Abstract
Background
The alkalistable and thermostable xylanases are in high demand for pulp bleaching in paper industry and generating xylooligosaccharides by hydrolyzing xylan component of agro-residues. The compost-soil samples, one of the hot environments, are expected to be a rich source of microbes with thermostable enzymes.
Methodology/Principal Findings
Metagenomic DNA from hot environmental samples could be a rich source of novel biocatalysts. While screening metagenomic library constructed from DNA extracted from the compost-soil in the p18GFP vector, a clone (TSDV-MX1) was detected that exhibited clear zone of xylan hydrolysis on RBB xylan plate. The sequencing of 6.321 kb DNA insert and its BLAST analysis detected the presence of xylanase gene that comprised 1077 bp. The deduced protein sequence (358 amino acids) displayed homology with glycosyl hydrolase (GH) family 11 xylanases. The gene was subcloned into pET28a vector and expressed in E. coli BL21 (DE3). The recombinant xylanase (rMxyl) exhibited activity over a broad range of pH and temperature with optima at pH 9.0 and 80°C. The recombinant xylanase is highly thermostable having T1/2 of 2 h at 80°C and 15 min at 90°C.
Conclusion/Significance
This is the first report on the retrieval of xylanase gene through metagenomic approach that encodes an enzyme with alkalistability and thermostability. The recombinant xylanase has a potential application in paper and pulp industry in pulp bleaching and generating xylooligosaccharides from the abundantly available agro-residues.
Introduction
Hemicellulose represents the second most abundant renewable polymer of plant cell walls after cellulose, and xylan is the main constituent in lignocellulosic agro-residues. The backbone of xylan is mainly composed of β-1,4-linked xylosyl residues along with various groups (arabinosyl, acetyl and glucuronosyl) in their side chains. These heterogeneous polysaccharides play a critical role in maintaining the cell wall integrity by making covalent and non-covalent bonds with cellulosic fibres and lignins [1], [2]. The heterogeneity in xylans necessitates multiplicity in the xylanolytic enzyme system that comprises endoxylanase (1,4-β-D-xylan xylanohydrolase; EC 3.2.1.8), β xylosidase (1,4 β-D-xylan xylohydrolase; EC 3.2.1.37) α-glucuronidase, α-L-arabinofuranosidase, and acetyl xylan esterase. The CAZY database (http://www.cazy.org/fam/acc_GH.html) categorized xylanases into six glycosyl hydrolase families GH5, GH8, GH10, GH11, GH30 and GH43 [3]. Among these, family 10 and 11 xylanases are widely distributed where GH10 xylanases share versatile substrate specificity with higher molecular weight, while GH11 xylanases are more stringent in their substrate specificity with low molecular weight and are considered as true xylanases [4], [5]. Hydrolysis of xylans by xylanases is of interest to many industries like ramie fibre degumming, food processing, textile, biofuel, feed and paper/pulp industries [6], [7], [8]. The xylanases that withstand extreme conditions prevailing in the industrial processes are in high demand in paper pulp processing and feed industries.
Although several xylanases have been reported from a number of microorganisms, most of them do not have adequate thermostability and alkalistability for their application in paper and pulp industries. Majority of xylanases have been obtained from the culturable 0.1 to 1% of the total microbial diversity existing in natural environments. The culture-independent metagenomic approaches permit retrieval of genes encoding useful enzymes from environmental samples without involving laborious and elaborate methods for the cultivation of microbes. The immense demand for alkalistable and thermostable xylanases encouraged us to adapt this innovative strategy for retrieving genes that encode thermo-alkali-stable xylanases. In this investigation, a metagenomic library was constructed and screened for clones with xylanase activity. Xylanase encoding gene (Mxyl) was subcloned and expressed, and the recombinant xylanase was purified and characterized. To the best of our knowledge, this is the first report on retrieving thermo-alkali-stable xylanase by a metagenomic approach.
Results
Construction of metagenomic library, DNA sequencing and bioinformatics analysis
When 5.0 µg of 20–30 kb of high molecular weight metagenomic DNA was digested with Sau3AI and the fragments were ligated into p18GFP vector with an efficiency of 3.6×104 clones per µg of DNA in constructing the library, the insert sizes were in the range of 3.0–8.0 kb with an average size of 5.5 kb. On screening, a clone having xylanase gene was spotted on RBB xylan containing LB-amp plate. The full sequence of the insert showed the size of 6.231 kbp that revealed its prokaryotic origin on blast analysis. The complete insert contained nine transcriptional units with a complete ORF of 1077 bp long xylanase gene. The sequence showed putative sequences of -35 (CACGCCA), -10 (TAAAAA) and ribosomal binding sites (AGGGG) at the upstream of xylanase gene followed by complete ORF having ATG and TAA as start and stop codons, respectively (Figure S1). The xylanase displayed five conserved regions (I-V) of GH11 xylanase having two catalytically important residues (Glu109 and Glu217) present in signature sequence II and V (Figure S2). Amino acid homology showed maximum identity (79%) with the xylanase gene of an uncultured bacterium and Actinoplanes sp. SE50/110 followed by a metagenomic GH 11 xylanase (71%). It shared 63–75% homology with xylanases produced by various species of Streptomyces. The xylanase retrieved in this investigation exhibits 75, 67 and 64% similarity with the endo-1,4 β-xylanases of Cellulomonas fimi, Micromonospora aurantiaca 27029 and Amycolatopsis mediterranei U32, respectively. It, however, has lower homology with the xylanases of Microbulbifer hydrolyticus (63%), Pseudomonas sp. ND137 (62%), uncultured Cellvibrio sp. (58%), Cellvibrio mixtus (57%) and Aspergillus fumigatus AF293 (52%) (Figure 1).
Figure 1. Phylogenetic tree of recombinant xylanase.
rMxyl showed highest homology with xylanase of Cellulomonas fimi ATCC 484 followed by uncultured microbial GH 11 xylanase. Neighbour Joining (NJ) Tree is constructed by using MEGA 4.0 software. Bootstrap values (n = 1000 replicates) are represented as percentage. The scale bar depicts the allowed changes per amino acid position.
Construction and expression of the xylanase gene in E. coli and localization of the encoding recombinant xylanase (rMxyl)
Xylanase gene was successfully cloned into pET28a(+) vector. It was confirmed by the release of insert from the vector on double digestion with NheI and XhoI. The sequencing results further confirmed the insert. The recombinant plasmid was expressed in E. coli BL21 (DE3) on induction with 1.0 mM IPTG at A600 of 0.6–0.7 and 30°C. At higher level of expression, it led to the formation of inclusion bodies, which could be solubilized using 6.0 M urea. The highest titre of the recombinant enzyme was achieved in 4–6 h. The construct (pET28a-Mxyl) expressed a high proportion of xylanase in cytoplasmic fraction (83%), followed by periplasmic (9%) and extracellular (8%) fractions after 4–5 h of induction. When xylanase gene was cloned and expressed in pET22b(+) vector, a high proportion of intracellular enzyme (>60%) was produced in the initial 3 h of induction, and thereafter, it declined. The periplamic xylanase was optimum at 12 h, while the extracellular fraction gradually increased and it reached a peak (29%) in 24 h.
Site directed mutagenesis
Muteins having Glu117Asp and Glu209Asp completely lost the activity. These two glutamates are highly conserved residues in the signature sequences LVEYYIVDN and MATEGY, and these are responsible for catalytic activity of GH 11 xylanase.
Purification, biochemical characterization and zymogram analysis of rMxyl
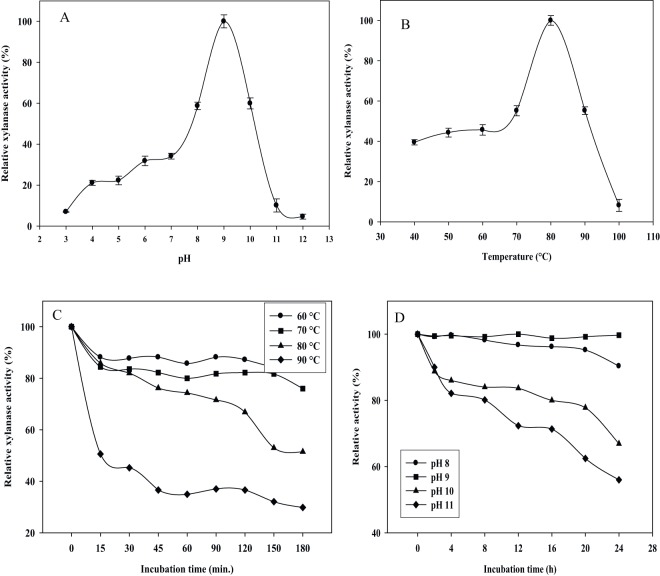
The recombinant xylanase was purified by Ni2+-NTA resin affinity chromatography and the purified recombinant protein could be eluted using imidazole (100–400 mM). The protein appeared as a single band of 40 kDa against the protein marker on 15% SDS-PAGE, and the recombinant xylanase revealed as a clear band of xylan hydrolysis by zymogram analysis (Figure 2). The effects of various physical and chemical parameters on the recombinant xylanase have been assessed. The xylanase exhibited broad range of pH (6.0–12.0) with optimum at 9.0, and it retained ∼55% residual activity at pH 10.0 (Figure 3A). The rMxyl is active in the temperature range between 40 and 100°C (Figure 3B) with optimum at 80°C, and retains more than 90–95% activity after exposure to 60 and 70°C for 3 h. The enzyme has a T1/2 of 2.0 h at 80°C and 15 min at 90°C (Figure 3C). The recombinant enzyme did not lose activity after 3 h exposure to pH 8.0 and 9.0, and thereafter, it declined (50% residual activity after 4 h). Approximately 20–45% loss in activity was recorded on either side of the pH optimum after 1 h incubation (Figure 3D). Mg2+, Sn2+ and Fe2+ stimulated rMxyl activity, while Hg2+ and Mn2+ significantly inhibited enzyme activity even at 1 mM. Other metal ions exerted inhibitory action on xylanase. More than 30% activity was lost in the presence of Mn2+ (Table 1). NBS and PMSF inhibited the activity to a significant extent even at 1 mM concentration. β-ME and DTT strongly inhibited enzyme activity. A stimulatory effect EDTA was recorded on xylanase activity.
Figure 2. Analysis of rMxyl using SDS–PAGE (15% polyacrylamide gel).
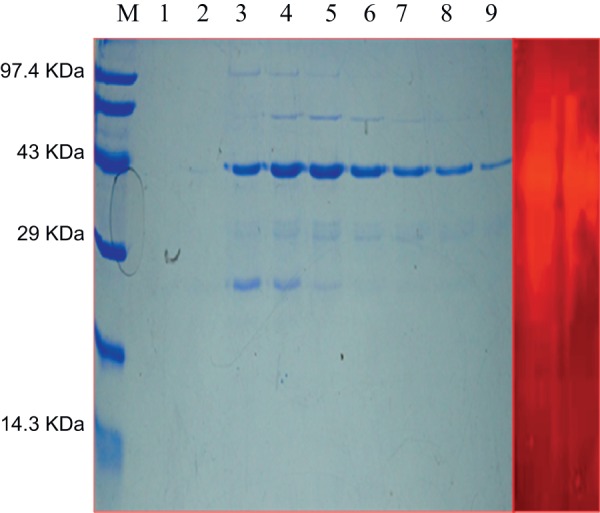
A. Lane 1 protein marker, Lane 2 and 3 are washes with 20 and 30 mM mM imidazole. Recombinanat xylanase was eluted using different concentrations of imidazole (100, 200, 250, 300, 400, 450, 500 mM). Purified xylanase showed molecular mass of ∼42 kDa on staining with Coomassie Brilliant Blue R-250. B. Zymogram analysis of purified xylanase using Congo red staining method.
Figure 3. Effect of pH and temperature on the activity and stability of rMxyl.
A and B: The recombinant xylanase incubated in various buffers (pH 3–12) and temperatures (40–100°C) and assayed for xylanase activity C: Recombinant xylanase was incubated in glycine-NaOH buffer without substrate and kept at various temperatures. Aliquots were collected at various time interval and store at 0°C for calculating residual activity. D: Similarly enzyme was incubated in various buffers (pH 8–11) and aliquots of different time intervals were used xylanase assays.
Table 1. Effect of modulators on rMxyl activity.
| Metal ions | 1 mM | 5 mM | 10 mM |
| Mg2+ | 106.45±1.05 | 99.65±0.98 | 87.38±0.45 |
| Fe2+ | 108.65±0.75 | 116.01±0.27 | 93.67±1.32 |
| Sn2+ | 110.43±0.67 | 76.12±0.44 | 45.17±0.63 |
| Ni2+ | 91.21±0.22 | 79.01±1.34 | 32.84±0.43 |
| Zn2+ | 91.67±0.32 | 76.64±0.78 | 32.89±0.89 |
| Pb2+ | 81.33±067 | 20.78±0.32 | 09.65±0.67 |
| K+ | 81.21±1.08 | 20.62±0.12 | 12.67 ±0.45 |
| Ag2+ | 73.48±0.53 | 54.55±0.69 | 27.83±0.98 |
| Ca2+ | 72.43±0.43 | 35.45±0.21 | 12.09±0.19 |
| Mn2+ | 71.76±0.63 | 27.34±1.32 | 09.67±0.27 |
| Ba2+ | 66.45±0.67 | 23.91±0.34 | 18.65±0.33 |
| Cd2+ | 54.67±0.43 | 29.33±0.49 | 12.87±0.65 |
| Co2+ | 59.15±1.23 | 29.63±0.65 | 12.54±1.12 |
| Na+ | 61.43±0.78 | 39.75±1.06 | 27.35±0.78 |
| Cu2+ | 29.12±0.18 | 15.76±0.76 | 10.09±0.87 |
| Hg2+ | 0 | 0 | 0 |
| Inhibitors | 1 mM | 5 mM | 10 mM |
| NBS | 46.66±0.12 | 35.67±0.09 | 20.12±0.11 |
| IAA | 103.45±0.54 | 89.75±0.32 | 69.85±1.56 |
| β-ME | 0 | 0 | 0 |
| DTT | 0 | 0 | 0 |
| EDTA | 105.65±1.23 | 107.19±1.01 | 89.98±0.56 |
| Detergents | 0.1% (v/v) | 0.5% (v/v) | |
| Tween 20 | 103.45±1.32 | 105.67±0.98 | |
| Triton X100 | 108.32±0.96 | 104.05±0.92 | |
| SDS | 97.34±1.32 | 65.89±0.19 | |
| Control | 100±0.12 | 100±0.23 | 100±0.67 |
Most of the metal ions were insignificant at 1 mM concentration; however xylanase activity was significantly inhibited at higher concentration by Pb2+, Ag2+, Ca2+, Mn2+, Ba2+, Cd2+ and Co2+. In the presence of Hg2+, enzyme lost completely its activity. Similarly, trace amounts of β-mercaptoethanol (β-ME) and dithiothreitol (DTT) completely inhibited the xylanase activity. Inhibition in the presence of N-bromosuccinimide (NBS) signifies the role of tryptophan in catalysis, while EDTA confirms it as a non-metallozyme.
Saccharification of agroresidues/hydrolysis of xylan
The rMxyl hydrolyzed xylan from various sources. The enzyme activity was very high in birchwood xylan (relative activity 100%) in comparison with that on xylan from beech wood (97%) and arabinoxylan (80%). There was no activity on carboxymethylcellulose (CMC) and other non-xylan polysaccharides (starch, pullulan and chitin). The Km and Vmax values of the enzyme on birchwood xylan are 8.0±1.21 mg/ml and 300±09.12 μmol/min/mg, respectively. The saccharification of wheat bran was high (15.2%) as compared to that of corncobs (9.89%) and sugarcane bagasse (4.71%). Various xylooligosaccharides were detected in the hydrolysates (Figure 4).
Figure 4. Profile of xylooligosaccharides liberated by the action of rMxyl.
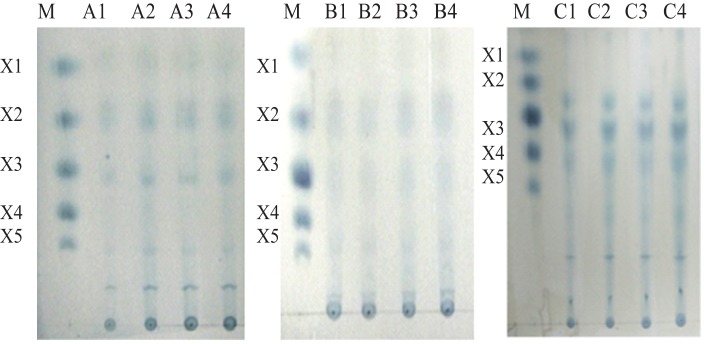
Lane (A1–A4)*: spots of X1, X2 and X3 were detected from wheat bran. Lane (B1–B4)*: hydrolysis from corncob that showed prominence of X2 and X3. While, X3, X4 and X5 were detected from hydrolysate of sugarcane bagasse (C1–C4)*. Lane M: Standards of various XOs (Sigma, USA). X1: xylose, X2: xylobiose; X3: xylotriose; X4: xyloptetraose; X5: xylopentaose *: 1/2/3/4 time intervals of 5, 15 and 30 min and 1 h, respectively.
Discussion
Although several xylanases have been reported from diverse microbiota using traditional culture dependent approaches [9], [10], [11], [12], majority of them do not endure the extreme temperature and alkaline conditions prevailing in industrial processes. An alternate strategy was, therefore, adapted to retrieve a thermo-alkali-stable xylanase gene (Mxyl) by culture-independent metagenomic approach. The metagenomic library constructed with DNA extracted from the compost-soil samples yielded a clone that produced xylanase. Although, the compost soils are in the acidic pH range, an alkalistable and thermostable endoglucanase had been reported from rice straw compost [13]. The culture independent approach has started yielding the useful biocatalysts from the hidden Pandora's Box [14], [15], [16]. A considerable success has also been achieved in obtaining xylanases with diverse attributes by using metagenomic approaches [17], [18], [19], [20], [21]. The protein encoded by xylanase gene comprises 358 amino acids, of which 16 are acidic and 21 basic. The predicted molecular weight, pI and instability index of recombinant xylanase are ∼40 kDa, 8.8 and 33.44 respectively. The xylanase contained a 43 amino acid long leader sequence at the N terminal region followed by a catalytic domain (44th–212th) of GH11 family interrupted by a short stretch of arginine and threonine rich non-catalytic region (WSVRQ2R2TG2TIT2). In addition, serine rich Q linker region (S2GS2DITVG2TS2G2TS2G2S3G2S10G4) has also been detected from amino acid 213 to 248 just after catalytic domain. Such repeated amino acids make linker regions that usually discriminate catalytic domain from carbohydrate binding domain [22], [23], [24]. Moreover, linkers have also been reported as integral parts of various xylanases that connect thermo-stabilizing domains, surface layer homology domains and dockerin domains which play a role in stabilizing the protein [22], . Amino acid homology and hydrophobic cluster analysis categorized this high molecular weight xylanase into GH 11 family. This is not unusual because xylanases of Clostridium stercorium also had been classified as a GH11 xylanase despite its high molecular mass (56 kDa) [29]. Metagenomic origin, distinct characteristics, lower homology and higher molecular weight (>30 kDa) make this a novel xylanase. The recombinant xylanases from Glacieola mesophila KMM241 and Geobacillus thermoleovorans displayed a similar pattern of recombinant protein localization [30], [31]. The profile of the distribution of the recombinant xylanase is similar to other xylanases cloned in pET28a(+) and pET22b(+) vectors as in G. thermoleovorans and B. halodurans [30], [32]. The integrated N- terminal pelb signal sequence in pET22b(+) directed the enzyme to periplasm that further led to secretion into the extracellular environment.
The site directed mutagenesis of two residues of glutamate to aspartate resulted in a complete loss of xylanase activity due to disruption in double displacement mechanism [33]. In order to take the advantage of thermostability of the recombinant xylanase, it was subjected to high temperature prior to purification by Ni2+-NTA agarose resins. This step reduced the extra load of non-His tagged, less thermostable and contaminant host proteins [30], [32].
The rMxyl exhibits optimum activity at higher temperature and pH which is similar to xylanases produced by Dictyoglomus thermolacticum, Thermotoga maritima, Bacillus acidicaldarius and Geobacillus thermoleovorans having optimal activity at or above 80°C [30], [34], [35], [36]. The activity and stability of rMxyl at higher pH are the crucial properties of xylanases for their applicability in paper processing industry. Xylanases of G. thermoleovorans, Bacillus firmus, B. stereothermophilus T-6 exhibited optimum activity at pH at 8.0 or above [30], [37], [39]. The shelf life of rMxyl is more than 3 months at 4°C, which retains greater than 90% activity. The recombinant xylanase is optimally active at 80°C and pH 9.0 that distinguishes it from already reported xylanases. The xylanase of Thermotoga maritima has Topt of 90°C, but it gets inactivated fast at pH 6.0 [35]. The highly thermostable xylanase with optimum temperature in the range between 80 and 105°C are available, but these xylanases exhibit their maxima either at acidic or neutral pH [35,35]. Similarly the alkalistability at higher pH is reported in many xylanases but are active at lower temperatures [37], [38]. The recombinant xylanase of GH10 family from Bacillus halodurans showed both properties together having optima at 75°C and pH 9.0, but it losses 50% activity at 65°C after 4 h and gets inactivated very fast at 80°C [32]. The metagenomic xylanase, on the other hand, has good thermostability at higher temperatures (60, 70 and 80°C) with only 20–30% loss after 3 h exposure. The most significant aspect of this investigation is obtaining a highly alkalistable (pHopt. 9.0) and thermostable (Topt. 80°C) xylanase from environmental samples by a metagenomic approach.
Cations (Mg2+, Sn2+ and Fe2+) stimulated the rMxyl activity like those of B. subtilis AMX-4 and an uncultured microbe [39], [40]. Even at 1 mM, Hg2+ and Mn2+ significantly inhibited the activity. The inhibition of xylanase by Hg2+ suggests the presence of tryptophan residues that oxidize indole ring, thereby inhibiting the xylanase activity [39], [41], [42], [43]. Xylanases from Streptomyces olivaceoviridis A1, Streptomyces sp. S27 and Bacillus subtilis strain R5 had been reported to be stimulated by Fe2+ and Mg2+ [19], [44], [45], [46], [47], and total loss of the enzyme in presence of Hg2+ and Mn2+ was reported in the xylanases from an uncultured microbe [19], [46], Penicillium sp. [48] and Plectosphaerella cucumerina [41]. The inhibition of xylanase activity by Cu2+ is similar to the majority of the xylanases [13], [39], [45], [49]. The stimulatory effect of EDTA could be due to a different catalytic mechanism as reported in xylanases of Aspergillus niger and Fusarium proliferatum [42], [49]. In Glaciecola mesophila KMM 241, EDTA caused ∼25% enhancement in activity [31]. N-BS inhibition suggests the involvement of tryptophan in xylanase activity [50], [51], [52]. Total loss of xylanase activity by β-ME and DTT suggests the distortion of disulfide linkages present between cystein residues [41], [49]. Detergents exerted a slight stimulatory effect on the recombinant xylanase which is a common feature of the other xylanases. However, rMxyl was inhibited by SDS as in other xylanases [39], [46], [49].
The rMxyl hydrolyzed birchwood and beechwood xylan efficiently. The structural similarity of beechwood and birchwood xylans may be the reason for the high activity [31]. The enzyme exhibited almost similar activities on oat spelt and arabinoxylan. Oat spelt xylan is a type of arabinoxylan very rich in arabinose (xylose/arabinose = 66∶34) [53], [54]. Interestingly the rMxyl liberated xylooligosaccaharides from xylan in just 5 min and it was sustainable on prolonged incubation. Several xylanases have been reported from various microorganisms that liberate xylooligosaccharides following xylan hydrolysis. Alkaline xylanases show better action on agro-residues by lowering the steric hindrance caused by cellulose and enhancing the solubility of hemicellulosic materials [54]. Xylanases from G. thermoleovorans [30], Bacillus halodurans [55] Thermomonospora fusca [56] have been reported to generate a similar profile of XOs. The metagenomic xylanase finds application in food industry for the production of xylooligosaccharides as prebiotics [57].
Conclusions
Most of the xylanases retrieved by culture-dependent and culture-independent approaches exhibit optimum activity in the pH and temperature range of 6.0–8.0 and 40–60°C, respectively. The xylanase obtained in this investigation through metagenomic approach (rMxyl) not only displays alkalistability and thermostability, but it also has a high thermostability. This is the first report of xylanase with twin stabilities obtained through culture-independent approach. A very low similarity in amino acid sequence of the enzyme with other known xylanases makes it a novel xylanase. The possibility of obtaining thermo-alkali-stable xylanase from composts may lead to an intense search for similar enzymes in this niche.
Materials and Methods
Ethics statement
No specific permits are required for collecting environmental samples used in this investigation. The samples did not have any animal or plant species because of the elevated temperature. The location is not privately owned or protected in any way and does not involve endangered and protected species.
Sample collection
The samples of compost-soil were collected in sterile polyethylene bags from the vicinity of a hot water spring near Fukuoka Japan and stored at 4°C. The pH of the samples is in the acidic range (3.0–4.5).
Strains and plasmids for DNA manipulations
Cloning vector p18 GFP was the gift from Dr. Taku Uchiyama, Japan. The pGEM-T Easy vector (Promega, USA), pET28a(+) and pET22b(+) of Novagen (Germany) were used for sequencing and expression of the xylanase gene, respectively. Escherichia coli DH10B, E. coli XL1 blue and E. coli BL21 (DE3) were used for the propagation of the plasmid. Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (UK).
Construction of metagenomic library
Soil DNA was extracted according to Verma and Satyanarayana (2011) [58]. Metagenomic DNA was processed for constructing the metagenomic library. Five μg of metagenomic DNA was partially digested with 0.5 U of restriction enzyme Sau3AI. The fragments of 3–12 Kb were eluted from agarose gel (1.2%, w/v) by gel extraction kit according to manufacturer's protocol (Macherey-Nagel, Germany). Hundred nanogram (ng) of insert DNA and 300 ng of BamH I digested and dephosphorylated p18 GFP vector were ligated by using T4 DNA ligase overnight at 16°C. The ligation mixture was transformed into competent E. coli DH10B cells by heat shock method. The metagenomic library was spread and screened for xylanase activity on 0.3% (w/v) RBB-xylan (4-O-methyl-D-glucurono-D-xylan-remazol brilliant blue R) (Sigma, St. Louis, MO, USA) LB-ampicillin agar plates. The transformants were grown at 37°C overnight and observed for the zone of xylan hydrolysis.
DNA sequencing and bioinformatics analysis
The pure clone (TSDV-MX1) showing clear zone of xylanase hydrolysis was sequenced using M13 forward and reverse primers followed by different internal primers using Applied Biosystem 373 stretch automated sequencer (Applied Biosystems, Foster City, CA, USA) at Nucleic acid sequencing facility of the University of Delhi South Campus, New Delhi (India) for obtaining full sequence of the insert. The ORFs were identified by using the NCBI's open reading frame (ORF) Finder tool (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). BLASTN and BLASTP of NCBI were used to align the nucleotide and amino acid sequences, respectively. Multiple alignments of the amino acids were carried out using the CLUSTALW programme (http://www.ebi.ac.uk/clustalW). The phylogenetic analysis was done using MEGA 2.1 with neighbour joining strategy.
Construction of expression plasmids pET28a-Mxyl and pET22b-Mxyl
The full length xylanase gene was pulled out from the clone (TSDV-MX1) by PCR using specific primers MxylF1 (CCCGCTAGCATGACAGCGAGTTTGAGGAAGA) and MxylR1 (CCCCTCGAGTTACGGCGTGTTTCCGTAGC) having the compatible NheI and XhoI restriction sites of pET28a(+) in the forward and reverse primers, respectively. Mxyl F2 (CCCGGATCCATGACAGCGAGTTTGAGGAAGA) and Mxyl R2 (CCCCTCGAGTTAC-GGCGTGTTTCCGTAGC) were used for cloning into pET22b(+) vector. The gene was amplified under the defined PCR conditions (initial denaturation 3 min at 95°C followed by 29 cycles of 10 sec at 98°C, 30 s at 59°C and 1 min at 68°C with a final extension step at 68 °C for 10 min). The PCR products pET28a(+) and pET22b(+) were digested with the restriction enzymes and ligated into the vectors followed by transformation into competent E. coli XL1 blue cells to obtain pET28-Mxyl and pET22-Mxyl.
Expression of xylanase (rMxyl) using pET28a-Mxyl and pET 22b-Mxyl
The recombinant constructs were confirmed by colony PCR followed by double digestion of the construct with restriction enzymes. These clones were processed for sequencing. The recombinant plasmid having the accurate sequence was then transformed into E. coli BL21 (DE3) competent cells for expression of the recombinant protein. The transformants were grown in kanamycin (50 µg/ml) containing LB medium at 37°C with 200 rpm in an incubator shaker to achieve an optical density of O.D. = 0.5 to 0.7. Afterwards the expression was induced by adding isopropyl-β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM and the culture was further cultivated at 30°C. The samples were collected at 1 h intervals for determining the enzyme titres.
Site directed mutagenesis
Multiple sequence alignment of recombinant xylanase with those of the known xylanases revealed Glu117 and Glu209 as catalytically important residues. Experimentally it has been proved by site directed mutagenesis using Geneart site directed mutagenesis kit (Invitrogen, Carsband, USA). Two point mutations (Glu117Asp and Glu209Asp) were created in the native xylanase gene and expressed in E. coli BL21 (DE3). The induced mutations were confirmed by sequencing.
Localization of rMxyl
Localization of the recombinant protein was determined by collecting the intracellular, extracellular and periplasmic fractions from the same culture. Extracellular, intracellular and periplasmic fractions were collected according to Verma and Satynarayana, 2012 [30].
Xylanase assay
Xylanase was assayed according to Archana and Satyanarayana (1997) [59]. The 0.5 ml of appropriately diluted enzyme was incubated at 80°C with 0.5 ml of 1% (w/v) birchwood xylan (Sigma, St. Louis, Mo.) in 100 mM glycine-NaOH buffer (pH 9.0) for 10 min. The reducing sugars were determined using 3, 5-dinitrosalicylic acid (DNSA) reagent [60]. One unit of xylanase is defined as the amount of enzyme required to liberate 1 μmol of reducing sugar as xylose ml−1 min−1 under the assay conditions.
Purification of recombinant xylanase and zymogram analysis
The rMxyl was purified by affinity chromatography using Ni2+-NTA agarose (Novagen, Germany) according to Verma and Satynarayana, 2012 [30]. For zymogram analysis, the recombinant protein was electrophoresed on native gel and the stripes of the gel were layered on 0.3% (w/v) xylan-agar plates of pH 9.0 at 70°C for 3 h. The gel was then removed from the plate and stained with Congo-red dye followed by destaining with 1 M NaCl for better appearance of the band showing clear zone of xylan hydrolysis.
Biochemical characterization of rMxyl
The kinetic characteristics of the recombinant xylanase like the effect of pH, temperature, metal ions, inhibitors and detergents on enzyme activity, thermostability and substrate specificity have been studied. The optimum pH was determined by incubating the recombinant enzyme in various buffers ranging from pH 3.0 to 12.0. Different buffers were used (pH 3.0 to 6.0, 0.1 M citrate buffer, for 7.0 and 8.0, 0.1 M sodium phosphate buffer and for 9.0–12.0, glycine-NaOH buffer). Similarly for optimum temperature, the recombinant enzyme was assayed over a range of temperatures from 40 to 100°C at pH 9.0. For thermostability, the residual xylanase activity was measured after incubating the recombinant enzyme at different temperatures (60–100) °C for 15 min to 3 h. The effects of various metal ions, inhibitors and detergents were determined by incorporating these into reaction mixtures followed by xylanase assay. The substrate specificity was quantitated by assaying the recombinant enzyme on various substrates (different sources of xylan as well as substrates other than xylan like starch, cellulose, chitin and pullulan in glycine-NaOH buffer). Kinetic properties of the recombinant enzyme (Km and Vmax) on different xylans from birchwood, beechwood and oat spelt were calculated from Lineweaver-Burk double reciprocal plots.
Saccharification of agroresidues/hydrolysis of xylan
One % (w/v) standards of xylooligosaccharides (X2–X6) and agro-residues (wheat bran, corn cobs and sugarcane bagasse) were treated with recombinant xylanase (10 U – 20 U/g) to find out the hydrolysis of XOs and lignocellulosic substrates. All the substrates (wheat bran, corn cobs and sugarcane bagasse) were suspended in glycine-NaOH buffer (pH 9.0) and incubated at 80°C. Aliquots at the desired intervals were collected and analyzed on silica based TLC plates (Merck, Germany) to determine the hydrolysis products. The saccharification of agro-residues was determined using DNSA reagent [60].
Nucleotide sequence accession numbers
The nucleotide sequence of the xylanase gene is deposited at NCBI GenBank (Accession no. AFP81696).
Supporting Information
Deduced amino acid sequence of recombinant xylanase (rMyl) and its nucleotide sequence. The red underlined region is leader sequence, cyan highlighted regions represents GH11 catalytic domain. Grey highlighted regions are compositionally biased regions that were not used in database search and proposed as linker regions. Bluish-green highlighted region depicts substrate binding domain.
(DOC)
Multiple sequence alignment of xylanase with other xylanases available in database. GenBank accession number and source of microorganisms were given as follows: 182406872 (glycosyl hydrolase family 11 precursor [uncultured bacterium]),17826947 (Pseudomonas sp. ND137), 29367333 (uncultured Cellvibrio sp.), 388259220 (Cellvibrio sp. BR), 302868167 (Micromonospora aurantiaca ATCC 27029), 386849796 (Actinoplanes sp. SE50/110), 194368056 (Streptomyces sp. S27). Five signature sequences: I (AYLTLYGW) II (VEYYIVDN), III (FWQYWSV), IV (HFDAWASLG) and V(MATEGY) of GH11 family are coloured. The two catalytically important residues (Glu 117 and Glu 209) are marked with black circle.
(DOC)
Funding Statement
This study was supported by the Department of Biotechnology, Govt. of India (BT/PR-8492/BCE/08/497/2006). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Coughlan MP, Hazlewood GP (1993) β-1,4 D-xylan-degrading enzyme systems: biochemistry, molecular biology and applications. Biotechnol Appl Biochem 17: 259–289. [PubMed] [Google Scholar]
- 2.Hori H, Elbein AD (1985) The biosynthesis of plant cell wall polysaccharides. In: Higuchi T. (Ed.), Biosynthesis and Biodegradation of Wood Components. Academic Press Inc., Orlando, FL, 109–135.
- 3. Collins T, Gerday C, Feller G (2005) Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol Rev 29: 3–23. [DOI] [PubMed] [Google Scholar]
- 4. Henrissat B (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Paes G, Berrin JG, Beaugrand J (2012) GH11 xylanases: Structure/function/properties relationships and applications. Biotechnol Adv 30: 564–592. [DOI] [PubMed] [Google Scholar]
- 6. Khandeparkar R, Bhosle NB (2007) Application of thermoalkalophilic xylanase from Arthrobacter sp. MTCC 5214 in biobleaching of kraft pulp. Biores Technol 98: 897–903. [DOI] [PubMed] [Google Scholar]
- 7. Menon V, Prakash G, Prabhune A, Rao M (2010) Biocatalytic approach for the utilization of hemicellulose for ethanol production from agricultural residue using thermostable xylanase and thermotolerant yeast. Biores Technol 101: 5366–5373. [DOI] [PubMed] [Google Scholar]
- 8. Woldesenbet F, Puneet A, Virk, Gupta N, Sharma P (2012) Effect of microwave irradiation on xylanase production from wheat bran and biobleaching of eucalyptus kraft pulp. Appl Biochem Biotechnol 167: 100–108. [DOI] [PubMed] [Google Scholar]
- 9. Sunna A, Bergquist PL (2003) A gene encoding a novel extremely thermostable 1, 4-β- xylanase isolated directly from an environmental DNA sample. Extremophiles 7: 63–70. [DOI] [PubMed] [Google Scholar]
- 10. Sunna A, Moracci M, Rossi M, Antranikian G (1997) Glycosyl hydrolases from hyperthermophiles. Extremophiles 1: 2–13. [DOI] [PubMed] [Google Scholar]
- 11. Kulkarni N, Shendye A, Rao M (1999) Molecular and biological aspects of xylanases. FEMS Microbiol Rev 23: 411–456. [DOI] [PubMed] [Google Scholar]
- 12. Sharma S, Adhikari S, Satyanarayana T (2007) Alkali-thermostable and cellulose free xylanase production by an extreme thermophile Geobacillus thermoleovorans . W J Microbiol Biotechnol 23: 483–490. [Google Scholar]
- 13. Son-Ng I, Li CW, Yeh Y, Chen PT, Chir JL, et al. (2009) A novel endoglucanase from the thermophilic bacterium Geobacillus sp. 70PC53 with high activity and stability over a broad range of temperatures. Extremophiles 13: 425–435. [DOI] [PubMed] [Google Scholar]
- 14. Yuhong Z, Shi P, Liu W, Meng K, Bai Y, et al. (2009) Lipase diversity in glacier soil based on analysis of metagenomic DNA fragments and cell Culture. J Microbiol Biotechnol 158: 200–212. [DOI] [PubMed] [Google Scholar]
- 15. Jungang L, Zhang K, Wenjun H (2010) Cloning and biochemical characterization of a novel lipolytic gene from activated sludge metagenome, and its gene product. Microbial cell factories. 9: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu EY, Kwon MA, Lee M, Oh JY, Choi JE, et al. (2011) Isolation and characterization of cold-active family VIII esterases from an arctic soil metagenome. Appl Microbiol Biotechnol 90: 573–581. [DOI] [PubMed] [Google Scholar]
- 17. Sunna A, Bergquist PL (2003) A gene encoding a novel extremely thermostable 1, 4-β- xylanase isolated directly from an environmental DNA sample. Extremophiles 7: 63–70. [DOI] [PubMed] [Google Scholar]
- 18. Lee CC, Kibblewhite-Accinelli RE, Wagschal K, Robertson GH, Wong DWS (2006) Cloning and characterization of a cold-active xylanase enzyme from an environmental DNA library. Extremophiles 10: 295–300. [DOI] [PubMed] [Google Scholar]
- 19. Hu Y, Zhang G, Li A, Chen J, Ma L (2008) Cloning and enzymatic characterization of a xylanase gene from a soil-derived metagenomic library with an efficient approach. Appl Microbiol Biotechnol 80: 823–830. [DOI] [PubMed] [Google Scholar]
- 20. Yamada K, Terahara T, Kurata S, Yokomaku T, Tsuneda S, et al. (2008) Retrieval of entire genes from environmental DNA by inverse PCR with pre amplification of target genes using primers containing locked nucleic acids. Environ Microbiol 10(4): 978–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang G, Wang Y, Yang P, Luo H, Huang H, et al. (2010) Molecular detection and diversity of xylanase genes in alpine tundra soil. Appl Microbiol Biotechnol 87: 1383–1393. [DOI] [PubMed] [Google Scholar]
- 22. Li R, Kibblewhite R, Orts WJ, Lee CC (2009) Molecular cloning and characterization of multidomain xylanase from manure library. World J Microbiol Biotechnol 25: 2071–2078. [Google Scholar]
- 23. Gilkes NR, Henrissat B, Kilburn DG, Miller RC Jr, Warren RA (1991) Domains in microbial 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev 55: 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brennan Y, Callen WN, Christoffersen L, Dupree P, Goubet F, et al. (2004) Unusual Microbial Xylanases from Insect Guts. Appl Environ Microbiol 70 (6): 3609–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shibuya H, Kaneko S, Hayashi K (2000) Enhancement of the thermostability and hydrolytic activity of xylanase by random gene shuffling. Biochem J 349: 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Notenboom V, Boraston AB, Williams SJ, Kilburn DG, Rose DR (2002) High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding. Biochemistry 41: 4246–4254. [DOI] [PubMed] [Google Scholar]
- 27. Winterhalter C, Liebl W (1995) Two extremely thermostable xylanases of the hyperthermophilic bacterium Thermotoga maritima MSB8. Appl Environ Microbiol 61: 1810–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fontes CM, Hazlewood GP, Morag E, Hall J, Hirst BH, et al. (1995) Evidence for a general role for non-catalytic thermostabilizing domains in xylanases from thermophilic bacteria. Biochem J 307: 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sakka K, Kojima Y, Kondo T, Karita S, Ohmiya K, et al. (1993) Nucleotide sequence of Clostridium stercorarium xynA gene encoding xylanase A: Identification of catalytic and cellulose binding domains. Biosci Biotechnol Biochem 57: 273–277. [DOI] [PubMed] [Google Scholar]
- 30. Verma D, Satyanarayana T (2012) Cloning, expression and applicability of thermo-alkali-stable xylanase of Geobacillus thermoleovorans in generating xylooligosaccharides from agro-residues. Biores Technol 107: 333–338. [DOI] [PubMed] [Google Scholar]
- 31. Guo B, Chen X, Sun C, Zhou B, Zhang Y (2009) Gene cloning, expression and characterization of a new cold-active and salt tolerant endo-β-1, 4-xylanase from marine Glaciecola mesophila KMM 241. Appl Microbiol Biotechnol 84: 1107–1115. [DOI] [PubMed] [Google Scholar]
- 32. Mamo G, Delgado O, Martinez A, Mattiasson B, Kaul RJ (2006) Cloning, sequencing analysis and expression of a gene encoding an endoxylanase from Bacillus halodurans S7. Mol Biotechnol 33: 149–159. [DOI] [PubMed] [Google Scholar]
- 33. Shi P, Tian J, Yuan T, Liu X, Huang H, et al. (2011) Paenibacillus sp. Strain E18 bifunctional xylanase-glucanase with a single catalytic domain. Appl Environ Microbiol 76(11): 3620–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yoon HS, Han NS, Kim CH (2004) Expression of Thermotoga maritima endo-β-1,4 xylanase gene in E. coli and characterization of the recombinant enzyme. Agric Chem Biotechnol 47: 157–160. [Google Scholar]
- 35. Mathrani IM, Ahring BK (1992) Thermophilic and alkaliphilic xylanase from several Dictyoglomus isolates. Appl Microbiol Biotechnol 38: 23–27. [Google Scholar]
- 36. Uchino F, Fukuda O (1983) Taxonomic characteristics of an acidophilic strain of Bacillus producing thermophilic acidophilic amylase and thermostable xylanase. Agric Biol Chem 47: 965–967. [Google Scholar]
- 37. Khasin A, Alchanati I, Shoham Y (1993) Purification and characterization of a thermostable xylanase from Bacillus stearothermophilus T-6. Appl Environ Microbiol 59: 1725–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chang WS, Tsai CL, Tseng MJ (2004) Cloning and characterization of two thermostable xylanases from an alkaliphilic Bacillus firmus . Biochem Biophys Res Commun 319: 1017–1025. [DOI] [PubMed] [Google Scholar]
- 39. Zhao Y, Luo H, Meng K, Shi P, Wang G, et al. (2011) A xylanase gene directly cloned from the genomic DNA of alkaline wastewater sludge showing application potential in the paper industry. Appl Biochem Biotechnol 165: 35–46. [DOI] [PubMed] [Google Scholar]
- 40. Yoon KH (2009) Cloning of the Bacillus subtilis AMX-4 xylanase gene and characterization of the gene product. J Microbiol Biotechnol 19: 1514–9. [DOI] [PubMed] [Google Scholar]
- 41. Maalej I, Belhaj I, Masmoudi NF, Belghith H (2009) Highly Thermostable Xylanase of the Thermophilic Fungus Talaromyces thermophilus: Purification and charact-erization. Appl Biochem Biotechnol. 158: 200–212. [DOI] [PubMed] [Google Scholar]
- 42. Zhang GM, Huang J, Huang GR, Ma LX, Zhang XE (2007) Molecular cloning and heterologous expression of a new xylanase gene from Plectosphaerella cucumerina . Appl Microbiol Biotechnol 74: 339–346. [DOI] [PubMed] [Google Scholar]
- 43. Liu L, Zhang G, Zhang Z, Wang S, Chen H (2011) Terminal amino acids disturb xylanase thermostability and activity. J Biol Chem. 286: 44710–44715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jalal A, Rashid N, Rasool N, Akhtar M (2009) Gene cloning and characterization of a xylanase from a newly isolated Bacillus subtilis strain R5. J Biosci Bioeng 107 (4): 360–365. [DOI] [PubMed] [Google Scholar]
- 45. Li N, Meng K, Wang Y, Shi P, Luo H, et al. (2008) Cloning, expression, and characterization of a new xylanase with broad temperature adaptability from Streptomyces sp. S9. Appl Microbiol Biotechnol 80: 231–240. [DOI] [PubMed] [Google Scholar]
- 46. Li R, Kibblewhite R, Orts WJ, Lee CC (2009) Molecular cloning and characterization of multidomain xylanase from manure library. World J Microbiol Biotechnol 25: 2071–2078. [Google Scholar]
- 47. Wang Y, Zhang H, He Y, Luo H, Yao B (2007) Characterization, gene cloning, and expression of a novel xylanase XYNB from Streptomyces olivaceoviridis A1. Aquaculture 267: 328–334. [Google Scholar]
- 48.Liu W, Shi P, Chen Q, Yang P, Wang G, et al. (2010) Gene cloning, over expression, characterization of a xylanase from Penicillium sp. CGMCC 1699. Appl. Biochem. Biotechnol. 162, 1–12. [DOI] [PubMed]
- 49. Matteotti C, Bauwens J, Brasseur C, Tarayre C, Thonart P, et al. (2012) Identification and characterization of a new xylanase from Gram-positive bacteria isolated from termite gut (Reticulitermes santonensis). Protein Expression and Purification 83: 117–127. [DOI] [PubMed] [Google Scholar]
- 50. Irena R, Jacek P, Stanislaw B (2006) Isolation and properties of Aspergillus niger IBT-90 xylanase for bakery. Appl Microbiol Biotechnol 69: 665–671. [DOI] [PubMed] [Google Scholar]
- 51. Yang Y, Zhang W, Huang J, Lin L, Lian H, et al. (2010) Purification and characterization of an extracellular xylanase from Aspergillus niger C3486. African J Microbiol Res 4(21): 2249–2256. [Google Scholar]
- 52. Vieira DS, Degreve L (2009) An insight into the thermostability of a pair of xylanases: the role of hydrogen bonds. Mol Phys 107: 59–69. [Google Scholar]
- 53. Kormelink FJM, Voragen AGJ (1993) Degradation of different [(glucurono)arabino] xylans by a combination of purified xylan-degrading enzymes. Appl Microbiol Biotechnol 38: 688–695. [Google Scholar]
- 54. Gruppen H, Hamer RJ, Voragen AGJ (1992) Water unextractable cell wall material from wheat flour. 2. Fractionation of alkali extracted polymers and comparison with water extractable arabinoxylans. J Cereal Sci 16: 53–67. [Google Scholar]
- 55. Mamo G, Hatti-Kaul R, Mattiasson B (2007) Fusion of carbohydrate binding modules from Thermotoga neapolitana with a family 10 xylanase from Bacillus halodurans S7. Extremophiles 11: 169–177. [DOI] [PubMed] [Google Scholar]
- 56. Sun JY, LU MQ, Weng XY, Qian LC, Gu SH (2007) Expression of recombinant Thermomonospora fusca xylanase A in Pichia pastoris and xylooligosaccharides released from xylan by it. Food Chem 104: 1055–1064. [Google Scholar]
- 57. Vazquez MJ, Alonso JL, Dominguez H, Parajo JC (2000) Xylooligosaccharides: manufacture and applications. Trends Food Sci Technol 11: 387–393. [Google Scholar]
- 58. Verma D, Satynarayana T (2011) An improved protocol for DNA extraction from alkaline soil and sediment samples for constructing metagenomic libraries. Appl Biochem Biotechnol 165: 454–464. [DOI] [PubMed] [Google Scholar]
- 59. Archana A, Satyanarayana T (1997) Xylanase production by thermophilic Bacillus licheniformis A99 in solid-state fermentation. Enzyme Microb Technol 21: 12–17. [Google Scholar]
- 60. Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugars. Anal Chem 31: 426–428. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Deduced amino acid sequence of recombinant xylanase (rMyl) and its nucleotide sequence. The red underlined region is leader sequence, cyan highlighted regions represents GH11 catalytic domain. Grey highlighted regions are compositionally biased regions that were not used in database search and proposed as linker regions. Bluish-green highlighted region depicts substrate binding domain.
(DOC)
Multiple sequence alignment of xylanase with other xylanases available in database. GenBank accession number and source of microorganisms were given as follows: 182406872 (glycosyl hydrolase family 11 precursor [uncultured bacterium]),17826947 (Pseudomonas sp. ND137), 29367333 (uncultured Cellvibrio sp.), 388259220 (Cellvibrio sp. BR), 302868167 (Micromonospora aurantiaca ATCC 27029), 386849796 (Actinoplanes sp. SE50/110), 194368056 (Streptomyces sp. S27). Five signature sequences: I (AYLTLYGW) II (VEYYIVDN), III (FWQYWSV), IV (HFDAWASLG) and V(MATEGY) of GH11 family are coloured. The two catalytically important residues (Glu 117 and Glu 209) are marked with black circle.
(DOC)