Background: Mitochondria send signals to the nucleus to modulate gene expression when mitochondrial function is perturbed.
Results: Cyclin-dependent kinase E1 (CDKE1) was identified as an essential component in regulation of responses to perturbation of mitochondrial electron transport.
Conclusion: Mitochondrial regulation is integrated with growth, energy, and other cellular stress signaling pathways.
Significance: The identification of a molecular link between mitochondrial retrograde regulation and growth and stress signaling pathways.
Keywords: Gene Transcription, Mitochondria, Plant Molecular Biology, Respiration, Signaling, Arabidopsis, Retrograde, Stress
Abstract
Plants must deal effectively with unfavorable growth conditions that necessitate a coordinated response to integrate cellular signals with mitochondrial retrograde signals. A genetic screen was carried out to identify regulators of alternative oxidase (rao mutants), using AOX1a expression as a model system to study retrograde signaling in plants. Two independent rao1 mutant alleles identified CDKE1 as a central nuclear component integrating mitochondrial retrograde signals with energy signals under stress. CDKE1 is also necessary for responses to general cellular stresses, such as H2O2 and cold that act, at least in part, via anterograde pathways, and integrates signals from central energy/stress sensing kinase signal transduction pathways within the nucleus. Together, these results place CDKE1 as a central kinase integrating diverse cellular signals and shed light on a mechanism by which plants can effectively switch between growth and stress responses.
Introduction
Mitochondria and plastids are semiautonomous organelles found in all plant cells. Although they contain a limited genetic coding capacity, encoding 57 and 128 proteins, respectively, in Arabidopsis thaliana (Arabidopsis) (1), greater than 95% of the 1500 and 2500 proteins estimated to be present in mitochondria and chloroplasts are encoded by genes located in the nucleus. The expression of nuclear located genes encoding mitochondrial and chloroplast proteins is controlled by either anterograde or retrograde pathways, depending on the source of the signal(s) regulating these sets of genes (2). Anterograde regulation refers to top-down regulatory pathways, where endogenous (e.g. hormones) or exogenous (e.g. light) signals directly impact nuclear gene expression. Retrograde regulation refers to signals (e.g. reactive oxygen species) that originate in mitochondria or plastids and modify or alter nuclear gene expression networks.
The concept of retrograde regulation for mitochondria and plastids is over 30 years old (3, 4). For plastid retrograde regulation, signals have been classified into five different groups, plastidial gene expression, pigment biosynthesis, reactive oxygen species, redox effectors, and metabolites. Plastidial retrograde regulation has been investigated with forward genetic approaches, and genome uncoupled (gun) mutants were identified. Five of the six GUN genes, GUN2–6, encode components closely associated with tetrapyrrole biosynthesis. GUN1 on the other hand, is a chloroplast localized pentatricopeptide repeat (2, 5). The report of a gain-of-function gun mutant, gun6, (5) strongly supports the role for heme as a signaling molecule and a mechanistic model for the Mg-ProtoIX-mediated plastid signal was recently presented (6). Additional factors or signals associated with plastid retrograde signaling include a plastid bound homeodomain transcription factor (9), the transcription factor ABI4 (7), PAP (8), and β-cyclocitral (9).
The characterization of mitochondrial retrograde signaling has been extensively carried out in Saccharomyces cerevisiae (yeast), where several retrograde pathways and the components involved have been identified and characterized (10). The most extensively characterized system is associated with inhibition or dysfunction of the TCA cycle, and three factors, called Rtg1, -2, and -3, have been characterized to mediate this response, in addition to a number of positive and negative regulators (10). Whereas Rtg1 and -3 are basic helix-loop-helix leucine transcription factors (10), Rtg2 contains a kinase domain that shares similarities with a variety of proteins, including cell cycle proteins (11). Furthermore, it has been demonstrated that the Rtg-regulated pathway interacts with the target of rapamycin pathway, involved in nitrogen metabolism in yeast, via Lst8p as a negative regulator of the Rtg pathway (12). In mammalian cells the master regulator NFκB links mitochondrial retrograde regulation with other regulatory pathways (13).
The induction of the alternative oxidase (AOX)2 at a transcript and protein level has been widely used as a model system to study mitochondrial retrograde signaling in plants (14), and a genetic system designed to identify components in these signal transduction pathways has been reported (15); but to date no components have been identified. Studies on mitochondrial retrograde signaling suggest converging or synergistic pathways with plastidial retrograde pathways. A study investigating the role of Arabidopsis prolyl-tRNA synthetase, a dual targeted protein required for both mitochondria and plastid translation, has shown that disruption of both mitochondrial and plastidial translation is required to elicit retrograde signals (16). At the receiver end, the transcription factor ABI4 has been shown to regulate targets of both plastidial and mitochondrial retrograde signaling pathways (17, 18).
Here, we describe the identification of the regulator of AOX1a 1 (rao1) mutant, encoding a nuclear localized cyclin-dependent kinase E1 (CDKE1), as an essential molecular component of mitochondrial retrograde signaling in Arabidopsis. Furthermore, we show that the induction of AOX in response to retrograde signals is likely under the control of both transcriptional and post-transcriptional regulation.
EXPERIMENTAL PROCEDURES
Col:LUC Construction and Line Selection
The −2000 bp AOX1a promoter region upstream of the translation start site, previously shown to be responsive to a variety of treatments that perturb mitochondrial function (17, 19), was cloned in front of the reporter gene LUCIFERASE (LUC). A Col:LUC line with a single T-DNA insertion was isolated by Southern blot analysis and hybridized with a 500-bp probe amplified from the inserted LUC gene (supplemental Fig. S1).
Mutagenesis and Mutant Screen
Approximately 30,000 homozygous Col:LUC seeds were mutagenized for 16 h in 100 ml of 0.25% (v/v) ethyl methane sulfonate, then washed in several volumes of water over a period of 6 h. Seeds were sown on soil and grown in growth chambers at 22 °C, 16 h, 120 μmol m−2 s−1 light/8 h dark. ∼20,000 M2 plants, representing 20 pooled family populations were screened for altered LUC reporter gene expression under mitochondrial stress. Plants were grown for 14 days on Gamborg's B5 growth media (PhytoTechnology) with 3% (w/v) sucrose and 0.8% agar (w/v). Stress treatments were applied 3 h into the light cycle by spraying plants with either 50 μm antimycin A (AA), 50 μm myxothiazol, 25 mm monofluoracetate, or 20 mm H2O2, as described previously (17, 20). Plants were returned to normal growth conditions for 6 h. Cold treatment was applied to 14-day-old plants by placing them at 4 °C for 16 h. Ultraviolet light (UV) was applied for 30 min and then plants were placed in normal growth conditions for 6 h. After 6 h (or 16 h for cold treatment), 2.5 mm luciferin (GoldBio) was applied to plants and luminescence was measured using a NightOWL bioluminescence imaging system (Berthold).
Genetic Mapping and Gene Identification
Transgenic Arabidopsis lines carrying the LUCIFERASE reporter gene system, linked to the 2000-bp AOX1a promoter region upstream of the translational start site (referred to as Col:LUC) were generated. A single insert homozygous line for the T-DNA insertion in the Columbia-0 (Col-0) background was mutagenized by ethane methyl sulfonate (supplemental Fig. 1, ColTL3). A similar line carrying a single, homozygous copy of the T-DNA construct was generated in Landsberg erecta ecotype (Ler:LUC) for mapping purposes. rao1–1 and rao1–2 mutants were crossed with Ler:LUC. Homozygous F2 rao1–1 and rao1–2 plants were selected and DNA extracted. Linkage between simple sequence length polymorphism markers (i.e. a set of insertion/deletion (Indel) polymorphism sites between Col-0 and Ler Arabidopsis ecotypes) (21) and a mutation was established based on a low recombination frequency. Markers were selected from Cereon database (supplemental Table S1 and Fig. S2). Initial mapping of a population of 50 homozygous F2 rao1–1 and rao1–2 plants all mapped at 25.478 Mb (MBK5-SSPL marker, supplemental Fig. S2). A second mapping was carried out with expanded small sequence length polymorphism markers in 50 homozygous F2 rao1–1 plants and 138 homozygous F2 rao1–2 plants and 50 homozygous F2 rao1–1 plants, recombinants were identified and genotyped. Both rao1–1 and rao1–2 mutations mapped to a 2.48-Mb region on chromosome 5 between marker CER457391 and the end of the chromosome.
Allelism between rao1–1 and rao1–2 was confirmed by the lack of phenotype complementation in F1 seeds resulting from cross between the 2 lines. rao1–1 and rao1–2 mutant plants were complemented with the full-length coding sequence of CDKE1 that was PCR amplified from Col-0 cDNA and cloned into pDONR201 (Invitrogen), and subsequently transferred to a binary vector pK7WG2 under the control of the constitutive Cauliflower Mosaic Virus 35S promoter. This binary vector was then transformed into Agrobacterium tumefaciens GV3130 strain. Floral dipping was performed with an inoculum medium containing 10% (w/v) sucrose and 0.05% (v/v) Silwet-77, as previously described (22). T1 transformants were screened on kanamycin antibiotic as described (23). The presence of the transgene in the complemented plants was confirmed by PCR analysis. The progeny of the primary transformants segregated for wild-type and rao1 mutant phenotypes; plants with wild-type induction responses all contained the transgene, whereas plants showing the rao1 phenotype did not.
Next Generation Sequencing and Data Analysis
50 ng of genomic DNA were isolated from pooled Arabidopsis seedlings for each line (Col:LUC (wild-type), rao1–1 backcrossed line 1, rao1–1 backcrossed line 2, rao1–1 backcrossed line 3, and rao1–2) using the DNeasy Plant Maxi kit (Qiagen), according to manufacturer's instructions. Genomic DNA was used as input in library generation using Nextera DNA sample prep kits (EpiCentre) according to the manufacturer's instructions. Library quality was checked using an Agilent bioanalyzer and quantified using a Nanodrop (Thermo Scientific) and Qubit (Invitrogen) before Next-Generation Sequencing with 2 × 50-bp paired-end read lengths on an Illumina HiSeq1000. Primary data processing was performed by RTA, .BCL conversion, and de-multiplexing was performed by CASAVA version 1.8.2. Alignments for Col:LUC read sequences were performed against TAIR9 reference sequences (arabidopsis.org) and a fasta reference genome sequence (Col:LUC reference) was generated for Col:LUC to ensure SNP calls present in the wild-type parental lineage could be excluded from rao1 sequence data analysis. Read sequences for rao1–1 and rao1–2 re-sequencing were aligned to the Col:LUC reference genome sequence using CASAVA version 1.8.2 and SNP calls generated using GATK software pipelines (www.broadinstitute.org/gsa/wiki/index.php/The_Genome_Analysis_Toolkit) and Best Practices (www.broadinstitute.org/gsa/wiki/index.php/Best_Practice_Variant_Detection_with_the_GATK_v3). Only homozygous SNP calls were used and SNPs were also filtered on base call quality, mapping quality scores, mutations occurring in annotated coding regions only and base changes generated by ethyl methane sulfonate mutagenesis. Furthermore, mutations were shortlisted only if all back-crossed lines contained the mutation, and there was a candidate mutation in the same gene in the allelic line. Sorted .bam and .gvf files were visualized in IGV (www.broadinstitute.org/igv/) and Tablet (bioinf.scri.ac.uk/tablet/) viewers to identify candidate SNPs in the pre-defined genomic region of interest at the base of chromosome 5.
Protein Sequence Alignment and Phylogenetic Tree Generation
The protein coding sequence for CDKE1 (AT5G63610) was downloaded from the TAIR website (www.arabidopsis.org). Using the CDKE1 sequence, a blast search for selected plant species was carried out using the Phytozome website (www.phytozome.net). The most related sequences were then aligned using the sequence alignment program MAFFT (24). Multiple sequence alignments were visualized using the program Multiple Align Show. Phylogeneitic trees were drawn using the Phylip (version 3.68) software package and visualized using A tree Viewer (25).
Quantitative RT-PCR Leaf Tissue from Col:LUC, rao1–1, and rao1–2-1—14-day-old seedlings treated with 50 μm AA, UV, or mock control treatments, as described above, were harvested in biological triplicate at 30 min, 1 h, and 3 h after treatment. RNA isolation, cDNA generation, and quantitative RT-PCR were performed as described previously (26). Primers and assay details for AOX1a have been described previously (26). LUC primers are listed in supplemental Table S1.
Global Transcript Analyses
Analysis of global changes in transcript abundance in response to AA treatment in the rao1 mutants and kin10 RNAi lines were carried out using Affymetrix ATH1 microarray genechips. For the rao1 mutant analysis, arrays were performed using RNA isolated in biological triplicate from Col:LUC, rao1–1, and rao1–2 14-day-old seedlings treated with either mock treatment or 3 h of 50 μm AA treatments. Similarly, for the kin10 RNAi expression analysis, arrays were performed using RNA isolated in biological triplicate from Ler, kin10 RNAi#1, and kin10 RNAi#7 14-day-old seedlings with either mock treatment or 3 h of 50 μm AA treatments. aRNA generation, hybridization, washing, and scanning the gene chips and all primary data analysis was performed according to the manufacturer's instructions. CEL files were MAS5 normalized to obtain present/absent calls across all Genechips. Probesets were defined as absent and removed from further analysis if they returned absent or marginal calls in 2 or more of the 3 biological replicates in all genotypes/conditions tested. Probesets relating to organelle transcripts and Affymetrix bacterial controls were also removed, leaving a present set of 16,607 probesets for the rao1 arrays and 16,423 for the kin10 arrays. GC-Robust Muliarray Average normalized gene expression values from the defined present set were analyzed to identify differentially expressed genes by a regularized t test based on a Bayesian statistical framework using the software program Cyber-T (27) (cybert.microarray.ics.uci.edu/) as described in Ref. 28. Microarray data for the two independent mutant lines (rao1–1 and rao1–2) were analyzed independently at first to ensure no probes displayed differential expression between the genotypes. Changes were considered significant at an FDR correction level of PPDE (>P) >0.95 and a fold-change greater than 1.5-fold (positive or negative). Protein-protein interaction networks were drawn using the online tools BAR Arabidopsis Interaction Viewer (29) and PAIR (30). Gene lists of stress responsive transcripts controlled via RAO1 activity were defined as shown in supplemental Fig. S8. Gene lists of KIN10-associated transcripts were utilized from Ref. 31 and hierarchical clusters of Log2 fold-changes were generated in Partek Genomics Suite version 6.5 (Partek) using average linkage and Euclidean distance parameters. The stress responsive network affected by mutation in CDKE1 is shown in supplemental Datasets S1-S6.
Western Blots
Mitochondria were isolated from 14-day-old plants treated with 50 μm AA or exposed with 30 min UV light, as described above. Western blot immunodetections were carried out as described previously (32) using antibodies to AOX (33) and TOM40 (32). Serial dilutions of total mitochondrial protein were used to ensure that the signal obtained was in the linear range related to protein levels.
GFP Localization
GFP localization of CDKE1 was performed as described previously, with mitochondrial mCherry utilized as a mitochondrial marker and DAPI staining (Invitrogen) to confirm nuclear localization (34).
Bimolecular Fluorescence Complementation
Bimolecular fluorescence complementation was carried out as outlined previously (35) in onion epithelial cells. DAPI staining (Invitrogen) was utilized as per the manufacturer's instructions, to confirm nuclear localization.
Phenotypic Characterization of rao1 and T-DNA Insert Lines
A full phenotypic characterization of wild-type (Col:LUC) and mutant lines (rao1–1, rao1–2, and GABI_564F11) analyzed in this study is shown in supplemental Fig. S9 and carried out as described (36). At least 15 plants per genotype were used for analysis. For plate-based phenotypic analysis, seedlings were grown on Gamborg's B5 agar medium supplemented with 3% (w/v) sucrose. For the quantification of maximum root length and maximum rosette leave area, photos were first taken of the plants, and the root length and leave area were then measured using Image Processing and Analysis tool in the software ImageJ64.
T-DNA Insertion Line
Four T-DNA insertion lines for cdke1 (SALK_072781, SALK_117306, SALK_138675, and GABI_564F11) were obtained from the NASC (European Arabidopsis Stock Centre). However, homozygous lines were only obtained from GABI_564F11 through PCR analyses. T-DNA insertion homozygous lines were confirmed by PCR using the gene-specific primers (LP and RP) and T-DNA-specific primer (LB). The precise location of the T-DNA insertion was determined by sequencing.
RESULTS
Identification of Regulators of Alternative Oxidase1a (rao) Mutants
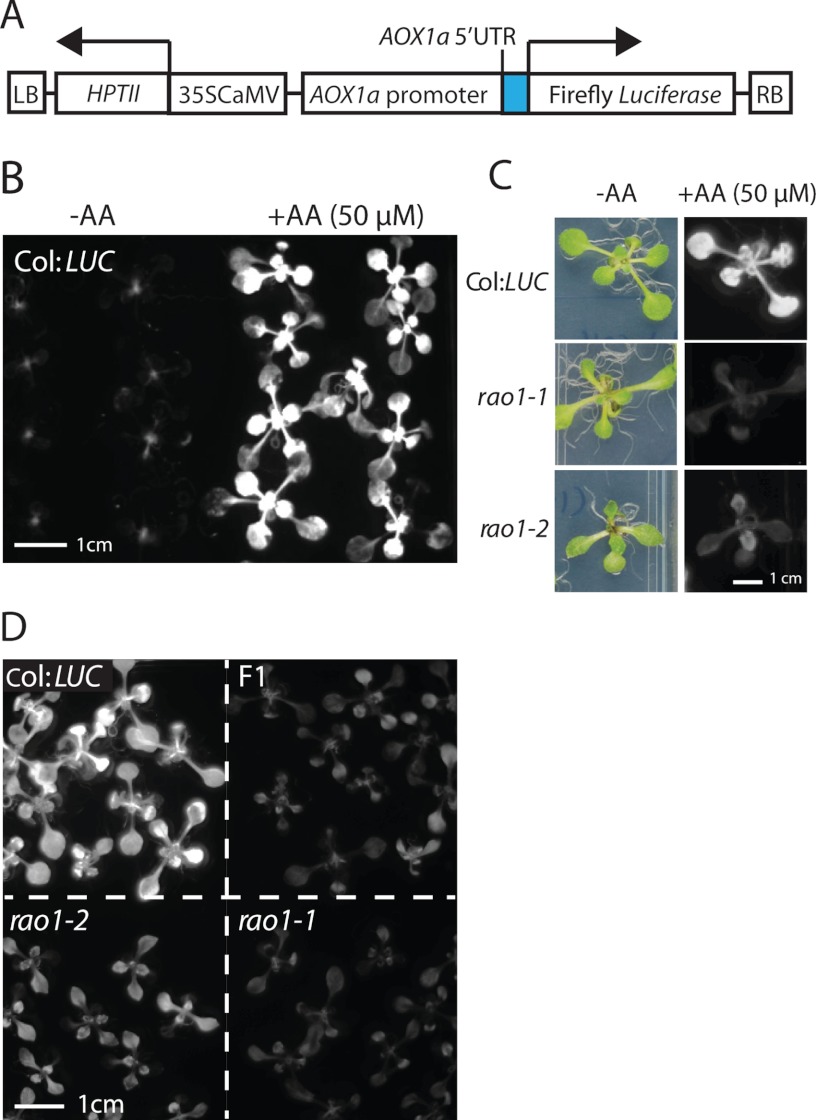
A genetic screen to identify the molecular components of mitochondrial retrograde signaling was developed using a transgenic Arabidopsis line carrying the LUCIFERASE reporter gene system, linked to the 2000-bp AOX1a promoter region upstream of the translational start site (AOX1a:LUC, Fig. 1A and supplemental Fig. S1, plants referred to as Col:LUC plants). Transgenic Col:LUC plants in the wild-type background showed low LUC expression under normal growth conditions (Fig. 1B, −AA) but this increased substantially after 6 h treatment with 50 μm AA, an inhibitor of Complex III of the mitochondrial electron transport chain (Fig. 1B, +AA, 50 μm). Mutagenized M2 progeny were screened for the inability to induce LUC in response to retrograde signals after treatment with AA. Two mutant lines, rao1–1 and rao1–2, were identified (Fig. 1C), both mapped to a 2.48-Mb region at the bottom of chromosome 5 (supplemental Fig. S2), and therefore, were crossed to test for allelism. rao1–1 and rao1–2 are recessive mutations and the first filial generation (F1) progeny from the cross, all F1 plants showed a rao1 mutant phenotype, confirming that rao1–1 and rao1–2 are allelic mutations in the same gene (Fig. 1D).
FIGURE 1.
Identification of two allelic mutants rao1–1 and rao1–2 as upstream retrograde regulators of AOX1a. A, a schematic diagram of the T-DNA insert containing ALTERNATIVE OXIDASE1a promoter driving a luciferase reporter gene (AOX1a:LUC). The AOX1a promoter consists of the region 2000 bp upstream of the translational start site of AOX1a, including the 99-bp AOX1a 5′ UTR. LB, T-DNA left border sequences; HPTII, hygromycin phosphotransferase II; 35S CaMV, 35S Cauliflower mosaic virus promoter; RB, T-DNA right border sequences. B, Col:LUC plants sprayed with deionized water (−AA) or with 50 μm antimycin A (AA) to elicit a mitochondrial retrograde signal. Luciferin was applied and LUC reporter activity was visualized in a NightOwl bioluminescence imager after 6 h treatment. C, rao1–1 and rao1–2 do not express AOX1a:LUC under AA treatment. D, allelism of rao1–1 and rao1–2. F1, the first filial generation from rao1–1 and rao1–2 crosses.
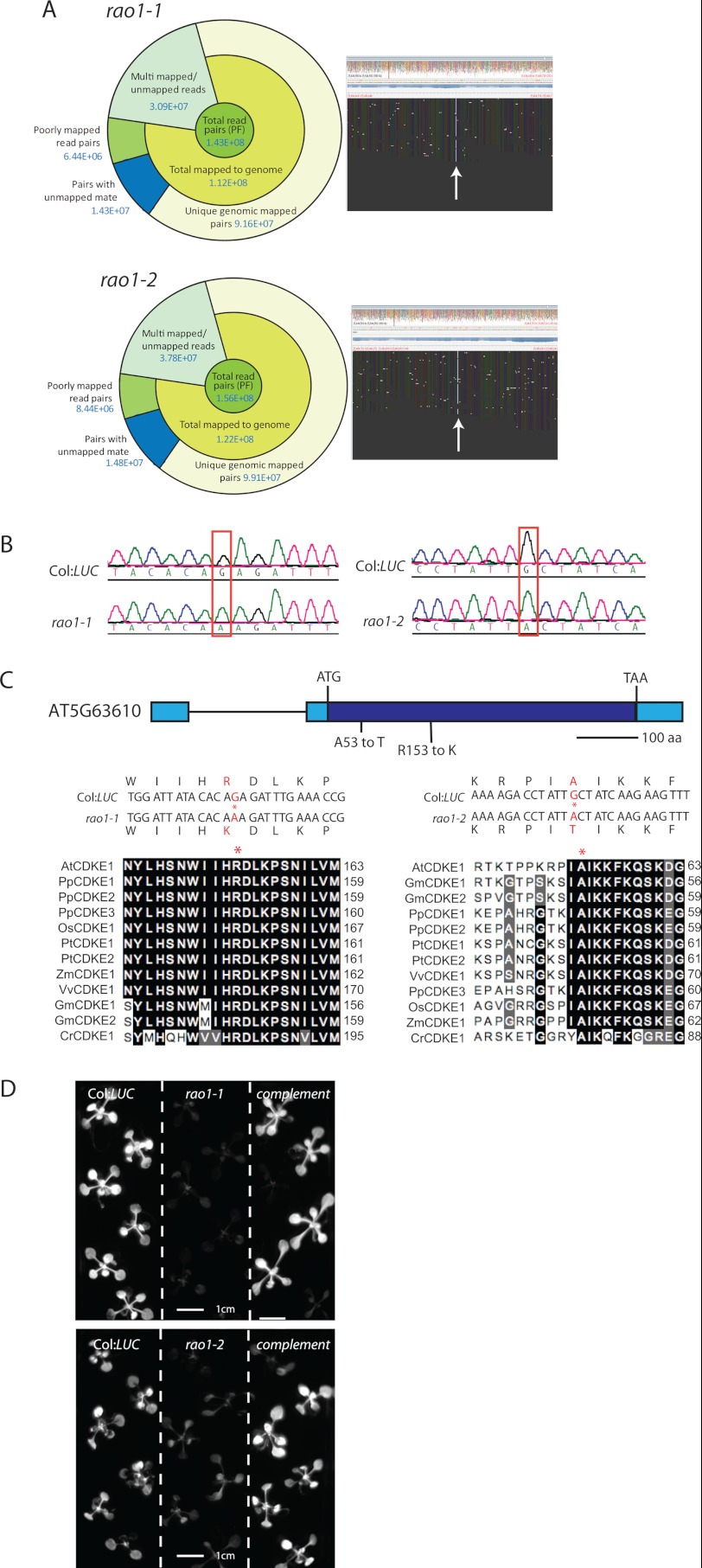
Next generation sequencing was used to catalog candidate mutations in common genes in these lines. Short sequence reads were aligned to a reference sequence generated from the re-sequencing of the Col:LUC line, and homozygous SNPs were called. rao1–1 and rao1–2 alignment coverage and details are shown in Fig. 2A. A candidate mutation was identified in AT5G63610, which encodes a CDKE1 (Fig. 2A, SNPs visualized in Tablet (http://bioinf.scri.ac.uk/tablet/); supplemental Fig. S3). Both rao1–1 and rao1–2 contain nonsynonymous mutations in the CDKE1 coding sequence as confirmed by Sanger sequence analysis (Fig. 2B). rao1–1 contains an amino acid change from an arginine to a lysine at position 153 within the serine/threonine protein kinase active site (Fig. 2, B and C). rao1–2 contains an amino acid change from an alanine to a threonine at position 53 in the protein kinase ATP binding site (Fig. 2, B and C). Both mutations occur in highly conserved regions of the coding sequence (Fig. 2C). In both rao1–1 and rao1–2 lines, the mutant phenotype could be rescued by complementation with constructs containing the wild-type CDKE1 coding sequence; as shown by fully restored, wild-type induction of LUC in response to AA treatment in the primary transformants (Fig. 2D). These results confirm that the rao1 phenotype is the result of a mutation in CDKE1.
FIGURE 2.
rao1–1 and rao1–2 encode a CDKE1 (AT5G63610). A, next generation sequencing analysis identified 2 candidate mutations in CDKE1 including a summary of alignment and .bam alignments for the region of interest visualized in Tablet (bioinf.scri.ac.uk/tablet). B, Sanger sequence chromatographs confirming nucleotide changes within the RAO1 coding sequence. C, a schematic model of the AT5G63610 gene structure with the relative position of amino changes shown in red for rao1–1 and rao1–2. Alignments of CDKE1 proteins from a range of plant species are shown. Asterisks indicate mutated amino acids in rao1–1 (left panel) and rao1–2 (right panel). At, Arabidopsis thaliana (Arabidopsis); Gm, Glycine max (soybean); Pp, Physcomitrella patens (moss); Pt, Populus trichocarpa (poplar); Os, Oryza sativa (rice); Vv, Vitis vinifera (grape); Zm, Zea mays (maize); Cr, Chlamydomonas reinhardtii (green algae). D, complementation of rao1–1 and rao1–2 with wild-type RAO1 results in restored wild-type induction of AOX1a:LUC under AA treatment.
Diverse Cellular Stress Signaling Pathways Are Regulated through RAO1
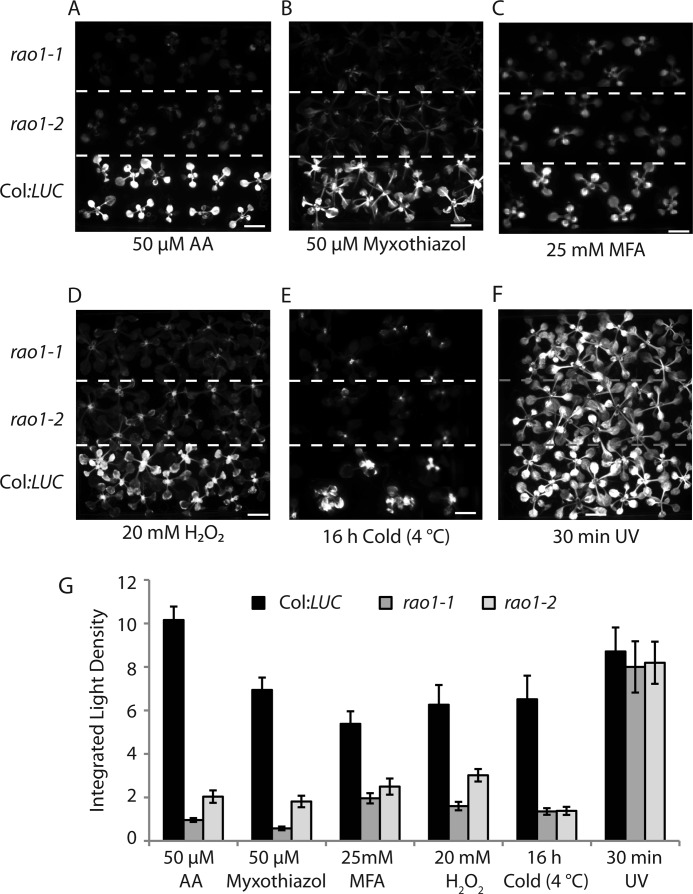
Although AA is a well defined inhibitor of Complex III, it also acts as an inhibitor to one pathway of cyclic electron transport (37). Therefore plants were screened for a retrograde response to 50 μm myxothiazol (which, like AA, inhibits Complex III in the mitochondrial respiratory chain but at a different site: AA inhibits at the N site and myxothiazol inhibits at the P site (38)). Neither rao1–1 nor rao1–2 could induce LUC expression in response to myxothiazol (Fig. 3, A, B, and G). rao1 plants were treated with monofluoracetate, an inhibitor of the TCA cycle at the site of aconitase (15). Although monofluoracetate induced LUC in Col:LUC plants, this induction was abolished in both rao1–1 and rao1–2 (Fig. 3, C and G). Therefore, the rao1 mutant plants failed to induce AOX1a in response to three different inhibitors affecting mitochondrial function. rao1 plants were also treated with 20 mm H2O2, an important reactive oxygen species signaling molecule (39, 40), to investigate RAO1 function in a wider stress signaling context. The induction of LUC observed in Col:LUC plants in response to H2O2 was abolished in the rao1 mutant background (Fig. 3, D and G). Thus, RAO1 is also involved in mediating the response of AOX1a to more general cellular oxidative stress signals. To investigate the extent of RAO1 regulation in general abiotic stress responses we also subjected plants to 16 h of cold treatment, which also failed to induce reporter gene expression in the mutant background compared with Col:LUC plants (Fig. 3, E and G). Finally, rao1 mutants were also exposed to ultraviolet (UV) light, to primarily target chloroplast function and, possibly generate alternative cellular reactive oxygen species (41). rao1–1 and rao1–2 lines showed no impairment of the observed wild-type induction of LUC in response to UV treatment, indicating that cellular responses to UV stress are signaled via pathway(s) independent of RAO1 function (Fig. 3, F and G). Previous studies also have shown that there are distinct pathways for induction of AOX1a, where mutants were identified that do not induce LUC expression with AA treatment, yet induce LUC with monofluoracetate treatment (13). Together, these screens demonstrate that rao1 mutants provide a powerful tool to investigate and dissect the stress signaling pathways operating in plants and are involved in both anterograde and retrograde signaling pathways.
FIGURE 3.
Characterization and quantification of AOX1a promoter inductions in rao1–1 and rao1–2 following different stress treatments. Col:LUC, rao1–1, and rao1–2 seedlings were treated with: A, 50 μm AA; B, 50 μm myxothiazol; C, 25 mm monofluoracetate (MFA); D, 20 mm H2O2; E, 16 h cold treatment (4 °C); F, 30 min ultraviolet (UV) light. Luciferase (LUC) reporter gene activity was visualized in a NightOwl bioluminescence imager after a 6-h treatment unless otherwise stated. Scale bars indicate 1 cm. G, processed images were analyzed for quantification of LUC expression using the Integrated Light Intensity from 8-bit images calculated in the Java software ImageJ. Error bars denote S.E. (n > 8 measurements).
RAO1 Is Responsible for Gating the Extent of AOX1a Induction in Response to Retrograde Signals
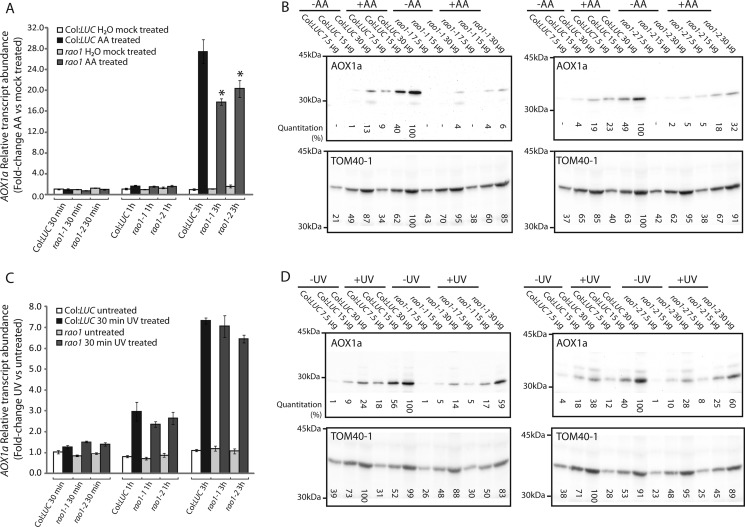
AOX1a transcript and protein abundances were measured over a 5-h time course during AA and UV treatments (Fig. 4). At 3 h of exposure to AA, the ∼28-fold significant induction (p < 0.001, Student's t test) of the AOX1a transcript that was observed in Col:LUC plants was significantly reduced (∼18-fold, p < 0.01, Student's t test) in both rao1–1 and rao1–2 lines (Fig. 4). As expected, the AOX protein abundance in Col:LUC plants increased substantially, but this was essentially absent in rao1 mutants (Fig. 4B). As rao1–1 and rao1–2 represented ethyl methane sulfonate mutants that resulted in a single amino acid change (Fig. 2, B and C), T-DNA insertions for this gene were analyzed. Of four T-DNA mutants annotated as containing an insertion in CDKE1 (At5g63610), only one (GABI_564F11) was found to be truly located in this gene. Analysis of the transcript and protein abundance for AOX in this T-DNA line revealed a similar pattern to that observed in rao1–1 and rao1–2, in that induction of transcript abundance by AA treatment is reduced by ∼50% and induction of protein abundance is reduced (supplemental Fig. S5, A and B).
FIGURE 4.
Characterization of AOX1a transcript and protein inductions in rao1–1 and rao1–2. A, AOX1a transcript abundance in Col:LUC, rao1–1, and rao1–2 lines after 30 min, 1 h, and 3 h AA treatment. Seventeen-day-old plants were sprayed with deionized water (−AA) or 50 μm AA. Relative transcript abundance fold-changes between mock treated and AA-treated plants for the respective genotypes are shown. B, immunodetection of AOX1a in isolated mitochondria after a 5-h AA treatment. Plants were sprayed with deionized water (−AA) or 50 μm AA. Relative protein abundance (quantification values indicated below each blot) was determined by quantitative densitometry. TOM40–1 protein abundance is also shown as a loading control. C, AOX1a transcript abundance in Col:LUC, rao1–1, and rao1–2 lines at 30 min, 1 h, and 3 h recovery after a 30-min UV treatment. Relative transcript abundance fold-changes between untreated and UV-treated plants are shown. D, immunodetection of AOX1a in isolated mitochondria from plants treated with UV for 30 min followed by 5 h recovery. TOM40–1 protein abundance is also shown as a loading control.
The results presented in Fig. 4, A and B, suggest that RAO1 can act at two levels to regulate the expression of AOX1a, at a transcript level, where transcript abundance was significantly reduced by between 30 and 50% (Fig. 4A, supplemental Fig. 5A), depending on the mutant examined, and at a post-transcriptional level, as a greater reduction in AOX1a protein was observed (Fig. 4B, supplemental Fig. 5B). To study this further we investigated conditions where induction of LUC was normal, and subsequently examined AOX1a transcript and protein levels in this situation. From the analysis of the LUC screening assay (Fig. 3), treatment with UV led to induction of luminescence in both Col:LUC and rao-1 mutant lines. Thus transcript and protein abundance for AOX1a was examined under UV treatment (Fig. 4, C and D). Induction of the AOX1a transcript was not significantly reduced in the rao-1 mutant lines compared with Col:LUC (Fig. 4C), however, the induction of protein was reduced by ∼40% (Fig. 4D). This was consistently observed in both rao-1 lines. Quantitative RT-PCR assays confirmed that the differences observed between the AOX1a transcript and protein responses to AA treatment are not due to a splicing defect or incomplete transcript processing in the rao1 background (supplemental Fig. S6).
Genomewide Transcriptome Responses in rao-1 Mutant Plants
To investigate the extent to which RAO1 is necessary to regulate genomewide responses to mitochondrial dysfunction, Affymetrix microarray analyses were performed on Col:LUC, rao1–1, and rao1–2 plants under untreated conditions and under 3 h stress treatment with AA (GEO accession GSE36011). Gene transcripts were classified into one of four gene lists: 1) not significantly changed under any condition comparison tested; 2) transcripts with altered expression in response to AA treatment, that are controlled independently of RAO1 function or activity; 3) gene targets positively regulated through RAO1; and 4) gene targets negatively regulated through RAO1 activity (supplemental Fig. S7 and Datasets S1–S6).
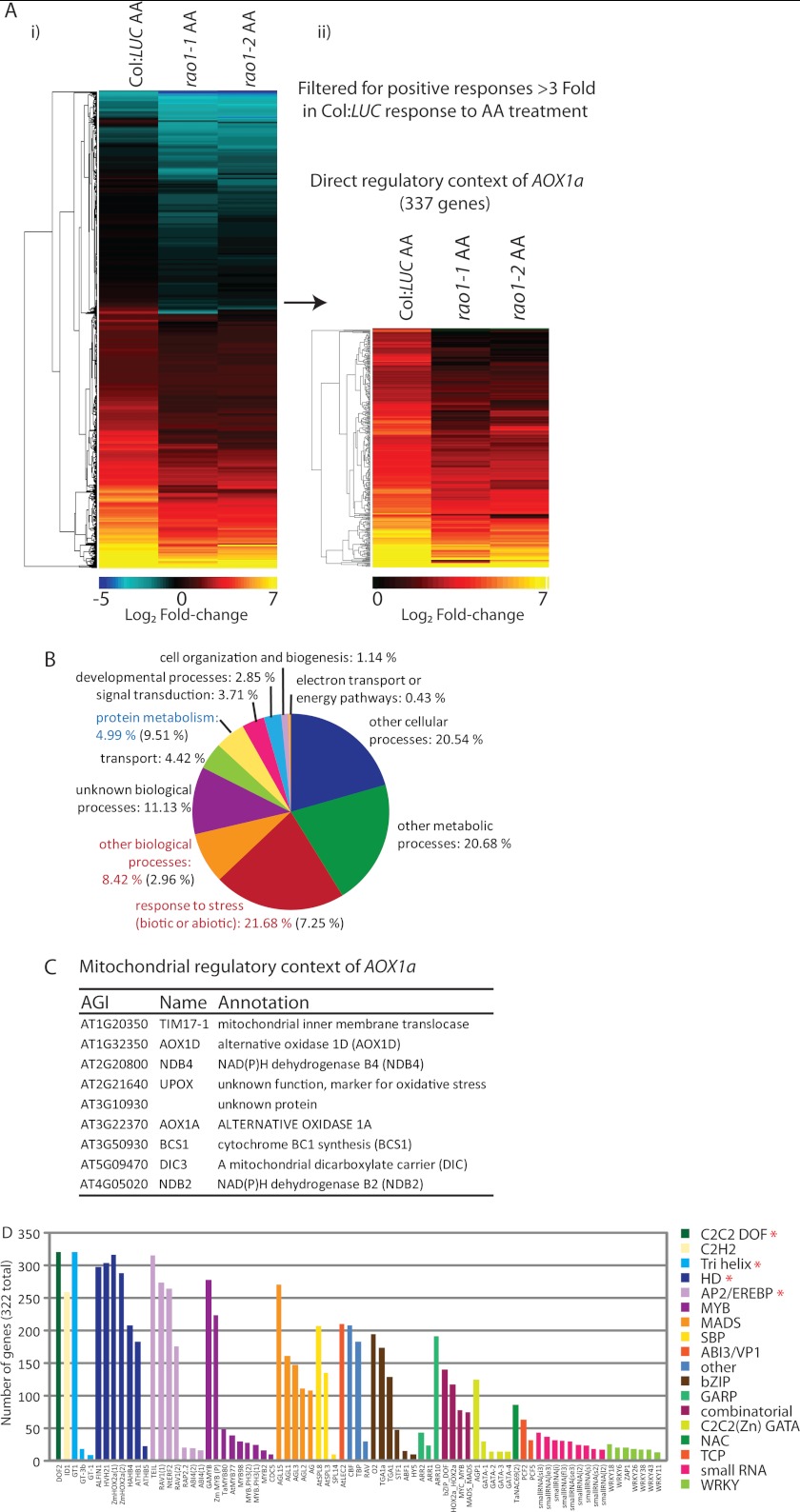
To identify early target genes regulated synergistically with AOX1a through RAO1, gene expression profiles for Geneset 3 were further filtered for positive wild-type responses greater than 3-fold (Log2 fold-change, untreated versus AA treated), which were significantly impaired by greater than 30% in both rao1–1 and rao1–2 under stress. This produced a list of 337 transcripts encoding proteins with diverse functions and cellular localizations (Fig. 5A). In this list, there is a significant over-representation of transcripts involved in stress responses to biotic and abiotic stimuli (22.68 versus 7.25% genomewide) and a significant under-representation of transcripts involved in protein metabolism and synthesis (4.99 versus 9.51% genomewide) (Fig. 5B). Within this list of core stress responsive transcripts regulated through RAO1 are numerous transcription factors, including AP2 domain factors (supplemental Dataset S5). Within this core list of 337 transcripts are those associated with energy status (Dark-induced 6 (DIN6) (31), numerous protein kinases, MAPK kinases, a predicted mitochondrial pentatricopeptide repeat protein (AT2G20720), and transmembrane receptor proteins, including one predicted to be localized to the chloroplast (AT2G32140) (supplemental Dataset S5). Nine mitochondrial components were identified, all of which have been previously defined as part of the mitochondrial stress response (42) (Fig. 5C). This indicates that the primary mitochondrial response to diverse stress conditions is mediated via RAO1 activity. Binding sites for a number of transcription factor families with proposed roles mediating plant abiotic stress responses and controlling mitochondrial energy metabolism were over-represented in this 337 Geneset. These included AP2/EREBP (ABI4 binding sequences (17)), MYB and bZIP (31), MADS, Homeodomain (HD factors), and C2C2 DOF and TCP factors (28) (Fig. 5D). Indeed, a predicted binding site for several of these classes of transcription factors were found in the upstream regions of all 337 transcripts in this stress responsive subnetwork (Fig. 5D, asterisks).
FIGURE 5.
Global identification of target genes regulated through RAO1 activity in response to mitochondrial retrograde stress signals. Affymetrix microarray analysis of global gene expression responses to AA treatment in Col:LUC, rao1–1, and rao1–2 plants was conducted as shown in supplemental Fig. S7 and Dataset S1. A Geneset was identified that contains transcripts that are positively regulated through RAO1 function in response to mitochondrial retrograde signals (Geneset 3). A, hierarchical cluster of fold-change responses to AA treatment for those transcripts in Geneset 3. This set was further filtered for positive responses >3-fold to give 337 genes. Scale bar shows the Log2 magnitude of fold-changes. B, functional categorization (GO Biological process, www.arabidopsis.org/tools/bulk/go) for the 337 genes defined as the direct regulatory context for AOX1a. Annotations in red are significantly over-represented and those in blue are significantly under-represented (p < 0.0001, χ-squared test). C, list of genes from the 337 genes that are localized in the mitochondria. D, occurrence of known transcription factor binding site sequences over-represented within the 1000-bp upstream regions for these genes. Binding sites are colored according to transcription factor family groups, specific family members with characterized binding sites are shown in the x axis. Asterisks represent binding sites that are present in all 337 upstream regions.
RAO1 Interacts with the Energy Sensing SnRK1 Kinase KIN10
As the core stress response mediated through RAO1 activity appears to include diverse functions in stress responses and energy state perception, the predicted protein-protein interactome for CDKE1 was defined (supplemental Fig. S8A). A protein kinase, KIN10, which has been shown previously to integrate stress, darkness, and energy signaling with growth was identified as having putative interactions with CDKE1 (Interlog confidence value 21 (31)) (supplemental Fig. S8A). Expression profiles for the KIN10 gene targets show dramatic overlap with the expression profiles observed for rao1 mutant plants under AA treatment (supplemental Fig. S8B).
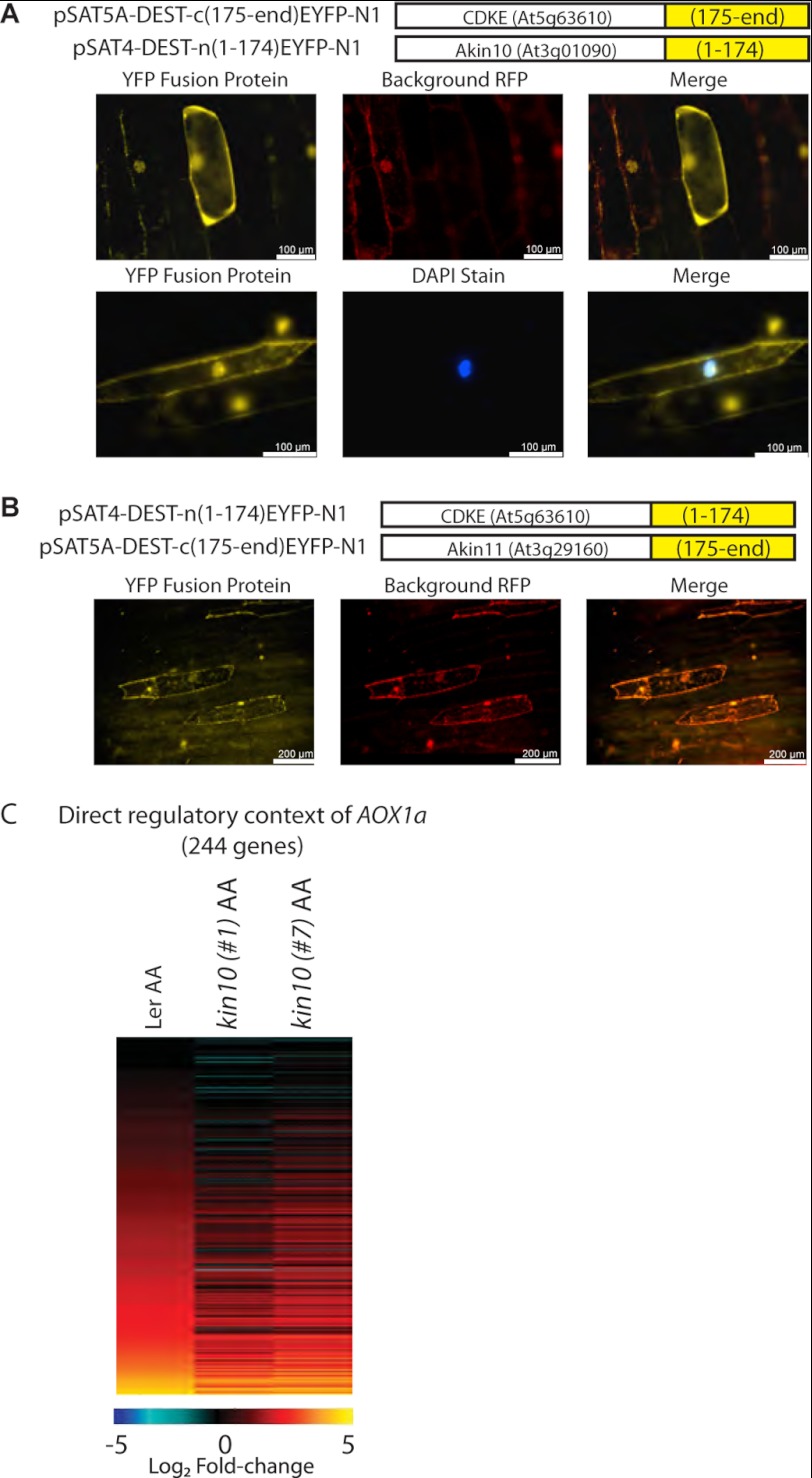
We sought to confirm the interaction between RAO1 and KIN10 using bimolecular fluorescence complementation assays (Fig. 6). RAO1 and KIN10 show a clear interaction within the nucleus, evidenced by a yellow fluorescent protein (YFP) signal distinct from autofluorescence (Fig. 6A). Interactions between RAO1 and KIN11 were also tested as a control but no YFP signal that was independent of autofluorescence could be detected (Fig. 6B). Analysis of the KIN10 coding sequence revealed a nuclear localization sequence and GFP localization shows clear nuclear targeting, as well as cytosolic targeting (Supplemental Fig. S4). Here, we confirm that KIN10 can be transported to the nucleus where it interacts with the exclusively nuclear localized RAO1 (Fig. 6, supplemental Fig. S4). To further explore the functional significance of the RAO1-KIN10 interaction, it was reasoned that the 337 genes that were identified as regulated by RAO1 (Fig. 5A, ii), should change in a similar manner in kin10 mutants under AA treatment. Although none of the T-DNA insertions annotated represent a knock-out, two RNAi lines, previously used to characterize KIN10 (31), were obtained and it was confirmed that transcript abundance for KIN10 was reduced to between 40 (kin10#1) and 20% (kin10#7) of wild-type levels, in Ler background (supplemental Fig. S5C). Of the 337 genes identified in the direct regulatory context of AOX1a by RAO1 (Fig 5A, ii), 279 were expressed in the Ler ecotype. Of these, 244 genes were induced in response to AA treatment in Ler plants, and 217 of these were also induced in response to AA in one or both kin10 RNAi lines (Fig. 6C). Notably, as was seen for the rao lines (Fig. 5A, ii), the magnitude of expression induction seen in the kin10 RNAi plants was smaller than the inductions seen in Ler plants (for 191 of the 217 genes) (Fig. 6C compare with Fig. 5A, ii; supplemental Dataset S5).
FIGURE 6.
Bimolecular fluorescence complementation of CDKE1 with KIN10 and KIN11. A, CDKE1, KIN10, and KIN11 were incorporated into a split-YFP system, with C- and N-terminal fusions to either the 1–174 amino acids (AA) or 175-end AA of YFP. These constructs were biolistically transformed into onion epidermal cells in a variety of construct combinations, to identify protein-protein interactions (see “Experimental Procedures”). To control for autofluorescence, the RFP channel was observed in addition to the YFP channel. A, for KIN10 and CDKE1, an interaction was observed, with a clear YFP signal in the nucleus and a diffused signal in the cytoplasm. B, for KIN11 and CDKE1, no independent YFP signal (differentiated from autofluorescence) could be observed. C, Affymetrix microarray analysis of the 337 genes defined as positively regulated by RAO1 were analyzed in response to AA treatment in Ler, kin10#1, and Kin10#2 plants. The genes are ordered in the same order as described in the legend to Fig. 4A, ii, showing that the induction in transcript abundance of genes is reduced in the kin10 RNAi lines compared with wild-type (Ler) (fold-changes are shown in supplemental Dataset 5). Scale bar shows the Log2 magnitude of fold-changes.
DISCUSSION
Although the induction of AOX in a variety of plant species has been used as a model system to study mitochondrial to nuclear communication for almost two decades (14), the molecular components that mediate this signaling are largely unknown. As so called end points of retrograde signaling, ABI4 and WRKY transcription factors have been previously shown to coordinate both mitochondrial and plastid retrograde signaling within the nucleus (17, 18, 43), but it is unclear how diverse hormone, energy, stress, and developmental signals concurrently influence transcription factor activity and transcription rates. The identification of CDKE1/RAO1 in this study provides a far greater understanding of how the induction of AOX is integrated into the general regulatory context of the cell. As the name indicates, cell cycle-dependent kinases are associated with cell division, and CDKE1/RAO1 is proposed to play a role in integrating environmental signals with plant development, but CDKE1/RAO1 does not directly interact with any other cyclin components (44). CDKE1/RAO1 is a component of the kinase module of the plant mediator complex that relays regulatory signals between specific transcription factors bound to the promoter and RNA polymerase II (45). Importantly, rao1 mutant plants do not show the transcriptional reprogramming of stress responsive components (such as ABI4) under normal conditions that is observed with complete aox1a knock-out plants (26). Indeed, only 119 transcripts genomewide are significantly altered under normal conditions in both rao1–1 and rao1–2 by greater than 2.5-fold compared with Col:LUC. Thus, the genomewide transcriptional response shown here represents the integrated network controlled through direct phosphorylation events associated with the mediator complex of RNA polymerase and not any secondary effect of mis-regulated central stress responsive transcription factors.
This particular kinase appears to have a dual role(s) central to the mediator complex of RNA polymerase II. First, CDKE1/RAO1 has a demonstrated role as a CDK, integrating cellular responses to environmental signals with cell division or elongation. Mutation in CDKE1/RAO1 has been identified previously in genetic screens for flowering abnormalities (hen3, HUA enhancer mutants) with a mild early flowering phenotype when mutated alone, and abnormalities in cell fate determination in a hua1–1/hua2–1 background (supplemental Fig. S9) (46). Second, from the results presented here CDKE1/RAO1 has a central role as a sensitive relay between specific stress-induced transcription factors actively bound to the promoter and RNA polymerase II, thereby directly affecting transcription under stress. Together, this suggests that CDKE1/RAO1 provides a mechanism by which plant cells can switch between growth and stress responses to changing environmental conditions. This is further supported by the results of the global transcriptional analyses, as transcripts relating to both growth and stress are affected in the rao1 mutant background, albeit in opposite ways (Fig. 4B, supplemental Fig. S9). Cell wide stress responses are initiated by CDKE1, whereas protein metabolism and protein synthesis (essentially the building blocks for cell division and growth) and photosynthetic components are switched off (Fig. 4B).
It is apparent from the results obtained that CDKE1/RAO1 acts both at a transcriptional and post-transcriptional level to regulate AOX1a expression. The phenomena of additional means of post-transcriptional control of AOX in response to stress has been observed or suggested independently in a number of different stress conditions in previous studies (19, 47, 48), however, without any likely explanation of the molecular mechanisms involved. In particular, induction of AOX1a transcript and protein takes place under phosphate starvation in plants, along with the synthesis of galactoglycerolipid, which occurs via three different pathways (47). A suppressor of one of these pathways identified a mitochondrial outer membrane protein that has an identical effect on the induction of AOX1a as rao1, namely digalactosyldiacylglycerol1 (dgd1) SUPPRESSOR1 (DGS1) (47). Under AA stimulation in this mutant while induction of transcript abundance for AOX1a appeared normal as determined by Northern blotting, induction of the AOX1a protein was essentially abolished. Furthermore, this mutant had the same root growth delayed phenotype as was observed with rao-1 mutants (supplemental Fig. S9B). Thus there appears to be a specific regulatory pathway that regulates the accumulation of AOX1a protein. It is interesting to note that the yeast orthologues of DGS1 on the mitochondrial outer membrane, called the nuclear control of ATPase 2 (NCA2), are involved in regulating mitochondrial gene expression in yeast (49, 50). Although it is not possible to determine whether accumulation of the AOX1a protein is due to a translational or post-translation effect, it is notable that there are a number of post-transcriptional control mechanisms that govern mRNA fate both prior to nuclear export and within the cytoplasm, which we know very little about in plants. A number of these processes are regulated by phosphorylation events. For example, in mammals, the 5′ capping events and cap-binding proteins that occur co-transcriptionally on the nascent pre-mRNA as it emerges from RNA polymerase II, are regulated in part through the activity of mammalian target of rapamycin kinases, with central roles in cell proliferation, growth, and energy sensing (51). The 5′ cap is postulated to have roles in mRNA protection, promoting downstream processing and, via target of rapamycin signaling, interacting with cap-binding proteins and other RNA binding complexes to preferentially stimulate synthesis of proteins from new transcripts (allowing quick responses under stress conditions with limited translational machinery) (51, 52). This function of target of rapamycin in Arabidopsis appears to be conserved and may be a downstream acceptor for KIN10 signaling in plants (53), perhaps also mediated through CDKE1 in the nucleus in a post-transcriptional manner.
Previously it has been shown that the SnRK1 protein kinase KIN10 in Arabidopsis acts as a central regulator or integrator of signals for growth, energy metabolism, and stress (31). Here we have shown that KIN10 can be targeted to the nucleus that is consistent with previous reports of nuclear targeting of KIN10 (54), with this report noting that KIN10 nuclear localization depends on the tissue and developmental stage, which suggests that its location in the nucleus may be regulated. The interaction with CDKE1/RAO1 in the nucleus provides evidence that CDKE1/RAO1 is the acceptor site for KIN10 signals. Consistent with the interaction demonstrated in this study between KIN10 and CDKE1/RAO1, it has been shown that yeast Snf1 interacts physically with proteins of the yeast mediator complex (55). Furthermore, analysis of changes in the transcript abundance in kin10 RNAi lines shows a large overlap with the genes identified as regulated by RAO1.
Consistent with this proposal of a switch between growth and stress response, genetic manipulation of AOX results in the altered growth of plants. Tobacco suspension cells unable to induce AOX continue to grow under severe nutrient limiting conditions in contrast to cells that can induce AOX and essentially stop growing (56). Furthermore, in tobacco cells and Chlamydomonas reinhardtii, a lack of AOX results in larger cells (57, 58). Thus the KIN10 and CDKE1/RAO1 interplay represents an efficient protein kinase signal cascade that essentially provides a mechanism by which plants can detect and integrate signals from multiple input sources, and switch between growth and stress responses. The results presented here suggest that anterograde and retrograde pathways interact at an early stage in the signaling process, i.e. KIN10, and that both transcriptional and post-transcriptional pathways operate, but the extent of each may differ under different treatments, as observed with the differences between UV and the other treatments used in this study. Combined these responses reveal how fine-tuned the signaling system is and how responses to different treatments can differ, even if some of the components used in the response to different treatments are the same.

This article contains supplemental Table S1, Figs. S1–S9, and Datasets S1–S6.
- AOX
- alternative oxidase
- AA
- antimycin A
- SNP
- single nucleotide polymorphism
- CDKE1
- cyclin-dependent kinase E1
- Ler
- Landsberg erecta
- PAP
- 3′-phosphoadenosine 5′-phosphate.
REFERENCES
- 1. Pfannschmidt T. (2010) Plastidial retrograde signalling. A true “plastid factor” or just metabolite signatures? Trends Plant Sci. 15, 427–435 [DOI] [PubMed] [Google Scholar]
- 2. Woodson J. D., Chory J. (2008) Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9, 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bradbeer J. W., Atkinson Y. E., Borner T., Hagemann R. (1979) Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid-synthesized RNA. Nature 279, 816–817 [Google Scholar]
- 4. Liao X. S., Small W. C., Srere P. A., Butow R. A. (1991) Intramitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Woodson J. D., Perez-Ruiz J. M., Chory J. (2011) Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr. Biol. 21, 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kindgren P., Norén L., López Jde D., Shaikhali J., Strand A. (2012) Interplay between HEAT SHOCK PROTEIN 90 and HY5 controls PhANG expression in response to the GUN5 plastid signal. Mol. Plant 5, 901–913 [DOI] [PubMed] [Google Scholar]
- 7. Sun X., Feng P., Xu X., Guo H., Ma J., Chi W., Lin R., Lu C., Zhang L. (2011) A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat. Commun. 2, 477 doi: 10.1038/ncomms1486 [DOI] [PubMed] [Google Scholar]
- 8. Estavillo G. M., Crisp P. A., Pornsiriwong W., Wirtz M., Collinge D., Carrie C., Giraud E., Whelan J., David P., Javot H., Brearley C., Hell R., Marin E., Pogson B. J. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23, 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ramel F., Birtic S., Ginies C., Soubigou-Taconnat L., Triantaphylidès C., Havaux M. (2012) Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. U.S.A. 109, 5535–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Z., Butow R. A. (2006) Mitochondrial retrograde signaling. Annu. Rev. Genet. 40, 159–185 [DOI] [PubMed] [Google Scholar]
- 11. Bork P., Sander C., Valencia A. (1992) An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. U.S.A. 89, 7290–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giannattasio S., Liu Z., Thornton J., Butow R. A. (2005) Retrograde response to mitochondrial dysfunction is separable from TOR1/2 regulation of retrograde gene expression. J. Biol. Chem. 280, 42528–42535 [DOI] [PubMed] [Google Scholar]
- 13. Jazwinski S. M., Kriete A. (2012) The yeast retrograde response as a model of intracellular signaling of mitochondrial dysfunction. Front. Physiol. 3, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Millar A. H., Whelan J., Soole K. L., Day D. A. (2011) Organization and regulation of mitochondrial respiration in plants. Annu. Rev. Plant Biol. 62, 79–104 [DOI] [PubMed] [Google Scholar]
- 15. Zarkovic J., Anderson S. L., Rhoads D. M. (2005) A reporter gene system used to study developmental expression of alternative oxidase and isolate mitochondrial retrograde regulation mutants in Arabidopsis. Plant Mol. Biol. 57, 871–888 [DOI] [PubMed] [Google Scholar]
- 16. Kleine T., Voigt C., Leister D. (2009) Plastid signalling to the nucleus. Messengers still lost in the mists? Trends Genet. 25, 185–192 [DOI] [PubMed] [Google Scholar]
- 17. Giraud E., Van Aken O., Ho L. H., Whelan J. (2009) The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 150, 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koussevitzky S., Nott A., Mockler T. C., Hong F., Sachetto-Martins G., Surpin M., Lim J., Mittler R., Chory J. (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719 [PubMed] [Google Scholar]
- 19. Ho L. H., Giraud E., Uggalla V., Lister R., Clifton R., Glen A., Thirkettle-Watts D., Van Aken O., Whelan J. (2008) Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in Arabidopsis. Plant Physiol. 147, 1858–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clifton R., Lister R., Parker K. L., Sappl P. G., Elhafez D., Millar A. H., Day D. A., Whelan J. (2005) Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol. Biol. 58, 193–212 [DOI] [PubMed] [Google Scholar]
- 21. Bell C. J., Ecker J. R. (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144 [DOI] [PubMed] [Google Scholar]
- 22. Clough S. J., Bent A. F. (1998) Floral dip. A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 23. Harrison S. J., Mott E. K., Parsley K., Aspinall S., Gray J. C., Cottage A. (2006) A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Katoh K., Misawa K., Kuma K., Miyata T. (2002) MAFFT. A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zmasek C. M., Eddy S. R. (2001) ATV. Display and manipulation of annotated phylogenetic trees. Bioinformatics 17, 383–384 [DOI] [PubMed] [Google Scholar]
- 26. Giraud E., Ho L. H., Clifton R., Carroll A., Estavillo G., Tan Y. F., Howell K. A., Ivanova A., Pogson B. J., Millar A. H., Whelan J. (2008) The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol. 147, 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baldi P., Long A. D. (2001) A Bayesian framework for the analysis of microarray expression data. Regularized t-test and statistical inferences of gene changes. Bioinformatics 17, 509–519 [DOI] [PubMed] [Google Scholar]
- 28. Giraud E., Ng S., Carrie C., Duncan O., Low J., Lee C. P., Van Aken O., Millar A. H., Murcha M., Whelan J. (2010) TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22, 3921–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Geisler-Lee J., O'Toole N., Ammar R., Provart N. J., Millar A. H., Geisler M. (2007) A predicted interactome for Arabidopsis. Plant Physiol. 145, 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin M., Shen X., Chen X. (2011) PAIR. The predicted Arabidopsis interactome resource. Nucleic Acids Res. 39, D1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baena-González E., Rolland F., Thevelein J. M., Sheen J. (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942 [DOI] [PubMed] [Google Scholar]
- 32. Lister R., Carrie C., Duncan O., Ho L. H., Howell K. A., Murcha M. W., Whelan J. (2007) Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell 19, 3739–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Elthon T. E., Nickels R. L., McIntosh L. (1989) Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol. 89, 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carrie C., Kühn K., Murcha M. W., Duncan O., Small I. D., O'Toole N., Whelan J. (2009) Approaches to defining dual-targeted proteins in Arabidopsis. Plant J. 57, 1128–1139 [DOI] [PubMed] [Google Scholar]
- 35. Citovsky V., Gafni Y., Tzfira T. (2008) Localizing protein-protein interactions by bimolecular fluorescence complementation in planta. Methods 45, 196–206 [DOI] [PubMed] [Google Scholar]
- 36. Boyes D. C., Zayed A. M., Ascenzi R., McCaskill A. J., Hoffman N. E., Davis K. R., Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis. A model for high throughput functional genomics in plants. Plant Cell 13, 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang R., Sharkey T. D. (2009) Photosynthetic electron transport and proton flux under moderate heat stress. Photosynth. Res. 100, 29–43 [DOI] [PubMed] [Google Scholar]
- 38. Busso C., Tahara E. B., Ogusucu R., Augusto O., Ferreira-Junior J. R., Tzagoloff A., Kowaltowski A. J., Barros M. H. (2010) Saccharomyces cerevisiae coq10 null mutants are responsive to antimycin A. FEBS J. 277, 4530–4538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mittler R., Vanderauwera S., Suzuki N., Miller G., Tognetti V. B., Vandepoele K., Gollery M., Shulaev V., Van Breusegem F. (2011) ROS signaling. The new wave? Trends Plant Sci. 16, 300–309 [DOI] [PubMed] [Google Scholar]
- 40. Suzuki N., Koussevitzky S., Mittler R., Miller G. (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35, 259–270 [DOI] [PubMed] [Google Scholar]
- 41. Jenkins G. I. (2009) Signal transduction in responses to UV-B radiation. Annu. Rev Plant Biol. 60, 407–431 [DOI] [PubMed] [Google Scholar]
- 42. Van Aken O., Zhang B., Carrie C., Uggalla V., Paynter E., Giraud E., Whelan J. (2009) Defining the mitochondrial stress response in Arabidopsis thaliana. Mol Plant 2, 1310–1324 [DOI] [PubMed] [Google Scholar]
- 43. Shang Y., Yan L., Liu Z. Q., Cao Z., Mei C., Xin Q., Wu F. Q., Wang X. F., Du S. Y., Jiang T., Zhang X. F., Zhao R., Sun H. L., Liu R., Yu Y. T., Zhang D. P. (2010) The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Boruc J., Van den Daele H., Hollunder J., Rombauts S., Mylle E., Hilson P., Inzé D., De Veylder L., Russinova E. (2010) Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network. Plant Cell 22, 1264–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mathur S., Vyas S., Kapoor S., Tyagi A. K. (2011) The mediator complex in plants. Structure, phylogeny and expression profiling of representative genes in a dicot (Arabidopsis thaliana) and a monocot (Oryza sativa) during reproduction and abiotic stress. Plant Physiol. 157, 1609–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang W., Chen X. (2004) HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 131, 3147–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moellering E. R., Benning C. (2010) Phosphate regulation of lipid biosynthesis in Arabidopsis is independent of the mitochondrial outer membrane DGS1 complex. Plant Physiol. 152, 1951–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rhoads D. M., McIntosh L. (1992) Salicylic acid regulation of respiration in higher plants. Alternative oxidase expression. Plant Cell 4, 1131–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Camougrand N., Pélissier P., Velours G., Guérin M. (1995) NCA2, a second nuclear gene required for the control of mitochondrial synthesis of subunits 6 and 8 of ATP synthase in Saccharomyces cerevisiae. J. Mol. Biol. 247, 588–596 [DOI] [PubMed] [Google Scholar]
- 50. Pélissier P., Camougrand N., Velours G., Guérin M. (1995) NCA3, a nuclear gene involved in the mitochondrial expression of subunits 6 and 8 of the Fo-F1 ATP synthase of S. cerevisiae. Curr. Genet. 27, 409–416 [DOI] [PubMed] [Google Scholar]
- 51. Topisirovic I., Svitkin Y. V., Sonenberg N., Shatkin A. J. (2011) Cap and cap-binding proteins in the control of gene expression. RNA 2, 277–298 [DOI] [PubMed] [Google Scholar]
- 52. Le Hir H., Séraphin B. (2008) EJCs at the heart of translational control. Cell 133, 213–216 [DOI] [PubMed] [Google Scholar]
- 53. Baena-González E., Sheen J. (2008) Convergent energy and stress signaling. Trends Plant Sci. 13, 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bitrián M., Roodbarkelari F., Horváth M., Koncz C. (2011) BAC-recombineering for studying plant gene regulation. Developmental control and cellular localization of SnRK1 kinase subunits. Plant J. 65, 829–842 [DOI] [PubMed] [Google Scholar]
- 55. Kuchin S., Treich I., Carlson M. (2000) A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. U.S.A. 97, 7916–7920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sieger S. M., Kristensen B. K., Robson C. A., Amirsadeghi S., Eng E. W., Abdel-Mesih A., Møller I. M., Vanlerberghe G. C. (2005) The role of alternative oxidase in modulating carbon use efficiency and growth during macronutrient stress in tobacco cells. J. Exp. Bot. 56, 1499–1515 [DOI] [PubMed] [Google Scholar]
- 57. Gérin S., Mathy G., Blomme A., Franck F., Sluse F. E. (2010) Plasticity of the mitoproteome to nitrogen sources (nitrate and ammonium) in Chlamydomonas reinhardtii. The logic of Aox1 gene localization. Biochim. Biophys. Acta 1797, 994–1003 [DOI] [PubMed] [Google Scholar]
- 58. Parsons H. L., Yip J. Y., Vanlerberghe G. C. (1999) Increased respiratory restriction during phosphate-limited growth in transgenic tobacco cells lacking alternative oxidase. Plant Physiol. 121, 1309–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]