Abstract
Imatinib has so far been the first-choice treatment in chronic myeloid leukemia with excellent results. However, only a proportion of patients achieve major molecular response – hence the need to find biological predictors of outcome to select the optimal therapeutic strategy now that more potent inhibitors are available. We investigated a panel of 20 polymorphisms in seven genes, potentially associated with the pharmacogenetics of imatinib, in a subset of 189 patients with newly diagnosed chronic myeloid leukemia enrolled in the TOPS trial. The analysis included polymorphisms in the transporters hOCT1, MDR1, ABCG2, OCTN1, and OATP1A2, and in the metabolizing genes CYP3A4 and CYP3A5. In the overall population, the OCTN1 C allele (rs1050152), a simple combination of polymorphisms in the hOCT1 gene and another combination in the genes involved in imatinib uptake were significantly associated with major molecular response. The combination of polymorphisms in imatinib uptake was also significantly associated with complete molecular response. Analyses restricted to Caucasians highlighted the significant association of MDR1 CC (rs60023214) genotype with complete molecular response. We demonstrate the usefulness of a pharmacogenetic approach for stratifying patients with chronic myeloid leukemia according to their likelihood of achieving a major or complete molecular response to imatinib. This represents an attractive opportunity for therapy optimization, worth testing in clinical trials.
Introduction
The introduction of the Bcr-Abl tyrosine kinase inhibitor imatinib mesylate in the first-line treatment of Philadelphia chromosome-positive (Ph+) chronic myeloid leukemia (CML) revolutionized patients' outcome. Given the excellent hematologic and cytogenetic response rates that can be obtained in the chronic phase of the disease,1 the goal of CML therapy has moved to the achievement of a major molecular response (MMR; defined as a 3-log reduction in Bcr-Abl transcript level from a standardized baseline value)2 and possibly a complete molecular response [CMR; defined as at least a 4-log reduction corresponding to undetectable Bcr-Abl transcript by realtime reverse transcription polymerase chain reaction (PCR)],2 which might represent an ‘operational cure’ as well as a prerequisite for discontinuation of imatinib.3
Research in CML has since focused on finding biological predictors of response allowing for treatment optimization. It is well recognized that interpatient variability in drug response reflects the systemic levels or intracellular concentrations of the drug, known to be associated with the pharmacokinetics (absorption, distribution, and metabolism) of the drug itself.4 Imatinib is metabolized by the cytochrome-P450 - mostly CYP3A4 and 3A5 isoforms.5 The active uptake of imatinib into cells is known to be mediated mainly by the hOCT1 transporter (encoded by the SLC22A1 gene),6-7 whereas its efflux is mediated by the ABC transporters, in particular ABCB1 (also known as MDR1) and, to a lesser extent, ABCG2.6,8-10 Besides these, other transporters may be important in the absorption, distribution, and elimination of imatinib, including the families of organic cation transporters (OCT) and organic anion transporters (OAT). In particular, a recent study by Hu et al.11 identified imatinib as a substrate of OATP1A2 (encoded by SLCO1A2), whereas the involvement of members of the OCTN family as imatinib transporters is still uncertain.
Interpatient variability in imatinib metabolism/transport is substantial and thus far unexplained. A possibility is that genetic polymorphisms in genes encoding imatinib-metabolizing enzymes and transporters may influence the extent to which imatinib is delivered to target cells. Accordingly, genetic polymorphisms of the candidate genes CYP3A4/3A5, MDR1, ABCG2, OATP1A2, OCTN1 (encoded by SLC22A4) and hOCT1 could affect expression of corresponding proteins and thus may predict differences in responses to imatinib. To explore this hypothesis, we selected a panel of polymorphisms in these genes and genotyped 189 newly diagnosed CML patients treated with imatinib in the framework of the Tyrosine kinase OPtimization and Selectivity (TOPS) phase III trial.12
Design and Methods
Study population
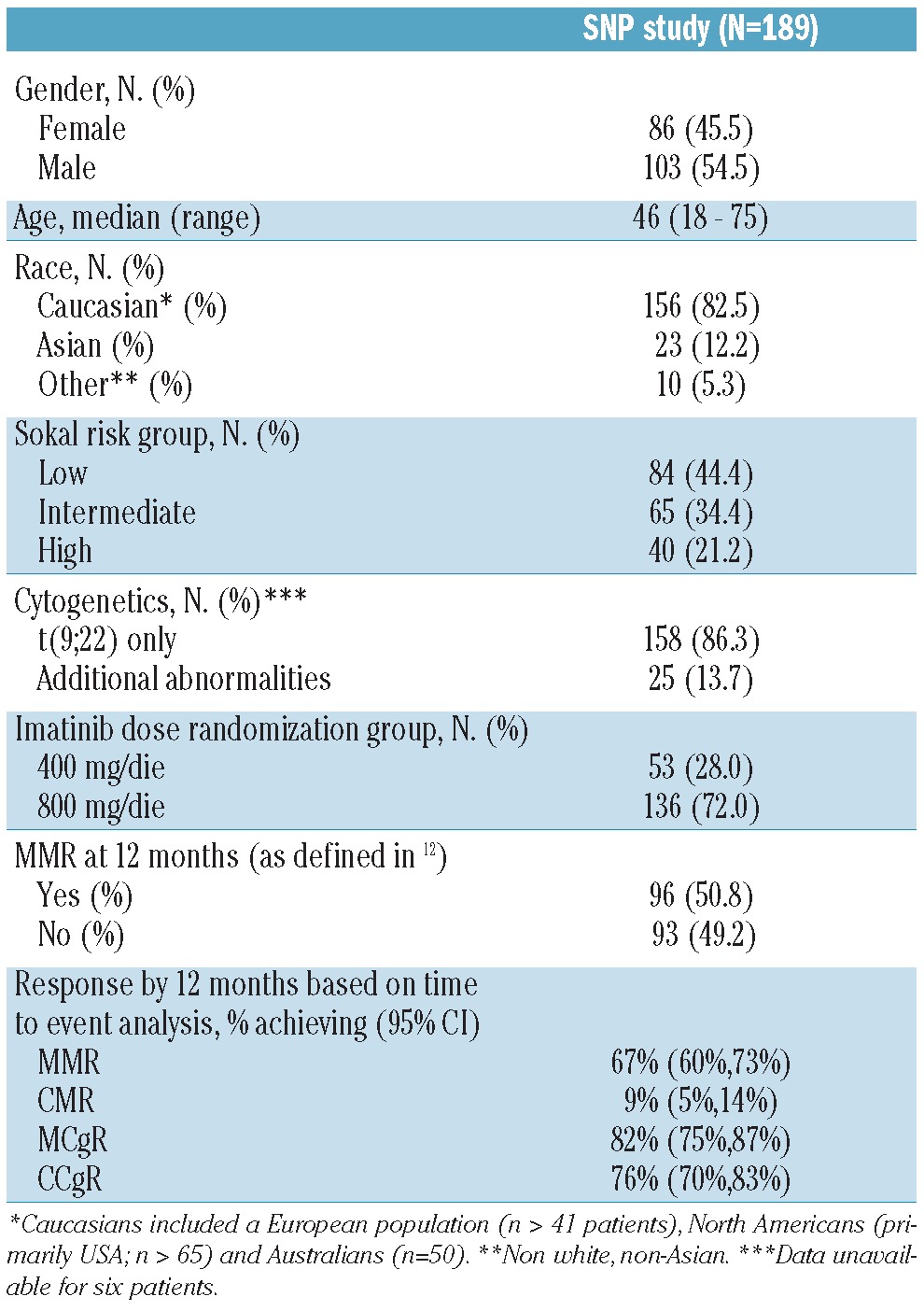
A total of 189 CML patients receiving imatinib were retrospectively enrolled in this pharmacogenetic study. These patients represented a subset of the population of the TOPS trial - a randomized, phase III study of imatinib 400 mg/day versus imatinib 800 mg/day in newly diagnosed and previously untreated CML patients in chronic phase.12 The patients' selection was based exclusively on availability of written informed consent for correlative sub-studies, according to the Helsinki Declaration and its later amendments, and sufficient amount of archived material. The study was approved by all local ethical committees. The patients' characteristics and treatment protocol have been described elsewhere.12 Table 1 summarizes selected demographic characteristics of our study population.
Table 1.
Patients' and disease characteristics of the study population.
Evaluation of response to imatinib
Molecular response (MR) was classified based on BCR-ABL to control gene transcript ratios, expressed on the International Scale.13 MMR and CMR were defined as ratios ≤0.1% and ≤0.0032%, respectively. Cytogenetic response (CgR) based on bone marrow assessment was classified as complete (CCgR; 0% Ph+ cells), partial (PCgR; >0 to 35% Ph+ cells), minor (>35 to 65% Ph+ cells), minimal (> 65 to 95% Ph+ cells), and none (>95 to 100% Ph+ cells). A major cytogenetic response (MCgR) was defined as achieving either a CCgR or a PCgR. Time to response (MMR, CMR, MCgR, CCgR) was defined as the interval between the date of randomization and the first date of achieving a response. Patients who progressed to accelerated phase or blast crisis were classified as non-responders at all time points.
Genotyping analysis
DNA was extracted from cryopreserved white blood cells using conventional methods. The characteristics of the studied polymorphisms (one insertion/deletion and 19 single nucleotide polymorphisms, from here on all referred to as SNP in the text) are reported in Online Supplementary Table S1. Genotypes were determined by PCR-based assays (restriction fragment length polymorphism and/or real-time) according to published methods,14-20 or as recommended by the manufacturer (Online Supplementary Table S1). Positive and negative controls were included in each reaction as quality controls. In addition, the accuracy of genotyping was confirmed by repetition of 100% of the samples. The replicates were 100% concordant.
Statistical analysis
Genotype distribution – overall and by race – was tested for Hardy-Weinberg equilibrium (HWE) with exact tests.21 The association of individual SNP with MMR, CMR, MCgR, and CCgR was assessed using the exact P-value based on a Cochran-Armitage trend test in an additive model with the number of minor alleles (0, 1, or 2) as the predictor. Survival analyses were also used to examine the relationship between SNP and MMR, CMR, MCgR, and CCgR. Time to response was defined as the interval between randomization and the date of first confirmed response for any of the given response variables under study. The follow-up time was censored at the date of last assessment for those not achieving a response. Hazard ratios were estimated with Cox proportional hazards models and evaluated with likelihood ratio tests. Dose of imatinib was included as a covariate in the models to adjust for possible differential response in the two imatinib dose groups. Statistically significant associations observed in the Cox regression analyses were further evaluated through plots based on the Kaplan-Meier method. The Kaplan-Meier curves were compared with log-rank tests of trend, assuming an additive genetic model. Haplotype frequencies were estimated using the expectation-maximization algorithm. Linkage disequilibrium between pairs of SP was estimated with Lewontin's D′, and likelihood ratio tests were performed to assess linkage disequilibrium among multiple loci. Empirical P-values obtained from permutation tests (n = 10,000 permutations) were used to evaluate the association between haplotypes and imatinib response at 12 months. All statistical tests were two-sided, with statistical significance defined as P<0.05. No adjustments were made for multiple comparisons. Statistical analyses were conducted using SAS version 9.1 (SAS Institute Inc., Cary, NC, USA).
The primary purpose of this study was to evaluate the effect of individual SNP on response to imatinib therapy. As an exploratory analysis, we also examined simple summary measures of functionally related (uptake and efflux) SNP. These were defined as the number of alleles, across the genes of interest, hypothesized or found in this study to be associated with a favorable response. According to the results of in vitro studies reported in the literature, we hypothesized a favorable response in the presence of hOCT1 rs4646277, rs4646278, and rs2282143, MDR1 rs1128503 (also known as C1236T), rs60023214 (also known as C3435T), rs1128501, and rs10245483 and OATP1A2 rs11568563 major alleles and ABCG2 rs2231137 and rs2231142 variant alleles.21 No functional knowledge was available for the remaining SNP and the major allele was thus hypothesized to be associated with favorable response unless the present study suggested a different relationship.
Results
Genotype distribution
Genotype distributions of the 20 candidate SNP are summarized in Online Supplementary Table S2A-C. Five SNP (hOCT1: rs4646277, rs4646278; MDR1: rs1128501; CYP3A4: rs28371759 and CYP3A5: rs28365083) were homozygous for the major allele for all patients and were excluded from the analyses. The comparison of genotype frequencies according to the patients' demographic characteristics revealed differences between Asian and Caucasian subgroups (Online Supplementary Table S2A, B). For this reason analyses of associations of SNP with response restricted to Caucasians were performed in addition to analyses based on all patients. A deviation from the HWE was observed for a single SNP (MDR1 rs10245483) in the Caucasian population; no departures from the HWE were observed for any other SNP, either in the overall population or in Caucasians. When restricting the analysis to the Caucasian population, the genotype distributions of MDR1 rs60023214 and rs2032582 (known as G2677T/A), CYP3A4 rs2740574 and CYP3A5 rs776746 differed significantly by geographical region. However, none of the four genotypes in the overall Caucasian group showed deviation from the HWE.
Outcomes of imatinib therapy
Treatment outcomes for all patients and only the Caucasians are presented in Figure 1. With a median total follow-up of 29 months among the entire study population, the 12-month post-randomization cumulative incidences of MMR, CMR, MCgR and CCgR were 67% (95% CI, 60%-73%), 9% (5%-14%), 82% (75%-87%), and 76% (70%-83%), respectively. Response rates in the Caucasian subgroup were similar.
Figure 1.
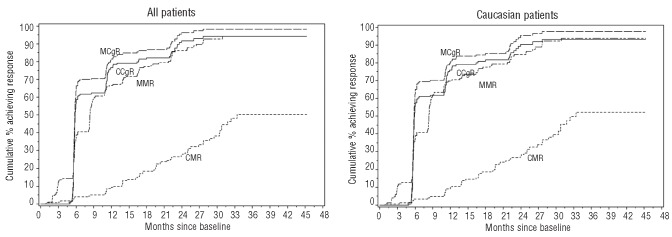
Rates of cytogenetic and molecular response to imatinib in the study population. Cumulative incidence of patients achieving major cytogenetic response (MCgR), complete cytogenetic response (CCgR), major molecular response (MMR) and complete molecular response (CMR) in the overall population (left panel) and in Caucasians (right panel).
Outcomes of imatinib therapy according to candidate genotypes
Treatment outcomes were compared according to: (i) each candidate genotype, (ii) summary measures based on combinations of SNP in the same gene, and (iii) summary measures based on combinations of SNP in functionally related (uptake and efflux) genes. Results of associations with CgR and MR, adjusted for imatinib dose, among all patients are grouped in Online Supplementary Table S3. All the most relevant results estimated with Cox proportional hazard models and evaluated with likelihood ratio tests are presented in Table 2.
Table 2.
Most relevant correlations between treatment outcomes and candidate genotypes or SNP combinations in the overall population.
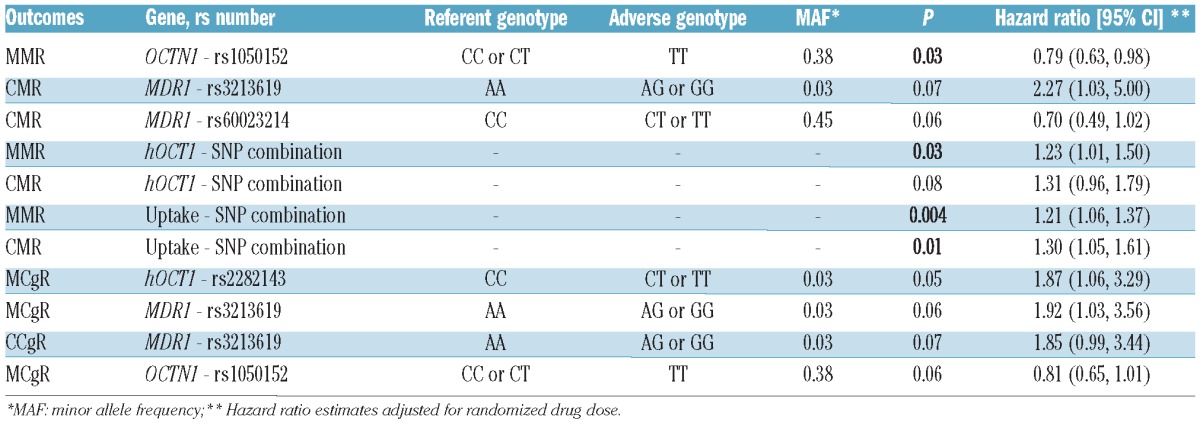
As far as MR was concerned, presence of the C allele in OCTN1 (rs1050152) had a favorable impact on MMR achievement (P=0.03). Nearly significant associations were observed for the presence of MDR1 A (rs3213619) and MDR1 C (rs60023214) alleles with achievement of CMR (P=0.07 and P=0.06 respectively). With respect to the summary measures, a combination of SNP in the hOCT1 gene (see Online Supplementary Table S4 for combination design and distribution in the study population) was significantly correlated with MMR (P=0.03). The same combination had a weak effect on CMR achievement (P=0.08). When considering summary measures of uptake (see Online Supplementary Table S5 for combination design and distribution in the study population) and efflux, only the former was associated with both MMR and CMR (P=0.004 and P=0.01, respectively).
As far as CgR was concerned, hOCT1 C (rs2282143) was significantly associated with MCgR (P=0.05); nearly significant associations were observed for the presence of MDR1 A (rs3213619) and OCTN1 C (rs1050152) alleles with achievement of MCgR (P=0.06 for both).
We also carried out analyses limited to Caucasian patients, our most representative sample set. All the results are presented in Online Supplementary Table S6, while the most relevant ones are grouped in Table 3. The analyses yielded results similar to those observed in the overall population. In particular, the presence of OCTN1 C (rs1050152) allele had a significantly favorable impact on achievement of MMR and a weak effect on CMR (P=0.03 and P=0.07, respectively). A combination of SNP in hOCT1 (Online Supplementary Table S4) was again associated with achievement of MMR (P=0.05). Similarly, achievement of MMR and CMR is associated with an increasing number of favorable alleles in genes functionally related to imatinib uptake (Table 3; P=0.01 for both). In addition to these results, the analyses in the Caucasian subgroup pointed out the influence of two additional SNP on the achievement of MR. The CC genotype in MDR1 (rs60023214) was significantly associated with MMR achievement (P=0.005), and a similar tendency was observed for the achievement of CMR (P=0.09). A nearly significant association between CYP3A4 GG genotype (rs270574) and MMR achievement was also observed (P=0.07). None of the analyzed SNP or combination of SNP was associated with MCgR or CCgR.
Table 3.
Most relevant correlations between treatment outcomes and candidate genotypes or SNP combinations among Caucasians.
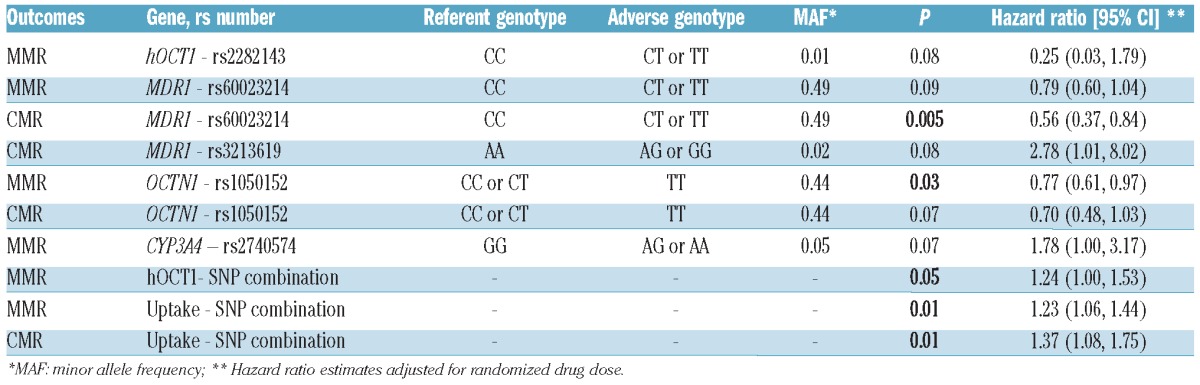
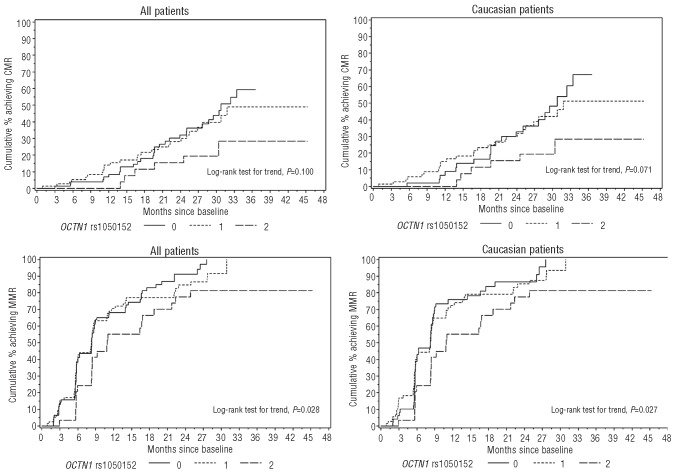
Plots of the estimated fraction of patients achieving response by time since treatment initiation based on the Kaplan-Meier method were analyzed in the overall population and separately for Caucasians. Presence of the minor allele in OCTN1 (TT rs1050152), was associated with decreased rate of MMR in the overall population and Caucasians (P=0.028 and P=0.027, respectively; Figure 2). Results were similar for CMR, and in general the curves separated at later times compared to those for MMR (Figure 2). Similarly, the combination of favorable alleles in the genes involved in imatinib uptake was associated with increased rates of both MMR and CMR (P=0.004 and P=0.015, respectively, in the overall population, and P=0.005 and P=0.009, respectively, in Caucasians). In addition, an increase in the number of favorable alleles in the hOCT1 gene was associated with an increased MMR rate (P=0.030 and P=0.043 in the overall population and Caucasians, respectively).
Figure 2.
Molecular response by OCTN1 genotype. Cumulative incidence of patients achieving MMR and CMR according to OCTN1 rs1050152 genotype in the overall population (left panels) and in Caucasians (right panels). Given that the major allele is C and the minor allele is T, CC and CT carriers had significantly higher MMR and CMR rates with respect to TT carriers.
In addition to these results, the analyses in the Caucasian subgroup confirmed the influence of two additional SNP on MR. The major allele in MDR1 (CCrs60023214) was significantly associated with a higher rate of CMR and had a weak effect on MMR (P=0.005 and P=0.081, respectively; Figure 3A). The major allele in CYP3A4 (AA rs2740574) was associated with a decreased rate of MMR (P=0.038; Figure 3B).
Figure 3.
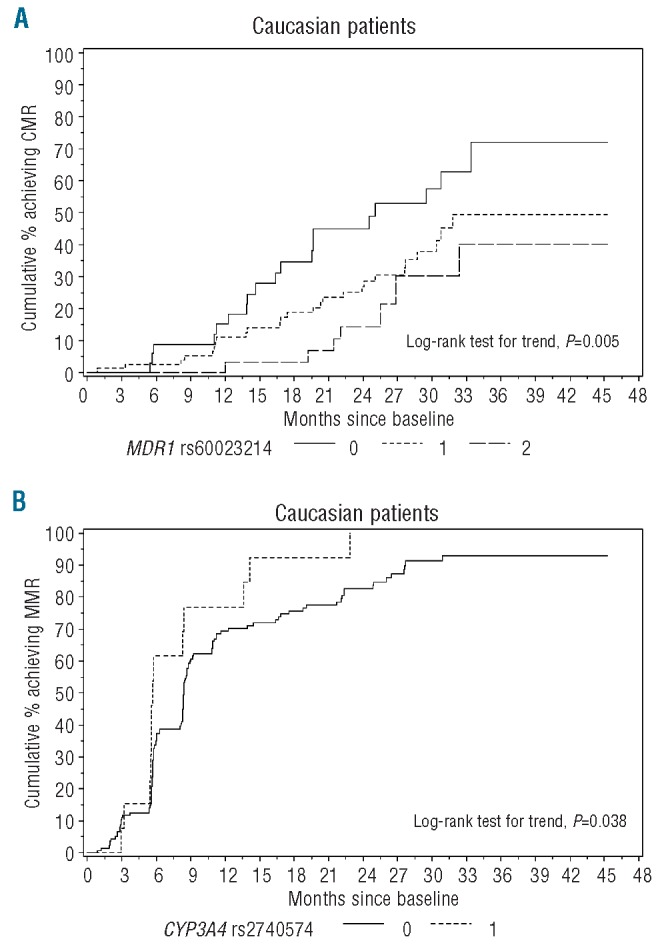
Molecular response in Caucasians according to MDR1 and CYP3A4 genotypes. (A) Cumulative incidence of patients achieving CMR according to MDR1 rs60023214 genotype. Given that for rs60023214 the major allele is C and the minor allele is T, CC carriers had a significantly higher CMR rate with respect to CT and TT carriers. (B) Cumulative incidence of patients achieving MMR according to CYP3A4 rs2740574 genotype. Given that for rs2740574 the major allele is A and the minor allele is G, AA carriers had a significantly higher MMR rate with respect to AG carriers (no GG carriers were detected in our study population).
As previously reported, the three MDR1 polymorphisms rs1129503 (C1236T), rs2032582 (G2677T/A), and rs60023214 (C3435T), were found to be in strong linkage disequilibrium (rs1129503 and rs2032582: D′=0.76; rs1129503 and rs60023214: D′=0.51; rs2032582 and rs60023214: D′=0.61; test for linkage disequilibrium among the three markers P<0.0001). However none of the resulting haplotypes (see Online Supplementary Table S7 for haplotype frequencies) was associated with response to imatinib.
Analogous analyses were performed for loss of MMR, MCgR, and CCgR. However, the loss of response rates was quite low and no suggestion of association could be detected.
Analyses were also performed for evidence of differences by drug dose (imatinib 400 mg versus 800 mg), but no statistically significant associations were observed.
Discussion
The spectrum of therapeutic options for CML patients has recently been enriched by second-generation tyrosine kinase inhibitors that are more potent and/or more selective than imatinib in Bcr-Abl inhibition. Nilotinib and dasatinib had already demonstrated remarkable efficacy in imatinib-resistant cases22,23 and phase II and III studies in newly diagnosed patients have shown even higher rates of responses, obtained earlier during therapy, than those achievable with imatinib.24-27 Paradoxically, the availability of multiple therapeutic options is not paralleled by the availability of biological predictors of outcome enabling identification, at the time of diagnosis, of those patients who are more likely to benefit from nilotinib or dasatinib rather than from imatinib - hence there is still a need for CML treatment optimization. To this end, a refined stratification of patients in terms of likelihood of achieving MMR, or even better CMR, would be the necessary starting point. Despite intensive research efforts, for years the Sokal score has been the only reliable factor for risk prediction at the time of diagnosis. Only recently has pretherapy hOCT1 transporter activity been demonstrated to correlate with MMR and to predict for long-term risk of resistance to imatinib.28,29
In several pathological conditions, pharmacogenetics has proven to be a potential source of biomarkers given the known influence of polymorphisms in key genes encoding drug transporters and metabolizing enzymes on intracellular drug delivery and, therefore, on the effectiveness of the drugs. In CML, only three studies had so far explored this field, but they all suffered from the limitation of being conducted in heterogeneous populations including patients at different stages of disease, not all treated with imatinib first-line30,31 and of having limited sample sizes.32,33
In the present study, we investigated a panel of polymorphisms that can be hypothesized to influence imatinib transport and metabolism in a large series of newly diagnosed, previously untreated CML patients receiving imatinib in the framework of the TOPS phase III trial. This is a worldwide trial that enrolled patients of different ethnic origin – which brings benefit to the search for common genetic traits of response. We found, using a multiple candidate gene-SNP approach, that there are genes in the imatinib metabolism and transport pathways that are associated with the degree of MR to imatinib therapy. Examining multiple candidate genes (20 SNP in 7 genes associated with imatinib metabolism and transport) also enables the potential effects of gene-gene interactions to be assessed.
In this study, a combination of SNP in hOCT1 was significantly associated with MMR, regardless of ethnicity. This finding is biologically plausible, especially in the light of the fact that all the studied variants are in the coding region and lead to an amino acid change, assigning a putative functional role to these SNP. It has been demonstrated that hOCT1 activity is an important determinant of MR to imatinib in CML patients.28,34 It is, therefore, reasonable to expect that a combination of SNP may influence hOCT1 expression and/or activity, thus playing a role in the achievement of MMR. It remains an open question whether hOCT1 genotypes determine the levels of hOCT1 mRNA transcript or alter the transporter activity/selectivity – thus further studies are warranted in order to elucidate the correlation of hOCT1 genotypes and activity. Another combination of SNP, all non-synonymous - in genes involved in imatinib uptake (hOCT1, OCTN1, OATP1A2) - was even more significantly associated with both MMR and CMR, regardless of ethnicity. This finding, also biologically plausible, shows the importance of individual pharmacokinetic differences for the response to imatinib therapy. Indeed, interindividual variation in imatinib uptake can be considered as a complex phenotypic trait, which can most probably be explained as a complex genotypic feature (i.e. one SNP versus a combination of SNP). However, little or no functional data are available in the literature for these SNP, so we can only hypothesize an effect on imatinib pharmacokinetics based on the fact that they all correspond to an amino acid variation in the protein sequence. An interesting finding was that the OCTN1 rs1050152-C allele was significantly associated with MMR in both the overall population and Caucasians. As far as we are aware, none of the pharmacokinetic studies of imatinib has pointed out an involvement of the OCTN1 protein in imatinib transport; however, it cannot be excluded that this transporter is directly or indirectly involved in imatinib uptake. It is also possible that this association occurred by chance - making it essential to replicate the observation in an independent dataset, as well as to carry out further in vitro studies assessing what role OCTN1 may play in the cellular uptake of imatinib.
Our analyses in the Caucasian population also ascribed a role to an MDR1 polymorphism in the achievement of CMR, confirming that this transporter may be a critical determinant of intracellular imatinib levels. Several pharmacogenetic association studies have focused on MDR1 variants. In particular the role of three variants: rs1128503, rs60023214, and rs2032582 – better known from published studies as C1236T, C3435T and G2677T/A – has been extensively studied, with strikingly contrasting results. Dulucq et al.30 were the first to report an association between MDR1 rs2032582 (TT), rs1128503 (TT/TA) and higher MMR rates. They also found that the haplotype rs1128503 C, rs2032582 G, rs60023214 C was statistically linked to less frequent MMR. A subsequent study by Kim et al. could not replicate any of these observations – no association emerged between any of the MDR1 gene polymorphisms analyzed and clinical outcome; however, no haplotype was assessed.31 More recently, Ni et al.32 reported that the rate of resistance to imatinib was higher in patients carrying the rs1128503 TT variant. Maffioli et al.33 reported a protective effect from primary failure for the rs2032582 T allele, however the rs1128503, rs2032582, rs60023214 TGC haplotype - differing only by one allele from the one reported by Dulucq et al.30 as favorable - was more frequently found in patients with primary resistance to imatinib.
In our data set, MDR1 haplotypes were not associated with imatinib response. On the other hand, we found a statistically significant correlation between MDR1 rs60023214 and CMR. Functional studies35,36 have shown a key role for rs60023214 in the altered expression and in vivo activity of P-glycoprotein, product of the MDR1 gene. However, our data showed a significantly higher CMR rate in carriers of the CC genotype than in carriers of the CT/TT genotypes, while results of these studies – the T allele has been reported to be associated with lower transcript levels as compared with the C allele – would suggest the opposite. Our observations are in agreement with the findings of Ni et al.32 who reported that the rate of resistance was higher in CT/TT genotype carriers than in CC genotype carriers, and in disagreement with Maffioli et al.33 In this latter study, the CC genotype was associated with a higher probability of primary treatment failure, but the results need to be considered with caution as only four patients experienced primary treatment failure. Another critical issue to bear in mind is that definitions of MR as well as of resistance to imatinib in CML have only recently been standardized; this unfortunately represents a confounding element, actually making it impossible to compare results in terms of associations across different pharmacogenetic studies. However, taken together these results suggest that MDR1 is, directly or indirectly, an important determinant of intracellular imatinib concentration.
Another interesting finding, restricted to the Caucasian group, is the significant association between the CYP3A4 rs270574-GG minor allele and increased MMR rate. Cytochrome P450 isoform 3A4 is generally regarded as the major enzyme responsible for the metabolism of imatinib.37,38 The rs270574 SNP is located in the regulatory region of the gene and might, therefore, alter transcriptional activity, ultimately influencing enzyme expression. To date, there have been no studies addressing the functional implications of this specific polymorphism in vivo; however, in vitro experiments have suggested that the variant allele may be associated with higher CYP3A4 expression.39,40 Pharmacokinetic and pharmacodynamic assessments in the phase I study of imatinib in CML proposed that variability in CYP3A4 activity might, at least in part, account for the observed interpatient variability in drug exposure and that co-administration of drugs inducing CYP3A4 could increase imatinib metabolism and lead to low/ineffective imatinib plasma levels.41 One would thus expect lower response rates to be associated with the rs270574 variant – which actually contrasts with our findings. On the other hand, a recent study in CML investigating the influence of in vivo CYP3A4 activity on the achievement of MR to imatinib found that higher activity significantly correlated with higher CMR rates at 12 months.42 This observation, in line with our findings, might have a rationale in the fact that the main imatinib metabolite formed by CYP3A4, the N-desmethylated piperazine derivative (CGP74588), is pharmacologically active, with a potency and selectivity similar to those of imatinib,37,38 and longer terminal half-life.43,44 It may be hypothesized that the presence of the variant allele leads to a larger amount of CGP74588. Another intriguing hypothesis is that CGP74588 can be subjected to a different transport process – which would be in line with the observation of higher cellular uptake of CGP74588 as compared to imatinib made by le Coutre et al.44 Obviously no definitive explanation can be given and further studies on the pharmacokinetics of CGP74588 are needed.
The study of combinations of SNP to assess response to imatinib therapy provides only a preliminary examination of the potential for creation of a panel of markers to drive treatment decisions. Creation of such a panel of markers will require larger sample sizes for development followed by testing in an independent set.
In conclusion, this study showed that genotyping should be taken into account in CML patients in an attempt to individualize treatment further, with the aim of enhancing efficacy in terms of achievement of MMR and CMR. On the basis of these findings, stratification of patients according to genotypes may be proposed for selection between imatinib and second-generation tyrosine kinase inhibitors, and represents an attractive opportunity for new clinical trials.
Acknowledgments
Funding: This work was supported by Novartis Oncology, Clinical Development, TOPS Correlative Studies Network, the University of Bologna (“Progetti strategici”), and the Ministry of Education, University and Research of Italy (MIUR, grant number 2009TNXL9P to SA). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures: Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Druker BJ, Guilhot F, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355(23):2408-17 [DOI] [PubMed] [Google Scholar]
- 2.Hughes TP, Branford S. Monitoring disease response to tyrosine kinase inhibitor therapy in CML. Hematology Am Soc Hematol Educ Program. 2009;477-87 [DOI] [PubMed] [Google Scholar]
- 3.Rousselot P, Huguet F, Rea D, Legros L, Cayuela JM, Maarek O, et al. Imatinib mesylate discontinuation in patients with chronic myelogenous leukemia in complete molecular remission for more than 2 years. Blood. 2007;109(1):58-60 [DOI] [PubMed] [Google Scholar]
- 4.Haiat S, Decleves X, Mittaine B, Legrand O, Francart S, Rio B, et al. Determination of imatinib plasma levels by high performance liquid chromatography (HPLC): evaluation of pharmacokinetic variability and haematological consequences. Blood. 2006;108 Abstract 1375. [Google Scholar]
- 5.Peng B, Lloyd P, Schran H. Clinical pharmacokinetics of imatinib. Clin Pharmacokinet. 2005;44(9):879-94 [DOI] [PubMed] [Google Scholar]
- 6.Thomas J, Wang L, Clark RE, Pirmohamed M. Active transport of imatinib into and out of cells: implications for drug resistance. Blood. 2004;104(12):3739-45 [DOI] [PubMed] [Google Scholar]
- 7.White DL, Saunders VA, Dang P, Engler J, Zannettino AC, cambareri AC, et al. OCT-1-mediated influx is a key determinant of the intracellular uptake of imatinib but not nilotinib (AMN107): reduced OCT-1 activity is the cause of low in vitro sensitivity to imatinib. Blood. 2006;108(2):697-704 [DOI] [PubMed] [Google Scholar]
- 8.Mahon FX, Belloc F, Lagarde V, Chollet C, Moreau-Gaudry F, Reiffers J, et al. MDR1 gene overexpression confers resistance to imatinib mesylate in leukemia cell line models. Blood. 2003;101(6):2368-73 [DOI] [PubMed] [Google Scholar]
- 9.Illmer T, Schaich M, Platzbecker U, Freiberg-Richter J, Oelschlägel U, von Bonin M, et al. P-glycoprotein-mediated drug efflux is a resistance mechanism of chronic myelogenous leukemia cells to treatment with imatinib mesylate. Leukemia. 2004;18(3):401-8 [DOI] [PubMed] [Google Scholar]
- 10.Burger H, Nooter K. Pharmacokinetic resistance to imatinib mesylate: role of the ABC drug pumps ABCG2 (BCRP) and ABCB1 (MDR1) in the oral bioavailability of imatinib. Cell Cycle. 2004;3(12):1502-5 [DOI] [PubMed] [Google Scholar]
- 11.Hu S, Franke RM, Filipski KK, Hu C, Orwick SJ, de Bruijn EA, et al. Interaction of imatinib with human organic ion carriers. Clin Cancer Res. 2008;14(10):3141-8 [DOI] [PubMed] [Google Scholar]
- 12.Cortes JE, Baccarani M, Guilhot F, Druker BJ, Branford S, kim DW, et al. Phase III, randomized, open-label study of daily imatinib mesylate 400 mg versus 800 mg in patients with newly diagnosed, previously untreated chronic myeloid leukemia in chronic phase using molecular end points: tyrosine kinase inhibitor optimization and selectivity study. J Clin Oncol. 2010;28(3):424-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes T, Deininger M, Hochhaus A, Branford S, Radich J, Kaeda J, et al. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendation for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108(1):28-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goreva OB, Grishanova AY, Domnikova NP, Mukhin OV, Lyakhovich W. MDR1 gene C1236T and C6+139T polymorphisms in the Russian population: associations with predisposition to lymphoproliferative diseases and drug resistance. Bull Exp Biol Med. 2004;138(4):404-6 [DOI] [PubMed] [Google Scholar]
- 15.Jamroziak K, Młynarski W, Balcerczak E, Mistygacz M, Trelinska J, Mirowski M, et al. Functional C3435T polymorphism of MDR1 gene: an impact on genetic susceptibility and clinical outcome of childhood acute lymphoblastic leukemia. Eur J Haematol. 2004;72(5):314-21 [DOI] [PubMed] [Google Scholar]
- 16.Kurzawski M, Pawlik A, Gornik W, DroŸdzik M. Frequency of common MDR1 gene variants in Polish population. Pharmacol Rep. 2006;58(1):35-40 [PubMed] [Google Scholar]
- 17.Hu LL, Wang XX, Chen X, Chang J, Li C, Zhang Y, et al. BCRP gene polymorphisms are associated with susceptibility and survival of diffuse large B-cell lymphoma. Carcinogenesis. 2007;28(8):1740-4 [DOI] [PubMed] [Google Scholar]
- 18.Radriguez-Antona C, Sayi JG, Gustafsson LL, Bertilsson L, Ingelman-Sundberg M. Phenotype-genotype variability in the human CYP3A locus as assessed by the probe drug quinine and analysis of variant CYP3A4 alleles. Biochem Biophys Res Comm. 2005;338(1):299-305 [DOI] [PubMed] [Google Scholar]
- 19.Hu YF, He J, Chen GL, Wang D, Liu ZQ, Zhang C, et al. CYP3A5*3 and CYP3A4*18 single nucleotide polymorphisms in a Chinese population. Clin Chim Acta. 2005; 353(1-2):187-92 [DOI] [PubMed] [Google Scholar]
- 20.Van Schaik RHN, van der Heiden IP, van den Anker JN, Lindemans J. CYP3A5 variant allele frequencies in Dutch Caucasians. Clin Chem. 2002;48(10):1668-71 [PubMed] [Google Scholar]
- 21.Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48(2):361-72 [PubMed] [Google Scholar]
- 22.Kantarjian HM, Giles F, Gattermann N, Bhalla K, Alimena G, Palandri F, et al. Nilotinib (formerly AMN107), a highly selective BCR-ABL tyrosine kinase inhibitor, is effective in patients with Philadelphia chromosome-positive chronic myelogenous leukemia in chronic phase following imatinib resistance and intolerance. Blood. 2007;110(10):3540-6 [DOI] [PubMed] [Google Scholar]
- 23.Hochhaus A, Kantarjian HM, Baccarani M, Lipton JH, Apperley J F,, Druker BJ, et al. Dasatinib induces notable hematologic and cytogenetic responses in chronic-phase chronic myeloid leukemia after failure of imatinib therapy. Blood. 2007;109(6):2303-9 [DOI] [PubMed] [Google Scholar]
- 24.Rosti G, Palandri F, Castagnetti F, Breccia M, Levato L, Gugliotta G, et al. Nilotinib for the frontline treatment of Ph(+) chronic myeloid leukemia. Blood. 2009;114(24): 4933-8 [DOI] [PubMed] [Google Scholar]
- 25.Cortes JE, Jones D, O'Brien S, Jabbour E, Konopleva M, Ferrajoli A, et al. Nilotinib as front-line treatment for patients with chronic myeloid leukemia in early chronic phase. J Clin Oncol. 2010;28(3):392-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kantarjian HM, Hochhaus A, Saglio G, De Souza C, Flinn IW, Stenke L, et al. Nilotinib versus imatinib for the treatment of patients with newly diagnosed chronic phase, Philadelphia chromosome-positive, chronic myeloid leukaemia: 24-month minimum follow-up of the phase 3 randomised ENESTnd trial. Lancet Oncol. 2011;12(9): 841-51 [DOI] [PubMed] [Google Scholar]
- 27.Kantarjian HM, Shah NP, Cortes JE, Baccarani M, Agarwal MB, Undurraga MS, et al. Dasatinib or imatinib in newly diagnosed chronic-phase chronic myeloid leukemia: 2-year follow-up from a randomized phase 3 trial (DASISION). Blood 2012; 119(5):1123-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White DL, Dang P, Engler J, frede A, Zrim S, Osborn M, et al. Functional activity of the OCT-1 protein is predictive of long-term outcome in patients with chronic-phase chronic myeloid leukemia treated with imatinib. J Clin Oncol. 2010;28(16):2761-7 [DOI] [PubMed] [Google Scholar]
- 29.White DL, Radich J, Soverini S, saunders VA, Frede A, Dang P, et al. Chronic phase chronic myeloid leukemia patients with low OCT-1 activity randomised to highdose imatinib achieve better responses, and lower failure rates, than those randomized to standard-dose. Haematologica. 2012;97(6):907-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dulucq S, Bouchet S, Turcq B, Lippert E, Etienne G, Reiffers J, et al. Multidrug resistance gene (MDR1) polymorphisms are associated with major molecular responses to standard-dose imatinib in chronic myeloid leukemia. Blood. 2008;112(5): 2024-7 [DOI] [PubMed] [Google Scholar]
- 31.Kim DH, Sriharsha L, Xu W, Kamel-Reid S, Liu X, Siminovitch K, et al. Clinical relevance of a pharmacogenetic approach using multiple candidate genes to predict response and resistance to imatinib therapy in chronic myeloid leukemia. Clin Cancer Res. 2009;15(14):4750-8 [DOI] [PubMed] [Google Scholar]
- 32.Ni LN, Li JY, Miao KR, Qiao C, Zhang SJ, Qiu HR, et al. Multidrug resistance gene (MDR1) polymorphisms correlate with imatinib response in chronic myeloid leukemia. Med Oncol. 2011;28(1):265-9 [DOI] [PubMed] [Google Scholar]
- 33.Maffioli M, Camós M, Gaya A, Hernández-Boluda Juan-Carlos, Álvarez-Larrán A, et al. Correlation between genetic polymorphims of the hOCT1 and MDR1 genes and the response to imatinib in patients newly diagnosed with chronicphase chronic myeloid leukemia. Leuk Res. 2011;35(8):1014-9 [DOI] [PubMed] [Google Scholar]
- 34.White DL, Saunders VA, Dang P, Engler J, Venables A, Zrim S, et al. Most CML patients who have a suboptimal response to imatinib have low OCT-1 activity: higher doses of imatinib may overcome the negative impact of low OCT-1 activity. Blood. 2007;110(12):4064-72 [DOI] [PubMed] [Google Scholar]
- 35.Hoffmeyer S, Burk O, von Richter O, Arnold H P,, Brockmöller J, Johne A, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci USA. 2000; 97(7):3473-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, et al. A “silent” polymorphisms in the MDR1 gene changes substrates specificity. Science. 2007;315(5811):525-8 [DOI] [PubMed] [Google Scholar]
- 37.Cohen MH, Williams G, Johnson JR, Duan J, Gobburu J, Rahman A, et al. Approval summary for imatinib mesylate capsules in the treatment of chronic myelogenous leukemia. Clin Cancer Res. 2002;8(5):935-42 [PubMed] [Google Scholar]
- 38.Frye RF, Fitzgerald SM, Lagattuta TF, Hruska MW, Egorin MJ. Effect of St John's wort on imatinib mesylate pharmacokinetics. Clin Pharmacol Ther. 2004;76(4):323-9 [DOI] [PubMed] [Google Scholar]
- 39.Peng B, Hayes M, Resta D, Racine-Poon A, Druker BJ, Talpaz M, et al. Pharmacokinetics and pharmacodynamics of imatinib in a phase I trial with chronic myeloid leukemia patients. J Clin Oncol. 2004;22(5):935-42 [DOI] [PubMed] [Google Scholar]
- 40.Gréen H, Skoglund K, Rommel F, Mirghani RA, Lotfi K. Cyp3A activity influences imatinib response in patients with chronic myeloid leukemia: a pilot study on in vivo CYP3A activity. Eur J Clin Parmacol. 2010; 66(4):383-6 [DOI] [PubMed] [Google Scholar]
- 41.Amirimani B, Walker AH, Weber BL, Rebbeck TP. Re: modification of clinical presentation of prostate tumors by a novel genetic variant in CYP3A4. J Natl Cancer Inst. 1999;91(18):1588-90 [DOI] [PubMed] [Google Scholar]
- 42.Amirimani B, Ning B, Deits AC, Weber BL, Kadlubar FF, Rebbeck TR. Increased transcriptional activity of the CYP3A4*1B promoter variant. Environ Mol Mutagen. 2003; 42(4):299-305 [DOI] [PubMed] [Google Scholar]
- 43.Larson RA, Druker BJ, Guilhot F, O'Brien SG, Riviere GJ, Krahnke T, et al. Imatinib pharmacokinetics and its correlation with response and safety in chronic-phase chronic myeloid leukemia: a subanalysis of the IRIS study. Blood. 2008;111(8):4022-8 [DOI] [PubMed] [Google Scholar]
- 44.le Coutre P, Kreuzer KA, Pursche S, Bonin M, Leopold T, Baskaynak G, et al. Pharmacokinetics and cellular uptake of imatinib and its main metabolite CGP74588. Cancer Chemother Pharmacol. 2004;53(4):313-23 [DOI] [PubMed] [Google Scholar]