Background: Radioresistance is a critical factor restricting efficacy of radiotherapy.
Results: Phosphorylation of both rpS3 and TRAF2 induces dissociation of rpS3-TRAF2 complex and influences radioresistance through activation of NF-κB pathway.
Conclusion: Phosphorylation of rpS3 and TRAF2 is a key control point of radioresistance in NSCLC cells.
Significant: Our findings reveal a novel radioresistance mechanism through functional orchestration of rpS3, TRAF2, and NF-κB in NSCLC cells.
Keywords: Lung Cancer, NF-kappa B (NF-KB), Phosphorylation, Radiation Biology, Serine Threonine Protein Kinase, TRAF2, Non-small Cell Lung Cancer, Radioresistance, rpS3
Abstract
Radioresistance is considered as a main factor restricting efficacy of radiotherapy. However, the exact molecular mechanism of radioresistance has not been explained yet. In this study, to elucidate radioresistance mechanism in lung cancer, we compared radiation responses in two types of non-small cell lung cancer (NSCLC) cells with different radiosensitivity and identified key molecules conferring radioresistance. In radioresistant NSCLC cells, ionizing radiation (IR) led to casein kinase 2α (CK2α)- and PKC-mediated phosphorylation of rpS3 and TRAF2, respectively, which induced dissociation of rpS3-TRAF2 complex and NF-κB activation, resulting in significant up-regulation of prosurvival genes (cIAP1, cIAP2, and survivin). Also, dissociated phospho-rpS3 translocated into nucleus and bound with NF-κB complex (p65 and p50), contributing to p65 DNA binding property and specificity. However, in radiosensitive NSCLC cells, IR-mediated rpS3 phosphorylation was not detected due to the absence of CK2α overexpression. Consequently, IR-induced rpS3-TRAF2 complex dissociation, NF-κB activation, and prosurvival gene expression were not presented. Taken together, our findings revealed a novel radioresistance mechanism through functional orchestration of rpS3, TRAF2, and NF-κB in NSCLC cells. Moreover, we provided the first evidence for the function of rpS3 as a new TRAF2-binding protein and demonstrated that phosphorylation of both rpS3 and TRAF2 is a key control point of radioresistance in NSCLC cells. These results suggest that regulation of rpS3 and TRAF2 in combination with radiotherapy could have high pharmacological therapeutic potency for radioresistance of NSCLC.
Introduction
Lung cancer is not only the leading cause of cancer death, but also the most commonly diagnosed cancer type worldwide. It is separated into two morphologic types: non-small cell lung cancer (NSCLC),3 making up the majority of lung cancer cases, and small cell lung cancer (SCLC). Most patients with lung cancer are diagnosed with inoperable and advanced stage disease (1, 2). Accordingly, radiotherapy performs a critical role in curative management of these inoperable NSCLC patients (3). However, therapeutic outcomes of radiotherapy are not fully satisfactory in many cases, and radioresistance is considered as a main factor that restricts successful treatment of radiotherapy (4).
Radioresistance is closely associated with alteration of radiation-induced DNA damage responses and enhanced repair capacity, resulting in resistance to apoptosis and tumor repopulation during radiation treatment (5). Upon irradiation, numerous genes and proteins are involved in shifting of balance between cell survival and death (6). Cancer cells may acquire resistance to radiation-induced apoptosis by dynamic interplay and regulation of multiple prosurvival factors (7). Nuclear factor-κB (NF-κB) is a typical intrinsic cellular suppressor of apoptosis. After exposure to both low and high doses of ionizing radiation (IR), DNA damage-induced NF-κB promotes antiapoptotic gene expression. Also, constitutive activation of NF-κB is associated with oncogenic transformation, chemo/radioresistance, and metastasis. Therefore, its activation has been reported to be a novel marker of tumor radioresistance, and researchers have focused on the mechanism of NF-κB-mediated radioresistance in radiosensitizer development (8, 9).
NF-κB activation is intimately involved in tumor necrosis factor receptors (TNFRs)-mediated signaling, which has been established in radiation-induced adaptive responses (10). Because TNFRs do not have any enzymatic properties, the signaling processes are achieved by recruitment of intracellular scaffold proteins including TNFR-associated factor 2 (TRAF2), TNFR1-associated death domain protein (TRADD), and receptor-interacting protein 1 (RIP1). In response to TNFα and other stress stimuli such as radiation, the cytoplasmic adaptor proteins regulate activation of inhibitor of κB (IκB) kinase (IKK) complex. IKK phosphorylation of IκB proteins leads to their Lys-48-linked polyubiquitination and subsequent degradation, allowing nuclear translocation of NF-κB and regulation of antiapoptotic gene expression (11, 12). Seven members of the mammalian TRAF family have been considered as one of the most important scaffold proteins. In particular, as a prototypical member of the TRAFs, TRAF2 plays a pivotal role in protection against apoptosis and cellular stress (13, 14). Post-translational modifications such as phosphorylation and ubiquitination are a major control mechanism of TRAF2 function. Recent studies indicate that knockdown of TRAF2 induces radiosensitization of cancer cells through inhibition of NF-κB signaling and suggest that TRAF2 could be an attractive candidate as a radiosensitizing target (15).
Ribosomal protein S3 (rpS3) is a component of the 40S ribosomal subunit and mainly involved in ribosomal maturation and translation initiation through association with the eukaryotic initiation factors, eIF2 and eIF3 (16). Moreover, rpS3 has been demonstrated to be capable of various extraribosomal functions including regulation of DNA repair, transformation, development, apoptosis, and transcription (17, 18). rpS3 also appears to possess a general DNA repair endonuclease activity that participates in the cleavage of damaged DNA lesions caused by UV irradiation and oxidative stress (19). Significantly, rpS3 has been reported as another essential non-Rel subunit of the native NF-κB complex (p65 homodimer or p65-p50 heterodimer) and cooperates with Rel dimmers to achieve full DNA binding, regulating the specificity of NF-κB transcriptional activation (20). However, the precise physiological functions of rpS3 and the regulation mechanism have remained unclear.
The purpose of this study was to elucidate radioresistance mechanism in NSCLC cells and to identify key molecules conferring radioresistance. We demonstrated that IR-dependent phosphorylation of rpS3 and TRAF2 by casein kinase 2α (CK2α) and protein kinase C (PKC), respectively, plays an important role in survival signal transduction pathway in NSCLC cells. Our findings provide a possible explanation of how NSCLC cells could acquire and regulate resistance to radiation. Furthermore, we suggest that regulation of rpS3 and TRAF2 could be an attractive pharmaceutical target for radioresistance of NSCLC and could ultimately contribute to effective radiation treatment of lung cancer.
EXPERIMENTAL PROCEDURES
Cell Culture and Irradiation
Human NSCLC cell lines, A549 and NCI-H460, were acquired from the American Type Culture Collection (ATCC). Cells were grown in RPMI 1640 medium supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in 95% air, 5% CO2. Cells were exposed to a single dose of γ-rays using the Gamma Cell 40 Exactor (Nordion International, Inc., Kanata, Ontario, Canada) at a dose rate of 0.81 grays/min.
siRNA Transfection
Following the manufacturer's instructions, cells were seeded on 6-well dishes and transfected with siRNA oligonucleotides (100 ng) against rpS3, CK2α, and PKCδ/ϵ (ON-TARGETplus SMARTpool, Dharmacon, Chicago, IL), using DharmaFECT 1 reagent.
Colony-forming Assay
Cells were plated at a density of 300 cells in 6-well dishes and transiently transfected with rpS3-siRNA. After 24 h, cells were exposed to specific dose of IR and subsequently grown for 14 days. Then, cells were fixed with 10% methanol, 10% acetic acid and stained with 1% crystal violet solution. Colonies containing more than 50 cells were scored as survivors by using densitometric software (21).
Northern Blot Analysis
After 2 h of irradiation, total cellular RNA (20 μg) was isolated from the cells (5 × 106 cells) using TRIzol (Invitrogen). Northern blotting was performed as described previously (22), using radiolabeled DNA probe targeting human rpS3 and CK2α cDNA.
Western Blot Analysis and Immunoprecipitation
Following the desired treatments, total cell lysates were prepared using RIPA lysis buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100, 25 mm NaF, 1 mm DTT, 20 mm EGTA, 1 mm Na3VO4, 0.3 mm PMSF, and 5 units/ml aprotinin). As occasion demanded, nuclear or ribosome fractionation was performed as described previously (18). For Western blot analysis, denaturated protein lysates (40 μg) were subjected to SDS-PAGE. Separated proteins were transferred to a nitrocellulose membrane followed by blocking with 5% skim milk in TBST (10 mm Tris, 100 mm NaCl, and 0.1% Tween 20) for 1 h at room temperature. Membranes were then probed with specific primary antibodies followed by peroxidase-conjugated secondary antibody and visualized using an ECL detection system. For immunoprecipitation, protein (200 μg) was immunoprecipitated overnight with specific primary antibody and protein A-agarose beads (50 μl, Santa Cruz Biotechnology, Santa Cruz, CA). They were washed three times with lysis buffer and boiled in 2× SDS sample buffer for 5 min followed by centrifugation. Resulting clear supernatants were subjected to SDS-PAGE. The following steps were taken using the same method applied for Western blotting. All antibodies used for Western blot analysis and immunoprecipitation were acquired from Santa Cruz Biotechnology and Cell Signaling Technology (Beverly, MA).
In Vitro and In Vivo Kinase Assay
For in vitro kinase assay, cell lysates (after 2 h of irradiation) were prepared using RIPA lysis buffer, immunoprecipitated with specific primary antibody overnight, and then incubated for 4 h with protein A-Sepharose beads (50 μl, Invitrogen). Beads were washed with RIPA lysis buffer and reacted with His-tagged rpS3 (WT or T221A) in the presence of [γ-32P]ATP (10 μCi) (PerkinElmer Life Sciences) and 30 μl of kinase buffer (20 mm Tris, pH 7.5, and 10 mm MgCl2) for 30 min. The reaction was stopped by the addition of 4× SDS sample buffer and boiling for 5 min. Samples were subjected to SDS-PAGE and analyzed by autoradiography to determine the phosphorylation status of rpS3 by CK2α. For the in vivo kinase assay, cells were transfected with FLAG-CK2α (WT or kinase-dead) or co-transfected with CK2α (WT) and HA-rpS3 (WT or T221A) for 24 h and then irradiated. Lysates were immunoprecipitated with specific primary antibody overnight. The Western blotting was conducted using phospho-Ser/Thr antibody.
In Vitro Pull-down Assay
Prepared TRAF2- (or rpS3)-bound resin was incubated at 4 °C for 8 h with 10 μg of purified GST-rpS3 (or GST-TRAF2) in 100 μl of the binding buffer (20 mm Tris-HCl, pH 7.9, 100 mm KCl, 2.5 mm CaCl2, 2.5 mm MgCl2, 1 mm DTT, and 0.1% Triton X-100). After a brief centrifugation, the beads were washed three times with washing buffer (50 mm imidazole, 500 mm NaCl, and 20 mm Tris-HCl, pH 7.9) and resuspended in elution buffer (1 m imidazole, 500 mm NaCl, and 20 mm Tris-HCl, pH 7.9). The samples were subjected to SDS-PAGE and immunoblotting with a primary anti-GST antibody and a peroxidase-conjugated secondary antibody.
Luciferase Reporter Gene Assay
Cells were transiently co-transfected with 3 μg of NF-κB luciferase reporter gene (NF-κB-Luc) plasmid, IκBα mutant plasmid (ΔIκBα), CK2α-siRNA, or PKCδ-siRNA. Following overnight transfection, luciferase reporter gene assay was carried out as described previously (22).
Chromatin Immunoprecipitation (ChIP) Assay
After 2 h of irradiation, HA-rpS3- (WT) and HA-rpS3 (T221A)-expressing cells (5 × 108) were cross-linked in 1% formaldehyde and quenched in 125 mm glycine followed by washing in PBS. Cells were lysed and sonicated. The extract was centrifuged, diluted in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1 mm EDTA, 20 mm Tris-HCl, pH 8.1, and 200 mm NaCl), and precleared using protein A/G-agarose and calf thymus DNA at 4 °C for 1 h. Next, immunoprecipitation was performed with anti-p65, -rpS3, or -IgG antibody. Immunoprecipitates were collected and washed in low salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1, and 150 mm NaCl), high salt buffer (as before, containing 500 mm NaCl), and LiCl wash buffer (0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mm EDTA, and 10 mm Tris-HCl, pH 8.1). Then, they were washed two times in 10 mm Tris, 5 mm EDTA. DNA was extracted from beads using 100 μl of elution buffer (1% SDS and 0.1 m NaHCO) and supplemented with 0.25 m NaCl. Following overnight incubation at 65 °C for reversal of cross-links, samples were incubated for an additional 1 h at 65 °C with 10 μm EDTA, 40 μm Tris, pH 6.8, and 2 μg of proteinase K. DNA was purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA). PCR was performed with the primers that encompass the human interleukin-8 (IL-8), IκBα, and β-actin promoter. Primers were as follows: IL-8, 5′-GGGCCATCAGTTGCAAATC-3′ and 5′-TTCCTTCCGGTGGTTTCTTC-3′; IκBα, 5′-GACGACCCCAATTCAAATCG-3′ and 5′-TCAGCTCGGGGAATTTCC-3′; β-actin, 5′-TGCACTGTGCGGCGAAGC-3′ and 5′-TCGAGCCATAAAAGGCAA-3′.
Real-time Reverse Transcription (RT)-PCR
cDNA was prepared following the protocol of the ImProm-IITM reverse transcription system (Promega, Madison, WI). Primers for cellular inhibitor of apoptosis 1 (cIAP1), cIAP2, and survivin were designed using Primer Express 4.0 (Applied Biosystems, Foster, CA). Real-time PCR was performed using an iCycler iQ multicolor real-time PCR detection system (Bio-Rad) as follows: initial denaturation at 94 °C for 5 min followed by 30 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 40 s, and final elongation at 72 °C for 10 min. The intensity of each gene was normalized against GAPDH expression.
DNA Fragmentation Assay
Cells were seeded at a density of 4 × 105 cells in 96-well plates, incubated overnight, and then exposed to the desired treatment of IR and/or siRNA. Apoptosis induction was determined by analysis of cytoplasmic histone-associated DNA fragmentation using a cell death detection kit (Roche Applied Science, Mannheim, Germany), according to the manufacturer's instructions.
Statistical Analysis
All numeric data were presented as mean ± S.D. and analyzed using the one-way analysis of variance on ranked data followed by a Tukey's honestly significant difference test and the two-way analysis of variance on ranked data and a Bonferroni's post test using Prism 4 (GraphPad Software, San Diego, CA). All results described in this study were confirmed by three independent experiments Supplemental Experimental Procedures.
RESULTS
Identification of rpS3 as a Target for Regulation of Radiosensitivity in NSCLC Cells
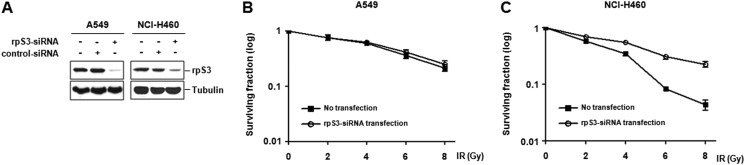
A549 and NCI-H460 cells can be used as models for radioresistant and radiosensitive NSCLC cells, respectively, due to their considerable difference in radiosensitivity (7, 23–25). Based on diverse extraribosomal functions of ribosomal proteins, we hypothesized that they could be associated with tumor radioresistance. To identify novel radiosensitizing target(s), we performed siRNA screening directed against the ribosomal proteins in irradiated NSCLC cells. The results yielded several hits of ribosomal protein that have different survival effects in A549 and NCI-H460 cells. Among these hits, we decided to validate rpS3 as a novel radiosensitizing target. To investigate whether rpS3 regulates radiosensitivity in NSCLC cells, rpS3-siRNA was prepared (Fig. 1A) and applied to a colony-forming assay (Fig. 1, B and C). As a result, transient rpS3 knockdown did not change radiosensitivity in A549 cells, whereas treatment of rpS3-siRNA caused a significant increase in proliferation of NCI-H460 cells. Furthermore, to examine whether or not our results are cell line-specific, we used other models for radioresistant and radiosensitive NSCLC cells, NCI-H1299 and NCI-H157, respectively. We reconfirmed that A549 and NCI-H1299 cells are more radioresistant than NCI-H460 and H157 cells in accordance with previous studies (7, 23–25)(supplemental Fig. S1A). The possible regulatory effect of rpS3 on radiosensitivity was also verified in NCI-H1299 and NCI-H157 cells (supplemental Fig. S1, B and C). Taken together, these results indicate that silencing of rpS3 shows different responses between radioresistant and radiosensitive NSCLC cells and suggest the possibility that rpS3 could be a regulator of radiosensitivity in NSCLC cells.
FIGURE 1.
Identification of rpS3 as a target for regulation of radiosensitivity in NSCLC cells. A, efficacy and specificity of rpS3-siRNA were confirmed by Western blot analysis. B and C, cell survival curves of rpS3-silenced A549 and NCI-H460 cells under irradiation were assessed by a colony-forming assay. Gy, grays.
IR-activated Phosphorylation of rpS3 by CK2α in Radioresistant NSCLC Cells
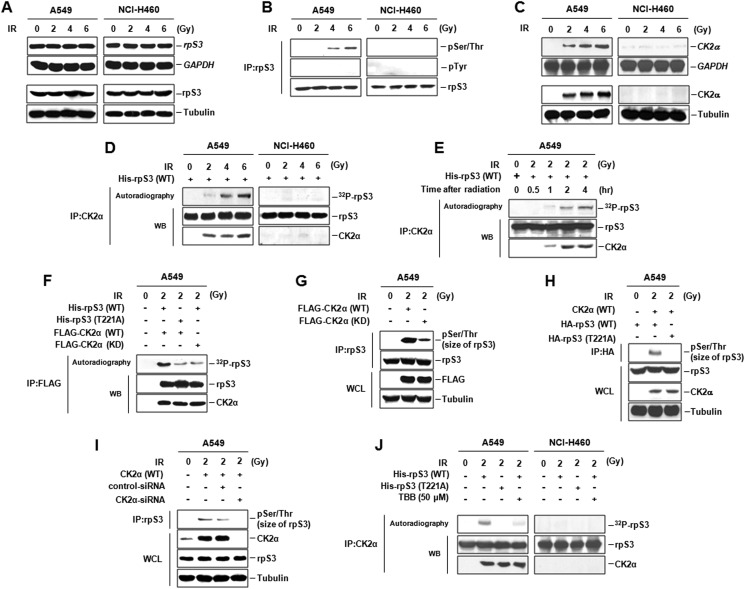
To investigate how rpS3 is associated with regulation of radiosensitivity in NSCLC cells, molecular modification and signaling involvement of rpS3 were monitored. rpS3 mRNA and protein were constitutively expressed without dependence on IR irradiation in both A549 and NCI-H460 cells (Fig. 2A). However, IR-induced rpS3 phosphorylation at Ser/Thr sites appeared only in radioresistant A549 cells (Fig. 2B). Next, we performed an in vitro kinase assay with purified recombinant rpS3 and several kinases immunoprecipitated by specific antibody from two NSCLC cell lines (data not shown). As a result, we found CK2α to be a candidate for an upstream kinase of rpS3. As shown in Fig. 2C, we identified a significant difference in IR-induced CK2α expression between radioresistant A549 and radiosensitive NCI-H460 cells. In NCI-H460 cells, CK2α mRNA and protein were expressed at extremely low levels irrelevant to the dose of IR. However, IR increased both mRNA and protein expression of CK2α in A549 cells. Similar results were observed in radioresistant NCI-H1299 and radiosensitive NCI-H157 cells (supplemental Fig. S2A). Interestingly, CK2α-mediated rpS3 phosphorylation was dose- and time-dependently induced only in radioresistant A549 cells (Fig. 2, D and E). To further investigate which residues are the target of CK2α in rpS3, based on the three-dimensional structure of rpS3 (Protein Data Bank (PDB) ID: 3KC4), we mutated several Ser/Thr residues to Ala and used this mutant as a substrate for FLAG-CK2α (WT). As shown in Fig. 2F, CK2α-induced rpS3 phosphorylation was eliminated by T221A mutation of rpS3, indicating that Thr-221 of rpS3 is required for CK2α-mediated phosphorylation in irradiated A549 cells. This specific phosphorylation event of rpS3 also occurs under in vivo conditions (Fig. 2, G and H). We verified that inhibition of CK2α expression led to complete blockage of the rpS3 phosphorylation in vivo in irradiated A549 cells (Fig. 2I). Direct association between IR-induced rpS3 phosphorylation at Thr-221 and CK2α was reconfirmed by treatment of CK2α inhibitor, 4,5,6,7-tetrabromobenzotriazole (TBB, Sigma-Aldrich) in A549 and NCI-H460 cells (Fig. 2J). Furthermore, CK2α-mediated phosphorylation of rpS3 was presented only in radioresistant NCI-H1299 cells under irradiation (supplemental Fig. S2B). According to these results, we propose the different radiation responses through rpS3 phosphorylation in NSCLC cells with different radiosensitivity and suggest that IR-activated CK2α directly phosphorylates rpS3 at Thr-221 under both in vitro and in vivo conditions in radioresistant NSCLC cells.
FIGURE 2.
IR-activated phosphorylation of rpS3 by CK2α in radioresistant NSCLC cells. A, IR-induced rpS3 mRNA and protein expression were detected by Northern blot analysis and immunoblotting, respectively. Gy, grays. B, IR-induced rpS3 phosphorylation was verified through immunoprecipitation (IP) of rpS3 followed by immunoblotting for phospho-Ser/Thr (pSer/Thr) and phospho-Tyr (pTyr) antibody. C, significant difference in IR-activated CK2α expression between A549 and NCI-H460 cells was detected by Northern blot analysis and immunoblotting. D and E, dose-dependent (D) and time-dependent (E) phosphorylation of rpS3 by CK2α was measured by in vitro kinase assay. WB, Western blot analysis. F, CK2α-mediated rpS3 phosphorylation at Thr-221 was confirmed by in vitro kinase assay with rpS3 T221A and CK2α (kinase-dead (KD)). G and H, phosphorylation of rpS3 by CK2α was also measured by in vivo kinase assay. WCL, whole cell lysates. I, direct association of IR-induced rpS3 phosphorylation with CK2α was confirmed by treatment of CK2α-siRNA. J, direct association between IR-induced rpS3 phosphorylation (Thr-221) and CK2α was reconfirmed by use of the rpS3 T221A and CK2α inhibitor, TBB, in both A549 and NCI-H460 cells.
IR-activated Nuclear Localization of Phospho-rpS3 in Radioresistant NSCLC Cells
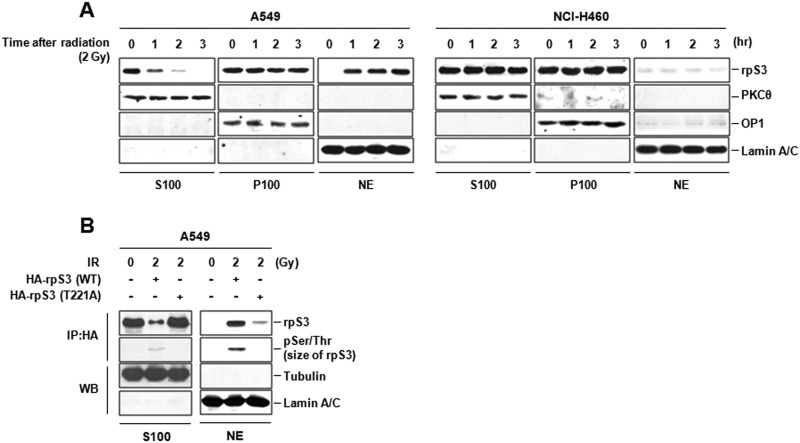
rpS3 critically regulates translation, and also possesses various extraribosomal functions involved in DNA repair, apoptosis, and transcriptional regulation (18). These nonribosomal multiple functions of rpS3 are thought to be possible according to its different post-translational modifications (26). To determine whether IR could change the subcellular localization of rpS3, the protein levels of rpS3 in nonribosomal, ribosomal, and nuclear region were investigated in irradiated A549 and NCI-H460 cells. As shown in Fig. 3A, IR caused a marked reduction of nonribosomal cytosolic rpS3 and concomitant increase of nuclear rpS3 with a time-dependent manner only in radioresistant A459 cells. To further examine whether IR-activated rpS3 phosphorylation affects nuclear localization of rpS3, we conducted immunoprecipitation from cytosol and nuclear fractions of irradiated A549 cells after transfection with HA-rpS3 (WT or T221A). We identified that IR-induced nuclear localization of rpS3 was significantly inhibited by transfection with rpS3 T221A mutant (Fig. 3B). Also, the involvement of IR-activated nuclear localization of rpS3 and its phosphorylation status was confirmed in NCI-H1299 and NCI-H157 cells. Under irradiation, nuclear localized rpS3 was observed only in radioresistant NCI-H1299 cells after transfection with HA-rpS3 (WT) (supplemental Fig. S3). Collectively, we suggest that IR-activated nuclear localization of nonribosomal rpS3 is phosphorylation (Thr-221)-dependent in NSCLC cells.
FIGURE 3.
IR-activated nuclear localization of phospho-rpS3 in radioresistant NSCLC cells. A, IR-induced translocation of rpS3 from cytosol into nucleus was assayed by Western blot analysis. The PKCθ oxidative phosphorylation complex 1 (OP1) and lamin A/C were used as markers for nonribosomal associated supernatant (S100), ribosomal pellet (P100), and nuclear extract (NE), respectively. 2 Gy, 2 grays. B, involvement of IR-activated nuclear localization of rpS3 and its phosphorylation status were determined by immunoprecipitation (IP) and Western blot analysis (WB). pSer/Thr, phospho-Ser/Thr.
IR-dependent Dissociation of rpS3-TRAF2 Complex in Radioresistant NSCLC Cells
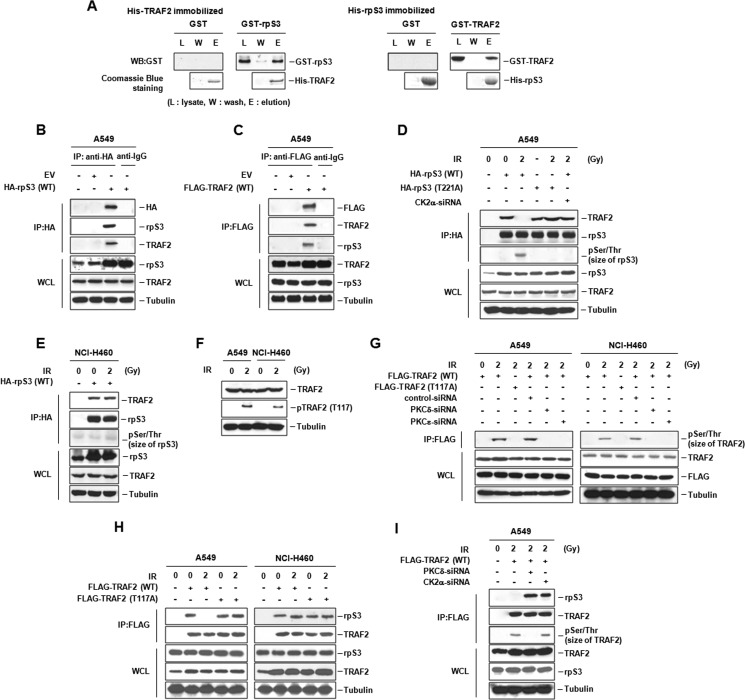
Next, we investigated IR-induced biochemical dynamic events. A yeast two-hybrid system was used to find proteins interacting with rpS3, and the results revealed several interacting partners of rpS3. TRAF2 was one of the most strongly and frequently interacting molecules for rpS3 in our assay system (data not shown). Based on regulatory properties of TRAF2 in tumorigenesis and biochemical relationships with various binding partners (14), we hypothesized that the relationship of rpS3 and TRAF2 could be functionally involved in the radioresistance of NSCLC cells. We examined the interaction of rpS3 with TRAF2 using in vitro pull-down assay and confirmed that rpS3 and TRAF2 can bind to each other (Fig. 4A). Also, the interaction of rpS3 (or TRAF2) with endogenous TRAF2 (or rpS3) was detected in vivo in A549 cells (Fig. 4, B and C). Interestingly, in radioresistant A549 cells, rpS3 was not able to form a complex with TRAF2 under irradiation, whereas rpS3-TRAF2 complex formation was detected in nonirradiated conditions. In rpS3 T221A or CK2a-siRNA transfected A549 cells, rpS3 formed a complex with TRAF2 under irradiation (Fig. 4D). However, unlike A549 cells showing IR-dependent rpS3-TRAF2 complex dissociation, in radiosensitive NCI-H460 cells, rpS3 bound with TRAF2 irrelevant to IR irradiation (Fig. 4E). Similar results were presented in radioresistant NCI-H1299 and radiosensitive NCI-H157 cells (supplemental Fig. S4A). These results suggest that IR-dependent dissociation of rpS3-TRAF2 complex is presented only in radioresistant NSCLC cells through CK2α-mediated rpS3 phosphorylation at Thr-221.
FIGURE 4.
IR-dependent dissociation of rpS3-TRAF2 complex in radioresistant NSCLC cells. A, the interaction of rpS3 with TRAF2 was verified by in vitro pull-down assay. Coomassie Blue staining showed that equal amounts of His fusion proteins were used. WB, Western blot analysis. B and C, in vivo binding of rpS3 and TRAF2 was measured by reciprocal immunoprecipitation (IP) assay. EV, empty vector. WCL, whole cell lysates. D, CK2α-activated rpS3 phosphorylation resulted in dissociation of rpS3-TRAF2 complex in irradiated A549 cells. Direct involvement of IR-induced rpS3 phosphorylation in the complex dissociation was verified by overexpression of rpS3 T221A and silencing of CK2α. Gy, grays; pSer/Thr, phospho-Ser/Thr. E, in irradiated NCI-H460 cells, rpS3-TRAF2 complex formation was shown through immunoprecipitation assay. F, phosphorylation status of TRAF2 (pTRAF2) was detected by immunoblotting in both A549 and NCI-H460 cells under irradiation. G, direct association of IR-activated TRAF2 phosphorylation with PKC was confirmed by use of TRAF2 T117A and PKCδ/ϵ-siRNA. H, in A549 and NCI-H460 cells, a critical role of TRAF2 phosphorylation in IR-dependent rpS3-TRAF2 complex dissociation was proved by overexpression of TRAF2 T117A. I, dissociation of rpS3-TRAF2 complex was dependent on phosphorylation status of rpS3 and TRAF2 by CK2α and PKC, respectively.
Post-translational modifications such as phosphorylation and ubiquitination are major regulation mechanisms of TRAF2 signaling including TNFα-mediated IKK activation. PKC phosphorylation of TRAF2 at Thr-117 is reported to promote IKK and NF-κB activation (12). To verify the effect of TRAF2 phosphorylation in formation/dissociation of rpS3-TRAF2 complex, we determined the phosphorylation status of TRAF2 in A549 and NCI-H460 cells. As shown in Fig. 4, F and G, IR-induced TRAF2 phosphorylation at Thr-117 was detected and abrogated by PKCδ/ϵ-siRNA in both A549 and NCI-H460 cells. These results indicated a direct involvement of PKC in IR-activated phosphorylation of TRAF2, suggesting that PKCδ/ϵ are upstream kinases of TRAF2. Next, we investigated whether or not TRAF2 phosphorylation actually influences the IR-dependent complex dissociation, and it was confirmed by use of T117A mutant of TRAF2. The results revealed that TRAF2 phosphorylation at Thr-117 is necessarily required for IR-dependent rpS3-TRAF2 complex dissociation in radioresistant A549 cells (Fig. 4H). As shown in supplemental Fig. S4B, although IR induced TRAF2 phosphorylation in both NCI-H1299 and NCI-H157 cells, dissociation of rpS3-TRAF2 complex was confirmed only in radioresistant NCI-H1299 cells. IR-induced dissociation of rpS3-TRAF2 complex was not found under treatment of CK2α-siRNA and PKCδ-siRNA in irradiated A549 cells (Fig. 4I). Taken together, these results indicate that rpS3-TRAF2 complex is dissociated only if both rpS3 and TRAF2 are phosphorylated by IR-activated CK2α and PKCδ, respectively. In other words, formation and dissociation of rpS3-TRAF2 complex are closely associated with phosphorylation status of rpS3 and TRAF2 in NSCLC cells.
NF-κB Activation by IR-induced Dissociation of rpS3-TRAF2 Complex in Radioresistant NSCLC Cells
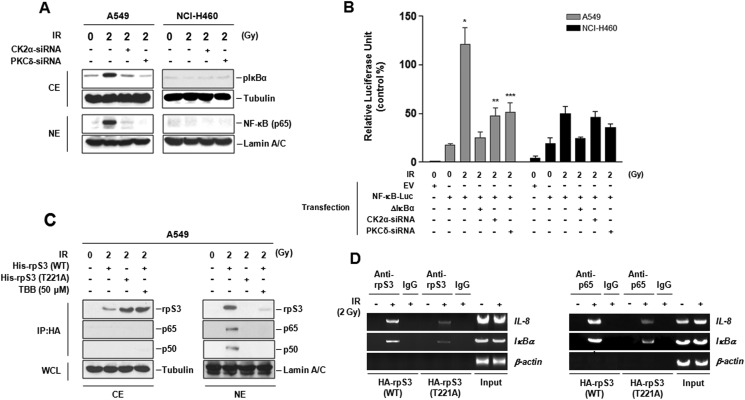
We investigated the relationship of IR-induced rpS3-TRAF2 complex dissociation and activation of NF-κB pathway in NSCLC cells. As shown in Fig. 5, A and B, IR-induced increase in IκBα phosphorylation, nuclear translocation of p65, and NF-κB transcriptional activation was detected only in radioresistant A549 cells. These increases were significantly suppressed through inhibition of rpS3-TRAF2 complex dissociation by CK2α- and PKCδ-siRNA treatment. These results were also confirmed in radioresistant NCI-H1299 and radiosensitive NCI-H157 cells (supplemental Fig. S5A). In addition, Thr-221 phosphorylation-dependent nuclear localized rpS3 bound with NF-κB complex (p65 and p50) in irradiated A549 cells (Fig. 5C). Direct association of these phosphorylation-dependent specific events of rpS3 with CK2α was reconfirmed by use of CK2α inhibitor, TBB. Taken together, these results indicate that activation of NF-κB pathway in NSCLC cells is intimately involved in dissociation of rpS3-TRAF2 complex and cellular post-translational modification (phosphorylation and localization) of rpS3 under irradiation.
FIGURE 5.
NF-κB activation by IR-induced dissociation of rpS3-TRAF2 complex in radioresistant NSCLC cells. A, effect of rpS3-TRAF2 complex dissociation on IR-induced IκBα phosphorylation and p65 nuclear translocation was confirmed by silencing of CK2α and PKCδ. CE, cytoplasm extract; NE, nuclear extract; Gy, grays. B, effect of rpS3-TRAF2 complex dissociation on IR-induced NF-κB transcriptional activity was measured by luciferase assay. Data represent the mean ± S.D. *, p < 0.05; NF-κB-Luc plasmid-transfected cells versus NF-κB-Luc plasmid-transfected irradiated cells, **, p < 0.05; NF-κB-Luc plasmid-transfected irradiated cells versus NF-κB-Luc plasmid and CK2α-siRNA-transfected irradiated cells, ***, p < 0.05; NF-κB-Luc plasmid-transfected irradiated cells versus NF-κB-Luc plasmid and PKCδ-siRNA-transfected irradiated cells. EV, empty vector. C, binding of IR-dependent nuclear localized phospho-rpS3 with NF-κB complex (p65 and p50) was shown by immunoprecipitation assay. D, recruitment of rpS3 and endogenous p65 to the κB site of IL-8 and IκBα promoters was measured by ChIP analysis. A549 cells were transfected with HA-rpS3 (WT) and HA-rpS3 (T221A), allowed to recover for 24 h, irradiated, and then harvested. Chromatin-bound DNA was immunoprecipitated with anti-rpS3 and anti-p65 antibody, respectively. 10% of the chromatin samples were used as positive control (Input).
Next, to verify the direct evidence for association between rpS3 and NF-κB activation, we performed additional experiments using a ChIP assay (Fig. 5D). We focused on specific κB sites within the promoter of IL-8 and IκBα as well known NF-κB target genes (20). In comparison with transfection of HA-rpS3 (WT) into A549 cells, transfection of HA-rpS3 (T221A) caused a significant decrease in the recruitment of rpS3 and p65 to IL-8 and IκBα promoter region. In radioresistant NCI-H1299 cells, it was reconfirmed that rpS3 phosphorylation critically affects the recruitment of p65 to IL-8 and IκBα promoter region (supplemental Fig. S5B). However, there was not much recruitment of rpS3 or p65 to the β-actin promoter, which does not have κB sites. These results indicated that the recruitment of rpS3 and p65 to the chromatin regions is specific for κB sites of target genes, and the p65 DNA binding property is closely associated with IR-dependent phosphorylation of rpS3 at Thr-221. Taken together, these results suggest that significant increase of NF-κB transcriptional activation in radioresistant NSCLC cells could be possible by elevating p65 DNA binding properties through IR-induced binding of phospho-rpS3 with p65 subunit of NF-κB complex. Ultimately, these events could promote NF-κB-mediated prosurvival gene expression.
Regulation of Radioresistance by IR-induced Modification of rpS3 and TRAF2 in NSCLC Cells
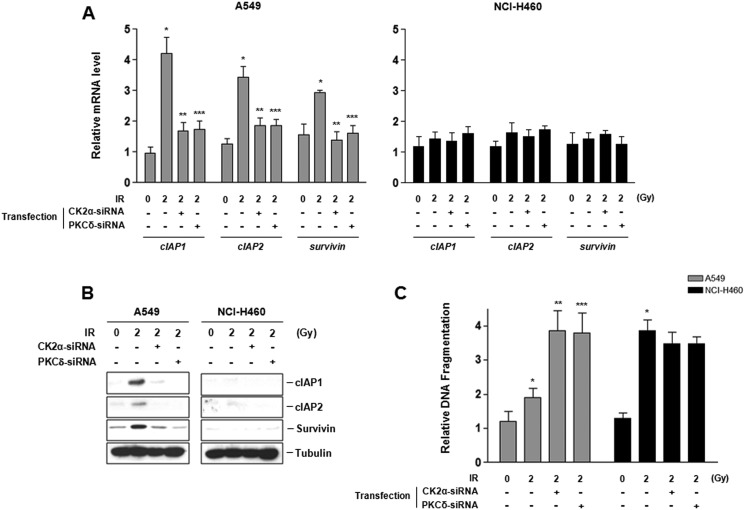
Next, we examined IR-induced transcriptional alterations in cIAP1, cIAP2, and survivin, which are well known as NF-κB-regulated prosurvival genes (27). Only in radioresistant A549 cells did significant induction of cIAP1, cIAP2, and survivin mRNA and concomitant overexpression of their proteins occur following irradiation. Expression of these genes was reduced by CK2α- and PKCδ-siRNA treatment (Fig. 6, A and B). Furthermore, we confirmed that phosphorylation of rpS3 and TRAF2 is involved in regulation of radiation-induced apoptosis using treatment of siRNA against their upstream kinase. When compared with NCI-H460 cells, A549 cells were more resistant to IR-mediated cytoplasmic histone-associated DNA fragmentation. However, silencing of CK2α and PKCδ caused a significant increase of radiosensitivity in irradiated A549 cells (Fig. 6C). Similar results were observed in radioresistant NCI-H1299 and radiosensitive NCI-H157 cells (supplemental Fig. S6). These results suggest that IR-induced phosphorylation of rpS3 and TRAF2 by CK2α and PKC, respectively, leads to dissociation of rpS3-TRAF2 complex. Then, activation of their downstream signal transduction process, such as NF-κB signaling, accelerates prosurvival gene expression, regulates radiation-induced apoptosis, and ultimately confers radioresistance in NSCLC cells.
FIGURE 6.
Regulation of radioresistance by IR-induced modification of rpS3 and TRAF2 in radioresistant NSCLC cells. A, quantitative analysis of mRNA levels of prosurvival genes (cIAP1, cIAP2, and survivin) in irradiated A549 and NCI-H460 was analyzed by real-time RT-PCR. Data represent the mean ± S.D. *, p < 0.05; irradiated cells versus control cells, **, p < 0.05; irradiated cells versus CK2α-siRNA-transfected irradiated cells, ***, p < 0.05; irradiated cells versus PKCδ-siRNA-transfected irradiated cells. Gy, grays. B, expression of cIAP1, cIAP2, and survivin in irradiated A549 and NCI-H460 was analyzed by Western blot analysis. C, functional involvement of phosphorylation of rpS3 and TRAF2 in IR-induced DNA damage response was measured by apoptosis assay. Data represent the mean ± S.D. *, p < 0.05; irradiated cells versus control cells, **, p < 0.05; irradiated cells versus CK2α-siRNA-transfected irradiated cells, ***, p < 0.05; irradiated cells versus PKCδ-siRNA-transfected irradiated cells.
DISCUSSION
IR increases intracellular free radical formation followed by DNA strand breaks and subsequent dysfunction of mitochondria, endoplasmic reticulum, and other organelles. These radiation-induced cellular events lead to activation of proapoptotic signaling, and eventually, tumor cell killing (28). At the same time, damage-induced cell cycle arrests provide necessary and sufficient time for repair, and then a large number of proteins are activated to enable repair of radiation-induced DNA damage. Abnormal activation of cell survival signaling and enhancement of repair capacity have been reported as one of main causes for resistance to radiation, and further, they induce elements of carcinogenesis such as repopulation and invasion of tumor cells (29). Some studies on the mechanism of radioresistance in NSCLC have suggested the potential involvement of p53 mutation, altered expression of survival proteins including X-linked inhibitor of apoptosis protein (XIAP) and survivin, or activation of phosphoinositide 3-kinase (PI3K)/AKT signaling (15). However, unfortunately, these studies have provided only limited understanding of the lung cancer radioresistance. At present, there are no molecular targeted approaches that are successfully combined with radiotherapy of NSCLC. Identification of molecular factors responsible for conferring radioresistance has been necessary to develop new drugs that increase sensitivity to radiation. Thus, an accurate knowledge of the functional mechanisms is urgently needed to overcome the radioresistance of NSCLC.
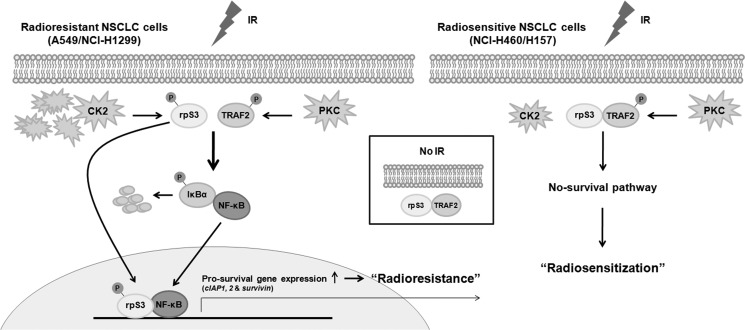
In this study, we sought to clarify radioresistance mechanism in two types of NSCLC cells, radioresistant (A549/NCI-H1299) and radiosensitive (NCI-H460/H157) NSCLC cells. We demonstrated that IR-dependent post-translational modifications of rpS3 and TRAF2 play a critical role in regulation of radioresistance in NSCLC cells. A schematic model based on our results is presented in Fig. 7. Several important findings include the following: (i) rpS3 directly interacts with TRAF2 under nonirradiated condition; (ii) in radioresistant NSCLC cells, IR leads to CK2α- and PKC-mediated phosphorylation of rpS3 and TRAF2, respectively, and then induces rpS3-TRAF2 complex dissociation, resulting in NF-κB activation and up-regulation of prosurvival gene expression (cIAP1, cIAP2, and survivin); (iii) the dissociated phospho-rpS3 translocates into the nucleus and binds with NF-κB complex (p65 and p50), inducing increase in NF-κB DNA binding activity; and (iv) in radiosensitive NSCLC cells, IR-mediated rpS3 phosphorylation is not detected due to the absence of CK2α overexpression. Also, concomitantly, activation of NF-κB and increase of prosurvival gene expression are not detected.
FIGURE 7.
Schematic diagrams illustrating the rpS3- and TRAF2-mediated radioresistance mechanism in NSCLC cells.
Ribosomal proteins are abundant in most cells and have been recently implicated in several nonribosomal functions. The multiple extraribosomal functions of rpS3 are closely associated with their post-translational modifications (26). Specifically, their phosphorylation status is crucial for the control of rpS3 functions under different physiological conditions including oxidative stress (18). Also, rpS3 switches from apoptosis to DNA repair through its phosphorylation (30). Regulatory roles of rpS3 have been mainly focused on the N-terminal region, particularly the KH domain, whereas those functions for the C-terminal region have been relatively unknown (31). Here, for the first time, we have indicated different radiation responses through rpS3 phosphorylation in radioresistant and radiosensitive NSCLC cells and demonstrated that IR-activated CK2α directly phosphorylates rpS3 at the C-terminal threonine residue (Thr-221) only in radioresistant NSCLC cells. Also, interestingly, the difference in rpS3 phosphorylation between the two types of NSCLC cells resulted from the existence of CK2α, an upstream kinase activator of rpS3 (Fig. 2 and supplemental Fig. S2).
CK2, a ubiquitous and constitutively active Ser/Thr protein kinase, is a heterotetramer composed of two catalytic subunits (α and α′) and two regulatory subunits (β). However, the individual subunits have been shown to act as free proteins. CK2 interacts with a large number (>300) of substrates in diverse locations as a master regulator and modulates cell cycle progression, proliferation, angiogenesis, and DNA repair (32). CK2 shows deregulated expression in most cancers, indicating increased protein expression and nuclear localization in comparison with their normal counterparts. siRNA-induced silencing of the CK2 catalytic subunits results in significant cell death (33). Given the essential roles for cell proliferation, CK2 has been proposed as a potentially important target for anticancer therapy (34). Based on structural studies (PDB ID: 1JWH), CK2 reveals a close interaction between the N-terminal region and activation segment, and this unique structure possibly contributes a constitutively active conformation of CK2 (35). Thus, activation of CK2 is intimately associated with its protein expression level. Interestingly, we found that IR-induced CK2α overexpression and subsequent CK2α-mediated rpS3 phosphorylation are presented only in radioresistant A549/NCI-H1299 cells (Fig. 2 and supplemental Fig. S2). In dividing cells, phosphorylation of rpS3 is involved in prevention of its proapoptotic function (30). As shown in Fig. 6C and supplemental Fig. S6, siRNA-mediated CK2α suppression, inducing subsequent inhibition of rpS3 phosphorylation, actually enhanced radiosensitivity only in irradiated radioresistant A549/NCI-H1299 cells. Taken together, these results suggest that CK2α could promote cell survival through regulation of rpS3 phosphorylation and allow cells to either live or die in response to radiation. Considering the meaningful relationship of radioresistance with CK2 overexpression, we suggest that CK2 expression in NSCLC or other cancer types could supply somewhat predictable information on efficacy of radiotherapy.
Also, we demonstrated that regulation of radioresistance through CK2α-mediated rpS3 phosphorylation is closely related with TRAF2 in irradiated NSCLC cells (Fig. 4 and supplemental Fig. S4). TRAF2, a versatile adaptor protein, has been reported to perform diverse biological functions including apoptosis, immune cell signaling, inflammation, organogenesis, and cell survival (36). TRAF2 is involved in activation of diverse protein kinase cascades and both canonical and noncanonical activation pathways of NF-κB (37). Moreover, inhibition of TRAF2 down-regulates G2-M phase control proteins, resulting in increased G2-M cell cycle arrest, growth inhibition, and radiosensitization (15). These regulatory effects of TRAF2 are mainly dependent on interactions with other proteins: TRADD, FAS-associated death domain protein (FADD), RIP1, TGFβ-associated kinase 1 (TAK1), TAK1-binding protein (TAB), NF-κB-inducing kinase (NIK), A20, cIAPs, and TRAF5 (14, 38). Thus, the functional complexity of TRAF2 signaling is considered to be associated with recruitment and the combination of the numerous TRAF2-interacting proteins. In this study, we showed the first evidence for the function of rpS3 as a new TRAF2-binding protein and demonstrated the formation of rpS3-TRAF2 complex under nonirradiated condition. Also, we proposed a novel radioresistance mechanism through IR-induced rpS3-TRAF2 complex dissociation. As shown in Fig. 4 and supplemental Fig. S4, in radiosensitive NCI-H460/H157 cells, rpS3 was always bound to TRAF2 irrelevant to IR irradiation. On the other hand, in radioresistant A549/NCI-H1299 cells, rpS3-TRAF2 complex was dissociated only if both rpS3 and TRAF2 were phosphorylated by IR-activated CK2α and PKC, respectively. Under phosphorylation of rpS3 at Thr-221, PKC-mediated TRAF2 phosphorylation (Thr-117) was necessarily required for rpS3-TRAF2 complex dissociation. Several previous studies have reported that TRAF2 phosphorylation at Thr-117 promotes recruitment of catalytic IKKα and IKKβ subunit. In addition, this phosphorylation determines Lys-63-linked polyubiquitination of TRAF2, which contributes association of TRAF2 with TAK1 binding TAB2/3 adaptors and subsequent activation of JNK, IKK, and NF-κB (12). In addition, we verified that rpS3 directly binds to C-terminal region of TRAF2 by using a yeast two-hybrid system (data not shown). Taken together, these results suggest that TRAF2 is sequestered by rpS3 under nonirradiated condition and that the rpS3-sequestering region of TRAF2 might coincide with the binding site of the TRAF2-associated proteins, TRADD and RIP. It appears as if binding of rpS3 with TRAF2 could inhibit the cell proliferation and survival ability of TRAF2. Thus, under irradiation, despite the PKC-mediated TRAF2 phosphorylation, the absence of CK2α in radiosensitive NCI-H460/H157 cells could result in the maintenance of rpS3-TRAF2 complex formation, which inhibits subsequent NF-κB activation. However, radiation-dependent phosphorylation of both rpS3 and TRAF2 affects dissociation of stable rpS3-TRAF2 complex and induces release of phospho-rpS3 and phospho-TRAF2, a functionally active form.
Nuclear accumulation of rpS3 is reported to increase DNA repair through their endonuclease activity, thereby facilitating neuronal survival from UV radiation (30). As shown in Fig. 3 and supplemental Fig. S3, we demonstrated that nonribosomal rpS3 is increased in nuclear translocation and accumulates only in radioresistant A549/NCI-H1299 cells under IR irradiation. IR-activated nuclear localization of rpS3 was phosphorylation-dependent. Furthermore, we indicated that nuclear-localized phospho-rpS3 interacts with NF-κB complex (p65 and p50) (Fig. 5C). According to these findings, we suggest that the rpS3 function as a target molecule of radioresistance in NSCLC cells may be conferred by its distinct subcellular localization through regulation of phosphorylation status. Interestingly, the results of Fig. 5D and supplemental Fig. S5B verified that p65 DNA binding is closely associated with IR-dependent phosphorylation of rpS3 at Thr-221. Collectively, we propose that IR-induced direct binding of phospho-rpS3 with p65 subunit in the nucleus could stabilize the NF-κB complex, enhance NF-κB DNA binding activity, and elevate NF-κB-mediated prosurvival gene expression in radioresistant A549/NCI-H1299 cells. Simultaneously, nuclear accumulation of rpS3 as an endonuclease might enhance the repair of IR-damaged DNA in A549/NCI-H1299 cells. These synergistic survival effects could raise the possibility that A549/NCI-H1299 cells acquire and regulate radioresistance in comparison with NCI-H460/H157 cells. Also, under irradiation, dissociation of phospho-TRAF2 from the complex could activate the IKK complex, leading to IκBα phosphorylation and NF-κB activation. Taken together, these results suggest that radioresistance in NSCLC cells is due to acceleration of cell survival property and repair capacity through functional association with rpS3, TRAF2, and NF-κB. Actually, CK2α- and PKCδ-mediated dissociation of rpS3-TRAF2 complex promoted radioresistance by inhibition of apoptosis in IR-irradiated radioresistant A549/NCI-H1299 cells (Fig. 6 and supplemental Fig. S6).
NSCLC reveals significant molecular heterogeneity, and four NSCLC cell lines used in this study have slightly different characterizations. Nevertheless, based on their degree of radioresistance/radiosensitivity, they showed the same radiation responses through our novel radioresistance mechanism. Accordingly, we determined that our results are not cell line-specific and that they could be fully extended into the entire scope of NSCLC cells.
Up to now, the single exact molecular mechanism of radioresistance in NSCLC cells was still unclear. In this study, we propose a novel radioresistance mechanism through functional orchestration of rpS3, TRAF2, and NF-κB in NSCLC cells and demonstrate that phosphorylation of both rpS3 and TRAF2 is a key control point of radioresistance in NSCLC cells. Despite excluding the potential role of the tumor microenvironment in radioresistance, our findings provide a possible explanation for how NSCLC cells could acquire and regulate resistance to radiation. Furthermore, we suggest that regulation of rpS3 and TRAF2 in combination with radiotherapy could have high pharmacological therapeutic potency for radioresistance, and eventually, lead to increased efficacy of radiation treatment in NSCLC.
This work was supported by the Nuclear R&D Program (Grants 2011-0030601 and 2011-0020777) and by the Basic Science Research Program (Grant 2011-0007625) through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology.

This article contains supplemental Experimental Procedures and Figs. S1–S6.
- NSCLC
- non-small cell lung cancer
- cIAP
- cellular inhibitor of apoptosis
- CK2
- casein kinase 2
- IKK
- inhibitor of κB (IκB) kinase
- IR
- ionizing radiation
- RIP
- receptor-interacting protein
- rpS3
- ribosomal protein S3
- TAK1
- TGFβ-associated kinase 1
- TNFR
- tumor necrosis factor receptor
- TRADD
- TNFR1-associated death domain protein
- TRAF
- TNFR-associated factor
- TBB
- 4,5,6,7-tetrabromobenzotriazole
- Luc
- luciferase.
REFERENCES
- 1. Baumann M., Krause M., Zips D., Petersen C., Dittmann K., Dörr W., Rodemann H. P. (2004) Molecular targeting in radiotherapy of lung cancer. Lung Cancer 45, Suppl. 2, S187–S197 [DOI] [PubMed] [Google Scholar]
- 2. Sause W. T. (1999) The role of radiotherapy in non-small cell lung cancer. Chest 116, 504S–508S [DOI] [PubMed] [Google Scholar]
- 3. Koh P. K., Faivre-Finn C., Blackhall F. H., De Ruysscher D. (2012) Targeted agents in non-small cell lung cancer (NSCLC): clinical developments and rationale for the combination with thoracic radiotherapy. Cancer Treat. Rev. 38, 626–640 [DOI] [PubMed] [Google Scholar]
- 4. Bussink J., van der Kogel A. J., Kaanders J. H. (2008) Activation of the PI3-K/AKT pathway and implications for radioresistance mechanisms in head and neck cancer. Lancet Oncol. 9, 288–296 [DOI] [PubMed] [Google Scholar]
- 5. Shimura T., Kakuda S., Ochiai Y., Nakagawa H., Kuwahara Y., Takai Y., Kobayashi J., Komatsu K., Fukumoto M. (2010) Acquired radioresistance of human tumor cells by DNA-PK/AKT/GSK3β-mediated cyclin D1 overexpression. Oncogene 29, 4826–4837 [DOI] [PubMed] [Google Scholar]
- 6. Igney F. H., Krammer P. H. (2002) Death and anti-death: tumour resistance to apoptosis. Nat. Rev. Cancer 2, 277–288 [DOI] [PubMed] [Google Scholar]
- 7. Kim W., Youn H., Seong K. M., Yang H. J., Yun Y. J., Kwon T., Kim Y. H., Lee J. Y., Jin Y. W., Youn B. (2011) PIM1-activated PRAS40 regulates radioresistance in non-small cell lung cancer cells through interplay with FOXO3a, 14-3-3, and protein phosphatases. Radiat. Res. 176, 539–552 [DOI] [PubMed] [Google Scholar]
- 8. Criswell T., Leskov K., Miyamoto S., Luo G., Boothman D. A. (2003) Transcription factors activated in mammalian cells after clinically relevant doses of ionizing radiation. Oncogene 22, 5813–5827 [DOI] [PubMed] [Google Scholar]
- 9. Yoshida K., Sasaki R., Nishimura H., Okamoto Y., Suzuki Y., Kawabe T., Saito M., Otsuki N., Hayashi Y., Soejima T., Nibu K., Sugimura K. (2010) Nuclear factor-κB expression as a novel marker of radioresistance in early-stage laryngeal cancer. Head Neck 32, 646–655 [DOI] [PubMed] [Google Scholar]
- 10. Murley J. S., Baker K. L., Miller R. C., Darga T. E., Weichselbaum R. R., Grdina D. J. (2011) SOD2-mediated adaptive responses induced by low-dose ionizing radiation via TNF signaling and amifostine. Free Radic. Biol. Med. 51, 1918–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wajant H., Pfizenmaier K., Scheurich P. (2003) Tumor necrosis factor signaling. Cell Death Differ. 10, 45–65 [DOI] [PubMed] [Google Scholar]
- 12. Li S., Wang L., Dorf M. E. (2009) PKC phosphorylation of TRAF2 mediates IKKα/β recruitment and K63-linked polyubiquitination. Mol. Cell 33, 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bradley J. R., Pober J. S. (2001) Tumor necrosis factor receptor-associated factors (TRAFs). Oncogene 20, 6482–6491 [DOI] [PubMed] [Google Scholar]
- 14. Arch R. H., Gedrich R. W., Thompson C. B. (1998) Tumor necrosis factor receptor-associated factors (TRAFs)–a family of adapter proteins that regulates life and death. Genes Dev. 12, 2821–2830 [DOI] [PubMed] [Google Scholar]
- 15. Zheng M., Morgan-Lappe S. E., Yang J., Bockbrader K. M., Pamarthy D., Thomas D., Fesik S. W., Sun Y. (2008) Growth inhibition and radiosensitization of glioblastoma and lung cancer cells by small interfering RNA silencing of tumor necrosis factor receptor-associated factor 2. Cancer Res. 68, 7570–7578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schäfer T., Maco B., Petfalski E., Tollervey D., Böttcher B., Aebi U., Hurt E. (2006) Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature 441, 651–655 [DOI] [PubMed] [Google Scholar]
- 17. Jang C. Y., Lee J. Y., Kim J. (2004) RpS3, a DNA repair endonuclease and ribosomal protein, is involved in apoptosis. FEBS Lett. 560, 81–85 [DOI] [PubMed] [Google Scholar]
- 18. Kim T. S., Kim H. D., Kim J. (2009) PKCδ-dependent functional switch of rpS3 between translation and DNA repair. Biochim. Biophys. Acta 1793, 395–405 [DOI] [PubMed] [Google Scholar]
- 19. Kim S. H., Lee J. Y., Kim J. (2005) Characterization of a wide range base-damage-endonuclease activity of mammalian rpS3. Biochem. Biophys. Res. Commun. 328, 962–967 [DOI] [PubMed] [Google Scholar]
- 20. Wan F., Anderson D. E., Barnitz R. A., Snow A., Bidere N., Zheng L., Hegde V., Lam L. T., Staudt L. M., Levens D., Deutsch W. A., Lenardo M. J. (2007) Ribosomal protein S3: a KH domain subunit in NF-κB complexes that mediates selective gene regulation. Cell 131, 927–939 [DOI] [PubMed] [Google Scholar]
- 21. Niyazi M., Niyazi I., Belka C. (2007) Counting colonies of clonogenic assays by using densitometric software. Radiat. Oncol. 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang H. J., Youn H., Seong K. M., Yun Y. J., Kim W., Kim Y. H., Lee J. Y., Kim C. S., Jin Y. W., Youn B. (2011) Psoralidin, a dual inhibitor of COX-2 and 5-LOX, regulates ionizing radiation (IR)-induced pulmonary inflammation. Biochem. Pharmacol. 82, 524–534 [DOI] [PubMed] [Google Scholar]
- 23. Guo W. F., Lin R. X., Huang J., Zhou Z., Yang J., Guo G. Z., Wang S. Q. (2005) Identification of differentially expressed genes contributing to radioresistance in lung cancer cells using microarray analysis. Radiat. Res. 164, 27–35 [DOI] [PubMed] [Google Scholar]
- 24. Das A. K., Sato M., Story M. D., Peyton M., Graves R., Redpath S., Girard L., Gazdar A. F., Shay J. W., Minna J. D., Nirodi C. S. (2006) Non-small-cell lung cancers with kinase domain mutations in the epidermal growth factor receptor are sensitive to ionizing radiation. Cancer Res. 66, 9601–9608 [DOI] [PubMed] [Google Scholar]
- 25. Kim W. Y., Oh S. H., Woo J. K., Hong W. K., Lee H. Y. (2009) Targeting heat shock protein 90 overrides the resistance of lung cancer cells by blocking radiation-induced stabilization of hypoxia-inducible factor-1α. Cancer Res. 69, 1624–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jang C. Y., Shin H. S., Kim H. D., Kim J. W., Choi S. Y., Kim J. (2011) Ribosomal protein S3 localizes on the mitotic spindle and functions as a microtubule associated protein in mitosis. Biochem. Biophys. Res. Commun. 429, 57–62 [DOI] [PubMed] [Google Scholar]
- 27. Li F., Sethi G. (2010) Targeting transcription factor NF-κB to overcome chemoresistance and radioresistance in cancer therapy. Biochim. Biophys. Acta 1805, 167–180 [DOI] [PubMed] [Google Scholar]
- 28. Koukourakis M. I. (2012) Radiation damage and radioprotectants: new concepts in the era of molecular medicine. Br. J. Radiol. 85, 313–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Longley D. B., Johnston P. G. (2005) Molecular mechanisms of drug resistance. J. Pathol. 205, 275–292 [DOI] [PubMed] [Google Scholar]
- 30. Lee S. B., Kwon I. S., Park J., Lee K. H., Ahn Y., Lee C., Kim J., Choi S. Y., Cho S. W., Ahn J. Y. (2010) Ribosomal protein S3, a new substrate of Akt, serves as a signal mediator between neuronal apoptosis and DNA repair. J. Biol. Chem. 285, 29457–29468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wan F., Weaver A., Gao X., Bern M., Hardwidge P. R., Lenardo M. J. (2011) IKKβ phosphorylation regulates RPS3 nuclear translocation and NF-κB function during infection with Escherichia coli strain O157:H7. Nat. Immunol. 12, 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meggio F., Pinna L. A. (2003) One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17, 349–368 [DOI] [PubMed] [Google Scholar]
- 33. Olsen B. B., Wang S. Y., Svenstrup T. H., Chen B. P., Guerra B. (2012) Protein kinase CK2 localizes to sites of DNA double-strand break regulating the cellular response to DNA damage. BMC Mol. Biol. 13, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trembley J. H., Chen Z., Unger G., Slaton J., Kren B. T., Van Waes C., Ahmed K. (2010) Emergence of protein kinase CK2 as a key target in cancer therapy. Biofactors 36, 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Niefind K., Guerra B., Ermakowa I., Issinger O. G. (2001) Crystal structure of human protein kinase CK2: insights into basic properties of the CK2 holoenzyme. EMBO J. 20, 5320–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Au P. Y., Yeh W. C. (2007) Physiological roles and mechanisms of signaling by TRAF2 and TRAF5. Adv. Exp. Med. Biol. 597, 32–47 [DOI] [PubMed] [Google Scholar]
- 37. Xia Z. P., Chen Z. J. (2005) TRAF2: a double-edged sword? Sci. STKE 2005, pe7. [DOI] [PubMed] [Google Scholar]
- 38. Blackwell K., Zhang L., Thomas G. S., Sun S., Nakano H., Habelhah H. (2009) TRAF2 phosphorylation modulates tumor necrosis factor α-induced gene expression and cell resistance to apoptosis. Mol. Cell Biol. 29, 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]