Background: T helper (Th) cell differentiation is a complex process regulated by multiple factors.
Results: PIM kinases promote Th1 differentiation through regulating the expression of genes important for this process.
Conclusion: PIM kinases were identified as new regulators of Th1 cell differentiation.
Significance: This study provides new insights into the mechanisms controlling Th cell differentiation.
Keywords: Differentiation, Gene Regulation, Immunology, siRNA, T Cell, Kinase
Abstract
The differentiation of human primary T helper 1 (Th1) cells from naïve precursor cells is regulated by a complex, interrelated signaling network. The identification of factors regulating the early steps of Th1 cell polarization can provide important insight in the development of therapeutics for many inflammatory and autoimmune diseases. The serine/threonine-specific proviral integration site for Moloney murine leukemia virus (PIM) kinases PIM1 and PIM2 have been implicated in the cytokine-dependent proliferation and survival of lymphocytes. We have established that the third member of this family, PIM3, is also expressed in human primary Th cells and identified a new function for the entire PIM kinase family in T lymphocytes. Although PIM kinases are expressed more in Th1 than Th2 cells, we demonstrate here that these kinases positively influence Th1 cell differentiation. Our RNA interference results from human primary Th cells also suggest that PIM kinases promote the production of IFNγ, the hallmark cytokine produced by Th1 cells. Consistent with this, they also seem to be important for the up-regulation of the critical Th1-driving factor, T box expressed in T cells (T-BET), and the IL-12/STAT4 signaling pathway during the early Th1 differentiation process. In summary, we have identified PIM kinases as new regulators of human primary Th1 cell differentiation, thus providing new insights into the mechanisms controlling the selective development of human Th cell subsets.
Introduction
The different subsets of human T helper (Th)3 cells play distinct roles in the immune system. Among these are Th1 and Th2 type cells, which originate from common naïve precursor cells and protect the host against intracellular and extracellular pathogens, respectively. Disturbances in the normal Th1 or Th2 cell generation or in the balance between the different subsets have been implicated in the development of many autoimmune and allergic diseases. Identification of the factors regulating Th1 and Th2 cell differentiation is an active area of investigation and may provide important insight in the development of better diagnostics and therapies for these diseases.
The most important stimuli for inducing Th cell differentiation from a naïve precursor cell are the T cell receptor (TCR)-mediated activation and the cytokine milieu surrounding the cell. Stimulation through TCR induces the activation of several transcription factors, such as nuclear factor of activated T cells (NFAT), activator protein-1, and nuclear factor κB (NF-κB), which are important inducers of several cytokine genes and have been implicated in the regulation of Th1/Th2 cell development (for reviews, see Refs. 1 and 2). The hallmark cytokine produced by Th1 cells, IFNγ, is induced upon the TCR activation by NFAT and NF-κB (3–7). Secreted IFNγ positively autoregulates Th1 differentiation by binding to its receptor and activating STAT1. TCR activation and STAT1 induce the expression of T box expressed in T cells (T-BET), a key transcription factor driving the Th1 polarization (8, 9). T-BET further induces the transcription of IFNγ and enhances the expression of interleukin-12 receptor β2 (IL-12Rβ2) (9–11). IL-12Rβ2 is not expressed in naïve T cells but is complementarily induced by TCR activation and T-BET and maintained by IFNγ (12, 13). Once the expression of IL-12Rβ2 is up-regulated, IL-12, a key cytokine driving Th1 differentiation, is able to activate STAT4, an important inducer of IFNγ and IL-12Rβ2 expression (11, 14–17). The Th1 cell differentiation is thus regulated by an interrelated signaling network with multiple factors positively regulating each other.
The proviral integration site for Moloney murine leukemia virus (PIM) family of serine/threonine-specific kinases consists of three members, PIM1, PIM2, and PIM3. These kinases are evolutionarily highly conserved and have largely overlapping functions but differ in their tissue distribution (18, 19). PIM1 and PIM2 have been reported to be predominantly expressed in hematopoietic cells where their expression is regulated by several cytokines and growth factors (19, 20). PIM3 has been suggested previously to be expressed mainly in brain, kidney, liver, and epithelia (18, 21) and very recently in mouse and human CD4+ T cells (22). PIM kinases are known regulators of cytokine-dependent proliferation and survival in hematopoietic cells (23–25). The substrates and binding partners identified for PIM kinases include Bcl2-associated death promoter (BAD) protein, a proapoptotic protein; p100, an activator of c-Myb transcription factor; and NFATc1, a mediator of TCR signaling as well as several factors involved in cell cycle regulation (20). PIM1 has also been shown to activate c-MYC target genes by chromatin phosphorylation (26). In addition, PIM kinases have been implicated in the inhibition of STAT5 and STAT6 signaling through the regulation of suppressor of cytokine signaling proteins SOCS1 and SOCS3 (27, 28).
We have previously reported that the expression of PIM family genes is up-regulated by Th1-polarizing cytokines (29), suggesting a role for PIM kinases in the regulation of Th1 cell differentiation. In this study, we have investigated the functional role of PIM family kinases in the regulation of human Th1 cell differentiation. We demonstrate that the PIM kinases promote the Th1 cell differentiation by inducing several factors critical for the Th1 polarization. Our RNA interference results depict that the PIM kinases are important for the up-regulation of two Th1-driving pathways, IFNγ/T-BET and IL-12/STAT4 pathways.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfections
Human mononuclear cells were isolated from the cord blood of healthy neonates using Ficoll-Paque isolation (Amersham Biosciences). CD4+ cells were further purified using DYNAL magnetic beads (Invitrogen). Cells from several individuals were pooled after the isolation. Cell cultures were made in Yssel's medium (Iscove's modified Dulbecco's medium (Invitrogen) supplemented with Yssel's medium concentrate, penicillin/streptomycin, and 1% AB serum). Cells were activated with plate-bound α-CD3 (0.5 μg/well) and soluble α-CD28 (0.5 μg/ml; both from Immunotech, Marseille, France) and at the same time polarized toward Th1 cells with 2.5 ng/ml IL-12 or toward Th2 cells with 10 ng/ml IL-4 (both from R&D Systems, Minneapolis, MN). IL-2 (40 units/ml; R&D Systems) was added into the cultures after 48 h of priming.
For PIM knockdown experiments, freshly isolated CD4+ cells were suspended in Opti-MEM I (Invitrogen) and transfected with siRNA oligonucleotides (Sigma-Aldrich) (see Table 1) using the nucleofection technique (Amaxa, Cologne, Germany). 4 × 106 cells were transfected with a total of 4.5 μg of siRNA (4.5 μg of non-targeting (NT) siRNA or 1.5 μg each of PIM1/2/3 siRNA). The nucleofected cells were allowed to rest for 20 h in RPMI 1640 medium (Sigma-Aldrich) supplemented with penicillin/streptomycin, 2 mm l-glutamine, and 10% FCS at 37 °C (2 × 106 cells/ml) and subsequently activated and cultured in Yssel's medium as described above. For STAT4 and STAT6 knockdown experiments, 1.5 μg of NT siRNA, STAT6 siRNA, or STAT4 siRNA (pool of two siRNAs; 1:1) was used. Nucleofections and culturing were performed similarly as in the PIM knockdown experiments except that the cells were rested for 24 h after nucleofection. Research involving the use of blood from unknown donors was permitted by the Finnish Ethics Committee.
TABLE 1.
Sequences of primers, probes, and siRNA oligonucleotides used
F, forward; R, reverse; FAM, carboxyfluorescein; TAMRA, tetramethylrhodamine.
| TaqMan RT-PCR | |
| EF1α-Probe | 5′-6(FAM)-AGCGCCGGCTATGCCCCTG-(TAMRA)-3′ |
| EF1α-F | 5′-CTGAACCATCCAGGCCAAAT-3′ |
| EF1α-R | 5′-GCCGTGTGGCAATCCAAT-3′ |
| IFNγ-Probe | 5′-6(FAM)-TGCTGGCGACAGTTCAGCCATCAC-(TAMRA)-3′ |
| IFNγ-F | 5′-TGTCCAACGCAAAGCAATACA-3′ |
| IFNγ-R | 5′-CTCGAAACAGCATCTGACTCCTT-3′ |
| IL-12Rβ2-Probe | 5′-6(FAM)-TGCATTGCTATCATCATGGTGGGCAT-(TAMRA)-3′ |
| IL-12Rβ2-F | 5′-CGTTTGTGGCACCAAGCA-3′ |
| IL-12Rβ2-R | 5′-GCTGGAAGTAATGCGTTGAGAA-3′ |
| STAT4-Probe | 5′-6(FAM)-AGTCTCGCAGGATGTCAGCGAATGG-(TAMRA)-3′ |
| STAT4-F | 5′-GCTGAGAGCTGTAGTGTTTACCGA-3′ |
| STAT4-R | 5′-AATAAAGGCCGGTTGTCTGCT-3′ |
| T-bet-Probe | 5′-6(FAM)-TCAGCATGAAGCCTGCATTCTTGCC-(TAMRA)-3′ |
| T-bet-F | 5′-ACAGCTATGAGGCTGAGTTTCGA-3′ |
| T-bet-R | 5′-GGCCTCGGTAGTAGGACATGGT-3′ |
| siRNA sequence | |
| NT | 5′-GCGCGCUUUGUAGGAUUCG-3′ |
| PIM1 | 5′-GAAGGUGAGCUCGGGUUUC-3′ |
| PIM2 | 5′-GUGGAGUUGUCCAUCGUGACA-3′ |
| PIM3 | 5′-GGCGTGCTTCTCTACGATA-3′ |
| STAT4 siRNA1 | 5′-GGUACAACGUGUCAACCAA-3′ |
| STAT4 siRNA2 | 5′-GGCAACGAUUCUUCUUCAA-3′ |
| STAT6 | 5′-AAGCAGGAAGAACUCAAGUUU-3′ |
Sorting and Analysis of Human Memory Th1 Cells
CD4-positive lymphocytes from buffy coats of healthy donors (Red Cross Finland Blood Service, Helsinki, Finland) were isolated as described under “Cell Culture and Transfections.” After isolation, CD4+ cells were stained with CD183 (CXCR3)-allophycocyanin and CD4-phycoerythrin (both from BD Pharmingen), and the CXCR3+CD4+ cell population was isolated using a FACSAria IIu cell sorter (BD Biosciences). A fraction of the CXCR3− CD4+ cell population was collected as control. Anti-mouse IgG1 κ-allophycocyanin and anti-mouse IgG1 κ-phycoerythrin (both from BD Pharmingen) were used as isotype controls. Sorted cell populations were activated with plate-bound anti-CD3 and soluble anti-CD28 and cultured in Yssel's medium (as described above). After 24 and 48 h of culturing, samples for Western blotting were collected. The intracellular IFNγ production of the CXCR3-positive and -negative populations was analyzed after 40 h of activation by intracellular cytokine staining as described below.
Real Time Quantitative RT-PCR
Total RNA samples were isolated and prepared for RT-PCR analysis as described previously (30). The gene expression levels were measured using the TaqMan ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA) (31). The primers and probes used (Oligomer, Helsinki, Finland; see Table 1) were designed using Primer Express software (Applied Biosystems). Universal Probe Library assays (Roche Applied Science) were designed using ProbeFinderTM software. The mRNA levels were normalized against the levels of a housekeeping gene, elongation factor 1α (EF1α) (31).
Western Blotting
Cells were lysed in Triton X-100 lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% Triton-X-100, 5% glycerol, 1% SDS, 1 mm Na3VO4, 10 mm NaF), boiled for 5 min, and sonicated, and equal amounts of protein were subsequently separated by SDS-PAGE and transferred to nitrocellulose membrane. Proteins were detected using the following antibodies: mouse α-PIM1 (12H8, Santa Cruz Biotechnology, Santa Cruz, CA), rabbit α-PIM2 (ATLAS Antibodies, Stockholm, Sweden or D1D2, Cell Signaling Technology, Beverly, MA), rabbit α-PIM3 (D17C9, Cell Signaling Technology), mouse α-T-BET (4B10, Santa Cruz Biotechnology), rabbit α-STAT1 p84/p91 (E-23, Santa Cruz Biotechnology), rabbit α-STAT4 (C-20, Santa Cruz Biotechnology), mouse α-STAT6 (BD Biosciences), mouse α-GAPDH (number 5G4, 6C5, HyTest, Turku, Finland), and mouse α-β-actin (Sigma-Aldrich). Horseradish peroxidase-conjugated goat α-mouse IgG (Santa Cruz Biotechnology) or α-rabbit IgG (BD Biosciences) were used as secondary antibodies. The proteins were visualized with enhanced chemiluminescence (GE Healthcare), quantified with a microcomputer imaging device (M5+, Imaging Research Inc., St. Catharines, Canada), and normalized against β-actin. For quantification, suitable non-overexposed film was used for each protein studied.
MilliplexTM Cytokine Assay
To measure IFNγ production from Th1-polarized cells, duplicate samples were stained on 96-well plates (Milliplex Map kit, Millipore, Billerica, MA) according to the manufacturer's instructions and measured using the Luminex® 100TM system (Luminex, Austin, TX). The cytokine concentrations of cell culture supernatants were normalized against relative cell counts obtained by flow cytometry.
Immunofluorescent Staining and Flow Cytometry
To measure the expression of IL-12Rβ2 on the surface of Th1-polarized cells, cells were first incubated with rat α-human IL-12Rβ2 (CD212) antibody or rat IgG2a (isotype control; both from BD Pharmingen) and then with biotin-conjugated α-rat antibody (BD Pharmingen) and finally labeled with streptavidin-peridinin-chlorophyll protein complex (BD Pharmingen). Samples were measured with the FACSCaliburTM system and analyzed with CellQuest Pro (both from BD Biosciences).
Intracellular Cytokine Staining
The flow cytometric analysis of intracellular cytokine staining of nucleofected Th1-polarized cells was performed as described previously (32). In short, the NT and PIM1 + PIM2 + PIM3 (PIM123) siRNA-nucleofected cells were activated and cultured under Th1-polarizing conditions for 6 days after which the cells were harvested and washed with PBS. Half of the cells were restimulated with 5 ng/ml phorbol 12-myristate 13-acetate (Calbiochem) and 0.5 pg/ml ionomycin (Sigma-Aldrich) in Yssel's medium, and the other half was incubated in Yssel's medium and used as an unstimulated control. After 2 h of incubation, 10 μg/ml brefeldin A (Alexis Biochemicals, Lausanne, Switzerland) was added, and incubation was continued for another 3 h. Cells were washed twice with 0.5% BSA, PBS (w/v), 0.01% NaN3; fixed with 4% paraformaldehyde, PBS; and permeabilized with 0.5% saponin, PBS. Anti-human IFNγ-FITC (Invitrogen) was used for staining of the intracellular IFNγ and anti-mouse IgG1-FITC (Invitrogen) was used as an isotype control. Cells were analyzed with the FACSCalibur system and analyzed with CellQuest Pro (both from BD Biosciences).
Microarray Studies
Nucleofected cells were harvested at 6 h after culturing them under Th1- or Th2-polarizing conditions from three independent biological replicate cultures. The subsequent sample treatments were performed at the Finnish Microarray and Sequencing Centre, Turku, Finland. Total RNA was isolated with an RNeasy Mini kit (Qiagen, Valencia, CA), and sample preparation for GeneChip (Affymetrix, Santa Clara, CA) oligonucleotide array hybridizations was performed according to the manufacturer's instructions using 250 ng of total RNA as starting material with a GeneChip 3′ IVT Express kit (Affymetrix). Samples were hybridized to HG-U219 arrays using a GeneTitan® instrument (both from Affymetrix). Microarray data were normalized using a robust multiarray average algorithm (33). Duplicate and unannotated probe sets were removed using the genefilter package in R.4 In the case of duplicates, the probe set with the highest interquartile range was retained. Present and absent calls for probe sets were generated by fitting the chip-wide expression data to a two-component Gaussian distribution function using the standard expectation-maximization algorithm implemented in the mixtools package in R (35). A probe set was defined to be present if it has an expression value higher than the threshold where the two components of the Gaussian distribution densities meet (36). ComBat with non-parametric empirical priors was used to correct for the batch effect (37). Differential expression analysis was done using the moderated paired t test as implemented in limma (38). The genes were considered as differentially expressed when the Benjamini-Hochberg adjusted false discovery rate was <0.1 and log -fold change was <−0.5 or >0.5. Overrepresentation analysis of transcription factor binding sites was done using GeneTrail (39).
RESULTS
Expression of PIM Kinases in Th Cells Is Regulated by the Knockdown of STAT4 and STAT6
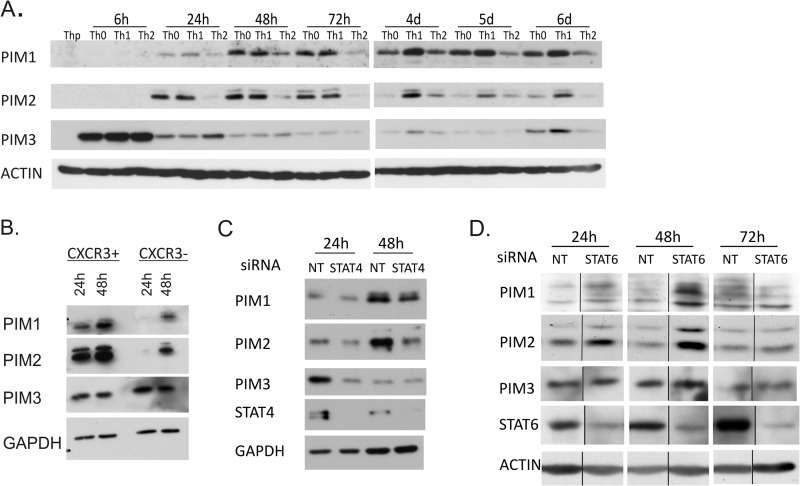
To analyze the expression levels of PIM family kinases during human T helper cell differentiation, naïve Th precursor (Thp) cells were activated and either cultured in neutral conditions without polarizing cytokines (Th0) or differentiated in the Th1 or Th2 direction. With the Western blotting measurement of the protein levels of PIM1, PIM2, and PIM3, enhanced expression was observed in all the treated cells relative to Thp cells at the time points studied, indicating that the PIM kinases are induced in response to TCR activation (Fig. 1A). The expression of PIM3 was much more rapidly induced than PIM1 or PIM2 and was strongly up-regulated already 6 h after activation. PIM2 was observed to be induced more rapidly than PIM1 after the primary activation and polarization and reached its maximum earlier (Fig. 1A). Importantly, however, the expression of PIM kinases was more pronouncedly induced in cells polarized in the Th1 direction than in the Th2 direction. Any Th1 preference of PIM3 could only be seen at later time points usually 72 h to 4 days after activation. These results are consistent with our previous data indicating that PIM1 is preferentially expressed by Th1 cells at both mRNA and protein levels (29). In this earlier study, PIM2 mRNA was also shown to be up-regulated in human Th1 cells.
FIGURE 1.
Expression of PIM kinases in Th cells is regulated by the knockdown of STAT4 and STAT6. A, expression of PIM1, PIM2, and PIM3 during human Th cell differentiation. Naïve CD4+ Thp cells isolated from cord blood were activated (Th0) or stimulated with IL-12 (Th1) or IL-4 (Th2). Samples were harvested for Western blotting at the indicated time points. The panels show representative data from four biological replicates. B, expression of PIM1, PIM2, and PIM3 in activated human peripheral CXCR3+ and CXCR3− CD4+ T cells. CD4+ cells were isolated from buffy coats, and the CXCR3-positive and -negative populations were isolated by flow cytometric cell sorting. Sorted cells were activated and harvested at the indicated time points. The panels show representative data of five biological replicates. C, CD4+ T cells were nucleofected with STAT4 (pooled siRNAs 1 + 2) or NT siRNA and polarized in the Th1 direction 20–24 h after nucleofection. Samples were harvested for Western blotting at the indicated time points. The panels show representative data from three biological replicates. D, CD4+ T cells were nucleofected with STAT6 or NT siRNA and polarized in the Th2 direction 24 h after nucleofection. Samples were harvested for Western blotting at the indicated time points. The panels show representative data from two biological replicates. Vertical lines represent repositioned gel lanes that are from the same blot and the same exposure. d, days.
To answer the question whether the observed up-regulation of PIM kinase expression in the in vitro polarized Th1 cells can also be seen in Th1 cells developed in vivo, we studied the expression of PIM kinases in CXCR3+CD4+ cells isolated from buffy coats. CXCR3 is a chemokine receptor exclusively expressed on memory Th1 cells distinguishing them from Th2 and Th17 cells (40, 41). The proportion of the CXCR3-positive cells of total CD4+ cells ranged from 10 to 25% depending on the donor. A fraction of the CXCR3-negative population was collected as a control. For Western blot analysis, the CXCR3-positive and -negative fractions were activated and cultured for 24 or 48h after which the samples were collected. For intracellular IFNγ staining, a proportion of cells was harvested after 40 h of activation and restimulated with phorbol 12-myristate 13-acetate and ionomycin, and intracellular IFNγ was measured by flow cytometry. The CXCR3-positive restimulated cells produced more IFNγ than the CXCR3-negative cells (average 16.7 versus 2.0%, respectively; p = 0.014). Furthermore, the Western blotting measurement of the protein levels of PIM1, PIM2, and PIM3 revealed an enhanced expression of PIM1 and PIM2 by the memory Th1 cells compared with CXCR3-negative cells (Fig. 1B). However, at the PIM3 protein level, no clear difference was observed between the CXCR3-positive and -negative cells.
To further study the regulation of PIM expression in Th cell differentiation, we investigated the involvement of STAT4- and STAT6-dependent pathways. For this purpose, human naïve CD4+ cells were nucleofected with NT, STAT4-, or STAT6-specific siRNA oligonucleotides (Table 1). The cells were then cultured in the presence of IL-12 (Th1) or IL-4 (Th2) for 24, 48, or 72 h. When the cells were cultured in Th1 conditions, the levels of PIM1, PIM2, and PIM3 were diminished in response to the depletion of STAT4 (Fig. 1C). On the other hand, the levels of PIM1 and PIM2 were increased in response to the depletion of STAT6 when cells were cultured in Th2 conditions (Fig. 1D). PIM3 was not affected by the STAT6 depletion. These results suggest that STAT4 is at least in part responsible for the up-regulation of PIM kinases during Th1 cell polarization, whereas during Th2 cell polarization, the expression of PIM1 and PIM2 is suppressed by STAT6-dependent signaling.
PIM Kinases Are Important for the Induction of IFNγ and T-BET during the Early Phases of Human Th1 Cell Development
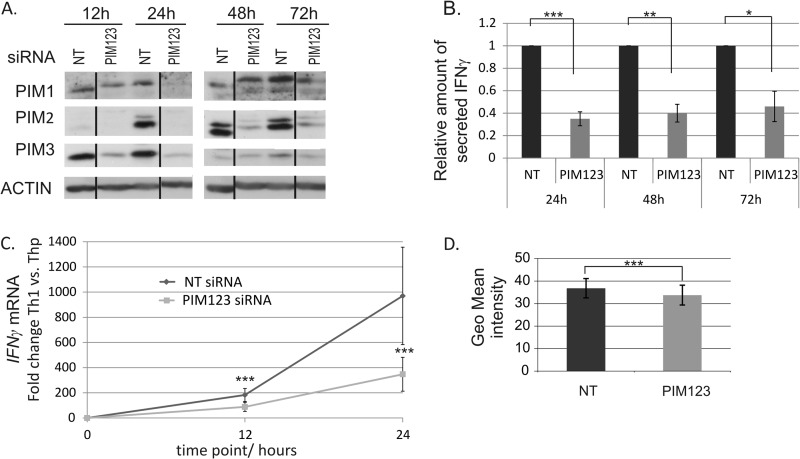
Having observed that PIM kinases were preferentially expressed by Th1 cells compared with Th2 cells, we chose to examine whether they have any role in the regulation of the Th1 differentiation process. Thus, the potential influence of the PIM knockdown on the expression of IFNγ and T-BET was analyzed. To address these questions, an siRNA approach was used to knock down the expression of PIM family members in human naïve CD4+ cells. The cells were nucleofected with NT or PIM-specific siRNA oligonucleotides (Table 1) and polarized in the Th1 direction. PIM1-, PIM2-, and PIM3-specific siRNAs were used simultaneously. The knockdown efficiencies of the siRNAs were controlled by measuring the PIM protein levels at the indicated time points (Fig. 2A). All PIM siRNAs knocked down their targets efficiently and specifically, and the triple knockdown was as efficient as the individual siRNAs separately (Fig. 2A and data not shown). The siRNA-mediated knockdown of three PIM kinases lasted up to 48–72 h after which its effect decreased most likely because of the proliferation of the cells (Fig. 2A). The knockdown data obtained by these siRNAs was also confirmed with two other siRNA sequences for each PIM kinase.
FIGURE 2.
Knockdown of PIM kinases down-regulates IFNγ at mRNA and protein levels during Th1 cell polarization. A, PIM1, PIM2, and PIM3 siRNAs efficiently inhibit the induction of PIM kinase expression in human CD4+ cells. Naïve CD4+ cells nucleofected with the indicated siRNAs were activated and polarized in the Th1 direction 20 h after nucleofection. Samples were harvested for Western blotting at the indicated time points. Representative data are shown from seven independent experiments. B, knockdown of PIM kinases inhibits IFNγ protein production. Cell culture supernatants from nucleofected and Th1-polarized cells were collected at the indicated time points, and the amount of IFNγ produced by the cells was measured by a cytokine assay. The bars represent the mean values ±S.E. (error bars) of the relative levels of IFNγ in PIM123 siRNA samples relative to the control samples. The values of the control samples were set as 1. Data collected from four to five independent experiments are shown. C, the induction of IFNγ mRNA is inhibited by PIM knockdown during early Th1 polarization. The levels of IFNγ mRNA in the nucleofected cells were analyzed by TaqMan RT-PCR. The graph represents the average -fold differences ±S.E. (error bars) of IFNγ mRNA levels in siRNA-treated Th1 cells compared with the average Thp value. D, PIM123 siRNA- or NT siRNA-nucleofected cells were cultured for 6 days under Th1-polarizing conditions. The cells where then harvested, and half of the cells were stimulated with phorbol 12-myristate 13-acetate and ionomycin, whereas the other half was left unstimulated. Cells were stained for intracellular IFNγ and analyzed with flow cytometry. Bars represent the average ±S.E. (error bars) geometrical (Geo) mean of fluorescence intensity calculated from four independent experiments. Vertical lines represent repositioned gel lanes that are from the same blot and the same exposure. Statistical significances were calculated using the two-tailed paired t test: *, p < 0.05; **, p < 0.02; ***, p > 0.01. siRNAs used were NT siRNA or PIM123 siRNAs.
Because the effects of siRNA oligos are diluted after a few days in culture, our experiments were carried out at the early phases of Th1 cell differentiation. The analyses of the cell culture supernatants by Milliplex cytokine assays indicated that IFNγ production was most prominently inhibited when all three PIM kinases were knocked down by siRNA (Fig. 2B and data not shown). This effect lasted for at least 72 h, and the PIM triple knockdown cells secreted less than 50% of the IFNγ secreted by the control cells. Importantly, TaqMan RT-PCR analysis revealed that the level of IFNγ mRNA was significantly down-regulated by PIM siRNAs already after 12 h of polarization, indicating that PIM kinases affect IFNγ production at the transcriptional level as well (Fig. 2C). Although the effects of siRNA oligos are diluted after a few days in culture, intracellular IFNγ staining of nucleofected cells revealed that there was still a small but statistically significant reduction in intracellular IFNγ after 6 days of culture and restimulation (Fig. 2D).
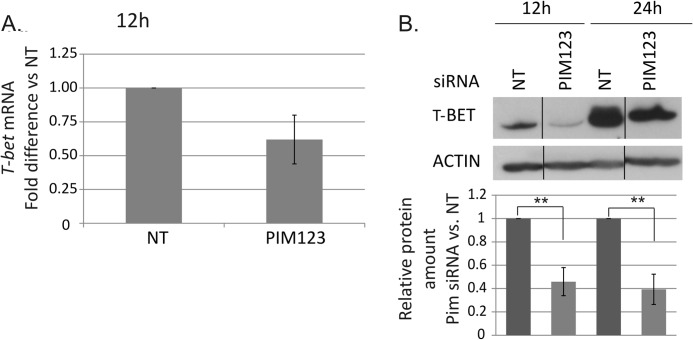
The measurements of T-BET mRNA indicated that it is rapidly and highly induced in response to the activation and IL-12 stimulation of naïve Thp cells. This early induction of T-BET mRNA was delayed by the PIM knockdown with the effects observed already 12 h after polarization although no longer after 24 h of culture (Fig. 3A and data not shown). Consistent with the mRNA data, Western blotting analysis of T-BET protein levels demonstrated that its expression was also down-regulated in response to the PIM depletion (Fig. 3B). Strong effects were seen after both 12 and 24 h of culture. The PIM triple knockdown down-regulated T-BET at the protein level more than 50% in comparison with control cells. The level of T-BET was highly up-regulated under Th1-polarizing conditions during the first 24 h of culture (Fig. 3B), which may have reduced the effects of PIM siRNAs after the 24-h time point. Taken together, our data indicate that the depletion of PIM kinases slows down the induction of IFNγ and T-BET at both the mRNA and protein levels.
FIGURE 3.
Depletion of PIM kinases down-regulates T-BET at mRNA and protein levels. A, the levels of T-BET mRNA in the nucleofected cells were analyzed by TaqMan RT-PCR. The graph represents the average -fold differences ±S.E. (error bars) of T-BET mRNA in PIM siRNA-treated Th1 cells compared with NT siRNA-treated Th1 cells. The value of the control cells was set as 1. The data were calculated from five of six independent experiments. B, the bars represent the mean values ±S.E. (error bars) of the relative levels of T-BET protein between the control and PIM siRNA samples after normalization against the levels of β-actin. The values of the control samples were set as 1. The Western blots are representatives of these experiments. The data were calculated from six of seven independent experiments. Vertical lines represent repositioned gel lanes that are from the same blot and the same exposure. Statistical significances were calculated using the two-tailed paired t test: **, p < 0.02. siRNAs used were NT siRNA or PIM123 siRNAs.
PIM Kinases Regulate the IL-12/STAT4 Signaling Pathway during Early Th1 Cell Polarization
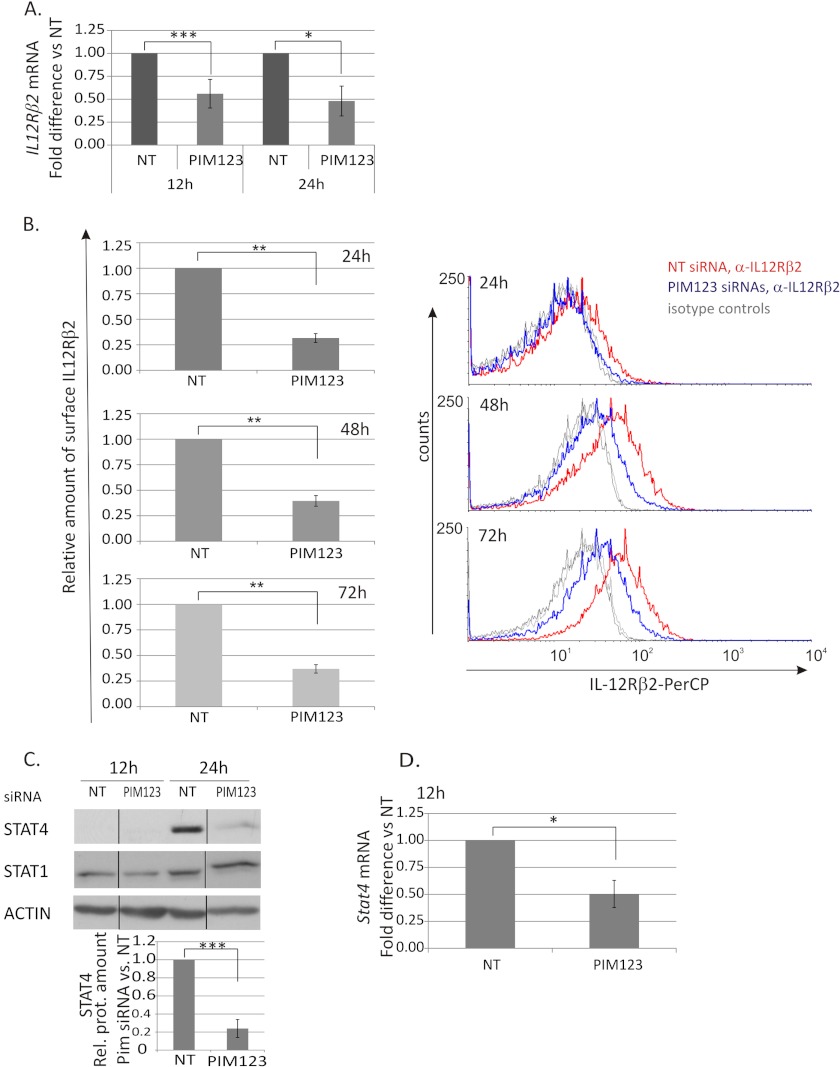
IL-12/STAT4 signaling is a critical pathway in the Th1 cell differentiation. Because PIM kinases were observed to regulate the expression of T-BET and IFNγ, it was of interest to study whether they have any influence on the expression of IL-12Rβ2 and/or STAT4, which have been shown to be direct or indirect targets of T-BET and IFNγ signaling (9, 11, 12, 42). We noticed that the depletion of PIM kinases from CD4+ cells significantly down-regulated the expression of IL-12Rβ2 at both the mRNA and protein levels (Fig. 4, A and B). The IL-12Rβ2 mRNA was down-regulated by the PIM siRNAs most efficiently at the 12-h time point, and this down-regulation by PIM triple knockdown was still more than 50% and statistically significant at the 24-h time point (Fig. 4A). At the protein level, the effect was seen already at the 24-h time point and started to slowly decrease thereafter. The triple knockdown still had a clear effect after 72 h of culturing as the relative amount of surface IL-12Rβ2 was 40% as compared with control (Fig. 4B).
FIGURE 4.
PIM kinases regulate the IL-12Rβ2/STAT4 pathway in Th1-polarized cells. CD4+ cells were nucleofected and cultured as in Fig. 2. A, samples were harvested at different time points during the culture for TaqMan RT-PCR analysis. These graphs represent the average -fold differences ±S.E. (error bars) of IL-12Rβ2 mRNA levels in PIM siRNA-treated Th1 cells compared with NT siRNA-treated Th1 cells. Data were calculated from five of six independent experiments. B, expression of the cell surface IL-12Rβ2 protein was analyzed by immunofluorescent staining and flow cytometry at the indicated time points. The bars represent the average -fold changes ±S.E. (error bars) of the geometric mean intensities of IL-12Rβ2 in PIM siRNA samples compared with control samples. The data were calculated from six independent experiments. The histograms show representative data from these experiments (red line, NT siRNA Th1; blue line, PIM123 siRNA Th1; gray lines, isotype controls). Samples were harvested for Western blotting (C) and TaqMan RT-PCR (D) analyses at the indicated time points. The bars represent the mean values ±S.E. (error bars) of the relative levels of STAT4 between control and PIM siRNA samples obtained by quantifying and normalizing against the levels of β-actin. The value of control samples was set as 1. The data were calculated from four independent experiments. The Western blot of STAT1 shows representative data from four independent experiments. Vertical lines represent repositioned gel lanes that are from the same blot and the same exposure. A–D, statistical significances were calculated using the two-tailed paired t test: *, p < 0.05; **, p < 0.02; ***, p > 0.01 (A, C, and D); *, p < 0.005; **, p < 0.0005 (B). NT indicates control siRNA. Rel. prot., relative protein; PerCP, peridinin-chlorophyll protein complex.
We also investigated the influence of PIM kinases on STAT4 and STAT1, which are important mediators of IL-12- and IFNγ-induced signaling pathways, respectively. The expression of STAT4 was transiently down-regulated by the triple knockdown of PIM kinases at 24 h after Th1 priming, whereas the expression of STAT1 was not influenced by the PIM siRNAs (Fig. 4C). Consistent with the protein data, STAT4 mRNA was down-regulated by the PIM knockdown at the 12-h time point, although the effect was no longer seen at the 24-h time point (Fig. 4D and data not shown). Taken together, our results show that the depletion of PIM kinases from human Th1-polarized cells down-regulates the expression of IL-12Rβ2 and has also a transient effect on STAT4.
Transcriptional Profiling of PIM Kinase Knockdown Th1 Cells Reveals Potential Downstream Transcriptional Regulators
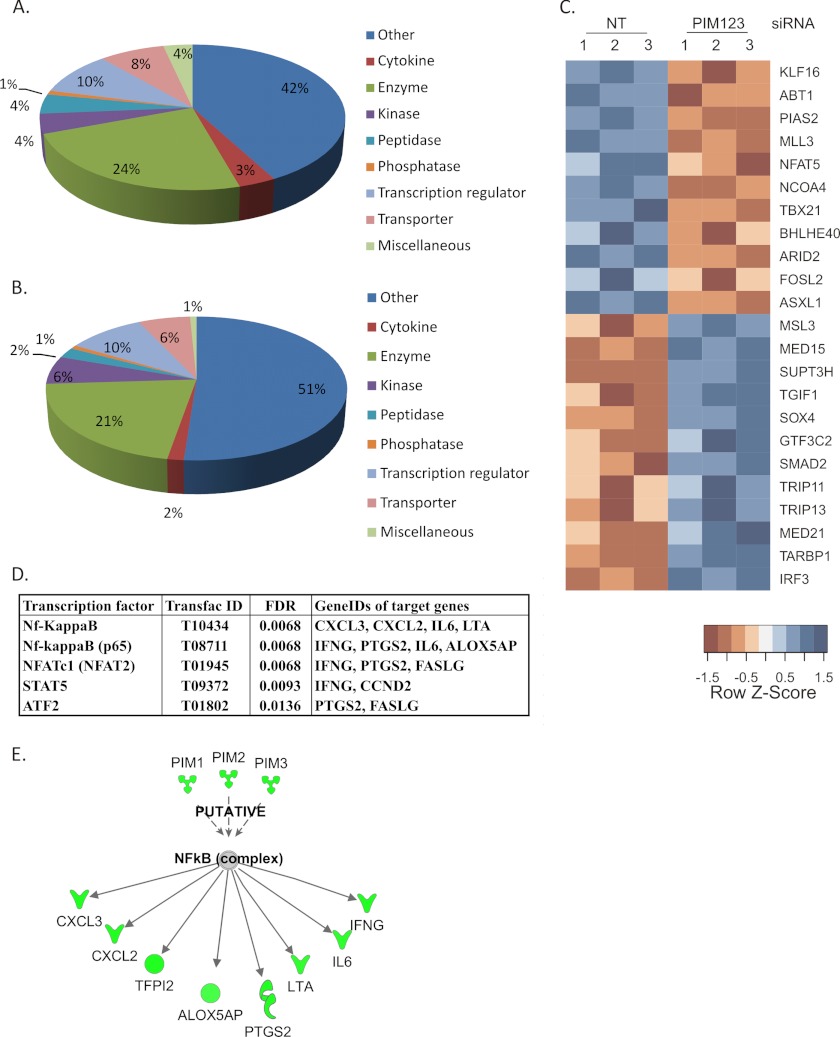
Having observed that the knockdown of PIM kinases impairs the IFNγ and T-BET expression as well as the IL-12/STAT4 pathway, we further studied the transcriptional regulation downstream of PIM kinases. We carried out a genome-wide transcriptional profiling of PIM knockdown T cells polarized in the Th1 and Th2 directions. For this purpose, cells were nucleofected with either NT siRNA or a mixture of PIM1, PIM2, and PIM3 siRNAs and cultured under Th1- or Th2-polarizing conditions. Six hours after cell activation, samples were collected for microarray analysis. This early time point was selected to avoid indirect influences of T-BET and STAT4 down-regulation induced by PIM knockdowns. The microarray analysis was performed from 3 independent replicate cultures and the PIM kinase knockdowns were confirmed by Western blot and/or TaqMan RT-PCR from samples collected at the 24-h time point (data not shown). The expected target genes of PIM kinase knockdown, IFNγ and T-BET, were both found to be down-regulated in the data set when PIM123 siRNA Th1 samples were compared with NT Th1 samples (supplemental Table S1). In cells polarized in the Th1 direction, altogether 241 genes were found to be differentially expressed (DE), and 114 of these were down-regulated. Similar analysis of the effect of PIM knockdown was performed with cells polarized in the Th2 direction. The DE genes of Th1 differentiated cells were compared with DE genes of Th2-polarized cells (supplemental Fig. S1). This comparison revealed that the majority of the DE genes of both Th1- and Th2-polarized cells were different, and only 24 genes were differentially expressed in both cell types. To further analyze the microarray results, the DE genes of Th1-polarized PIM knockdown cells were classified into functional classes using annotations given by the Ingenuity Pathway Analysis system (Fig. 5, A and B). The distribution of both up- and down-regulated genes into functional classes was found to be similar. Among the down-regulated genes, 11 were transcriptional regulators, including T-BET, protein inhibitor of activated STAT 2 (PIAS2), and NFAT5 (Fig. 5C). On the other hand, 12 transcription factors, including TGFβ signaling-related proteins TGFB-induced factor homeobox 1 (TGIF1), SMAD family member 2 (SMAD2), and SRY (sex-determining region Y) box 4 (SOX4), were up-regulated when PIM kinases were knocked down. In addition to these differentially expressed transcription factors, we further investigated which transcription factors could be responsible for the observed transcriptional changes using the whole list of differentially expressed genes. For this purpose, an overrepresentation analysis of transcription factor binding motifs on gene promoters was performed using the GeneTrail tool. By this analysis, we identified four transcription factors whose target genes were enriched among the differentially regulated genes (Fig. 5E). These transcription factors were NFATc1, Nf-κB, STAT5, and activating transcription factor 2 (ATF2). Both Nf-κB complex and p65 subunit were among these transcription factors, and altogether, seven Nf-κB target genes were found to be down-regulated. Further on, Ingenuity Pathway Analysis was used to analyze the putative transcription factors downstream of PIM kinases. In addition to the above mentioned seven Nf-κB target genes, the Ingenuity Pathway Analysis resulted in one more Nf-κB-regulated gene, TFPI2, which was also down-regulated (Fig. 5D and supplemental Table S1). In summary, these results indicate that down-regulation of PIM kinases influences many other genes in addition to IFNγ, IL12Rβ2, STAT4, and T-BET during the early stages of Th1 cell differentiation.
FIGURE 5.
Down-regulation of PIM kinases results in transcriptional changes. Human naïve CD4+ cells were isolated and nucleofected with NT or PIM123 siRNAs and cultured for 6 h after which the RNA samples were collected for microarray analysis. Down-regulated (A) and up-regulated (B) genes (NT siRNA versus PIM123 siRNA; false discovery rate (FDR), <0.1; log -fold change, <−0.5/>0.5) were classified into functional categories according to Ingenuity Pathway Analysis annotations. C, PIM kinase knockdown altered the expression of 23 transcriptional regulators. Expression values are standardized using the z-scores for visualization purposes. Red color indicates a low level of expression, whereas blue color indicates a high level of expression. Biological replicates are indicated by numbers 1, 2, and 3. D, overrepresentation analysis of transcription factor binding motifs on gene promoters revealed four transcription factors whose target genes were enriched. All of the genes except FAS ligand (FASLG) were down-regulated. E, overrepresentation analysis of transcription factor binding motifs on gene promoters by the GeneTrail tool and Ingenuity Pathway analysis revealed altogether eight Nf-κB target genes that were down-regulated. NT indicates control siRNA. IFNG, IFNγ. LTA, lymphotoxin alpha.
DISCUSSION
In this study, we have identified for the first time the role of the serine/threonine kinases PIM1, PIM2, and PIM3 as regulators of human Th1 cell differentiation. These results demonstrate that all three PIM kinases are expressed more in Th1 than Th2 cells and that both STAT4 and STAT6 regulate their expression. In Th2-polarizing conditions, STAT6 appears to down-regulate PIM1 and PIM2 expression, whereas in Th1 conditions, the depletion of STAT4 leads to down-regulation of all three PIM kinases. Our results suggest that in addition to up-regulation in Th1 conditions the expression of PIM kinases is also negatively regulated in Th2 cells. Very recently it was reported that all PIM kinases are up-regulated in response to activation in murine and human CD4+ T cells (22). Similarly, as in our results, a more pronounced and faster up-regulation of PIM3 was observed in comparison with PIM1 and PIM2 in activated murine CD4+ T cells. However, in contrast to our data, no difference in PIM1 expression was found among Th1, Th2, and Th17 cells. Notably, however, they used murine CD4+ cells in which the expression of Pim kinases may differ from that observed in human cells.
In vivo differentiated Th1 cells express high levels of CXCR3 chemokine receptor, which distinguishes them from Th2 and Th17 cells (40, 41). CXCR3 is essential for effector and memory Th1 cell migration, and its expression is regulated by T-BET (41). Similarly as in the in vitro polarized Th1 cells, we found that the CXCR3-positive cells expressed more PIM1 and PIM2 than the CXCR3-negative cells. Although our results did not show any difference in PIM3 expression between these two cell populations, these results indicate that at least PIM1 and PIM2 might have a role in in vivo developed Th1 cells.
During the early differentiation of Th1 cells, PIM kinases positively influence the production of IFNγ, the hallmark cytokine produced by Th1 cells. Consistent with this, our data show these kinases to be important for the expression of T-BET and the IL-12/STAT4 signaling pathway, which are critical driving factors during the early Th1 cell polarization. The effects of PIM knockdown on T-BET and IFNγ were already seen 12 h after activation. T-BET was down-regulated at the protein level by the PIM siRNAs until 24 h after activation, whereas the PIM triple knockdown Th1 cells secreted less IFNγ than the control Th1 cells for at least 72 h. Although the effect seen on the secretion of IFNγ decreased to some extent toward the 72-h time point, a slightly impaired Th1 response was observed in intracellular cytokine staining of restimulated Th1 cells at day 6 in PIM knockdown cells compared with control. In line with these results, the PIM depletion had a strong effect on surface IL-12Rβ2 at all the time points studied until 72 h. IL-12Rβ2 has been shown to be regulated by T-BET (9, 11), although contradictory data also exist (17, 42). It is possible that the influence of PIM kinases on IL-12Rβ2 expression is at least partly mediated via the early regulation of T-BET. STAT4 mRNA expression was transiently down-regulated by PIM siRNAs at the 12-h time point and at the protein level at the 24-h time point. Because T-BET has been reported to induce STAT4 expression (42), it is possible that PIM kinases influence STAT4 at least partly via T-BET.
Our PIM siRNA experiments have shown that the PIM triple knockdown has a more prominent effect than any of the PIM siRNAs individually. This supports the idea that PIM kinases can substitute each other's functions in cellular processes, and it is in line with mouse PIM triple knock-out studies (19, 20). The PIM triple knock-out mice displayed reduced body mass and moderate defects in T cell proliferation, whereas Pim1−/− knock-out mice did not have any pronounced phenotype (19, 20). Our own individual PIM siRNAs, however, affected the Th1 cell differentiation, and the simultaneous use of all three PIM siRNAs enhanced these effects on all of the factors studied. The RNA interference data obtained by the individual PIM siRNAs was also confirmed by using two other siRNA sequences for each PIM kinase. Similar effects on IFNγ secretion were obtained with all three siRNA sequences, thus further supporting the hypothesis.
The knockdown effects of the PIM kinases were generally transient. This is best explained by the fact that the siRNA oligonucleotides are diluted after the cells start to proliferate. Although the early induction of the PIM proteins in Th1 cells was almost completely inhibited by the siRNAs, their amounts began to increase 48–72 h after the initiation of Th1 polarization, thus explaining why the effects of the knockdown were reversed relatively quickly. However, the intracellular cytokine staining showed that the PIM knockdown Th1 cells still had at day 6 a reduced amount of intracellular IFNγ compared with control cells. This indicates that although the effects of PIM knockdown are transient the Th1 differentiation process is impaired by PIM kinase knockdown. In addition, T-BET and IFNγ themselves are very highly induced during the first hours after the Th1 priming, which can also reduce the effects of the PIM knockdown. By contrast, the expression of IL-12Rβ2 was still strongly inhibited by the PIM triple knockdown at the 72-h time point. Because STAT4 and IFNγ are known to induce and maintain the expression of IL-12Rβ2 (12, 13, 15, 17), it is possible that the long term influence of PIM kinases on IL-12Rβ2 is mediated via its effects on IFNγ and STAT4.
Taken together, our results show that PIM kinases induce the early stages of human Th1 cell differentiation. They positively regulate several factors and signaling pathways critical for Th1 polarization. The mechanism through which PIM kinases regulate T-BET and IFNγ transcription during the early phases of Th1 polarization is currently unknown. The expression of T-BET has also been shown to be controlled by TCR activation and the IFNγ/STAT1 signaling pathway (8, 9). In addition, IL-12 and STAT4 have been reported to induce T-BET at the transcriptional level (10, 42, 43), although this mechanism has been shown to take place in the late phase of Th1 cell differentiation (44). The expression of IFNγ is also regulated by TCR activation and STAT4 (11, 14). The possible role of PIM kinases in promoting the activity of STAT1 or STAT4 is not currently known. Our unpublished data5 suggest that PIM2 does not influence the serine phosphorylation of STAT1 at Ser727 or STAT4 at Ser721, sites important for their transcriptional activities (45). In addition, there are no conserved substrate recognition sequences for PIM kinases in STAT1 or STAT4 (46, 47). Therefore, it is not very likely that PIM1, PIM2, or PIM3 would directly regulate the activities of STAT1 or STAT4 by serine phosphorylation.
Because PIM kinase knockdown has an effect on important Th1 differentiation regulators, we performed a genome-wide analysis of transcriptional changes caused by the PIM triple knockdown 6 h after induction of Th1 differentiation. This analysis showed that altogether 241 genes were differentially expressed and that the majority of the differentially expressed genes found in Th1 cells were not affected by the PIM knockdown in Th2 cells. Among the genes found differentially expressed in Th1 cells were TGFβ signaling-related transcription factors SMAD2, TGIF1, and SOX4, which were all up-regulated. Although important for Th17 and regulatory T cell differentiation, TGFβ represses the differentiation of other Th subtypes (48–51). Smad2 knock-out mice studies have also shown that Smad2 inhibits IFNγ production by Th1 cells (52). The up-regulation of these genes under Th1-polarizing conditions is in line with the impaired Th1 response seen when the expression of PIM kinases have been down-regulated. Interestingly, NFATc1, Nf-κB, STAT5, and ATF2 target genes were enriched among the differentially expressed genes. In addition, all the target genes of these transcription factors except FAS ligand (FASLG) were down-regulated. Moreover, Ingenuity Pathway Analysis revealed an additional Nf-κB target gene, TFIP2, down-regulated in response to PIM silencing. Two of these transcription factors, NFAT and NF-κB, induced upon TCR activation contribute to the early induction of T-BET and IFNγ expression (3, 4, 53). Interestingly, both NFAT and NF-κB have been reported to be targets of PIM kinases. PIM1 phosphorylates NFATc1 on serine residues and thereby enhances its transcriptional activity (54). On the other hand, PIM2 stimulates NF-κB-dependent transcription through the phosphorylation of oncogenic serine/threonine kinase Cot, which activates NF-κB (55). ATF2, has both serine and threonine phosphorylation sites that have been reported to be phosphorylated by several different kinases, such as p38 and c-JUN N-terminal kinase (JNK) (56). In addition, ATF-c-JUN binds to IFNγ promoter (57, 58). Taken together, these results support the idea that the effects of PIM kinases on the expression of T-BET and IFNγ as well as other genes are mediated at least in part through enhanced activation of NFAT and/or NF-κB.
On the basis of earlier observations (59), it is possible that PIM kinases contribute to the regulation of Th1 cell polarization by regulating the Runt-related transcription factor (RUNX) family of transcription factors. PIM1 has been shown to phosphorylate RUNX proteins and enhance their transcriptional activity. RUNX proteins in turn have been implicated in the regulation of Th1/Th2 differentiation. RUNX1 has been shown to repress GATA-binding protein 3 (GATA3) expression and to inhibit Th2 differentiation (60), whereas RUNX3 promotes Th1 polarization in cooperation with T-BET. T-BET induces the expression of RUNX3, and they are both required for the maximal production of IFNγ in Th1 cells (34). Thus, taken together, it is possible that PIM kinases promote Th1 polarization by several mechanisms, i.e. by enhancing the early induction of T-BET and IFNγ as well as by increasing the activities of RUNX proteins.
The identification of factors and mechanisms influencing the early steps of human Th cell differentiation is important for the development of interventions and therapeutics for many autoimmune and allergic diseases. In this study, we report a new function for PIM kinases, namely the regulation of Th1 cell development, thus providing new insights into the mechanisms controlling the selective development of human Th cell subsets.
Acknowledgments
We are grateful to Sanna Edelman, Antti Ellonen, Marjo Hakkarainen, Mirkka Heinonen, Sarita Heinonen, Outi Melin, Anu Neuvonen, Johanna Myllyviita, Robert Molder, and Eeva Rainio for contributions and technical assistance in this work. We thank Dr. Anjana Rao for a careful review and comments on the manuscript.
This work was supported by The Academy of Finland, European Union FP7 Grant “Systems Biology of T-cell activation in health and disease” (SYBILLA), the Academy of Finland (Centre of Excellence in Molecular Systems Immunology and Physiology Research, 2012–2017, Decision 250114), The Sigrid Jusélius Foundation, The Department of Biotechnology, The Government of India, The National Technology Agency of Finland (TEKES), The Finnish Cultural Foundation, The Turku University Hospital Research Fund (EVO), The Väinö and Laina Kivi Foundation, The Foundation for Finnish Society against Tuberculosis, Oskar Öflund Foundation, The Research Foundation of the Pulmonary Diseases, and The Ida Montin Foundation.

This article contains supplemental Fig. S1 and Table S1.
The microarray data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO accession number GSE40542.
R. Gentleman, V. Carey, W. Huber, and F. Hahne, genefilter: methods for filtering genes from microarray experiments, R package version 1.38.0 online manual.
J. Tahvanainen and R. Lahesmaa, unpublished data.
- Th
- T helper
- NFAT
- nuclear factor of activated T cells
- NF-κB
- nuclear factor κB
- NT
- non-targeting
- PIM
- proviral integration site for Moloney murine leukemia virus
- RUNX
- Runt-related transcription factor
- SMAD2
- SMAD family member 2
- SOCS
- suppressor of cytokine signaling
- SOX4
- SRY (sex-determining region Y) box 4
- T-BET
- T box expressed in T cells
- TCR
- T cell receptor
- TGIF1
- TGFB-induced factor homeobox 1
- IL-12Rβ2
- interleukin-12 receptor β2
- Thp
- Th precursor
- DE
- differentially expressed
- ATF
- activating transcription factor
- PIM123
- PIM1 + PIM2 + PIM3.
REFERENCES
- 1. Murphy K. M., Ouyang W., Farrar J. D., Yang J., Ranganath S., Asnagli H., Afkarian M., Murphy T. L. (2000) Signaling and transcription in T helper development. Annu. Rev. Immunol. 18, 451–494 [DOI] [PubMed] [Google Scholar]
- 2. Rautajoki K. J., Kylaniemi M. K., Raghav S. K., Rao K., Lahesmaa R. (2008) An insight into molecular mechanisms of human T helper cell differentiation. Ann. Med. 40, 322–335 [DOI] [PubMed] [Google Scholar]
- 3. Sica A., Dorman L., Viggiano V., Cippitelli M., Ghosh P., Rice N., Young H. A. (1997) Interaction of NF-κB and NFAT with the interferon-γ promoter. J. Biol. Chem. 272, 30412–30420 [DOI] [PubMed] [Google Scholar]
- 4. Kiani A., García-Cózar F. J., Habermann I., Laforsch S., Aebischer T., Ehninger G., Rao A. (2001) Regulation of interferon-γ gene expression by nuclear factor of activated T cells. Blood 98, 1480–1488 [DOI] [PubMed] [Google Scholar]
- 5. Peng S. L., Gerth A. J., Ranger A. M., Glimcher L. H. (2001) NFATc1 and NFATc2 together control both T and B cell activation and differentiation. Immunity 14, 13–20 [DOI] [PubMed] [Google Scholar]
- 6. Porter C. M., Clipstone N. A. (2002) Sustained NFAT signaling promotes a Th1-like pattern of gene expression in primary murine CD4+ T cells. J. Immunol. 168, 4936–4945 [DOI] [PubMed] [Google Scholar]
- 7. Aronica M. A., Mora A. L., Mitchell D. B., Finn P. W., Johnson J. E., Sheller J. R., Boothby M. R. (1999) Preferential role for NF-κB/Rel signaling in the type 1 but not type 2 T cell-dependent immune response in vivo. J. Immunol. 163, 5116–5124 [PubMed] [Google Scholar]
- 8. Lighvani A. A., Frucht D. M., Jankovic D., Yamane H., Aliberti J., Hissong B. D., Nguyen B. V., Gadina M., Sher A., Paul W. E., O'Shea J. J. (2001) T-bet is rapidly induced by interferon-γ in lymphoid and myeloid cells. Proc. Natl. Acad. Sci. U.S.A. 98, 15137–15142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Afkarian M., Sedy J. R., Yang J., Jacobson N. G., Cereb N., Yang S. Y., Murphy T. L., Murphy K. M. (2002) T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat. Immunol. 3, 549–557 [DOI] [PubMed] [Google Scholar]
- 10. Szabo S. J., Kim S. T., Costa G. L., Zhang X., Fathman C. G., Glimcher L. H. (2000) A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100, 655–669 [DOI] [PubMed] [Google Scholar]
- 11. Mullen A. C., High F. A., Hutchins A. S., Lee H. W., Villarino A. V., Livingston D. M., Kung A. L., Cereb N., Yao T. P., Yang S. Y., Reiner S. L. (2001) Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science 292, 1907–1910 [DOI] [PubMed] [Google Scholar]
- 12. Szabo S. J., Dighe A. S., Gubler U., Murphy K. M. (1997) Regulation of the interleukin (IL)-12R β2 subunit expression in developing T helper 1 (Th1) and Th2 cells. J. Exp. Med. 185, 817–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang J. T., Shevach E. M., Segal B. M. (1999) Regulation of interleukin (IL)-12 receptor β2 subunit expression by endogenous IL-12: a critical step in the differentiation of pathogenic autoreactive T cells. J. Exp. Med. 189, 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barbulescu K., Becker C., Schlaak J. F., Schmitt E., Meyer zum Büschenfelde K. H., Neurath M. F. (1998) IL-12 and IL-18 differentially regulate the transcriptional activity of the human IFN-γ promoter in primary CD4+ T lymphocytes. J. Immunol. 160, 3642–3647 [PubMed] [Google Scholar]
- 15. Lawless V. A., Zhang S., Ozes O. N., Bruns H. A., Oldham I., Hoey T., Grusby M. J., Kaplan M. H. (2000) Stat4 regulates multiple components of IFN-γ-inducing signaling pathways. J. Immunol. 165, 6803–6808 [DOI] [PubMed] [Google Scholar]
- 16. Nishikomori R., Usui T., Wu C. Y., Morinobu A., O'Shea J. J., Strober W. (2002) Activated STAT4 has an essential role in Th1 differentiation and proliferation that is independent of its role in the maintenance of IL-12R β2 chain expression and signaling. J. Immunol. 169, 4388–4398 [DOI] [PubMed] [Google Scholar]
- 17. Letimier F. A., Passini N., Gasparian S., Bianchi E., Rogge L. (2007) Chromatin remodeling by the SWI/SNF-like BAF complex and STAT4 activation synergistically induce IL-12Rβ2 expression during human Th1 cell differentiation. EMBO J. 26, 1292–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eichmann A., Yuan L., Bréant C., Alitalo K., Koskinen P. J. (2000) Developmental expression of pim kinases suggests functions also outside of the hematopoietic system. Oncogene. 19, 1215–1224 [DOI] [PubMed] [Google Scholar]
- 19. Mikkers H., Nawijn M., Allen J., Brouwers C., Verhoeven E., Jonkers J., Berns A. (2004) Mice deficient for all PIM kinases display reduced body size and impaired responses to hematopoietic growth factors. Mol. Cell. Biol. 24, 6104–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bachmann M., Möröy T. (2005) The serine/threonine kinase Pim-1. Int. J. Biochem. Cell Biol. 37, 726–730 [DOI] [PubMed] [Google Scholar]
- 21. Feldman J. D., Vician L., Crispino M., Tocco G., Marcheselli V. L., Bazan N. G., Baudry M., Herschman H. R. (1998) KID-1, a protein kinase induced by depolarization in brain. J. Biol. Chem. 273, 16535–16543 [DOI] [PubMed] [Google Scholar]
- 22. Jackson L. J., Pheneger J. A., Pheneger T. J., Davis G., Wright A. D., Robinson J. E., Allen S., Munson M. C., Carter L. L. (2012) The role of PIM kinases in human and mouse CD4+ T cell activation and inflammatory bowel disease. Cell. Immunol. 272, 200–213 [DOI] [PubMed] [Google Scholar]
- 23. Wang Z., Bhattacharya N., Weaver M., Petersen K., Meyer M., Gapter L., Magnuson N. S. (2001) Pim-1: a serine/threonine kinase with a role in cell survival, proliferation, differentiation and tumorigenesis. J. Vet. Sci. 2, 167–179 [PubMed] [Google Scholar]
- 24. White E. (2003) The pims and outs of survival signaling: role for the Pim-2 protein kinase in the suppression of apoptosis by cytokines. Genes Dev. 17, 1813–1816 [DOI] [PubMed] [Google Scholar]
- 25. Fox C. J., Hammerman P. S., Thompson C. B. (2005) The Pim kinases control rapamycin-resistant T cell survival and activation. J. Exp. Med. 201, 259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zippo A., De Robertis A., Serafini R., Oliviero S. (2007) PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat. Cell Biol. 9, 932–944 [DOI] [PubMed] [Google Scholar]
- 27. Peltola K. J., Paukku K., Aho T. L., Ruuska M., Silvennoinen O., Koskinen P. J. (2004) Pim-1 kinase inhibits STAT5-dependent transcription via its interactions with SOCS1 and SOCS3. Blood 103, 3744–3750 [DOI] [PubMed] [Google Scholar]
- 28. Chen X. P., Losman J. A., Cowan S., Donahue E., Fay S., Vuong B. Q., Nawijn M. C., Capece D., Cohan V. L., Rothman P. (2002) Pim serine/threonine kinases regulate the stability of Socs-1 protein. Proc. Natl. Acad. Sci. U.S.A. 99, 2175–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aho T. L., Lund R. J., Ylikoski E. K., Matikainen S., Lahesmaa R., Koskinen P. J. (2005) Expression of human pim family genes is selectively up-regulated by cytokines promoting T helper type 1, but not T helper type 2, cell differentiation. Immunology 116, 82–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lund R. J., Ylikoski E. K., Aittokallio T., Nevalainen O., Lahesmaa R. (2003) Kinetics and STAT4- or STAT6-mediated regulation of genes involved in lymphocyte polarization to Th1 and Th2 cells. Eur. J. Immunol. 33, 1105–1116 [DOI] [PubMed] [Google Scholar]
- 31. Hamalainen H. K., Tubman J. C., Vikman S., Kyrölä T., Ylikoski E., Warrington J. A., Lahesmaa R. (2001) Identification and validation of endogenous reference genes for expression profiling of T helper cell differentiation by quantitative real-time RT-PCR. Anal. Biochem. 299, 63–70 [DOI] [PubMed] [Google Scholar]
- 32. Tahvanainen J., Pykäläinen M., Kallonen T., Lähteenmäki H., Rasool O., Lahesmaa R. (2006) Enrichment of nucleofected primary human CD4+ T cells: a novel and efficient method for studying gene function and role in human primary T helper cell differentiation. J. Immunol. Methods 310, 30–39 [DOI] [PubMed] [Google Scholar]
- 33. Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 34. Djuretic I. M., Levanon D., Negreanu V., Groner Y., Rao A., Ansel K. M. (2007) Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 8, 145–153 [DOI] [PubMed] [Google Scholar]
- 35. Benaglia T., Chaveau D., Hunter D. R., Young D. S. (2009) Mixtools: an R package for analyzing mixture models. J. Stat. Software 32, 1–29 [Google Scholar]
- 36. Lee H. J., Suk J. E., Patrick C., Bae E. J., Cho J. H., Rho S., Hwang D., Masliah E., Lee S. J. (2010) Direct transfer of α-synuclein from neuron to astroglia causes inflammatory responses in synucleinopathies. J. Biol. Chem. 285, 9262–9272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson W. E., Li C., Rabinovic A. (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 [DOI] [PubMed] [Google Scholar]
- 38. Smyth G. K. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- 39. Backes C., Keller A., Kuentzer J., Kneissl B., Comtesse N., Elnakady Y. A., Müller R., Meese E., Lenhof H. P. (2007) GeneTrail—advanced gene set enrichment analysis. Nucleic Acids Res. 35, W186–W192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yamamoto J., Adachi Y., Onoue Y., Adachi Y.S., Okabe Y., Itazawa T., Toyoda M., Seki T., Morohashi M., Matsushima K., Miyawaki T. (2000) Differential expression of the chemokine receptors by the Th1- and Th2-type effector populations within circulating CD4+ T cells. J. Leukoc. Biol. 68, 568–574 [PubMed] [Google Scholar]
- 41. Groom J. R., Luster A. D. (2011) CXCR3 in T cell function. Exp. Cell Res. 317, 620–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Usui T., Preiss J. C., Kanno Y., Yao Z. J., Bream J. H., O'Shea J. J., Strober W. (2006) T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription. J. Exp. Med. 203, 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ylikoski E., Lund R., Kyläniemi M., Filén S., Kilpeläinen M., Savolainen J., Lahesmaa R. (2005) IL-12 up-regulates T-bet independently of IFN-γ in human CD4+ T cells. Eur. J. Immunol. 35, 3297–3306 [DOI] [PubMed] [Google Scholar]
- 44. Schulz E. G., Mariani L., Radbruch A., Höfer T. (2009) Sequential polarization and imprinting of type 1 T helper lymphocytes by interferon-γ and interleukin-12. Immunity 30, 673–683 [DOI] [PubMed] [Google Scholar]
- 45. Chen W., Daines M. O., Khurana Hershey G. K. (2004) Turning off signal transducer and activator of transcription (STAT): the negative regulation of STAT signaling. J. Allergy Clin. Immunol. 114, 476–489 [DOI] [PubMed] [Google Scholar]
- 46. Palaty C. K., Clark-Lewis I., Leung D., Pelech S. L. (1997) Phosphorylation site substrate specificity determinants for the Pim-1 protooncogene-encoded protein kinase. Biochem. Cell Biol. 75, 153–162 [PubMed] [Google Scholar]
- 47. Peng C., Knebel A., Morrice N. A., Li X., Barringer K., Li J., Jakes S., Werneburg B., Wang L. (2007) Pim kinase substrate identification and specificity. J. Biochem. 141, 353–362 [DOI] [PubMed] [Google Scholar]
- 48. Bettelli E., Carrier Y., Gao W., Korn T., Strom T. B., Oukka M., Weiner H. L., Kuchroo V. K. (2006) Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 [DOI] [PubMed] [Google Scholar]
- 49. Veldhoen M., Hocking R. J., Atkins C. J., Locksley R. M., Stockinger B. (2006) TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24, 179–189 [DOI] [PubMed] [Google Scholar]
- 50. Li M. O., Flavell R. A. (2008) TGF-β: a master of all T cell trades. Cell 134, 392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lund R., Aittokallio T., Nevalainen O., Lahesmaa R. (2003) Identification of novel genes regulated by IL-12, IL-4, or TGF-β during the early polarization of CD4+ lymphocytes. J. Immunol. 171, 5328–5336 [DOI] [PubMed] [Google Scholar]
- 52. Takimoto T., Wakabayashi Y., Sekiya T., Inoue N., Morita R., Ichiyama K., Takahashi R., Asakawa M., Muto G., Mori T., Hasegawa E., Saika S., Shizuya S., Hara T., Nomura M., Yoshimura A. (2010) Smad2 and Smad3 are redundantly essential for the TGF-β-mediated regulation of regulatory T plasticity and Th1 development. J. Immunol. 185, 842–855 [DOI] [PubMed] [Google Scholar]
- 53. McCracken S. A., Hadfield K., Rahimi Z., Gallery E. D., Morris J. M. (2007) NF-κB-regulated suppression of T-bet in T cells represses Th1 immune responses in pregnancy. Eur. J. Immunol. 37, 1386–1396 [DOI] [PubMed] [Google Scholar]
- 54. Rainio E. M., Sandholm J., Koskinen P. J. (2002) Cutting edge: transcriptional activity of NFATc1 is enhanced by the Pim-1 kinase. J. Immunol. 168, 1524–1527 [DOI] [PubMed] [Google Scholar]
- 55. Hammerman P. S., Fox C. J., Cinalli R. M., Xu A., Wagner J. D., Lindsten T., Thompson C. B. (2004) Lymphocyte transformation by Pim-2 is dependent on nuclear factor-κB activation. Cancer Res. 64, 8341–8348 [DOI] [PubMed] [Google Scholar]
- 56. Gozdecka M., Breitwieser W. (2012) The roles of ATF2 (activating transcription factor 2) in tumorigenesis. Biochem. Soc. Trans. 40, 230–234 [DOI] [PubMed] [Google Scholar]
- 57. Jones B., Chen J. (2006) Inhibition of IFN-γ transcription by site-specific methylation during T helper cell development. EMBO J. 25, 2443–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Filén S., Ylikoski E., Tripathi S., West A., Björkman M., Nyström J., Ahlfors H., Coffey E., Rao K. V., Rasool O., Lahesmaa R. (2010) Activating transcription factor 3 is a positive regulator of human IFNG gene expression. J. Immunol. 184, 4990–4999 [DOI] [PubMed] [Google Scholar]
- 59. Aho T. L., Sandholm J., Peltola K. J., Ito Y., Koskinen P. J. (2006) Pim-1 kinase phosphorylates RUNX family transcription factors and enhances their activity. BMC Cell Biol. 7, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Komine O., Hayashi K., Natsume W., Watanabe T., Seki Y., Seki N., Yagi R., Sukzuki W., Tamauchi H., Hozumi K., Habu S., Kubo M., Satake M. (2003) The Runx1 transcription factor inhibits the differentiation of naive CD4+ T cells into the Th2 lineage by repressing GATA3 expression. J. Exp. Med. 198, 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]