Background: The p12 subunit of DNA polymerase δ is degraded in response DNA damage.
Results: RNF8 was identified as a ubiquitin ligase that targets p12 for degradation.
Conclusion: RNF8 is involved in the DNA damage-induced degradation of p12 in vivo.
Significance: RNF8 integrates signaling for p12 destruction with the cellular processes of DNA repair and translesion synthesis.
Keywords: DNA damage response, DNA polymerase, DNA repair, DNA replication, Protein degradation, Ubiquitin ligase, Ubiquitination, RNF8
Abstract
DNA polymerase δ consists of four subunits, one of which, p12, is degraded in response to DNA damage through the ubiquitin-proteasome pathway. However, the identities of the ubiquitin ligase(s) that are responsible for the proximal biochemical events in triggering proteasomal degradation of p12 are unknown. We employed a classical approach to identifying a ubiquitin ligase that is involved in p12 degradation. Using UbcH5c as ubiquitin-conjugating enzyme, a ubiquitin ligase activity that polyubiquitinates p12 was purified from HeLa cells. Proteomic analysis revealed that RNF8, a RING finger ubiquitin ligase that plays an important role in the DNA damage response, was the only ubiquitin ligase present in the purified preparation. In vivo, DNA damage-induced p12 degradation was significantly reduced by shRNA knockdown of RNF8 in cultured human cells and in RNF8−/− mouse epithelial cells. These studies provide the first identification of a ubiquitin ligase activity that is involved in the DNA damage-induced destruction of p12. The identification of RNF8 allows new insights into the integration of the control of p12 degradation by different DNA damage signaling pathways.
Introduction
DNA polymerase δ plays a key role in eukaryotic DNA replication and is also involved in gap-filling in DNA repair processes (1, 2). Pol δ2 was originally identified in rabbit reticulocytes (3) and was first purified from calf thymus (4, 5). Calf thymus and human Pol δ are the best studied examples of Pol δ in higher eukaryotes, whereas yeast Pol δ has been extensively studied in lower eukaryotes (1). Human Pol δ is a heterotetramer (Pol δ4) composed of p125 (the catalytic subunit), p50, p68, and p12 and has been extensively studied as the reconstituted complex expressed in insect cells (2, 6–8). Recent work has revealed that human Pol δ4 is tightly regulated in response to DNA damage and during cell cycle progression (Ref. 9), and reviewed in Ref. 2). The p12 subunit of Pol δ is degraded in response to UV, alkylating agents, and replication stress under the control of ATR, resulting in the conversion of Pol δ4 to Pol δ3, the heterotrimer lacking p12 (10). There is emerging evidence that the switchover to Pol δ3 in response to genotoxic agents serves a genoprotective role. Pol δ3 has altered properties that endow it with a greater level of proofreading activity (11, 12). In addition, Pol δ3 is likely to be the primary form of Pol δ activity present after DNA damage so that it is the form involved in gap-filling reactions during DNA repair and also during translesion synthesis in S phase cells (9).
The degradation of p12 is dependent on an intact ubiquitination system (10), but the proximal ubiquitination enzymes that are responsible for targeting p12 for destruction have not been identified. In this study, we provide evidence that the RING finger ubiquitin ligase, RNF8, participates in the regulation of p12. RNF8 was discovered over a decade ago (13, 14) and was shown to be able to cooperate with E2s that could catalyze the formation of both Lys-48-linked and Lys-63-linked ubiquitin chains (15). RNF8 has an FHA domain near its N terminus and a RING finger domain at its C terminus; this domain structure is shared with the mitotic checkpoint regulator CHFR (checkpoint with forkhead and ring finger) (16).
RNF8 plays a prominent role in the assembly of the DNA damage response (DDR) complexes at double-stranded DNA breaks induced by IR (17–21). RNF8 is recruited to MDC1, which is phosphorylated by ATM via its FHA domain (22). The ubiquitination of histones H2A and H2AX leads to the recruitment of 53BP1 (p53-binding protein 1) and BRCA1 (breast cancer susceptibility protein 1) to the damage sites and plays a key role in coordinating the DNA damage response in the assembly of repair factors and chromatin remodeling (23–25). The Lys-63-linked polyubiquitination of histones by RNF8 also requires the action of the downstream ubiquitin ligase, RNF168, which extends and maintains the ubiquitination of H2A/X histones (26). In addition to its recruitment to IR-induced foci (IRIF), RNF8 is also recruited to sites of UV damage in a cell cycle-independent manner following ATR and MDC1 recruitment to NER generated ssDNA (27). Thus, RNF8 is recruited via independent pathways to IRIF and NER repair foci to induce histone ubiquitination. RNF8 is also capable of mono- and polyubiquitination of PCNA, implicating it as a factor in the activation of translesion synthesis (28). RNF8 interacts with Class III E2s (14) and with UbcH5c to generate Lys-48-linked polyubiquitin chains as well as with UBC13 to produce Lys-63-linked polyubiquitin chains (15, 28, 29). Recent studies support a role for both Lys-48-linked and Lys-63-linked ubiquitination of histones in the DDR (30).
In this study, we provide evidence that RNF8 is involved in targeting of the p12 subunit of Pol δ for degradation in response to DNA damage. We discuss how these findings place the regulation of p12 degradation within the current framework of the DNA damage signaling pathways of the DDR, NER, and the intra-S phase checkpoint.
EXPERIMENTAL PROCEDURES
Proteins and Antibodies
Recombinant human proteins (E1 ubiquitin-activating enzyme, ubiquitin, ubiquitin K48R, ubiquitin K63R, UbcH5c, Cdc34, and UbcH2), ATP-regenerating solution, ubiquitin aldehyde, and HeLa S100 fraction were obtained from Boston Biochem (Cambridge, MA). Antibodies against individual Pol δ subunits, γH2AX, and cyclopyrimidine dimers (CPDs) were obtained as described previously (9). RNF8 antibody was obtained from Abcam Inc. (Cambridge, MA). His-tagged RNF8 was expressed in Sf9 cells and purified as described previously (28). The GFP-RNF8 plasmid was constructed for expression in mammalian cells in the pEGFP vector as described (31) and used for the construction of the RING-deleted mutant (Δ-RING) (19) and R42A FHA-defective mutant (R42A) (20). GST-p12 was expressed in Escherichia coli and purified as described previously (7).
In Vitro Ubiquitination Assay for Ubiquitin Ligase Activity against p12
The column fractions to be tested for ubiquitin ligase activity were added to reaction mixtures containing 40 mm Tris-HCl, pH 7.5, 2 mm dithiothreitol, 5 mm MgCl2, 10 μg of ubiquitin, 30 nm E1 enzyme, 500 nm UbcH5c, 20 ng of ubiquitin aldehyde, 1× energy-regenerating solution, and 300 ng of GST-p12 in a total volume of 15 μl. The reaction mixtures were incubated at 30 °C for 1 h and terminated by the addition of 0.8 ml of PBS, 0.05% Nonidet P-40. The GST-p12 and its ubiquitinated products were pulled down by the addition of 15 μl of glutathione-Sepharose-4B beads. The mixtures were rotated at 4 °C for 60 min. The beads were then washed six times with PBS, 0.05% Nonidet P-40. The bound proteins were extracted by boiling in 30 μl of SDS-PAGE sample buffer for 5 min. The proteins were then resolved by SDS-PAGE on 12 or 10% acrylamide gels and immunoblotted with anti-ubiquitin.
Purification of Ubiquitin Ligase Activity
Sephacryl S-300 HR, phenyl-Sepharose CL-4B, Superdex S200 HR 10/30, and Mono Q 5/5 columns were obtained from GE Healthcare Life Sciences.
Step 1: Phenyl-Sepharose Chromatography
HeLa cells (2.1 × 109 cells, National Cell Culture Center, Minneapolis, MN) were suspended in 20 ml of TGEE buffer (20 mm Tris-HCl, pH 7.8, 10% glycerol, 0.5 mm EGTA, 1 mm EDTA, 1 mm MgCl2) containing 200 mm NaCl. The cells were disrupted by passage through a French press and centrifuged (10,000 × g, 60 min). The supernatant was diluted 4-fold to 50 mm NaCl with TGEE buffer and passed through an anti-p125 immunoaffinity column (7 ml) to remove most of the DNA polymerase δ (and endogenous p12) as described previously (10). The flow-through from the column was dialyzed against TGEED buffer (TGEE plus 2 mm dithiothreitol) containing 500 mm ammonium sulfate and then loaded onto a phenyl-Sepharose CL-4B column (11 ml). The column was washed with 5 bed volumes of TGEED, 20 mm potassium phosphate, 500 mm ammonium sulfate and then eluted with a 10-bed volume linear gradient from 500 to 0 mm ammonium sulfate in TGEED. Fractions of 1.5 ml were collected. The fractions were analyzed for ubiquitin ligase activity with UbcH5c added as the E2 ligase and GST-p12 as the substrate. Fractions (30–68) containing ubiquitin ligase activity were pooled and concentrated to 2.5 ml by centrifugation through 30,000 molecular weight cutoff Centricon tubes (Millipore)
Step 2: Sephacryl S300 HR Gel Filtration
The pooled fractions were loaded onto a Sephacryl S300 HR gel filtration column (140 ml) and eluted with TGEED, 150 mm NaCl. Fractions of 2 ml were collected. The fractions were analyzed for ubiquitin ligase activity with UbcH5c added as the E2 ligase. Fractions 46–61 were pooled.
Step 3: Mono Q Anion Exchange Chromatography
The pooled fractions were dialyzed against HEPES buffer (40 mm HEPES, pH 7.9, 0.1 mm EDTA, 2 mm dithiothreitol, 10% glycerol) containing 50 mm KCl and loaded on a 1-ml Mono Q column equilibrated with HEPES buffer. The column was washed with 2 bed volumes of HEPES buffer, and the bound proteins were eluted with a 20-bed volume linear gradient from 0 to 500 mm KCl in HEPES buffer. Fractions of 0.5 ml were collected. The fractions were analyzed for ubiquitin ligase activity with UbcH5c added as the E2 ligase. Fractions 7–14 were pooled and concentrated to 250 μl as described above.
Step 4: Superdex S200 HR 10/30 Chromatography
The concentrated fractions (250 μl) were chromatographed on a Superdex S200 HR 10/30 FPLC column and eluted with TGEED/150 mm NaCl. Fractions of 0.25 ml were collected. The fractions were analyzed for ubiquitin ligase activity with UbcH5c added as the E2 ligase.
Mass Spectrometry Analysis
An aliquot of fraction 39 from the Superdex 200 chromatography was subjected to Multidimensional Protein Identification Technology (MudPIT) and tandem mass spectrometry (LC/LC/MS/MS) at the Scripps Research Institute essentially as described by Ref. 32.
shRNA Knockdown of RNF8 and Mouse Epithelial RNF8−/− Cells
Stable A549 cells in which RNF8 was depleted by shRNA against RNF8 were isolated using four different shRNAs as described previously (28). HuSH 29-mer shRNA constructs against human RNF8 and control shRNA (noneffective shGFP) were synthesized by OriGene Technologies, Inc. (Rockville, MD). The RNF8-shRNA target sequences were as follows: shR1, GTGACCATGTCCAGGATTCTGAGGCTCAA; shR2, GGACGAGGATTTGGTGTCACATACCAACT; shR3, GCAGAAAGCCTCAAACTCTTCAGCATCTC; and shR4, GAGTGGAGCAACTAGAGAAGACTTTCCAG.
A549 cells in which RNF8 was knocked down were selected according to the protocol supplied by OriGene Technologies. Cells were maintained in RPMI medium 1640 supplemented with 10% fetal bovine serum, plated on 6-well dishes, and transfected with the control and RNF8 shRNA plasmids using Lipofectamine 2000 (Invitrogen). The growth medium was replaced after 24 h with fresh medium containing 2 μg/ml puromycin, and selection pressure was maintained for 2–3 weeks to obtain single clonal populations. Clonal populations of cells against RNF8 shRNA or control shRNA were taken from each plate for further culture and analysis. Stable cells were then maintained in 0.5 μg/ml puromycin. Mouse epithelial fibroblast (MEF) RNF8−/− cells derived from mouse RNF8 knockouts were a generous gift from Dr. Junjie Chen and maintained as described (29).
Immunofluorescence Localization Studies
Immunofluorescence localization and UV irradiation through polycarbonate filters with 5-μm pores were performed as described previously (9). HEK293T cells were maintained and grown on glass-chambered slides, fixed, and stained for p125, p68, p50, PCNA, and γH2AX as described previously (9). Fluorescence images were taken using a Zeiss Axiovert 200M microscope with a black-and-white CCD AxioCam and pseudo-colored with AxioVision 4.6 software. Unless otherwise stated, most images were taken at a magnification of 40×.
RESULTS
In Vitro Screening Assay for Ubiquitin Ligase Activities That Polyubiquitinate p12
We set up a screening assay for ubiquitin ligase activity by using GST-p12 as the substrate. In this protocol, ubiquitinated GST-p12 was recovered by pulldown with glutathione beads and analyzed by Western blotting for visualization (see “Experimental Procedures”). Because the ubiquitin ligases require the presence of specific E2s, several recombinant E2 enzymes were tested for their abilities to stimulate GST-p12 ubiquitination using commercially available HeLa S100 fraction as a source of ubiquitin ligase activity. The HeLa S100 fraction itself exhibited low but detectable activity for the ubiquitination of GST-p12 (Fig. 1, lane 2). The E2s tested were UbcH5c, which is active in concert with a number of ubiquitin ligases from both the RING and the HECT domain families (33) and has been widely used for in vitro studies (34). Two other E2s tested were Cdc34 and UbcH2. Cdc34 (UbcH3) is involved in the regulation of the G1/S cell cycle transition as the E2 for the Skp1-Cullin 1-F-box protein-ROC1 ubiquitin ligase complex (35, 36). UbcH2, the human homolog of yeast Rad6, is involved in the Rad6-Rad18 monoubiquitination of PCNA that triggers translesion bypass (37). UbcH5c induced a robust stimulation of the ubiquitination of GST-p12 using HeLa extract as the ubiquitin ligase source (Fig. 1, lane 4), whereas Cdc34 exhibited a much weaker reaction and UbcH2(Rad6) had almost no activity (Fig. 1, lanes 6 and 8, respectively). Note that the products formed are predominantly high molecular weight polyubiquitinated species.
FIGURE 1.
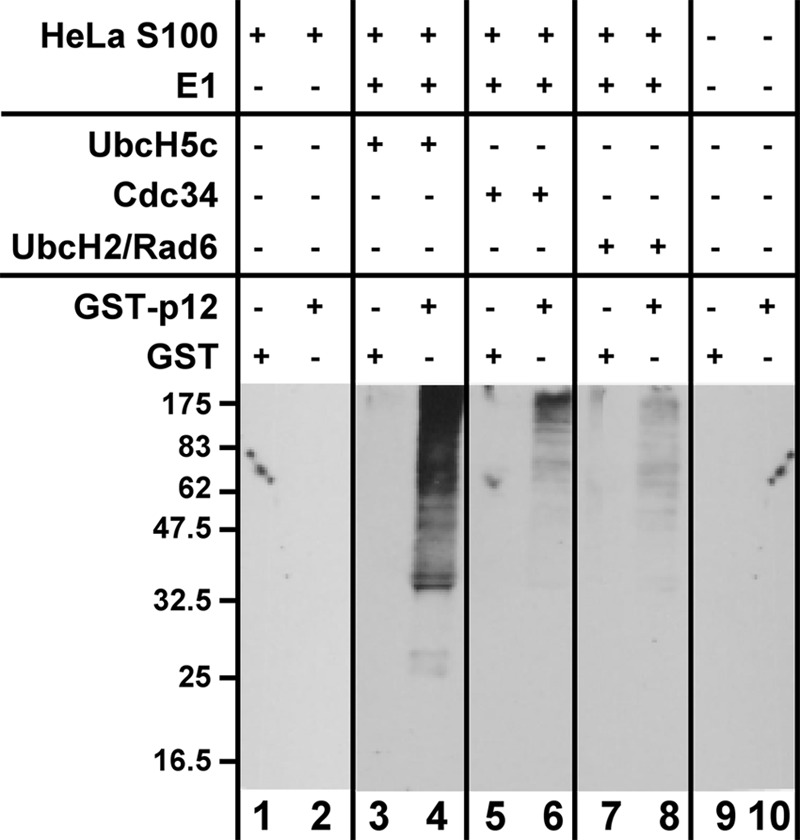
In vitro assay for the ubiquitination of GST-p12. The in vitro assay was performed as described under ”Experimental Procedures.“ The reaction mixtures contained E1, ubiquitin, an ATP-generating system, HeLa S100 lysate as a source of ubiquitin ligase, UbcH5c, Cdc34, or UbcH2 as the E2, and GST-p12 as the substrate. The reactions were incubated for 1 h at 30 °C. The products were pulled down with glutathione beads and analyzed by SDS-PAGE followed by Western blotting for ubiquitin (see ”Experimental Procedures“).
Purification of a UbcH5c-dependent Ubiquitin Ligase for p12
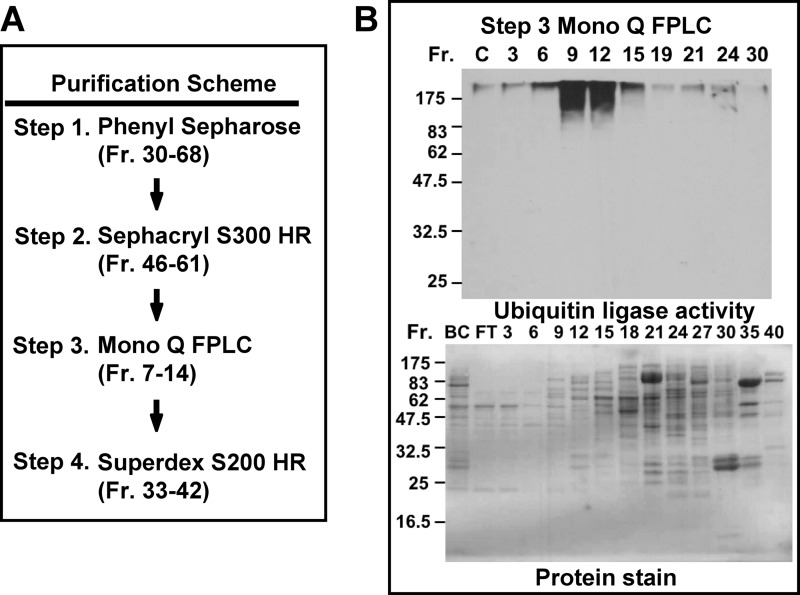
A HeLa cell lysate was first passed through an immunoaffinity column consisting of immobilized antibody against the p125 subunit of Pol δ to remove endogenous Pol δ as described previously (10). The flow-through fraction after the immunoaffinity chromatography step was then subjected to four sequential chromatography purifications on phenyl-Sepharose, Sephacryl S300 HR, Mono Q FPLC ion exchange, and Superdex S200 HR as schematically shown in Fig. 2A. The fractions from each column were assayed for ubiquitination of GST-p12 in the presence of UbcH5c, and those displaying activity were pooled for the subsequent steps (see ”Experimental Procedures“). The analysis for ubiquitin ligase activity for the Mono Q FPLC (Step 3) is shown in Fig. 2B (upper panel). The major peak of ubiquitin ligase activity was eluted between fractions 9 and 12 with a minor peak eluting at fraction 24. The protein stain for the SDS-PAGE of the column fractions is shown in Fig. 2B (lower panel). Fractions 9–12 contained significantly less protein than the later fractions, indicating that the ubiquitin ligase activity was likely to be considerably enriched.
FIGURE 2.
Purification of a ubiquitin ligase that ubiquitinates p12 from HeLa cells. A, purification scheme for the identification of ubiquitin ligase activity against GST-p12. Details are given under ”Experimental Procedures.“ The starting material was the lysate from 2.1 × 109 HeLa cells. Fractions (Fr.) from each column were assayed for ubiquitin ligase activity in the presence of UbcH5c as the E2 ubiquitin-conjugating enzyme (Fig. 1). Fractions containing ubiquitin ligase activity were pooled (fraction numbers are given in parentheses). B, upper panel, assay of the column fractions from Step 3 (Mono Q chromatography) for ubiquitin ligase activity. Lower panel, Ponceau Red stain for proteins in the same column fractions. “C” is a control without added enzyme; “BC” refers to the pooled enzyme before chromatography; “FT” refers to the pooled flow through fractions.
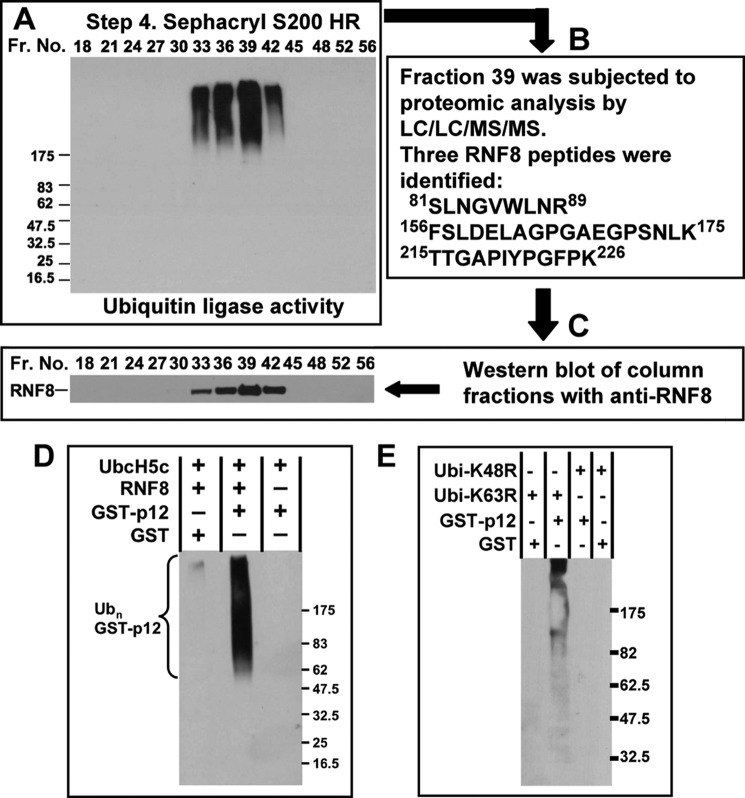
The pooled fractions from the Mono Q column (Step 3) were then subjected to Superdex S200 HR gel filtration chromatography (Step 4). The assays of the column fractions showed that there was a single peak between fractions 33 and 42 (Fig. 3A). Fraction 39 was subjected to proteomic analysis by LC/LC/MS/MS. Peptides for 240 proteins were identified in the proteomic analyses, only one of which was involved in ubiquitination reactions. This protein was identified as RNF8, for which three peptides were found (81SLNGVWLNR89, 156FSLDELAGPGAEGPSNLK175, and 215TTGAPIYPGFPK226, Fig. 3B). To confirm the identification of RNF8 as the ubiquitin ligase in the column fractions, the latter were Western blotted with anti-RNF8 (Fig. 3C). The Western blots for RNF8 corresponded with the ubiquitin ligase activity (Fig. 3A), both in position and in relative intensity. These results show that the combination of RNF8 and UbcH5c is effective in the polyubiquitination of p12 and confirm the data showing that HeLa cell lysates contain an active UbcH5c-dependent ubiquitination system for p12.
FIGURE 3.
Identification of the ubiquitin ligase present in the Superdex S200 fractions. A, assay for ubiquitin ligase activity in the fractions (Fr.) from Superdex S200 HR chromatography (Fig. 2A). Fractions 7–14 from the Mono Q column (Fig. 2B) were concentrated and applied to a Superdex S200 HR 10/30 FPLC column. B, an aliquot of fraction 39 was subjected to proteomic analysis by LC/LC/MS/MS. The three RNF8 peptides that were identified are shown. C, the fractions shown were further analyzed by Western blotting for RNF8. D, in vitro ubiquitination of GST-p12 by purified recombinant His-RNF8 and UbcH5c. Reactions were performed as described under ”Experimental Procedures“ and contained GST-p12, His-RNF8, E1, UbcH5c, and ubiquitin. Reaction products were pulled down with glutathione-Sepharose and Western blotted with anti-ubiquitin antibody. E, RNF8 synthesizes Lys-48-linked polyubiquitin chains on GST-p12. Reactions were performed as for D, except that the K63R or K48R ubiquitin (Ubi) mutants were used instead of wild type ubiquitin.
To confirm that RNF8 and UbcH5c can polyubiquitinate p12, we purified recombinant RNF8 and UbcH5c expressed in E. coli and used these to ubiquitinate GST-p12 (Fig. 3D). The results show that RNF8 polyubiquitinates GST-p12 readily in the presence of UbcH5c in vitro. To confirm that the polyubiquitin chains formed were linked via canonical Lys-48 isopeptide linkages between the ubiquitin moieties, and not the Lys-63-linked chains associated with RNF8 signaling in the DDR (23–25), the assay was also performed using the K48R and K63R ubiquitin mutants. No reaction products were observed when the K48R ubiquitin was used (Fig. 3E), confirming that the polyubiquitination occurs via Lys-48 linkages. The K63R ubiquitin mutant supported the polyubiquitination of GST-p12, showing that the polyubiquitination did not involve Lys-63 isopeptide linkages.
Effects of RNF8 Depletion on the Degradation of p12 in Response to DNA Damage in Vivo
The demonstration that RNF8 ubiquitinates p12 in vitro provided the impetus to investigate its role in vivo. To address this question, we utilized shRNA knockdowns of RNF8 in cultured cells using four shRNAs against RNF8 (shR1–shR4, see ”Experimental Procedures“). The analyses demonstrate that shR2 was the most efficient of the four tested for the depletion of RNF8 (Fig. 4A), and shR2-derived cells were therefore used in subsequent experiments. The degradation of p12 with increasing UVC dose was examined in control and RNF8 knockdown cells (Fig. 4B). The lysates were also Western blotted for Chk1-Ser(P)-345 and γH2AX to monitor the activation of DNA damage signaling. The Western blots for p12 were quantified and plotted against UVC dose (Fig. 4C). The results show that the sensitivity of p12 degradation to UV dose was significantly reduced by RNF8 knockdown.
FIGURE 4.
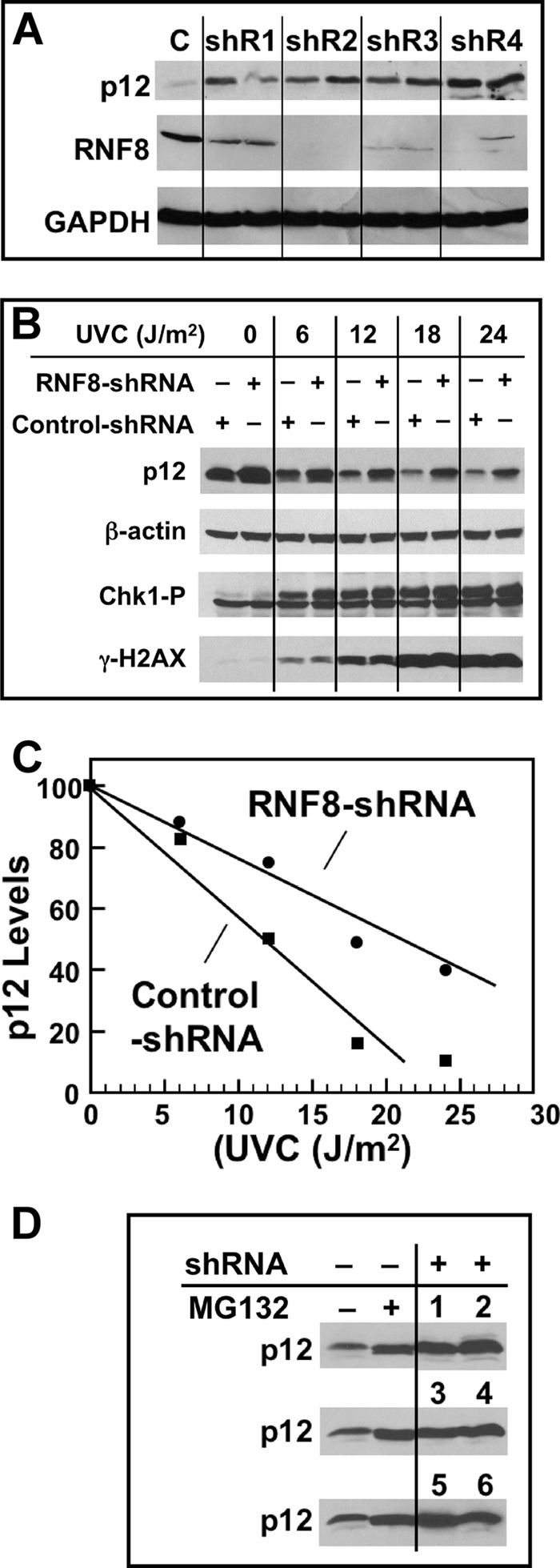
Effects of RNF8 depletion on p12 degradation by UV. A, four different shRNAs (shR1–shR4) and a control shRNA (C) were used to produce respective stable A549 cells (see ”Experimental Procedures“). Each sample was treated in duplicate with UVC (20 J/m2) and Western blotted for RNF8 and p12 after 4 h. GAPDH was used as a loading control. B, UV dose dependence of p12 degradation in RNF8-depleted A549 cells; shR2 was used in these and the following experiments. Cell cultures were treated with UVC (0, 6, 12, 18, and 24 J/m2) and analyzed 4 h later by Western blotting for p12, Chk1-p, γH2AX, and β-actin (loading control). C, the blots for p12 were quantified by densitometry and normalized against the loading control. The relative levels of p12 were then plotted against UVC dose. D, comparison of p12 levels in unperturbed RNF8 knockdown cells. The two left lanes show p12 levels in control shRNA-treated cells in the absence and presence of the proteasome inhibitor MG132 (10 μm), which was added 30 min prior to analysis. The two right lanes with the three rows of numbers show p12 levels in six individual shR2 RNF8 clonal isolates without MG132.
In addition, we had noted that the levels of p12 were generally elevated in the RNF8 knockdown cells when compared with control cells (Fig. 4B). We compared the levels of p12 in cells treated with the proteasome inhibitor MG132 (10) with the levels of p12 in RNF8-depleted cells (Fig. 4D). The results show that RNF8 depletion raises p12 levels in a manner similar to MG132 treatment, indicating that RNF8 participates in the normal cellular process of p12 turnover, as well as in the DNA damage-induced degradation of p12.
The half-lives of p12 in untreated and UVC-treated control and RNF8 shRNA knockdown cells were then determined in the presence of cycloheximide to inhibit protein synthesis (Fig. 5, A and B). The levels of p12 were determined by densitometry and plotted on a semi-log plot against time (Fig. 5C). The half-lives of the control and RNF8-shRNA cells were both estimated to be 16.5 h. After UVC treatment, the half-life of p12 was estimated to be ∼1.0 h in the control cells and 3.0 h in the RNF8 knockdown cells (Fig. 5C). Thus, these results show that RNF8 is involved in the UV-induced targeting of p12 for degradation in vivo. It is noted that complete stabilization of p12 was not observed. This indicates that p12 degradation is only partly dependent on RNF8 and that there are likely to be other redundant mechanisms that contribute to p12 degradation. In this situation, it is expected that complete degradation of p12 will be delayed when one of these mechanisms is disabled, as evidenced by the increased half-life.
FIGURE 5.
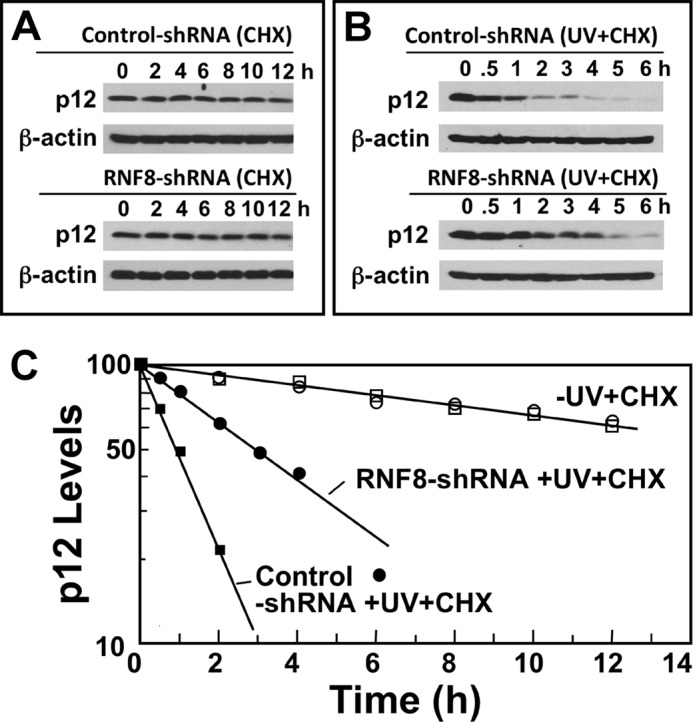
Determination of the half-lives of p12 in control RNF8 knockdown cells. A, control and shR2 RNF8 cells were transferred to fresh medium containing 10 μg/ml cycloheximide (CHX). Cells were harvested at the indicated times (0–12 h) after treatment and analyzed by Western blotting for p12. B, control and shR2 RNF8 cells were treated with UVC (10 J/m2) and transferred to fresh medium containing 10 μg/ml cycloheximide. Cells were harvested at the indicated times (0–6 h) after treatment and analyzed by Western blotting for p12. β-Actin was used as the loading control. C, the relative amounts of p12 for the Western blots shown in A and B were determined by densitometry and were plotted on a semi-log plot against time. Data for control shRNA in the presence of cycloheximide or cycloheximide + UV are shown as open or closed squares, respectively; data for RNF8-shRNA cells in the presence of cycloheximide or cycloheximide + UV are shown as open or closed circles, respectively.
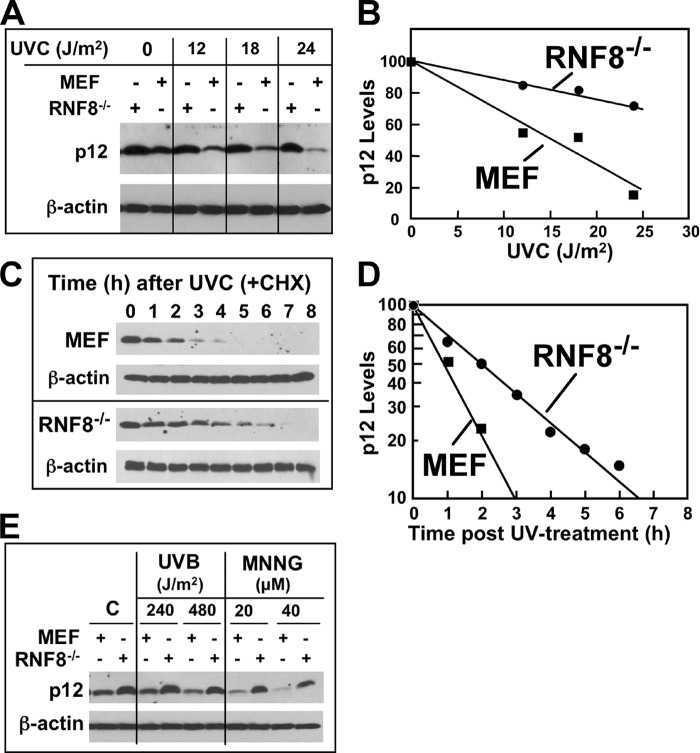
To further confirm the effects observed on the reduction of p12 degradation in RNF8-depleted cells, we examined the effects of UV on p12 degradation in mouse epithelial fibroblasts derived from RNF8 knock-out mice (29). The degradation of p12 was significantly retarded in RNF8−/− cells by comparison with control cells in response to increasing UV dose (Fig. 6, A and B). From the densitometry data, we estimated that the degradation of p12 was decreased by ∼50–60% by knock-out of RNF8 (Fig. 6B). The time course of degradation of p12 after UV was examined in the presence of cycloheximide and showed that the rate of degradation of p12 was significantly reduced (Fig. 6, C and D). The half-life of p12 in the RNF8−/− cells estimated from the semi-log plots of p12 levels against time was more than doubled from that of the control MEF cells (0.9–2.1 h). These results are consistent with those obtained from the RNF8 shRNA knockdown cells (Fig. 5C). These results show that RNF8 is involved in the UV-induced degradation of p12 in vivo and also confirm that it is not the sole regulatory system that targets p12 for proteasomal degradation.
FIGURE 6.
UV-induced p12 degradation is impaired in mouse RNF8 knock-out cells. A, control MEF or their RNF8 knock-out cells (RNF8−/−) were treated with 0, 12, 18, and 24 J/m2 UVC, allowed to recover for 2 h, and analyzed by Western blotting for p12. β-Actin was used as a loading control. B, the relative amounts of p12 for the Western blots shown in panel A were determined by densitometry and corrected for loading against β-actin. Data were normalized to the p12 levels with no UV treatment and plotted against UV dose. C, time dependence of p12 degradation in RNF8−/− cells. Control MEF and RNF8−/− cells were treated with UVC (8 J/m2) and transferred to fresh medium containing 10 μg/ml cycloheximide (CHX). Cells were harvested at the indicated times (0–8 h) after treatment and analyzed by Western blotting for p12. D, the relative amounts of p12 for the Western blots shown in panel C were determined by densitometry as for panel B and were plotted on a semi-log plot against time. E, RNF8−/− and control MEF were treated with UVB (240 or 480 J/m2) and Western blotted for p12 after 4 h or treated with 20 or 40 μm MNNG and examined by Western blotting after 4 h.
The effects of RNF8 knock-out on p12 degradation induced by UVB and an alkylating agent, MNNG, were examined (Fig. 6E). The degradation of p12 by both these agents was significantly retarded in RNF8−/− cells.
Subcellular Recruitment of RNF8 and Pol δ to Local Areas of UV-induced Damage
We have recently shown that Pol δ is efficiently recruited to local areas of UV damage produced by irradiation through UV-opaque polycarbonate membranes with 5-μm pores (9). Thus, it was important to establish whether RNF8 is recruited to sites of UV damage together with Pol δ to provide the necessary spatial evidence that it might be involved in p12 degradation. We utilized GFP-RNF8 to observe the recruitment of RNF8 to DNA damage foci. We also tested the GFP-RNF8 mutants in which the RING domain was deleted, and the R42A mutant in which the FHA domain was inactivated, to confirm that GFP-RNF8 was recruited to DNA damage like the endogenous RNF8 (27) (supplemental Fig. S1). Treatment of stably transfected cells showed that GFP-RNF8 was recruited to IRIF (22), to areas of local UV-induced irradiation (27), and also to damage foci induced by MNNG, which has not been previously observed (supplemental Fig. S1). The deletion of the RING domain did not affect the recruitment of RNF8, whereas inactivation of the FHA domain led to loss of recruitment; both are consistent with the mechanism for the recruitment of RNF8 via its FHA domain to phosphorylated MDC1 in response to IR and UV damage (27). These results validate the use of GFP-RNF8 for the examination of the co-localization of RNF8 with Pol δ and PCNA.
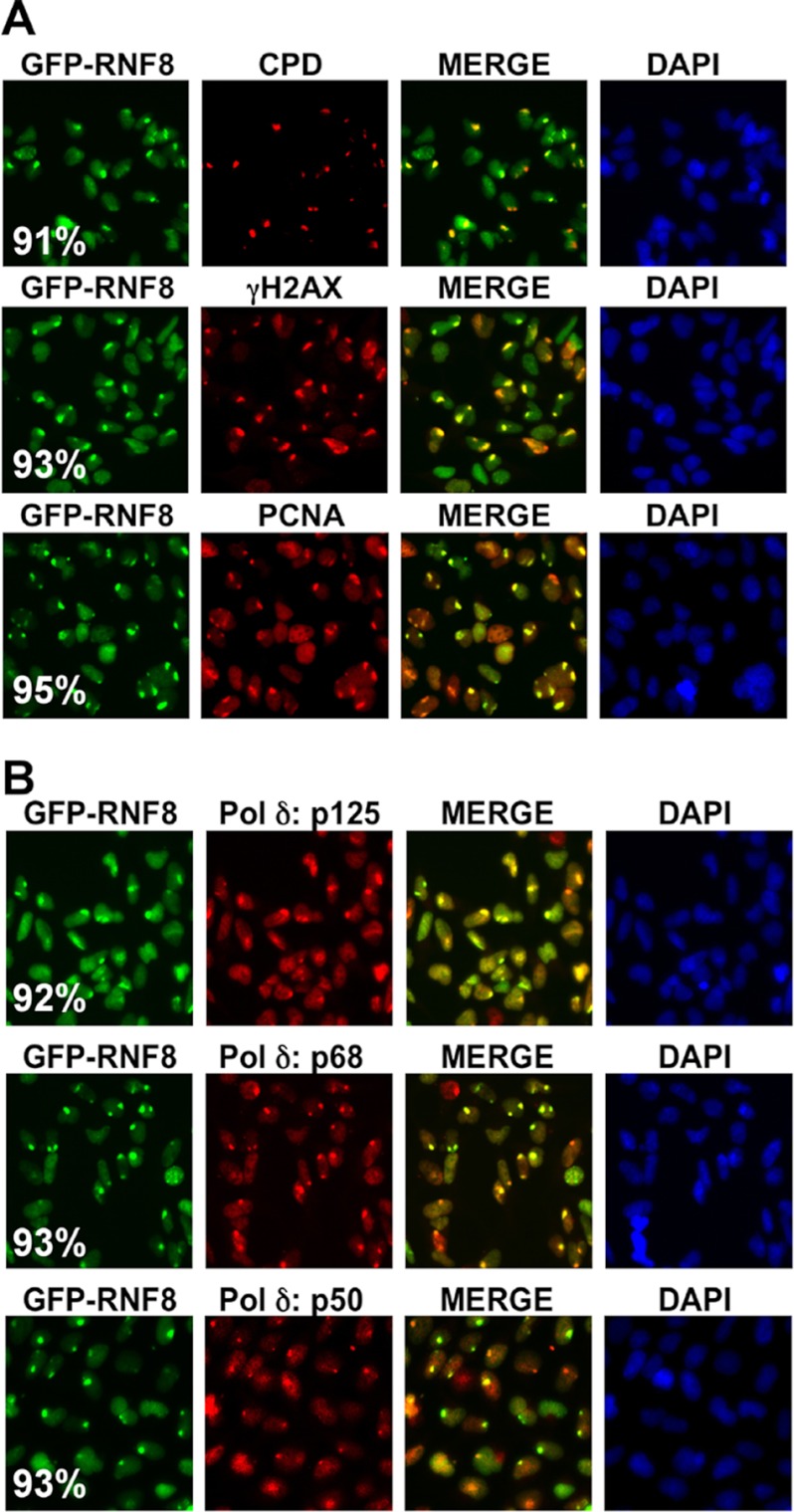
We irradiated cells through UV-opaque polycarbonate filters with 5-μm pores to produce local areas of UV irradiation (9). GFP-RNF8 was co-localized with CPDs, γH2AX, and PCNA to these areas of UV damage (Fig. 7A). Next we showed that RNF8 is co-localized with the p125, p68, and p50 subunits of Pol δ to these areas of local UV irradiation (Fig. 7B). Quantitation of the extent of co-localization gave values >90% in all cases (Fig. 7, A and B). These results show that RNF8 is recruited together with PCNA and Pol δ to sites of UV damage in asynchronous cell populations.
FIGURE 7.
Recruitment of GFP-RNF8, PCNA, and Pol δ to sites of UV damage as shown by co-localization with CPDs and γH2AX. A, co-localization of GFP-RNF8 to local areas of UV-induced damage as shown by co-localization with CPDs, γH2AX, and PCNA. HEK293T cells expressing GFP-RNF8 were treated with 75 J/m2 UVC through 5-μm pore polycarbonate filters and fixed after 4 h. GFP-RNF8 was visualized by green fluorescence; CPDs, γH2AX, and PCNA were visualized by red fluorescence by the use of Rhodamine Red-X-conjugated secondary antibodies. B, co-localization of GFP-RNF8 and the p125, p68, and p50 subunits of Pol δ to sites of local irradiation by UVC. p125, p68, and p50 were visualized by red fluorescence as for panel A. The extent of co-localization was quantified by counting >100 foci and expressed as a percentage as described previously (9). Values are shown in white lettering in the left-hand panels.
DISCUSSION
Evidence for the role of ubiquitination in the UV-induced degradation of p12 comes from its dependence on the presence of ubiquitin-activating enzyme, the polyubiquitination of p12, and its inhibition by proteasome inhibitors (10). The identity of the ubiquitin ligase(s) was, however, previously unknown. In this study, we took a classical approach of protein purification followed by proteomic analysis to identify RNF8 as a ubiquitin ligase that is able to polyubiquitinate p12 in vitro in concert with UbcH5c. Further analysis of the effects of UV on p12 degradation in RNF8 knockdown cells and in mouse RNF8−/− knock-out cells demonstrated that p12 degradation is significantly impaired, providing evidence that RNF8 is physiologically involved in the targeting of p12 for degradation in response to UV damage. Our data show that p12 degradation is not completely prevented, supporting the conclusion that p12 degradation is likely under the control of more than one ubiquitination pathway. In addition, we observed that RNF8 knockdown or RNF8−/− cells exhibited higher basal levels of p12, suggesting that the RNF8 is also involved in the normal cellular turnover of p12. Our studies now reveal the identity of one of the ubiquitin ligases involved in targeting p12 for degradation.
Our in vitro assays of ubiquitin ligase activity are based on the use of UbcH5c as the E2 enzyme that partners with RNF8 for the polyubiquitination of p12. Although it has yet to be determined whether UbcH5c is the partner for p12 degradation by RNF8 in vivo, we have shown that UbcH5c is the most effective of several E2s we tested for the in vitro PCNA ubiquitination by RNF8 (28) in addition to the p12 ubiquitination presented here. We have previously shown that RNF8 also utilizes Ubc13-Uev1a for the noncanonical Lys-63 polyubiquitination of PCNA (28). Studies have also shown that RNF8 uses Ubc13 to ubiquitinate histones H2A and H2AX, a process that plays a crucial role in the recruitment of downstream effectors such as p53BP1 and BRCA1 to double strand breaks during the DNA damage response (17–21). Much of the previous focus of RNF8 ubiquitination has involved studies of the noncanonical Lys-63 ubiquitination of DDR intermediates and histones (38) so that the degradation of p12 represents a new example of the involvement of RNF8 in degradation of a key protein in the DDR. Two other recent examples of RNF8 involvement in Lys-48-linked ubiquitination of substrates have been reported, that of DNA damage-induced degradation of the lysine demethylase KDM4A(JMJD2A) (39) and that of the degradation of the nonhomologous end-joining protein, KU80, as well as Chk2, the downstream kinase associated with ATM (30).
The identification of RNF8 as a ubiquitin ligase that regulates p12 is highly significant in the context of the developing concept that Pol δ3 is formed in response to DNA damage to function as the cellular form of Pol δ activity that participates in various DNA repair processes (reviewed in Ref. 2). The finding that p12 degradation is regulated, at least in part, by RNF8 is a major step in the assessment of Pol δ3 formation as a component of the cellular DNA damage responses.
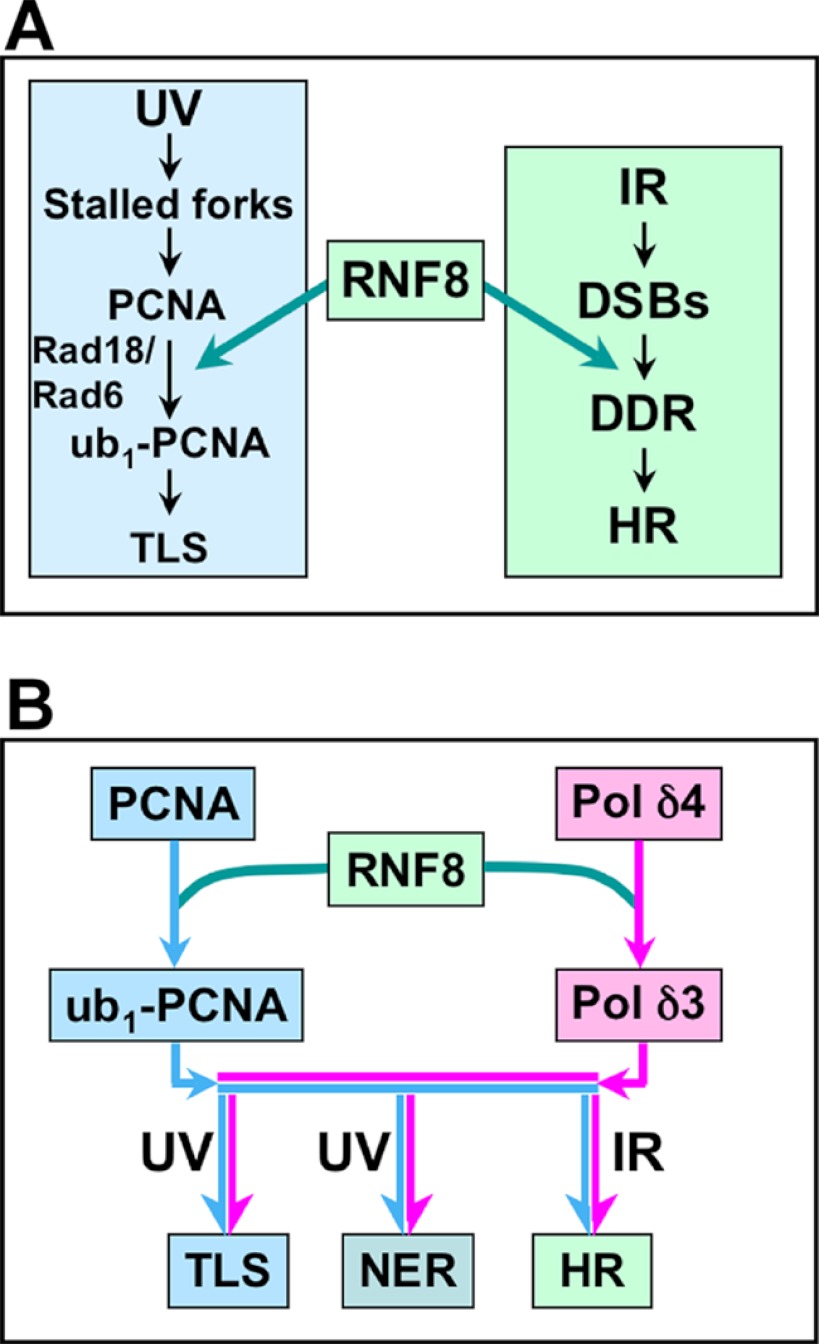
Our original investigations of the DNA damage-induced degradation of p12 were made in the context of the intra-S phase checkpoint triggered by UV for which ATR is the apical signaling kinase (10–12). In S phase cells, UV damage triggers translesion synthesis, for which monoubiquitination of PCNA plays a key role, and has been extensively investigated as part of the DNA damage tolerance pathway (37, 40, 41). In this system, the yeast Rad6-Rad18 and its human homologs have been shown to be the E3/E2 enzymes that lead to monoubiquitination of PCNA to trigger the so-called error-prone pathway involving translesion synthesis by TLS Pols as exemplified by Pol η. It is noted that the functions of Rad18/Rad6 have been largely associated with TLS in S phase cells. (We have not been able to determine whether Rad18 is involved in p12 degradation, and this remains an open possibility.) It is of significant interest that our previous studies have shown that RNF8 is also able to ubiquitinate PCNA (28), providing an alternative regulatory pathway for PCNA ubiquitination that might not be constrained to S phase cells (Fig. 8A).
FIGURE 8.
Overview of the recruitment and modification of PCNA and Pol δ by RNF8 involving different DNA damage pathways. A, RNF8 involvement in the ubiquitination of PCNA. The left panel shows the basic scheme of the activation of TLS in response to UV damage in S phase cells where CPDs block replication fork progression. The current well established scheme in which Rad6-Rad18 triggers monoubiquitination of PCNA to initiate TLS by recruitment of Pol η is shown (37, 54). The right panel indicates the response to IR, which triggers the DDR, where RNF8 plays a key role in the assembly of the complex of DDR proteins (17–19, 21). Shown here is the finding that RNF8 also ubiquitinates PCNA (28). DSBs, double strand breaks. B, a proposed role for RNF8 in integrating the formation of Pol δ3 with PCNA ubiquitination. This diagram shows that RNF8 could act to generate Pol δ3 and ub-PCNA in response to UV to act in TLS as well as in NER and in response to IR to act in HR. The light blue arrows show the pathway for ubiquitination of PCNA, and the pink arrows show the generation of Pol δ3 via the degradation of p12. Implicit in this pathway is that the formation of monoubiquitination of PCNA leads to the recruitment of TLS polymerases.
The ubiquitination of p12 and PCNA may be coordinated at the molecular level, given that they are present in complex at the primer termini, generating Pol δ3 and ub-PCNA. This possibility is consistent with models we have proposed for the switching of Pol η with Pol δ on ub-PCNA, in which formation of Pol δ3 is a key step. In these models, the loss of p12 releases a binding site for a PCNA interacting protein box, thereby facilitating the exchange of Pol η with Pol δ3 (2, 42).
The identification of RNF8 as an E3 ligase that targets p12 is significant from several additional aspects. The degradation p12 is global in that it occurs in all phases of the cell cycle (9), which implies that Pol δ3 is involved in multiple forms of DNA repair including NER. RNF8, which has a preeminent role in the DDR under the control of ATM, would appear on the surface to be an unexpected candidate for the regulation of p12 degradation. However, RNF8 has been shown to be recruited to sites of NER in an MDC1- and ATR-dependent manner (27). Thus, RNF8, unlike Rad6-Rad18, might provide a more global regulation for p12 degradation and has the potential for generating Pol δ3 for functions in NER and HR as well as TLS, as shown in Fig. 8B. There is evidence that Pol δ participates in the resynthesis step of NER (43–45). Although the role of Pol δ activity in double strand break repair by HR is still incompletely understood, it has been implicated in the process of heteroduplex extension in yeast (46, 47). It should be emphasized that the pathways shown in Fig. 8B provide a working hypothesis for which much further investigation is needed. One other aspect, not shown in Fig. 8B, that needs to be taken into account is that we have found that p12 degradation is dependent on ATR activation, which is normally associated with activation of UV damage response signaling. Thus, it is relevant to note that ATR activation can also result from the generation of ssDNA during resection of double strand break ends in HR (48, 49) and during NER (50–52). We have examined the co-localization of ATR, Pol η, and Pol δ to sites of UV damage (CPDs) and found that all three are recruited but with different kinetics, that of ATR being completed first followed by Pol η and then Pol δ (9).
The connection between p12 degradation and PCNA ubiquitination also brings into view the potential significance of the generation of ub-PCNA in contexts other than the TLS at stalled replication forks. The generation of ub-PCNA is associated with recruitment of TLS Pols and translesion synthesis, and recent studies have shown that Pol κ (as well as Pol δ) may be involved in the resynthesis step of NER (44, 53).
There remain many unanswered questions regarding the details of the regulation of Pol δ that leads to the conversion of Pol δ4 to Pol δ3 in response to genotoxic stress. Included among these are the identities of the other ubiquitin ligase(s) that participate in the targeting of p12 for degradation. These ubiquitin ligases could provide redundancy for the degradation of p12 and provide for conversion of Pol δ4 to Pol δ3 in response to multiple signaling inputs generated by genotoxic stresses. There are also questions regarding the identity of the E2 enzymes that cooperate with RNF8 to target p12 in vivo.
In summary, we have identified RNF8 as an E3 ligase that targets p12 for degradation in response to UV damage. The involvement of RNF8 integrates the regulation of Pol δ into the major DNA response system, the DDR, and provides further support for the concept that the formation of Pol δ3 is a significant event during DNA damage.
Acknowledgment
We thank Dr. Junjie Chen, University of Texas MD Anderson Cancer Center, for the generous gift of MEF and RNF8−/− cell lines.
This work was supported, in whole or in part, by National Institutes of Health Grants GM31973 and ES14737 (to M. Y. W. T. L.) and P41 GM103533 (to J. R. Y.).
This article was selected as a Paper of the Week.

This article contains supplemental Fig. S1.
- Pol δ
- DNA polymerase δ
- ATR
- ATM- and Rad3-related
- ATM
- ataxia telangiectasia mutated
- CPDs
- cyclobutane pyrimidine dimers
- DDR
- DNA damage response
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin-conjugating enzyme
- E3
- ubiquitin ligase
- FHA
- forkhead-associated
- IR
- ionizing radiation
- IRIF
- IR-induced foci
- MDC1
- mediator of DNA damage checkpoint 1
- MEF
- mouse epithelial fibroblast(s)
- HR
- homologous recombination
- MNNG
- N-methyl-N′-nitroso-N-nitrosoguanidine
- NER
- nucleotide excision repair
- PCNA
- proliferating cell nuclear antigen
- ub-PCNA
- ubiquitinated PCNA
- TLS
- translesion synthesis.
REFERENCES
- 1. Garg P., Burgers P. M. (2005) DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 40, 115–128 [DOI] [PubMed] [Google Scholar]
- 2. Lee M. Y., Zhang S., Lin S. H., Chea J., Wang X., Leroy C., Wong A., Zhang Z., Lee E. Y. (2012) Regulation of human DNA polymerase δ in the cellular responses to DNA damage. Environ. Mol. Mutagen. 53, 683–698 [DOI] [PubMed] [Google Scholar]
- 3. Byrnes J. J., Downey K. M., Black V. L., So A. G. (1976) A new mammalian DNA polymerase with 3′ to 5′ exonuclease activity: DNA polymerase δ. Biochemistry 15, 2817–2823 [DOI] [PubMed] [Google Scholar]
- 4. Lee M. Y., Tan C. K., So A. G., Downey K. M. (1980) Purification of deoxyribonucleic acid polymerase δ from calf thymus: partial characterization of physical properties. Biochemistry 19, 2096–2101 [DOI] [PubMed] [Google Scholar]
- 5. Lee M. Y., Tan C. K., Downey K. M., So A. G. (1984) Further studies on calf thymus DNA polymerase δ purified to homogeneity by a new procedure. Biochemistry 23, 1906–1913 [DOI] [PubMed] [Google Scholar]
- 6. Zhou Y., Chen H., Li X., Wang Y., Chen K., Zhang S., Meng X., Lee E. Y., Lee M. Y. (2011) Production of recombinant human DNA polymerase δ in a Bombyx mori bioreactor. PLoS ONE 6, e22224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li H., Xie B., Zhou Y., Rahmeh A., Trusa S., Zhang S., Gao Y., Lee E. Y., Lee M. Y. (2006) Functional roles of p12, the fourth subunit of human DNA polymerase δ. J. Biol. Chem. 281, 14748–14755 [DOI] [PubMed] [Google Scholar]
- 8. Xie B., Mazloum N., Liu L., Rahmeh A., Li H., Lee M. Y. (2002) Reconstitution and characterization of the human DNA polymerase δ four-subunit holoenzyme. Biochemistry 41, 13133–13142 [DOI] [PubMed] [Google Scholar]
- 9. Chea J., Zhang S., Zhao H., Zhang Z., Lee E. Y., Darzynkiewicz Z., Lee M. Y. (2012) Spatiotemporal recruitment of human DNA polymerase δ to sites of UV damage. Cell Cycle 11, 2885–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang S., Zhou Y., Trusa S., Meng X., Lee E. Y., Lee M. Y. (2007) A novel DNA damage response: rapid degradation of the p12 subunit of DNA polymerase δ. J. Biol. Chem. 282, 15330–15340 [DOI] [PubMed] [Google Scholar]
- 11. Meng X., Zhou Y., Lee E. Y., Lee M. Y., Frick D. N. (2010) The p12 subunit of human polymerase δ modulates the rate and fidelity of DNA synthesis. Biochemistry 49, 3545–3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meng X., Zhou Y., Zhang S., Lee E. Y., Frick D. N., Lee M. Y. (2009) DNA damage alters DNA polymerase δ to a form that exhibits increased discrimination against modified template bases and mismatched primers. Nucleic Acids Res. 37, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seki N., Hattori A., Sugano S., Suzuki Y., Nakagawara A., Ohhira M., Muramatsu M., Hori T., Saito T. (1998) Isolation, tissue expression, and chromosomal assignment of a novel human gene which encodes a protein with RING finger motif. J. Hum. Genet. 43, 272–274 [DOI] [PubMed] [Google Scholar]
- 14. Ito K., Adachi S., Iwakami R., Yasuda H., Muto Y., Seki N., Okano Y. (2001) N-Terminally extended human ubiquitin-conjugating enzymes (E2s) mediate the ubiquitination of RING-finger proteins, ARA54 and RNF8. Eur. J. Biochem. 268, 2725–2732 [DOI] [PubMed] [Google Scholar]
- 15. Plans V., Scheper J., Soler M., Loukili N., Okano Y., Thomson T. M. (2006) The RING finger protein RNF8 recruits UBC13 for lysine 63-based self polyubiquitylation. J. Cell. Biochem. 97, 572–582 [DOI] [PubMed] [Google Scholar]
- 16. Brooks L., 3rd, Heimsath E. G., Jr., Loring G. L., Brenner C. (2008) FHA-RING ubiquitin ligases in cell division cycle control. Cell. Mol. Life Sci. 65, 3458–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang B., Elledge S. J. (2007) Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. U.S.A. 104, 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kolas N. K., Chapman J. R., Nakada S., Ylanko J., Chahwan R., Sweeney F. D., Panier S., Mendez M., Wildenhain J., Thomson T. M., Pelletier L., Jackson S. P., Durocher D. (2007) Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huen M. S., Grant R., Manke I., Minn K., Yu X., Yaffe M. B., Chen J. (2007) RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900 [DOI] [PubMed] [Google Scholar]
- 21. Harper J. W., Elledge S. J. (2007) The DNA damage response: ten years after. Mol. Cell. 28, 739–745 [DOI] [PubMed] [Google Scholar]
- 22. Bekker-Jensen S., Mailand N. (2010) Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair 9, 1219–1228 [DOI] [PubMed] [Google Scholar]
- 23. Wu J., Huen M. S., Lu L. Y., Ye L., Dou Y., Ljungman M., Chen J., Yu X. (2009) Histone ubiquitination associates with BRCA1-dependent DNA damage response. Mol. Cell. Biol. 29, 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim H., Chen J. (2008) New players in the BRCA1-mediated DNA damage responsive pathway. Mol. Cells 25, 457–461 [PMC free article] [PubMed] [Google Scholar]
- 25. Luijsterburg M. S., van Attikum H. (2012) Close encounters of the RNF8th kind: when chromatin meets DNA repair. Curr. Opin. Cell Biol. 24, 439–447 [DOI] [PubMed] [Google Scholar]
- 26. Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D. H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., Lukas J., Lukas C. (2009) RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 [DOI] [PubMed] [Google Scholar]
- 27. Marteijn J. A., Bekker-Jensen S., Mailand N., Lans H., Schwertman P., Gourdin A. M., Dantuma N. P., Lukas J., Vermeulen W. (2009) Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol. 186, 835–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang S., Chea J., Meng X., Zhou Y., Lee E. Y., Lee M. Y. (2008) PCNA is ubiquitinated by RNF8. Cell Cycle 7, 3399–3404 [DOI] [PubMed] [Google Scholar]
- 29. Huen M. S., Huang J., Yuan J., Yamamoto M., Akira S., Ashley C., Xiao W., Chen J. (2008) Noncanonical E2 variant-independent function of UBC13 in promoting checkpoint protein assembly. Mol. Cell. Biol. 28, 6104–6112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Feng L., Chen J. (2012) The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat. Struct. Mol. Biol. 19, 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Plans V., Guerra-Rebollo M., Thomson T. M. (2008) Regulation of mitotic exit by the RNF8 ubiquitin ligase. Oncogene 27, 1355–1365 [DOI] [PubMed] [Google Scholar]
- 32. Bonner M. K., Poole D. S., Xu T., Sarkeshik A., Yates J. R., 3rd., Skop A. R. (2011) Mitotic spindle proteomics in Chinese hamster ovary cells. PLoS ONE 6, e20489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brzovic P. S., Klevit R. E. (2006) Ubiquitin transfer from the E2 perspective: why is UbcH5 so promiscuous? Cell Cycle 5, 2867–2873 [DOI] [PubMed] [Google Scholar]
- 34. Lorick K. L., Jensen J. P., Weissman A. M. (2005) Expression, purification, and properties of the Ubc4/5 family of E2 enzymes. Methods Enzymol. 398, 54–68 [DOI] [PubMed] [Google Scholar]
- 35. Sarikas A., Hartmann T., Pan Z. Q. (2011) The cullin protein family. Genome Biol. 12, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Plon S. E., Leppig K. A., Do H. N., Groudine M. (1993) Cloning of the human homolog of the CDC34 cell cycle gene by complementation in yeast. Proc. Natl. Acad. Sci. U.S.A. 90, 10484–10488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen J., Bozza W., Zhuang Z. (2011) Ubiquitination of PCNA and its essential role in eukaryotic translesion synthesis. Cell Biochem. Biophys. 60, 47–60 [DOI] [PubMed] [Google Scholar]
- 38. Campbell S. J., Edwards R. A., Leung C. C., Neculai D., Hodge C. D., Dhe-Paganon S., Glover J. N. (2012) Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation. J. Biol. Chem. 287, 23900–23910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mallette F. A., Mattiroli F., Cui G., Young L. C., Hendzel M. J., Mer G., Sixma T. K., Richard S. (2012) RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 31, 1865–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sale J. E., Lehmann A. R., Woodgate R. (2012) Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 13, 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Andersen P. L., Xu F., Xiao W. (2008) Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res 18, 162–173 [DOI] [PubMed] [Google Scholar]
- 42. Zhang Z., Zhang S., Lin S. H., Wang X., Wu L., Lee E. Y., Lee M. Y. (2012) Structure of monoubiquitinated PCNA: implications for DNA polymerase switching and Okazaki fragment maturation. Cell Cycle 11, 2128–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Overmeer R. M., Gourdin A. M., Giglia-Mari A., Kool H., Houtsmuller A. B., Siegal G., Fousteri M. I., Mullenders L. H., Vermeulen W. (2010) Replication factor C recruits DNA polymerase δ to sites of nucleotide excision repair but is not required for PCNA recruitment. Mol. Cell. Biol. 30, 4828–4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ogi T., Limsirichaikul S., Overmeer R. M., Volker M., Takenaka K., Cloney R., Nakazawa Y., Niimi A., Miki Y., Jaspers N. G., Mullenders L. H., Yamashita S., Fousteri M. I., Lehmann A. R. (2010) Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell 37, 714–727 [DOI] [PubMed] [Google Scholar]
- 45. Mocquet V., Lainé J. P., Riedl T., Yajin Z., Lee M. Y., Egly J. M. (2008) Sequential recruitment of the repair factors during NER: the role of XPG in initiating the resynthesis step. EMBO J. 27, 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maloisel L., Fabre F., Gangloff S. (2008) DNA polymerase δ is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Mol. Cell. Biol. 28, 1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li X., Stith C. M., Burgers P. M., Heyer W. D. (2009) PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase δ. Mol. Cell 36, 704–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shiotani B., Zou L. (2009) Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol. Cell 33, 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tomimatsu N., Mukherjee B., Deland K., Kurimasa A., Bolderson E., Khanna K. K., Burma S. (2012) Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair 11, 441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lagerwerf S., Vrouwe M. G., Overmeer R. M., Fousteri M. I., Mullenders L. H. (2011) DNA damage response and transcription. DNA Repair 10, 743–750 [DOI] [PubMed] [Google Scholar]
- 51. Vrouwe M. G., Pines A., Overmeer R. M., Hanada K., Mullenders L. H. (2011) UV-induced photolesions elicit ATR-kinase-dependent signaling in non-cycling cells through nucleotide excision repair-dependent and -independent pathways. J. Cell Sci. 124, 435–446 [DOI] [PubMed] [Google Scholar]
- 52. Matsumoto M., Yaginuma K., Igarashi A., Imura M., Hasegawa M., Iwabuchi K., Date T., Mori T., Ishizaki K., Yamashita K., Inobe M., Matsunaga T. (2007) Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells. J. Cell Sci. 120, 1104–1112 [DOI] [PubMed] [Google Scholar]
- 53. Lehmann A. R. (2011) DNA polymerases and repair synthesis in NER in human cells. DNA Repair 10, 730–733 [DOI] [PubMed] [Google Scholar]
- 54. Waters L. S., Minesinger B. K., Wiltrout M. E., D'Souza S., Woodruff R. V., Walker G. C. (2009) Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 73, 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]