Background: Expression of ΔEGFR, a mutant of EGFR in gliomas, correlates with poor prognosis.
Results: Access to the nucleus is required for full oncogenicity of ΔEGFR, and nuclear ΔEGFR regulates transcription of target genes via c-Myc.
Conclusion: Functional association of nuclear ΔEGFR with c-Myc is necessary for ΔEGFR-induced oncogenicity.
Significance: These data show a novel activity of ΔEGFR and offer new opportunities for therapeutic intervention.
Keywords: ChIP, Glioblastoma, Myc, Nuclear Translocation, Receptor Tyrosine Kinase, ChIP-Seq, EGFRvIII, Nuclear RTK, Transcription
Abstract
ΔEGFR is a potent glioblastoma oncogene which has been studied primarily as a plasma membrane kinase. Using intracranial xenograft studies in mice, we show that blocking ΔEGFR access to the nucleus attenuates its tumorigenicity and, conversely, that promoting nuclear accumulation enhances this, providing the first in vivo evidence that the nuclear actions of ΔEGFR contribute strongly to its oncogenic function. Nuclear actions of ΔEGFR include regulation of gene expression by participation in chromatin-bound complexes, and genome-wide mapping of these sequences by chromatin immunoprecipitation and massively parallel sequencing identified 2294 peaks. Bioinformatic analysis showed enrichment of the E-box motif in the dataset, and c-Myc and ΔEGFR were corecruited to the promoters of and transcriptionally activated a subset of nuclear ΔEGFR chromatin targets. Knockdown of c-Myc decreased the expression of these targets and diminished ΔEGFR-stimulated anchorage-independent colony formation. We conclude that transcriptional regulation of target genes by association with gene regulatory chromatin in cooperation with c-Myc by nuclear ΔEGFR makes a unique contribution to its oncogenicity and propose that this venue provides new targets for therapeutic intervention.
Introduction
The EGF receptor (EGFR)2 gene is frequently amplified, overexpressed, and mutated in glioblastoma multiforme, the most aggressive form of primary brain tumor, which is also refractory to therapy. Expression of ΔEGFR (also called EGFRvIII), the most common mutation of EGFR (1), correlates with advanced disease and resistance to therapy (2). Recent work from several laboratories has indicated that EGFR can shuttle directly into the nucleus in normal proliferating and cancerous cells. The functions of nuclear EGFR include regulating gene transcription via physical interaction with transcription factors (3–10) and DNA damage repair (11). Correlative studies suggest an association of high nuclear EGFR and poor clinical outcome in breast (12), oral (13, 14), bladder (15), and ovarian cancer (12, 16), allowing that nuclear EGFR may also be significant in the behavior of gliomas. Nuclear localization for ΔEGFR has been reported recently in the context of association with STAT5 (10) and STAT3 (8, 9). These interactions were shown to be important for ΔEGFR regulation of genes, including the Bcl-XL (10) and Cox2 gene (9). The functional role of the nuclear ΔEGFR is currently unknown. Here we investigate the biological consequence of translocation of ΔEGFR to the nucleus and show that nuclear localization of ΔEGFR is associated with increased tumorigenicity in glioma cells both in vitro and in vivo.
To uncover molecular interactions between ΔEGFR and DNA in vivo on a genome-wide scale, we used ChIP-Seq in conjunction with bioinformatic analysis. We identified that the E-box motif, which is the binding motif of the c-Myc/Max obligatory heterodimer (17), is enriched in ΔEGFR targeted promoters. The c-Myc oncogene is a helix-loop-helix leucine zipper transcription factor that is involved in a myriad of cellular events including cell cycle progression, cellular growth and metabolism, differentiation, and apoptosis (18). Alterations in c-Myc that have been detected in numerous solid tumors, including glioma and blood malignancies, are often associated with cancer aggressiveness and poor treatment prognosis (19). We show that ΔEGFR associates with c-Myc in the nucleus and is corecruited to target genes. Ablation of c-Myc by RNAi decreases both expression of ΔEGFR/c-Myc target genes and ΔEGFR-induced anchorage-independent colony formation in glioma cells.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
FuGENE HD (Roche) was used for transient transfections. Cell lines were fingerprinted for identity using a PCR-based analysis (GenomeLab Human STR Primer set from Beckman Coulter) that interrogates a set of 12 short tandem repeats (supplemental Table S1) (20). AG1478 and Leptomycin B were from Calbiochem.
Plasmid Construction, Mutagenesis, and RNAi
EGFR and ΔEGFR fragments were PCR-cloned from pcDNA 3.1/EGFR and pcDNA 3.1/ΔEGFR, respectively, into the pENTR/TEV/D-TOPO vector (Invitrogen) using primers EGFR-F and EGFR-R. Mutagenesis of NES and NLS constructs was performed using the QuikChange mutagenesis kit (Stratagene) using appropriate primers (supplemental Table S2) according to the instructions of the manufacturer. All the constructs were subcloned into pcDNA-DEST47 (Invitrogen) to generate C-terminal GFP-tagged protein. To generate C-terminal V5-tagged proteins, the pENTR/TEV/D-TOPO constructs were used to introduce a V5 tag using EGFR-F and EGFR-V5-R or EGFR-NLS-V5-R (for generating the SV mutant). The constructs were subcloned into the 1726zeoG retrovirus (a derivative of 1726zeo (20) with a Gateway destination vector (Invitrogen) cassette in the unique EcoRI site) (21). Importinβ, CRM1, as well as control RNAis were purchased from Dharmacon RNAi Technologies (Lafayette, CO). The c-Myc shRNA construct was obtained from Addgene.
Immunofluorescence Confocal Assays
The cellular location of proteins was determined using indirect immunofluorescence as follows. Cells were grown on glass coverslips in 6-well culture plates. When the cells were 50% confluent, they were rinsed with PBS, fixed in phosphate-buffered 4% paraformaldehyde, and then processed for immunofluorescence staining. The primary antibodies used for immunofluorescence were EGFR (Calbiochem), phospho-EGFR (Tyr-1068) (Cell Signaling), V5 (Invitrogen), Importinβ (Santa Cruz Biotechnology, Inc.), and CRM1 (BD Biosciences). Secondary antibodies from Molecular Probes were either Alexa Fluor 488- or Alexa Fluor 546-labeled goat antibodies. Nuclei were visualized using TOPRO3 (blue color, Molecular Probes) or DAPI. Slides were further processed for imaging and confocal analysis using an Olympus LSM 510 or Olympus Fluoview FV1000 microscope and a ×60 objective.
Cellular and Biochemical Assays
Cell extraction, immunoprecipitation, Western blot analysis, fractionation (22), biotinylation experiment, and quantitative real-time PCR (23) were performed as described earlier.
Soft Agar Assay
A three-dimensional agarose colony formation assay was performed as described earlier (24). Briefly, single cells (1200 cells/well in a 12-well plate) suspended in 0.5% agarose were overlaid on a solidified 0.7% agarose matrix and allowed to grow for up to 16 days. Starting on day10, colonies were counted using the GelCounttm technology (Oxford Optronix, Inc., Oxford, United Kingdom). The GelCounttm scan and image acquisition used a Microsoft Windows-based program. GelCounttm software was developed by Oxford Optronix, Inc.
Animal Experiment
All animal procedures were approved by the Institutional Animal Care and Use Committee of the M.D. Anderson Cancer Center. For in vivo studies, either 100,000 or 35,000 cells were inoculated intracranially into nude mice (10 animals/group) using a guide screw. The animals were sacrificed when moribund. The formalin-fixed tumor mass was processed for immunohistochemistry as described.
Immunohistochemistry
Immunohistochemistry was performed in the Division of Surgery histology core facility using the LabVision Autostainer 360. Mouse brains were paraffin-embedded, and 4-μm brain sections were used for immunohistochemical analysis. Antigen retrieval was done on the LabVision PTmodule with citrate buffer (pH 6.0), and the Biocare Mouse on Mouse HRP polymer kit was used for detection of the signal. Immunoreactive staining was visualized using an avidin-biotin complex technique with diaminobenzidine (Invitrogen) as the chromogenic substrate and hematoxylin as the counterstain. The primary antibody used was against V5 (1:500 dilution).
ChIP and ChIP Sequencing (ChIP-Seq) Experiment
ChIP (V5 antibody (Invitrogen)) and sequential ChIP (V5/c-Myc antibody (Cell Signaling Technology) in U87 cells were done as described earlier (25). The quantitative PCR was performed on ChIP DNA using FastStart SYBR Green Master (Roche). Primers sequences for the ChIP are listed in supplemental Table S2. For ChIP-Seq, DNA from ChIP using V5 antibody was submitted to the DNA Analysis Facility at the M.D. Anderson Cancer Center for next-generation sequencing on Illumina GAIIx. The raw sequencing image data were analyzed by the Illumina analysis pipeline to generate sequence reads of 36 bps aligned to the unmasked human reference genome (NCBI v36, hg18) using the ELAND software (Illumina). ChIP-enriched binding peaks were identified by Model-based analysis of ChIP-Seq (MACS) (26). Analysis of ChIP-Seq peaks for annotation was done using the single nearest gene approach using the GREAT software (27).
RESULTS
Nuclear Localization of ΔEGFR Is Regulated by Sequences in the Juxtamembrane Region
In agreement with previous reports (8–10), analysis of glioma cell lines overexpressing the EGFR, ΔEGFR, or ΔEGFR kinase-inactive mutant (ΔEGFRki) to levels similar to those encountered in human tumors consistently demonstrated their presence in the nuclear fraction (supplemental Fig. S1A). This was also confirmed by confocal analysis (supplemental Fig. S1, B and C). Further, as shown for EGFR (supplemental Fig. S2) (28), nuclear ΔEGFR was derived from the plasma membrane, and the nuclear movement of ΔEGFR was dependent on Importinβ1 and CRM1, suggesting that it uses the same molecular machinery as EGFR (supplemental Fig. S3).
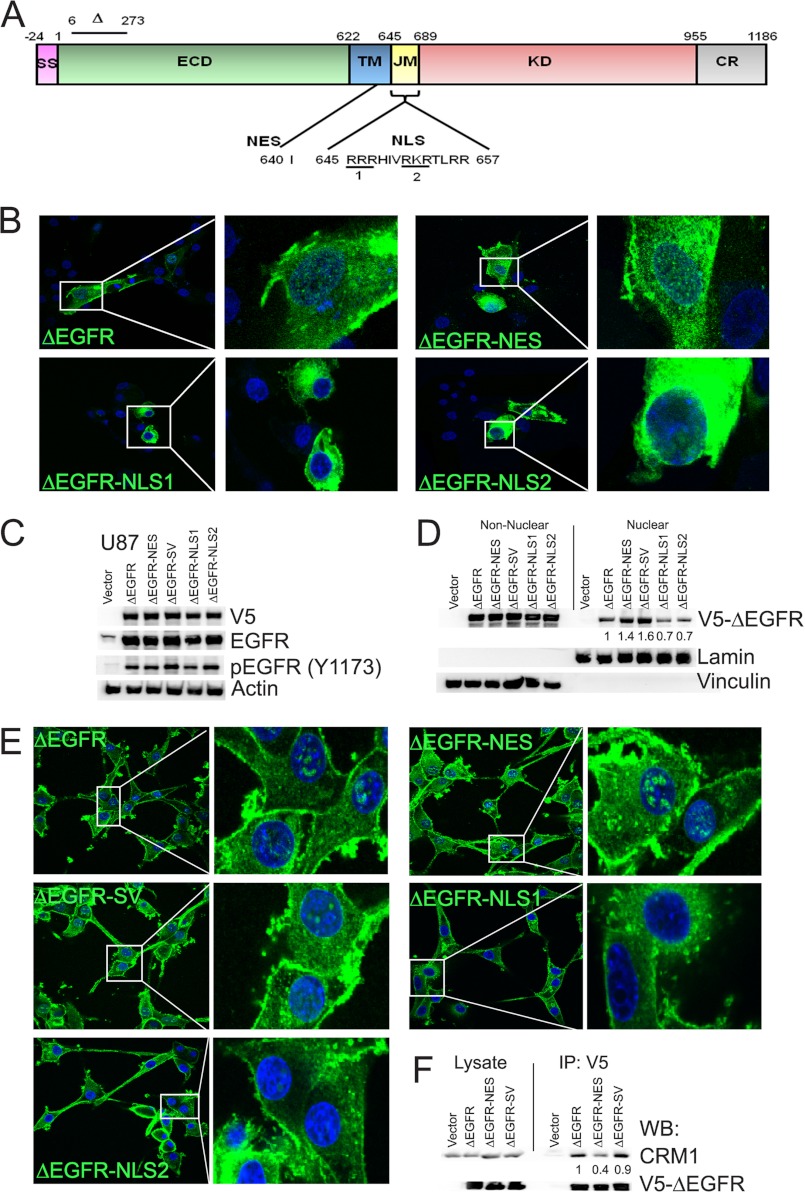
Although nuclear localization sequence (NLS) has been described for EGFR, no export sequences have been identified. By in silico analysis of ΔEGFR sequence using the program NetNES 1.1 (29), we identified a NES (Ile-640) (Fig. 1A) and generated a NES mutant in which isoleucine was mutated to alanine (ΔEGFR-NES) (supplemental Table S3). The NLS of EGFR (Fig. 1A) is responsible for its nuclear localization (22). To investigate whether the same sequence plays a role in ΔEGFR functions, we created ΔEGFR mutants where the basic residues of NLS1 and NLS2 were mutated to alanine either individually (ΔEGFR-NLS1 and ΔEGFR-NLS2) or together (ΔEGFR-NLS-DM). We also created a mutant in which the NLS of SV-40 LT antigen, 126PKKKRKV132, was tagged to the C terminus of ΔEGFR (ΔEGFR-SV). All of the above-mentioned mutations were also made in EGFR for comparison. Confocal analyses of the mutants in U87 cells showed that the NES mutants for both ΔEGFR and EGFR (Fig. 1B and supplemental Fig. S5A, respectively) were enriched in the nucleus and the perinuclear region, whereas the NLS mutants were excluded from the nuclear compartment. These data provide support for the idea that access to the nucleus by EGFR and its mutant forms is a regulated process of biological significance and that the same sequences regulate this process in wild-type and mutant EGFR.
FIGURE 1.
ΔEGFR contains NES and NLS signals that are functional. A, schematic of EGFR depicting the deleted region of ΔEGFR as well as the location of the NLS and NES signals. SS, signal peptide; ECD, extracellular domain; TM, transmembrane domain; KD, kinase domain; CR, C-terminal regulatory region. B, confocal analysis of GFP-ΔEGFR and its mutants (green) in U87 cells. C, stable expression of V5-ΔEGFR and the mutants in U87 cells. D, biochemical analysis of U87 stable cells for nuclear V5-ΔEGFR. Quantitation and statistical analysis of Western blotting are shown in supplemental Fig. S4A. E, confocal analyses of U87 stable cells using V5 antibody (green). Nuclei were counterstained using DAPI (blue) for all confocal analyses. F, coimmunoprecipitation (IP) and Western blot analysis (WB) showing decreased association of ΔEGFR and ΔEGFR-NES with CRM1. Quantitation and statistical analysis of Western blotting are shown in supplemental Fig. S4B.
To delineate the consequence of restricting ΔEGFR to the nucleus or the cytoplasm, we generated U87 glioma lines stably expressing these mutants. As shown in Fig. 1C, all the cell lines express the V5-tagged proteins. Biochemical fractionation demonstrated that the ΔEGFR-NES and ΔEGFR-SV lines exhibit increased nuclear ΔEGFR compared with U87-ΔEGFR (Fig. 1D). Conversely, the ΔEGFR-NLS1 and ΔEGFR-NLS2 mutants had decreased nuclear localization as compared with ΔEGFR (Fig. 1D). Confocal analysis also confirmed that ΔEGFR-NES and ΔEGFR-SV levels were increased in the nucleus, whereas ΔEGFR-NLS1 and ΔEGFR-NLS2 had reduced localization (Fig. 1E). To further define the NES sequence, we determined the association of ΔEGFR, ΔEGFR-NES, and ΔEGFR-SV (as a non-mutated but nuclear-enriched control) with CRM1. As shown in Fig. 1F, the robust association of CRM1 with ΔEGFR was decreased upon mutation of the NES signal that we created in ΔEGFR-NES. The association was maintained in the ΔEGFR-SV mutant, which has the SV40-NLS sequence attached to ΔEGFR and, thus, has the intact NES signal. Analogous results were observed in stable cell lines of EGFR and the respective mutants in U87 cells by microscopy and biochemical fractionation (supplemental Fig. S5).
Modulation of the NLS Sequence Suppresses Nuclear Localization Independent of ΔEGFR Activity
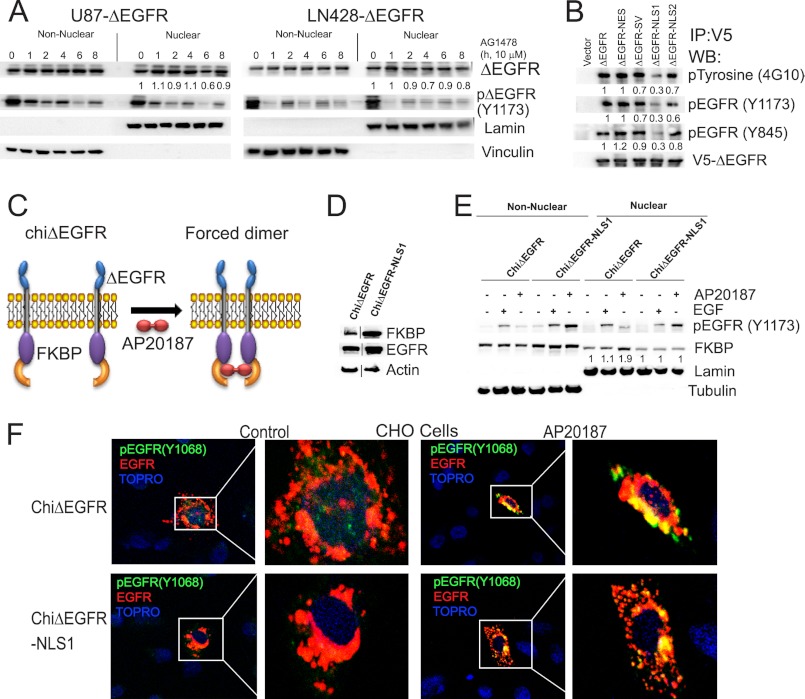
To define the requirement of ΔEGFR activity for its nuclear translocation, we treated U87-ΔEGFR and LN428-ΔEGFR cells with AG1478 (an EGFR tyrosine kinase inhibitor) and determined the levels of nuclear ΔEGFR. Inhibition of activity had no effect on the amount of ΔEGFR in the nucleus in both the cell lines (Fig. 2A). This is consistent with our earlier data that ΔEGFRki can translocate to the nucleus in glioma cells (supplemental Fig. S1A). Efficacy of AG1478 was determined by assaying for tyrosine phosphorylation on the Tyr-1173 residue, which is a major phosphorylation site, accounting for about 35% of the autophosphorylation observed in ΔEGFR (30). Recent findings have delineated an important role for the juxtamembrane domain on the catalytic activity of EGFR, wherein the juxtamembrane domain acts as an activation domain in the EGFR dimer (31). As the mutations in the NES and NLS are in the juxtamembrane, we examined whether this altered the kinase activity. In agreement with published results (31), mutations on R646A and R647A found in ΔEGFR-NLS1 (Fig. 1C), EGFR-NLS1 and EGFR-NLS-DM (supplemental Fig. S5E), resulted in reduced activity as compared with the non-mutated receptors. Immunoprecipitation studies also showed that the NLS1 and NLS-DM mutants had diminished activity, whereas the other mutants were similar to the non-mutated receptors (Fig. 2B for ΔEGFR and supplemental Fig. S5F for EGFR).
FIGURE 2.
Forced dimerization of chiΔEGFR-NLS1 restores activity of ΔEGFR-NLS1. A, biochemical fractionation and Western blot analysis of U87-ΔEGFR and LN428-ΔEGFR cells after AG1478 treatment. Quantitation and statistical analysis of Western blotting are shown in supplemental Fig. S4, C and D. B, immunoprecipitation (IP) followed by Western blot (WB) analysis in U87 stable cells for total phosphotyrosine as well as EGFR-specific phosphoresidues. Quantitation and statistical analysis of Western blotting are shown in supplemental Fig. S4, E–G. C, schematic representation showing the mode of action of chiΔEGFR. D, stable expression of chiΔEGFR and chiΔEGFR in U87 cells. Actin was used as a loading control. E, nuclear and non-nuclear fractions from U87 stable cells after stimulation with either EGF or AP20187 were analyzed by Western blotting for FK506 binding protein (FKBP). Lamin B and tubulin served as markers for nuclear and non-nuclear fractions, respectively. Quantitation and statistical analysis of Western blotting are shown in supplemental Fig. S4H. F, confocal analyses of U87 stable cells stimulated with either EGF or AP20187. Cells were immunostained using pEGFR (Tyr-1068) antibody (green) and EGFR antibody (red). Nuclei were counterstained using Topro3 (blue). CHO, Chinese hamster ovary.
The observation that some mutations in the juxtamembrane domain reduced the activation of ΔEGFR raised the possibility that the reduced nuclear localization on the NLS mutants was primarily a consequence of reduced activity. To test this directly, we used a chimeric ΔEGFR (chiΔEGFR) that can be artificially activated by dimerization in response to a chemical dimerizer, AP20187 (23) (Fig. 2C). We engineered the NLS1 mutation in chiΔEGFR (chiΔEGFR-NLS1) and generated U87 stable clones that overexpress either chiΔEGFR or chiΔEGFR-NLS1 (Fig. 2D). AP20187 treatment resulted in increased phosphorylation for both chiΔEGFR and chiΔEGFR-NLS1 (Fig. 2E). Importantly, the levels of nuclear chiΔEGFR-NLS1 did not change in response to the treatment (Fig. 2E), showing that the alteration of the NLS sequence suppresses nuclear localization independently of the activity state. We confirmed these observations using confocal microscopy where AP20187 treatment induced increased translocation of chiΔEGFR but not chiΔEGFR-NLS1 (Fig. 2F). This demonstrates that exclusion of ΔEGFR from the nuclear compartment by the NLS1 mutation is not due to loss of kinase activity, as restoration of activity did not induce its translocation to the nucleus.
Nuclear ΔEGFR Drives Tumor Formation in Glioma Cells
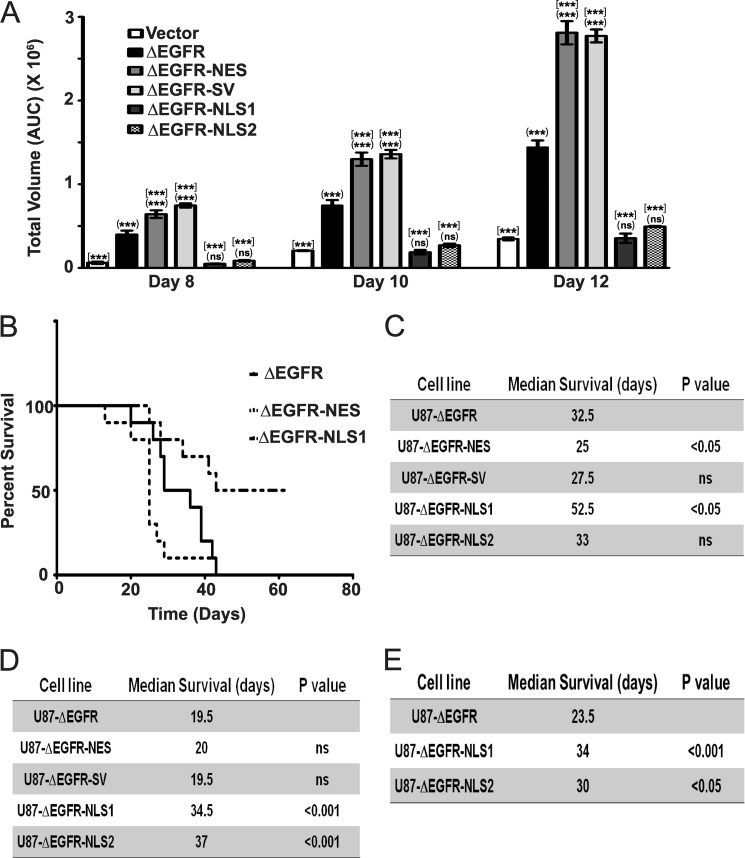
To determine whether nuclear localization affects the ability of ΔEGFR to promote cellular transformation, we analyzed anchorage-independent colony formation. U87-ΔEGFR-NES and U87-ΔEGFR-SV cells showed increased colony formation than U87-ΔEGFR, whereas U87-ΔEGFR-NLS1 and U87-ΔEGFR-NLS2 cells had diminished colony formation (Fig. 3A, representative images shown in supplemental Fig. S6A). We also assessed the impact of altered localization on the growth of U87 tumors in an intracranial xenograft model. In the first experiment, in which animals bearing ΔEGFR cells had a median survival time of 32.5 days, ΔEGFR-NES cells led to more rapid tumor growth in vivo with a reduced median survival time of 25 days (p < 0.05; Fig. 3, B and C), demonstrating that promoting nuclear localization of ΔEGFR also accelerated in vivo growth. ΔEGFR-SV cells also showed a trend toward more rapid tumor growth with a median survival of 27.5 days (Fig. 3C). Conversely, ΔEGFR-NLS1 tumors resulted in an extended median survival time of 54 days (p < 0.05), suggesting that the reduction of nuclear ΔEGFR reduces its oncogenic potential (Fig. 3, B and C). In this experiment, ΔEGFR-NLS2 cells yielded a survival time indistinguishable from ΔEGFR (33 days), raising the possibility that it is the attenuated kinase activity of the NLS1 mutant that is primarily responsible for its reduced tumor formation. Therefore, we assessed tumor growth in a second experiment in which the median survival of animals bearing ΔEGFR cells was significantly shorter (19.5 days). Tumor formation by ΔEGFR-NES and ΔEGFR-SV were not accelerated, possibly because tumor formation was already occurring at a near-maximal rate. However, both U87-ΔEGFR-NLS1 and U87-ΔEGFR-NLS2 showed significantly increased median survival rates of 34.5 and 37 days, respectively (p < 0.01; Fig. 3D and supplemental Fig. S6B). This suggests that both NLS mutants limit the oncogenic potential of ΔEGFR in intracranial xenografts and that these limitations are revealed at different rates of progression to morbidity. To clarify the role of U87-ΔEGFR-NLS2, we repeated the xenografts studies using U87-ΔEGFR-NLS1 and U87-ΔEGFR-NLS2 as well as U87-ΔEGFR cells. In this third experiment, we observed significantly increased median survival rates of 34 and 30 days for U87-ΔEGFR-NLS1 and U87-ΔEGFR-NLS2, respectively (Fig. 3E and supplemental Fig. S6C). This suggests that results from the very first animal experiment was an outlier for U87-ΔEGFR-NLS2 cells and confirms our conclusions that restriction of ΔEGFR to the cytoplasm compromises its oncogenic capabilities. Sustained expression of the ΔEGFR proteins was confirmed by immunohistochemical analysis of tumor sections with V5 antibody (supplemental Fig. S6, D–F). To further rule out the possibility that decreased kinase activity is responsible for decreased oncogenic activity of ΔEGFR-NLS1, we used U87-chiΔEGFR-NLS1 for intracranial xenograft studies, with half the group receiving 2 weeks of infusion with AP20187 (to induce dimerization) while the rest were treated with vehicle. There was no significant difference between the median survival time of the two groups (control, 32 days; AP20187, 33 days; supplemental Fig. S7, A and B), suggesting that loss of kinase activity is not responsible for the decreased tumorigenic potential of ΔEGFR-NLS1. Western blotting for phospho-Tyr-1173 in the tumors confirmed the effectiveness of AP20187 in vivo (supplemental Fig. S7C). We have shown earlier that forced dimerization of chiΔEGFR accelerates tumor growth (23). These experiments together support the conclusion that promoting nuclear localization of ΔEGFR promotes transformation and tumor growth in vivo and that suppressing access to the nucleus suppresses these manifestations of oncogenicity.
FIGURE 3.
Nuclear ΔEGFR promotes an aggressive oncogenic phenotype in glioma cells. A, effect of ΔEGFR and the mutants on anchorage-independent colony formation of U87 cells. (***), comparison between U87–1726zeo and other cell lines. [***], comparison between U87-ΔEGFR and other cell lines. ***, p < 0.001; ns, not significant; n = 3 ± S.E.; at least triplicate samples/experiment. AUC, area under the curve. B, Kaplan-Meier survival curves for mice (10 animals/group) injected intracranially with U87-ΔEGFR, U87-ΔEGFR-NES, and U87-ΔEGFR-NLS1 cells. Lines for U87-ΔEGFR-NLS2 and U87-ΔEGFR-SV that were not significantly different from the U87 control are omitted for clarity. C, median survival times for mice carrying U87-ΔEGFR and the mutants. Statistical significance was assessed by log-rank test. D, median survival times for mice carrying U87-ΔEGFR and the mutants in a repeat experiment (10 animals/group). Statistical significance was assessed by log-rank test. E, median survival times for mice carrying U87-ΔEGFR and the NLS mutants (8 animals/group). Statistical significance was assessed by log-rank test.
Genome-wide Target Analysis of ΔEGFR in Glioblastoma Cells
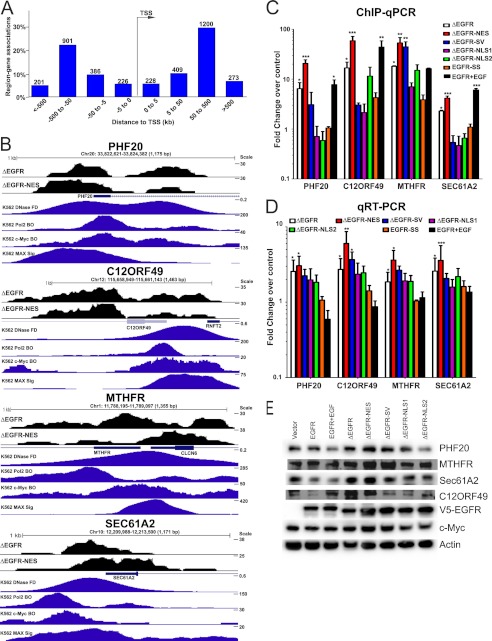
Because nuclear EGFR is implicated in regulation of transcription (3, 4, 6–9), we performed a ChIP-Seq analysis to identify genomic targets of ΔEGFR. We first validated our ChIP-Seq samples by quantitative PCR of the known ΔEGFR target promoter, Aurora A (supplemental Fig. S8A) (10). From our ChIP-Seq experiment, altogether about 2.5 million unique tags and 4857 and 6355 peaks were identified for ΔEGFR and ΔEGFR-NES, respectively. Each peak was assigned to the closest gene as described under “Methods.” The complete list for peak annotation in terms of the closest gene is provided in supplemental Tables S4 & S5. About 23% of ΔEGFR peaks and 21% of ΔEGFR-NES peaks overlapped with promoters (Fig. S8, B and C) (5% of peaks would be located in promoters by chance). The peaks present in both ΔEGFR and ΔEGFR-NES (2294 peaks) datasets were used to identify the probable targets of ΔEGFR (supplemental Fig. S8D). We performed a functional analysis of the genes associated with the promoters to which the ChIP-Seq peaks mapped using Ingenuity Pathway Analysis (Ingenuity® Systems). The analysis classified the genes into 28 categories, of which the most significant ones were Cell-to-Cell Signaling and Interaction, Cellular Assembly and Organization, Cellular Function and Maintenance, and Molecular Transport (supplemental Table S6). Further, evaluation of the distance to the nearest transcription start site (TSS) showed a tendency for the ΔEGFR-binding loci to be observed distal to the TSS (Fig. 4A and supplemental Fig. S8E). Fig. 4B depicts the promoter region occupancy for some of the genes that were found in both ΔEGFR and ΔEGFR-NES. We also show an in silico analysis where we used previously published ChIP-Seq data for Pol2, c-Myc, and open chromatin in K562 cells to demonstrate that these could be potential regions of active transcription. For further validation, we performed ChIP quantitative PCR at predicted loci in cells expressing the different ΔEGFR localization mutants. Of the 14 regions that we examined, nine showed the following pattern and were considered bona fide targets of ΔEGFR: increased recruitment of ΔEGFR over control, increased recruitment of ΔEGFR-NES as compared with ΔEGFR, and diminished recruitment of ΔEGFR-NLS1 and ΔEGFR-NLS2 as compared with ΔEGFR (Fig. 4C and supplemental Table S7). To determine whether recruitment of ΔEGFR had an effect on transcription of the downstream target gene, we analyzed the mRNA levels by quantitative real-time PCR (Fig. 4D) and protein by Western blotting (Fig. 4E) in U87-ΔEGFR and the various mutant lines. Supplemental Table S7 provides a comprehensive list of the genes that we tested for the validation studies and shows that we could validate four genes from the initial 14-gene list as targets of ΔEGFR at both the transcript and protein levels.
FIGURE 4.
ΔEGFR binding to human glioma genome. A, the distance of each peak in both directions from the TSS. B, transformed ChIP-Seq data for ΔEGFR and ΔEGFR-NES in the UCSC genome browser. Shown are the loci PHF20, C12ORF49, MTHFR, and SEC61A2. C, ChIP quantitative PCR analyses of selected genes in U87 stable cell lines. D, Real-time quantitative PCR analyses of selected genes in U87 stable cell lines. For all quantitative PCR experiments, values in the stable cell lines are shown relative to those in vector control cells (normalized to 1), and error bars represent mean ± S.E. from three biological replicates. E, Western blot analyses of selected genes in U87 stable cell lines. Quantitation and statistical analysis of Western blotting are shown in supplemental Fig. S8F. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Analysis of ChIP-Seq Regions for Consensus Transcription Factor Binding Sites
To identify DNA-binding partners of ΔEGFR, we looked for enrichment of transcription factor binding sites in DNA sequences surrounding the peaks (1 kb) that were localized in promoter and upstream regions using TRANSFAC (Biobase biological databases) and used MAST (33) to identify the presence of the motif of interest. We found a significant overrepresentation of the E-box motif in the gene regions bound by ΔEGFR (supplemental Table S8). Of 559 regions analyzed, about 55 E-Box motif-containing regions were identified. We validated the top 14 regions that had an e-value cutoff of 10 from this list, as shown earlier (Fig. 4 and supplemental Table S7).
Nuclear ΔEGFR and c-Myc Are Found to Be Associated on E-box-containing Promoters
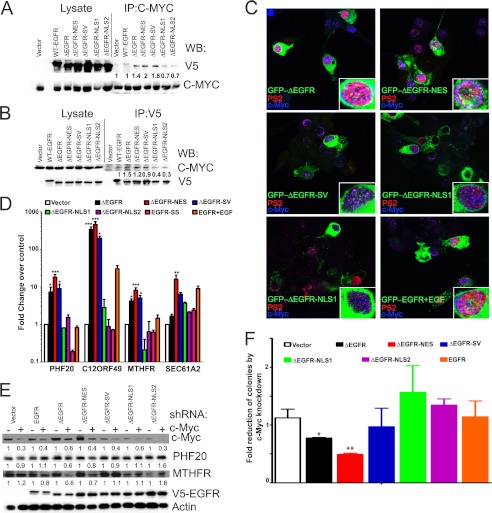
To explore whether c-Myc, an E-Box motif binding transcription factor, could be a binding partner through which ΔEGFR associates with the chromatin, we asked whether ΔEGFR and c-Myc could interact with each other in a physiological setting. c-Myc associated robustly with ΔEGFR as well as with the ΔEGFR-NES and ΔEGFR-SV mutants (Fig. 5A), as determined by c-Myc immunoprecipitation. The association was faintly discernible in ΔEGFR-NLS1 and ΔEGFR-NLS2 as well as in EGF-stimulated EGFR and absent in the control cells (Fig. 5A). The reciprocal immunoprecipitation of EGFR, ΔEGFR, and the mutants showed similar results, with robust levels of c-Myc detected in association with ΔEGFR and ΔEGFR-NES and decreased amounts with ΔEGFR-NLS1 and ΔEGFR-NLS2 (Fig. 5B). Furthermore, cotransfected GFP-ΔEGFR, GFP-ΔEGFR-NES, and, to a lesser extent, GFP-ΔEGFR-SV (green) and c-Myc (blue) were distinctly colocalized (cyan) in the nucleus, by confocal microscopy (Fig. 5C). This colocalization was not obvious in EGF-stimulated GFP-EGFR-, GFP-ΔEGFR-NLS1, and GFP-ΔEGFR-NLS2-transfected cells. Further, GFP-ΔEGFR, GFP-ΔEGFR-NES, and GFP-ΔEGFR-SV also colocalized (yellow) with phospho-Pol2 (PS2, red), a marker of active transcription (Fig. 5C). Thus, ΔEGFR and c-Myc interact physically with each other in the nucleus and seem to associate in areas of active transcription. To determine whether this association was reflected as corecruitment to the target gene promoters that harbor the E-boxes, we performed sequential ChIP/quantitative PCR analysis for the known ΔEGFR target genes. Both c-Myc and ΔEGFR were significantly corecruited onto the promoters of the target genes (Fig. 5D). To validate the role of c-Myc in ΔEGFR-induced transcription of target genes, we depleted c-Myc in U87 stable cell lines cells and found a significant down-regulation of protein levels of the target genes (Fig. 5E). Further, knockdown of c-Myc compromised the growth of the cells on soft agar only in ΔEGFR- and ΔEGFR-NES-overexpressing cells but did not affect cells that were not driven by nuclear ΔEGFR (Fig. 5F), indicating that c-Myc is involved in the oncogenic functions of nuclear ΔEGFR.
FIGURE 5.
Nuclear ΔEGFR associates with c-Myc to promote oncogenic phenotype in glioma cells. A, coimmunoprecipitation (IP) and Western blot (WB) analysis showing c-Myc association with ΔEGFR. Quantitation and statistical analysis of Western blotting are shown in supplemental Fig. S9A. B, coimmunoprecipitation and Western blot analysis showing ΔEGFR association with c-Myc. Quantitation and statistical analysis of Western blotting are shown in supplemental Fig. S9B. C, localization of GFP-EGFR, GFP-ΔEGFR, and GFP-tagged ΔEGFR mutants (green), phosphoPol2 (PS2, red), and c-Myc (blue) in U87 cells. The insets show magnification of a single cell for better visualization. D, sequential ChIP quantitative PCR analysis for c-Myc and the EGFR, ΔEGFR, and ΔEGFR mutants in stable U87 cells. The results have been normalized to the vector control. E, Western blot analysis of U87 stable cells after knockdown of c-Myc by shRNA. One representative blot of three replicates is shown. Quantitation and statistical analysis of Western blotting are shown in supplemental Fig. S9C. F, anchorage-independent colony formation analysis of U87 stable cells after knockdown of c-Myc by shRNA. Values are relative to vector control cells without c-Myc knockdown (normalized to 1). Error bars represent mean ± S.E. from three biological replicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
DISCUSSION
Although there have been reports of nuclear ΔEGFR (8–10), we provide here the first demonstration of its specific oncogenic contribution and mode of action via regulation of gene expression in cooperation with transcription factors, including c-Myc. Central to our findings are the observations in three-dimensional colony formation and in intracranial xenografts that access to the nucleus is a determinant of oncogenic function, making the analysis of this minority component of ΔEGFR important.
We found that the nuclear movement of ΔEGFR is independent of kinase activity in glioblastoma cells. Even when the lower levels of activity of the NLS mutants were overcome by forced dimerization of chiΔEGFR-NLS1, the increased kinase activity was not sufficient to overcome either the reduced ability of chiΔEGFR-NLS1 to enter the nuclear compartment or the decreased tumorigenic potential that results from the altered localization. This activity independence of nuclear localization is in contrast to the majority of previous studies of nuclear EGFR (9, 11) that used either breast cancer or epidermoid carcinoma cells, and so cellular context and mutational status is likely a determining factor for whether translocation of ΔEGFR is activity-dependent. One aspect of this cell type specificity may be the signaling pathways that are deregulated, and this aspect of the biology of nuclear ΔEGFR is worth further investigation, as it may offer points of intervention.
We characterize an NES in EGFR and ΔEGFR for the first time. Enrichment of the NES mutants in the nucleus and loss of interaction with CRM1 (Fig. 1 and supplemental Fig. S5) also indicates that the NES is likely to be a functional export signal in both ΔEGFR and EGFR. The NLS described for EGFR earlier (22) has equivalent functions in ΔEGFR. We were, however, unable to express ΔEGFR-NLS-DM. In contrast, the EGFR-NLS-DM mutant showed robust expression (supplemental Fig. S5), suggesting a potentially interesting difference between the wild-type and mutant EGFRs. The detailed characterization of this difference and whether it affects the functioning of the molecules remains to be undertaken. It is of interest that up to 2-fold enrichment of ΔEGFR (Fig. 1) in the nucleus was able to accelerate tumor formation only to a limited extent, whereas a decrease in nuclear ΔEGFR was able to consistently diminish the tumorigenicity of ΔEGFR (Fig. 3). This suggests that although a threshold amount of nuclear ΔEGFR is required for its oncogenic functions, ΔEGFR does not need to localize to the nucleus at high concentrations to bring about its oncogenic nuclear functions. In fact, our data indicate that a minority of ΔEGFR is in the nucleus but that this relatively small portion makes a unique contribution to the potency of the oncogene.
A key contribution of our data is the comprehensive analysis of chromatin sequences associated with ΔEGFR-containing complexes by ChIP-Seq, and we provide a catalogue of 2294 potential target sequences. Analysis of distances between ΔEGFR-associated regions from the common dataset and TSSs revealed that although 19% of ΔEGFR binding sites are associated with promoters, 28, 47, and 61% are located within 5–50 kb, 50–500 kb, and > 500 kb, respectively (supplemental Fig. S8E). Enrichment at such large distances from the TSS is not without precedent in global ChIP studies of bona fide transcription factors, suggesting that the sequences we have obtained represent a biologically relevant map of ΔEGFR complex-targeted sequences and so regulated genes. The global chromatin binding sites for various transcription factors such as GATA1, GATA2, FLI1, SCL (34), Prdm14 (35), SMAD3, SMAD4, and FOXH1 (36), as well as the recent analysis of 88 nuclear receptors and 121 collaborating factors (37), are also highly enriched at introns and regions distal to the TSS. In this study, our focus for follow-up was on target regions found in promoters of genes. Analysis of the other regions that were found is the subject of an ongoing study.
c-Myc is known to bind to the E-box motifs enriched in our dataset (17) and has been implicated in gliomagenesis (38). Down-regulation of the expression of ΔEGFR/c-Myc target genes coupled with decreased tumorigenic potential of U87-ΔEGFR upon c-Myc depletion suggests the importance of c-Myc in ΔEGFR-induced tumorigenicity. Curiously, in transiently expressing GFP-ΔEGFR, GFP-ΔEGFR-NES, and GFP-ΔEGFR-SV cells, there was increased staining for both PS2 and c-Myc (supplemental Fig. S10). Increased c-Myc protein was also seen in Western blot analyses (Fig. 4E). It has been demonstrated that nuclear EGFR, STAT3, and Src form a trimeric complex on c-Myc promoter and control its expression in pancreatic cancer cells (32). A quick scan of our ChIP-Seq data showed that neither the promoter nor intronic regions of the c-Myc gene were identified as targets. This suggests that control of c-Myc gene expression by ΔEGFR might be either indirect or at the posttranscriptional level.
We showed recently that STAT5b is a nuclear interacting partner of ΔEGFR and that the two molecules are recruited to and regulate target genes, including Aurora A and Bcl-XL (10). Two previous studies have also shown that ΔEGFR can interact with STAT3 in the nucleus (8, 9). Our data show for the first time the nuclear interaction of ΔEGFR with a protein (c-Myc) outside of the STAT family members. Whether there is convergence in targets of ΔEGFR/c-Myc and ΔEGFR/STAT family members remains to be determined.
In summary, our data demonstrate that a minority proportion of ΔEGFR found in the nucleus of glioblastoma cells is regulated by sequences in the juxtamembrane region of the receptor, that nuclear access is important for the full oncogenic activity of ΔEGFR, and that nuclear ΔEGFR associates with c-Myc to elicit its oncogenic functions. In addition to delineating the mechanism of nuclear translocation of ΔEGFR for the first time, we show the role of nuclear ΔEGFR in ΔEGFR-induced tumorigenesis. This assumes importance in the context of glioblastoma multiforme because ΔEGFR is highly tumorigenic in glioma cells (as compared with EGFR) and is overexpressed in about 30% of glioblastoma multiforme. Because restricting ΔEGFR from entry into the nucleus decreased oncogenicity of ΔEGFR, it is likely that a significant extent of ΔEGFR oncogenic functions is exerted via the nuclear-specific function of ΔEGFR. Therefore, understanding the nuclear functions of ΔEGFR offers the opportunity for the discovery of new targets for the development of anti-glioma strategies. Further, identification of c-Myc, an oncogenic transcription factor as an important mediator of nuclear transcriptional functions of ΔEGFR, an oncogenic receptor tyrosine kinase connects two central nodes of signaling in tumors. This novel connection can be potentially exploited in a therapeutic setting.
Acknowledgments
We thank Dr. Mien-Chie Hung for helpful discussions that supported our studies. We also thank Verlene Henry and Lindsay Holmes of the Department of Neurosurgery, University of Texas M.D. Anderson Cancer Center, for help in carrying out animal experiments.
This work was supported, in whole or in part, by grants from the National Cancer Institute National Institutes of Health Grants RO1CA108500 (to O. B.) and P50CA127001 (to O. B. and A. E. G.). This work was also supported by M.D. Anderson Cancer Center Core Grant NCI CA016672, including the DNA Analysis facility.

This article contains supplemental Figs. S1–S10 and Tables S1–S8.
- EGFR
- EGF receptor
- NES
- nuclear export signal
- NLS
- nuclear localization signal
- ChIP-Seq
- ChIP coupled with parallel sequencing
- TSS
- transcription start site
- STAT
- signal transducer and activator of transcription.
REFERENCES
- 1. Nishikawa R., Ji X. D., Harmon R. C., Lazar C. S., Gill G. N., Cavenee W. K., Huang H. J. (1994) A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc. Natl. Acad. Sci. U.S.A. 91, 7727–7731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Furnari F. B., Fenton T., Bachoo R. M., Mukasa A., Stommel J. M., Stegh A., Hahn W. C., Ligon K. L., Louis D. N., Brennan C., Chin L., DePinho R. A., Cavenee W. K. (2007) Malignant astrocytic glioma. Genetics, biology, and paths to treatment. Genes Dev. 21, 2683–2710 [DOI] [PubMed] [Google Scholar]
- 3. Hanada N., Lo H. W., Day C. P., Pan Y., Nakajima Y., Hung M. C. (2006) Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol. Carcinog. 45, 10–17 [DOI] [PubMed] [Google Scholar]
- 4. Hung L. Y., Tseng J. T., Lee Y. C., Xia W., Wang Y. N., Wu M. L., Chuang Y. H., Lai C. H., Chang W. C. (2008) Nuclear epidermal growth factor receptor (EGFR) interacts with signal transducer and activator of transcription 5 (STAT5) in activating Aurora-A gene expression. Nucleic Acids Res. 36, 4337–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huo L., Wang Y. N., Xia W., Hsu S. C., Lai C. C., Li L. Y., Chang W. C., Wang Y., Hsu M. C., Yu Y. L., Huang T. H., Ding Q., Chen C. H., Tsai C. H., Hung M. C. (2010) RNA helicase A is a DNA-binding partner for EGFR-mediated transcriptional activation in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 107, 16125–16130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin S. Y., Makino K., Xia W., Matin A., Wen Y., Kwong K. Y., Bourguignon L., Hung M. C. (2001) Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 3, 802–808 [DOI] [PubMed] [Google Scholar]
- 7. Lo H. W., Hsu S. C., Ali-Seyed M., Gunduz M., Xia W., Wei Y., Bartholomeusz G., Shih J. Y., Hung M. C. (2005) Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell 7, 575–589 [DOI] [PubMed] [Google Scholar]
- 8. de la Iglesia N., Konopka G., Puram S. V., Chan J. A., Bachoo R. M., You M. J., Levy D. E., Depinho R. A., Bonni A. (2008) Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 22, 449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lo H. W., Cao X., Zhu H., Ali-Osman F. (2010) Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol. Cancer Res. 8, 232–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Latha K., Li M., Chumbalkar V., Gururaj A., Hwang Y., Dakeng S., Sawaya R., Aldape K., Cavenee W. K., Bogler O., Furnari F. B. (2013) Nuclear EGFRvIII-STAT5b complex contributes to glioblastoma cell survival by direct activation of the Bcl-XL promoter. Int. J. Cancer 132, 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang S. C., Hung M. C. (2009) Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin. Cancer Res. 15, 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lo H. W., Xia W., Wei Y., Ali-Seyed M., Huang S. F., Hung M. C. (2005) Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 65, 338–348 [PubMed] [Google Scholar]
- 13. Psyrri A., Egleston B., Weinberger P., Yu Z., Kowalski D., Sasaki C., Haffty B., Rimm D., Burtness B. (2008) Correlates and determinants of nuclear epidermal growth factor receptor content in an oropharyngeal cancer tissue microarray. Cancer Epidemiol. Biomarkers Prev. 17, 1486–1492 [DOI] [PubMed] [Google Scholar]
- 14. Psyrri A., Yu Z., Weinberger P. M., Sasaki C., Haffty B., Camp R., Rimm D., Burtness B. A. (2005) Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clin. Cancer Res. 11, 5856–5862 [DOI] [PubMed] [Google Scholar]
- 15. Kim J., Jahng W. J., Di Vizio D., Lee J. S., Jhaveri R., Rubin M. A., Shisheva A., Freeman M. R. (2007) The phosphoinositide kinase PIKfyve mediates epidermal growth factor receptor trafficking to the nucleus. Cancer Res. 67, 9229–9237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xia W., Wei Y., Du Y., Liu J., Chang B., Yu Y. L., Huo L. F., Miller S., Hung M. C. (2009) Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Mol. Carcinog. 48, 610–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pelengaris S., Khan M., Evan G. (2002) c-MYC. More than just a matter of life and death. Nat. Rev. Cancer 2, 764–776 [DOI] [PubMed] [Google Scholar]
- 18. Soucek L., Evan G. I. (2010) The ups and downs of Myc biology. Curr. Opin. Genet. Dev. 20, 91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Albihn A., Johnsen J. I., Henriksson M. A. (2010) MYC in oncogenesis and as a target for cancer therapies. Adv. Cancer Res. 107, 163–224 [DOI] [PubMed] [Google Scholar]
- 20. Shingu T., Chumbalkar V. C., Gwak H. S., Fujiwara K., Kondo S., Farrell N. P., Bogler O. (2010) The polynuclear platinum BBR3610 induces G2/M arrest and autophagy early and apoptosis late in glioma cells. Neuro. Oncol. 12, 1269–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen B., Borinstein S. C., Gillis J., Sykes V. W., Bogler O. (2000) The glioma-associated protein SETA interacts with AIP1/Alix and ALG-2 and modulates apoptosis in astrocytes. J. Biol. Chem. 275, 19275–19281 [DOI] [PubMed] [Google Scholar]
- 22. Hsu S. C., Hung M. C. (2007) Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J. Biol. Chem. 282, 10432–10440 [DOI] [PubMed] [Google Scholar]
- 23. Hwang Y., Chumbalkar V., Latha K., Bogler O. (2011) Forced dimerization increases the activity of ΔEGFR/EGFRvIII and enhances its oncogenicity. Mol. Cancer Res. 9, 1199–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kajiwara Y., Panchabhai S., Levin V. A. (2008) A new preclinical 3-dimensional agarose colony formation assay. Technol. Cancer Res. Treat. 7, 329–334 [DOI] [PubMed] [Google Scholar]
- 25. Gururaj A. E., Singh R. R., Rayala S. K., Holm C., den Hollander P., Zhang H., Balasenthil S., Talukder A. H., Landberg G., Kumar R. (2006) MTA1, a transcriptional activator of breast cancer amplified sequence 3. Proc. Natl. Acad. Sci. U.S.A. 103, 6670–6675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., Lowe C. B., Wenger A. M., Bejerano G. (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lo H. W., Ali-Seyed M., Wu Y., Bartholomeusz G., Hsu S. C., Hung M. C. (2006) Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin β1 and CRM1. J. Cell Biochem. 98, 1570–1583 [DOI] [PubMed] [Google Scholar]
- 29. la Cour T., Kiemer L., Mølgaard A., Gupta R., Skriver K., Brunak S. (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 17, 527–536 [DOI] [PubMed] [Google Scholar]
- 30. Huang H. S., Nagane M., Klingbeil C. K., Lin H., Nishikawa R., Ji X. D., Huang C. M., Gill G. N., Wiley H. S., Cavenee W. K. (1997) The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J. Biol. Chem. 272, 2927–2935 [DOI] [PubMed] [Google Scholar]
- 31. Red Brewer M., Choi S. H., Alvarado D., Moravcevic K., Pozzi A., Lemmon M. A., Carpenter G. (2009) The juxtamembrane region of the EGF receptor functions as an activation domain. Mol. Cell 34, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jaganathan S., Yue P., Paladino D. C., Bogdanovic J., Huo Q., Turkson J. (2011) A functional nuclear epidermal growth factor receptor, SRC and Stat3 heteromeric complex in pancreatic cancer cells. PloS ONE 6, e19605. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33. Bailey T. L., Gribskov M. (1998) Combining evidence using p-values. Application to sequence homology searches. Bioinformatics 14, 48–54 [DOI] [PubMed] [Google Scholar]
- 34. Tijssen M. R., Cvejic A., Joshi A., Hannah R. L., Ferreira R., Forrai A., Bellissimo D. C., Oram S. H., Smethurst P. A., Wilson N. K., Wang X., Ottersbach K., Stemple D. L., Green A. R., Ouwehand W. H., Göttgens B. (2011) Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev. Cell 20, 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma Z., Swigut T., Valouev A., Rada-Iglesias A., Wysocka J. (2011) Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat. Struct. Mol. Biol. 18, 120–127 [DOI] [PubMed] [Google Scholar]
- 36. Kim S. W., Yoon S. J., Chuong E., Oyolu C., Wills A. E., Gupta R., Baker J. (2011) Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev. Biol. 357, 492–504 [DOI] [PubMed] [Google Scholar]
- 37. Tang Q., Chen Y., Meyer C., Geistlinger T., Lupien M., Wang Q., Liu T., Zhang Y., Brown M., Liu X. S. (2011) A comprehensive view of nuclear receptor cancer cistromes. Cancer Res. 71, 6940–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jensen N. A., Pedersen K. M., Lihme F., Rask L., Nielsen J. V., Rasmussen T. E., Mitchelmore C. (2003) Astroglial c-Myc overexpression predisposes mice to primary malignant gliomas. J. Biol. Chem. 278, 8300–8308 [DOI] [PubMed] [Google Scholar]