Background: VERNALIZATION1 (VRN1) binds DNA sequence nonspecifically in vitro, and its loss causes vernalization defects in plants.
Results: Dominant suppression of VRN1 targets was lethal, and a triple mutant of the VRN1 B3 domain ablated DNA binding.
Conclusion: The DNA-interacting surface of VRN1 B3 is defined.
Significance: B3 domains are ubiquitous in plants, and learning how VRN1 B3 binds DNA is important for understanding B3-mediated processes.
Keywords: Arabidopsis, DNA-binding Protein, DNA-Protein Interaction, Nuclear Magnetic Resonance, Protein Crystallization, B3 Domain, Vernalization
Abstract
The B3 DNA-binding domain is a plant-specific domain found throughout the plant kingdom from the alga Chlamydomonas to grasses and flowering plants. Over 100 B3 domain-containing proteins are found in the model plant Arabidopsis thaliana, and one of these is critical for accelerating flowering in response to prolonged cold treatment, an epigenetic process called vernalization. Despite the specific phenotype of genetic vrn1 mutants, the VERNALIZATION1 (VRN1) protein localizes throughout the nucleus and shows sequence-nonspecific binding in vitro. In this work, we used a dominant repressor tag that overcomes genetic redundancy to show that VRN1 is involved in processes beyond vernalization that are essential for Arabidopsis development. To understand its sequence-nonspecific binding, we crystallized VRN1(208–341) and solved its crystal structure to 1.6 Å resolution using selenium/single-wavelength anomalous diffraction methods. The crystallized construct comprises the second VRN1 B3 domain and a preceding region conserved among VRN1 orthologs but absent in other B3 domains. We established the DNA-binding face using NMR and then mutated positively charged residues on this surface with a series of 16 Ala and Glu substitutions, ensuring that the protein fold was not disturbed using heteronuclear single quantum correlation NMR spectra. The triple mutant R249E/R289E/R296E was almost completely incapable of DNA binding in vitro. Thus, we have revealed that although VRN1 is sequence-nonspecific in DNA binding, it has a defined DNA-binding surface.
Introduction
As its name suggests, the protein VERNALIZATION1 (VRN1) was discovered for its role in Arabidopsis vernalization (1), a physiological process by which flowering accelerates in response to prolonged cold (i.e. winter). This process is essential for many crops to flower and is a key agronomic trait. The reason for the failure of vrn1 mutants to vernalize is their inability during this cold treatment to suppress a key regulator of flowering, FLC (2, 3).
Although vrn1 mutants appear to have only a single phenotype, there are clues that VRN1 has much broader roles. For example, VRN1 is highly expressed throughout Arabidopsis and associates with transcriptionally active regions of nuclei (4). Furthermore, overexpression of VRN1 causes early flowering, morphological defects, and inappropriate gene expression (1).
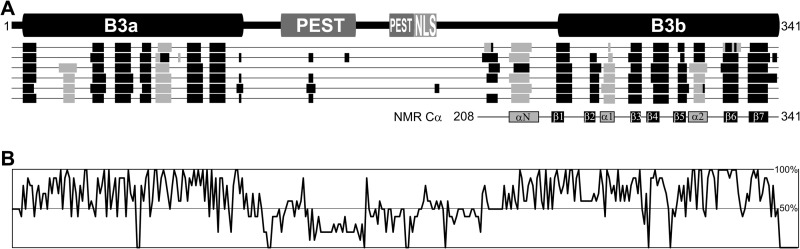
Almost two-thirds of the VRN1 protein sequence is composed of two predicted B3 DNA-binding domains, named by the discovery of the DNA binding ability of a protein region termed B3 in the maize protein VIVIPAROUS1 (5). VRN1 also has two putative PEST protein turnover domains and a putative nuclear localization sequence (Fig. 1A) (1). A region upstream of the second B3 domain is predicted to be helical, and its Cα shifts in NMR (6) agreed with this prediction. The ∼100-residue region linking the first B3 domain to the helix before the second B3 domain is predicted to be unstructured (Fig. 1A), and among the proteins most similar to VRN1, this region is the most poorly conserved in sequence (Fig. 1B and supplemental Fig. S1).
FIGURE 1.
Schematic of VRN1. A, VRN1 consists of two B3 domains of ∼100 residues each. Between them are two putative PEST-rich protein turnover domains and a Lys-rich nuclear localization sequence (NLS). Below VRN1 (from top to bottom) are the outputs from secondary structure prediction programs jalign, jfreq, jhmm, jnet, jpssm, and jpred, which suggest that only the B3 domains and a region preceding B3b are structured. The VRN1(208–341) protein has been studied in solution by NMR, and the Cα shift data (6) correlated well with these predictions. The secondary structural elements are numbered for later reference. B, summary of the percentage conservation within an alignment of VRN1 and its 10 most similar proteins from the GenBankTM Data Bank. Conservation is high at the B3 domains but also in the region preceding B3b. For the full sequence alignment this panel was based on, see supplemental Fig. S1.
B3 domains are found throughout the plant kingdom and have been classified into four families (ARF, RAV, LAV, and REM) that bind the DNA sequences TGTCTC (ARF), CACCTG (RAV), and CATGCA (LAV) in vitro (7). VRN1 is the only member of the REM B3 domain-containing protein class whose DNA binding has been studied, and it exhibits strong sequence-nonspecific DNA binding in vitro (1).
Arabidopsis encodes over 100 B3 domain-containing proteins that are implicated by genetics in processes as diverse as seed development, drought tolerance, hormone response, and flowering time (7). Most are thought to be sequence-specific DNA-binding proteins with cis-elements characteristic for each B3 class.
Here, we structurally and functionally characterize VRN1, presenting chimeric VRN1 repressor data that strongly support a broader role for VRN1 in processes essential to Arabidopsis development. This highlights the fundamental importance of VRN1 in plant development. We generated recombinant protein from a construct of VRN1 comprising residues 208–341 (VRN1(208–341), representing one of the two B3 domains) and show that like the full-length protein, this single B3 domain displays sequence-nonspecific DNA binding properties. We determined a high resolution crystal structure for VRN1(208–341) and compared it with the NMR structures of two B3 domain proteins that have not been assigned in planta functions: RAV1 is a sequence-specific DNA-binding protein (8, 9), whereas At1g16640 is reported not to bind DNA (10). We used NMR to identify putative DNA-binding sites, which we tested with a series of 16 mutations in VRN1(208–341) that included a triple Arg-to-Glu change that abolished in vitro DNA-binding activity. These data suggested which residues were most important but also showed that the VRN1(208–341) DNA-binding surface was large.
EXPERIMENTAL PROCEDURES
Dominant Suppression of in Vivo VRN1 Targets
The VRN1 (At3g18990) ORF was amplified using sense and antisense primers (5′-GAT GCC ACG CCC TTT CTT CCA TAA GTT GAT-3′ and 5′-GAT GCC ACG CCC TTT CTT CCA TAA GTT GAT-3′) and ligated into the SmaI site of the p35SSRDXG vector to produce a chimeric repressor gene fused with the SRDX repression domain (GLDLDLELRLGFA), and the region corresponding to each transgene was transferred into the pBCKH plant expression vector using the Gateway system (Invitrogen) (11). C-terminal placement of the SRDX tag was ideal, as VRN1 retains its native function with C-terminal fusions to the much larger GFP (4). The binary construct was transferred to Agrobacterium tumefaciens by electroporation and transformed into wild-type Columbia A. thaliana plants by floral dip (12). T1 transgenic plants were selected on Murashige and Skoog medium-agar plates containing 3% sucrose and 30 μg/ml hygromycin. Subsequently, T0/T1 seeds were sown without the pressure of hygromycin selection and screened for small deformed plants.
VRN1(208–341) Expression, Purification, and Selenomethionine Labeling
Native VRN1(208–341) protein was produced for crystallization as described previously (13). To produce selenomethionine (SeMet)4-labeled protein, Escherichia coli cells containing a pQE30-VRN1(208–341) construct (13) were grown in minimal M63 medium containing seleno-dl-methionine (Sigma S3875) and standard amino acids before expression was induced by isopropyl β-d-thiogalactopyranoside as described previously (14). SeMet-VRN1(208–341) was purified and concentrated for crystallization as described (13).
VRN1(208–341) Crystal Structure Determination
Native VRN1(208–341) was crystallized at 4 °C using the hanging drop vapor diffusion method (13). SeMet-labeled protein was crystallized under similar conditions using a protein concentration of 19 mg/ml and 0.1 m MES (pH 6.0), 1.4 m sodium chloride, and 10 mm manganese chloride as the well solution. Crystals of SeMet-VRN1(208–341) appeared after 20 days, whereas native crystals formed in 6 days. Crystals of SeMet-VRN1(208–341) grew to a maximum size of ∼200 × 50 × 50 μm.
Crystals were harvested by transferring them into a solution containing 0.1 m MES (pH 6.0), 2 m sodium chloride, 10 mm manganese chloride, and 20% (v/v) ethylene glycol as a cryopreservative. Crystals were then flash-cooled in liquid nitrogen and stored under liquid nitrogen. These were transferred to a dry shipper for transport to the Australian Synchrotron, where diffraction data were measured on beamline MX1 at 0.956 Å (15). A single-wavelength anomalous diffraction data set was measured from the SeMet-VRN1(208–341) crystal, and a high resolution native data set was measured from the native VRN1(208–341) crystal. Crystals of VRN1(208–341) contain two molecules in the crystallographic asymmetric unit.
Data were indexed, integrated, and scaled using HKL2000 (16). Heavy atom positions were identified using ShelxD (17). Phasing was performed with PHENIX Autosol, and PHENIX Autobuild (18) was used to generate an initial model (R = 22.9%/Rfree = 27.8%) This was further refined with rounds of manual model building in Coot (19) and refinement in PHENIX Refine (20). Of the 133 residues in the designed construct, 117 residues were modeled in each of the two molecules in the asymmetric unit. In both molecules, residues 208–220 and 339–341 of VRN1(208–341) were not modeled due to poor electron density at the N and C termini, indicating disorder. Four chloride ions were included in the model at crystal contact points where water molecules were not sufficient to explain the electron density.
Cross-validation Rfree calculation was performed with 5% of the data. The quality of the model was monitored throughout refinement using MolProbity (21). Data collection and refinement statistics are given in Table 1. Raw data images will be made available on the Diffraction IMage Experiment Repository (DIMER) website.
TABLE 1.
Data collection, phasing, and refinement statistics
Values in parentheses are for the top shell of data.
| Native VRN1(208–341) | SeMet VRN1(208–341) | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.956 | 0.956 |
| Space group | C2 | C2 |
| Cell parameters | ||
| a, b, and c (Å) | 106.2, 47.3, 61.4 | 105.9, 47.9, 61.3, |
| α, β, γ | 90.0°, 115.0°, 90.0° | 90.0°, 114.5°, 90.0° |
| Resolution range (Å) | 50.0–1.6 (1.66–1.6) | 31.3–2.7 (2.8–2.7) |
| No. of observations | 139,720 | 114,844 |
| No. of unique reflections | 34,680 (3439) | 7689 (766) |
| Multiplicity | 4.0 (4.0) | 14.9 (15.2) |
| Completeness (%) | 94.6 (93.7) | 99.0 (98.6) |
| 〈I/σI〉 | 23 (4.6) | 23.8 (21.5) |
| Rmerge (%) | 5.3 (29.8) | 8.8 (15.9) |
| Phasing | ||
| Resolution range (Å) | 31.3–2.7 | |
| No. of selenium atoms/asymmetric unit | 2 | |
| Figure of merit (before/after density modification) | 0.32/0.72 | |
| Refinement | ||
| Resolution range (Å) | 23.65–1.60 (1.65–1.60) | |
| No. of reflections | 34,661 | |
| R-factor | 0.120 (0.142) | |
| Free R-factor | 0.166 (0.234) | |
| No. of atoms | ||
| Protein | 1962 | |
| Chloride ion | 4 | |
| Water | 366 | |
| Wilson B-factor | 14 | |
| Average B-values (Å2) | ||
| Protein | 19 | |
| Chloride ion | 19 | |
| Water | 34 | |
| r.m.s.d. in ideal | ||
| Bond lengths (Å) | 0.011 | |
| Bond angles | 1.3° | |
| Ramachandran plot (%) | ||
| Favored | 98.3 | |
| Outliers | 0.0 | |
| MolProbity score | 0.68, 100th percentile | |
B3a Modeling by I-TASSER
To generate a model structure for B3a using the VRN1(208–341) structure, we entered the first 104 amino acids of VRN1 (Met-1–Asn-104) into I-TASSER (22) and, as a restraint, uploaded the Protein Data Bank file for VRN1(208–341) in “specify template without alignment” with otherwise default parameters used.
NMR Titration Experiments with VRN1(208–341) and dsDNA
15N-Labeled VRN1(208–341) was produced as described (6), and after size exclusion, its final concentration was 0.74 mg/ml (43.6 μm). An 18-bp fragment of DNA was made by heating a mixture of 0.25 ml of 1 mm ssDNA oligonucleotide JM121 (5′-AAA AAT TGC ATG TCA TTC-3′) and 0.25 ml of 1 mm ssDNA oligonucleotide JM122 (5′-GAA TGA CAT GCA ATT TTT-3′) to 95 °C (368 K) for 5 min and allowing the solution to cool to room temperature (294 K). This 18-bp region corresponds to a specific region of FLC where VRN1 was suspected to bind in vivo, but the short repeating AGTC probes we later used in fluorescence polarization and the previously demonstrated sequence nonspecificity of VRN1 would suggest that any DNA sequence would give a similar result in NMR DNA binding experiments.
The 18-bp fragment of DNA was purified by size exclusion chromatography using a Sephacryl S75 column (GE Healthcare) in 50 mm sodium phosphate (pH 7.0), 150 mm NaCl, and 1 mm DTT. After size exclusion, the concentration of the DNA was 0.46 mg/ml (42 μm).
To avoid precipitation when combining the 18-bp DNA with 15N-labeled VRN1(208–341), we found it necessary to combine them diluted and concentrate them after they were combined. Each NMR DNA binding experiment began by layering in an Amicon Ultra-4 concentrator with a 5-kDa cutoff (Millipore UFC800524) the following solutions: 1.85 ml of 38 μm 15N-labeled VRN1(208–341), a separating “cushion” of 2 ml of size exclusion buffer (50 mm sodium phosphate (pH 7.0), 150 mm sodium chloride, and 1 mm DDT), and either 0.1 or 0.2 m eq of 42 μm DNA. A no-DNA control containing only 15N-labeled VRN1(208–341) was also prepared. The mixture was concentrated to 0.5 ml (0.14 mm VRN1(208–341)) before 50 μl of D2O was added, and a heteronuclear single quantum correlation (HSQC) spectrum was acquired as described (6). With 0.2 m eq of DNA added, the number of scans was increased to 96 to obtain similar peak resolution (64 scans were performed for the no-DNA control and when 0.1 m eq of DNA had been added).
VRN1(208–341) Mutagenesis
Mutations were introduced into the pQE30-VRN1(208–341) construct (13) by PCR with complementary primer pairs. For brevity, the mutation will be followed only by the forward primer sequence with the mutagenic codon underlined: R249A, 5′-C AGA GTG GTT CTG GCG CCA TCC TAT CTA TAC-3′; Y254A, 5′-CCA TCC TAT CTA GCG AGA GGT TGC ATC ATG-3′; Y260A, 5′-GGT TGC ATC ATG GCG CTT CCT TCT GGG-3′; R289A, 5′-CAA TGG CCT GTT GCG TGT CTC TAC AAA GCC-3′; L291A, 5′-CCT GTT CGA TGT GCG TAC AAA GCC GGG-3′; and R296A, 5′-CTC TAC AAA GCC GGG GCG GCC AAA TTC AGT C-3′. VRN1(208–341) mutants were 15N-labeled during expression and purified as described (6) and concentrated to ∼0.3 mm. A two-dimensional 1H-15N HSQC spectrum was acquired for each mutant to ascertain the effect of each mutation on the protein. Subsequent to these six mutants, another group of six mutants were made with less conservative changes, including G295E (5′-CTC TAC AAA GCC GAG AGA GCC AAA TTC-3′), R289E (5′-CAA TGG CCT GTT GAG TGT CTC TAC AAA-3′), and R296E (5′-TAC AAA GCC GGG GAG GCC AAA TTC AGT-3′. To make the R289E/R296E double mutant, the single mutations were introduced sequentially. To make the R289A/R296A double mutant, the aforementioned R289A plasmid was mutagenized by PCR with a shorter primer for R296A (5′-TAC AAA GCC GGG GCG GCC AAA TTC AGT-3′). To replace the loop of VRN1 with the sequence RPSYLYRGCI with the equivalent At1g16640-derived loop sequence ISEKSSKS, PCR was done using phosphorylated primers JM563 (5′-tct agt aaa tcc ATG TAT CTT CCT TCT GGG-3′) and JM564 (5′-ttt ctc aga gat CAG AAC CAC TCT GAA GAA-3′). The bases encoding ISEKSSKS are shown in lowercase letters and underlined. The PCR product was ligated to create the mutant referred to hereafter as loopΔ. A final set of four mutants were made by incorporating R249E as a single, second, or third mutation with the appropriate template using primer JM577 (5′-AGA GTG GTT CTG GAG CCA TCC TAT CTA-3′) and its complement. All proteins were 15N-labeled, and an HSQC spectrum was acquired for each as previously described (6).
Fluorescence Polarization DNA Binding Assay
The aforementioned mutants and wild-type VRN1(208–341) were assayed for in vitro DNA-binding activity using a fluorescence polarization (FP) assay that measured fluorescence upon binding of each protein to 6-carboxyfluorescein (FAM)-labeled dsDNA probes. The dsDNA probes were made by combining JM565 (FAM-AGT CAG TCA GTC AGT-3′) with JM566 (FAM-ACT GAC TGA CTG ACT-3′) to a final concentration of 0.5 μm in water, placing them in boiling water for 5 min, and allowing the solution to cool at room temperature. An unlabeled probe for competition was made in a similar way using JM569 (5′-AGT CAG TCA GTC AGT-3′) and JM570 (5′-ACT GAC TGA CTG ACT-3′) diluted to 50 μm in water. The FAM-labeled dsDNA probe was then diluted to 25 nm with FP buffer (30 mm HEPES, 50 mm potassium chloride, 10 mm guanidinium chloride, 2 mm magnesium chloride, 0.5 mm EDTA, and 0.01% Nonidet P-40 (pH 7.5)). The protein concentration was quantified using A280 (NanoDrop), and purity was confirmed by SDS-PAGE. All FP assays were conducted in FP buffer.
The 16 mutant proteins were separated into four independent sets of experiments, each including the wild-type protein as a control. For binding reactions, 20 μl of 25 nm labeled probe was added to 80 μl of protein in FP buffer. The final protein concentrations were 1.5, 3, 5, 10, 20, 30, 50, 75, 100, 150, 300, and 600 nm, and the final probe concentration was 5 nm. Three 25-μl aliquots of each reaction mixture were transferred to a black opaque 384-well plate and incubated for 30 min at room temperature in the dark before measurement. FP was measured with a PerkinElmer EnVision multi-label plate reader (485 nm excitation and 520 nm emission). A saturation binding curve was fitted using GraphPad Prism, and the Kd value of each mutant was determined with an average Bmax of 125 nm found for the wild-type protein. To demonstrate that fluorescence was probe-specific, 2.5 μl of 50 μm unlabeled probe (1000-fold over the labeled probe) was added to those wells containing the highest concentration of labeled probe, and FP was re-assayed to demonstrate competitive displacement of the labeled probe.
RESULTS
Constitutively Expressed VRN1-SRDX Is Lethal
Arabidopsis vrn1 mutants have only one phenotype: their reduced vernalization response (1). This specificity seems at odds with the abundance of VRN1 and its mRNA expression in all plant tissues except pollen (4). A fully functional VRN1-GFP fusion associates throughout the nucleus (4). Furthermore, VRN1 overexpression causes accelerated flowering and pleiotropic phenotypes (1). These features suggest that VRN1 has more widespread roles in maintaining gene expression.
We tested the hypothesis that VRN1 function is protected by redundancy with chimeric repressor silencing technology (23). This approach arose from the observation that the essential repression domain of class II ERF (ethylene-responsive element-binding factor) transcription factors and of several TFIIIA-type zinc-finger transcription factors is located at the C terminus of these proteins, in a conserved sequence termed the EAR (ERF-associated amphiphilic repression) motif. When the EAR-like motif from SUPERMAN designated as “SRDX” was fused to DNA-binding proteins, the chimeras acted as strong repressors (24, 25). Constitutive expression of SRDX fusions has been heavily used in Arabidopsis to overcome genetic redundancy. The ability of the SRDX repression domain to overcome genetic redundancy has been demonstrated for a number of transcription factors, where each mimics the phenotypes of loss-of-function genetic mutations for the same transcription factor (26), e.g. 35S:AP3:SRDX and 35S:CUC1:SRDX (27). Thus, a VRN1-SRDX fusion is likely to act as a dominant repressor wherever VRN1 binds in planta, even at locations where other proteins share its roles.
We initially failed to isolate transformants containing the 35S:VRN1:SRDX construct (Fig. 2A) when the seeds of transformed plants were sown on Murashige and Skoog medium-agar containing hygromycin selection, probably due to lethality or a severe defect in growth or development. We then grew the T0/T1 seeds on Murashige and Skoog medium-agar without hygromycin but supplemented with glucose and found three small plants with the same deformed phenotype (Fig. 2, B and C). These deformed plants developed roots and true leaves but died before they could set seed. These three deformed plants along with 12 aphenotypic plants were tested by PCR with 35S and nos or VRN1 primers to show that the pleiotropy correlated with the VRN1 transgene (Fig. 2D). Sequencing the 35S/nos PCR product from these three lines confirmed that each contained a proper VRN1-SRDX fusion. Our results suggest that VRN1 regulates essential developmental processes in addition to regulating vernalization. We suspect that genetic redundancy is masking the role of VRN1 in essential developmental processes.
FIGURE 2.
Phenotype of VRN1-SRDX fusion lines. A, the VRN1-SRDX constructs consist of the constitutive CaMV35S promoter (p35S), a VRN1 ORF and SRDX fusion, and the nopaline synthase (nos) terminator. B, magnified images of three T1 VRN1-SRDX transgenic plants grown on glucose-containing medium without hygromycin selection showing their small size and deformed phenotype. C, the same three T1 plants next to non-transgenic neighboring plants. D, results from PCR analysis of genomic DNA extracted from transgenic and non-transgenic lines showing that the deformed plants contain the VRN1-SRDX transgene. The smaller VRN1 band corresponds to the non-intronic VRN1 ORF used in the VRN1-SRDX transgene.
A Structured and Conserved Region in VRN1 Orthologs
We next investigated the structure of VRN1 to gain insight into its function at a molecular level. Secondary structure analysis of VRN1 suggested that it consists of two predicted B3 domains, B3a and B3b, separated by an unstructured region of ∼100 residues (Fig. 1A). A region of ∼20 residues preceding B3b is also predicted to be helical. An alignment of the 10 proteins with the highest sequence similarity to VRN1 showed that this region preceding the second B3 domain is conserved (Fig. 1B and supplemental Fig. S1). The predictions showed good correlation with the secondary structure of VRN1(208–341) determined experimentally from the Cα chemical shift data acquired by solution NMR (Fig. 1A) (6).
VRN1(208–341) Adopts a β-Barrel Fold
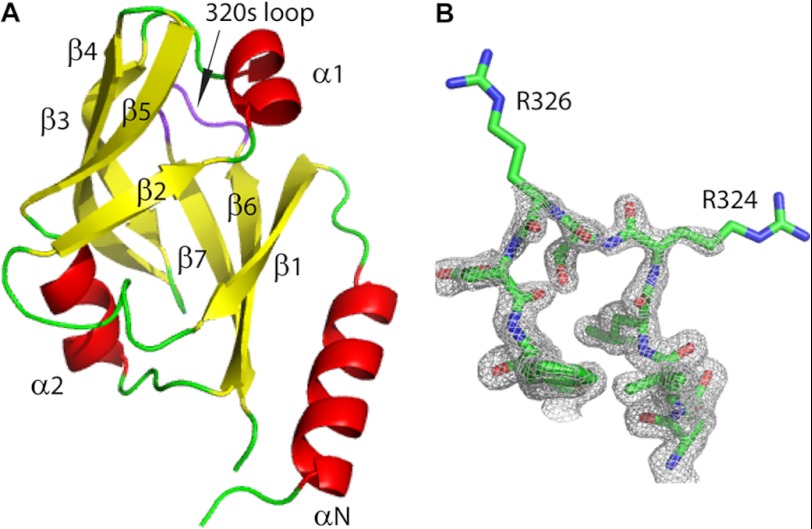
The crystal structure of VRN1(208–341) was solved by single-wavelength anomalous diffraction methods and refined to high resolution with excellent geometry (Table 1). The structure of the B3 domain of VRN1(208–341) reveals a seven-stranded β-barrel capped at either edge of the barrel by a short helix. As indicated above, the VRN1 B3b domain is preceded by a sequence of 35 residues conserved among VRN1 orthologs. The crystal structure shows that this sequence forms a long helix (αN) (Fig. 3A), as predicted by bioinformatics (Fig. 1A) and NMR secondary structure studies (6).
FIGURE 3.
Crystal structure of VRN1(208–341). A, structural model of VRN1(208–341) showing that its overall fold is a barrel of β-strands (yellow) capped at each end by short α-helices (red). The 320s loop, which could not be assigned by NMR (potentially due to flexibility), is shown in purple. The conserved N-terminal sequence of VRN1 proteins that is absent in other B3 domains forms a long α-helix (α-N, bottom right of the structure). B, the loop region Glu-321–Val-329 showing 2Fo − Fc electron density contoured at the 1σ level. Images were created in PyMOL (33).
The two molecules of VRN1(208–341) in the asymmetric unit are essentially identical (0.6 Å root mean square deviation (r.m.s.d.) for all 117 Cα atoms). A loop region comprising Arg-324–Val-329, which we refer to as the 320s loop, was predicted to exhibit conformational exchange based on an inability to assign Arg-324–Val-329 using NMR (6). In the crystal structure reported here, the main chain of this loop is well ordered and adopts similar structures in both molecules, though the side chains of Arg-324 and Arg-326 are disordered (Fig. 3B).
Comparison of VRN1 Structure with RAV1 and At1g16640
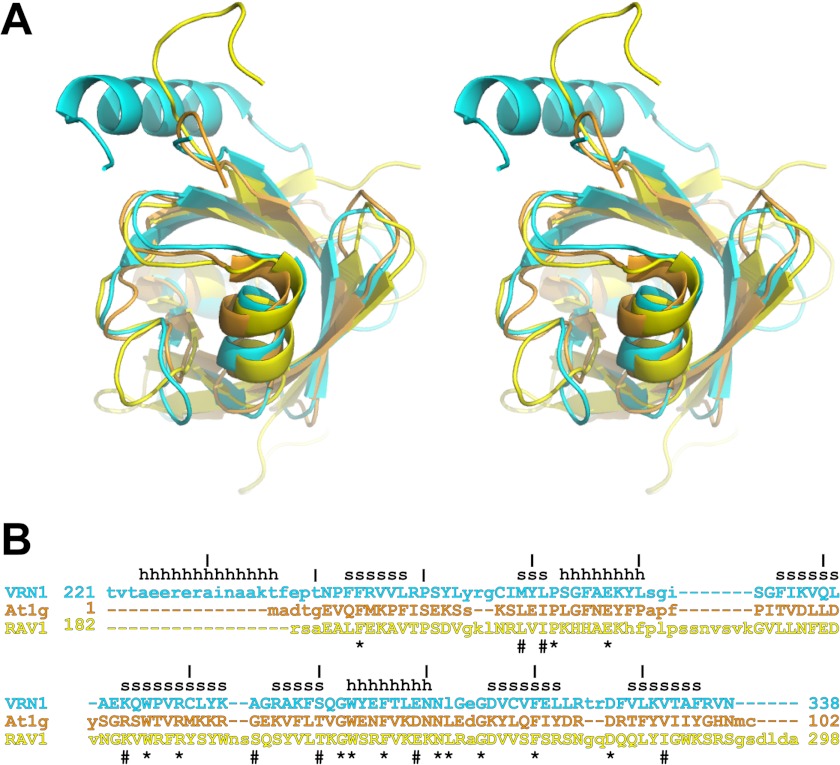
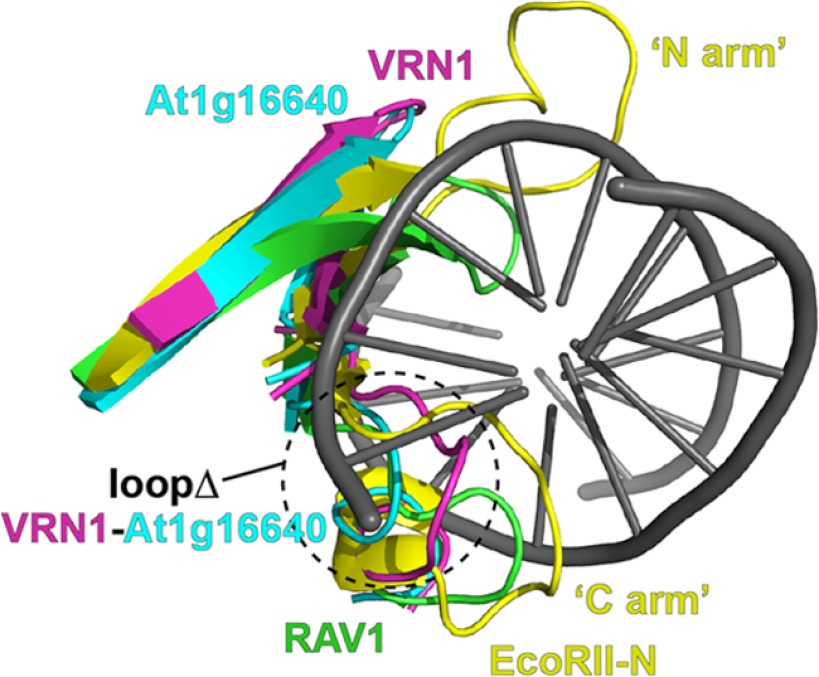
VRN1(208–341) is the first crystal structure of a B3 domain protein, although NMR structures of two plant B3 domain proteins, RAV1 (Protein Data Bank code 1WID) (9) and At1g16640 (code 1YEL) (10), have been reported previously. These three proteins share very low sequence identity (21–27%) yet have similar architectures, differing only in the addition of the preceding N-terminal helix in VRN1(208–341) (Fig. 4). Indeed, a DALI search (28) found RAV1 and At1g16640 structures to be the top hits when VRN1(208–341) was used as a search model: At1g16640 gives a Z-score of 12.6, with 21% sequence identity and a r.m.s.d. of 1.8 Å for 94 Cα atoms, and RAV1 gives a Z-score of 9.6, with 23% sequence identity and a r.m.s.d. of 2.6 Å for 98 Cα atoms. By comparison, DALI similarity analysis of the At1g16640 and RAV1 structures gives a Z-score of 10.1, with 27% sequence identity and a r.m.s.d. of 2.8 Å for 94 Cα atoms. Thus, of the three pairs of structures, VRN1(208–341) and At1g16640 are the most similar to each other using DALI for comparison. This structural similarity is reflected to some degree in the function of these three proteins. RAV1 binds a specific DNA sequence (8), VRN1 binds DNA sequence nonspecifically, and At1g16640 has been reported to not bind DNA (10).
FIGURE 4.
Structural superimposition and structure-based sequence alignment. A, the structures of VRN1(208–341) (cyan), At1g16640 (orange), and RAV1 (yellow) were aligned with SSM in Coot. The resulting structural superimposition is shown in stereo. B, the structure-based sequence alignment of these three proteins (same color scheme as in A) shows structurally aligned residues in uppercase letters. Residues that are conserved in all three structures are shown by asterisks, and residues that are highly conserved are shown by hash symbols. Secondary structure elements in the VRN1(208–341) structure are indicated by h for helix and s for strand. At1g, At1g16640.
How can we rationalize the in vitro nonspecific DNA binding of VRN1(208–341) and the lack of DNA binding in At1g16640? Comparison of the structures and sequences of VRN1(208–341) and At1g16640 revealed several features in the former that could contribute to DNA binding and that are absent in the latter. Thus, VRN1(208–341) is a more basic protein overall, with 17 basic residues and 12 acidic residues, whereas At1g16640 has an equal number of basic and acidic residues (14 each). Furthermore, the stretch of residues encompassing strands 4 and 5 and the beginning of helix 2 forms a contiguous surface on all three proteins. In RAV1, this region contributes the DNA-binding loop 5 and in total has four basic and no acidic residues. By comparison, this region of VRN1(208–341) lacks the DNA-binding residues of RAV1 but does comprise five basic residues and no acidic residues.
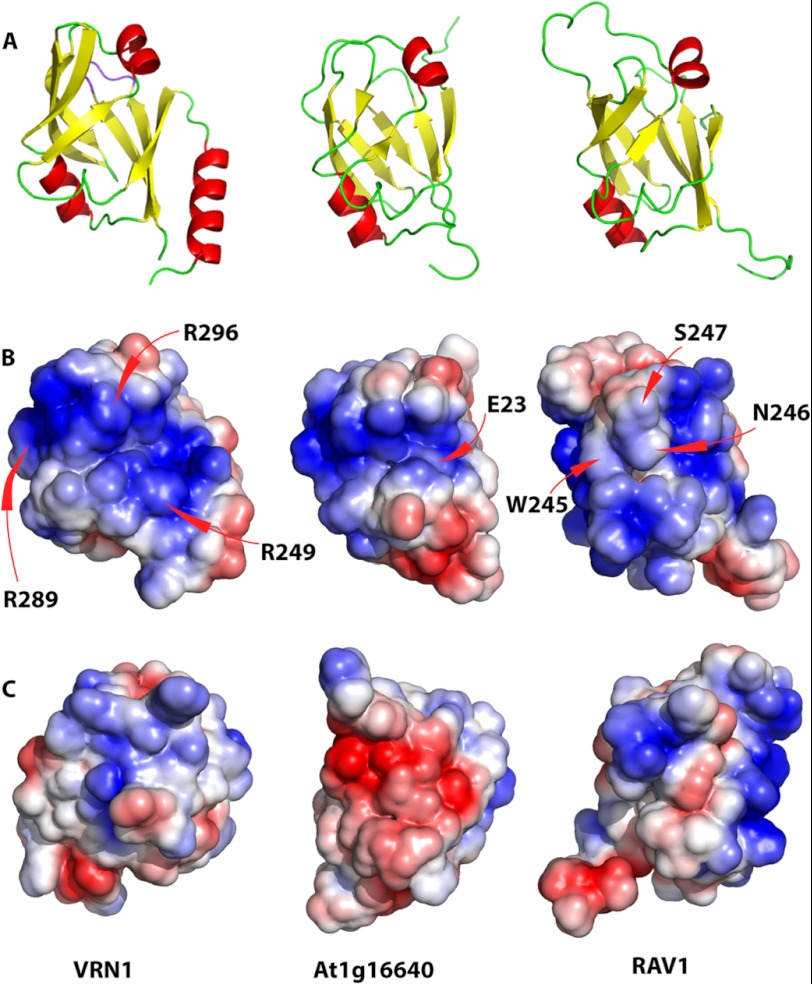
The electrostatic surface representations of the three B3 domains support these observations (Fig. 5). Trp-245, Asn-246, Ser-247, and Gln-249 of RAV1 are located in a region that separates two highly basic regions; these residues are proposed to interact directly with DNA bases (9). In both VRN1 and At1g16640, these residues are absent: in VRN1, this region is replaced by a broad continuous basic groove, whereas in At1g16640, a less expansive basic groove is apparent. In At1g16640, Glu-23 is located in the middle of this basic groove, although this is not apparent in the electrostatic surface. Finally, the greater overall basicity of RAV1 and VRN1 compared with At1g16640 is evident in the electrostatic surfaces.
FIGURE 5.
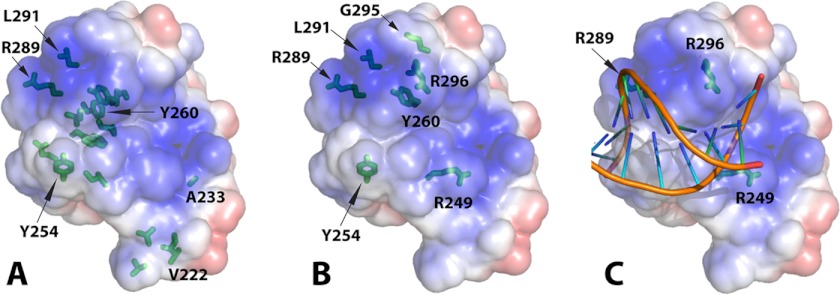
Structural alignment of VRN1, At1g16640, and RAV1 B3 domains. A, structural alignment showing the putative DNA-binding surface of VRN1(208–341) and RAV1. B, electrostatic surfaces of the three domains in the same orientation as in A. Arg residues contributing to the conspicuous basic patch on VRN1(208–341) (R249, R289, and R296) are labeled. C, electrostatic surfaces of the three domains rotated 180° around the vertical axis relative to the surface in B. The structures were first aligned with SSM in Coot and then oriented to show the putative DNA-binding region of RAV1 (9), with the surface charge ranging from −3 to 3 kbT/ec(J/C), where k is the Boltzmann constant (1.380 × 10−23 J/K), T is in Kelvin, and e is the charge on an electron (1.6 × 10−19 C).
We investigated the structures further to identify features that might contribute to the sequence-nonspecific DNA binding properties of VRN1(208–341). The largest differences between the B3 domains of VRN1(208–341) and RAV1 are localized to the loop regions that join the strands of the β-barrel. In general, the loops of RAV1 are longer than those of VRN1(208–341) and At1g16640 (Fig. 5). Thus, of the seven loops in the B3 fold, three of the VRN1(208–341) loops are shorter than those of RAV1 (by seven, two, and one residues). For At1g16640, four loops are shorter than those of RAV1 (by seven, two, two, and two residues).
Three of the seven RAV1 loops incorporate its DNA-binding residues (Lys-199 and Leu-200 in loop 1; Lys-207 in loop 2; and Trp-245, Asn-246, Ser-247, and Gln-249 in loop 5) (9), but these residues are generally not conserved in VRN1(208–341) or in At1g16640, and two of the DNA-binding residues in loop 5 are deleted in both VRN1(208–341) and At1g16640. These mutations and deletions may explain why VRN1(208–341) and At1g16640 do not exhibit the same DNA binding specificity of RAV1.
Model for the First B3 Domain of VRN1
There are two B3 domains in VRN1, B3a and B3b (Fig. 1), and both bind DNA (Ref. 1 and this work) and share a sequence identity of ∼18%. Here, we have focused on the structure and DNA-binding surface of B3b. Although 18% sequence identity may be considered low, the structural similarity of B3 domains despite similarly low sequence identity (Fig. 4) suggests that these two domains may share a similar fold. The notable lack in B3a of the N-terminal helix in VRN1(208–341) is because this region is not part of any B3 domain; rather, it is a specific and conserved region found to precede the second B3 domain of plant homologs of VRN1.
Like B3b, B3a is a highly basic domain (15 Arg/Lys residues and 11 Asp/Glu residues) consistent with DNA binding. We generated a homology model of B3a from the crystal structure of B3b and found that the modeled B3a structure overlaid well with B3b (r.m.s.d. of 1.3 Å) (supplemental Fig. S2) as well as the B3 domains of At1g16640 (r.m.s.d. of 1.8 Å) and RAV1 (r.m.s.d. of 2.5 Å). The B3a model also has a basic patch in a similar location as B3b, suggesting that the same surface may bind DNA. B3a is known to bind DNA in vitro (1), but further work is required to confirm that the binding faces are similarly located for both B3 domains and to establish whether the two domains bind DNA simultaneously in vivo and, if so, where along the DNA strand each binds.
Comparison with Bacterial Restriction Endonucleases
The next five highest hits in the DALI analysis are for three proteins: EcoRII (Protein Data Bank code 1NA6, chains A and B) (29), EcoRII (code 3HQF) (30), and BfiI (code 2C1L, chains A and B) (31), giving Z-scores ranging from 6.3 and 6.9, with sequence identities between 10 and 16% and r.m.s.d. between 2.6 and 3.1 Å for 93–96 Cα atoms. All three proteins are restriction endonucleases in which a DNA-binding effector domain contains a fold very similar to that found in the B3 family (9).
Identification of the VRN1 DNA-binding Surface
Previous work used HSQC NMR with the RAV1 B3 domain and DNA to predict the binding surface and propose a model for the complex (9). We monitored the VRN1(208–341) HSQC spectra after the addition of an 18-bp dsDNA probe. In almost all cases, the peaks corresponding to the responsive residues broadened and disappeared. Mapping these onto the structure showed that the binding residues could be mapped to one face of VRN1(208–341) (Fig. 6A and supplemental Fig. S3). These data were consistent with the electrostatic surface most favorable for VRN1(208–341)-DNA binding and the DNA-binding surface previously identified for the RAV1 B3 domain (9).
FIGURE 6.
VRN1(208–341)-DNA interaction. A, VRN1(208–341) in the same orientation as Fig. 3A and Fig. 5A displaying its electrostatic surface and marked with the residues most sensitive to NMR. The side chains of all residues in intermediate exchange at 0.2 m eq are shown in stick format, and those of the most sensitive residues in intermediate exchange at 0.1 m eq of DNA are also labeled textually. The figure with a slight rotation in supplemental Fig. S3 is better to show the “sidedness” of the DNA interaction. B, the side chains of VRN1(208–341) residues mutated preceding DNA binding studies are shown in stick format. C, the structure of VRN1(208–341) was superimposed onto the structure of EcoRII from the crystal structure of the EcoRII-DNA complex (Protein Data Bank code 3HQF) (30) using SSM in Coot. The structure of EcoRII was then removed, leaving this VRN1(208–341)-DNA overlay. No further manipulation was performed. PyMOL was used to produce the electrostatic surface of VRN1(208–341). The positions of three Arg residues (R249, R289, and R296) that, when mutated to Glu, abolished DNA binding are shown.
To confirm the VRN1(208–341) DNA-binding surface and to identify the most important regions, we produced protein mutants and assessed their in vitro DNA-binding activities using FP assays (Fig. 6B and Table 2). We used HSQC spectra to ensure that the fold of each protein was maintained (supplemental Figs. S4–S6). Initially, we created a series of six single Ala mutants targeting Tyr and Arg residues, but these changes had weak or no effect on DNA binding. The most severe were R249A and R296A, which decreased binding with a 4-fold change in Kd. We then made a second series of six mutants, including an R289A/R296A double mutant, a loop swap from At1g16640, three single Glu mutants, and an R289E/R296E double mutant. The single Glu changes R249E and R296E had the greatest effect (8- and 12-fold change in Kd). The effect on the binding strength of these single Glu mutations was similar to that of the R289A/R296A double mutant. The R289E/R296E double mutant showed a dramatic (50-fold) change in DNA binding Kd. We then created a third round of four mutants in which the R249E mutation was introduced singly or added to existing single or double Glu mutations. The R249E/R289E/R296E triple mutation almost completely abolished DNA binding (Table 2).
TABLE 2.
Estimated DNA-binding Kd values for VRN1(208–341) and mutants
To demonstrate accurate protein quantification, an equal loading of protein was run on an SDS-polyacrylamide gel and Coomassie Blue-stained (supplemental Fig. S7).
| Protein | Kd | Kd -fold decrease |
|---|---|---|
| nm | ||
| WT | 10.3 ± 1.1 | 1.0 |
| Y254A | 9.1 ± 1.2 | 0.9 |
| Y260A | 13.1 ± 1.0 | 1.3 |
| L291A | 13.2 ± 1.2 | 1.3 |
| R289A | 18.8 ± 1.8 | 1.8 |
| G295E | 25.5 ± 2.4 | 2.5 |
| LoopΔ | 26.5 ± 1.4 | 2.6 |
| R289E | 36.4 ± 4.9 | 3.5 |
| R249A | 37.1 ± 2.4 | 3.6 |
| R296A | 40.7 ± 3.5 | 4.0 |
| R249E | 86.3 ± 5.0 | 8.4 |
| R289A/R296A | 102.4 ± 8.0 | 9.9 |
| R296E | 121.9 ± 8.0 | 11.8 |
| R289E/R296E | 533.9 ± 44.1 | 51.8 |
| R249E/R296E | 601.2 ± 112.7 | 58.4 |
| R249E/R289E | 604.1 ± 105.3 | 58.7 |
| R249E/R289E/R296E | 1548.0 ± 721.5 | 150.3 |
Of the single mutations, the R296E substitution had the most deleterious effect on DNA binding, closely followed by R249E. Although R296E and R249E most strongly affected DNA binding, all three double Glu mutants had similar decreases in DNA-binding activity (52–59-fold change in Kd). The DNA binding of the triple mutant R249E/R289E/R296E was similar to that of the negative control, suggesting that DNA binding was completely abrogated. The fact that three mutations are required to abolish DNA binding suggests that the nonspecific DNA-binding surface of the VRN1 B3b domain is large or that there is redundancy in the DNA-binding residues.
Model of DNA Binding by the B3 Domain of VRN1
We generated a model for DNA binding by VRN1(208–341) by superimposing the VRN1(208–341) structure onto the EcoRII-DNA complex from Protein Data Bank code 3HQF (30) (Fig. 6C) using SSM in Coot (19). The DNA appears to fit within a groove of VRN1(208–341), and the phosphate backbone appears to be well placed to make contact with basic patches on the VRN1 surface.
DISCUSSION
Arabidopsis vrn1 mutants have only one phenotype: their reduced vernalization response (1). This specificity seems at odds with VRN1 abundance and mRNA expression in all plant tissues except pollen (4). A fully functional VRN1-GFP fusion associates throughout the nucleus (4). VRN1 overexpression also causes accelerated flowering and pleiotropic phenotypes (1). These features suggest that VRN1 has more widespread roles than just FLC regulation. Our finding that the expression of a 35S:VRN1:SRDX construct induced pleiotropic phenotypes and was eventually lethal is the first experimental evidence that VRN1 has roles that are essential to development but that are protected by genetic redundancy. The 35S:VRN1:SRDX construct did not induce embryo lethality in transgenic plants, which were still able to develop leaves and roots, albeit deformed. This finding indicates that despite the vernalization- and FLC-specific phenotype of vrn1 genetic mutants, VRN1 has more widespread roles in essential processes that remain to be revealed.
VRN1 has been shown to bind strongly to DNA but in a sequence-nonspecific manner (1). NMR analysis and its pattern of surface charge suggest that VRN1(208–341) has a definite DNA-binding surface. We therefore compared its structure with those of related proteins with known DNA-binding modes.
The structural similarity between bacterial endonucleases and B3 domain proteins has been noted previously (9) and led to the suggestion that the B3 domain-containing plant proteins may have evolved from a bacterial endonuclease (32). Comparison of the N-terminal domain from E. coli restriction endonuclease EcoRII (designated as EcoRII-N) with Arabidopsis RAV1 and At1g16640 B3 domains led Golovenko et al. (30) to claim that the structure of EcoRII-N in complex with DNA was the first structure of a B3 family protein with DNA. They compared RAV1 and At1g16640 structures with EcoRII-N and suggested that the B3 family proteins bind DNA in a similar fashion, which is perpendicular to the orientation proposed for the model of the RAV1-B3 DNA complex by Yamasaki et al. (9). More recently, Yamasaki et al. (32) performed modeling experiments that altered the helical axis of the DNA in their original model by 90°. Our study of the structure and DNA binding properties of VRN1 provides further evidence to support the DNA-binding models of both groups (30, 32).
EcoRII-N and RAV1 are reported to bind DNA in a sequence-specific manner, whereas VRN1 binds DNA in a sequence-nonspecific manner. This may be explained by the observation that RAV1 possesses two loops in positions similar to those in EcoRII that make intimate contact with the EcoRII recognition sequence within the DNA major groove, whereas VRN1 has only one such loop (Fig. 7). It appears that VRN1 may bind DNA in a manner that is mostly dependent on interaction with the phosphate groups of the DNA backbone. Golovenko et al. (30) reported that three EcoRII residues involved in binding the DNA phosphate backbone are conserved in RAV1 and At1g16640. We found that two of these residues are also conserved in VRN1: EcoRII Arg-81 (VRN1 Arg-289) and EcoRII Thr-100 (VRN1 Ser-300). However, EcoRII Lys-23 appears to be replaced by Val-246 in VRN1. When the crystal structures of VRN1 and EcoRII are aligned, a number of residues that make contacts with bases or phosphate groups in the EcoRII-DNA complex are structurally conserved in VRN1; these include EcoRII Arg-98/VRN1 Lys-298, EcoRII Tyr-85/VRN1 Tyr-292, EcoRII Tyr-41/VRN1 Tyr-260, EcoRII Arg-81/VRN1 Arg-289, and EcoRII Thr-100/VRN1 Ser-300.
FIGURE 7.
Structural alignment of EcoRII-N in complex with DNA (Protein Data Bank code 3HQF) with RAV1, At1g16640, and VRN1 B3 domains to highlight the two loops of EcoRII that interact with DNA. EcoRII-N is yellow, with residues 27–42 (bottom) and residues 78–101 (top) shown and referred to by Golovenko et al. (30) as the “N arm” and the “C arm,” respectively. RAV1 is green, with residues 194–206 (bottom) and residues 238–255 (top) shown. VRN1 is magenta, with residues 250–266 (bottom) and residues 287–302 (top) shown. At1g16640 is aqua, with residues 15–23 (bottom) and residues 50–66 (top) shown. The VRN1 and At1g16640 loops swapped in the VRN1 loopΔ mutant are marked.
The At1g16640 B3 domain was reported not to bind DNA (10). This lack of DNA binding could be explained by the more acidic surface of At1g16640 compared with RAV1 and VRN1. Curiously, like VRN1, At1g16640 has only one of the two loops of RAV1 and EcoRII. We replaced the loop of VRN1 (RPSYLYRGCI, VRN1(249–258)) with that of At1g16640 (ISEKSSKS, At1g16640(14–21)) expecting that this would abolish DNA binding. However, we were surprised to find that DNA binding by this loop substitution mutant (loopΔ) was only slightly reduced (2.6-fold change in Kd) (Table 2).
When first characterized, VRN1 was thought not to bind DNA based on initial electrophoretic mobility shift assays containing poly(dI-dC) (a nonspecific DNA-binding competitor). Only when the VRN1 protein concentration was increased or poly(dI-dC) was removed was DNA binding apparent. Surface plasmon resonance, which does not include a nonspecific DNA-binding competitor, confirmed the strong sequence-nonspecific DNA binding of VRN1 (1). Given the structural similarity between VRN1 B3b and At1g16640, it may be worth reinvestigating its DNA binding ability using these approaches.
Our mutagenesis experiments demonstrated that to disrupt DNA binding in the VRN1 B3b domain, charge reversal mutations were required (Arg to Glu). To completely abolish binding required three such charge reversal mutations. These findings suggest that the DNA-binding surface of VRN1(208–341) is large and involves potentially dozens of residues or that the binding surface is large but that there may be redundancy, so when one residue or region is removed, another can compensate. On the basis of our VRN1 structure, its comparison with Protein Data Bank structures of restriction enzyme-DNA complexes, and the effect of mutations on DNA binding, we have proposed a model for the binding of DNA by VRN1(208–341). This model could potentially explain how other B3 domains interact with dsDNA and provides a basis for future structure-function studies with Arabidopsis VRN1.
Acknowledgments
We thank X. Jia for help with NMR, F. Fontaine (Centre for Drug Discovery and Design, The University of Queensland) for help with fluorescence polarization DNA binding assays, Q. Kaas for help with PyMOL, and M. François and A. Argyros for technical assistance. We are grateful for access to The University of Queensland Remote Operation Crystallization and X-ray (ROCX) Diffraction Facility and the Australian Synchrotron.
This work was supported in part by a University of Queensland early career research award and Australian Research Council Queen Elizabeth II Fellowship DP0879133 (to J. S. M.), a National Health and Medical Research Council R. D. Wright Fellowship 401748 (to J. M. H.), and by Australian Research Council Australian Laureate Fellowship FL0992138 and Honorary National Health and Medical Research Council Research Fellowship 455829 (to J. L. M.).

This article contains supplemental Figs. S1–S7.
The atomic coordinates and structure factors (code 4I1K) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SeMet
- selenomethionine
- HSQC
- heteronuclear single quantum correlation
- FP
- fluorescence polarization
- FAM
- 6-carboxyfluorescein
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Levy Y. Y., Mesnage S., Mylne J. S., Gendall A. R., Dean C. (2002) Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297, 243–246 [DOI] [PubMed] [Google Scholar]
- 2. Michaels S. D., Amasino R. M. (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sheldon C. C., Burn J. E., Perez P. P., Metzger J., Edwards J. A., Peacock W. J., Dennis E. S. (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mylne J. S., Barrett L., Tessadori F., Mesnage S., Johnson L., Bernatavichute Y. V., Jacobsen S. E., Fransz P., Dean C. (2006) LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc. Natl. Acad. Sci. U.S.A. 103, 5012–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Suzuki M., Kao C. Y., McCarty D. R. (1997) The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA-binding activity. Plant Cell 9, 799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mylne J. S., Mas C., Hill J. M. (2012) NMR assignment and secondary structure of the C-terminal DNA binding domain of Arabidopsis thaliana VERNALIZATION1. Biomol. NMR Assign. 6, 5–8 [DOI] [PubMed] [Google Scholar]
- 7. Swaminathan K., Peterson K., Jack T. (2008) The plant B3 superfamily. Trends Plant Sci. 13, 647–655 [DOI] [PubMed] [Google Scholar]
- 8. Kagaya Y., Ohmiya K., Hattori T. (1999) RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamasaki K., Kigawa T., Inoue M., Tateno M., Yamasaki T., Yabuki T., Aoki M., Seki E., Matsuda T., Tomo Y., Hayami N., Terada T., Shirouzu M., Osanai T., Tanaka A., Seki M., Shinozaki K., Yokoyama S. (2004) Solution structure of the B3 DNA binding domain of the Arabidopsis cold-responsive transcription factor RAV1. Plant Cell 16, 3448–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Waltner J. K., Peterson F. C., Lytle B. L., Volkman B. F. (2005) Structure of the B3 domain from Arabidopsis thaliana protein At1g16640. Protein Sci. 14, 2478–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mitsuda N., Hiratsu K., Todaka D., Nakashima K., Yamaguchi-Shinozaki K., Ohme-Takagi M. (2006) Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol. J. 4, 325–332 [DOI] [PubMed] [Google Scholar]
- 12. Bechtold N., Ellis J., Pelletier G. (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. III Sci. Vie 316, 1194–1199 [Google Scholar]
- 13. King G., Hill J. M., Martin J. L., Mylne J. S. (2009) Expression, purification and preliminary X-ray diffraction studies of VERNALIZATION1208–341 from Arabidopsis thaliana. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 291–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shouldice S. R., Heras B., Jarrott R., Sharma P., Scanlon M. J., Martin J. L. (2010) Characterization of the DsbA oxidative folding catalyst from Pseudomonas aeruginosa reveals a highly oxidizing protein that binds small molecules. Antioxid. Redox. Signal. 12, 921–931 [DOI] [PubMed] [Google Scholar]
- 15. McPhillips T. M., McPhillips S. E., Chiu H.-J., Cohen A. E., Deacon A. M., Ellis P. J., Garman E., Gonzalez A., Sauter N. K., Phizackerley R. P., Soltis S. M., Kuhn P. (2002) Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 9, 401–406 [DOI] [PubMed] [Google Scholar]
- 16. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 17. Sheldrick G. M. (2010) Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D 66, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 20. Afonine P. V., Grosse-Kunstleve R. W., Adams P. D. (2005) The Phenix refinement framework. CCP4 Newsl. Protein Crystallogr. 42, 8 [Google Scholar]
- 21. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739 [DOI] [PubMed] [Google Scholar]
- 24. Hiratsu K., Ohta M., Matsui K., Ohme-Takagi M. (2002) The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 514, 351–354 [DOI] [PubMed] [Google Scholar]
- 25. Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kagale S., Links M. G., Rozwadowski K. (2010) Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 152, 1109–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oshima Y., Mitsuda N., Nakata M., Nakagawa T., Nagaya S., Kato K., Ohme-Takagi M. (2011) Novel vector systems to accelerate functional analysis of transcription factors using chimeric repressor gene-silencing technology (CRES-T). Plant Biotechnol. 28, 201–210 [Google Scholar]
- 28. Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou X. E., Wang Y., Reuter M., Mücke M., Krüger D. H., Meehan E. J., Chen L. (2004) Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold. J. Mol. Biol. 335, 307–319 [DOI] [PubMed] [Google Scholar]
- 30. Golovenko D., Manakova E., Tamulaitiene G., Grazulis S., Siksnys V. (2009) Structural mechanisms for the 5′-CCWGG sequence recognition by the N- and C-terminal domains of EcoRII. Nucleic Acids Res. 37, 6613–6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grazulis S., Manakova E., Roessle M., Bochtler M., Tamulaitiene G., Huber R., Siksnys V. (2005) Structure of the metal-independent restriction enzyme BfiI reveals fusion of a specific DNA-binding domain with a nonspecific nuclease. Proc. Natl. Acad. Sci. U.S.A. 102, 15797–15802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamasaki K., Kigawa T., Seki M., Shinozaki K., Yokoyama S. (2012) Trends Plant Sci., in press [DOI] [PubMed] [Google Scholar]
- 33. Schrödinger, LLC (2010) The PyMOL Molecular Graphics System, Version 1.4, Schrödinger, LLC, San Diego, CA [Google Scholar]