Background: Cyclotides are biologically active, plant-derived macrocyclic peptides. All cyclotides and their acyclic variants have been isolated from dicots.
Results: We characterized nine novel linear cyclotides from monocot plant Panicum laxum.
Conclusion: Our study provides the first evidence of linear cyclotides at the protein level in the Poaceae.
Significance: Ancient linear cyclotide analogs may have existed before the divergence of dicots and monocots.
Keywords: Genomics, Peptides, Plant, Plant Defense, Proteomics, Cyclotides, Monocot Plant, Poaceae, Linear Cyclotides, Panitides
Abstract
Cyclotides are disulfide-rich macrocyclic peptides that display a wide range of bioactivities and represent an important group of plant defense peptide biologics. A few linear variants of cyclotides have recently been identified. They share a high sequence homology with cyclotides but are biosynthetically unable to cyclize from their precursors. All hitherto reported cyclotides and their acyclic variants were isolated from dicot plants of the Rubiaceae, Violaceae, Cucurbitaceae, and recently the Fabaceae and Solanaceae families. Although several cyclotide-like genes in the Poaceae family were known from the data mining of the National Center for Biotechnology Information (NCBI) nucleotide database, their expression at the protein level has yet to be proven. Here, we report the discovery and characterization of nine novel linear cyclotides, designated as panitides L1–9, from the Panicum laxum of the Poaceae family and provide the first evidence of linear cyclotides at the protein level in a monocot plant. Disulfide mapping of panitide L3 showed that it possesses a cystine knot arrangement similar to cyclotides. Several panitides were shown to be active against Escherichia coli and cytotoxic to HeLa cells. They also displayed a high stability against heat and proteolytic degradation. Oxidative folding of the disulfide-reduced panitide L1 showed that it can fold efficiently into its native form. The presence of linear cyclotides in both dicots and monocots suggests their ancient origin and existence before the divergence of these two groups of flowering plants. Moreover, the Poaceae family contains many important food crops, and our discovery may open up new avenues of research using cyclotides and their acyclic variants in crop protection.
Introduction
Cyclotides are cysteine-rich miniproteins consisting of 28–37 residues with a cyclized backbone formed by an end-to-end amide bond (1). Their structures are stabilized by three disulfide bonds arranged in a cystine knot topology with the pattern of Cys I–IV, II–V, and III–VI (2). These structural features provide cyclotides an extraordinary stability to heat, chemical, and enzymatic degradation (3). Cyclotides can be further categorized into two major subfamilies, the Möbius and bracelet, which differ from each other by the presence or absence of a cis-Pro residue in loop 5, respectively (1).
Cyclotides exhibit a diverse range of biological activities, including insecticidal (4), antimicrobial (5), anti-HIV (6), neurotensin inhibition (7), and cytotoxicity (8, 9). They are found in a variety of dicot plants, including the Cucurbitaceae (10), Rubiaceae (11), Violaceae (12, 13), Fabaceae (9, 14), and Solanaceae families (15). They are ribosomally synthesized as linear precursors, excised at the N and C termini of their mature cyclotide domains, and then head-to-tail ligated to produce mature cyclic peptides (4). Their encoding genes were first characterized from the Rubiaceae plant Oldenlandia affinis (4) and subsequently in several other plant species (9, 15–19). The predicted precursors of cyclotides display low sequence homology among the known cyclotide-producing families, and sometimes even between species of the same genus. Except for the Fabaceae and Cucurbitaceae, the general gene arrangement is similar among the cyclotide-producing plants in Rubiaceae and Violaceae. A typical cyclotide gene encodes a precursor with an endoplasmic reticulum (ER)2 signal sequence, an N-terminal propeptide (NTPP), a mature cyclotide domain, and a C-terminal propeptide (CTPP) (20, 21). In some instances, the cyclotide genes encode for multiple mature cyclotide domains with each domain separated by a short N-terminal repeat region (4). In the Cucurbitaceae, the cyclotide genes called TIPTOP have recently been characterized from Momordica cochinchinensis (19). The TIPTOP genes are somewhat similar to a multidomain cyclotide gene but are distinguished by having both cyclic and acyclic knottin (cystine knot) domains in the same precursor. In the Fabaceae, the architecture of the cyclotide-containing precursor is distinctly different. The N-terminal proto-domain is totally absent in their precursors, which contain only the ER signal sequence followed directly by a mature cyclotide domain, a short linker region, and an albumin-1 chain a domain (9, 17).
Recently, a few acyclic variants of cyclotides have been identified. They share the same cystine arrangement and high sequence homology with cyclotides, but are biosynthetically unable to cyclize (18, 21, 22). They have been referred as “uncyclotides” by Nguyen et al. (21) or “acyclotides” by Poth et al. (15). In this study, the term linear cyclotides or acyclotides will be used. A common characteristic shared by the acyclic variants is the absence, in the cyclotide domains of their precursors and the processed linear cyclotide sequences, of the “essential” C-terminal Asn residue that serves as the recognition site for the putative thiol protease processing enzyme in the excision and cyclization of a cyclotide.
The occurrence of linear cyclotides was once thought to be rare with violacin A isolated from Viola odorata by Ireland et al. (22) in 2006 as the sole example. Since then, additional linear cyclotides have been identified from various plants, including hedyotide B2 from Hedyotis biflora (21), chassatide C7 and C8 from Chassalia chartacea (18), psyle C from Psychotria leptothyrsa (23), kalata B20-lin from O. affinis (24), and Phyb M from Petunia hybrida (15). Analysis of their genetic structures revealed that they contain, at the mature cyclotide domain, either a truncated precursor sequence without the essential C-terminal Asn residue and the CTPP or a mutation to a stop codon prior to the CTPP. Both mutations can account for the failure in the biosynthesis of cyclotide-containing precursors to produce the characteristic cyclic peptide backbone of cyclotides. Although the functional correlation between the circular structure and bioactivity remains controversial, several linear cyclotides have been shown to retain the bioactivities comparable with their cyclic counter parts (18, 23).
The present study of novel linear cyclotides in the Poaceae arose from our interest in identifying proteinaceous natural products in traditional medicines and important food crops. Recently, Mulvenna et al. (25) reported the existence of cyclotide-like genes from data mining of nucleotide databases in several Poaceae plants such as Oryza sativa, Zea mays, Triticum aestivum, and Hordeum vulgare. However, there is no evidence for their expression at the protein level. Following this lead, we screened six Poaceae species and found that linear variants of cyclotides but not cyclotides were abundantly expressed in Panicum laxum with the total peptide yield ∼0.25 mg/g of plant materials (wet weight). P. laxum, also known as lax panicgrass, is a perennial tropical grass, about 30–60 cm high, commonly found near roadsides and open space areas. It has value as cattle fodder and as an outstanding grass for sheep grazing in rubber plantations in tropic and subtropic regions (26, 27).
Our finding of linear cyclotides in the Poaceae (grass family) is significant. It provides new insights in understanding their distribution and evolution in plants, particularly in agronomically important food crops. This finding is also timely because of the recent discovery of cyclotides in other food crops such as Fabaceae (legume) and Solanaceae (potato) families (9, 15). The Poaceae is the fifth largest plant family with over 10,000 species divided into 12 subfamilies and over 700 genera (28). It is host to four of the most widely cultivated crops in the world and numerous other economically and agriculturally important plant species (29). Here, we are the first to report the isolation and characterization of linear cyclotides in P. laxum of the Poaceae family. It is also the first monocot plant found to produce linear cyclotides. We also characterized the expression profiles as well as the functional properties of these peptides. Our study confirms their presence in monocots and contributes to our understanding of cyclotides and their acyclic variants in the Poaceae family.
EXPERIMENTAL PROCEDURES
Isolation and Purification of Novel Panitides
Aerial parts of P. laxum (60 g) were extracted with 500 ml of 10% ethanol. After filtration, the aqueous extract was concentrated by rotary evaporation and subsequently fractionated by preparative HPLC using a C18 Vydac column (250 × 21 mm) on a Shimadzu system at a flow rate of 8 ml/min. A linear gradient of 1%/min of 0–80% buffer B was applied. Buffer A contains 0.05% (v/v) trifluoroacetic acid (TFA) in HPLC grade water, and buffer B contains 0.05% (v/v) TFA and 99.5% (v/v) acetonitrile. The fractions obtained were repurified to separate individual peptides by using a semipreparative C18 Vydac column (250 × 10 mm) at a flow rate of 3 ml/min with the same gradient.
Sequence Determination
10 μg of each peptide was dissolved in 30 μl of 100 mm ammonium bicarbonate buffer (pH 7.8), containing 20 mm dithiothreitol (DTT), and incubated for 1 h at 37 °C. The S-reduced peptides were either S-alkylated first with iodoacetamide (IAM) followed by enzymatic digestion or directly digested with endoproteinase Glu-C, trypsin, or chymotrypsin. The obtained peptide fragments were sequenced by MALDI-TOF MS/MS with nitrogen as the collision gas and applied collision energy of 1 keV. Assignments of isobaric residues Ile/Leu and Lys/Gln of panitides L1, L2, L4, L6, L7, and L8 were based on the cDNA sequences. For panitides L3 and L5, they were assigned based on digestion patterns by chymotrypsin and homology to other panitides and known cyclotides.
Cloning of Panitide Genes
RNA was prepared from fresh leaves and converted to single-stranded cDNA. Partial encoding genes of panitides were amplified by 3′ RACE PCR (Invitrogen) using degenerate forward primers encoding the ICGETCV sequence (5′-ATATGTGGNGARACNTGYGT-3′) and AFCGETC sequence (5′-GCATTATGTGGNGARACNTG-3′). The remaining encoding genes were obtained by 5′ RACE PCR (Invitrogen) using reverse primers based on the cDNA sequences obtained from 3′ RACE PCR. To identify whether panitide L4 gene contains intron, genomic DNA from fresh leaves was extracted. PCRs on DNA templates were then conducted with specific primers designed against 5′- and 3′-untranslated regions of panitide L4.
Disulfide Mapping
Panitide L3 (0.5 mg) was partially reduced in 2 ml of 100 mm citrate buffer, pH 3.0, containing 20% acetonitrile and 20 mm tris(2-carboxyethyl)phosphine at 37 °C for 40 min. Subsequently, N-ethylmaleimide (NEM) was added to a final concentration of 50 mm and incubated at 37 °C for another 15 min. The reaction was quenched by immediate injection of samples into a Vydac C18 column (250 × 4.6 mm) at a flow rate of 1 ml/min. Intermediate species were separated by RP-HPLC and analyzed by mass spectrometry to verify the number of NEM-alkylated cysteines. S-NEM intermediate species with one (1SS) or two (2SS) intact disulfide bonds were fully S-reduced with 20 mm DTT and S-alkylated with 40 mm IAM. S-alkylated peptides were analyzed by MS/MS.
Oxidative Folding of Panitide L1
Native panitide L1 was S-reduced with DTT and subsequently purified by RP-HPLC. After lyophilization, S-reduced panitide L1 at the final peptide concentration of 30 μm was refolded in 50% isopropyl alcohol (v/v) containing 100 mm ammonium bicarbonate, 1 mm reduced glutathione, pH 8.0. Aliquots at various time points were removed and monitored by RP-HPLC.
Antibacterial Assay
Two bacterial and two fungal strains from the American Type Culture Collection (ATCC) were used including Staphylococcus aureus ATCC 12600, Escherichia coli ATCC 25922, Candida albicans 11006, and Candida tropicalis 750. All the microbes were cultured in trypticase soy broth. The antimicrobial activities of the novel panitides were examined using the radial diffusion assay as described previously (9, 30). The synthetic dendrimeric peptide (RLYR)4-(K2K) was used as the positive control (31).
Cytotoxicity Assay
HeLa cells cultured in Dulbecco's modified Eagle's medium were seeded onto 96-well plate (Nunc) and incubated overnight. Peptides were then added to each well at the final concentration of 1–10 μm. After a 48-h incubation, cell viability was evaluated by using PrestoBlue cell viability reagent (Invitrogen). 1% Triton X-100 solution was used as positive control. The cytotoxicities of panitides were indicated by IC50 values (concentration that gives a survival index of 50%).
Heat Stability
10 μg of each peptide was added to 100 μl of distilled water and incubated at 100 °C for 1 h. A replicate was done as a control and placed at room temperature. The RP-HPLC profiles of the heated and control samples were compared with evaluate their stability.
Enzymatic Stability
10 μg of each peptide was added to 100 μl of 100 mm ammonium bicarbonate buffer (pH 7.8). 1 μl of 0.5 μg/μl trypsin was added and incubated at 37 °C for 3 h. A replicate was done as a control without adding trypsin. The RP-HPLC profiles of the treated and control samples were compared with evaluate their stability.
Accession Numbers
GenBankTM accession numbers of novel cyclotide-like sequences identified by BLAST searches are as follows: Agrostis stolonifera A (JU116752), B (DV868577), and C (DV868380); T. aestivum F (HX171398); H. vulgare B (EX575351); and Z. mays K (CF061604). There are no expressed sequence tags identified for Setaria italica genes as they were identified upon TBLASTN search of the whole genome shotgun contigs database.
RESULTS
Screening for Cyclotide-like Peptides in Poaceae Species
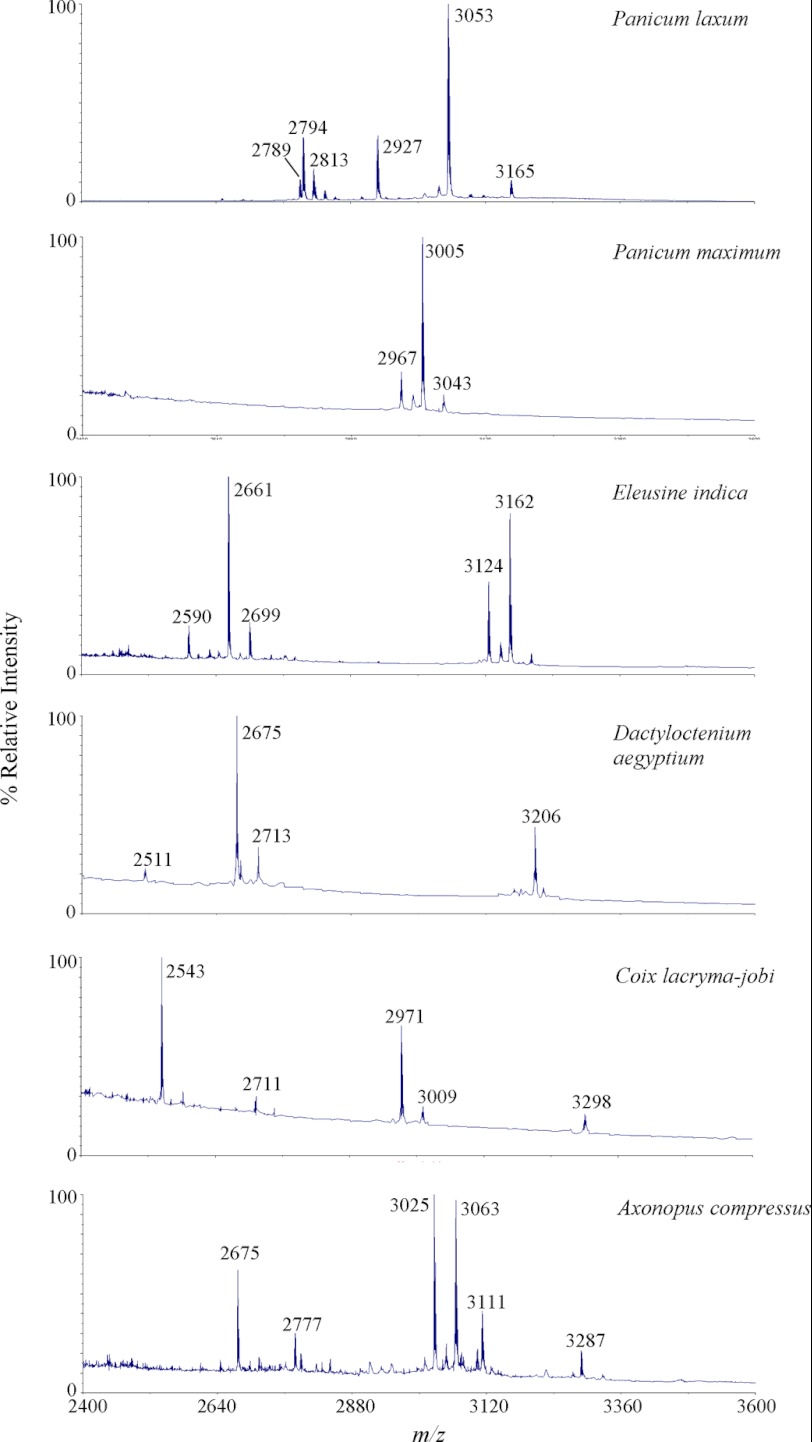
Six Poaceae species readily found in our campus in Singapore were screened for the presence of cyclotide-like peptides. They included P. laxum, Panicum maximum, Eleusine indica, Dactyloctenium aegyptium, Coix lacryma-jobi, and Axonopus compressus. Plant materials of each species (0.1 g) were ground in liquid nitrogen, extracted with 50% acetonitrile, and profiled by mass spectrometry. All tested plant species were found to express peptides around the cyclotide mass range of 2.5–4 kDa (Fig. 1) with six Cys after S-reduction and S-alkylation. To show that they are cyclotide-like peptides, P. laxum was selected as a model species for further investigation. ∼0.5 g of aerial tissues of P. laxum was extracted with 3 ml of water and semipurified on a C18 solid phase extraction column. The extraction column was washed with 20% acetonitrile and eluted with 80% acetonitrile. The eluted peptides were S-reduced with DTT and S-alkylated with IAM. Most of the eluted peptides displayed a mass increase of 348 Da, suggesting the presence of three disulfide bonds consistent with a cyclotide-like peptide.
FIGURE 1.
MS profiling of Poaceae species for the presence of cyclotide-like masses. All six plant samples including P. laxum, P. maximum, E. indica, D. aegyptium, C. lacryma-jobi, and A. compressus were found to produce peptides within the known cyclotide mass range of 2.5–4 kDa.
Isolation and Sequence Determination of Novel Linear Cyclotides from P. laxum
To determine the sequences of the putative cyclotide-like peptides in the Poaceae family and to provide sufficient samples for sequencing, a preparative-scale extraction of 60 g of aerial tissues of P. laxum was performed. After several rounds of repetitive RP-HPLC, eight novel peptides were isolated and designated as panitides L1–8. To determine their primary sequences, they were S-reduced with DTT. The S-reduced peptides were either S-alkylated with IAM or directly digested with trypsin, chymotrypsin, or endoproteinase Glu-C without S-alkylation. Enzyme-generated peptide fragments were sequenced by tandem mass spectrometry. All the novel peptides have acyclic structures and were found to incorporate pyroglutamic acid at their N termini (Table 1). They all have a Möbius-like primary structure and share a high sequence homology with the prototypic cyclotide kalata B1.
TABLE 1.
Novel linear cyclotides in Panicum laxum
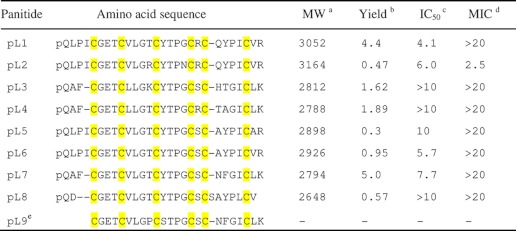
a Molecular weights (MW) are shown in monoisotopic masses.
b Yield of linear cyclotides (mg) were obtained from 60 g of plant materials.
c IC50 (μm), concentration that gives a 50% survival of HeLa cells.
d MIC (μm), minimal inhibitory concentration against E. coli.
e Panitide L9 (abbreviated as pL) is predicted from the gene sequence. Cys residues are highlighted in yellow.
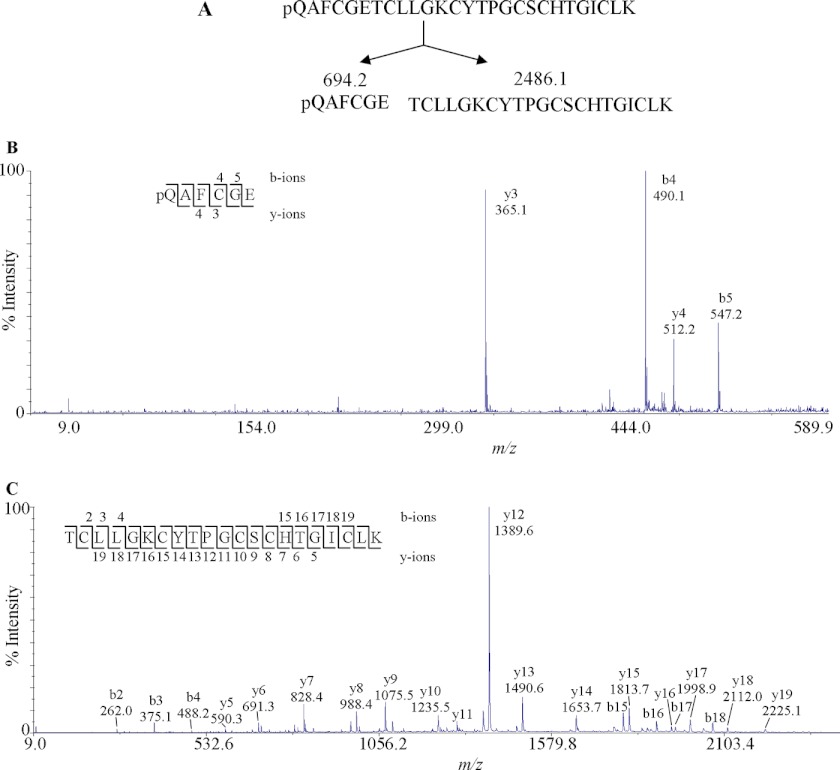
As an example, the MS/MS sequencing of panitide L3 is shown in Fig. 2. Panitide L3 had an m/z value of 2813, which became 3161 upon S-alkylation. The mass gain of 348 Da after S-alkylation indicates the presence of three disulfide bonds in the peptide structure. The linear backbone of panitide L3 was supported by two lines of evidence. First, the S-alkylated panitide L3 was extensively fragmented by MS/MS, which indicated its linear structure because cyclic peptides normally do not fragment efficiently under our MS/MS conditions. Second, endoproteinase Glu-C digestion of S-alkylated panitide L3 generated two fragments with m/z values of 2486 and 694. The total molecular mass of these two fragments is 18 Da bigger than the original S-alkylated panitide L3, corresponding to the addition of a single water molecule. This indicates that panitide L3 contains only one enzymatic susceptible site. If the peptide had a cyclic backbone, a single fragment with the m/z value of 3179 Da would be expected. Instead, two fragments were observed, suggesting that panitide L3 has a linear backbone. De novo sequencing of the digested fragments gave the full sequence of panitide L3 as pQAFCGETCLLGKCYTPGCSCHTGICLK with a pyroglutamyl residue at its N terminus. Using this approach, the amino acid sequences of other panitides were also determined.
FIGURE 2.
MS/MS analysis of panitide L3. A, S-alkylated panitide L3 was digested with endoproteinase Glu-C, generating two fragments with m/z values of 694.2 and 2486.1. B, MS/MS spectra of 694.2 Da fragment. C, MS/MS spectra of 2486.1 Da fragment. The peptides were digested with three different enzymes, endoproteinase GluC, trypsin, and chymotrypsin, as indicated under “Experimental Procedures.” The endoproteinase GluC fragments are shown here. The remaining sequence was obtained from other proteolytic fragments.
cDNA Cloning of Novel Panitides
Encoding cDNAs of the novel panitides were cloned by 3′ RACE PCR using degenerated primers targeting the ICGETCV and AFCGETC sequences. Full-length genes were subsequently obtained by 5′ RACE PCR. As a result, we obtained four full-length clones encoding panitides L1, L2, L4, and L6, and three partial clones encoding panitides L7, L8, and a novel sequence panitide L9. The supplemental material contains partial sequences for panitides L7, L8, and L9.
To compare the genetic structures of panitides at both DNA and mRNA levels, the encoding gene of panitide L4 was cloned from the leaf DNA using cDNA-derived sequences as primers. The DNA clone revealed that panitide L4 gene has no intron, which is similar to cyclotide genes of the Violaceae family.
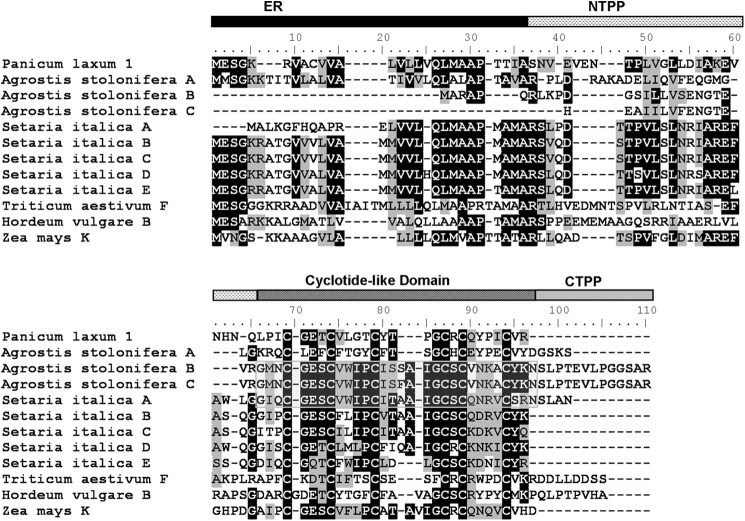
Translated precursors of panitide genes are shown in Fig. 3. Their primary sequences were aligned with the precursors of other cyclotide-like genes from the Poaceae including Z. mays A, T. aestivum A, Sorghum bicolor, H. vulgare, and Pennisetum glaucum. The panitide precursors display only a moderate sequence homology with other Poaceae cyclotide-like genes and even among themselves. None of the panitide precursors contain the essential C-terminal Asn residue that is commonly found in cyclotides and necessary for their biosynthesis into a cyclic form. They also lack the CTPP domain with a stop codon found immediately after the mature panitide domain.
FIGURE 3.
Translated sequences of panitide precursors. The primary sequences of the precursors were aligned with other cyclotide-like genes from the Poaceae including Z. mays (GenBank accession number CF060985), T. aestivum (CA617438), S. bicolor (BE125990), P. glaucum (CD725989), and H. vulgare (AL450615). The precursor numbers are labeled according to the corresponding mature panitides. The putative ER domain is colored in black, NTPP domain is indicated with a dashed horizontal pattern, and cyclotide-like domain is colored in dark gray.
Expression Profiles of Panitides
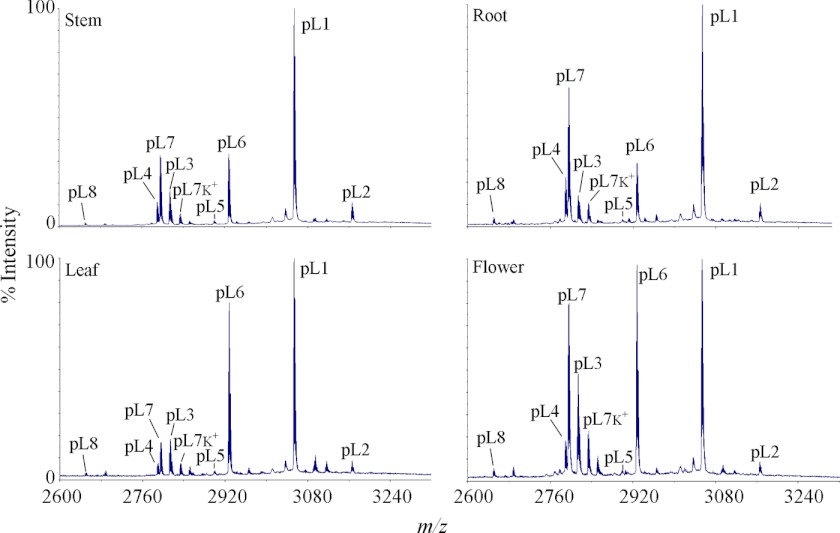
To determine whether the novel panitides are expressed universally or in a tissue-specific manner, four different plant parts including leaves, stems, roots, and flowers were extracted separately and profiled by mass spectrometry. As shown in Fig. 4, most of the panitides were found to be expressed in all the tissues examined, but their relative abundances are slightly variable in different plant parts. For example, panitide L1 is the most abundant panitide in all tissues followed by panitides L6 and L7. The relative abundances of panitides L6 and L7 vary among the four tissues examined. Panitide L6 has a higher expression level than panitide L7 in leaves and flowers, but a similar expression level in stems and lower in roots.
FIGURE 4.
MS profiles of panitides in different tissues of P. laxum. Four different plant parts were collected including leaf, stem, flower, and root. Their extracts were profiled by mass spectrometry. The peaks are labeled according to the masses of the corresponding panitides, which are abbreviated as pL followed by their numbers. Peaks labeled K+ indicate the potassium adducts (+38 Da).
Disulfide Mapping of Panitide L3
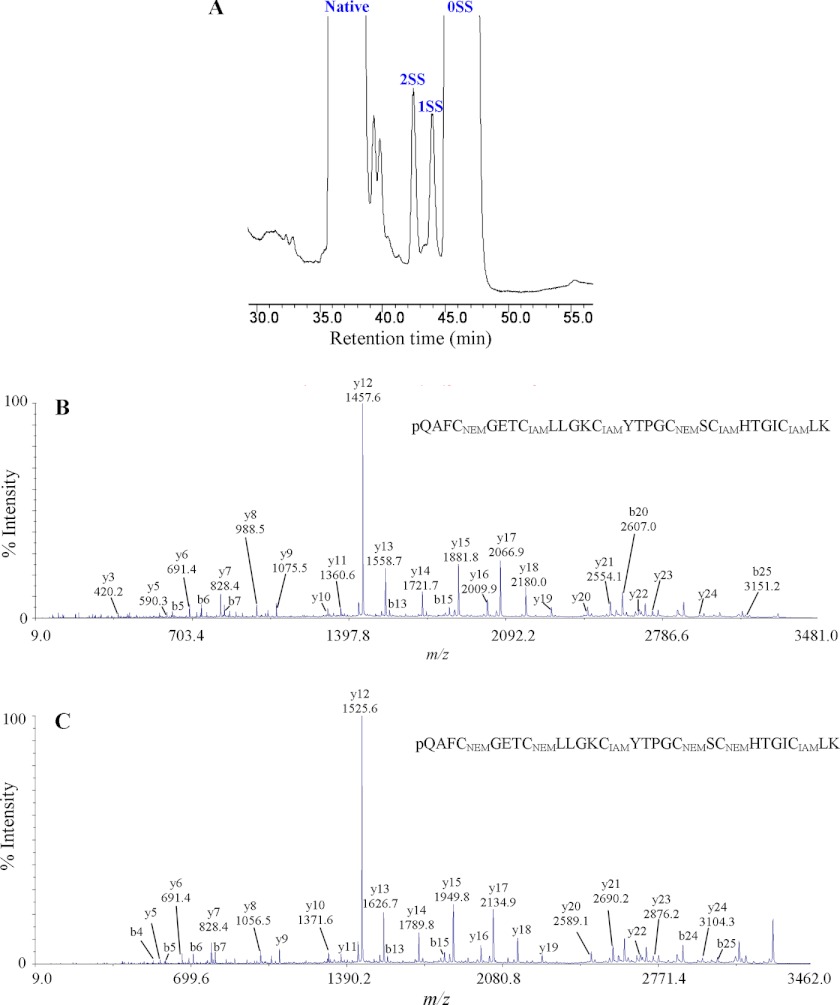
Because the novel panitides are isolated from a new plant family (the Poaceae), their disulfide linkages were determined to confirm whether they share the same cystine knot arrangement as other cyclotides. Panitide L3 was selected as a representative. Its connectivity was determined by a sequential S-tagging strategy with two different alkylation reagents, first with NEM and then with IAM. This approach was previously used for the disulfide mapping of kalata B1 (2), hedyotide B2 (21), and cliotide T2 (9).
Native panitide L3 was first partially S-reduced with tris(2-carboxyethyl)phosphine followed by S-alkylation with excess NEM. Two S-NEM labeled intermediates with one (1SS) or two (2SS) intact disulfide bonds were collected (Fig. 5A). These two intermediates were fully S-reduced with DTT, S-tagged with IAM, and sequenced by tandem mass spectrometry. The information from the differentially S-labeled panitide L3 was used to deduce the disulfide bond connectivity. As shown in Fig. 5B, the sequence of 2SS species after S-IAM alkylation was determined to contain two S-NEM residues on Cys I and IV and S-IAM on the remaining Cys residues. This established the connectivity as Cys I–IV. Similarly, the sequence of 1SS species after S-IAM alkylation was determined to contain two S-IAM residues on Cys III and VI and S-NEM residues on the remaining cysteines, establishing the connectivity as Cys III–VI (Fig. 5C). The connectivity of the third disulfide linkage was obtained by deduction as Cys II–V. Our results provide chemical evidence that panitide L3 possesses the same cystine knot arrangement as known cyclotides and acyclic variants in other plant families (2, 9, 21).
FIGURE 5.
Disulfide mapping of panitide L3. A, RP-HPLC profile of partially S-reduced and S-NEM-labeled panitide L3. 1SS and 2SS are intermediate species with one and two intact disulfide bonds, respectively. 0SS indicates the fully S-NEM-labeled panitide L3. The 1SS and 2SS intermediates were fully S-reduced with DTT and S-alkylated with IAM. B and C, MS/MS spectra of double S-tagged panitide L3. Their interpreted sequences are shown at the tops of panels B and C.
Oxidative Folding of Panitide L1
To study whether the reduced panitides can be efficiently refolded into the native conformation, we selected panitide L1 to probe its oxidative folding pathway. Panitide L1 was first reduced with excess DTT, and the reduced peptide was purified by RP-HPLC. After lyophilization, the oxidative refolding reaction was performed in 50% isopropyl alcohol (v/v) containing 100 mm ammonium bicarbonate, 1 mm reduced glutathione, pH 8.0, and the reduced peptide at the final concentration of 30 μm. Aliquots at selected time points were removed and analyzed by RP-HPLC. Correct folding was confirmed by mass spectrometry and coelution with the native peptide under RP-HPLC.
Fig. 6 shows the RP-HPLC traces of the folding reaction over a time course of 18 h. The native oxidized panitide L1 eluted significantly later than the reduced peptide, an RP-HPLC feature that is similar to the prototypic cyclotide kalata B1 (32). The reduced peptide was slowly converted into the native oxidized form with the correctly folded peptide starting to appear at 30 min. The folding reaction was nearly complete after 18 h.
FIGURE 6.
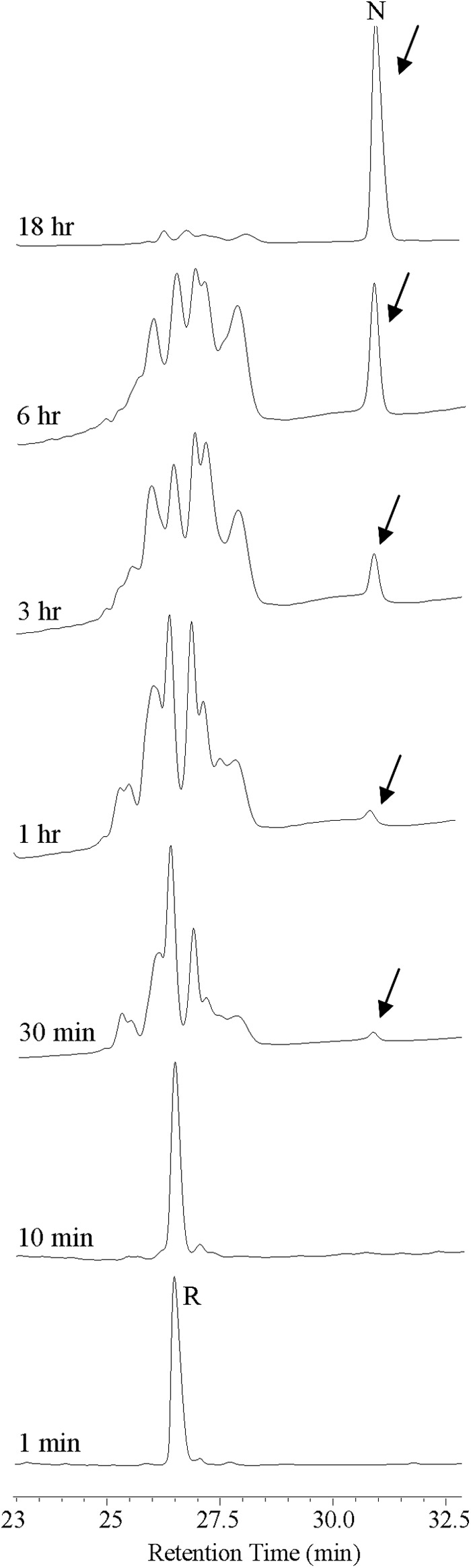
Oxidative folding pathway of panitide L1. Oxidation was performed in 50% isopropyl alcohol, 100 mm ammonium bicarbonate (pH 8.0), 1 mm reduced glutathione at room temperature. Aliquots were removed at various intervals, quenched with 1% trifluoroacetic acid, and analyzed by RP-HPLC. The time points are labeled on the HPLC traces. R and N are the reduced and native peptide, respectively. Arrows indicate the position of the native peptide.
Heat and Enzymatic Stability of Novel Panitides
To examine whether the novel panitides are stable to heat and enzymatic degradation, panitides L1 and L2 were subjected to heat treatment and tryptic digestion. As shown in Fig. 7, both peptides were stable to heat at 100 °C for 1 h. They were also resistant against tryptic digestion up to 3 h with more than 90% of the peptides remaining intact.
FIGURE 7.
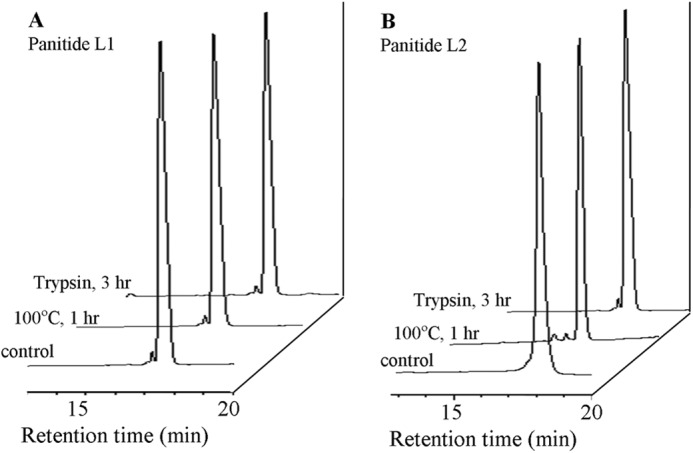
Thermal and enzymatic stability of panitides. Each peptide was incubated at 100 °C for 1 h or treated with trypsin for 3 h at 37 °C. A and B, RP-HPLC profiles of panitide L1 and L2 after treatment.
Antimicrobial Activity
Antimicrobial activities of novel panitides were tested against four different strains including E. coli, S. aureus, C. albicans, and C. tropicalis. No antimicrobial activity was detected for any of the tested panitides except for panitide L2, which was active against E. coli with a minimal inhibitory concentration value of 2.5 μm (Table 1).
Cytotoxicity Assay
Cytotoxicities of novel panitides against HeLa cells were evaluated using PrestoBlue reagent. Panitides L1, L2, L5, L6, and L7 exhibited cytotoxicity at a similar range as cyclotides with minimal inhibitory concentration values of 4.1, 6.0, 10, 5.7, and 7.7 μm, respectively. Panitide L3, L4, L5, and L8 did not show any inhibitory effect on cell growth at up to 10 μm (Table 1).
DISCUSSION
In this study, we have analyzed six species belonging to the Poaceae family and provided the first evidence at the protein level for the expression of linear cyclotides in a monocot plant. We isolated from P. laxum eight novel linear cyclotides at the protein level and characterized seven cDNA clones, one of which encodes a novel linear cyclotide. Our study provides new understanding about the distribution and evolution of cyclotides and their acyclic variants in plants.
Existence of Linear Cyclotides in the Poaceae Family
Several cyclotide-like genes have been identified in various species of the Poaceae, but their expression at the protein level has yet to be proven (25). The isolation of eight novel linear cyclotides in P. laxum confirms their existence as expressed proteins and thus expands the distribution of linear cyclotides to a new plant family, the Poaceae of the monocot flowering plants. Furthermore, screening of six grass species found in Singapore showed that they produced peptides with six cysteine residues and at the 2.5–4 kDa mass range, suggesting the putative occurrence of cyclotide-like peptides in the Poaceae family. However, additional proof of their cyclotide-like sequences in these plant species will be required. Nevertheless, the presence of linear cyclotides in both monocots and dicots suggests their ancient origin prior to the divergence of these two groups of flowering plants.
The discovery of novel linear cyclotides in the Poaceae family may impact on agriculture management. Poaceae is the most economically important of all plant families. It includes cereal grains, which account for more than 50% of food energy and proteins for human consumption (33), crops and grassland for cattle fodder, poultry, and animal husbandry, sugar cane for sugar production, and bamboo for timber and paper production. The ecological role of grasses is equally important as grasslands are estimated to comprise 20% of the vegetation cover (34), which provides a habitat and food for numerous species of animals. Our finding may also have an impact on the cattle industry as it has been reported that grasses account for 75% of food consumption by cattle, and P. laxum was among the species with highest dry weights in their diet (35). The presence of bioactive, thermally and proteolytically stable proteins in animal feed may stimulate an interest in the potential effects of diets containing cyclotides and cyclotide-like peptides on animal husbandry. Considering the potential antimicrobial (5), nematicidal (36), and oxytocin properties of cyclotides (37), a diet rich in cyclotide-producing plants may have a positive impact on animal health and aid regurgitation and food digestion of cattle. However, this is just a speculation based on known biological functions of cyclotides, and future study is required to confirm this speculation.
Biosynthesis Pathway of Linear Cyclotides in the Poaceae Family
Genetic characterization revealed that the novel panitide genes encode three major domains including an ER signal sequence, an NTPP domain, and a mature linear cyclotide domain followed directly by a stop codon. Similar to cyclotide genes of the Violaceae family, they do not contain an intron in the ER signal region (38). The ER signal sequence is likely to play a role in directing the protein into the ER lumen for oxidative folding and subsequently entering the secretory pathway. The function of the NTPP domain is still poorly understood. They are hypervariable in both sequence and length even among species of the same family. The NTPPs of panitide precursors consist of 23–35 residues as compared with 65 residues in kalata B1 (4) and 15–20 residues in chassatides and Phybs (15, 18), and are totally absent in cliotides (9). The low sequence conservation suggests that this domain may not be essential for the biosynthesis of cyclotides but plays other physiological functions in plants.
None of the novel panitides contain in their sequences the essential C-terminal Asn residue in loop 6, which is required for the backbone cyclization in cyclotides. The essential Asn residue is also absent in their precursors, which are found to be truncated with the putative Asn position occupied by a stop codon. This provides a biosynthetic explanation for the linear structure of panitides and reinforces the essential role of the C-terminal Asn residue for a head-to-tail backbone ligation. The biosynthesis of panitides is likely similar to other linear cyclotides such as chassatide C7 (18), hedyotide B2 (21), kalata B20-lin (24), Phyb M (15), and violacin A (22), which also lack the essential Asn residue in their sequences and precursors.
The novel linear cyclotides in P. laxum also share two other characteristics. First, all panitides were found to contain a post-translational pyroglutamyl modification at their N terminus. No peptide masses corresponding to non-pyroGlu panitides were detected, suggesting the conversion from Gln to pyroGlu residue during the biosynthesis of panitides is highly efficient. It is uncertain whether this N-terminal modification occurs spontaneously or is catalyzed by a specific glutamyl cyclase. Similar modifications have been identified in a number of plant peptides such as Phyb M isolated from P. hybrida (15), squash trypsin inhibitors from Cucurbitaceae plants (19, 39), and Ib-AMP from Impatiens balsamina (40). The incorporation of an N-terminal pyroGlu residue in panitides may be an alternative strategy to the backbone cyclization to provide protection against exopeptidase degradation. Second, although the novel panitides are Möbius-like, with five of them containing the typical Pro residue in loop 5, four were found to replace the highly conserved Pro by a Gly residue. Both Pro and Gly residues are frequently found in the turn structures of proteins, and it would be interesting to find out the structure-activity consequences of this replacement in a future study.
Analysis of the panitide precursors also revealed that the N-terminal cleavage site occurs between Asn and Gln residues. In other cyclotide-producing plant families such as the Rubiaceae, Violaceae, and Solanaceae, the N-terminal cleavage usually occurs after the dipeptide motif Val/Leu-Xaa or Xaa-Asn (18, 41). The former is catalyzed by an unknown protease, whereas the latter is likely to be catalyzed by an asparaginyl endopeptidase. The N-terminal processing of panitides probably follows the latter, which might be mediated by an asparaginyl endopeptidase enzyme.
Linear Cyclotides but Not Cyclotides Are Likely to Be More Common in the Poaceae Family
Thus far, all cyclotide-producing plants reported in the Rubiaceae, Violaceae, Fabaceae, and Solanaceae produced either cyclotides only or cyclotides together with a few acyclic variants (1, 9, 15, 18). Intriguingly, P. laxum expressed exclusively linear cyclotides without any cyclic counterparts detected under our experimental conditions. This suggests that the cyclic backbone may be a dispensable requirement for panitides. Our findings are consistent with previous work of Mulvenna et al. (25) that the cyclotide-like genes of various Poaceae species reported in the NCBI nucleotide database prior to 2006 lack the C-terminal Asn residue, leading to the prediction that the corresponding mature peptides are not cyclic. Our work supports this hypothesis, and it appears that linear cyclotides could be biologically relevant in the Poaceae family.
This prompts a follow-up question. Do cyclotides or the cyclic variants exist in the Poaceae family? The answer is likely yes. Since 2006, numerous new cyclotide-like genes have been deposited into the nucleotide database (25). TBLASTN search of novel cyclotide-like sequences showed that A. stolonifera contains three, Setaria italica contains five, and there is one each in T. aestivum, Z. mays, and H. vulgare. The predicted precursor sequences of these genes are summarized in Fig. 8. Of the predicted novel sequences, three are found to contain the essential Asn residue including two sequences in A. stolonifera and one in S. italica. They also contain the typical processing sites found in the Rubiaceae and Violaceae families with the N-terminal processing occurring between the Val/Leu-Xaa dipeptide motifs and the Gly residue and between the Asn and Ser-Leu motif at the C-terminal processing site, suggesting that the expressed mature products could be head-to-tail cyclic peptides.
FIGURE 8.
Novel sequences in the Poaceae family identified by database searching. The putative ER domain is colored in black, the NTPP domain is indicated with a dashed horizontal pattern, the cyclotide-like domain is colored in dark gray, and the CTPP domain is colored in light gray. The putative cyclotide sequences are boxed. Panitide L1 precursor was aligned together for comparison.
Novel Linear Cyclotides in the Poaceae Preserve the Knotted Arrangement
Panitides share similar cysteine spacing as both linear and cyclic forms of cyclotides. Disulfide mapping of panitide L3 showed that it possesses a cystine knot arrangement similar to known cyclotides (2, 21). Although there is little sequence homology of cyclotide precursors in different plant families, cyclotides of the Poaceae, Rubiaceae, Violaceae, and Fabaceae were all found to preserve the cystine knot arrangement, suggesting its structural and functional importance. The cystine knot arrangement is found in a variety of unrelated protein families of diverse species such as fungi, cone snails, snake venoms, insects, spiders, and plants (42). In plants, cystine knot peptides with molecular mass <4 kDa are known to be stable to heat and enzymatic degradation, and the cystine knot arrangement is considered to be a major contributing factor (3). Consistently, we showed that panitides are highly stable to heat and enzymatic degradation although the circular backbone feature has been abolished in these peptides. Furthermore, we also showed that panitide L1 can readily fold into its native form within 18 h. The structural stability of linear cyclotides together with an efficient folding pathway make them a potentially attractive natural scaffold for peptide-based drug design and grafting of bioactive epitopes.
Bioactivities
The diverse biological activities of cyclotides have been proposed to arise from their ability to target, interact, and disrupt cell membranes (43–46). In this work, we indirectly assessed the membrane-active properties of the novel panitides by using antimicrobial and cytotoxicity assays. Most of the tested peptides were found to be ineffective against our selected panel of bacteria. Only panitide L2 showed antibacterial activity against E. coli with the minimal inhibitory concentration value of 2.5 μm. This could be attributed to the high positive net charge of panitide L2 (+2) as compared with other panitides, which carry a +1, neutral, or negative net charge. This suggests that the electrostatic interaction may play an important role in the antimicrobial activity of panitides. For cytotoxicity, several panitides display activities ranging from 4.1 to 10 μm, which is comparable with the reported values of Möbius cyclotides such as kalata B1, varv A, varv E, and cliotide T3 (8, 9, 47). These results suggest that linear cyclotides in the Poaceae retained certain bioactivity profiles similar to their cyclotide counterparts.
In conclusion, our study reports the discovery of nine new linear cyclotides, panitides L1–9, in P. laxum. This finding expands their occurrence to a new plant family, the Poaceae, of which many species have great economic values to humans. Our finding is also of interest from the evolutionary point of view as linear cyclotides are now found in both dicot and monocot plants. We also predict from the NCBI nucleotide database the likely existence of cyclotides in two species, A. stolonifera and S. italica, of the grass family. The presence of cyclotides, cyclic or acyclic variants, in the Poaceae family may open up new avenues of research for crop protection considering their defensive roles in plants.
This work was supported by A*STAR Biomedical Research Council Grant 09/1/22/19/612 and the Competitive Research Grant by National Research Foundation in Singapore.

This article contains a supplemental file with partial sequences for panitides L7, L8, and L9.
- ER
- endoplasmic reticulum
- NTPP
- N-terminal propeptide
- CTPP
- C-terminal propeptide
- IAM
- iodoacetamide
- RACE
- rapid amplification of cDNA ends
- NEM
- N-ethylmaleimide
- RP-HPLC
- reverse phase-HPLC
- contig
- group of overlapping clones.
REFERENCES
- 1. Craik D. J., Daly N. L., Bond T., Waine C. (1999) Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J. Mol. Biol. 294, 1327–1336 [DOI] [PubMed] [Google Scholar]
- 2. Göransson U., Craik D. J. (2003) Disulfide mapping of the cyclotide kalata B1: chemical proof of the cystic cystine knot motif. J. Biol. Chem. 278, 48188–48196 [DOI] [PubMed] [Google Scholar]
- 3. Colgrave M. L., Craik D. J. (2004) Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: the importance of the cyclic cystine knot. Biochemistry 43, 5965–5975 [DOI] [PubMed] [Google Scholar]
- 4. Jennings C., West J., Waine C., Craik D., Anderson M. (2001) Biosynthesis and insecticidal properties of plant cyclotides: the cyclic knotted proteins from Oldenlandia affinis. Proc. Natl. Acad. Sci. U.S.A. 98, 10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tam J. P., Lu Y. A., Yang J. L., Chiu K. W. (1999) An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc. Natl. Acad. Sci. U.S.A. 96, 8913–8918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gustafson K. R., McKee T. C., Bokesch H. R. (2004) Anti-HIV cyclotides. Curr. Protein Pept. Sci. 5, 331–340 [DOI] [PubMed] [Google Scholar]
- 7. Witherup K. M., Bogusky M. J., Anderson P. S., Ramjit H., Ransom R. W., Wood T., Sardana M. (1994) Cyclopsychotride A, a biologically active, 31-residue cyclic peptide isolated from Psychotria longipes. J. Nat. Prod. 57, 1619–1625 [DOI] [PubMed] [Google Scholar]
- 8. Svangård E., Göransson U., Hocaoglu Z., Gullbo J., Larsson R., Claeson P., Bohlin L. (2004) Cytotoxic cyclotides from Viola tricolor. J. Nat. Prod. 67, 144–147 [DOI] [PubMed] [Google Scholar]
- 9. Nguyen G. K., Zhang S., Nguyen N. T., Nguyen P. Q., Chiu M. S., Hardjojo A., Tam J. P. (2011) Discovery and characterization of novel cyclotides originated from chimeric precursors consisting of albumin-1 chain a and cyclotide domains in the Fabaceae family. J. Biol. Chem. 286, 24275–24287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hernandez J. F., Gagnon J., Chiche L., Nguyen T. M., Andrieu J. P., Heitz A., Trinh Hong T., Pham T. T., Le Nguyen D. (2000) Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry 39, 5722–5730 [DOI] [PubMed] [Google Scholar]
- 11. Gustafson KR, S. R., Henderson LE, Parsons IC, Kashman Y, Cardellina JH, McMahon JB, Buckheit RB, Pannell LK, Boyd MR. (1994) Circulins A and B. Novel human immunodeficiency virus (HIV)-inhibitory macrocyclic peptides from the tropical tree Chassalia parvifolia. J. Am. Chem. Soc. 116, 9337–9338 [Google Scholar]
- 12. Hallock Y. F., Sowder R. C., 2nd, Pannell L. K., Hughes C. B., Johnson D. G., Gulakowski R., Cardellina J. H., 2nd, Boyd M. R. (2000) Cycloviolins A–D, anti-HIV macrocyclic peptides from Leonia cymosa. J. Org. Chem. 65, 124–128 [DOI] [PubMed] [Google Scholar]
- 13. Broussalis A. M., Göransson U., Coussio J. D., Ferraro G., Martino V., Claeson P. (2001) First cyclotide from Hybanthus (Violaceae). Phytochemistry 58, 47–51 [DOI] [PubMed] [Google Scholar]
- 14. Poth A. G., Colgrave M. L., Philip R., Kerenga B., Daly N. L., Anderson M. A., Craik D. J. (2011) Discovery of cyclotides in the Fabaceae plant family provides new insights into the cyclization, evolution, and distribution of circular proteins. ACS Chem. Biol. 6, 345–355 [DOI] [PubMed] [Google Scholar]
- 15. Poth A. G., Mylne J. S., Grassl J., Lyons R. E., Millar A. H., Colgrave M. L., Craik D. J. (2012) Cyclotides associate with leaf vasculature and are the products of a novel precursor in Petunia (Solanaceae). J. Biol. Chem. 287, 27033–27046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Simonsen S. M., Sando L., Ireland D. C., Colgrave M. L., Bharathi R., Göransson U., Craik D. J. (2005) A continent of plant defense peptide diversity: cyclotides in Australian Hybanthus (Violaceae). Plant Cell 17, 3176–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Poth A. G., Colgrave M. L., Lyons R. E., Daly N. L., Craik D. J. (2011) Discovery of an unusual biosynthetic origin for circular proteins in legumes. Proc. Natl. Acad. Sci. U.S.A. 108, 10127–10132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nguyen G. K., Lim W. H., Nguyen P. Q., Tam J. P. (2012) Novel cyclotides and uncyclotides with highly shortened precursors from Chassalia chartacea and effects of methionine oxidation on bioactivities. J. Biol. Chem. 287, 17598–17607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mylne J. S., Chan L. Y., Chanson A. H., Daly N. L., Schaefer H., Bailey T. L., Nguyencong P., Cascales L., Craik D. J. (2012) Cyclic peptides arising by evolutionary parallelism via asparaginyl-endopeptidase-mediated biosynthesis. Plant Cell 24, 2765–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dutton J. L., Renda R. F., Waine C., Clark R. J., Daly N. L., Jennings C. V., Anderson M. A., Craik D. J. (2004) Conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins. J. Biol. Chem. 279, 46858–46867 [DOI] [PubMed] [Google Scholar]
- 21. Nguyen G. K., Zhang S., Wang W., Wong C. T., Nguyen N. T., Tam J. P. (2011) Discovery of a linear cyclotide from the bracelet subfamily and its disulfide mapping by top-down mass spectrometry. J. Biol. Chem. 286, 44833–44844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ireland D. C., Colgrave M. L., Nguyencong P., Daly N. L., Craik D. J. (2006) Discovery and characterization of a linear cyclotide from Viola odorata: implications for the processing of circular proteins. J. Mol. Biol. 357, 1522–1535 [DOI] [PubMed] [Google Scholar]
- 23. Gerlach S. L., Burman R., Bohlin L., Mondal D., Göransson U. (2010) Isolation, characterization, and bioactivity of cyclotides from the Micronesian plant Psychotria leptothyrsa. J. Nat. Prod. 73, 1207–1213 [DOI] [PubMed] [Google Scholar]
- 24. Mylne J. S., Wang C. K., van der Weerden N. L., Craik D. J. (2010) Cyclotides are a component of the innate defense of Oldenlandia affinis. Biopolymers 94, 635–646 [DOI] [PubMed] [Google Scholar]
- 25. Mulvenna J. P., Mylne J. S., Bharathi R., Burton R. A., Shirley N. J., Fincher G. B., Anderson M. A., Craik D. J. (2006) Discovery of cyclotide-like protein sequences in graminaceous crop plants: ancestral precursors of circular proteins? Plant Cell 18, 2134–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ng K. F., Stur W. W., Shelton H. M. (1997) New forage species for integration of sheep in rubber plantations. J. Agric. Sci. 128, 347–355 [Google Scholar]
- 27. Firth D. J., Jones R. M., McFadyen L. M., Cook B. G., Whalley R. D. B. (2002) Selection of pasture species for groundcover suited to shade in mature macadamia orchards in subtropical Australia. Trop. Grasslands 36, 1–12 [Google Scholar]
- 28. Campbell C. S. (1985) The subfamilies and tribes of Gramineae (Poaceae) in the Southeastern United-States. J. Arnold Arboretum 66, 123–199 [Google Scholar]
- 29. Paterson A. H., Freeling M., Sasaki T. (2005) Grains of knowledge: Genomics of model cereals. Genome Res. 15, 1643–1650 [DOI] [PubMed] [Google Scholar]
- 30. Lehrer R. I., Rosenman M., Harwig S. S., Jackson R., Eisenhauer P. (1991) Ultrasensitive assays for endogenous antimicrobial polypeptides. J. Immunol. Methods 137, 167–173 [DOI] [PubMed] [Google Scholar]
- 31. Tam J. P., Lu Y. A., Yang J. L. (2002) Antimicrobial dendrimeric peptides. Eur. J. Biochem. 269, 923–932 [DOI] [PubMed] [Google Scholar]
- 32. Daly N. L., Clark R. J., Craik D. J. (2003) Disulfide folding pathways of cystine knot proteins: tying the knot within the circular backbone of the cyclotides. J. Biol. Chem. 278, 6314–6322 [DOI] [PubMed] [Google Scholar]
- 33. Cordain L. (1999) Cereal grains: humanity's double-edged sword. World Rev. Nutr. Diet. 84, 19–73 [DOI] [PubMed] [Google Scholar]
- 34. Shantz H. L. (1954) The Place of Grasslands in the Earths Cover of Vegetation. Ecology 35, 143–145 [Google Scholar]
- 35. Desbiez A. L. J., Santos S. A., Alvarez J. M., Tomas W. M. (2011) Forage use in domestic cattle (Bos indicus), capybara (Hydrochoerus hydrochaeris), and pampas deer (Ozotoceros bezoarticus) in a seasonal neotropical wetland. Mamm. Biol. 76, 351–357 [Google Scholar]
- 36. Colgrave M. L., Kotze A. C., Huang Y. H., O'Grady J., Simonsen S. M., Craik D. J. (2008) Cyclotides: natural, circular plant peptides that possess significant activity against gastrointestinal nematode parasites of sheep. Biochemistry 47, 5581–5589 [DOI] [PubMed] [Google Scholar]
- 37. Gran L., Sandberg F., Sletten K. (2000) Oldenlandia affinis (R&S) DC. A plant containing uteroactive peptides used in African traditional medicine. J. Ethnopharmacol. 70, 197–203 [DOI] [PubMed] [Google Scholar]
- 38. Gruber C. W., Elliott A. G., Ireland D. C., Delprete P. G., Dessein S., Göransson U., Trabi M., Wang C. K., Kinghorn A. B., Robbrecht E., Craik D. J. (2008) Distribution and evolution of circular miniproteins in flowering plants. Plant Cell 20, 2471–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mar R. I., Carver J. A., Sheil M. M., Boschenok J., Fu S. L., Shaw D. C. (1996) Primary structure of trypsin inhibitors from Sicyos australis. Phytochemistry 41, 1265–1274 [DOI] [PubMed] [Google Scholar]
- 40. Tailor R. H., Acland D. P., Attenborough S., Cammue B. P. A., Evans I. J., Osborn R. W., Ray J. A., Rees S. B., Broekaert W. F. (1997) A novel family of small cysteine-rich antimicrobial peptides from seed of Impatiens balsamina is derived from a single precursor protein. J. Biol. Chem. 272, 24480–24487 [DOI] [PubMed] [Google Scholar]
- 41. Trabi M., Mylne J. S., Sando L., Craik D. J. (2009) Circular proteins from Melicytus (Violaceae) refine the conserved protein and gene architecture of cyclotides. Org. Biomol. Chem. 7, 2378–2388 [DOI] [PubMed] [Google Scholar]
- 42. Gracy J., Le-Nguyen D., Gelly J. C., Kaas Q., Heitz A., Chiche L. (2008) KNOTTIN: the knottin or inhibitor cystine knot scaffold in 2007. Nucleic Acids Res. 36, D314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Henriques S. T., Huang Y. H., Castanho M. A., Bagatolli L. A., Sonza S., Tachedjian G., Daly N. L., Craik D. J. (2012) Phosphatidylethanolamine binding is a conserved feature of cyclotide-membrane interactions. J. Biol. Chem. 287, 33629–33643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang Y. H., Colgrave M. L., Daly N. L., Keleshian A., Martinac B., Craik D. J. (2009) The biological activity of the prototypic cyclotide kalata b1 is modulated by the formation of multimeric pores. J. Biol. Chem. 284, 20699–20707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang C. K., Colgrave M. L., Ireland D. C., Kaas Q., Craik D. J. (2009) Despite a conserved cystine knot motif, different cyclotides have different membrane binding modes. Biophys. J. 97, 1471–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kamimori H., Hall K., Craik D. J., Aguilar M. I. (2005) Studies on the membrane interactions of the cyclotides kalata B1 and kalata B6 on model membrane systems by surface plasmon resonance. Anal. Biochem. 337, 149–153 [DOI] [PubMed] [Google Scholar]
- 47. He W., Chan L. Y., Zeng G., Daly N. L., Craik D. J., Tan N. (2011) Isolation and characterization of cytotoxic cyclotides from Viola philippica. Peptides 32, 1719–1723 [DOI] [PubMed] [Google Scholar]