Abstract
The symbiosis between marine bioluminescent Vibrio bacteria and the sepiolid squid Euprymna is a model for studying animal–bacterial Interactions. Vibrio symbionts native to particular Euprymna species are competitively dominant, capable of outcompeting foreign Vibrio strains from other Euprymna host species. Despite competitive dominance, secondary colonization events by invading nonnative Vibrio fischeri have occurred. Competitive dominance can be offset through superior nonnative numbers and advantage of early start host colonization by nonnatives, granting nonnative vibrios an opportunity to establish beachheads in foreign Euprymna hosts. Here, we show that nonnative V. fischeri are capable of rapid adaptation to novel sepiolid squid hosts by serially passaging V. fischeri JRM200 (native to Hawaiian Euprymna scolopes) lines through the novel Australian squid host E. tasmanica for 500 generations. These experiments were complemented by a temporal population genetics survey of V. fischeri, collected from E. tasmanica over a decade, which provided a perspective from the natural history of V. fischeri evolution over 15,000–20,000 generations in E. tasmanica. No symbiont anagenic evolution within squids was observed, as competitive dominance does not purge V. fischeri genetic diversity through time. Instead, abiotic factors affecting abundance of V. fischeri variants in the planktonic phase sustain temporal symbiont diversity, a property itself of ecological constraints imposed by V. fischeri host adaptation.
Keywords: Adaptation, experimental evolution, fitness, mutualism, sepiolid squid, Vibrio
The symbiosis between marine bioluminescent Vibrio fischeri and their sepiolid squid hosts (Cephalopoda: Sepiolidae) has revolutionized the study of animal–bacteria interactions, serving as a model for the last two decades, because both bacteria and squids can be maintained independently of each other in the laboratory (Nishiguchi 2000). Symbiotic bacteria can be grown in pure culture and the hosts raised axenically. Squid hatchlings emerge from their eggs with gnotobiotic light organs and subsequently are colonized within hours by free-living planktonic V. fischeri from seawater (McFall-Ngai and Ruby 1991; Nyholm and Nishiguchi 2008). Adult animals seed the water column via venting 90–95% of the bacteria at dawn, when the squid bury in sand. The remaining 5–10% of the symbionts grow throughout the day and reestablish a full light organ population during this time (Ruby 1996). Vibrio fischeri inhabit the mantle cavity of the squid in a specialized, complex, morphological structure termed the light organ (McFall-Ngai and Ruby 1991; Nyholm and Nishiguchi 2008). Research suggests squid utilize the bacterial bioluminescence for counter-illumination concealment during its nocturnal activity (Jones and Nishiguchi 2004), and squid aid the bacteria by providing a microhabitat rich in nutrients (Soto et al. 2009).
The sepiolid squid genera Euprymna and Sepiola reside in the Indo-West Pacific Ocean and Mediterranean Sea, respectively (Soto et al. 2009). Euprymna species distributions are allopatric, whereas those of Sepiola co-occur (Nesis 1982). Hybridization among sepiolid squid species is exceedingly rare (Zamborsky and Nishiguchi 2011). Vibrio symbionts indigenous to Euprymna species are host specialists and exhibit competitive dominance, outcompeting allochthonous isolates. This implies acquired tradeoffs during symbiont adaptation to regional Euprymna hosts, and these fitness costs manifest themselves when colonizing other sepiolid squids (Nishiguchi 2002; Elena and Lenski 2003). Vib-rio symbionts in Sepiola are host generalists and display no competitive dominance. Despite this competitive dominance of symbionts within Euprymna species, previous research suggests secondary colonization events have occurred, whereby nonnative Vibrio genotypes have invaded new geographical areas to ultimately become established and simultaneously displace native symbionts from their former Euprymna hosts (Jones et al. 2006).
Interestingly, V. fischeri also forms light organ mutualisms with monocentrid fishes, and these fish isolates do not colonize squid well (Nishiguchi 2002). Recent work demonstrated that change in a single regulatory gene (rscS) was sufficient to dramatically improve colonization capability (Mandel et al. 2009). Host switching is common in the natural history of Vibrionaceae symbioses, regardless of whether the interactions were mutualisms, host–pathogen relationships, or commensalisms. Additionally, obligately free-living V. fischeri strains exist as bacterioplankton community members, unable to colonize monocentrid fish or sepiolid squid to initiate light organ symbioses. This abandonment and reacquisition of the symbiont lifestyle from an obligately free-living one has occurred numerous times based on phylogenetic analysis of closely related Vibrio species (Nishiguchi and Nair 2003). Vibrio fischeri is capable of persisting as a native part of marine sediment and sand biofilm microflora, skin and gut commensals of marine animals, and attached to floating debris, particles, zooplankton, phytoplankton, and carrion (Nishiguchi and Jones 2004; Soto et al. 2010).
Previous work has suggested abiotic factors, microbial allelopathy, and social cooperative behavior can significantly impact growth and abundance of V. fischeri symbiont populations existing as part of the free-living bacterioplankton (Soto et al. 2009). For instance, V. fischeri symbionts from Euprymna differ more from each other than those of Sepiola in how they grow over abiotic factor gradients (e.g., salinity) spanning their fundamental niche breadth, including lower and upper growth limits. Hence, abiotic factors provide a mechanism that may allow non-native V. fischeri from foreign geographical areas to invade a particular Euprymna host range and extirpate native symbionts through selective amplification of nonnative V. fischeri numbers in the oceanic water column while in the free-living phase. Additionally, abiotic factors, allelopathy, and microbial sociality may facilitate the intrusion of nonnative symbionts into a region to the detriment of autochthonous ones by providing the advantage of an early start during squid host colonization (i.e., “running start” or “headstart” colonization; Soto et al. 2009). Moreover, evolution of host specificity by V. fischeri may be drastically influenced by founder effects (Soto et al. 2009; Wollenberg and Ruby 2009).
Consequently, competitively dominant V. fischeri, locally adapted and specialized to a definitive Euprymna squid species, could be outflanked by foreign, more maladapted and less fit isolates during host colonization. Phenomena such as environmental stress, fluctuating abiotic factors, microbial allelopathy, quorum-sensing systems, and tight genetic bottlenecks may grant nonnative V. fischeri an opportunity to establish footholds in novel Euprymna hosts, despite the continued presence of competitively dominant isolates (Nyholm and Nishiguchi 2008). From these host beachheads, exotic V. fischeri could potentially adapt to new and alien Euprymna light organ habitats (Soto et al. 2009). Therefore, to test the hypothesis that nonnative strains make headway against competitive dominance, host colonization experiments were implemented in Euprymna scolopes and E. tasmanica where a non-native V. fischeri strain was given a numerical or running start advantage over an endemic one. In addition, we tested the hypothesis that foreign V. fischeri were evolutionarily amenable to expeditious host shifts. V. fischeri ES114 from the Hawaiian bobtail squid (E. scolopes) was serially passaged through the novel host E. tasmanica, the Australian dumpling squid. The goal was to use experimental evolution to adapt nonnative symbiont V. fischeri ES114 from Hawaiian E. scolopes to Australian host E. tasmanica for 500 generations to increase the competitive ability of a derived nonnative symbiont relative to the ancestral clone (Soto et al. 2010). This approach gave the nonnative symbiont a chance to become adapted to the novel host.
Previous research suggests approximately 500–750 generations is sufficient time for bacteria to respond to selection in a novel environment (Lenski et al. 1991). In the following experiments, relative fitness is defined as the proportion of symbionts that have successfully colonized a squid light organ (Nishiguchi et al. 1998; Nishiguchi 2002). Animal colonization experiments with the nonnative lines, evolved in E. tasmanica, were conducted in the ancestral Hawaiian squid host E. scolopes to detect possible trade-offs. Also, the accompanying effect of host adaptation to E. tasmanica on V. fischeri microbial physiology was examined to determine the relationship between symbiosis evolution and bacterial growth response to an abiotic factor ecologically important to V. fischeri during the free-living phase (e.g., salinity), including under environmentally stressful conditions (Nyholm and Nishiguchi 2008; Soto et al. 2009, 2010). Thus, the growth of the derived nonnative lines along a sodium chloride (NaCl) gradient, spanning the entire saline niche breadth, was compared to the ancestral state.
To complement the study of V. fischeri in squid E. tasmanica for 500 generations using microbial experimental evolution, symbiotic V. fischeri were isolated over a time span close to a decade from E. tasmanica animals (a duration representing 15,000–20,000 generations of V. fischeri evolution in this squid host) from wild ocean populations inhabiting a single locality in Australia (Botany Bay, New South Wales). Spatial population genetics studies with V. fischeri in allopatric and sympatric squid host populations have revealed high levels of symbiont biodiversity (Boettcher and Ruby 1994; Kimbell et al. 2002; Jones et al. 2006; Zamborsky and Nishiguchi 2011). Because elevated levels of genetic diversity exist in the geographical landscape of V. fischeri symbionts, we examined whether this diversity was maintained over temporal scales. Haplotype network analysis was used to examine any fluxes that may be occurring in V. fischeri genetic diversity over the sampled time period. Published work in temporal population genetics of natural bacterial populations is scant relative to literature existing on spatial genetic structure (Ramette and Tiedje 2007). Some of these examples include Escherichia coli from human hosts over an 11-month period (Caugant et al. 1981) and a Burkholderia cepacia stream study over 32 days (Wise et al. 1996). This article is the first to simultaneously integrate both microbial experimental evolution and temporal population genetics, uniquely providing microevolution and macroevolution viewpoints for a marine bioluminescent bacteria-squid symbiosis.
Methods and Materials
IN VIVO ANIMAL EXPERIMENTS
Animal experiments were completed in a 12:12 h dark–light cycle at 25°C in 10-mL scintillation vials containing 5-mL artificial seawater (34 parts per thousand [ppt]). Fresh seawater changes were made every 12 h and accompanied with bioluminescence measurements on a TD-20/20 luminometer (Turner Scientific, Sunnyvale, CA). Axenic squid hatchlings were inoculated with 1 × 103 V. fischeri colony forming units (CFUs)/mL for monoculture and competitive colonization experiments. This cell density is sufficient to ensure squid hatchling inoculation with V. fischeri. After 3 h of inoculation, animals were rinsed with sterile seawater, allowing colonization to be synchronized within this window (McCann et al. 2003). When the animals were sacrificed, squid light organs were homogenized, serially diluted, and plated onto 70% seawater tryptone (SWT) agar plates for enumeration (Nishiguchi et al. 1998). Symbionts from squid light organ homogenates have nearly 100% plating efficiency (Ruby 1996).
“SUPERIOR NUMBERS” AND “HEADSTART” COLONIZATION
For “superior numbers” and “headstart” colonization experiments, V. fischeri strains ES114 (native to Hawaiian E. scolopes) and ET401 (native to E. tasmanica) were used (Nishiguchi et al. 1997). The two strains are genetically distinct (Nishiguchi and Nair 2003). Vibrio fischeri ES114 was the nonnative against V. fischeri ET401 in E. tasmanica, and V. fischeri ET401 was the nonnative to V. fischeri ES114 for the cross-experiments performed in E. scolopes. Numerical (2:1, 5:1, and 10:1, n = 20) and running start (12 h, n = 20) advantages were given to the foreign strain. A running start time of 12 h, along with the ratios 2:1, 5:1, and 10:1, were chosen because these nonnative advantages seemed ecologically realistic and form a reasonable simulation of what may happen in nature. Additionally, venting 95% of non-native symbionts inhabiting the squid light organ after the 12-h headstart provided the most optimal condition for the native strain to infiltrate, since this was when nonnatives were most susceptible and their numbers decimated. Initiation of headstart colonization began once animals hatched in the dark. Sterile (axenic) animals (n = 20) served as negative controls. Pure culture inoculations of squid with only native or nonnative symbionts were positive controls (n = 20). Animals inoculated 1:1 with native to nonnative V. fischeri were competitive dominance controls. For headstart colonization experiments, a 12-h running start advantage was also given to the native strain as an additional control. These experiments were conducted through 48 h, at which point animals were sacrificed (see section “In vivo animal experiments” for further details). Vibrio fischeri ET401 is visibly luminescent whereas ES114 is not, and their relative numbers from platings of light organ homogenates containing both can be determined by counting the number of luminescent or bright colonies in the dark (“ET401” colonies; Nishiguchi et al. 1997). Subtracting this previous number from the total colonies on the plate counted (ET401 + ES114) yields the number of “ES114” colonies. This method of strain discrimination and enumeration during competitive co-inoculation experiments of sepiolid squid hosts with symbionts has been published previously (Nishiguchi et al. 1997, 1998). No unusual interactions occur between V. fischeri ET401 and ES114 with regard to visible bioluminescence on seawater high nutrient agar plates, and this phenotypic difference is 100% stable between these two specific strains, exhibiting no phase variation since penetrance is complete (Nishiguchi et al. 1997, 1998). Additionally, there is virtually no mutation from one phenotype to another relating to visible luminescence in V. fischeri ET401 and ES114.
IN VIVO EXPERIMENTAL EVOLUTION
Experimental evolution was conducted by serially passaging V. fischeri JRM200 (isogenic to V. fischeri ES114, see below) through axenic E. tasmanica hatchlings (n = 24) for 96 h, when animals were sacrificed. Axenic animals remained uninoculated, serving as negative controls (n = 24). Light organ homogenate fractions were used to continuously serial passage V. fischeri JRM200 through hatchlings for additional 96-h rounds. Homogenates were frozen (20% glycerol final concentration) every five serial passages (25 total) and stored at −80°C to create a “frozen fossil record.” After every fifth serial passage, competitive colonization experiments were completed with derived V. fischeri JRM200 and ancestral ES114 in E. tasmanica (n = 24). This assay determines the degree of competitiveness between different strains of symbiotic bacteria (Nishiguchi 2002). The “more fit” competitor will be the symbiont strain that comprises a significantly higher proportion of the final light organ population at 96 h. Vibrio fischeri JRM200 is a chloramphenicol-resistant isogenic derivative of V. fischeri ES114 (chloramphenicol sensitive; McCann et al. 2003). The spontaneous mutation rate of chloramphenicol resistance was negligible in V. fischeri ES114 (~1 × 10−10 CFUs/mL) and no reversion of the marker phenotype to wild-type was observed in V. fischeri JRM200 (Soto 2009). Axenic animals (n = 24) served as negative controls. Positive control animals (n = 24) were exclusively infected with only one of two competitors. Animals were sacrificed, light organs homogenized at 96 h and plated on SWT agar (Ruby 1996). The proportion of light organ colonization by each competitor was then quantified by screening 100 random colonies for chloramphenicol resistance (McCann et al. 2003). Neutral marker status of chloramphenicol resistance in E. tasmanica was assessed similarly in two different experiments (n = 13, n = 20). Chloramphenical resistance has already been shown to be a neutral marker in E. scolopes (McCann et al. 2003). Competitive colonization experiments between V. fischeri JRM200 and ES114 were also executed in E. scolopes (Hawaiian host) over 48 h using the same methodology conducted in E. tasmanica (Australian host) for all 24 of the 500-generation derived lines (n = 24). Animal experiments with E. scolopes were not completed through 96 h because this squid species is more fragile and short lived than E. tasmanica.
EFFECTS OF SQUID HOST EVOLUTION ON V. FISCHERI GROWTH ALONG A SALINITY GRADIENT
Correlated responses of V. fischeri adaptation to novel host E. tasmanica on this marine microbe’s ability to grow along a wide salinity range were studied using an assay previously published (Soto et al. 2009). Individual colonies from SWT plates for the 24 derived lines, ancestor V. fischeri ES114, and unevolved V. fischeri JRM200 were used to inoculate 18 × 150 mm test tubes containing 5-mL SWT. These test tubes served as starter cultures for the experiment. Tubes were incubated at 28°C while shaking at 225 rpm for 16 h. Thereafter, 10 μl of each overnight starter culture was used to inoculate test tubes containing 5 mL of fresh SWT liquid media. The subsequent cultures were incubated at 28°C and shaken at 225 rpm for 3 h. After 3 h of growth, a Uvikon XL spectrophotometer (Bio Tek Instruments, Winooski, VT) was used to measure optical density (OD600) of all cultures. Cultures were then inoculated into test tubes containing 5-mL LBS with salinities spanning 0.0–7.0% NaCl. Vibrio fischeri does not grow above 7.0% NaCl LBS (Soto et al. 2009). All cultures began at the same initial cell density of 5 × 105 CFU/mL. NaCl concentrations were increased by 0.1% increments between 0.0% and 7.0%. Test tubes were placed in a shaker for 24 h at 28°C and 225 rpm. Optical density (OD600) readings of each culture were measured at each NaCl concentration (n = 5).
COLLECTION, ISOLATION, AND CULTURE METHODS FOR HAPLOTYPE NETWORK ANALYSIS
Details of specimen collection and sampling strategy of bacterial symbionts from squid hosts have been described previously (Jones et al. 2006). With the exception of 2001 and 2002, E. tasmanica specimens were collected along the eastern Australian (Botany Bay, New South Wales) coast between 2000 and 2009. Squid light organs from adult animals were dissected, rinsed with sterile 34 ppt artificial seawater, homogenized aseptically with a pestle, and serially diluted in sterile 34 ppt seawater, and plated (100 μl) onto SWT agar plates (Nealson 1978; Nishiguchi et al. 1998). Plates were incubated for 24–48 h at 28°C. Ten to 20 colonies per animal for each year were identified using 10 μg Diagnostic Discs with the vibriostatic agent 0129 (Oxoid, Basingstoke, Hampshire, England) on SWT agar plates following manufacturer’s instructions, tested for bioluminescence using a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA), and sequenced at the 16S rRNA locus for verification. Positive isolates were then streaked for isolation onto SWT agar plates and incubated for 24–48 h at 28°C. Single colonies from these plates were used to inoculate 5 mL of SWT liquid media in test tubes for a period of 12–18 h at 28°C at 200 rpm. These pure cultures were then frozen as stocks in a final concentration of 20% glycerol and kept at –80°C until needed.
DNA EXTRACTION, POLYMERASE CHAIN REACTION (PCR) AMPLIFICATION, AND SEQUENCING OF BACTERIAL ISOLATES
Frozen stocks of V. fischeri were recovered by streaking onto SWT agar plates and incubating for 24–48 h at 28°C. Single colonies were picked from these cultures and inoculated into 5 mL of SWT liquid media in test tubes, which were then incubated at 28°C while shaking at 200 rpm for 12–18 h. Bacterial cells were then harvested by centrifugation at 7500 rpm for 10 min. The resulting pellet was resuspended in 180 μl of Buffer ATL (Qiagen DNeasy Blood and Tissue extraction kit, Qiagen Sciences, MD). The bacterial cells were lysed by addition of 20 μl Proteinase K and heated at 56°C for 1–3 h while shaking at 80 rpm. Once lysed, the cells were washed and DNA was extracted according to total DNA bacterial isolation protocols using the Qiagen DNeasy Blood and Tissue extraction kit (Qiagen Sciences) with manufacturer’s recommendations for Gram-negative bacteria. The DNA was eluted and stored in 200 μl Buffer AE. The quality and concentration of the extracted DNA was determined using a nanodrop spectrophotometer (Thermo Scientific, Waltham, MA).
A total of 5–10 ng of purified DNA was used as a template in the PCR amplification of a ~350-bp fragment of the glyceraldehyde phosphate dehydrogenase subunit A locus (gapA) using the primers: gapAfischeriVFF (5′-TCGAATTGTTTCTAATGGGTCA-3′) and gapAVFR (5′-AGCGGCGCTTCAGTATAGTC-3′; Jones et al. 2006). PCR reactions to amplify the gapA locus from V. fischeri were performed as 25 μl reactions containing: 5–10 ng of extract DNA, 0.2 μM of both forward and reverse primers, 0.2 U of Taq polymerase (GoTaq® Flexi DNA polymerase, Promega, San Luis Obispo, CA), 2.5 mM of magnesium chloride (MgCl2), a final concentration of 200 μM of dGTP, dATP, dCTP, and dTTP (50 μM each), and 1 × buffer (Tris-HCl, KCl, and 0.1% Triton X-100). PCR reactions were completed in a MJ Research Dyad Disciple thermal cycler (Waltham, MA). Samples were amplified using the following PCR cycle: initial denaturation at 94°C for 5 min, followed by 30 cycles at 94°C for 15 sec, 55°C for 15 sec, and 72°C for 30 sec for 32 cycles with one final termination step at 72°C for 7 min. PCR products were cleaned using the Qiagen PCR purification kit (Qiagen Sciences, Valencia, CA).
The gapA locus was sequenced according to BigDye Terminator version 3.1 Sequencing kit protocol (Applied Biosystems, Foster City, CA). PCR products were combined in a presequencing reaction mixture containing 3–10 ng clean PCR product, forward or reverse primer (10 mM), Big Dye combined with SeqSaver (Sigma, St. Louis, MO) in a 1:1 dilution for a 20% final volume, sequencing buffer (1× final concentration), and PCR H2O for a final volume of 10 μl. Presequencing reactions were run in an MJ Research Dyad Disciple thermal cycler. The presequenced gapA product was spin column purified (Edge Biosystems, Gaithersburg, PA), and mixed with 3–5 μl Hi-Di Formamide prior to sequencing to bring the final reaction volume over the minimum of 10 μl. Samples were sequenced on a 3100 ABI capillary sequencer (Applied Biosystems). Independent forward and reverse sequences were subsequently combined and edited using Sequencher v 4.6 (Gene Codes™, Ann Arbor, MI), and compared against the NCBI database through the Basic Local Alignment Search Tool (BLAST) to confirm gene identity with V. fischeri ES114. Sequences were exported in FASTA format and aligned with Clustal W2 (Thompson et al. 1994). Analysis of the sequencedata for building haplotype networks was completed in Arlequin v3.1 (Excoffier et al., 2005) and then used in TCS 1.21 (Clement et al. 2000) to determine genetic variance between the haplotypes and to create a haplotype network, respectively.
DNASP AND TCS ANALYSES
To measure variation within and among populations, genotype data were imported in PHYLIP sequential format into the computer program DnaSP version 4.10.9 (Rozas and Rozas 1999). For each individual population (now defined as comprising all the bacterial isolates from a single year), Arlequin generated measurements of DNA divergence by computing haplotype diversity (Nei 1987). To quantify genetic variation among populations (e.g., 2000 vs. 2007), DnaSP computed Nei’s coefficient of gene variation (Gst), which varies between 0 and 1 (Nei 1973, 1975). Gst approaching 0 infers the majority of variation is found within populations, whereas Gst approaching 1 infers the majority of variation is found between populations. The program TCS v. 1.21 (Clement et al. 2000) was utilized to generate a haplotype network depicting symbiont genotype distribution through time for V. fischeri isolated from E. tasmanica inhabiting Botany Bay, Australia. TCS created the haplotype network from symbiont genotype data by telescoping sequences into haplotypes and calculating the frequency of the haplotypes using a statistical parsimony algorithm (Templeton et al. 1992). Under this method, the probability of parsimony is calculated until it exceeds 0.95. The maximum number of steps connecting parsimoniously two haplotypes is the number of mutational differences just before this 95% cutoff (Clement et al. 2000). The program was run using default settings with gaps treated as a fifth state. A haplotype network was created and displayed as a graph with separate circles representing separate haplotypes, the size of each circle representing the number of isolates in each haplotype, and dissimilar colors signifying different years.
Results and Discussion
“SUPERIOR NUMBERS” AND “HEADSTART” COLONIZATION
Earlier work has addressed conceivable mechanisms of how competitive dominance can be countered (Soto et al. 2009). For example, severe genetic bottlenecks and founder effects inherent during symbiont colonization may place an upper asymptotic limit on the advantage that indigenous V. fischeri can gain over nonnatives during host adaptation due to extensive genetic drift (Novella et al. 1995; Wollenberg and Ruby 2009). Gaining superior numbers or a headstart (Tables 1A, B, 2A, B) in host squid colonization by nonnative V. fischeri compared to native strains may also offset and outflank competitive dominance. The possibility remains that any native V. fischeri remaining in the light organ, after these initial advantages to nonnatives dissipate, will eventually outcompete inhabiting nonnatives and retake the squid host, especially if natives continue preferential host occupancy during repeated venting events. However, these data (Tables 1A, B, 2A, B) clearly demonstrate that nonnatives can make some headway against competitive dominance with superior numbers and the advantage of “running start” host colonization. Additionally, distinct morphological changes arise in Euprymna hatchlings during initial phases of symbiosis consequent to incipient colonization by Vibrio symbionts, and these dramatic physical changes make perpetual and successive serial recolonizations from free-living Vibrio cells less probable upon maturation of the association. This transformation enables invading nonnative Vibrio symbionts to retain inhabitance of foreign Euprymna, despite continued existence of competitive dominant strains in seawater. Salinity and temperature have also demonstrated striking and sudden effects on Vibrio symbionts isolated from Euprymna, which may lead to countervailing of competitive dominance (Soto et al. 2009). Remarkably, slight alterations in salinity (e.g., Δ0.1% NaCl≈Δ1 ppt) can lead to abrupt changes in microbial growth. For this reason, the strain predominating within a given sphere at a particular time can be kaleidoscopic, depending on the irregularity of salinity occurring in a regional area due to local water currents and seasonal changes. In some instances, precipitous saline demarcations exist, where a specific strain thrives and languishes, suggesting that gradual zones of decreased microbial growth are not always evident (Soto et al. 2009). This is especially true as the boundaries of saline tolerance are approached for V. fischeri. A single fluctuation in 0.1% NaCl or 1 ppt at the extreme ends of saline niche breath (lower and upper salinity limits of growth) can determine whether a certain V. fischeri isolate will grow or not. Hence, minute salinity deviations in the marine environment can profoundly affect host squid and V. fischeri symbiotic relationships by enabling nonnative V. fischeri, a superior numbers advantage (or a headstart) over native isolates during host squid colonization, even at moderate oceanic salinities typical of most marine waters (Soto et al. 2009). Incorporating temperature into this sodium chloride scheme only enriches the ecological complexity and the evolutionary dynamics for the sepiolid squid–V. fischeri mutualism.
Table 1.
(A) Colonization experiments with superior numbers of nonnative Vibrio fischeri ET401 (2:1, 5:1, and 10:1) over native V. fischeri ES114 in Euprymna scolopes. (B) Headstart advantage (12 h) of nonnative V. fischeri ET401 over native V. fischeri ES114. Total inoculum of both strains in artificial seawater is 1 × 103 CFUs/mL. Squid hatchlings were sacrificed at 48 h for enumeration of V. fischeri. Letters indicate significant differences using fixed-treatment analysis of variance (P < 0.05, α = 0.05).
| Treatment (n=20) | Percent Vibrio fischeri ET401 (nonnative) |
|---|---|
| (A) Nonnative superior numbers in Euprymna scolopes. | |
| Axenic squid | 0 |
| ES114 positive control | 0 |
| ET401 positive control | 100 |
| 1:1 ET401:ES114 | 2.7A |
| 2:1 ET401:ES114 | 11.9B |
| 5:1 ET401:ES114 | 42.6C |
| 10:1 ET401:ES114 | 53.8D |
| (B) Nonnative headstart in E. scolopes. | |
| Axenic squid | 0 |
| ES114 positive control | 0 |
| ET401 positive control | 100 |
| 1:1 ET401:ES114 | 6.3A |
| ES114+12 h ET401 | 0 |
| ET401+12 h ES114 | 80.3B |
Table 2.
(A) Colonization experiments with superior numbers of nonnative Vibrio fischeri ES114 (2:1, 5:1, 10:1) over native V. fischeri ET401 in Euprymna tasmanica. (B) Headstart advantage (12 h) of nonnative V. fischeri ES114 over native V. fischeri ET401. Total inoculum of both strains in artificial seawater is 1 × 103 CFUs/mL. Squid hatchlings were sacrificed at 48 h for enumeration of V. fischeri. Letters indicate significant differences using fixed-treatment analysis of variance (P < 0.05, α = 0.05).
| Treatment (n=20) | Percent Vibrio fischeri ES114 (nonnative) |
|---|---|
| (A) Nonnative superior numbers in Euprymna tasmanica. | |
| Axenic squid | 0 |
| ES114 positive control | 100 |
| ET401 positive control | 0 |
| 1:1 ES114:ET401 | 5.4A |
| 2:1 ES114:ET401 | 15.2B |
| 5:1 ES114:ET401 | 65.7C |
| 10:1 ES114:ET401 | 98.2D |
| (B) Nonnative headstart in E. tasmanica. | |
| Axenic squid | 0 |
| ES114 positive control | 100 |
| ET401 positive control | 0 |
| 1:1 ES114:ET401 | 7.6A |
| ET401+12 h ES114 | 0 |
| ES114+12 h ET401 | 100B |
MICROBIAL EXPERIMENTAL EVOLUTION
If advantage of superior numbers or headstart colonization allow nonnative V. fischeri to successfully establish an initial beachhead in novel animal hosts, the question is raised whether this new invasive population can adapt to these new hosts, setting the stage for a study with microbial experimental evolution. These experiments also address the extent to which niche-invasion mutations and host range expansion are possible within V. fischeri (Cohan 2002). For instance, do evolutionary transitions or mutations exist that would permit a nonnative symbiont to invade a novel host and displace a native symbiont population that is already established? We estimated that V. fischeri populations undergo ~10 generations during the first 12 h of growth and colonization inside juvenile squid light organs. Vibrio strains reach a carrying capacity of approximately 105–106 CFUs per E. scolopes hatchling (Ruby 1996) and up to 107 CFUs per juvenile in larger E. tasmanica. Thereafter, the symbiont light organ population passes through ~5 generations per day, as 5% of the population remains after expulsion (Ruby 1996). Over a four-day period (96 h), V. fischeri experiences 20–25 generations in the squid host. Independent empirical and theoretical work from other laboratories support these estimates (Schuster et al. 2010). To differentiate V. fischeri serially transferred through E. tasmanica from antecedent V. fischeri ES114, V. fischeri JRM200 (isogenic to V. fischeri ES114 and chloramphenicol resistant) was utilized for all experiments. Chloramphenicol resistance was a neutral marker and did not affect symbiosis competency in E. tasmanica (Table 3) nor in E. scolopes (McCann et al. 2003). OD600 versus logarithmic cell density curves were used to achieve ratios of V. fischeri ES114 and JRM200 at 50:50 in seawater for all colonization experiments. Results were confirmed with agar plate (viable cell) enumeration and live cell counts using a cytometer under light microscopy. The proportion of V. fischeri ES114 and JRM200 observed in E. tasmanica after 96 h was not significantly different from the inoculation ratios in artificial seawater.
Table 3.
Chloramphenicol resistance in Vibrio fischeri strain JRM200 during Euprymna tasmanica colonization (two-tailed t-test α = 0.05).
ns, not significantly different (P > 0.05).
Vibrio fischeri JRM200 was serially passaged (1 × 103 CFUs/mL artificial seawater inoculum) through E. tasmanica 25 times and demonstrated an ever increasing proportion of the symbiont light organ population relative to the ancestor at consecutive evolutionary time points in E. tasmanica. At 400 and 500 generations, this value was significantly different compared to the ancestor (Table 4). This increase reinforces previous population genetic surveys suggesting allochthonous V. fischeri invasions transpire and can become established, with concurrent displacement of native ones in different Euprymna species distributions (Jones et al. 2006). Therefore, V. fischeri certainly are evolutionarily fluid and capable of swift adaptation to new hosts. Host adaptation by V. fischeri JRM200 to E. tasmanica is reminiscent to the step model evolution that has been previously reported in E. coli experimental evolution studies (Table 4; Lenski et al. 1991). Relative to ancestral V. fischeri ES114, no significant differences in bioluminescence, growth rate in squid light organs, or symbiont population carrying capacity occurred in the novel squid host by V. fischeri JRM200 as a result of adapting to E. tasmanica.
Table 4.
Competitive colonization experiments between Vibrio fischeri strains ES114 (ancestor) and JRM200 (derived) at different evolutionary time points in the novel squid host Euprymna tasmanica.
| Evolutionary time point (Generations) | Expected ES114:JRM200 (Percentage ancestor: percentage evolved) |
Observed ES114:JRM200 (Percentage ancestor: percentage evolved) |
|---|---|---|
| 0 (n=33) | 50:50 | 46:54 |
| 100 (n=24) | 50:50 | 47:53 |
| 200 (n=24) | 50:50 | 41:59 |
| 300 (n=24) | 50:50 | 41:59 |
| 400 (n=24) | 50:50 | 35:651 |
| 500 (n=24) | 50:50 | 36:641 |
Significantly different two-tailed t-test and sign test (P < 0.05, α = 0.05).
Subsequently, we investigated possible correlated responses in the ancestral host E. scolopes as a result of 400 (n = 6) and 500 generations (n = 24) of symbiont evolution in E. tasmanica. No significant differences were observed in light organ carrying capacity between V. fischeri JRM200 and ES114 at 48 h in ancestral E. scolopes, either in competition or monoculture experiments. Derived V. fischeri JRM200 was consistently less bioluminescent than progenitor V. fischeri ES114 over 48 h of colonization in the ancestral host environment, and at 12, 24, and 36 h this disparity was significantly different at 400 generations (Table 5) and over the entire 48 h at 500 generations (Table 6). These observations are consistent with previous findings (Schuster et al. 2010) and are congruent with competitive dominance resulting from host specialization and local adaptation of symbionts to Euprymna species in the environment, with the simultaneous acquisition of antagonistic pleiotropy or mutation accumulation. These factors may subsequently lead to trade-offs in other host squid species (Elena and Lenski 2003). For example, a general tendency exists of V. fischeri isolated from Australian E. tasmanica to be visibly luminescent on seawater nutrient agar plates, whereas those from Hawaiian E. scolopes are nonvisibly luminescent (Nishiguchi et al. 1997). Previous results presume nonvisibly luminescent V. fischeri become brighter as a result of host evolution in E. tasmanica, yet this study advocates that increased dimness (e.g., secondary loss of luminosity) in E. scolopes is perhaps a more likely scenario.
Table 5.
Vibrio fischeri JRM200 (six randomly selected clones of 24) reduced bioluminescence in ancestral host Euprymna scolopes after 400 generations of evolution in novel host E. tasmanica (±SE). Standard error (SE) bars were calculated using the unbiased estimator for the mean.
| Hours postinoculation | Mean axenic squid bioluminescence relative light units (n=5) | Mean ES114 bioluminescence relative light units (n=6) | Mean JRM200 bioluminescence relative light units (n=6) |
|---|---|---|---|
| 0 | 0.041 (±0.006) | 0.033 (±0.005) | 0.031 (±0.007) |
| 12 | 0.042 (±0.005) | 2.8091 (±0.736) | 0.3951 (±0.161) |
| 24 | 0.034 (±0.007) | 13.941 (±3.667) | 6.5111 (±2.940) |
| 36 | 0.032 (±0.010) | 11.461 (±2.867) | 2.9661 (±0.990) |
| 48 | 0.037 (±0.004) | 4.212 (±1.275) | 3.703 (±1.188) |
Significantly different analysis of variance (P < 0.05, α = 0.05).
Table 6.
Vibrio fischeri JRM200 reduced bioluminescence in ancestral host Euprymna scolopes after 500 generations of evolution in novel host E. tasmanica (±SE). SE bars were calculated using the unbiased estimator for the mean. “Unevolved” refers to V. fischeri JRM200 before experiencing any serial transfers through E. tasmanica (0 generations), whereas “Evolved” signifies V. fischeri JRM200 after undergoing serial passage through E. tasmanica for 500 generations.
| Hours postinoculation | Mean axenic squid (Negative control) bioluminescence relative light units (n=24) | Mean ES114 (Ancestor) bioluminescence relative light units (n=24) | Mean JRM200 (Unevolved) bioluminescence relative light units (n=24) | Mean JRM200 (Evolved) bioluminescence relative light units (n=24) |
|---|---|---|---|---|
| 0 | 0.021 (±0.004) | 0.026 (±0.006) | 0.025 (±0.005) | 0.033 (±0.004) |
| 12 | 0.031 (±0.006) | 10.161 (±1.043) | 10.561 (±0.915) | 0.6891 (±0.079) |
| 24 | 0.021 (±0.005) | 38.811 (±3.070) | 36.811 (±3.450) | 15.441 (±0.668) |
| 36 | 0.027 (±0.006) | 11.561 (±1.343) | 11.581 (±1.286) | 6.9581 (±0.812) |
| 48 | 0.034 (±0.005) | 39.681 (±2.044) | 39.471 (±3.845) | 29.781 (±2.313) |
Significantly different analysis of variance (P < 0.05, α = 0.05).
CORRELATED RESPONSES ALONG SALINITY GRADIENTS AFTER 500 GENERATIONS IN E. TASMANICA
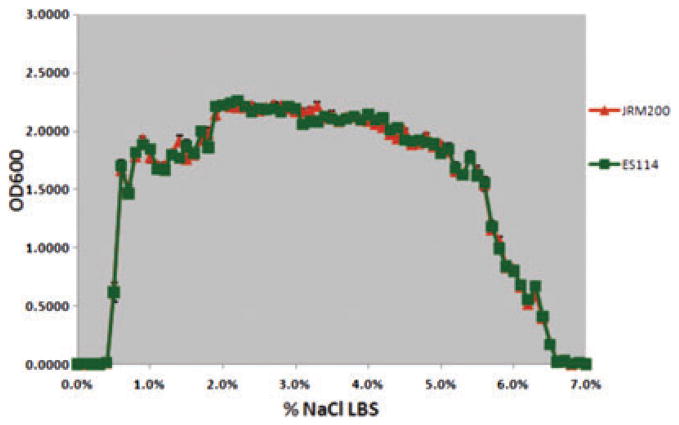
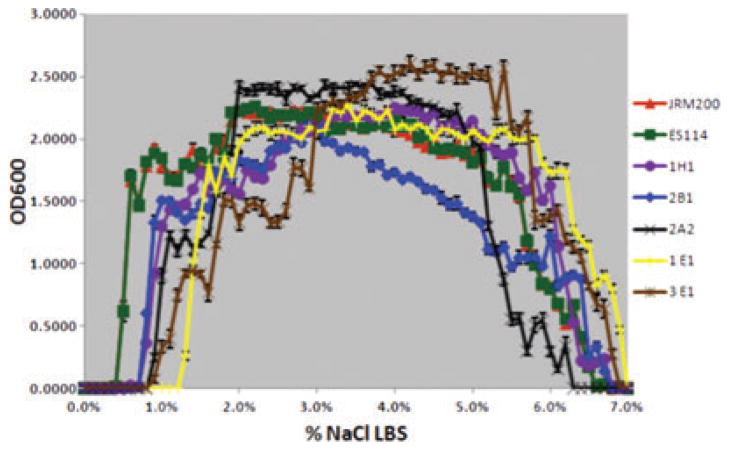
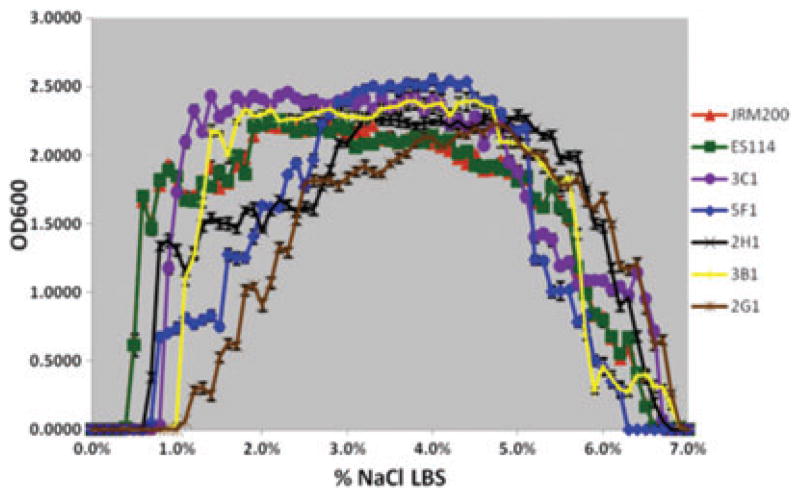
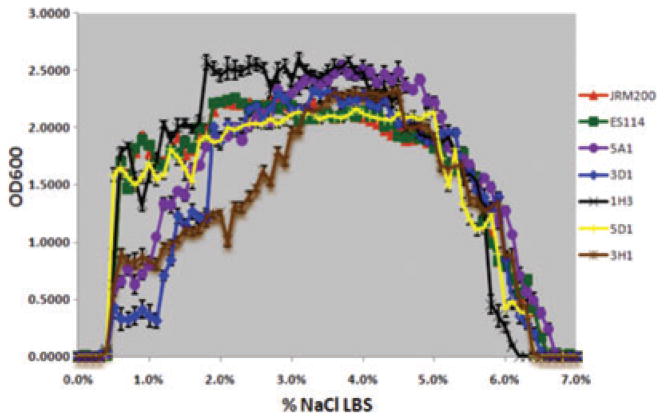
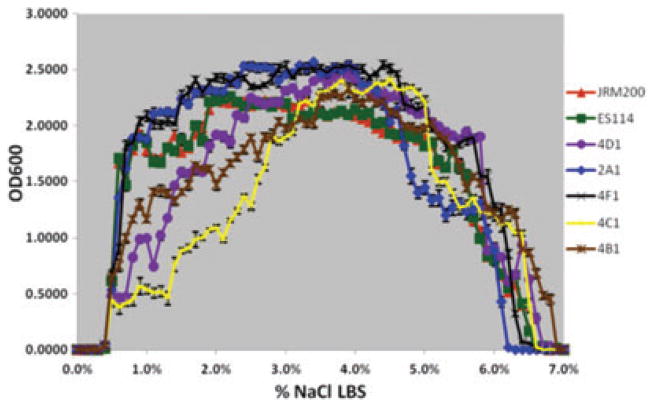
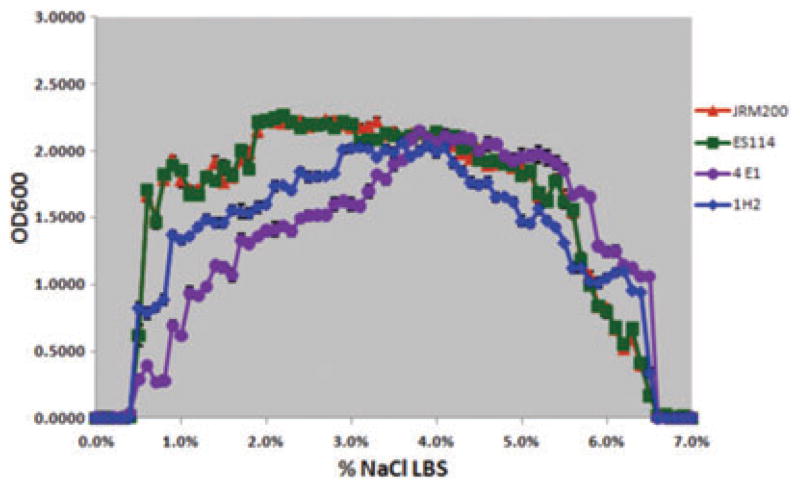
The ancestral and 24 V. fischeri lines serially passaged through E. tasmanica for 500 generations were grown along a salinity gradient to note correlated responses in this environment as a result of evolution in a novel squid host. Chloramphenicol resistance has no effect on V. fischeri growth along a salinity gradient, as V. fischeri ES114 and unevolved JRM200 were indistinguishable (Fig. 1). The ancestral lower and upper limits of microbial growth are 0.4% (SE = ±6.0×10−4) and 6.9% (SE = ±9.9×10−4) NaCl, respectively. Ten derived lines serially passaged in E. tasmanica for 500 generations exhibited a shift to the right in percent NaCl (increase), where microbial growth first occurred (minimum 0.01 OD600) along a salinity gradient relative to ancestral V. fischeri ES114 and unevolved JRM200 (Figs. 2, 3). Another 10 derived lines showed no shift where microbial growth first occurred but demonstrated a shift in percent NaCl where microbial growth last occurred (minimum 0.01 OD600) relative to the ancestor (Figs. 4, 5). The correlated responses of these 24 lines, as a result of undergoing novel host evolution in E. tasmanica, were the most common patterns observed. No single evolved line ever simultaneously expanded its lower and upper limit of growth relative to the ancestral osmolar niche as a result of host evolution. Apparently this was an evolutionary genetic or physiological constraint. Perhaps a trade-off exists in V. fischeri osmoregulation in its ability to grow at extremely low and high salinities. Two derived lines displayed no shift in percent NaCl where microbial growth initially or finally occurred relative to the ancestor (Fig. 6). However, these two lines still exhibited changes in growth abundance and biomass (i.e., amplitude) along the gradient—characteristics observed in all the derived lines. An additional two lines showed a shift to the left in percent NaCl (decrease) where microbial growth first occurred (0.2% NaCl, minimum 0.01 OD600) relative to the ancestral osmolar niche (Fig. 7); this was the rarest and most unique (8.3%) correlated response recorded. Presumably, acquiring the capacity to grow at ever lower salinities has physiological limitations (V. fischeri is not capable of growth in 0% NaCl culture media or distilled water).
Figure 1.
Growth curves of ancestral Vibrio fischeri ES114 and unevolved V. fischeri JRM200 along an increasing salinity gradient. No differences were observed between ancestral and unevolved strains used in the study.
Figure 2.
Five derived lines (1H1, 2B1, 2A2, 1E1, 3E1) serially passaged in Euprymna tasmanica for 500 generations and showing a shift to the right in percent NaCl where microbial growth first occurs (minimum 0.01 OD600) along a salinity gradient relative to ancestral Vibrio fischeri ES114 and unevolved JRM200.
Figure 3.
Five derived lines (3C1, 5F1, 2H1, 3B1, 2G1) serially passaged in Euprymna tasmanica for 500 generations exhibiting a shift to the right in percent NaCl where microbial growth first occurs (minimum 0.01 OD600) along a salinity gradient relative to ancestral Vibrio fischeri ES114 and unevolved JRM200.
Figure 4.
Five derived lines (5A1, 3D1, 1H3, 5D1, 3H1) serially passaged in Euprymna tasmanica for 500 generations and exhibiting a shift in percent NaCl where microbial growth last occurs (minimum 0.01 OD600) along a salinity gradient relative to ancestral Vibrio fischeri ES114 and unevolved JRM200.
Figure 5.
Five derived lines (4D1, 2A1, 4F1, 4C1, 4B1) serially passaged in Euprymna tasmanica for 500 generations and exhibiting a shift in percent NaCl where microbial growth last occurs (minimum 0.01 OD600) along a salinity gradient relative to ancestral Vibrio fischeri ES114 and unevolved JRM200.
Figure 6.
Two derived lines (4E1, 1H2) serially passaged in Euprymna tasmanica for 500 generations and exhibiting no shift in percent NaCl where microbial growth initially or finally occurs (minimum 0.01 OD600) along a salinity gradient relative to ancestral Vibrio fischeri ES114 and unevolved JRM200. However, both 4E1 and 1H2 still exhibited changes in growth abundance and biomass (i.e., amplitude) along the gradient.
Figure 7.
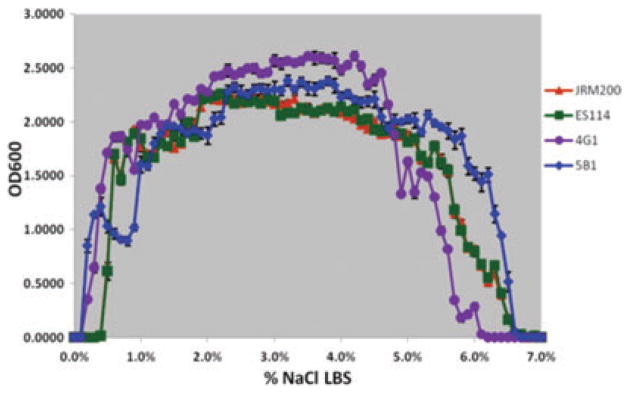
Two derived lines (4G1, 5B1) serially passaged in Euprymna tasmanica for 500 generations and exhibiting a shift to the left in percent NaCl where microbial growth first occurs (minimum 0.01 OD600) along a salinity gradient relative to ancestral Vibrio fischeri ES114 and unevolved JRM200.
Vibrio fischeri osmoregulation and osmolar niche breadth were clearly affected by evolution through a novel squid host. Although the ancestral osmolar niche breadth was considerably altered as a byproduct of adaptation to E. tasmancia, no obvious trends in the correlated responses of the derived lines were noted. Seemingly, the derived V. fischeri osmolar niche breadth has been “randomized” from the ancestral state, due to 500 generations of serial transfer in the Australian dumpling squid. Because the crypt spaces in the squid light organ are continuous with the mantle cavity, which itself is extracellular space that merges and is perpetually bathed with the ocean water environment through the siphon during squid ventilation, salinity within the chamber fluids of the squid light organ can only logically be considered as equal to that of the surrounding marine water. Possible complications include exudates or secretions by mucosal cells lining the crypts, and the existence of crypt microenvironments where limited or no mixing occurs. Adequate turbulent and laminar mixing are genuine concerns in batch cultures and bioreactors (Kresta and Brodkey 2004; Szalai et al. 2004), yet the roles of fluid blending and hydraulic principles play in shaping microcosm (squid light organ) salinities remain obscure and merit closer scrutiny. Undoubtedly, V. fischeri growth responses and physiological ranges of tolerances (i.e., niche breadths) to other abiotic factors have also been impacted. These results imply that bacterial stress responses are pleiotropically and epistatically influenced by host evolution. Previous studies in microbial experimental evolution investigating stress physiology has been substantial in demonstrating such effects in E. coli (Bennett and Lenski 1999; Bennett and Hughes 2009). Additionally, earlier work strongly suggested variable environments and symbiosis were correlated in their effects on V. fischeri microbial growth (Soto et al. 2009), providing evidence that stress on microbial physiology induced by abiotic factors and host colonization (e.g., immune defenses) may be coupled (Soto et al. 2010). How bacterial stress responses of host-associated prokaryotes react to challenges imposed by abiotic factors versus host immunity is poorly studied. For instance, do bacterial stress responses compartmentalize and specialize in how they maintain homeostasis against these two origins of stress, or is there cross-reactivity and pathway generalization? Future work is aimed at further phenotypic characterization and genome sequencing of the ancestral and derived V. fischeri lines to identify traits and loci underpinning adaptation to E. tasmanica.
TEMPORAL HAPLOTYPE NETWORK
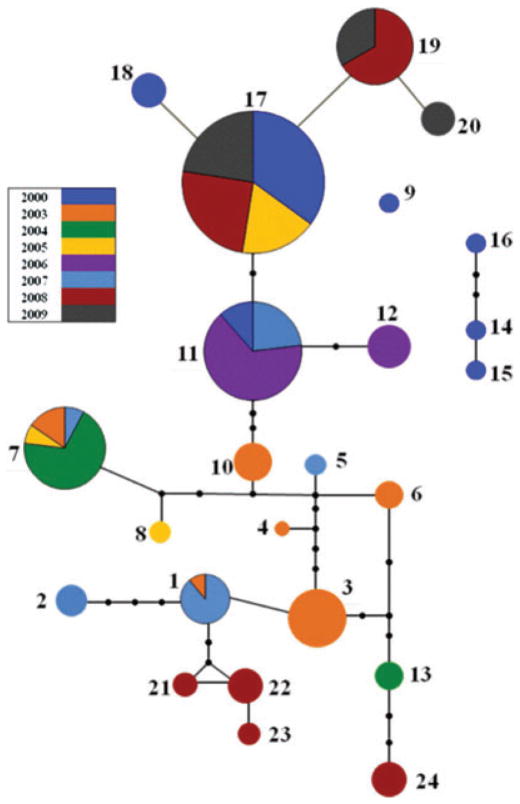
Variation produced by the in vivo experimental evolution study (artificial selection) among 24 lines was compared to wild V. fischeri isolates (natural history) procured from field-caught E. tasmanica specimens. The biodiversity of natural V. fischeri from Euprymna squid light organ populations was examined through a period of 10 years, a period of 15,000–20,000 generations in Vibrio symbiont evolution (Ruby and Asato 1993; Ruby 1996). The V. fischeri experimental evolution study performed inside a Euprymna squid host for 500 generations investigated microevolutionary processes (e.g., point mutations). Temporal analysis of diversity among symbiotic V. fischeri from E. tasmanica supplements experimental evolution studies by observing similar and other processes (e.g., horizontal gene transfer) at a macroevolutionary timescale, showing a haplotype network with extensive genetic diversity through time at the gapA locus (Fig. 8). Different colors in the haplotype network represent different years in which E. tasmanica were sampled, and the size of the circles signify number of isolates in each haplotype. A total of 24 different gapA haplotypes (alleles) were sampled through the years. Genetic distances between dissimilar haplotypes are represented by missing transitional forms, which are illustrated by black dots. For instance, Haplotypes 3 and 5 differ by six base changes, whereas Haplotypes 17 and 19 vary by one.
Figure 8.
Temporal haplotype network of Vibrio fischeri strains from the same Euprymna tasmanica population (Botany Bay NSW, Australia) over 10 years (2000–2009) using the gapA locus. Two major haplotypes have dominated during this time: haplotype 17, which has the largest sample size (represented by the size of the circle) over the entire span (11 years) and haplotype 11 (second largest representative haplotype). Other related haplotypes (i.e., 2, 8, 10, 20) were found only in one year, and were never recovered during the course of this study. Haplotypes that genetically were not connected to the rest of the network (9, 14, 15, 16) were all from the initial collection time point (2000), and were never recovered in subsequent years (Soto 2009).
Vibrio fischeri haplotypes colonizing E. tasmanica between the years 2000–2009 (except 2001 and 2002) can be considered as a rising and ebbing tide of biodiversity comprised by multiple variants that populate the squid hosts through successive years (Table 7). Annual diversity values (□k) have been corrected for uneven number of isolates and are directly comparable. Distinct haplotypes independently arose several times on separate occasions over a decade, signifying secondary colonization events are occurring. For example, Haplotype 17 was observed in 2000, 2005, 2008, and 2009, and Haplotype 11 is present in 2000, 2006, and 2007 (Fig. 8). Patterns of reemergence in E. tasmanica were not consistent for each haplotype; some haplotypes arose in successive years whereas others only recurred years later. Likewise, the prevalence of specific haplotypes at any given year varied greatly. No year existed where only a single haplotype was detected in E. tasmanica from the Botany Bay population. Peak diversity was recorded in 2003, whereas the minimum was in 2006 (Table 7). Closed loop V. fischeri Haplotypes 1, 21, and 22 represent unresolved ambiguities. Haplotypes 9, 14, 15, and 16 are 2000 V. fischeri haplotypes that were never resampled in subsequent years and are therefore disconnected to the main network. These haplotypes may represent locally extirpated, if not extinct, haplotypes or lineages lying in reservoirs that have yet to resurface. Populations of V. fischeri are vented daily from E. tasmanica, and these vented populations are potential inoculums for subsequent squid hatchlings. Therefore, it could be interpreted that the above patterns are indicative of expelled isolates thriving as opportunistic free-living bacteria colonizing squid hosts when environmental conditions favor their emergence. The gapA haplotypes most represented throughout the years (Haplotypes 7, 11, and 17) could represent environmental generalists. Haplotype 17 has also been discovered in a spatial population genetic survey of V. fischeri inhabiting light organs of E. scolopes, the Hawaiian bobtail squid (Jones et al. 2006), and may represent a V. fischeri generalist or biotype more able to shuttle reversibly between Hawaiian and Australian squid host distributions than other haplotypes (Soto 2009). The geographical range of E. scolopes relative to E. tasmanica is quite restricted in terms of inhabited area and range of environmental factors tolerated (Soto et al. 2009). For instance, E. scolopes is restricted to the coast of the Hawaiian archipelago, whereas E. tasmanica lives circum-continental to Australia. As a result, V. fischeri colonizing E. scolopes experience more constant habitats. Average annual temperature and salinity ranges for the marine realms of E. scolopes and E. tasmanica suggest that such spatial and temporal changes in salinity exist for both host habitats (Soto et al. 2009).
Table 7.
Vibrio fischeri haplotype diversity (□k value) sampled from Euprymna tasmanica cyclically increases and decreases through the years. □k values have been corrected for the different number of symbiont isolates per year.
| Year | □k value | No. of Haplotypes | No. of isolates |
|---|---|---|---|
| 2000 | 1.6635 | 7 | 23 |
| 2003 | 3.2005 | 6 | 15 |
| 2004 | 0.4297 | 2 | 10 |
| 2005 | 0.6898 | 3 | 22 |
| 2006 | 0.3190 | 2 | 20 |
| 2007 | 2.0074 | 5 | 17 |
| 2008 | 1.1521 | 6 | 22 |
| 2009 | 0.8909 | 3 | 13 |
| Combined | 4.8331 | 24 | 142 |
This work examining a decade of symbiont evolution associated with E. tasmanica is consistent with V. fischeri population genetics through space spanning diverse squid host species and oceans from across the globe (Kimbell et al. 2002; Jones et al. 2006; Zamborsky and Nishiguchi 2011), demonstrating similarity in V. fischeri population dynamics and diversity on both spatial and temporal scales. Whether oceanic currents or distributions of animal host metapopulations are chiefly responsible for V. fischeri dissemination in the South Pacific is unclear, as unknown Euprymna populations scattered amid Polynesian islands could be serving as stepping stone or corridor hosts. Sepiolid squids may be host habitat islands whose environmental complexity and heterogeneity (Travisano and Rainey 2000) cultivate and maintain Vibrio symbiont diversity. The V. fischeri fish host Cleidopus gloriamaris also co-occurs in Australian waters (Nishiguchi and Nair 2003), which may provide an additional source of symbiotic V. fischeri; however, prior work demonstrated that fish V. fischeri symbionts are quite different in their colonization capabilities than those found in sepiolid squid (Mandel et al. 2009). Vibrio fischeri haplotypes colonizing E. tasmanica are not continuously supplanted by V. fischeri haplotypes appearing in subsequent years. That is, there is an overall absence of anagenic symbiont evolution in sepiolid squid hosts and no direct evidence that V. fischeri fitness continuously improves in colonizing sepiolid squid hosts over time. Therefore, competitive dominance (Nishiguchi et al. 1998; Nishiguchi 2002) does not appear to sustain the same genetically distinct V. fischeri in Euprymna squid hosts over long evolutionary time periods, suggesting other determinants such as abiotic factors, ecological interactions between symbionts (competition, allelopathy, and social cooperative behavior), and founder effects have a strong influence in the sepiolid squid–Vibrio symbiosis (Nyholm and Nishiguchi 2008; Wollenberg and Ruby 2009). Host adaptation demonstrated by microbial experimental evolution does not transcend or upscale to long time intervals, however, experimental evolution does explain competitive dominance. Apparently, the same ecological processes fostering competitive dominance also nurture diversifying selection in V. fischeri populations in free-living and host-associated phases. Accordingly, competitive dominance does not purge V. fischeri haplotype diversity through time in the sepiolid squid–Vibrio symbiosis.
As Darwin did in his contrast between artificially selected pigeons and wild populations of rock doves to understand natural history and phenotypic variation of birds (Darwin 1859), contemplating genetic variation of experimentally evolved V. fischeri through a novel squid host to that of wild isolates procured from natural squid host populations can provide illumination to the evolution of these bioluminescent marine bacteria. To this end, the correlated responses documented in the salinity gradient by the derived lines from the experimental evolution study may enlighten why competitive dominance of native V. fischeri over nonnative isolates in Euprymna squid is an illusory barrier to secondary colonization. Vibrio fischeri do indeed adapt and specialize to the regional Euprymna species, yet this local host adaptation leads to “randomization” of the symbiont osmolar niche breadth (along with those of other abiotic factors). Thus, symbiotic V. fischeri vented back into the ocean by squid hosts every dawn are never able to optimize or trek their physiology to environmental conditions of the free-living phase, an especially important detriment to V. fischeri when environmental conditions themselves change. This phenomenon could account why no single V. fischeri haplotype predominates and persists through time in E. tasmanica. Periodic selection and selective sweeps may never be permitted in symbiotic V. fischeri, as specific symbiont haplotypes adapted to squid hosts of one generation may fail to consistently colonize hosts of the next generation due to the inability to thrive in the free-living phase under fluctuating environments. Instead alternate V. fischeri haplotypes (e.g., nonnatives) more numerous in the ocean under the particular circumstances are the genotypes to colonize the animal hosts. What V. fischeri haplotypes colonize Euprymna squid from one generation to next from the ocean may be a fortuitous or stochastic process. The existence of obligately free-living V. fischeri as bacterioplankton may be a stable evolutionary strategy that evades the physiological constraints imposed by squid host evolution, permitting local adaptation to abiotic factors of the free-living phase. Further mathematical elaboration of “symbiotic” versus “oceanic” V. fischeri ecotypes with game theory modeling is a reasonable next step.
Future work in spatial and temporal population genetics surveys of free-living bacterioplanktonic V. fischeri in the open ocean should be explored to determine if biodiversity differences exist between the free-living fraction and those colonizing light organ hosts. An important next step is plasmid population genetics and plasmid ecology, unaddressed since Boettcher and Ruby (1994). Examining symbiont plasmid diversity, possible coevolution with chromosomal loci, and how these attributes vary from bacterioplanktonic versus host-associated V. fischeri remain relatively unexplored topics in the sepiolid squid–Vibrio symbiosis. For microorganisms that alternate between free-living and host-affiliated phases (e.g., Rhizobium, V. fischeri), identifying sources of host shifts in associations between microbial symbionts and eukaryotic hosts, how these processes translate into patterns through space and time, and which mechanisms predominate at various scales are integral for more fully understanding interactions between hosts and their symbiotic microorganisms. Discerning the consequences of abiotic (salinity, temperature, environmental stress) versus host (immunity) factors on microbial ecology, biogeography, and diversity is necessary for coordinating all of these processes into a manageable scenario for microbial evolution. These perspectives of mutualisms have implications for medical microbiology and infectious disease, as many microbial pathogens of eukaryotic hosts (including humans) cycle between free-living and symbiotic phases.
Acknowledgments
Funding was provided by NSF-IOS0744498 and NIH-NIAID 1SC1A1081659–01 to MKN. W. Soto was supported by the NMSU RISE for the doctorate (NIH NIGMS R25GM061222). The authors would also like to thank M. McGrouther, P. Steinberg, N. Wilson, and G. Woolcott for help with squid collecting in Australia.
LITERATURE CITED
- Bennett AF, Hughes BS. Microbial experimental evolution. Am J Physiol Regul Integr Comp Physiol. 2009;297:17–25. doi: 10.1152/ajpregu.90562.2008. [DOI] [PubMed] [Google Scholar]
- Bennett AF, Lenski RE. Experimental evolution and its role in evolutionary physiology. Am Zool. 1999;39:346–362. [Google Scholar]
- Boettcher KJ, Ruby EG. Occurrence of plasmid DNA in the sepiolid squid symbiont Vibrio fischeri. Curr Microbiol. 1994;29:279–286. [Google Scholar]
- Caugant DA, Levin BR, Selander RK. Genetic diversity and temporal variation in the E. coli population of a human host. Genetics. 1981;98:467–490. doi: 10.1093/genetics/98.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Cohan FM. What are bacterial species? Annu Rev Microbiol. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin version 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Jones BW, Nishiguchi MK. Counterillumination in the bobtail squid, Euprymna scolopes (Mollusca: Cephalopoda) Mar Biol. 2004;144:1151–1155. [Google Scholar]
- Jones BW, Lopez JE, Huttenburg J, Nishiguchi MK. Population structure between environmentally transmitted vibrios and bobtail squids using nested clade analysis. Mol Ecol. 2006;15:4317–4329. doi: 10.1111/j.1365-294X.2006.03073.x. [DOI] [PubMed] [Google Scholar]
- Kimbell JR, McFall-Ngai MJ, Roderick GK. Two genetically distinct populations of bobtail squid, Euprymna scolopes, exist on the island of O’ahu. Pac Sci. 2002;56:347–355. [Google Scholar]
- Kresta SM, Brodkey RS. Turbulence in mixing applications. In: Paul EL, Atiemo-Obeng VA, Kresta SM, editors. Handbook of industrial mixing: science and practice. John Wiley & Sons, Inc; Hoboken, NJ: 2004. pp. 19–88. [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138:1315–1341. [Google Scholar]
- Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009;458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann J, Stabb EV, Millikan DS, Ruby EG. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl Environ Microbiol. 2003;69:5928–5934. doi: 10.1128/AEM.69.10.5928-5934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ, Ruby EG. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- Nealson KH. Isolation, identification, and manipulation of luminous bacteria. Meth Enzymol. 1978;57:153–165. [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proc Nat Acad Sci USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular population genetics and evolution. North Holland; Oxford, U.K: 1975. [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. Columbia Univ. Press; New York: 1987. [Google Scholar]
- Nesis KN. Cephalopods of the world. T.F.H; Neptune City, NJ: 1982. [Google Scholar]
- Nishiguchi MK. Temperature affects species distribution in symbiotic populations of Vibrio spp. Appl Environ Microbiol. 2000;66:3550–3555. doi: 10.1128/aem.66.8.3550-3555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi MK. Host recognition is responsible for symbiont composition in environmentally transmitted symbiosis. Microb Ecol. 2002;44:10–18. doi: 10.1007/BF03036870. [DOI] [PubMed] [Google Scholar]
- Nishiguchi MK, Jones BW. Microbial diversity within the Vibrionaceae. In: Seckbach J, editor. Origins, evolution, and biodiversity of microbial life. Cole-Kluwer Academic Publishers; Dordrecht, The Netherlands: 2004. pp. 531–548. [Google Scholar]
- Nishiguchi MK, V, Nair S. Evolution of symbiosis in the Vibrionaceae: a combined approach using molecules and physiology. Int J Syst Evol Microbiol. 2003;53:2019–2026. doi: 10.1099/ijs.0.02792-0. [DOI] [PubMed] [Google Scholar]
- Nishiguchi MK, Ruby EG, McFall-Ngai MJ. Phenotypic bioluminescence as an indicator of competitive dominance in the Euprymna-Vibrio symbiosis. In: Hastings JW, Kricka LJ, Stanley PE, editors. Bioluminescence and chemiluminescence. John Wiley and Sons; New York: 1997. pp. 123–126. [Google Scholar]
- Nishiguchi MK, Ruby EG, McFall-Ngai MJ. Competitive dominance during colonization is an indicator of coevolution in an animal-bacterial symbiosis. Appl Environ Microbiol. 1998;64:3209–3213. doi: 10.1128/aem.64.9.3209-3213.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novella I, Elena S, Moya A, Domingo E, Holland J. Size of genetic bottlenecks leading to virus fitness loss is determined by mean initial population fitness. J Virol. 1995;69:2869–2872. doi: 10.1128/jvi.69.5.2869-2872.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Nishiguchi MK. The evolutionary ecology of a sepiolid squid-Vibrio association: from cell to environment. Vie et Milieu. 2008;58:175–184. [PMC free article] [PubMed] [Google Scholar]
- Ramette A, Tiedje JM. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb Ecol. 2007;53:197–207. doi: 10.1007/s00248-005-5010-2. [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Ruby EG. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri–Euprymna scolopes light organ symbiosis. Annu Rev Microbiol. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- Ruby EG, Asato LM. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- Schuster BM, Perry LA, Cooper VS, Whistler CA. Breaking the language barrier: experimental evolution of non-native Vibrio fischeri in squid tailors luminescence to the host. Symbiosis. 2010;51:85–96. [Google Scholar]
- Soto W. Evolutionary ecology of symbionts within free-living and Host environments in the sepiolid squid-Vibrio symbiosis. Biology Department, New Mexico State University; Las Cruces: 2009. p. 208. [Google Scholar]
- Soto W, Gutierrez J, Remmenga MD, Nishiguchi MK. Salinity and temperature effects on physiological responses of Vibrio fischeri from diverse ecological niches. Microb Ecol. 2009;57:140–150. doi: 10.1007/s00248-008-9412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto W, Lostroh CP, Nishiguchi MK. Physiological responses to stress in the Vibrionaceae. In: Seckback J, Grube M, editors. Cooperation and stress in biology. Springer; New York, NY: 2010. pp. 1–16. [Google Scholar]
- Szalai ES, Alvarez MM, Muzzio FJ. Laminar mixing: a dynamical systems approach. In: Paul EL, Atiemo-Obeng VA, Kresta SM, editors. Handbook of industrial mixing: science and practice. John Wiley & Sons, Inc; Hoboken, NJ: 2004. pp. 89–144. [Google Scholar]
- Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travisano M, Rainey PB. Studies of adaptive radiation using model microbial systems. Am Nat. 2000;156:S35–S44. doi: 10.1086/303414. [DOI] [PubMed] [Google Scholar]
- Wise MG, McArthur JV, Wheat C, Shimkets LJ. Temporal variation in genetic diversity and structure of a lotic population of Burkholderia (Pseudomonas) cepacia. Appl Environ Microbiol. 1996;62:1558–1562. doi: 10.1128/aem.62.5.1558-1562.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg MS, Ruby EG. Population structure of Vibrio fischeri within the light organs of Euprymna scolopes from two Oahu populations. Appl Environ Microbiol. 2009;75:193–202. doi: 10.1128/AEM.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamborsky DJ, Nishiguchi MK. Phylogeographical patterns among sympatric populations of sepiolid squids and their Vibrio symbionts in the Mediterranean Sea. Appl Environ Microbiol. 2011;77:642–649. doi: 10.1128/AEM.02105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]