Background: Small molecule MEK1/2 inhibitors are designed to block ERK1/2-mediated signaling and inhibit proliferation.
Results: Novel instrumentation which measures cellular respiratory chain function found that four structurally distinct MEK1/2 inhibitors acutely affected mitochondrial bioenergetics.
Conclusion: The anti-mitochondrial effects of MEK1/2 inhibitors determined proliferative potential.
Significance: Impaired mitochondrial metabolism following MEK1/2 inhibition must be considered when using small molecule inhibitors to define this signaling pathway.
Keywords: ERK, MAP Kinases (MAPKs), Mitochondria, Proliferation, Respiratory Chain, MEK1/2, PD198306, PD98059, U0126, Complex I
Abstract
The Ras-MEK1/2-ERK1/2 kinase signaling pathway regulates proliferation, survival, and differentiation and, because it is often aberrant in tumors, is a popular target for small molecule inhibition. A novel metabolic analysis that measures the real-time oxidation state of NAD(H) and the hemes of the electron transport chain and oxygen consumption within intact, living cells found that structurally distinct MEK1/2 inhibitors had an immediate, dose-dependent effect on mitochondrial metabolism. The inhibitors U0126, MIIC and PD98059 caused NAD(H) reduction, heme oxidation, and decreased oxygen consumption, characteristic of complex I inhibition. PD198306, an orally active MEK1/2 inhibitor, acted as an uncoupler. Each MEK1/2 inhibitor depleted phosphorylated ERK1/2 and inhibited proliferation, but the most robust antiproliferative effects always correlated with the metabolic failure which followed mitochondrial inhibition rather than inhibition of MEK1/2. This warrants rethinking the role of ERK1/2 in proliferation and emphasizes the importance of mitochondrial function in this process.
Introduction
Most cancers develop autonomy from growth factor signaling often through mutations or changes in protein expression (1). The mitogen-activated protein kinase (MAPK) cascades have been identified as key regulators of cellular proliferation, differentiation, and survival, and one of these cascades, Raf-MEK1/2-ERK1/2, has been the target of many pharmaceutical endeavors (2–7). Processes as diverse as survival following oxidant injury, steroid biosynthesis, VEGF release, neuronal development, and cell cycle progression are believed to be regulated via this cascade where the end point obtained depends not only on the phosphorylation event but also on factors such as the duration and cellular location of the signal (8–15).
Because RAS structural alterations occur in nearly 30% of all cancers and BRAF mutations have been found in up to 60% of certain cancers, small molecule inhibitors of this signaling pathway have been widely developed and tested (10, 16, 17). Several MEK1/2 inhibitors including selumetinib, MEK162, GSK1120212, CI-1040, PD0325901, and XL518 have either been or are currently being evaluated in phase I/II clinical trials, and still others are in preclinical development (3–5). The survival of many myeloid leukemia cells, both in vitro and in vivo, depends on the activation of this MAPK pathway, and various MEK1/2 inhibitors have been used to successfully inhibit leukemic proliferation (18, 19). In this study we tested four MEK1/2 inhibitors in HL-60 myeloid leukemia cells which harbor an N-RAS mutation and demonstrate constitutive MAPK activation (18, 20).
The most effective preclinical compounds targeting the Raf-MEK1/2-ERK1/2 pathway are against MEK1/2. Because ERK1/2 are the only known substrates of MEK1/2, the proliferative inhibition and reduced survival seen following MEK1/2 inhibition are attributed to ERK1/2-mediated factors (4, 7). PD98059 and U0126 are the most popular preclinical MEK1/2 inhibitors used to study this pathway, and the results obtained with these compounds in cell culture have been used to justify the development of clinical inhibitors. Here we show that these structurally distinct MEK1/2 inhibitors and a newer inhibitor, MEK inhibitor I (MIIC),2 not only block ERK1/2 phosphorylation but also cause acute alterations of mitochondrial electron transport chain (ETC) function.
The ETC is composed of four protein complexes containing electron carriers embedded in the inner mitochondrial membrane and cytochrome c (Cytc) present in the intermembrane space. Complex III, Cytc, and cytochrome oxidase (complex IV) (CytOX) contain heme electron carriers which can either be oxidized (not carrying electrons) or reduced (carrying electrons), and their oxidation state can be measured with multiwavelength cell spectroscopy. Inhibition of the mitochondrial complexes results in an upstream reduction and a downstream oxidation of the electron carriers as well as decreasing oxygen consumption allowing multiwavelength cell spectroscopy, combined with NADH spectroscopy, to pinpoint the site of mitochondrial inhibition in living cells (21).
In this paper, we show that PD98059, U0126, and MIIC potently inhibit the ETC at complex I at concentrations routinely used in the literature for inhibition of MEK1/2, and we investigate the role of this inhibition on human leukemia cell proliferation. Furthermore, we show that PD198306, a newer and more potent MEK1/2 inhibitor, acts as a robust mitochondrial protonophore and uncouples oxidative phosphorylation at higher concentrations. In short, we show the strong mitochondrial effects of these compounds provide a new mechanism by which these inhibitors regulate cell proliferation.
EXPERIMENTAL PROCEDURES
Cell Culture
HL-60 cells were cultured at 37 °C in spinner flasks in phenol red-free RPMI 1640 medium (Invitrogen) containing antibiotic/antimycotic (Sigma) and 10% fetal bovine serum (Invitrogen) in a 95% air and 5% CO2 incubator. Cell density and viability were determined via trypan blue exclusion using the Countess® Automated Cell Counter (Invitrogen). Proliferation was defined as the increase in viable (trypan blue-excluding) cells over time compared with vehicle control-treated cells. This definition takes into account changes due to cell division, differentiation, and death.
Inhibitors
U0126 (C18H16N6S2) and PD198306 (C18H16F3IN2O2) were purchased from Tocris (Ellisville, MO). PD98059 (C16H13NO3) was from Promega. MEK inhibitor I designated MIIC (C21H18N4OS) was from EMD/Calbiochem. Rotenone and pyridaben were from Sigma. Rotenone was dissolved in ethanol. Pyridaben and the MEK1/2 inhibitors were dissolved in DMSO, and controls were treated with a correlating concentration of DMSO. Inhibitors were added as indicated in the figure legends.
Cell Spectroscopy and Analysis
Cells were spun down at 500 × g for 5 min and then resuspended in at a density of 2.0 × 107 cells/ml in RPMI 1640 medium and placed in a custom-built 5-ml chamber that consisted of a 17-mm inside-diameter quartz crucible embedded in an aluminum block maintained at 37.0 °C by a thermoelectric element. The oxygen concentration within the chamber was measured from the fluorescence lifetime of a phosphorescent membrane inserted through a 3-mm-diameter hole in the side of the crucible, and the top of the chamber was sealed with a stainless steel plunger. The stir bar was made of glass rather than Teflon, and all of the seals were made of Viton in accordance with good respirometry practice (22). The cells were oxygenated and deoxygenated under computer control by exchange of oxygen across 80 mm of oxygen-permeable silicone tubing immersed in the cell suspension using a feedback circuit to adjust the oxygen tension within the tubing to maintain constant oxygenation within the chamber; the tubing always contained 5% CO2 to maintain intracellular pH. Oxygen consumption was measured from the difference between the oxygen delivery to the cell suspension by the tubing and the rate change of the oxygen concentration of the cell suspension. The oxygen delivery was calculated from the oxygen gradient across the wall of tubing and the oxygen permeability of the tubing which was measured prior to each study.
Spectroscopy and Spectral Analysis
Heme attenuation spectra and NADH fluorescence spectra were measured with two separate CCD-spectrograph systems working in time-multiplexed mode at 50 Hz using a 6-ms on/4-ms off duty cycle. Contiguous spectra were averaged to give a temporal resolution of 0.5 s. A warm white light emitting diode (LED) was used for the attenuation spectra illumination which was mounted 10 mm below a bundle of three NA0.37 1-mm optical fibers. One fiber was used for attenuation spectra detection, one for fluorescence spectra detection and one was coupled to a 365-nm UV LED for fluorescence excitation. The two detection fibers were F-matched onto the slits of two 0.3-mm spectrographs (Triax 320; Horiba, Edison, NJ), each equipped with a 1024 × 128-pixel back-thinned CCD camera (DV401BV; Andor Technology, South Windsor, CT). The attenuation spectrograph was equipped with a 600 g/mm grating blazed at 500 nm, which provided complete spectra between 508 and 640 nm with a pixel bandpass of 0.16 nm. The slits were set to give a spectral resolution of 1 nm. The NADH fluorescence spectrograph was equipped with a 300 g/mm grating blazed at 500 nm, which provided complete spectra between 400 and 670 nm with a pixel bandpass of 0.33 nm. The slits were set to give a spectral resolution of 20 nm.
Heme oxidation changes were calculated by fitting a linear combination of model spectra to the change in attenuation spectrum (23) over the wavelength range 520–630 nm. The model spectra were: bH, bL, c1 hemes of complex III, Cytc, a605, a602, and a3 spectra from CytOx (23) and a quadratic background to account for any base-line drift. All model spectra were measured from the isolated enzymes except a602 (see Ref. 23). The b560 heme of complex II is not included in the model because it has a midpoint potential of −185mV (24) and so is unlikely to be reduced in the absence of exogenous reductants. The oxidized fraction of each heme (oxidation state) was calculated from the oxidation changes assuming full reduction during anoxia and full oxidation at high oxygen tension after addition of 1 μm complex I inhibitor rotenone.
NADH oxidation changes were calculated by fitting a linear combination of model spectra to the fluorescence intensity spectrum over the wavelength range 410–610 nm. The model spectra were the fluorescence spectrum of NADH, the fluorescence spectrum of the RPMI 1640 medium and a linear background. The NADH signal originates from both NADH and NADPH in both the cytosol and the mitochondria. The oxidation state of mitochondrial NADH was calculated from oxidation changes assuming full reduction of mitochondrial NADH during anoxia and full oxidation of mitochondrial NADH after 1 mm complex II inhibitor 3-nitropropionic acid, which inhibits the TCA cycle, and 1 μm protonophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP), assuming that NADPH and cytosolic NADH did not change with these interventions.
Quantitative Western Blotting
Aliquots containing of 2.0 × 106 cells were removed from the chamber and immediately combined with 500 μl of cold PBS, centrifuged at 4.3 × g for 90 s, and the resulting pellet was resuspended in Laemmli buffer heated to 75 °C and heated for 5 min. The proteins were separated on a 14% SDS-polyacrylamide gel (Novex) and transferred to an Immobilon P membrane. Membranes were blocked in Tris-buffered saline containing 0.05% Tween 20 (TBST) and 5% milk. Primary antibodies were diluted in TBST and 5% milk or BSA and exposed to the blot overnight at 4 °C. Primary antibodies ERK and phosphor-ERK (Thr-202, Tyr-204) were purchased from Cell Signaling. Blots were washed in TBST three times and exposed to fluorescent-conjugated secondary antibodies, mouse IgG IRDye800 (Rockland), and rabbit IgG Alexa Fluor 680, for 45 min. Blots were washed in TBST three times and once in TBS before being read on an Odyssey Imaging System (LiCor). Blots were analyzed and quantitated using custom software that corrects the fluorescence intensity of a protein band for the fluorescence intensity above and below the band. The amount of phosphorylated ERK1/2 fluorescence per total ERK1/2 fluorescence is expressed in the data, but it should be noted that this value is only proportional to the pERK1/2/total ERK1/2 due to differences in the efficiency of detection.
RESULTS
Dose-dependent Inhibition of Proliferation and Phosphorylation of ERK1/2 by Various MEK1/2 Inhibitors
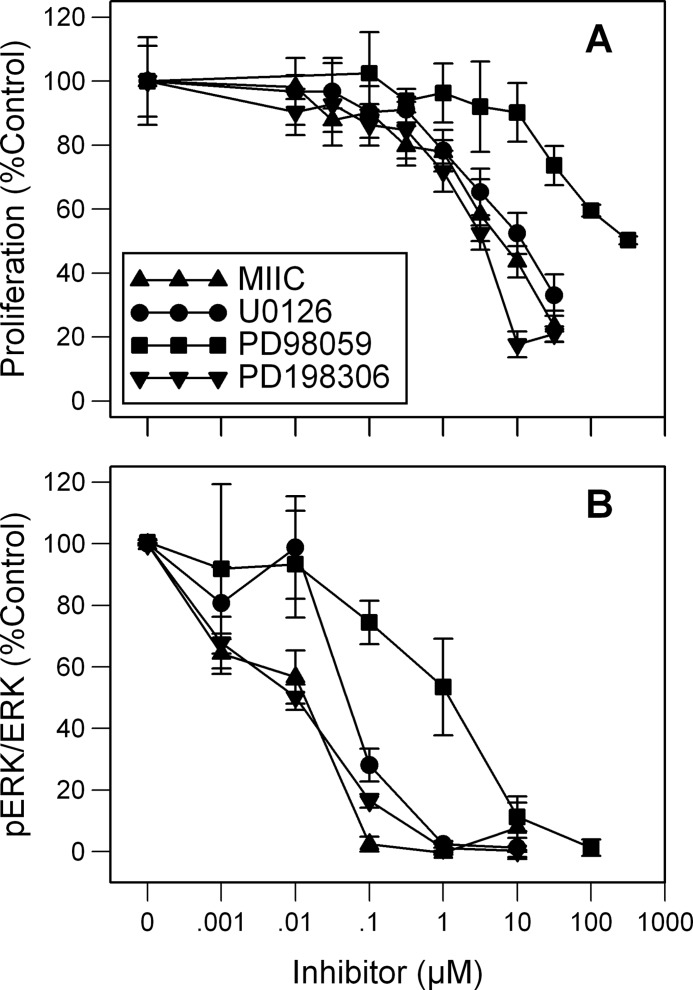
MEK inhibitors slowed the proliferation of HL-60 cells in a dose-dependent manner that varied with inhibitor (Fig. 1A). Exponentially growing HL-60 cells were treated with 1 nm-300 μm inhibitor for 48 h, and the number of viable cells was calculated and related to the proliferation of vehicle (DMSO)-treated cells. PD198306 showed a slightly stronger inhibition of proliferation per dose than MIIC and U0126. All three of these drugs were more potent at inhibiting proliferation than PD98059, a first generation MEK1/2 inhibitor. A 50% inhibition of proliferation was detected with 300 μm PD98059, the highest concentration tested (Fig. 1A). U0126 and MIIC, at 30 μm, inhibited the proliferation of HL-60 cells by 67 and 77%, respectively. Phosphorylation of ERK1/2, the only known substrates of MEK1/2, is the standard assay of MEK1/2 activity. To determine the MEK1/2 activity of inhibitor-treated HL-60 cells, total cell extracts were made 2 min after exposure to 1–100 μm inhibitor. Western analysis indicated that little or no pERK1/2 was found following 1 μm MIIC, U0126, and PD198306 and 100 μm PD98059 (Fig. 1B).
FIGURE 1.
Differential effects of MEK1/2 inhibitors on proliferation and pERK/ERK. A, dose response of MIIC, U0126, PD98059, and PD19830 on proliferation and phosphorylated ERK1/2 per ERK1/2 is shown. B, HL-60 cells were incubated with 1 nm--300 μm inhibitor for 48 h. The number of live cells was determined by trypan blue exclusion, and results are expressed as a ratio of vehicle-control-treated cell proliferation (A). Differences are significantly less than control (p < 0.05) at concentrations of 0.1 μm PD198306, 0.3 μm MIIC, 1 μm U0126, and 30 μm PD98059 and greater. Phosphorylated ERK1/2 per total ERK1/2 was determined using Western blot analysis of lysates taken 2 min after exposure to 1 nm--100 μm inhibitor. Results are expressed as mean ± S.D. (error bars; n = 3) and are significantly less than base line (p < 0.05) at concentrations of 0.001 μm PD198306 and MIIC, 0.01 μm U0126, and 1 μm PD98059 and greater.
Direct Effects of MEK1/2 Inhibitors on the ETC and Evidence for Complex I Inhibition
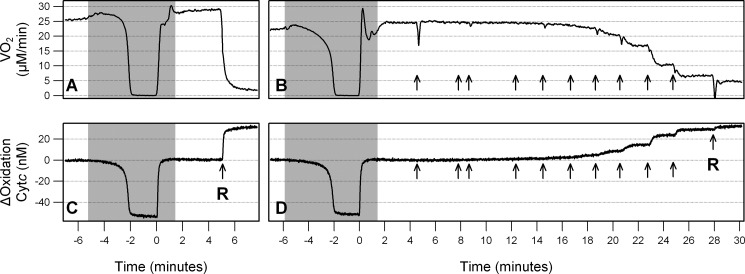
Multiwavelength cell spectroscopy was used to quantitate oxidation changes of the ETC hemes simultaneously with oxygen consumption (VO2). A typical study is shown in Fig. 2 where a downward deflection in Cytc denotes reduction and an upward deflection denotes oxidation. Oxygen concentration within the chamber was maintained at 100 μm (∼10% O2) except in the gray shaded area where the O2 concentration was altered under computer control. [O2] was decreased to zero (anoxia) and maintained there for 2 min then returned to 100 μm. Under anoxic conditions, electrons are not able to leave the chain, and all of the upstream redox centers, including Cytc, become fully reduced (Fig. 2, A and B). The addition of 1 μm rotenone, a complex I inhibitor, blocked entry of electrons into the chain, resulting in a complete oxidation of Cytc and blocked all mitochondrial VO2 (Fig. 2, A and B). The heme oxidation states were subsequently calculated assuming that the hemes were fully reduced and fully oxidized under anoxia and rotenone, respectively. The addition of U0126 indicated by the arrows, to final concentrations of 0.001, 0.003, 0.01, 0.03, 0.1, 0.3, 1, 3, 10, and 30 μm, resulted in a concentration-dependent decrease in VO2 and oxidation of Cytc (Fig. 2, B and D).
FIGURE 2.
U0126 acutely inhibits VO2 and Cytc oxidation in a concentration-dependent manner. Representative data were obtained from the multiwavelength cell spectroscopy system. Gray denotes areas where [O2] is <100 μm, and arrows show where inhibitor, either U0126 or 1 μm rotenone (R), was added. Oxygen consumption (VO2) is shown in A and B. Change in Cytc oxidation from base line is shown in C and D. A and C show a simple anoxia and rotenone study to fully reduce and fully oxidize the hemes. B and D show an anoxia, addition of U0126 to a final concentration of 0.001, 0.003, 0.01, 0.03, 0.1, 0.3, 1, 3, 10, and 30 μm, and finally rotenone.
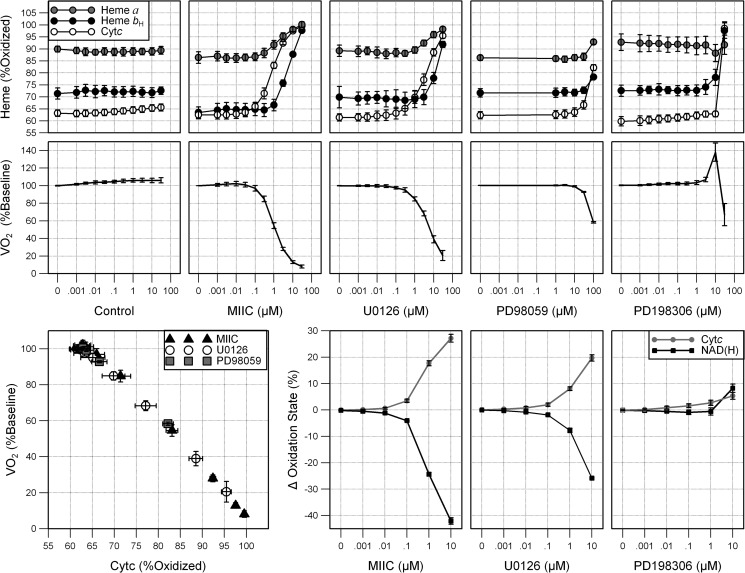
At base line 62 ± 1% of Cytc, 69 ± 4% of bH of the bc1 complex (complex III) and 89 ± 3% of CytOx were oxidized (Fig. 3, top). DMSO, given as a vehicle control, slightly increased VO2 but did not appreciably change the cytochrome oxidation state (Fig. 3, left). The addition of MIIC and U0126 caused a dose-dependent oxidation of Cytc, bH, and CytOx (Fig. 3, top). Cytc was 97.5 ± 0.3% and 88.5 ± 0.2% oxidized after 10 μm MIIC and U0126, respectively, and nearly fully oxidized after 30 μm. The bL and c1 hemes of the bc1 complex also oxidized in concert with the other hemes (data not shown). The oxidation was rapid and stabilized within 90 s (Fig. 2D). PD98059 also oxidized the hemes but required more drug, 30 μm and higher, to do so. PD198306, a newer generation MEK1/2 inhibitor, had little effect on the heme oxidation state until the highest concentration, 30 μm. A dose-dependent decrease in VO2 was noted following treatment with MIIC or U0126 (Fig. 3, bottom). Similarly, PD98059 decreased VO2 but required a greater concentration of drug. In contrast, PD198306 first increased VO2 at moderate concentrations (3 and 10 μm) and then decreased VO2 at the highest concentration (30 μm), where it also oxidized Cytc and bH. U0124, sold as a small molecule control for U0126 studies, had no effect VO2 or cytochrome oxidation state (data not shown).
FIGURE 3.
Concentration-dependent inhibition of ETC by structurally distinct MEK1/2 inhibitors. Oxidation state of heme bH of complex III, Cytc, and heme a of CytOx (upper panels), and oxygen consumption as a percentage of base line (lower panels) 2 min after treatment with 1 nm–30 μm MIIC, U0126, and PD198306, 1–100 μm PD98059, or equivalent amounts of vehicle-control (DMSO). Results are expressed as mean ± S.D. (error bars; n = 6). Cytc changes were significant (p < 0.005) at concentrations of 0.03 μm and higher for MIIC and U0126 (p < 0.05) at and above 0.01 μm PD198306 and 3 μm PD98059. VO2, CytOx, and CytbH changes were significant (p < 0.05), respectively, at the following concentrations: MIIC 0.3, 0.3, 1; U0126 0.1, 3, 10; PD98059 10, 100, 30; PD198306 0.03, 10, 10. Direct linear correlation between Cytc oxidation and VO2 was found for MIIC, U0126, and PD98059. Cytc oxidation and VO2 were measured 2 min after treatment with varying doses of inhibitor. The linear regression for each inhibitor was R2 > 0.99. Changes in Cytc oxidation state (upper panels) and NAD(H) oxidation state (lower panels) were measured following treatment with 0.001–10 μm MIIC, U0126, or PD198306. Measurements were taken 2 min after each dose. Results are expressed as mean ± S.D. (n = 3). p < 0.01 at concentrations of 1 μm U0126, 0.1 μm MIIC, and 10 μm PD198306 and higher.
A direct linear correlation, R>0.99, between Cytc oxidation and VO2 was found for MIIC, U0126, and PD98059 (Fig. 3, lower left). As each of these compounds was added Cytc oxidation increased in concert with a decrease in VO2. The results suggest that these inhibitors have the same mechanism of action on the mitochondria.
The oxidation of all ETC hemes along with inhibition of VO2 indicated that the MEK1/2 inhibitors were disrupting the ETC upstream of complex III. Similar to the hemes, NAD+ and NADH are a redox couple where NADH is the reduced form (carrying two electrons) and NAD+ is the oxidized form. To determine whether the inhibition was occurring at complex I or upstream of complex I, the oxidation state of NAD(H) was measured fluorometrically. Treatment with MIIC and U0126 resulted in a concentration-dependent reduction in NAD(H) which was significant starting at 0.1 μm and 1 μm (p < 0.005), respectively (Fig. 3, lower right). The effect of PD98059 could not be determined because the drug itself fluoresced. U0126 also fluoresced, but it could be spectrally resolved. PD198306 resulted in an oxidation in NAD(H) at 10 μm.
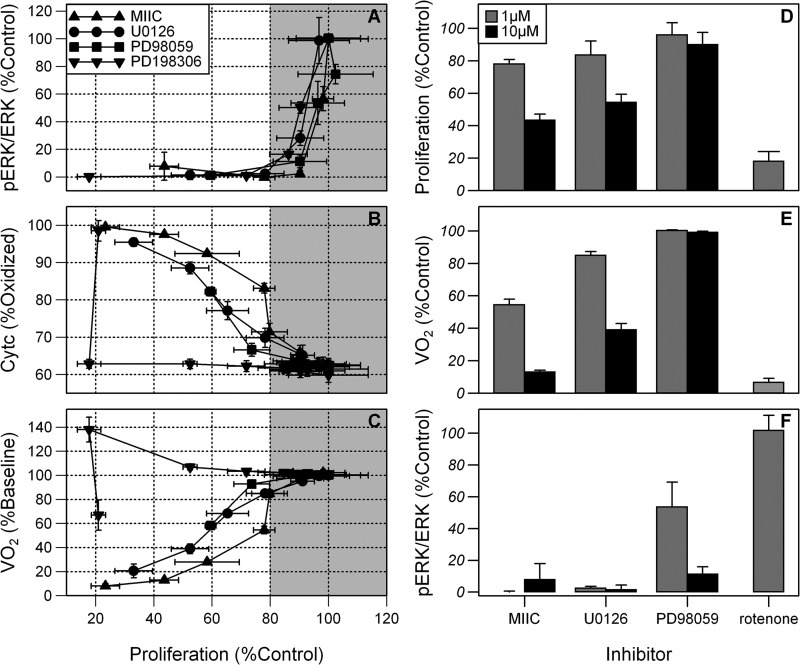
Complex I and Proliferative Inhibition Independent of MEK1/2 Inhibition
The concentration of each inhibitor required to reduce phosphorylation of ERK1/2 to ≤5% of base line produced only a 10–25% decrease in proliferation compared with vehicle-treated control cells at 48 h (Fig. 4A, gray section). Improved proliferative inhibition to 80% of control occurred at higher doses, with little or no further decrease in pERK1/2 levels. For the inhibitors MIIC, U0126, and PD98059 this improved proliferative inhibition strongly correlated with the oxidation of Cytc and a decrease in VO2 (Fig. 4, B and C). These changes are indicative of inhibition of the ETC. Similarly, PD198306 inhibits proliferation by just 25% when pERK1/2 is <5% of control. A greater proliferative inhibition was noted at higher doses, which did not decrease pERK1/2 levels further but did alter mitochondrial function.
FIGURE 4.
Antiproliferative effects of MEK1/2 inhibitors depend on mitochondrial inhibition. A–C, fraction of control proliferation versus the loss of pERK1/2/ERK1/2 (A), Cytc oxidation state (B), and VO2 (C) in HL-60 cells. Measurements of Cytc and VO2 were taken 2 min after each dose and are expressed as mean ± S.D. (error bars; n = 6) and cell lysates were obtained for pERK1/2 and ERK1/2 determination after 5 min and are expressed as mean ± S.D. (n = 3). MIIC, U0126, and PD198306 were used at concentrations of 0.001–30 μm and PD98059 at 0.001–100 μm. D–F, comparison of the effects of 1 μm and 10 μm MIIC, U0126, PD98059, and 1 μm rotenone on proliferation (D), VO2 (E), and pERK1/2/ERK1/2 expression (F) relative to control. Proliferation was measured at 48 h and VO2, and pERK1/2/ERK1/2 expression was measured at 2 and 5 min, respectively.
Rotenone, an established complex I inhibitor, caused no decrease in pERK1/2/ERK1/2 although there was significant inhibition of proliferation at 48 h to 18 ± 6% of vehicle-treated control cells, suggesting loss of pERK1/2 is not necessary for complex I inhibition (Fig. 4, D and F). As expected, 1 μm rotenone fully oxidized Cytc and inhibited VO2 to 6.2 ± 2.4% of control; the residual VO2 was nonmitochondrial in origin (Fig. 4E). Another known complex I inhibitor, pyridaben (1 μm), also oxidized Cytc, abolished mitochondrial VO2, and inhibited proliferation to 44 ± 4% of control (data not shown), showing that complex I inhibition robustly affects proliferation.
At 1 μm, MIIC and U0126 had only modest effects on proliferation despite lowering pERK1/2/ERK1/2 to <2.5% of control levels. This concentration caused partial inhibition of complex I as indicated by the intermediate inhibition of VO2 and oxidation of ETC cytochromes (Figs. 4E and 3). At a higher concentration of 10 μm, MIIC and U0126 further decreased VO2 and further inhibited proliferation but caused no detectable change in pERK1/2/ERK1/2 (Fig. 4, D–F), suggesting that the robust inhibition of proliferation is dependent on complex I inhibition. In contrast, the MEK1/2 inhibitor PD98059 at 10 μm inhibited pERK1/2/ERK1/2 to 11.3 ± 4.6% of control but had little effect on VO2, Cytc oxidation, or on proliferation, again suggesting that lowering pERK1/2/ERK1/2 alone has little effect on proliferation. Together these data support the requirement of complex I activity over pERK1/2 expression for maintenance of proliferation.
DISCUSSION
Using a new technology that provides a real-time depiction of mitochondrial metabolism in intact cells via measurements of NAD(H) oxidation state, heme oxidation states, and oxygen consumption, we found that several structurally distinct MEK1/2 inhibitors acutely and effectively altered mitochondrial metabolism. Three of the inhibitors tested, U0126, MIIC, and PD98059, had dose-dependent immediate effects on complex I of the ETC. Another MEK1/2 inhibitor, PD198306, acted acutely as a protonophore. As expected, all of the MEK1/2 inhibitors blocked phosphorylation of ERK1/2 in a dose-dependent manner, and all slowed the proliferation of HL-60 human leukemia cells. Notably, the depletion of pERK1/2 to levels <5% of control correlated with just a 10–25% decrease in proliferation compared with control. Further proliferative inhibition, down to 80% of control, occurred at higher doses of inhibitor and correlated strongly with the effects on the mitochondrial electron transport.
The immediate, dose-dependent reduction in NAD(H) and oxidation in heme bH of complex III, Cytc, and heme a of CytOx, as well as the corresponding decrease in VO2 caused by MIIC, U0126, and PD98059 are hallmarks of complex I inhibition. Such inhibition would explain the increase in the ADP:ATP ratio found in HEK293 cells treated with U0126 and PD98059 (25). U0126 also caused Cytc oxidation and inhibited VO2 in a normal mouse macrophage cell line, RAW264.7, and in two other human myeloid leukemia cell lines, ML-1 and AML-3 (data not shown), indicating that the inhibition of complex I is not specific to HL-60 cells. In addition, we found that U0126 oxidized Cytc and inhibited VO2 in isolated HL-60 mitochondria (data not shown), suggesting that U0126 acts to inhibit complex I independently of cytoplasmic MEK1/2. At least 60 different families of compounds have been found to inhibit complex I, highlighting its sensitivity to a vast variety of structurally unrelated natural and artificial compounds (26). The oxidation of Cytc in HL-60 cells was maintained for at least 2 days following a single dose of U0126 (data not shown), suggesting that the inhibition is prolonged.
It is possible that the structurally distinct inhibitors U0126, MIIC, and PD98059 bind directly to complex I. Alternatively, inhibition could be occurring through a signaling event. Such a signaling cascade would require a terminal signaling protein with access to complex I and therefore present in either the mitochondrial intermembrane space or the mitochondrial matrix. There have been several reports of the presence of ERK1/2 in mitochondria, but it is difficult to reconcile the mitochondrial location of ERK1/2 with the impermeability of the outer mitochondrial membrane to proteins lacking a mitochondrial import sequence (27–29). In any case, inhibition of complex I by MEK1/2 or ERK1/2 can be ruled out because low dose PD198306 resulted in the profound loss of pERK1/2, presumably indicating complete inhibition of MEK1/2 kinase activity, with no observable inhibition of complex I. Kinase inhibitors are notoriously nonspecific, and it remains possible that these compounds are targeting a kinase within the mitochondria whose physiological substrate is complex I. The mammalian complex I contains 45 subunits (30), and some of the subunits reportedly contain phosphorylation sites (31, 32) although consensus has not been reached as to their functionality and relevance (33).
Low concentrations of a protonophore decrease the membrane potential and stimulate respiration whereas high concentrations cause a decrease in respiration concomitant with a failure in the TCA cycle to reduce NADH (34). Our previous work with the protonophore CCCP has shown the Cytc oxidation state changes very little at low concentrations, where VO2 increases, but becomes highly oxidized at higher concentrations where VO2 decreases (35). PD198306 produced exactly this pattern of oxidation and electron flux changes consistent with it acting as a protonophore. Increases in VO2 can also arise if ATP consumption is increased, but the changes observed with PD198306 also occurred in the presence of the ATP synthase inhibitor oligomycin, indicating that ATP consumption is not driving the increased VO2 (data not shown). Similar to CCCP and another protonophore, carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), PD198306 contains a secondary amine group which, in this case, bridges two benzene rings. Loss of the proton on the amine group would lead to a negative charge that would be delocalized over the two benzene rings, thus explaining the protonophore activity at higher concentrations. It should be noted that PD198306 is a structural analog of the MEK1/2 inhibitors, CI-1040 and MEK162, used in clinical trials.
In summary, the role of the MAPK signaling pathway in cellular proliferation is well established, and the overexpression or activation of the upstream components of this pathway in certain cancers is undeniable; however, the use of MEK1/2-specific inhibitors to define the function of this pathway must be accepted with caution. As we have clearly shown, MEK1/2 inhibitors of diverse structure can alter mitochondrial metabolism in addition to depleting pERK1/2. Because the reduction of pERK1/2 to <5% of base line corresponded to only a 10–25% decrease in proliferation compared with control regardless of the inhibitor used and because the mitochondrial inhibition continued to correlate with enhanced proliferative inhibition, we propose a prominent role for mitochondrial-mediated proliferative control by these inhibitors. The concentrations used to obtain this level of proliferative inhibition were consistent with those published in the majority of literature related to these MEK1/2 inhibitors (36–41). Therefore, it is likely that many of the antiproliferative effects associated with these inhibitors are due to inhibition of mitochondrial function rather than strictly being mediated by the loss of pERK1/2.
This work was supported, in whole or in part, by National Institutes of Health Grants 5R21RR25803 and R01NS054298.
- MIIC
- MEK inhibitor I
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- Cytc
- cytochrome c
- CytOX
- cytochrome oxidase
- DMSO
- dimethyl sulfoxide
- ETC
- electron transport chain.
REFERENCES
- 1. Hanahan D., Weinberg R. A. (2000) The hallmarks of cancer. Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 2. Shaw R. J., Cantley L. C. (2006) Ras, PI(3)K and mTOR signalling control tumour cell growth. Nature 441, 424–430 [DOI] [PubMed] [Google Scholar]
- 3. Frémin C., Meloche S. (2010) From basic research to clinical development of MEK1/2 inhibitors for cancer therapy. J. Hematol. Oncol. 3, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roberts P. J., Der C. J. (2007) Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 26, 3291–3310 [DOI] [PubMed] [Google Scholar]
- 5. Sebolt-Leopold J. S. (2008) Advances in the development of cancer therapeutics directed against the RAS-mitogen-activated protein kinase pathway. Clin. Cancer Res. 14, 3651–3656 [DOI] [PubMed] [Google Scholar]
- 6. Pagès G., Lenormand P., L'Allemain G., Chambard J. C., Meloche S., Pouysségur J. (1993) Mitogen-activated protein kinases p42MAPK and p44 MAPK are required for fibroblast proliferation. Proc. Natl. Acad. Sci. U.S.A. 90, 8319–8323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Osborne J. K., Zaganjor E., Cobb M. H. (2012) Signal control through Raf: in sickness and in health. Cell Res. 22, 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paz C., Poderoso C., Maloberti P., Cornejo Maciel F., Mendez C., Poderoso J. J., Podestá E. J. (2009) Detection of a mitochondrial kinase complex that mediates PKA-MEK-ERK-dependent phosphorylation of mitochondrial proteins involved in the regulation of steroid biosynthesis. Methods Enzymol. 457, 169–192 [DOI] [PubMed] [Google Scholar]
- 9. Liu X., Yan S., Zhou T., Terada Y., Erikson R. L. (2004) The MAP kinase pathway is required for entry into mitosis and cell survival. Oncogene 23, 763–776 [DOI] [PubMed] [Google Scholar]
- 10. Steelman L. S., Franklin R. A., Abrams S. L., Chappell W., Kempf C. R., Bäsecke J., Stivala F., Donia M., Fagone P., Nicoletti F., Libra M., Ruvolo P., Ruvolo V., Evangelisti C., Martelli A. M., McCubrey J. A. (2011) Roles of the Ras/Raf/MEK/ERK pathway in leukemia therapy. Leukemia 25, 1080–1094 [DOI] [PubMed] [Google Scholar]
- 11. Berra E., Pagès G., Pouysségur J. (2000) MAP kinases and hypoxia in the control of VEGF expression. Cancer Metastasis Rev. 19, 139–145 [DOI] [PubMed] [Google Scholar]
- 12. Meloche S., Pouysségur J. (2007) The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1 to S phase transition. Oncogene 26, 3227–3239 [DOI] [PubMed] [Google Scholar]
- 13. Klesse L. J., Meyers K. A., Marshall C. J., Parada L. F. (1999) Nerve growth factor induces survival and differentiation through two distinct signaling cascades in PC12 cells. Oncogene 18, 2055–2068 [DOI] [PubMed] [Google Scholar]
- 14. Newbern J. M., Li X., Shoemaker S. E., Zhou J., Zhong J., Wu Y., Bonder D., Hollenback S., Coppola G., Geschwind D. H., Landreth G. E., Snider W. D. (2011) Specific functions for ERK/MAPK signaling during PNS development. Neuron 69, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guyton K. Z., Liu Y., Gorospe M., Xu Q., Holbrook N. J. (1996) Activation of mitogen-activated protein kinase by H2O2. Role in cell survival following oxidant injury. J. Biol. Chem. 271, 4138–4142 [DOI] [PubMed] [Google Scholar]
- 16. Davies H., Bignell G. R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M. J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B. A., Cooper C., Shipley J., Hargrave D., Pritchard-Jones K., Maitland N., Chenevix-Trench G., Riggins G. J., Bigner D. D., Palmieri G., Cossu A., Flanagan A., Nicholson A., Ho J. W., Leung S. Y., Yuen S. T., Weber B. L., Seigler H. F., Darrow T. L., Paterson H., Marais R., Marshall C. J., Wooster R., Stratton M. R., Futreal P. A. (2002) Mutations of the BRAF gene in human cancer. Nature 417, 949–954 [DOI] [PubMed] [Google Scholar]
- 17. Brose M. S., Volpe P., Feldman M., Kumar M., Rishi I., Gerrero R., Einhorn E., Herlyn M., Minna J., Nicholson A., Roth J. A., Albelda S. M., Davies H., Cox C., Brignell G., Stephens P., Futreal P. A., Wooster R., Stratton M. R., Weber B. L. (2002) BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 62, 6997–7000 [PubMed] [Google Scholar]
- 18. Morgan M. A., Dolp O., Reuter C. W. (2001) Cell-cycle-dependent activation of mitogen-activated protein kinase kinase (MEK1/2) in myeloid leukemia cell lines and induction of growth inhibition and apoptosis by inhibitors of RAS signaling. Blood 97, 1823–1834 [DOI] [PubMed] [Google Scholar]
- 19. Milella M., Kornblau S. M., Estrov Z., Carter B. Z., Lapillonne H., Harris D., Konopleva M., Zhao S., Estey E., Andreeff M. (2001) Therapeutic targeting of the MEK/MAPK signal transduction module in acute myeloid leukemia. J. Clin. Invest. 108, 851–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Collins S. J. (1987) The HL-60 promyelocytic leukemia cell line: proliferation, differentiation, and cellular oncogene expression. Blood 70, 1233–1244 [PubMed] [Google Scholar]
- 21. Hollis V. S., Palacios-Callender M., Springett R. J., Delpy D. T., Moncada S. (2003) Monitoring cytochrome redox changes in the mitochondria of intact cells using multi-wavelength visible light spectroscopy. Biochim. Biophys. Acta 1607, 191–202 [DOI] [PubMed] [Google Scholar]
- 22. Haller T., Ortner M., Gnaiger E. (1994) A respirometer for investigating oxidative cell metabolism: toward optimization of respiratory studies. Anal. Biochem. 218, 338–342 [DOI] [PubMed] [Google Scholar]
- 23. Kim N., Ripple M. O., Springett R. (2011) Spectral components of the α-band of cytochrome oxidase. Biochim. Biophys. Acta 1807, 779–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu L., Xu J. X., Haley P. E., Yu C. A. (1987) Properties of bovine heart mitochondrial cytochrome b560. J. Biol. Chem. 262, 1137–1143 [PubMed] [Google Scholar]
- 25. Dokladda K., Green K. A., Pan D. A., Hardie D. G. (2005) PD98059 and U0126 activate AMP-activated protein kinase by increasing the cellular AMP:ATP ratio and not via inhibition of the MAP kinase pathway. FEBS Lett. 579, 236–240 [DOI] [PubMed] [Google Scholar]
- 26. Degli Esposti M. (1998) Inhibitors of NADH-ubiquinone reductase: an overview. Biochim. Biophys. Acta 1364, 222–235 [DOI] [PubMed] [Google Scholar]
- 27. Wortzel I., Seger R. (2011) The ERK cascade: distinct functions within various subcellular organelles. Genes Cancer 2, 195–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nowak G., Clifton G. L., Godwin M. L., Bakajsova D. (2006) Activation of ERK1/2 pathway mediates oxidant-induced decreases in mitochondrial function in renal cells. Am. J. Physiol. Renal. Physiol. 291, F840–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Becker T., Böttinger L., Pfanner N. (2012) Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem. Sci. 37, 85–91 [DOI] [PubMed] [Google Scholar]
- 30. Efremov R. G., Baradaran R., Sazanov L. A. (2010) The architecture of respiratory complex I. Nature 465, 441–445 [DOI] [PubMed] [Google Scholar]
- 31. Papa S., Sardanelli A. M., Scacco S., Petruzzella V., Technikova-Dobrova Z., Vergari R., Signorile A. (2002) The NADH: ubiquinone oxidoreductase (complex I) of the mammalian respiratory chain and the cAMP cascade. J. Bioenerg. Biomembr. 34, 1–10 [DOI] [PubMed] [Google Scholar]
- 32. Chen R., Fearnley I. M., Peak-Chew S. Y., Walker J. E. (2004) The phosphorylation of subunits of complex I from bovine heart mitochondria. J. Biol. Chem. 279, 26036–26045 [DOI] [PubMed] [Google Scholar]
- 33. Schilling B., Bharath M. M. S., Row R. H., Murray J., Cusack M. P., Capaldi R. A., Freed C. R., Prasad K. N., Andersen J. K., Gibson B. W. (2005) Rapid purification and mass spectrometric characterization of mitochondrial NADH dehydrogenase (complex I) from rodent brain and a dopaminergic neuronal cell line. Mol. Cell. Proteomics. 4, 84–96 [DOI] [PubMed] [Google Scholar]
- 34. Steinlechner-Maran R., Eberl T., Kunc M., Margreiter R., Gnaiger E. (1996) Oxygen dependence of respiration in coupled and uncoupled endothelial cells. Am. J. Physiol. 271, C2053–2061 [DOI] [PubMed] [Google Scholar]
- 35. Kim N., Ripple M. O., Springett R. (2012) Measurement of the mitochondrial membrane potential and pH gradient from the redox Poise of the hemes of the bc1 complex. Biophys. J. 102, 1194–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ripple M. O., Kalmadi S., Eastman A. (2005) Inhibition of either phosphatidylinositol 3-kinase/Akt or the mitogen/extracellular-regulated kinase, MEK/ERK, signaling pathways suppress growth of breast cancer cell lines, but MEK/ERK signaling is critical for cell survival. Breast Cancer Res. Treat. 93, 177–188 [DOI] [PubMed] [Google Scholar]
- 37. DeSilva D. R., Jones E. A., Favata M. F., Jaffee B. D., Magolda R. L., Trzaskos J. M., Scherle P. A. (1998) Inhibition of mitogen-activated protein kinase kinase blocks T cell proliferation but does not induce or prevent anergy. J. Immunol. 160, 4175–4181 [PubMed] [Google Scholar]
- 38. Bost F., McKay R., Dean N., Mercola D. (1997) The JUN kinase/stress-activated protein kinase pathway is required for epidermal growth factor stimulation of growth of human A549 lung carcinoma cells. J. Biol. Chem. 272, 33422–33429 [DOI] [PubMed] [Google Scholar]
- 39. Hotokezaka H., Sakai E., Kanaoka K., Saito K., Matsuo K., Kitaura H., Yoshida N., Nakayama K. (2002) U0126 and PD98059, specific inhibitors of MEK, accelerate differentiation of RAW264.7 cells into osteoclast-like cells. J. Biol. Chem. 277, 47366–47372 [DOI] [PubMed] [Google Scholar]
- 40. Gysin S., Lee S. H., Dean N. M., McMahon M. (2005) Pharmacologic inhibition of RAF→MEK→ERK signaling elicits pancreatic cancer cell cycle arrest through induced expression of p27Kip1. Cancer Res. 65, 4870–4880 [DOI] [PubMed] [Google Scholar]
- 41. Beloueche-Babari M., Jackson L. E., Al-Saffar N. M., Workman P., Leach M. O., Ronen S. M. (2005) Magnetic resonance spectroscopy monitoring of mitogen-activated protein kinase signaling inhibition. Cancer Res. 65, 3356–3363 [DOI] [PubMed] [Google Scholar]