Abstract
Understanding more about the host's immune response to different Cryptococcus spp. will provide additional insight into the pathogenesis of cryptocococcis. We hypothesized that the ability of C. gattii to cause disease in immunocompetent humans depends on a distinct innate cytokine response of the host to this emerging pathogen. In the current study we assessed the cytokine profile of human peripheral blood mononuclear cells (PBMCs) of healthy individuals, after in vitro stimulation with 40 different well-defined heat-killed isolates of C. gattii, C. neoformans and several hybrid strains. In addition, we investigated the involvement of TLR2, TLR4 and TLR9 in the pro-inflammatory cytokine response to C. gattii. Isolates of C. gattii induced higher concentrations of the pro-inflammatory cytokines IL-1β, TNF-α and IL-6 and the Th17/22 cytokine IL-17 and IL-22 compared to C. neoformans var neoformans and C. neoformans var grubii. In addition, clinical C. gattii isolates induced higher amounts of cytokines than environmental isolates. This difference was not observed in C. neoformans var. grubii isolates. Furthermore, we demonstrated a likely contribution of TLR4 and TLR9, but no role for TLR2, in the host's cytokine response to C. gattii. In conclusion, clinical heat-killed C. gattii isolates induced a more pronounced inflammatory response compared to other Cryptococcus species and non-clinical C. gattii. This is dependent on TLR4 and TLR9 as cellular receptors.
Introduction
The incidence of cryptococcosis has increased dramatically over the past decades, due in a large part to the global HIV pandemic. More than 600,000 deaths are estimated to occur each year as a result of cryptococcal meningoencephalitis [1]. The species C. neoformans is an opportunistic pathogen mainly affecting immunocompromised hosts. In contrast, C. gattii mainly causes disease in apparently immunocompetent hosts at lower incidence [2], [3]. C. gattii is emerging over the past decade as a pathogen in the Pacific North-West of North America and has caused a large outbreak on Vancouver Island [4], [5]. This outbreak was mainly caused by a single, hypervirulent genotype of C. gattii, namely AFLP6A/VGIIa [6].
Cells of the innate immune system are important for initial defense against pathogens. Upon contact with pathogens, they produce pro-inflammatory cytokines such as tumor necrosis factor (TNF)-α, Interleukin (IL)-1β and IL-6, thereby initiating a specific adaptive cellular immune response. Anti-inflammatory cytokines such as IL-1RA are also produced and act as downregulators of this immune response. Of particular interest for fungal infections, the cytokines IL-1β and IL-6 in the presence of IL-23 induce the development of T-helper (Th)17 cells. IL-17 and IL-22, the major cytokines excreted by Th17 cells, have several pro-inflammatory functions, one of which is eliciting defensin production by epithelial cells [7]. Previous studies have shown a crucial role of Th17 cells in human antifungal defense against mucosal Candida albicans infections [8]–[10]; but the role of this particular Th-lymphocyte subset in anti-cryptococcal defense is not clear.
Which cytokines are released depends on recognition of microbial components by pattern recognition receptors (PRRs) on the cells of the innate immune system. Toll-like receptors (TLRs), a well-defined set of PRRs, are expressed on a variety of cells and are important mediators of pro-inflammatory cytokine release. However, their role in mediating cytokine response to Cryptococcus spp. is being debated [11]–[15].
Understanding more about the host's immune response to different Cryptococcus spp, will provide additional insight into the pathogenesis of cryptocococcis. We hypothesized that the ability of C. gattii to cause disease in immunocompetent humans depends on a distinct innate cytokine response of the host to this emerging pathogen. Therefore, in the current study we assessed the cytokine profile of human peripheral blood mononuclear cells (PBMCs) of healthy individuals, after in vitro stimulation with well-defined heat-killed isolates of C. gattii, C. neoformans and several hybrids. In addition, we investigated the involvement of TLR2, TLR4 and TLR9 in the pro-inflammatory cytokine response to C. gattii.
Results
Quantitative comparison of cytokine induction between different Cryptococcus spp
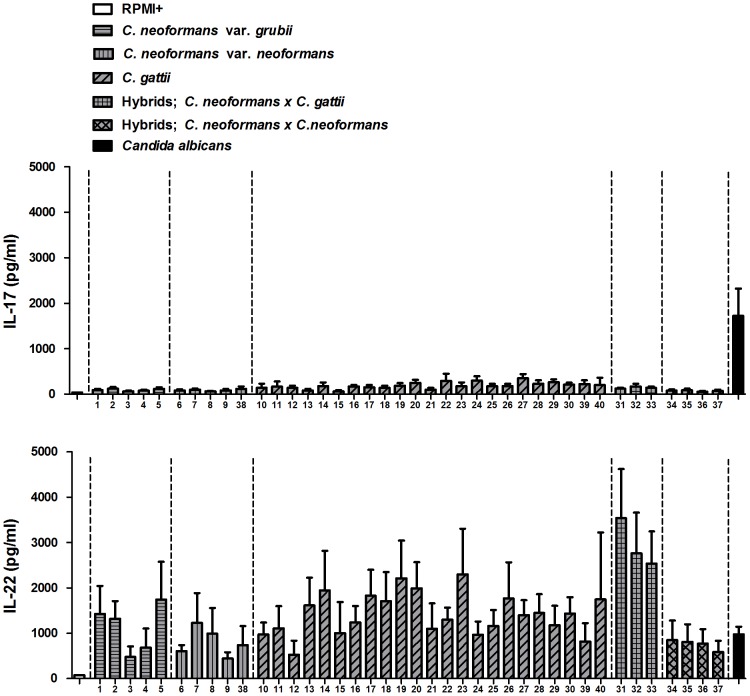
We determined the concentration of several cytokines produced by PBMCs upon stimulation with 40 different heat-killed Cryptococcus species complex isolates in order to elucidate the cytokine milieu in cryptococcal infection and to explore differences between the species. In preliminary experiments, we determined that the minimal concentration of yeasts necessary to induce cytokine production is 107 microorganisms/mL (data not shown). There was substantial inter-strain variation in the production of the pro-inflammatory cytokines IL-1β, TNF-α, IL-6 and the anti-inflammatory cytokine IL-1Ra. TNF-α and IL-1β were induced in low amounts (up to 300 pg/mL). Interestingly, production of these cytokines using a 100-fold lower concentration of Candida albicans was much higher (data not shown). Results for the induction of T-cell derived cytokines IL-17 and IL-22 after 7 days of incubation are shown in Figure 1. It appeared that the studied Cryptococcus strains induce low amounts of IL-17 but substantial quantities of IL-22, again with significant inter-strain variation in the production of these cytokines.
Figure 1. All forty Cryptococcus strains induce low amounts of IL-17, but high amounts of IL-22.
IL-17 and IL-22 production after 7 d by PBMCs stimulated with RPMI+, either one of 40 different heat-killed Cryptococcus strains [107 microorganisms/mL] or heat-killed Candida albicans [105 microorganisms/mL] is shown respectively. Mean values ± SE (n = 5) of three independent experiments are presented.
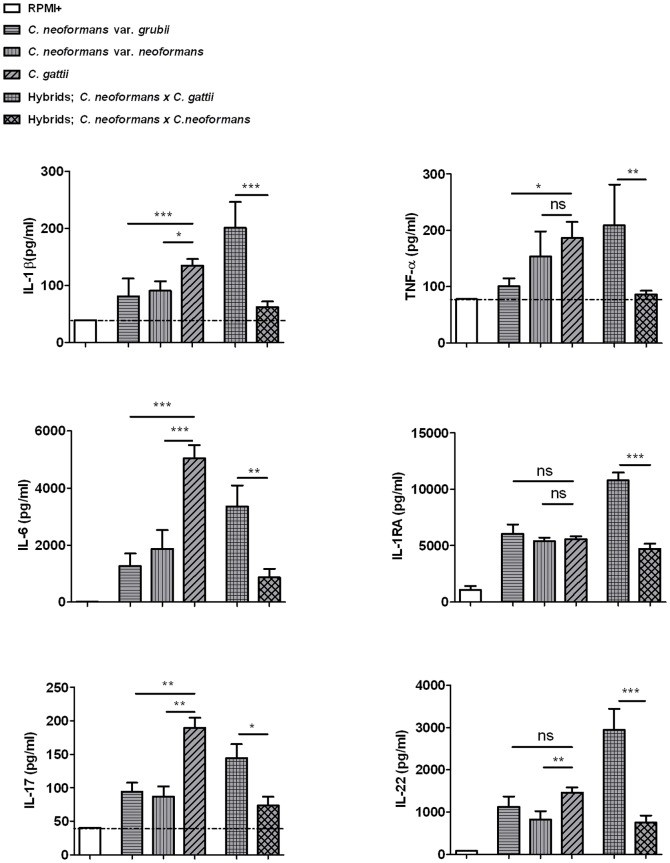
Figure 2 shows a quantitative comparison of cytokine induction between two varieties of C. neoformans, C. gattii and various hybrid isolates. C. gattii was a more potent inducer of the pro-inflammatory cytokines TNF-α, IL-1β, IL-6 and the T-cell cytokines IL-17 and IL-22, compared to both C. neoformans varieties. The different species did not differ with regard to IL-1Ra induction. Interestingly, the interspecies hybrids containing C. gattii as a partner of the mating pair induced significantly higher cytokine production than hybrids which were the result of mating between the two varieties of C. neoformans. This suggests that an inheritable factor is responsible for the difference in cytokine production.
Figure 2. Comparison of C. gattii isolates and interspecies hybrids with C. neoformans isolates and hybrids between both C. neoformans varieties.
The forty heat-killed Cryptococcus isolates are grouped according to (sub)species. Cytokine production by human PBMCs after 24 h (IL-1β, TNF-α, IL-6 and IL-1Ra) and 7 d (IL-17 and IL-22) incubation is shown. Mean values (n = 5 to 7) ± SE of three independent experiments are presented. *, p 0.01 to 0.05; **, p 0.001 to 0.01; ***, p<0.001. The horizontal line represents the lower detection limit.
Quantitative comparison of cytokine induction between environmental and clinical strains within the Cryptococcus species complex
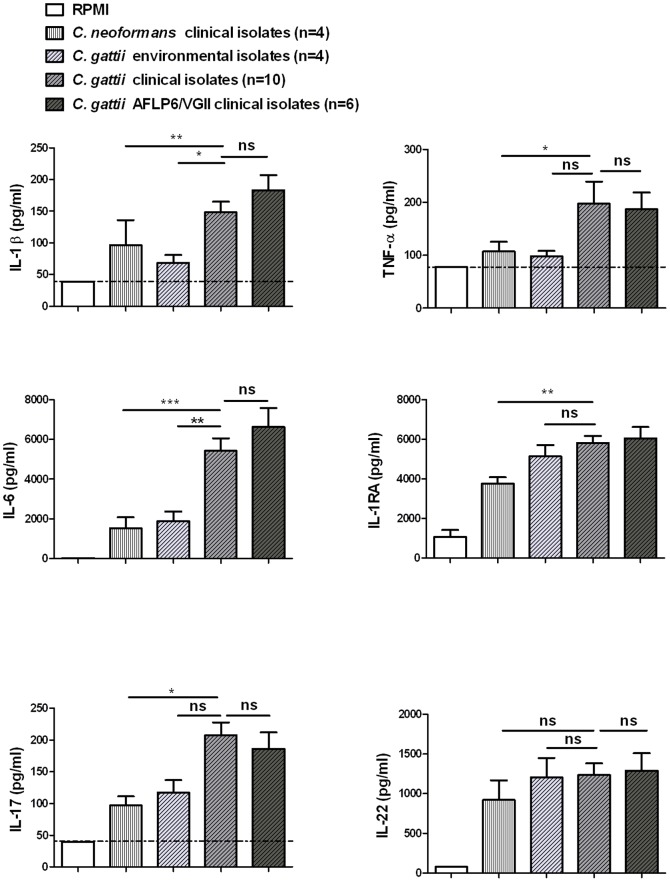
Sixteen clinical C. gattii isolates (isolates 10,12,14,18,19–21,23–29,39,40), of which six isolates belonging to the genotype AFLP6/VGII which was involved in the Vancouver Island outbreak, were compared to four environmental C. gattii isolates (isolates 13,15,16,17), as well as to four clinical C. neoformans isolates (isolates 1,4,5,9), with regard to the cytokine induction (Figure 3). Clinical C. gattii isolates induced significantly higher IL-1β and IL-6 amounts compared to environmental isolates. Moreover, clinical C. gattii isolates also induced higher IL-1β, IL-6, TNF-α, IL-1Ra and IL-17 than clinical C. neoformans isolates. The C. gattii genotype AFLP6/VGII, however, induced no higher amounts of other cytokines compared to the other clinical C. gattii isolates.
Figure 3. Comparison of cytokine production by PBMCs induced by clinical or environmental cryptococcal isolates.
Heat killed clinical isolates of C. gattii are compared to environmental C. gattii isolates and to clinical isolates of C. neoformans. The clinical isolates of C. gattii genotype AFLP6/VGII are depicted separately. Mean values (n = 5 to 7) ± SE values of three independent experiments are presented. *, p 0.01 to 0.05; **, p 0.001 to 0.01; ***, p<0.001. The horizontal line represents the lower detection limit.
In a different panel of Cuban C. neoformans var grubii isolates, comparison of clinical with environmental isolates showed no significant difference (P value for IL-6 and IL-22: 0.19 and 0.07 respectively) in cytokine production (Figure 4). The induction of low levels of cytokines by C. neoformans var grubii isolates, as seen in the panel of 40 isolates, was confirmed.
Figure 4. Comparison of cytokine production by PBMCs induced by clinical or environmental C. neoformans var grubii isolates.
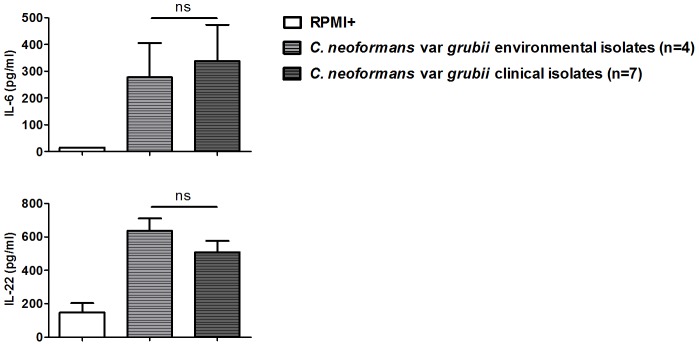
Cytokine production by human PBMCs after 24 h (IL-6) and 7 d (IL-22) incubation with heat-killed isolates is shown. Mean values (n = 7) ± SE of three independent experiments are presented. ns, not significant.
Involvement of different Pattern Recognition Receptors (PRRs) in cytokine production induced by C. gattii
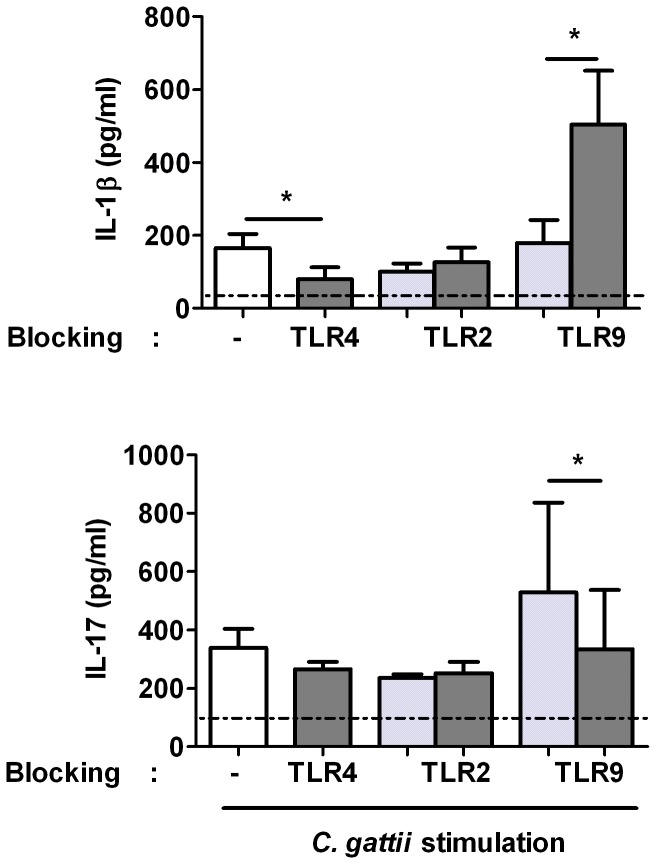
To assess which PRRs are involved in recognizing C. gattii, we performed experiments in which PBMCs were preincubated for one hour with specific PRR blocking reagents prior to stimulation with heat-killed C. gattii or, as a control, culture medium. Stimulation with culture medium showed undetectable levels for all cytokines (not shown). Blocking TLR2 had no effect on cytokine production by C. gattii, whereas this antibody significantly inhibited IL-1beta production after stimulation with Pam3cys (a known TLR-2 ligand) (Figure S1). Blocking TLR4 significantly diminished IL-1β induction by C. gattii, with a trend towards significance for TNF-α (P = 0.06). Interestingly, blocking TLR9 led to significantly higher concentrations of IL-1β induced by C. gattii compared to its control, and a trend towards significance (P = 0.06) was found for TNF-α. Blocking TLR9 had a negative effect (P = 0.03) on IL-17 production induced by C. gattii (Figure 5 for the effect on IL-1β and IL-17).
Figure 5. The role of TLR2, TLR4 and TLR9 in IL-1β and IL-17 induction by C. gattii.
Cytokine production by human PBMCs preincubated for one hour with culture medium (white bar) or PRR blocking reagents (dark gray bars) or their control (light gray bar) prior to stimulation with heat-killed C. gattii (strain B5742) [107/ml]. IL-1β is determined after 24 h incubation, IL-17 is determined after 7 d incubation. Mean values ± SE of eight individuals in 4 independent experiments (IL-17) or six individuals in 5 independent experiments (IL-1β) (with exclusion of additional four individuals with undetectable cytokine induction by C. gattii) are presented. *, p 0.01 to 0.05. The horizontal line represents the lower detection limit.
We performed these experiments also with C. neoformans var grubii (H99). The latter isolate did not elicit a substantial pro-inflammatory cytokine response in PBMCs, as shown in previous experiments with other strains. Moreover, we did not observe an increase in IL-1β and TNF-α production induced by C. neoformans var grubii when blocking TLR9 (results not shown).
Discussion
In the present study we investigated the in-vitro cytokine production of human PBMCs incubated with 40 different heat-killed isolates of the Cryptococcus neoformans species complex. We demonstrate that C. gattii isolates induces higher concentrations of pro-inflammatory and Th17/22 cytokines compared to C. neoformans var. neoformans and C. neoformans var. grubii. In addition, we found that clinical C. gattii isolates were able to induce higher amounts of cytokines than environmental isolates or clinical C. neoformans isolates. Furthermore, we demonstrated a contribution of TLR4 and TLR9, but no role for TLR2, in the host's cytokine response to C. gattii.
Our results indicate that Cryptococcus neoformans species complex seems to induce mainly a IL-22 response, with surprisingly low IL-17 production. This argues against a Th17 response to cryptococcal infection as we hypothesized, but rather to an exclusively IL-22 producing subset of Th-cells. A candidate for this areTh22 cells. These cells have not been found in mice so far; therefore only studies with human cells can be used to determine the role of this Th subset in host defense against cryptococcal infections. The aryl hydrocarbon receptor is identified to mediate IL-22 production without mediating IL-17 and seems to be critical in differentiation of naïve T cells to Th22 cells [16]. IL-22 is a unique cytokine in that it acts only on non-immune cells including keratinocytes, myofibroblasts and epithelial cells in tissues of the respiratory system, skin and digestive tract, which express receptors for this cytokine [17]. IL-22 promotes the production of antimicrobial agents called β-defensins by epithelial cells and serves in mucosal defenses against pathogens. It is tempting to speculate that IL-22 is important for the initial anti-cryptococcal defense because it has a function at the place of entrance of this yeast, namely the epithelial surface of the respiratory system. However, to confirm these speculations, further research should be attempted to identify the role of IL-22 and Th22 cells in clinical patients with cryptococcosis.
Higher amounts of the pro-inflammatory cytokines IL-1β, TNFα, IL-6, IL-17 and IL-22 by human PBMCs were induced by C. gattii compared to both varieties of C. neoformans, indicating that certain (virulence) factors of C. gattii are responsible for a more pronounced inflammatory reaction. This finding is strengthened as the same trend was seen in the hybrids containing C. gattii as a partner of the mating pair. Therefore we suggest that an inheritable factor is responsible for the difference in cytokine production.
Our finding that C. gattii induces a more powerful pro-inflammatory response aimed at more efficient defense against the pathogen is supported by the work of Ngamskulrungroj et al [18]. The authors compared the pathogenesis of the two Cryptococcus species in mice using an inhalation model and they found that in naive mice, C. gattii grew significantly slower in blood than C. neoformans. Infection with C. gattii was restricted to the lungs, while C. neoformans dissimenated to the brain causing meningoencephalitis. When mice were infected intravenously with low inoculums of yeast, C. neoformans was more virulent than C. gattii. Apparently, in this murine model the host's peripheral immune cells are able to clear C. gattii infection more efficiently, probably by a more adequate cytokine response. However, in humans, C. gattii species seem to be more virulent, as they are able to cause disease in apparently immunocompetent hosts. Large-scale environmental colonization for C. gattii was found during the Vancouver Island outbreak, whereas only relatively few people developed overt disease [6]. It can be hypothesized that a specific defect in the innate immune system of affected hosts predisposes them to infection with C. gattii. Furthermore, other factors such as intracellular survival, outgrowth or dissemination may also be important for virulence of C. gattii, independent of the initial pro-inflammatory cytokine response [19]. In our experiments we used PBMCs of healthy individuals who are expected to have an adequate immune response to C. gattii. These cells reflect the second line of defense when the yeast enters the host after inhalation. Our results showed a less optimal recognition and initial cytokine induction of C. neoformans var. grubii and var. neoformans, which suggests that in a host with inadequate cellular immunity this less optimal innate cytokine response leads more easily to infection with C. neoformans var. grubii and var. neoformans compared to C. gattii. Clinical data support this, since infections of immunocompromised hosts with C. neoformans var grubii is far more prevalent than infection with C. gattii [20].
A potential limitation of our study is that heat-killed instead of live cryptococci were used. However, at the temperatures used for heat-killing, most virulence factors (capsular polysaccharide, lipoproteins) are retained. Moreover, in number of previous studies, heat-killed cryptococci were used and significant inflammatory responses specific for capsulated and unencapsulated cryptococci were found [21], [22]. One study investigated lymphocyte proliferation after stimulation with live and heat-killed cryptococci and found no difference [23]. Thus, we feel that in this study, the use of heat-killed crytococci is justified.
Our experiments using a virulent C. gattii strain in stimulating PBMCs that were pre-incubated with specific PRR-blocking reagents indicate a role for TLR4 and TLR9 in recognizing Cryptococcus and subsequently modulation of the pro-inflammatory cytokine response. TLR4 seemed to be involved in mounting a pro-inflammatory cytokine response. Previous studies suggest that glucuronoxylomannan, the major capsular component [15] or other cryptococcal cell wall elements [24] are involved in binding to TLR4. In this study we did not design experiments in order to identify which cell wall components are involved in the initial cytokine response. Cytokine responses appeared to be independent of TLR2 recognition, since blocking of this receptor had no effect on cytokine concentrations. This contrasted with what is found in mice by Biondo et al. who demonstrated a key role of TLR2, but not of TLR4 [12]. Other studies, however, found no major role for TLR2 in survival of cryptococcal infections in a murine model [11], [13].
Based on our results, a special role in Cryptococcus recognition can be ascribed to TLR9. Unmethylated CpG-rich DNA is the best-known ligand for this receptor. Nakamura et al. have shown that TLR9 recognizes cryptococcal DNA [14]. We found that this receptor mediates IL-17 production, without any effect on IL-22. Conversely, blockade of TLR9 resulted in increased IL-1β production in response to C. gattii. The latter effect opposes the possible effect of TLR4. However, a specific combination of PRRs that bind available fungal PAMPs lead to pathways that interact with each other because of a limited set of shared adaptor molecules and transcription factors, and converge to a tailored response [25]. Likely, TLR9 and TLR4 work together in recognizing Cryptococcus and their signaling pathways interact downstream. Interestingly, we did not see TLR9 dependent negative modulation of C. neoformans var. grubii, indicating that the TLR9 dependent recognition of Cryptococcus is species-dependent. Negative modulation of immune responses to fungal pathogens mediated by TLR9 have been observed in other studies [26]. As the host's response to C. gattii relies on an initial pro-inflammatory cytokine response more than in C. neoformans infections, it can be speculated that susceptibility to C. gattii is influenced by subtle TLR polymorphisms and not necessarily by a defective adaptive immune response.
In the present study we investigated the in-vitro cytokine production of human PBMCs incubated with 40 different heat-killed isolates of Cryptococcus neoformans species complex. We demonstrated that isolates of C. gattii induce higher concentrations of the pro-inflammatory cytokines IL-1β, TNF-α and IL-6 and the Th17/22 cytokines IL-17 and IL-22 compared to C. neoformans var neoformans and C. neoformans var grubii. In addition, we found that clinical C. gattii isolates induced higher amounts of IL-1beta and IL-6 than environmental isolates. Furthermore, we demonstrated a likely contribution of TLR4 and TLR9, but no role for TLR2, in the host's cytokine response to C. gattii. In conclusion, clinical C. gattii isolates induced a more pronounced inflammatory cytokine response compared to other Cryptococcus species and non-clinical C. gattii that is dependent on TLR4 and TLR9 as cellular receptors.
Materials and Methods
Cryptococcal strains
Forty cryptococcal isolates from the CBS Fungal Biodiversity Centre (Utrecht, the Netherlands) were used in this study. These isolates were obtained from laboratory, clinical, environmental and veterinary sources. A detailed overview of the origin, sero- and AFLP genotype of these isolates is provided in Table 1. Twenty-three isolates were identified as C. gattii, 5 C. neoformans var. neoformans, 5 C. neoformans var. grubii and 7 hybrids, 3 of which were interspecies hybrids between C. gattii and C. neoformans var. neoformans and 4 hybrids between both C. neoformans varieties. In addition, 11 Cuban isolates were used in separate experiments, all identified as C. neoformans var grubii (Table 2).
Table 1. Details of the 40 cryptococcal isolates.
| No. in experiment | Isolate | Other specification | Species and varieties | Sero-type | AFLP-genotype | Origin | Reference/Source |
| 1 | 125.91 | CBS10512 | C. neoformans var. grubii | A | 1 | Cryptococcal meningitis patient, Tanzania | Lengeler et al., 2002 |
| 2 | CBS8336 | C. neoformans var. grubii | A | 1 | Decaying wood of Cassia tree, Brazil | Boekhout et al., 2001 | |
| 3 | CBS8710 | CBS10515, H99 | C. neoformans var. grubii | A | 1 | Subculture of type strain of Cryptococcus neoformans var. grubii (H99) | Boekhout et al., 2001 |
| 4 | CBS996(T) | C. neoformans var. grubii | A | 1 | Clinical isolate, Argentina | Boekhout et al., 1997 | |
| 5 | P152 | C. neoformans var. grubii | A | 1 | AIDS patient, Zimbabwe | Boekhout et al., 2001 | |
| 6 | B-3501 | CBS6900 | C. neoformans var. neoformans | D | 2 | Genetic offspring of CBS6885×CBS7000 ( = NIH12×NIH433) | Boekhout et al., 2001 |
| 7 | JEC20 | CBS10511, NIH-B4476 | C. neoformans var. neoformans | D | 2 | Congenic pair with JEC21 that differs only in mating type | Kwon-Chung et al., 1992a |
| 8 | JEC21 | CBS10513, NIH-B4500 | C. neoformans var. neoformans | D | 2 | Congenic pair with JEC20 that differs only in mating type | Kwon-Chung et al., 1992a |
| 9 | WM629(R) | CBS10079 | C. neoformans var. neoformans | D | 2 | HIV positive human, reference strain of molecular type VNIV, Melbourne, Australia. | Meyer et al., 1999 |
| 10 | CBS6998 | NIH365 | C. gattii VGI | B | 4 | Human, Thailand | Boekhout et al., 1997 |
| 11 | CBS8273 | CBS6289, RV20186, NIH-B-3939 | C. gattii VGI | B | 4 | Subculture of type strain of Cryptococcus gattii (RV 20186) | Boekhout et al., 1997 |
| 12 | WM179(R) | CBS10078 | C. gattii VGI | B | 4 | Immunocompetent human, reference strain of molecular type VGI, Sydney, Australia | Meyer et al., 2003 |
| 13 | WM276 | CBS10510 | C. gattii VGI | B | 4 | Eucalyptus tereticornis, Mt. Annan, New South Wales, Australia | Kidd et al., 2005 |
| 14 | CN043 | C. gattii VGIII | B | 5 | Human, Auckland, New Zealand | Katsu et al., 2004 | |
| 15 | CBS8755 | HOO58-I-682 | C. gattii VGIII | C | 5A | Detritus of almond tree, Colombia | Boekhout et al., 2001 |
| 16 | WM161(R) | CBS10081 | C. gattii VGIII | B | 5B | Eucalyptus camaldulensis wood from hollow, reference strain of molecular type VGIII, San Diego, USA | Meyer et al., 2003 |
| 17 | WM728 | C. gattii VGIII | B | 5B | Eucalyptus sp. debris from car park of zoo, San Diego, USA | Boekhout et al., 2001 | |
| 18 | CBS6955(T) | NIH191, ATCC32608 | C. gattii VGIII | C | 5C | Human, type strain of Cryptococcus bacillisporus, California, USA | Boekhout et al., 1997 |
| 19 | CBS6993 | NIH18 | C. gattii AFLP5 = VGIII | C | 5C | Human, California, USA | Boekhout et al., 1997 |
| 20 | A1M R265 | CBS10514 | C. gattii VGII | B | 6 | Immunocompetent male, Duncan, Vancouver Island, Canada | Kidd et al., 2004 |
| 21 | A1M R368 | A1M-R376 | C. gattii VGII | B | 6 | Immunocompetent male, Victoria, Canada | Kidd et al., 2004 |
| 22 | CBS1930 | C. gattii VGII | B | 6 | Sick goat, Aruba | Boekhout et al., 1997 | |
| 23 | CBS6956 | NIH444, ATCC32609 | C. gattii VGII | B | 6 | Immunocompetent human, Seattle, USA, | Boekhout et al., 1997 |
| 24 | WM178(R) | IFM50894, CBS10082 | C. gattii VGII | B | 6 | Immunocompetent human, lung, reference strain of molecular type VGII, Sydney, Australia | Meyer et al., 2003 |
| 25 | AV55 | CBS10090 | C. gattii VGII | B | 6A | HIV-negative human, Greece | Hagen et al. 2012 |
| 26 | AV54 | CBS10089 | C. gattii VGII | B | 6B | HIV-negative human, Greece | Hagen et al. 2012 |
| 27 | B5742 | C. gattii VGIV | C | 7 | Human, Punjab, India | Katsu et al., 2004 | |
| 28 | B5748 | C. gattii VGIV | B | 7 | HIV positive patient, India | Diaz and Fell, 2005 | |
| 29 | M27055 | C. gattii VGIV | C | 7 | Clinical, Johannesburg, South Africa | Latouche et al., 2002 | |
| 30 | WM779(R) | IFM50896, CBS10101 | C. gattii VGIV | C | 7 | Cheetah, reference strain of molecular type VGIV, Johannesburg, South Africa | Meyer et al., 2003 |
| 31 | CBS10488 | AMC770616 | C. gattii AFLP4×C. neoformans AFLP2 | BD | 8 | HIV-negative human, The Netherlands | Bovers et al., 2006 |
| 32 | CBS10489 | AMC2010404 | C. gattii AFLP4×C. neoformans AFLP2 | BD | 8 | HIV-positive human, The Netherlands | Bovers et al., 2006 |
| 33 | CBS10496 | LSPQ#308 | C. gattii AFLP4×C. neoformans AFLP1 | BD | 9 | HIV-positive human, Canada, visited Mexico | Bovers et al., 2008 |
| 34 | CBS132 | - | AD | 3 | Type strain C. neoformans, peach, Italy | Boekhout et al., 2001 | |
| 35 | NYJ40 | - | AD | 3 | - | Boekhout et al., 2001 | |
| 36 | RV52733 | - | AD | 3 | - | Boekhout et al., 2001 | |
| 37 | RV52755 | - | AD | 3 | - | Boekhout et al., 2001 | |
| 38 | CBS5467 | C. neoformans var. neoformans | D | 2 | Milk from mastitic cow, Switzerland | Boekhout et al., 1997 | |
| 39 | IHEM14941 Slimy | RV 63979, IHEM14941, CBS11687 | C. gattii | B | 10 | HIV- patient from Mexico, Spain | Hagen et al. 2012 |
| 40 | IHEM14941 White | RV 63979, IHEM14941 | C. gattii | B | 10 | HIV- patient from Mexico, Spain | Hagen et al. 2012 |
Table 2. Details of 11 additional C. neoformans var grubii isolates, arranged by Microsatellite Complex (MC) [29].
| Number in experiment | Isolate | Other specification | Species | Serotype | MC | Origin | Reference/Source |
| I | 37-07-17 | Cuba 617-05 | C. neoformans var grubii | A | MC1 | Clinical | Illnait Zaragozi et al., 2010 |
| II | 44-08-52 | Cuba CA 1-5 | C. neoformans var grubii | A | MC1 | Environmental | Illnait Zaragozi et al., 2010 |
| III | 37-07-03 | Cuba 24-2b | C. neoformans var grubii | A | MC1 | Environmental | Illnait Zaragozi et al., 2010 |
| IV | 36-10-01 | Cuba CH-2 | C. neoformans var grubii | A | MC2 | Environmental | Illnait Zaragozi et al., 2010 |
| V | 44-08-16 | Cuba 569-06 | C. neoformans var grubii | A | MC2 | Clinical | Illnait Zaragozi et al., 2010 |
| VI | 36-09-16 | Cuba 225-99 | C. neoformans var grubii | A | MC2 | Clinical | Illnait Zaragozi et al., 2010 |
| VII | 36-09-32 | Cuba 227-01 | C. neoformans var grubii | A | MC3 | Clinical | Illnait Zaragozi et al., 2010 |
| VIII | 36-09-57 | Cuba 0119 | C. neoformans var grubii | A | MC3 | Clinical | Illnait Zaragozi et al., 2010 |
| IX | 36-10-46 | Cuba 30-2D | C. neoformans var grubii | A | MC3 | Environmental | Illnait Zaragozi et al., 2010 |
| X | 36-10-56 | Cuba 315-01 | C. neoformans var grubii | A | MC4 | Clinical | Illnait Zaragozi et al., 2010 |
| XI | 36-09-53 | Cuba 098 | C. neoformans var grubii | A | MC6 | Clinical | Illnait Zaragozi et al., 2010 |
Prior to the experiments, the strains were freshly grown on Sabouraud dextrose agar plates. A suspension of each strain was prepared in sterile phosphate buffered saline (PBS), heat-killed overnight at 56°C and quantified by spectrophotometry at a wavelength of 530 nm. The suspensions were checked for fungal and bacterial growth on a Sabouraud dextrose agar plate and a blood agar plate respectively. No growth was observed after 5 days. All strains were stored at 4°C until used.
Candida strain
Heat-killed Candida albicans ATCC MYA-3573 (UC 820), a well described clinical isolate, suspended in sterile PBS, was used as a positive control.
Reagents and antibodies
Bartonella LPS, a penta-acylated LPS which is an antagonist of TLR4-dependent signaling, was obtained as previously described [27]. An anti-TLR2 monoclonal antibody from eBioscience (San Diego, CA, USA) was used, and an irrelevant isotype-matched murine IgG1 κ isotype (Biolegend, San Diego, CA, USA) as control. TLR9 inhibitory oligonucleotides ODN TTAGGG (anti TLR9) [28] and its negative control were obtained from InvivoGen (San Diego, CA, USA).
Isolation and stimulation of PBMCs
Human peripheral blood mononuclear cells (PBMCs) were collected from buffy coats of healthy donors after written informed consent had been obtained. PBMCs were isolated using density gradient centrifugation on Ficoll-Hypaque (GE Healthcare, Uppsala, Sweden). The cells from the interphase were aspirated and washed three times in sterile PBS and resuspended in culture medium RPMI 1640 Dutch modification (Sigma-Alderich, St Louis, MO, USA) supplemented with 1% L-glutamine, 1% pyruvate and 1% gentamicin. Cells were counted in a Coulter Counter Z® (Beckman Coulter, Fullerton, CA, USA), and adjusted to 5×106 cells/ml. Thereafter, they were incubated in a round-bottom 96-wells plate (volume 200 µl/well) at 37°C and 5% CO2 with either one of the heat-killed cryptococcal strains (final concentration of 107/ml), or heat-killed C. albicans (final concentration of 105/mL, which is known to induce substantial amounts of cytokines) or culture medium alone. After 24 hours or 7 days (in the presence of 10% human pool serum) supernatants were collected and stored at −20°C until being assayed.
In a subsequent experiment, PBMCs were preincubated for one hour with inhibitory ligand for TLR4 (Bartonella quintana LPS (200 ng/ml) or culture medium as control, anti-TLR2 or control antibody (10 µg/ml), TLR9 inhibitory oligonucleotides and its negative control (25 µg/ml). After preincubation, C. gattii B5742, isolate 27 in the previous experiment, or specific TLR ligands were added, such as Pam3cys or E.coli LPS (10 µg/ml and 10 ng/ml respectively] and PBMCs were incubated as described.
Cytokine assays
Tumor necrosis factor-α (TNF-α), Interleukin-1β (IL-1β), IL-6 and IL-1 receptor antagonist (IL-1Ra) concentrations were determined from the culture supernatant after 24 hours of incubation using commercially available ELISA kits (TNF-α, IL-1β and IL-1Ra: R&D systems, Minneapolis, MN, USA. IL-6: Sanquin,Amsterdam, the Netherlands) according to the manufacturer's instructions. T-cell derived cytokines IL-17 and IL-22 concentrations were determined in the supernatant after 7 days of incubation using ELISA kits (R&D systems). Lower detection limits were 78 pg/ml, 39 pg/ml, 15 pg/ml, 200 pg/ml, 40 pg/ml and 78 pg/ml for TNF-α, IL-1β, IL-6, IL-1Ra, IL-17 and IL-22 respectively.
Ethics statement
Written informed consent of healthy donors was provided. The study was approved by the Medical Ethical Committee Arnhem-Nijmegen in the Netherlands.
Statistical analysis
Results from at least three different experiments with a range of 5–7 donors were pooled and analyzed using GraphPad Prism 5 software (GraphPad, San Diego, CA). Data are given as mean ± SE. The Mann-Whitney U-test for unpaired, nonparametrical data was used to compare differences in cytokine production between two groups. The Kruskal-Wallis test with Dunn's multiple comparison test was used when more than two groups were compared. The Wilcoxon matched-pairs signed rank test was used to analyze differences in cytokine production between inhibitors and their controls in the inhibition experiments. The level of significance was set at p<0.05.
Supporting Information
IL-1β induction by Pam3cys and E. coli LPS after blocking of TLR2 and TLR4 respectively. IL-1β production by human PBMCs is shown (A) induced by pam3cys [10 µg/ml] after preincubated for one hour with anti-TLR2 or control antibody [10 µg/ml] and (B) by E. coli LPS [10 ng/ml] after preincubation for one hour with TLR4 antagonist Bartonella quintana LPS [200 ng/ml] or culture medium. Mean values (n = 10) ± SE of five independent experiments are presented.
(TIF)
Acknowledgments
The authors thank Ferry Hagen for providing cryptococcal strains.
Funding Statement
M.G.N. was supported by a Vici Grant of the Netherlands Organization for Scientific Research. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, et al. (2009) Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23: 525–530. [DOI] [PubMed] [Google Scholar]
- 2. Speed B, Dunt D (1995) Clinical and host differences between infections with the two varieties of Cryptococcus neoformans . Clin Infect Dis 21: 28–34 discussion 35-26. [DOI] [PubMed] [Google Scholar]
- 3. Chayakulkeeree M, Perfect JR (2006) Cryptococcosis. Infect Dis Clin North Am 20: 507–544, v–vi. [DOI] [PubMed] [Google Scholar]
- 4. Byrnes EJ, Li W, Lewit Y, Ma H, Voelz K, et al. (2010) Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog 6: e1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chaturvedi V, Chaturvedi S (2011) Cryptococcus gattii: a resurgent fungal pathogen. Trends Microbiol 19: 564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, et al. (2004) A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A 101: 17258–17263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eyerich S, Eyerich K, Cavani A, Schmidt-Weber C (2010) IL-17 and IL-22: siblings, not twins. Trends Immunol 31: 354–361. [DOI] [PubMed] [Google Scholar]
- 8. van de Veerdonk F, Gresnigt M, Kullberg B, van der Meer J, Joosten L, et al. (2009) Th17 responses and host defense against microorganisms: an overview. BMB Rep 42: 776–787. [DOI] [PubMed] [Google Scholar]
- 9. van de Veerdonk FL, Kullberg BJ, Netea MG (2010) Pathogenesis of invasive candidiasis. Curr Opin Crit Care 16: 453–459. [DOI] [PubMed] [Google Scholar]
- 10. Eyerich K, Foerster S, Rombold S, Seidl HP, Behrendt H, et al. (2008) Patients with chronic mucocutaneous candidiasis exhibit reduced production of Th17-associated cytokines IL-17 and IL-22. J Invest Dermatol 128: 2640–2645. [DOI] [PubMed] [Google Scholar]
- 11. Yauch LE, Mansour MK, Shoham S, Rottman JB, Levitz SM (2004) Involvement of CD14, toll-like receptors 2 and 4, and MyD88 in the host response to the fungal pathogen Cryptococcus neoformans in vivo. Infect Immun 72: 5373–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biondo C, Midiri A, Messina L, Tomasello F, Garufi G, et al. (2005) MyD88 and TLR2, but not TLR4, are required for host defense against Cryptococcus neoformans . Eur J Immunol 35: 870–878. [DOI] [PubMed] [Google Scholar]
- 13. Nakamura K, Miyagi K, Koguchi Y, Kinjo Y, Uezu K, et al. (2006) Limited contribution of Toll-like receptor 2 and 4 to the host response to a fungal infectious pathogen, Cryptococcus neoformans . FEMS Immunol Med Microbiol 47: 148–154. [DOI] [PubMed] [Google Scholar]
- 14. Nakamura K, Miyazato A, Xiao G, Hatta M, Inden K, et al. (2008) Deoxynucleic acids from Cryptococcus neoformans activate myeloid dendritic cells via a TLR9-dependent pathway. J Immunol 180: 4067–4074. [DOI] [PubMed] [Google Scholar]
- 15. Shoham S, Huang C, Chen JM, Golenbock DT, Levitz SM (2001) Toll-like receptor 4 mediates intracellular signaling without TNF-alpha release in response to Cryptococcus neoformans polysaccharide capsule. J Immunol 166: 4620–4626. [DOI] [PubMed] [Google Scholar]
- 16. Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H (2009) Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat Immunol 10: 864–871. [DOI] [PubMed] [Google Scholar]
- 17. Eyerich S, Eyerich K, Pennino D, Carbone T, Nasorri F, et al. (2009) Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J Clin Invest 119: 3573–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ngamskulrungroj P, Chang Y, Sionov E, Kwon-Chung KJ (2012) The primary target organ of Cryptococcus gattii is different from that of Cryptococcus neoformans in a murine model. MBio 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma H, Hagen F, Stekel DJ, Johnston SA, Sionov E, et al. (2009) The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc Natl Acad Sci U S A 106: 12980–12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown GD, Netea MG, SpringerLink (Online service) (2007) Immunology of Fungal Infections. Dordrecht: Springer.
- 21. Levitz SM, Farrell TP, Maziarz RT (1991) Killing of Cryptococcus neoformans by human peripheral blood mononuclear cells stimulated in culture. J Infect Dis 163: 1108–1113. [DOI] [PubMed] [Google Scholar]
- 22. Siddiqui AA, Shattock RJ, Harrison TS (2006) Role of capsule and interleukin-6 in long-term immune control of Cryptococcus neoformans infection by specifically activated human peripheral blood mononuclear cells. Infect Immun 74: 5302–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mody CH, Syme RM (1993) Effect of polysaccharide capsule and methods of preparation on human lymphocyte proliferation in response to Cryptococcus neoformans . Infect Immun 61: 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Doering TL (2009) How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans . Annu Rev Microbiol 63: 223–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van de Veerdonk F, Kullberg B, van der Meer J, Gow N, Netea M (2008) Host-microbe interactions: innate pattern recognition of fungal pathogens. Curr Opin Microbiol 11: 305–312. [DOI] [PubMed] [Google Scholar]
- 26. Kasperkovitz PV, Khan NS, Tam JM, Mansour MK, Davids PJ, et al. (2011) Toll-like receptor 9 modulates macrophage antifungal effector function during innate recognition of Candida albicans and Saccharomyces cerevisiae . Infect Immun 79: 4858–4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Popa C, Abdollahi-Roodsaz S, Joosten LA, Takahashi N, Sprong T, et al. (2007) Bartonella quintana lipopolysaccharide is a natural antagonist of Toll-like receptor 4. Infect Immun 75: 4831–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stunz LL, Lenert P, Peckham D, Yi AK, Haxhinasto S, et al. (2002) Inhibitory oligonucleotides specifically block effects of stimulatory CpG oligonucleotides in B cells. Eur J Immunol 32: 1212–1222. [DOI] [PubMed] [Google Scholar]
- 29. Illnait-Zaragozi MT, Martínez-Machín GF, Fernández-Andreu CM, Boekhout T, Meis JF, et al. (2010) Microsatellite typing of clinical and environmental Cryptococcus neoformans var. grubii isolates from Cuba shows multiple genetic lineages. PLoS One 5: e9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
IL-1β induction by Pam3cys and E. coli LPS after blocking of TLR2 and TLR4 respectively. IL-1β production by human PBMCs is shown (A) induced by pam3cys [10 µg/ml] after preincubated for one hour with anti-TLR2 or control antibody [10 µg/ml] and (B) by E. coli LPS [10 ng/ml] after preincubation for one hour with TLR4 antagonist Bartonella quintana LPS [200 ng/ml] or culture medium. Mean values (n = 10) ± SE of five independent experiments are presented.
(TIF)