A.Z and Pol II binding sites around the integrations, using the seqMINER platform [41]. Previously generated MLV integrations and random control sequences datasets [4] were also re-annotated on the UCSC hg19 genome. For all pairwise comparisons we applied a 2-sided Fisher's exact test. The threshold for statistical significance was set at a P value<0.05.
Supporting Information
Association between MLV and SIN-MLV integration sites and CpG islands, CNCs, PolII and histone modifications. Distribution of the distance of SIN-MLV (green bars) and MLV wt (red bars) integrations from the midpoint of CpG islands (A) or CNCs (B) in a 20 kb window. In the y axis is plotted the percentage of the total number of CpG islands or CNCs located at ±50 kb distance from the integrations. The black line indicates the distribution of random control sites. (C) The distribution of epigenetic marks in a 10 kb window around vector integration sites (IS) shown for H3K27me3 (top panels), H2A.Z (middle panels), PolII (lower panels) with respect to MLV integrations (left panels) or SIN-MLV integrations (right panels). See legend of Figure 1C for explanation of the graphs.
(PDF)
MLV and SIN-MLV integration sites and clusters in CD34+ HSPC-specific loci. Distribution of MLV (red) and SIN-MLV (green) integration clusters (horizontal solid bars) and integrations (vertical marks) in the CD34, ELF1, NFE2, and RUNX2, MECOM and PRDM16 loci as displayed by the UCSC Genome Browser. The base position feature at the top (scale bar and chromosome number) identifies the genomic coordinates of the displayed region.
(PDF)
Acknowledgments
A.M. is a student of the Ph.D. Program in Cellular and Molecular Biology of the Vita-Salute San Raffaele University, Milan, Italy.
Funding Statement
This work was supported by grants from the European Commission (PERSIST), the European Research Council (GT-SKIN), and the Italian National Research Council (CNR – EPIGEN). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Coffin JM, Huges SH, Varmus HE (1997) Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [PubMed]
- 2. Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, et al. (2005) Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol 3: 848–858. [DOI] [PubMed] [Google Scholar]
- 3. Cattoglio C, Facchini G, Sartori D, Antonelli A, Miccio A, et al. (2007) Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood 110: 1770–1778. [DOI] [PubMed] [Google Scholar]
- 4. Cattoglio C, Pellin D, Rizzi E, Maruggi G, Corti G, et al. (2010) High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood 116: 5507–5517. [DOI] [PubMed] [Google Scholar]
- 5. Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD (2007) HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res 17: 1186–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aiuti A, Cattaneo F, Galimberti S, Benninghoff U, Cassani B, et al. (2009) Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med 360: 447–458. [DOI] [PubMed] [Google Scholar]
- 7. Hacein-Bey-Abina S, Le Deist F, Carlier F, Bouneaud C, Hue C, et al. (2002) Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med 346: 1185–1193. [DOI] [PubMed] [Google Scholar]
- 8. Boztug K, Schmidt M, Schwarzer A, Banerjee PP, Diez IA, et al. (2010) Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N Engl J Med 363: 1918–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mavilio F, Pellegrini G, Ferrari S, Di Nunzio F, Di Iorio E, et al. (2006) Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat Med 12: 1397–1402. [DOI] [PubMed] [Google Scholar]
- 10. Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, et al. (2009) Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 326: 818–823. [DOI] [PubMed] [Google Scholar]
- 11. Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, et al. (2008) Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest 118: 3132–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, et al. (2008) Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest 118: 3143–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krause D (2011) Gene Therapy for Wiskott-Aldrich Syndrome: Benefits and Risks. The Hematologist 8: 10. [Google Scholar]
- 14. Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, et al. (2006) Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med 12: 401–409. [DOI] [PubMed] [Google Scholar]
- 15. Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, et al. (2010) Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med 16: 198–204. [DOI] [PubMed] [Google Scholar]
- 16. Zychlinski D, Schambach A, Modlich U, Maetzig T, Meyer J, et al. (2008) Physiological Promoters Reduce the Genotoxic Risk of Integrating Gene Vectors. Mol Ther [DOI] [PubMed] [Google Scholar]
- 17. Maruggi G, Porcellini S, Facchini G, Perna SK, Cattoglio C, et al. (2009) Transcriptional Enhancers Induce Insertional Gene Deregulation Independently From the Vector Type and Design. Mol Ther 17: 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Modlich U, Navarro S, Zychlinski D, Maetzig T, Knoess S, et al. (2009) Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol Ther 17: 1919–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, et al. (2009) The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest 119: 964–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Felice B, Cattoglio C, Cittaro D, Testa A, Miccio A, et al. (2009) Transcription factor binding sites are genetic determinants of retroviral integration in the human genome. PLoS ONE 4: e4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thornhill SI, Schambach A, Howe SJ, Ulaganathan M, Grassman E, et al. (2008) Self-inactivating gammaretroviral vectors for gene therapy of X-linked severe combined immunodeficiency. Mol Ther 16: 590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim SY, Pritchard JK (2007) Adaptive evolution of conserved noncoding elements in mammals. PLoS Genet 3: 1572–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cattoglio C, Maruggi G, Bartholomae C, Malani N, Pellin D, et al. (2010) High-definition mapping of retroviral integration sites defines the fate of allogeneic T cells after donor lymphocyte infusion. PLoS One 5: e15688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cui K, Zang C, Roh TY, Schones DE, Childs RW, et al. (2009) Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4: 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang GP, Levine BL, Binder GK, Berry CC, Malani N, et al. (2009) Analysis of lentiviral vector integration in HIV+ study subjects receiving autologous infusions of gene modified CD4+ T cells. Mol Ther 17: 844–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Engelman A, Cherepanov P (2008) The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog 4: e1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, et al. (2006) Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog 2: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, et al. (2008) H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135: 649–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Studamire B, Goff SP (2008) Host proteins interacting with the Moloney murine leukemia virus integrase: multiple transcriptional regulators and chromatin binding factors. Retrovirology 5: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kustikova O, Fehse B, Modlich U, Yang M, Dullmann J, et al. (2005) Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science 308: 1171–1174. [DOI] [PubMed] [Google Scholar]
- 31. Modlich U, Kustikova OS, Schmidt M, Rudolph C, Meyer J, et al. (2005) Leukemias following retroviral transfer of multidrug resistance 1 (MDR1) are driven by combinatorial insertional mutagenesis. Blood 105: 4235–4246. [DOI] [PubMed] [Google Scholar]
- 32. Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, et al. (2006) Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol 24: 687–696. [DOI] [PubMed] [Google Scholar]
- 33. Modlich U, Bohne J, Schmidt M, von Kalle C, Knoss S, et al. (2006) Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood 108: 2545–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bosticardo M, Ghosh A, Du Y, Jenkins NA, Copeland NG, et al. (2009) Self-inactivating retroviral vector-mediated gene transfer induces oncogene activation and immortalization of primary murine bone marrow cells. Mol Ther 17: 1910–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kustikova O, Brugman M, Baum C (2010) The genomic risk of somatic gene therapy. Semin Cancer Biol 20: 269–278. [DOI] [PubMed] [Google Scholar]
- 36. Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, et al. (2010) Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature 467: 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Almarza D, Bussadori G, Navarro M, Mavilio F, Larcher F, et al. (2011) Risk assessment in skin gene therapy: viral-cellular fusion transcripts generated by proviral transcriptional read-through in keratinocytes transduced with self-inactivating lentiviral vectors. Gene Ther 18: 674–681. [DOI] [PubMed] [Google Scholar]
- 38. Cesana D, Sgualdino J, Rudilosso L, Merella S, Naldini L, et al. (2012) Whole transcriptome characterization of aberrant splicing events induced by lentiviral vector integrations. J Clin Invest 122: 1667–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heckl D, Schwarzer A, Haemmerle R, Steinemann D, Rudolph C, et al. (2012) Lentiviral vector induced insertional haploinsufficiency of Ebf1 causes murine leukemia. Mol Ther 20: 1187–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moiani A, Paleari Y, Sartori D, Mezzadra R, Miccio A, et al. (2012) Lentiviral vector integration in the human genome induces alternative splicing and generates aberrant transcripts. J Clin Invest 122: 1653–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ye T, Krebs AR, Choukrallah MA, Keime C, Plewniak F, et al. (2010) seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res 39: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Association between MLV and SIN-MLV integration sites and CpG islands, CNCs, PolII and histone modifications. Distribution of the distance of SIN-MLV (green bars) and MLV wt (red bars) integrations from the midpoint of CpG islands (A) or CNCs (B) in a 20 kb window. In the y axis is plotted the percentage of the total number of CpG islands or CNCs located at ±50 kb distance from the integrations. The black line indicates the distribution of random control sites. (C) The distribution of epigenetic marks in a 10 kb window around vector integration sites (IS) shown for H3K27me3 (top panels), H2A.Z (middle panels), PolII (lower panels) with respect to MLV integrations (left panels) or SIN-MLV integrations (right panels). See legend of Figure 1C for explanation of the graphs.
(PDF)
MLV and SIN-MLV integration sites and clusters in CD34+ HSPC-specific loci. Distribution of MLV (red) and SIN-MLV (green) integration clusters (horizontal solid bars) and integrations (vertical marks) in the CD34, ELF1, NFE2, and RUNX2, MECOM and PRDM16 loci as displayed by the UCSC Genome Browser. The base position feature at the top (scale bar and chromosome number) identifies the genomic coordinates of the displayed region.
(PDF)