Abstract
Conventional clinical and pathologic risk factors in stage II colon cancer provide limited prognostic information and do not predict response to adjuvant 5-fluorouracil-based chemotherapy. New prognostic and predictive biomarkers are needed to identify patients with highest recurrence risk who will receive the greatest absolute risk reduction from adjuvant chemotherapy. We review below the evidence for conventional risk factors in node-negative colon cancer patients, followed by a discussion of promising new molecular and genetic markers in this malignancy. Gene expression profiling is an emerging tool with both prognostic and predictive potential in oncology. For stage II colon cancer patients, the Oncotype DX Colon Cancer test is now commercially available as a prognostic marker, and the ColoPrint assay is expected to be released later this year. Current evidence for both of these assays is described below, concluding with a discussion of potential future directions for gene expression profiling in colon cancer risk stratification and treatment decision-making.
Keywords: Stage II colon cancer, gene signature, gene expression profile, biomarker, recurrence
Introduction
Among the estimated 106,100 new cases of colon cancer diagnosed in the United States in 2009, approximately 25%-30% will have node-negative (stage II) disease.1-3 With surgery alone, the overall survival at five years for unselected patients with stage II colon cancer is about 80%.4 Adjuvant chemotherapy offers a minimal incremental benefit tothe average stage II patient, with improvement in survival less than 5% at five years.4,4 In other words, for every one hundred patients with stage II colon cancer exposed to the risks of chemotherapy, eighty would have survived anyway, sixteen would have cancer recurrence regardless of treatment, and only about four patients would derive benefit. To distinguish high from low risk stage II patients, clinical and pathologic features are currently used even though there is minimal prospective data defining the relative impact of these conventional risk factors on an individual patient’s risk of recurrence or likelihood of benefit from chemotherapy.5-14 Better tools are needed to identify the stage II patients at highest risk of recurrence who stand to get the most benefit from adjuvant chemotherapy.
Prognostic factors are characteristics associated with a patient’s clinical outcome, such as likelihood of survival or relapse. For example, in colon cancer, nodal involvement is a poor prognostic factor in that it portends a shorter survival but does not predict for or against response to treatment.5 Predictive factors are characteristics that correlate with likelihood of response to therapy. Borrowing an example from breast cancer, the presence or absence of the estrogen receptor on tumor cells predicts whether hormone therapy will be effective in a given patient; this same factor also is prognostic of improved outcomes, independent of the use of hormonal therapy. In stage II colon cancer, additional factors to identify patients at the highest risk for recurrence (prognostic factors) as well as to predict those most likely to benefit from chemotherapy (predictive factors) are needed to refine the selection of patients for adjuvant chemotherapy.
Gene expression profiling (GEP) enables the screening of thousands of genes in patients with distinct clinical characteristics (such as cancer remission versus recurrence) in order to identify subsets of genes with differential expression between patient groups. This powerful technique is prognostic as well as predictive of treatment response in patients with early-stage breast cancers.15-18 GEP is now under study in early-stage colon cancer patients as a potential means to improve our ability to identify those patients most likely to recur and to predict benefit from adjuvant therapy.
Conventional risk assessment in patients with resected colon cancer is reviewed below. We will then present the current data for GEP as a prognostic factor in this malignancy, focusing upon two new assays for risk assessment in patients with stage II colon cancer, the Oncotype DX Colon Cancer test (Genomic Health, Inc., Redwood City, CA) and ColoPrint (Agendia BV, Amsterdam, The Netherlands). We conclude with a discussion of potential future directions for GEP in colon cancer.
Prognostic Import of Stage in Colon Cancer
Survival rates in colon cancer are strongly influenced by stage at diagnosis, underscoring the prognostic relevance of the American Joint Committee on Cancer (AJCC) staging system in this disease. Although the overall survival at 5 years is 65.2% overall, the differential is dramatic between stages, with five-year survival of 90.8% for localized disease (stages I and II), 69.5% for stage III, and 11.3% for stage IV.1 In the QUASAR study, a large, randomized phase III study of adjuvant chemotherapy in patients with predominantly stage II colon cancer, the overall survival rate at five years was approximately 80% with surgery alone.4 This is corroborated by a meta-analysis of seven adjuvant studies in patients with stages II and III colon cancer randomized to surgery alone or adjuvant fluoropyrimidine therapy.19 In contrast, for patients with stage III disease treated with surgery alone, the overall survival rate at five years is approximately 50%.19-22
Risk Stratification in Treatment Decisions
The mainstay of treatment for stages II and III colon cancers is surgical resection. In stage III colon cancer, postsurgical adjuvant fluoropyrimidine-based chemotherapy has been the standard of care since the 1980s when two landmark studies demonstrated that fluorouracil plus levamisole reduced mortality by approximately 30% in lymph node-positive (stage III) patients.22,23 In 2004, the MOSAIC trial showed that the addition of oxaliplatin to 5-fluorouracil and leucovorin (FOLFOX) as postsurgical adjuvant therapy for stage III patients reduced relapse by comparison with fluorouracil and leucovorin alone with hazard ratio (HR) 0.76 (95% CI 0.62-0.92).14 Based upon the MOSAIC trial, six months of combination chemotherapy with the FOLFOX regimen is now the recommended adjuvant treatment for patients with stage III colon cancer.12
Studies and subset analyses of patients with stage II colon cancer, however, have historically demonstrated minimal (if any) survival or recurrence benefit from the use of postsurgical adjuvant chemotherapy.13,20,22,24,25 Because of the high survival rates in patients treated with surgery alone coupled with modest rates of risk reduction from chemotherapy, it is estimated that large sample sizes (over 8,000 patients) would be required to demonstrate with statistical significance a 2% survival difference between treatment and control arms in this disease.24,26,27 To date, the largest study of stage II colon cancer patients was the randomized QUASAR study (N = 3239 patients with colorectal cancer, 91% with stage II disease, and 2291 with tumors of the colon).4 QUASAR demonstrated a small but significant improvement in overall survival at five years of approximately 3.6% (95% CI 1.0-6.0) with the use of postsurgical adjuvant fluorouracil plus leucovorin by comparison with no adjuvant therapy.
Current consensus guidelines for the adjuvant treatment of stage II colon cancer patients require careful assessment of recurrence risk, based upon the presence of clinical and pathologic risk factors, discussed further below, to guide decisions regarding postsurgical adjuvant therapy. The presence of high risk features is considered an indication to treat with adjuvant chemotherapy following an algorithm similar to that for stage III disease.12 For average risk stage II patients, however, individualized decision-making between patient and physician is recommended given the unclear benefit of postsurgical chemotherapy over observation alone.
Conventional Risk Factors in Stage II Colon Cancer
High risk features included in the current guidelines of the National Comprehensive Cancer Network (NCCN) are listed in Table 1.5-10, 12-13, 19. Incidence and impact information about these risk factors derives from subgroup analyses of the large adjuvant trials in colorectal cancer. In INT-0035, there were 318 stage II colon cancer patients enrolled; among these, perforation was present in 6%, obstruction in approximately 16%, tumor T4 stage in approximately 18%, and poorly differentiated histology in approximately 11%.13 Retrospective analysis of the pooled data from the 403 stage II patients in INT-0035 and the NCCTG adjuvant trial showed perforation and obstruction to be significantly associated with increased recurrence risk, though only perforation was independently prognostic in multivariate analysis (P ≤ .01).13 In MOSAIC, 576 of the 899 stage II colon cancer patients included in the study were classified as “high risk” based upon the presence of one or more high risk features; these patients demonstrated shorted disease free survival than low risk stage II patients in both treatment arms.28 A preliminary subset analysis of conventional risk factors in 1436 patients with stage II colon cancer enrolled in the QUASAR study was presented at the American Society of Clinical Oncology (ASCO) Annual Meeting in 2009.29 Among these patients, T4 tumors and fewer than 12 lymph nodes sampled were features associated with significantly increased recurrence risk, with HRs of 1.83 (P = .005) and 1.47 (P = .040), respectively. Lymphovascular invasion was not significantly associated with recurrence, while high tumor grade was significantly associated with a lower risk of recurrence (HR 0.62, P = .026), counter to standard definitions of risk. Mismatch repair protein deficiency, which is associated with microsatellite instability (MSI), was also associated with significantly lower risk of recurrence (HR 0.32, P < .001) and is discussed further below. See Table 2 for the relative impacts of different high risk features in subset analyses of stage II patients in the QUASAR study.29
Table 1. Stage II Colon Cancer High Risk Features in NCCN Guidelines1.
| Bowel obstruction | Grade 3-4 histology |
|---|---|
| T3 tumors with localized perforation |
Lymphatic or vascular invasion |
| T4 tumors (penetrating to surface of visceral> peritoneum or directly invading or adhering to other organs or structures) |
Close, indeterminate or positive margins |
| < 12 lymph nodes examined |
NCCN Practice Guidelines in Oncology–v.2.2010 Colon Cancer, National Comprehensive Cancer Network, 2010
Table 2. Impact of Conventional High Risk Factors in Stage II Colon Cancer Patients in QUASAR1.
| Risk Factor (Incidence) | Hazard Ratio for Recurrence at 3 Years (95% CI) |
P value |
|---|---|---|
| Mismatch repair deficiency (13%) |
0.32 (0.15, 0.69) | < 0.001 |
| T 4 tumor stage (15%) | 1.83 (1.23, 2.75) | 0.005 |
| High grade tumor (29%) | 0.62 (0.40, 0.96) | 0.026 |
| < 12 lymph nodes examined (62%) |
1.47 (1.01, 2.14) | 0.040 |
| Lymphovascular invasion (13%) | 1.40 (0.88, 2.23) | 0.175 |
(Is permission needed to present this data? It was an oral presentation at ASCO 2009, available online if one has an ASCO membership.)
Kerr D, Gray R, Quirke P, Watson D, Yothers G, et al.: A quantitative multigene RT-PCR assay for prediction of recurrence in stage II colon cancer: Selection of the genes in four large studies and results of the independent, prospectively designed QUASAR validation study, American Society of Clinical Oncology Annual Meeting. Orlando, FL, J Clin Oncol, 2009
It is noteworthy that these conventional prognostic factors for increased recurrence risk have not been validated as predictive factors for chemotherapy benefit. In stage II patients with high risk criteria included in the MOSAIC trial, disease-free survival at five years was 82.1% with the use of FOLFOX by comparison with 74.9% with fluorouracil plus leucovorin, a nonsignificant finding with HR 0.74 and 95% CI 0.52-1.06.28 Low risk patients received no recurrence or survival benefit with the use of FOLFOX by comparison with fluorouracil plus leucovorin alone. Among the 403 stage II patients in pooled analysis of the Intergroup 0035 (INT-0035) and the North Central Cancer Treatment Group (NCCTG) trials, there was no significant interaction with treatment benefit for any of the prognostic factors evaluated.13
Ultimately, these conventional clinical and pathologic risk factors in stage II colon cancer do not predict response to therapy and do not clearly discriminate between high and low risk stage II patients. With a high likelihood of cure after surgery alone contrasting with the poor prognosis of those patients who do experience recurrence, better prognostic and predictive tools are in great need.
Molecular and Genetic Markers in Colorectal Cancer
Multiple molecular and genetic markers have been studied in colon cancer patients with the goal of better characterizing stage II and III patients’ risk of recurrence and likelihood of benefit from chemotherapy.30,31 Among these many markers, however, only a small subset demonstrates clear prognostic significance, with even fewer markers proving to be predictive of treatment response. A selection of the markers with the greatest level of evidence is presented below.
Microsatellite instability (MSI) refers to mutations within DNA microsatellites, repetitive DNA sequences of one to six base pairs located throughout the genome.30,32 These mutations are due to deficiency in one or more mismatch repair proteins, sporadic or inherited, and occur with high frequency (MSI-high) in approximately 15% of colorectal cancer.33,34 Tumors from patients with the Lynch syndrome (hereditary nonpolyposis colorectal cancer) demonstrate MSI due to germline mutations in one or more mismatch repair genes, most commonly MLH1, MSH2, MSH6, and PMS2.35 In contrast, tumors with sporadic MSI exhibit deficient mismatch repair primarily due to hypermethylation of a mismatch repair gene promoter.36 Multiple studies suggest that tumors with microsatellite instability are associated with significantly better overall survival than microsatellite-stable tumors.29-31,34,37-46 In a meta-analysis of 32 studies, the HR for survival was 0.65 (95% CI 0.59-0.71) for tumors with microsatellite instability.34 Interestingly, pooled analyses also suggest that, in addition to being a positive prognostic factor as above, MSI may be a predictive marker for lower responsive rates to fluorouracil-based chemotherapy.42,43 Other studies, however, offer conflicting data.47-49
In contrast to MSI, chromosomal instability (CIN) refers to structural chromosomal abnormalities or abnormalities in chromosome number within tumor cells, often suggested by the finding of aneuploidy or polyploidy.50 CIN is present in approximately 65%-70% of colorectal cancers and is associated with a worse prognosis.31,50 Meta-analysis data from 63 studies (N = 10,126 patients) showed that CIN conferred a HR for overall survival of 1.45 (P < .001).50 Assessment of CIN is hampered by a lack of consistent methodology. Flow cytometry is the most common technique used to identify CIN but is unable to identify the specific chromosomal abnormalities present. Deletion or loss of heterozygosity (LOH) of chromosome 18q is a chromosomal abnormality frequently associated with CIN and is the most common known cytogenetic abnormality in colorectal cancer, identified in approximately 70% of colorectal cancer in multiple studies.31,38,51 Although individual studies and meta-analysis data have suggested that tumor LOH at 18q may be associated with poor prognosis, this finding is not universal and may differ between stage II and stage III colon cancer patients.38,46,51,52 The Eastern Cooperative Oncology Group (ECOG) trial 5202 is currently underway to prospectively evaluate the benefit of adjuvant chemotherapy in patients with LOH at 18q. Figure 1[FS1] displays the ECOG 5202 study schema.
Figure 1.
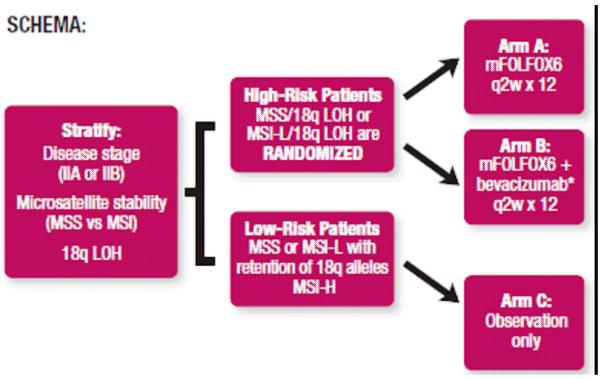
ECOG 5202 Study Schema (Is permission needed? This is from the ECOG flyer for the trial.)
Recently, mutation of the Kirsten ras gene (KRAS) has been validated as a predictive factor for nonresponse to therapies directed at the epidermal-growth factor receptor (EGFR) in patients with metastatic colorectal cancer.53-55. KRAS mutation, present in 30%-40% of tumors, does not appear to be an independent negative prognostic factor, in that patients in the control arms of studies of EGFR inhibitors in colorectal cancer demonstrate similar outcomes regardless of KRAS mutational status.53-55 Interestingly, the V600E mutation in BRAF, another gene in the KRAS signal transduction pathway, is also associated with nonresponse to EGFR-targeted therapy based upon early data.56-58 Subset analyses from the CAIRO-2, CRYSTAL, and PETACC-3 trials suggest that the BRAF V600E mutation, unlike KRAS mutation, is associated with poor prognosis which may confound interpretation of its predictive value.59-61 BRAF mutations are present in approximately 5% to 10% of colorectal tumors and appear to be mutually exclusive with KRAS mutations.56,59,62,63 BRAF mutation may be more common in patients with the CpG island methylator phenotype (CIMP), which itself has been associated with a positive prognosis, highlighting the complexity of these molecular pathways.64
Gene Expression Profiling as a Prognostic and Predictive Tool
Gene expression profiling (GEP) is an emerging tool. This methodology attempts to identify differentially expressed subsets of genes (gene signatures) in groups of patients with distinct clinical outcomes. In the example of breast cancer, a reverse-transcriptase-polymerase-chain-reaction (RT-PCR) assay is now commercially available (Oncotype DX, Genomic Health Inc., Redwood City, CA) to refine assessment of recurrence risk in lymph node-negative, estrogen receptor-positive breast cancer patients.15,65,66 The Oncotype DX assay generates a 21-gene signature from paraffin-embedded tumor specimens. In a study evaluating the prognostic role of this gene signature, 668 tumor blocks from patients enrolled in National Surgical Adjuvant Breast and Bowel Project (NSABP) trial B-14 were analyzed, with 51% being identified as low risk, 22% intermediate risk, and 27% high risk for recurrence, based upon reference-normalized expression measurements from selected cancer-related genes in the signature.15 A low recurrence score was associated with recurrence rate of 6.8% at 10 years, by comparison with 14.3% and 30.5% in the intermediate and low recurrence score groups, respectively. A retrospective case-control study of 4,964 Kaiser Permanente patients demonstrated a significant association between the Oncotype DX 21-gene recurrence score and breast cancer-associated death.67 The 21-gene signature also was predictive of extent of chemotherapy benefit when retrospectively applied to 651 patients with lymph node-negative, estrogen receptor-positive breast cancer randomized to tamoxifen versus tamoxifen plus adjuvant chemotherapy in the NSABP B-20 trial.16 In these patients, a high recurrence score was associated with a mean absolute decrease in 10-year distant recurrence rate of 27.6% with the use of chemotherapy (relative risk 0.26, 95% CI 0.13-0.53), while those patients with a low recurrence score did not appear to derive any reduction in recurrence risk. The test for interaction between chemotherapy treatment and recurrence score was significant (P = .038).
Consensus guidelines now include the use of Oncotype DX for risk stratification and chemotherapy decision-making in lymph node-negative, estrogen-receptor positive breast cancer patients.65,66 Another commercially-available assay, MammaPrint (Agendia BV, Amsterdam, the Netherlands), uses DNA microarray analysis of frozen tissue specimens to identify a 70-gene signature for risk stratification in lymph-node negative patients with breast cancer.17,18,65,68 This 70-gene signature has demonstrated prognostic value independent of conventional clinicopathologic features but has not yet been validated as a predictive marker for chemotherapy benefit. Two large, randomized trials, TAILORx and MINDACT, are ongoing to prospectively evaluate the prognostic and predictive value of Oncotype DX and MammaPrint, respectively, in lymph node-negative breast cancer patients.
Gene Expression Profiling in Colon Cancer
In colon cancer, multiple small studies have evaluated gene expression profiles derived from RT-PCR or DNA microarray analysis for potential prognostic value.63,69-80 Interpretation of these individual studies is limited by small sample sizes, heterogeneous patient populations, unclear significance of the genes included in each signature, and minimal (if any) external validation. Furthermore, the prognostic value of GEP has not been carefully compared with that of the conventional clinical and pathologic risk factors discussed previously.81 Despite these limitations, a meta-analysis of studies of various gene expression assays in eight cohorts of patients with stage II colorectal cancer (N = 271 patients overall) showed a likelihood ratio of 4.7 (95% CI 3.2-6.8) for endpoints of either recurrence or death within three years.82 Cumulatively, these early studies and meta-analysis data suggest promise for GEP as a means to discriminate recurrence risk in patients with early-stage colon cancer, though minimal data exists to demonstrate any predictive value of GEP for chemotherapy response in colon cancer to date.83
Two new gene expression signatures have recently shown promise as potential prognostic markers for recurrence risk in stage II and III colon cancers.29,84 Kerr et al presented results of a quantitative, multigene RT-PCR assay (the Oncotype DX Colon Cancer test.) using a validation set of 1,436 tumor specimens from stage II colon cancer patients in the QUASAR study.29 Glas et al presented results of a DNA microarray gene signature (ColoPrint) developed in 180 patients with stages I, II, and III colorectal cancer with a validation set of 178 stage II and III colon cancer samples.84 These tests are discussed in further detail below and summarized in Table 3.
Table 3. Features of the Oncotype DX Colon Cancer Test and ColoPrint Assay.
| Oncotype DX Colon Cancer Test1 |
ColoPrint2 | |
|---|---|---|
| Tissue specimen required |
Formalin-fixed, paraffin- embedded tumor specimens |
Fresh frozen tumor specimens |
| Type of assay | RT-PCR | High density Agilent 44K oligonucleotide array |
| Validation Set | Stage II colon cancer patients in QUASAR trial |
|
| Primary Endpoint | Recurrence risk (RR) at 3 years |
Distant-metastasis- free survival (DMFS) |
| Risk groups (incidence) |
Low (43.7% of patients) | Low (61% of patients) |
| Intermediate (30.7% of patients) |
High (39% of patients) | |
| High (25.6% of patients) | ||
| Risk according to group |
Low risk: 12% RR at 3 years | Low risk: 89% DMFS at 5 years |
| Intermediate risk: 18% RR at 3 years |
High risk: 62% DMFS at 5 years |
|
| High: 22% RR at 3 years |
Kerr D, Gray R, Quirke P, Watson D, Yothers G, et al.: A quantitative multigene RT-PCR assay for prediction of recurrence in stage II colon cancer: Selection of the genes in four large studies and results of the independent, prospectively designed QUASAR validation study, American Society of Clinical Oncology Annual Meeting. Orlando, FL, J Clin Oncol, 2009
Glas AM RP, Salazar R, Capella G, Moreno V, et al.: Development and validation of a robust prognostic and predictive signature for colorectal cancer (CRC) patients., American Society of Clinical Oncology Annual Meeting. Orlando, FL, J Clin Oncol, 2009
Oncotype DX Colon Cancer Test
Following upon their development of Oncotype DX for breast cancer patients, researchers at Genomic Health, Inc. selected 761 genes with potential significance in colon cancer.29 Candidate gene expression was assayed by RT-PCR in a development set of fixed, paraffin-embedded tissue samples from patients with surgically resected colorectal cancers from one of four adjuvant studies: NSABP C-01/C-02 (N = 270), Cleveland Clinic Foundation (N = 765), NSABP C-04 (N = 308), and NSABP C-06 (N = 508). Based upon the differential gene expression in these 1,851 patient tumor samples, 48 genes significantly associated with recurrence risk and 66 genes significantly associated with treatment benefit were identified. The final assay, branded as the Oncotype DX Colon Cancer test, was developed using the 7 genes most strongly associated with recurrence, the 6 genes most strongly identified with treatment benefit, and 5 reference genes for standardization.
The resulting 7-gene recurrence score (0-100) and 6-gene treatment score (0-100) were then applied to tissue blocks from patients enrolled in the landmark QUASAR study as a clinical validation set.4 Among the 3,239 patients enrolled in QUASAR overall, evaluable tissue blocks were available from 1,436 patients with confirmed stage II colon cancer (excluding patients with rectal cancer), of whom 711 were treated in the surgery alone arm and 725 in the surgery plus chemotherapy arm. Conventional risk factors were balanced between the two groups. It is noteworthy that fewer than 12 lymph nodes were sampled in over 60% of the study population overall, a conventional high risk feature.6-9
Among the 711 patients enrolled in the surgery-alone arm of the study, the 7-gene recurrence score was significantly associated with recurrence risk at three years after surgery (P = .004). Based upon the recurrence score, 43.7% of patients were classified as low risk (score < 30), 30.7% as intermediate risk (score 30-40), and 25.6% as high risk (score ≥ 41). Within each group, Kaplan-Meier estimates of recurrence at three years were 12%, 18%, and 22%, respectively; the HR for recurrence between the low and high risk groups was 1.47 (P = .046) using the Cox model.
In multivariate analyses, the recurrence score retained prognostic significance independent of conventional prognostic factors including mismatch repair status, tumor T stage, number of lymph nodes examined, grade, and presence of lymphovascular invasion. Tumor T4 stage also was an independent negative prognostic factor (HR 1.83, P = .005), while mismatch repair protein deficiency was an independent positive prognostic factor (HR 0.32, P < .001). One of these two findings was present in approximately 25% of patients.
Of note, neither the treatment score nor the recurrence score was predictive of treatment benefit in the 725 patients treated with fluorouracil and leucovorin. There are no published studies evaluating the predictive value of the Oncotype DX Colon Cancer test recurrence or treatment scores in patients treated with FOLFOX. Preliminary data presented at the Gastrointestinal Cancers Symposium in 2010 showed similar gene expression and recurrence scores between stage II and III patients in the development sets for the Oncotype DX Colon Cancer test, though there was no clear interaction between the recurrence score and stage.85 The Oncotype DX Colon Cancer test was released for commercial use in January of 2010.
ColoPrint
Another GEP assay expected to be available within the year is ColoPrint, a 38-gene signature developed by Agendia, following upon its predecessor microarray assay in breast cancer, MammaPrint.86 The ColoPrint signature was developed using a training set of fresh frozen tumor specimens from 180 patients with stages I, II, and III colorectal cancer, using high density Agilent 44K oligonucleotide arrays.84 The development studies identified three main molecular subclasses with good, intermediate, and poor prognosis. The gene signature from the 104 patients characterized as intermediate risk was then analyzed with cross validation for association with five-year distant metastasis free survival (DMFS). The resulting signature was applied to a validation set of 178 tumor samples from patients with stages II and III colon cancer. A set of 38 genes were significantly associated with prognosis; the specific genes identified have yet to be presented. According to preliminary results presented at the ASCO Annual Meeting in 2009 and the Gastrointestinal Cancers Symposium in 2010, the 38-gene signature classified 61% of samples as low risk and 39% as high risk, with a HR for DMFS of 3.19 between groups.84,87 DMFS at five years was 89% in the low risk group and 62% in the high risk group. The signature retained prognostic significance when applied separately to stage II and stage III samples, as well as to patients treated with or without adjuvant therapy. The majority of patients with microsatellite instability were classified as low-risk by this assay.87
Current Limitations of GEP in Colon Cancer
Will recurrence scores yield useful prognostic information for the majority of patients?
In the QUASAR subset of evaluable stage II colon patients treated with surgery alone, the Oncotype DX Colon Cancer test distinguished patients as high, intermediate, and low risk based upon recurrence risk at three years.29 High risk patients had a 22% chance of recurrence, by comparison with 12% in the low risk patients, as described above. This represents a much smaller degree of discrimination between risk groups by comparison with the test in breast cancer patients, in whom high risk corresponds with a 30.5% risk of recurrence at ten years, by comparison with 6.8% in low risk patients.15 The presence of either T4 tumor stage or mismatch repair protein deficiency (one of which was present in approximately 25% of patients in this study) generally outweighed the prognostic significance of the Oncotype DX Colon Cancer test recurrence score, also limiting the number of patients for whom this assay will provide useful prognostic information. Longitudinal data will be required to determine whether the recurrence score maintains prognostic value over a longer follow up period.
Although the preliminary results of the ColoPrint gene signature validation set initially suggest a greater ability to discriminate risk, the findings are confounded by inclusion of both stage II and stage III patients. In the validation set of 178 samples, 61% of patients were assessed as low risk and 39% as high risk, corresponding to recurrence rates of 11% and 38% at five years, respectively, as above.84 The authors state that the profile was significant within stage II and stage III patients (with P = .0058 and P = .036, respectively) but do not provide stage-specific recurrence rates for each risk group, nor any information about distribution of stage II and III patents within each risk group. Thus, it is unknown at present whether ColoPrint provides relevant prognostic information beyond standard tumor, node, and metastasis (TNM) staging. Its performance will require validation in larger samples with stratification by TNM stage. Agendia is now conducting the Prospective Study for the Assessment of Recurrence Risk in Stage II Colon Cancer (PARSC) to better define the performance of ColoPrint in stage II patients.86
It is important to note that both of these assays have been validated in colon cancer patients only, and do not apply to rectal cancer. It is unknown whether these gene signatures are universal across races and ages.
Does lack of predictive value limit application?
While preliminary results suggest that both the Oncotype DX Colon and ColoPrint gene signatures may provide prognostic information about recurrence risk, neither assay has demonstrated the ability to predict which patients will benefit from adjuvant chemotherapy, a primary goal of personalized medicine in oncology. Unlike the Oncotype DX breast cancer assay which does predict which patients will benefit from adjuvant chemotherapy, the Oncotype DX Colon Cancer test did not predict treatment benefit from fluorouracil and leucovorin in the QUASAR stage II colon cancer patients as discussed above.16,29 Data regarding predictive value of the ColoPrint assay has not been presented.
The absence of predictive value of the Oncotype DX Colon gene signature in stage II colon cancer patients treated with adjuvant fluorouracil and leucovorin raises several interesting questions. The findings may be confounded by the presence of other high risk features such as inadequate lymph node sampling, which was present in over 60% of patients in both arms of the QUASAR study. It may be worthwhile to analyze subset results from conventionally low risk patients in QUASAR separately. Another question that surfaces pertains to the limited efficacy of fluorouracil monotherapy: Would the recurrence score or treatment score be predictive of benefit from FOLFOX in patients assessed as high risk by this assay?
Ultimately, although Oncotype DX Colon does not predict which patients will derive disproportionate chemotherapy benefit, the relative benefit from fluorouracil and leucovorin chemotherapy is proportional across risk groups. Therefore, the patients at highest risk of recurrence derive the greatest absolute risk reduction with the use of chemotherapy. Identifying these patients is a very worthwhile outcome.
Are these tests feasible Outside of clinical trials?
The Oncotype DX Colon Cancer test requires fixed, paraffin-embedded tumor tissue, specimens which are generally readily available from patients with resected colon cancers. Genomic Health’s predecessor RT-PCR assay, Oncotype DX for breast cancer, has been widely adopted, demonstrating the feasibility of this approach. In contrast, however, ColoPrint requires fresh frozen tissue for its microarray assay, a requirement which may limit uptake of this assay. Cost effectiveness analysis for these tests has yet to be performed.
Conclusions
Gene expression profiling may provide prognostic value independent of conventional clinical and pathologic risk factors in stage II colon cancer, a disease in which the overall favorable prognosis after surgery alone conflicts with the poor prognosis of the subset who do experience recurrence. Given the marginal benefit of chemotherapy in unselected patients with stage II colon cancer, the ideal test would identify the patients most likely to benefit from adjuvant treatment – a so-called predictive test. In the absence of predictive tests, however, a test which better refines prognosis is critical to more clearly identify patients who will derive the greatest absolute reduction in recurrence risk, even though the relative reduction may be uniform across risk groups.
Preliminary results of the Oncotype DX Colon Cancer test recurrence score in the large, randomized, QUASAR validation set are provocative, suggesting independent prognostic value superior to the majority of conventional risk factors currently used to determine which stage II colon cancer patients receive adjuvant chemotherapy. If confirmed in the final, peer-reviewed publication of this study, these results support further investigation of this gene signature as a means to improve risk stratification in stage II patients. How best to integrate the Oncotype DX Colon Cancer test recurrence score results with conventional risk factors, however, remains unknown. In the QUASAR validation set for this assay, the recurrence score along with tumor T4 stage and mismatch repair protein deficiency were the strongest independent prognostic factors in multivariate analysis; inadequate lymph node sampling was also a significant poor prognostic factor, while high grade of tumor, paradoxically, was a positive prognostic factor, perhaps due to its strong association with mismatch repair protein deficiency. Based upon the results of this single study, a combination of Oncotype DX Colon Cancer test recurrence score, tumor T4 stage, the number of lymph nodes sampled, and mismatch repair status emerges as a potential prognostic algorithm for stage II colon cancer patients. This combined testing algorithm would, of course, require validation in an external data set to demonstrate superiority to conventional risk stratification alone. For example, it would be of great interest to test this algorithm in specimens obtained from patients in the ongoing ECOG 5202 trial for whom tumor T stage, nodal status, and mismatch repair protein testing are readily available.
Although prediction of chemotherapy benefit is the ultimate goal of personalized cancer care, the lack of predictive value of GEP in colon cancer thus far should not devalue the prognostic significance of these tests. It is important to note that there is minimal (if any) evidence for standard clinical and pathologic features as predictors of chemotherapy benefit, yet these features are widely used in stage II colon cancer treatment algorithms. Moreover, unlike the majority of conventional risk factors, GEP is a dynamic tool with significant, untapped potential. Ultimately, the significant association between these gene signatures and recurrence risk offers an important proof of principle supporting the ongoing study of gene expression profiling as a means to personalize colon cancer care.
Acknowledgments
This study was supported in part by a grant from the National Cancer Institute (P01CA130818) through the Center for Translational and Policy Research on Personalized Medicine (TRANSPERS) at the University of California, San Francisco.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
Dr. Kelley has received an honorarium for speaker training from Genomic Health, Inc. Dr. Venook has received an honorarium from and served on the advisory board to Genomic Health, Inc.
References
- 1.Horner MJRL, Krapcho M, Neyman N, et al. SEER Cancer Statistics Review, 1975-2006. National Cancer Institute; 2009. FS2. [Google Scholar]
- 2.Cancer in California . Department of Health Services; State of California: Dec, 2005. 1988-2002. 2005.[FS3] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 4.Quasar Collaborative G. Gray R, Barnwell J, et al. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370:2020–9. doi: 10.1016/S0140-6736(07)61866-2. [FS4][FS5] [DOI] [PubMed] [Google Scholar]
- 5.Compton CC, Fielding LP, Burgart LJ, et al. Prognostic factors in colorectal cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med. 2000;124:979–94. doi: 10.5858/2000-124-0979-PFICC. [DOI] [PubMed] [Google Scholar]
- 6.Bilimoria KY, Palis B, Stewart AK, et al. Impact of tumor location on nodal evaluation for colon cancer. Dis Colon Rectum. 2008;51:154–61. doi: 10.1007/s10350-007-9114-2. [DOI] [PubMed] [Google Scholar]
- 7.Johnson PM, Porter GA, Ricciardi R, Baxter NN. Increasing negative lymph node count is independently associated with improved long-term survival in stage IIIB and IIIC colon cancer. J Clin Oncol. 2006;24:3570–5. doi: 10.1200/JCO.2006.06.8866. [DOI] [PubMed] [Google Scholar]
- 8.Wong SL, Ji H, Hollenbeck BK, Morris AM, Baser O, Birkmeyer JD. Hospital lymph node examination rates and survival after resection for colon cancer. JAMA. 2007;298:2149–54. doi: 10.1001/jama.298.18.2149. [DOI] [PubMed] [Google Scholar]
- 9.Sarli L, Bader G, Iusco D, et al. Number of lymph nodes examined and prognosis of TNM stage II colorectal cancer. Eur J Cancer. 2005;41:272–9. doi: 10.1016/j.ejca.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Steinberg SM, Barkin JS, Kaplan RS, Stablein DM. Prognostic indicators of colon tumors. The Gastrointestinal Tumor Study Group experience. Cancer. 1986;57:1866–70. doi: 10.1002/1097-0142(19860501)57:9<1866::aid-cncr2820570928>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 11.Wanebo HJ, Rao B, Pinsky CM, et al. Preoperative carcinoembryonic antigen level as a prognostic indicator in colorectal cancer. N Engl J Med. 1978;299:448–51. doi: 10.1056/NEJM197808312990904. [DOI] [PubMed] [Google Scholar]
- 12.Oncology–v.2.2009 Colon Cancer. National Comprehensive Cancer Network; [Accessed: July 10, 2009]. 2009. Practice Guidelines NCCN. 2009, at http://www.nccn.org/professionals/physician_gls/PDF/colon.pdf. [Google Scholar]
- 13.Moertel CG, Fleming TR, Macdonald JS, et al. Intergroup study of fluorouracil plus levamisole as adjuvant therapy for stage II/Dukes’ B2 colon cancer. J Clin Oncol. 1995;13:2936–43. doi: 10.1200/JCO.1995.13.12.2936. [DOI] [PubMed] [Google Scholar]
- 14.Andre T, Boni C, Mounedji-Boudiaf L, et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med. 2004;350:2343–51. doi: 10.1056/NEJMoa032709. [DOI] [PubMed] [Google Scholar]
- 15.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 16.Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–34. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 17.van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 18.van ’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 19.Gill S, Loprinzi CL, Sargent DJ, et al. Pooled analysis of fluorouracil-based adjuvant therapy for stage II and III colon cancer: who benefits and by how much? J Clin Oncol. 2004;22:1797–806. doi: 10.1200/JCO.2004.09.059. [DOI] [PubMed] [Google Scholar]
- 20.Douillard JY, Bennouna J. Adjuvant chemotherapy for colon cancer: a confusing area! Ann Oncol. 2005;16:1853–4. doi: 10.1093/annonc/mdi413. [DOI] [PubMed] [Google Scholar]
- 21.Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476–87. doi: 10.1056/NEJMra040958. [DOI] [PubMed] [Google Scholar]
- 22.Moertel CG, Fleming TR, Macdonald JS, et al. Levamisole and fluorouracil for adjuvant therapy of resected colon carcinoma. N Engl J Med. 1990;322:352–8. doi: 10.1056/NEJM199002083220602. [DOI] [PubMed] [Google Scholar]
- 23.Laurie JA, Moertel CG, Fleming TR, et al. The North Central Cancer Treatment Group and the Mayo Clinic Surgical adjuvant therapy of large-bowel carcinoma: an evaluation of levamisole and the combination of levamisole and fluorouracil. J Clin Oncol. 1989;7:1447–56. doi: 10.1200/JCO.1989.7.10.1447. [DOI] [PubMed] [Google Scholar]
- 24.Wolpin BM, Meyerhardt JA, Mamon HJ, Mayer RJ. Adjuvant treatment of colorectal cancer. CA Cancer J Clin. 2007;57:168–85. doi: 10.3322/canjclin.57.3.168. [DOI] [PubMed] [Google Scholar]
- 25.Schippinger W, Samonigg H, Schaberl-Moser R, et al. A prospective randomised phase III trial of adjuvant chemotherapy with 5-fluorouracil and leucovorin in patients with stage II colon cancer. Br J Cancer. 2007;97:1021–7. doi: 10.1038/sj.bjc.6604011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benson AB, 3rd, Schrag D, Somerfield MR, et al. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22:3408–19. doi: 10.1200/JCO.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 27.Buyse M, Piedbois P. Should Dukes’ B patients receive adjuvant therapy? A statistical perspective. Semin Oncol. 2001;28:20–4. doi: 10.1016/s0093-7754(01)90247-7. [DOI] [PubMed] [Google Scholar]
- 28.de Gramont ABC, Navarro M, Tabernero J, et al. Oxaliplatin/5FU/LV in adjuvant colon cancer: Updated efficacy results of the MOSAIC trial, including survival, with a median follow-up of six years. J Clin Oncol; American Society of Clinical Oncology Annual Meeting; Chicago, IL. 2007; 2007. FS6. [Google Scholar]
- 29.Kerr D, Gray R, Quirke P, et al. A quantitative multigene RT-PCR assay for prediction of recurrence in stage II colon cancer: Selection of the genes in four large studies and results of the independent, prospectively designed QUASAR validation study. J Clin Oncol; American Society of Clinical Oncology Annual Meeting; Orlando, FL. 2009.2009. [Google Scholar]
- 30.Shankaran V, Wisinski KB, Mulcahy MF, Benson AB., 3rd The role of molecular markers in predicting response to therapy in patients with colorectal cancer. Mol Diagn Ther. 2008;12:87–98. doi: 10.1007/BF03256274. [DOI] [PubMed] [Google Scholar]
- 31.Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9:489–99. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 32.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–61. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 33.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57. [PubMed] [Google Scholar]
- 34.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–18. doi: 10.1200/JCO.2005.01.086. FS7. [DOI] [PubMed] [Google Scholar]
- 35.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–32. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 36.Bettstetter M, Dechant S, Ruemmele P, et al. Distinction of hereditary nonpolyposis colorectal cancer and sporadic microsatellite-unstable colorectal cancer through quantification of MLH1 methylation by real-time PCR. Clin Cancer Res. 2007;13:3221–8. doi: 10.1158/1078-0432.CCR-06-3064. [DOI] [PubMed] [Google Scholar]
- 37.French AJ, Sargent DJ, Burgart LJ, et al. Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin Cancer Res. 2008;14:3408–15. doi: 10.1158/1078-0432.CCR-07-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halling KC, French AJ, McDonnell SK, et al. Microsatellite instability and 8p allelic imbalance in stage B2 and C colorectal cancers. J Natl Cancer Inst. 1999;91:1295–303. doi: 10.1093/jnci/91.15.1295. [DOI] [PubMed] [Google Scholar]
- 39.Hemminki A, Mecklin JP, Jarvinen H, Aaltonen LA, Joensuu H. Microsatellite instability is a favorable prognostic indicator in patients with colorectal cancer receiving chemotherapy. Gastroenterology. 2000;119:921–8. doi: 10.1053/gast.2000.18161. [DOI] [PubMed] [Google Scholar]
- 40.Jensen SA, Vainer B, Kruhoffer M, Sorensen JB. Microsatellite instability in colorectal cancer and association with thymidylate synthase and dihydropyrimidine dehydrogenase expression. BMC Cancer. 2009;9:25. doi: 10.1186/1471-2407-9-25. FS8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanza G, Gafa R, Santini A, Maestri I, Guerzoni L, Cavazzini L. Immunohistochemical test for MLH1 and MSH2 expression predicts clinical outcome in stage II and III colorectal cancer patients. J Clin Oncol. 2006;24:2359–67. doi: 10.1200/JCO.2005.03.2433. [DOI] [PubMed] [Google Scholar]
- 42.Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–57. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sargent DJMS, Thibodeau SN, Labianca R, et al. Confirmation of deficient mismatch repair (dMMR) as a predictive marker for lack of benefit from 5-FU based chemotherapy in stage II and III colon cancer (CC): A pooled molecular reanalysis of randomized chemotherapy trials. J Clin Oncol; American Society of Clinical Oncology Annual Meeting; Chicago. 2008.2008. [Google Scholar]
- 44.Sinicrope FA, Rego RL, Foster N, et al. Microsatellite instability accounts for tumor site-related differences in clinicopathologic variables and prognosis in human colon cancers. Am J Gastroenterol. 2006;101:2818–25. doi: 10.1111/j.1572-0241.2006.00845.x. [DOI] [PubMed] [Google Scholar]
- 45.Sinicrope FA, Rego RL, Halling KC, et al. Prognostic impact of microsatellite instability and DNA ploidy in human colon carcinoma patients. Gastroenterology. 2006;131:729–37. doi: 10.1053/j.gastro.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe T, Wu TT, Catalano PJ, et al. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med. 2001;344:1196–206. doi: 10.1056/NEJM200104193441603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim GP, Colangelo LH, Wieand HS, et al. Prognostic and predictive roles of high-degree microsatellite instability in colon cancer: a National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project Collaborative Study. J Clin Oncol. 2007;25:767–72. doi: 10.1200/JCO.2006.05.8172. [DOI] [PubMed] [Google Scholar]
- 48.Tejpar SBF, Delorenzi M, Fiocca R, et al. Microsatellite instability (MSI) in stage II and III colon cancer treated with 5FU-LV or 5FU-LV and irinotecan (PETACC 3-EORTC 40993-SAKK 60/00 trial). J Clin Oncol; American Society of Clinical Oncology Annual Meeting; Orlando, FL. 2009; 2009. [DOI] [PubMed] [Google Scholar]
- 49.Bertagnolli MM, Niedzwiecki D, Compton CC, et al. Microsatellite instability predicts improved response to adjuvant therapy with irinotecan, fluorouracil, and leucovorin in stage III colon cancer: Cancer and Leukemia Group B Protocol 89803. J Clin Oncol. 2009;27:1814–21. doi: 10.1200/JCO.2008.18.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walther A, Houlston R, Tomlinson I. Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut. 2008;57:941–50. doi: 10.1136/gut.2007.135004. [DOI] [PubMed] [Google Scholar]
- 51.Popat S, Houlston RS. A systematic review and meta-analysis of the relationship between chromosome 18q genotype, DCC status and colorectal cancer prognosis. Eur J Cancer. 2005;41:2060–70. doi: 10.1016/j.ejca.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 52.Roth ADTS, Yan P, Fiocca R, et al. Correlation of molecular markers in colon cancer with stage-specific prognosis: Results of the translational study on the PETACC 3 - EORTC 40993-SAKK 60-00 trial; Gastrointestinal Cancers Symposium; San Francisco, CA. 2009.2009. [Google Scholar]
- 53.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–34. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 54.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 55.van Cutsem ELI, D’Haens G, Moiseyenko V, et al. KRAS status and efficacy in the first-line treatment of patients with metastatic colorectal cancer treated with FOLFIRI with or without cetuximab: The CRYSTAL experience. Journal of Clinical Oncology; American Society of Clinical Oncology Annual Meeting; Chicago, IL. 2008.2008. p. 2. [Google Scholar]
- 56.Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–12. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 57.Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27:5924–30. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 58.Loupakis F, Ruzzo A, Cremolini C, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101:715–21. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tol J, Nagtegaal ID, Punt CJ. BRAF mutation in metastatic colorectal cancer. N Engl J Med. 2009;361:98–9. doi: 10.1056/NEJMc0904160. [DOI] [PubMed] [Google Scholar]
- 60.Van Cutsem ELI, Folprecht G, Nowacki M, et al. Cetuximab plus FOLFIRI in the treatment of metastatic colorectal cancer (mCRC): The influence of KRAS and BRAF biomarkers on outcome: Updated data from the CRYSTAL trial; Gastrointestinal Cancers Symposium; Orlando, FL. 2010.2010. [Google Scholar]
- 61.Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466–74. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 62.Bamford S, Dawson E, Forbes S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–8. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramos FJCG, Tabernero JM, Salazar R, et al. Genomic profiling for prognosis prediction and KRas, BRaf, and Phosphatidylinositol-3-OH kinase (PI3K) mutations prediction in stage II and III colorectal cancer (CRC) patients (pts); Gastrointestinal Cancers Symposium; San Francisco, CA. 2009.2009. [Google Scholar]
- 64.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–6. doi: 10.1136/gut.2008.155473. FS9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.NCCN Practice Guidelines in Oncology–v.1.2009 Breast Cancer. National Comprehensive Cancer Network; [Accessed: August 10, 2009]. 2009. 2009, at. [Google Scholar]
- 66.Harris L, Fritsche H, Mennel R, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 67.Habel LA, Shak S, Jacobs MK, et al. A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast Cancer Res. 2006;8:R25. doi: 10.1186/bcr1412. FS10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buyse M, Loi S, van’t Veer L, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98:1183–92. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 69.Garman KS, Acharya CR, Edelman E, et al. A genomic approach to colon cancer risk stratification yields biologic insights into therapeutic opportunities. Proc Natl Acad Sci USA. 2008;105:19432–7. doi: 10.1073/pnas.0806674105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Jiang Y, Casey G, Lavery IC, et al. Development of a clinically feasible molecular assay to predict recurrence of stage II colon cancer. J Mol Diagn. 2008;10:346–54. doi: 10.2353/jmoldx.2008.080011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Connell MJPS, Yothers G, Constantino JP, et al. Relationship between tumor gene expression and recurrence in stage II/III colon cancer: Quantitative RT-PCR assay of 757 genes in fixed paraffin-embedded (FPE) tissue. J Clin Oncol; American Society of Clinical Oncology Annual Meeting; Orlando, FL. 2006.2006. [Google Scholar]
- 72.Barrier A, Boelle PY, Roser F, et al. Stage II colon cancer prognosis prediction by tumor gene expression profiling. J Clin Oncol. 2006;24:4685–91. doi: 10.1200/JCO.2005.05.0229. [DOI] [PubMed] [Google Scholar]
- 73.Cavalieri D, Dolara P, Mini E, et al. Analysis of gene expression profiles reveals novel correlations with the clinical course of colorectal cancer. Oncol Res. 2007;16:535–48. doi: 10.3727/096504007783438376. [DOI] [PubMed] [Google Scholar]
- 74.Croner RS, Fortsch T, Bruckl WM, et al. Molecular signature for lymphatic metastasis in colorectal carcinomas. Ann Surg. 2008;247:803–10. doi: 10.1097/SLA.0b013e31816bcd49. [DOI] [PubMed] [Google Scholar]
- 75.Eschrich S, Yang I, Bloom G, et al. Molecular staging for survival prediction of colorectal cancer patients. J Clin Oncol. 2005;23:3526–35. doi: 10.1200/JCO.2005.00.695. [DOI] [PubMed] [Google Scholar]
- 76.Munoz Llarena AGA, Suarez B, Jangi M, et al. Gene expression profile of human colorectal cancer using oligonucleotide microarray. J Clin Oncol; American Society of Clinical Oncology Annual Meeting; Orlando, FL. 2009.2009. [Google Scholar]
- 77.Koehler A, Bataille F, Schmid C, et al. Gene expression profiling of colorectal cancer and metastases divides tumours according to their clinico pathological stage. J Pathol. 2004;204:65–74. doi: 10.1002/path.1606. [DOI] [PubMed] [Google Scholar]
- 78.Watanabe T, Kobunai T, Sakamoto E, et al. Gene expression signature for recurrence in stage III colorectal cancers. Cancer. 2009;115:283–92. doi: 10.1002/cncr.24023. [DOI] [PubMed] [Google Scholar]
- 79.Bandres E, Malumbres R, Cubedo E, et al. A gene signature of 8 genes could identify the risk of recurrence and progression in Dukes’ B colon cancer patients. Oncol Rep. 2007;17:1089–94. [PubMed] [Google Scholar]
- 80.Johnston PGMK, Kay E, Black J, et al. A genetic signature of relapse in stage II colorectal cancer derived from formalin fixed paraffin embedded tissue (FFPE) using a unique disease specific colorectal array. J Clin Oncol; American Society of Clinical Oncology Annual Meeting; 2006.2006. [Google Scholar]
- 81.Cascinu S, Zaniboni A, Scartozzi M, Meriggi F. Molecular biology for stage II colorectal cancer: the jury is still out. J Clin Oncol. 2007;25:2861. doi: 10.1200/JCO.2006.10.0966. author reply 2-3. [FS11] [DOI] [PubMed] [Google Scholar]
- 82.Lu AT, Salpeter SR, Reeve AE, et al. Gene expression profiles as predictors of poor outcomes in stage II colorectal cancer: A systematic review and meta-analysis. Clin Colorectal Cancer. 2009;8:207–14. doi: 10.3816/CCC.2009.n.035. [DOI] [PubMed] [Google Scholar]
- 83.Del Rio M, Molina F, Bascoul-Mollevi C, et al. Gene expression signature in advanced colorectal cancer patients select drugs and response for the use of leucovorin, fluorouracil, and irinotecan. J Clin Oncol. 2007;25:773–80. doi: 10.1200/JCO.2006.07.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Glas AMRP, Salazar R, Capella G, et al. Development and validation of a robust prognostic and predictive signature for colorectal cancer (CRC) patients. J Clin Oncol; American Society of Clinical Oncology Annual Meeting; Orlando, FL. 2009.2009. [Google Scholar]
- 85.O’Connell MJLI, Gray RG, Quirke P, et al. Comparison of molecular and pathologic features of stage II and stage III colon cancer in four large studies conducted for development of 12-gene colon cancer recurrence score; Gastrointestinal Cancers Symposium; Orlando, FL. 2010.2010. [Google Scholar]
- 86.ASCO Studies Bolster Agendia’s Marketing Plans for ColoPrint and MammaPrint. Pharmacogenomics Reporter. 2009 [FS12][FS13] [Google Scholar]
- 87.Salazar RBR, Bruin S, Capella G, et al. Development and validation of a robust high-throughput gene expression test (ColoPrint) for risk stratification of colon cancer patients; Gastrointestinal Cancers Symposium; Orlando, FL. 2010.2010. [Google Scholar]


