Abstract
When establishing the most appropriate cells from the huge numbers of a cell library for practical use of cells in regenerative medicine and production of various biopharmaceuticals, cell heterogeneity often found in an isogenic cell population limits the refinement of clonal cell culture. Here, we demonstrated high-throughput screening of the most suitable cells in a cell library by an automated undisruptive single-cell analysis and isolation system, followed by expansion of isolated single cells. This system enabled establishment of the most suitable cells, such as embryonic stem cells with the highest expression of the pluripotency marker Rex1 and hybridomas with the highest antibody secretion, which could not be achieved by conventional high-throughput cell screening systems (e.g., a fluorescence-activated cell sorter). This single cell-based breeding system may be a powerful tool to analyze stochastic fluctuations and delineate their molecular mechanisms.
Cell-based technology is sustained by a wide variety of cell species, such as bacteria, yeast, insect and plant, as well as animal and human cells, for research and industrial use. In particular, cells utilized in regenerative medicine and producing various biopharmaceuticals, such as cytokines, antibodies, enzymes, proteins, peptides and metabolites, have significantly contributed toward human welfare. For the practical use of biopharmaceuticals, it is essential to obtain the most appropriate cells from a candidate cell population. Thus far, conventional cell screening systems have been based on colony isolation by assuming all cells in a colony possess homogeneous characteristics1,2. However, recent single cell-based analyses have revealed that each cell in an isogenic cell population shows diverse and heterogeneous gene expression, morphology and/or cell proliferation3,4,5. For example, each cell of a mouse embryonic stem (ES) cell colony shows heterogeneous expression of the ES marker protein Rex16. Humanized immunoglobulin G (IgG)-producing Chinese hamster ovary (CHO) cells are a mixture of clones showing stochastic fluctuations in IgG production7. Thus, a more rational approach has been considered necessary to isolate and culture the most suitable cells from a huge number of cell candidates by single-cell isolation and expansion (i.e., single cell-based breeding). Although we have examined the use of a fluorescence-activated cell sorter (FACS) as a practical approach, the recovered cells suffered from mechanical stresses probably associated with high voltage and pressure, as well as chemical stress from the sheath solution for cell suspension. A conventional FACS system requires cell sample consisting of ~1 × 105 cell population and containing more than 0.1% positive cells8. Considering the cell sample may be limited, a large portion of the sample is required for FACS optimization, in which cell sample is not recovered. These current issues of FACS technology prompted us to develop a novel system capable of isolating single positive cells from less than 1 × 105 cells under undisruptive conditions. In this study, we have developed an automated single-cell analysis and isolation system to facilitate high-throughput isolation of fluorescently labeled mammalian cells in an undisruptive and single cell-based manner. The robot is a stand-alone machine with a microchamber array chip (containing ~2.5 × 105 cells) and an automated micromanipulator with a glass capillary and fluorescence microscope system, which isolates single positive cells from ~2.5 × 105 cells under undisruptive conditions. Here, we report single cell-based selection and expansion of mouse ES cells with a homogeneous genetic background for the pluripotency marker gene Rex1 and hybridomas that highly secrete antibody, using our automated single-cell isolation system.
Results
Development of the automated single-cell analysis and isolation system
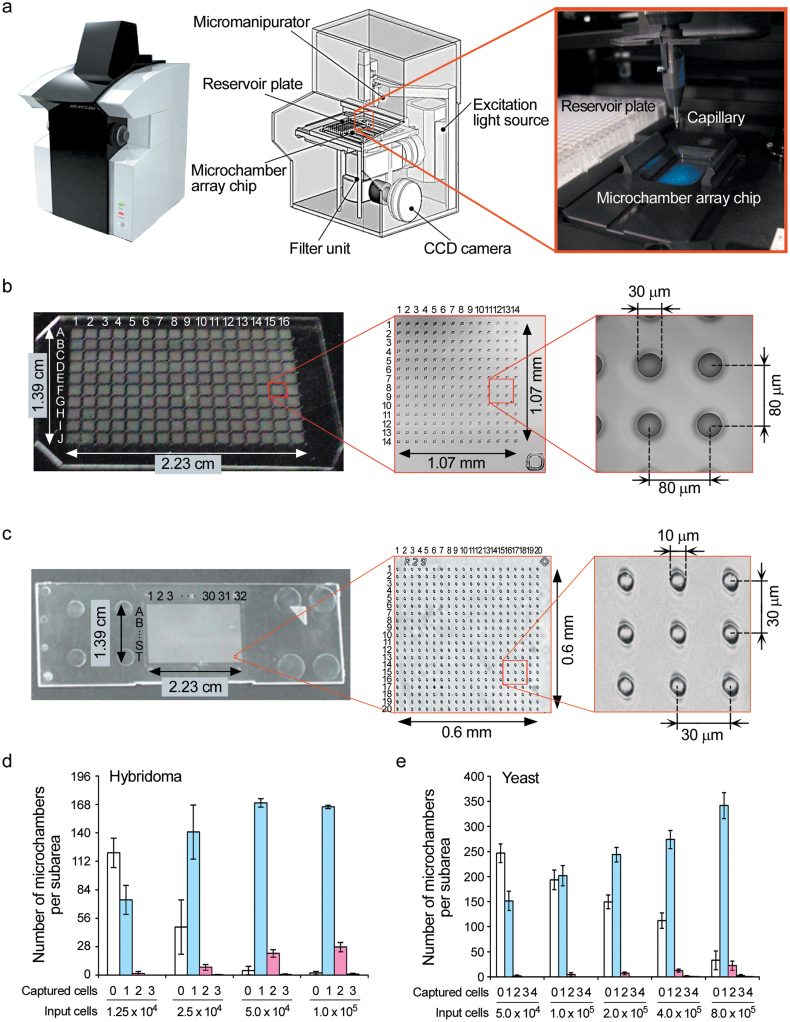
Recently, a microchamber array chip was developed, which allows single-cell microarray analysis of a large cell population (~2.0 × 105 cells)8. Each microchamber (10 μm in diameter) is designed to accommodate only one cell and enables weak fluorescence detection at a high signal to noise ratio by excluding noise signals from negative cells. However, both the fluorescence analysis and single-cell isolation of on-chip cells have been carried out manually. A fully automated single-cell isolation system may improve screening efficiency for the most appropriate cell from a candidate cell population. Therefore, we constructed a robot that executed the following procedures automatically. First, acquisition of the fluorescent intensity of each cell on the microchamber array chip. Second, permutation of cells by the order of their fluorescent intensities. Third, physical retrieval of desired cells with a glass capillary attached to a micromanipulator. Fourth, movement and release of isolated cells to the reservoirs of a multi-well plate (Figure 1a). Detailed specifications of the robot were described in “Methods section.” Two types of microchamber array chips were fabricated with polydimethylsiloxane (PDMS) for mammalian cells (31,360 wells on a 1.39 × 2.23 cm2 total area; 30 μm diameter, 80 μm well-to-well pitch and 30 μm well depth) (Figure 1b), and with polystyrene (PS) for small mammalian or yeast cells (256,000 wells on a 1.39 × 2.23 cm2 total area; 10 μm diameter, 30 μm well-to-well pitch and 10 μm well depth) (Figure 1c). Various numbers of hybridomas were introduced into 30-μm microchambers by brief centrifugation. Approximately 80 ~ 90% of microchambers were occupied by single hybridomas (Figure 1d). Similarly, yeast cells were introduced into 10-μm microchambers at comparable efficiency (Figure 1e).
Figure 1. Overview of the automated single-cell analysis and isolation system.
(a) Automated single-cell analysis and isolation system. (b) Microchamber array chip made of polydimethylsiloxane (PDMS, 31,360 wells, 30 μm diameters). Left, overview (10 × 16 subareas); center, subarea (14 × 14 wells); right, wells. (c) Microchamber array chip made of polystyrene (PS, 256,000 wells, 10 μm diameters). Left, overview (20 × 32 subareas); center, subarea (20 × 20 wells); right, wells. (d) Cell numbers of hybridoma in each 30-μm PDMS microchamber. Error bars = SD (n = 6). (e) Cell numbers of yeast in each 10-μm PS microchamber. Cell numbers of hybridoma in each 30-μm PDMS microchamber. Error bars = SD (n = 6).
Flowchart of the automated single-cell analysis and isolation system
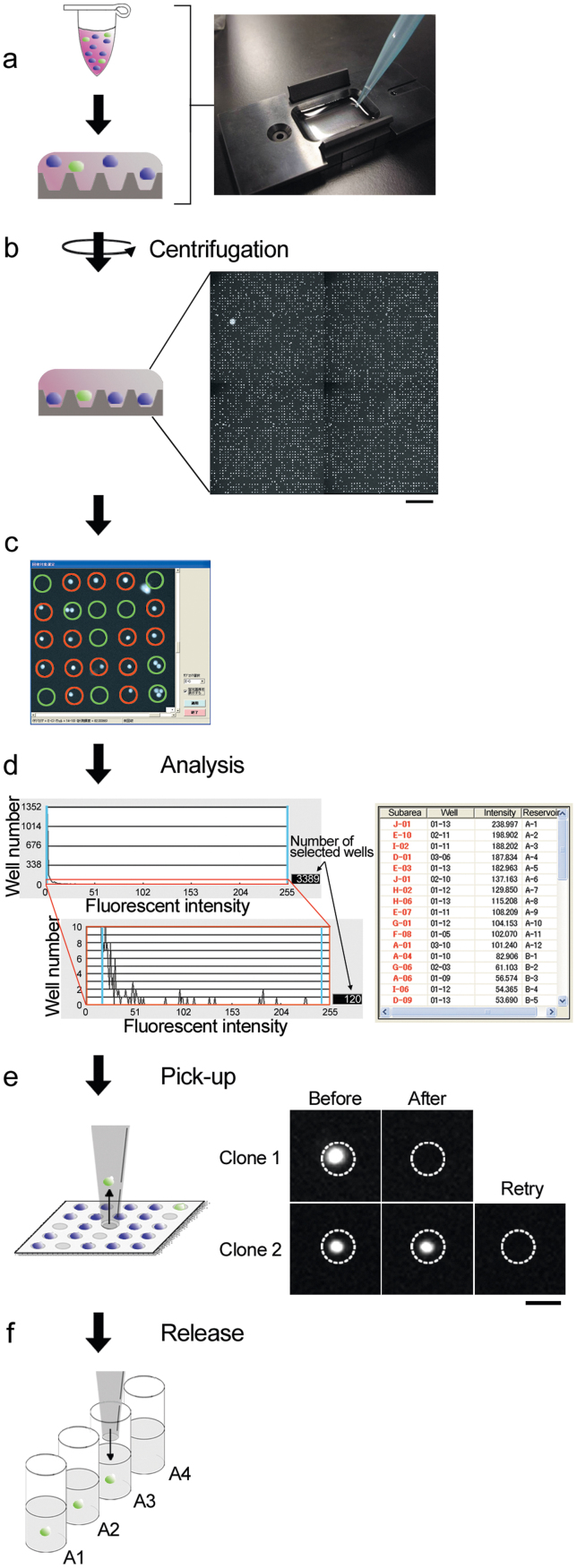
Cell manipulation by the robot was carried out as follows. Cells were introduced into microchambers by brief centrifugation (Figure 2, steps a and b) and covered with culture medium, which could be cultured for at least 24 h. The fluorescent intensities of 9,600 microchambers on a chip were measured by the robot for 30 s (14 min for a 256,000 microchamber array chip) (step b; Supplementary video S1 online). Microchambers containing no or more than 2 fluorescent particles were excluded from further analyses (step c). Finally, a histogram together with a list of correlations between the position and fluorescent intensity of each cell was generated (step d). Cells of interest could be virtually marked in a descending/ascending/random order of fluorescent intensity. Marked cells were automatically collected with a glass capillary attached to the micromanipulator of the robot, which were confirmed by elimination of fluorescence in the target microchamber (step e). Upon failure, the robot automatically repeated the collection process. Each cell was transferred and released into the culture medium of an assigned well in 96- or 384-well plates (step f). The reciprocal movement of the glass capillary required 15 s for each cell (Supplementary video S2 online).
Figure 2. Flow chart of the automated single-cell analysis and isolation system.
Approximately 5.0 × 104 cells in culture medium were added to the microchamber array chip equipped with an aluminum frame (step (a)) and then introduced into 30-μm PDMS microchambers by brief centrifugation (50 × g, at room temperature for 1 min) (step (b)). The microchamber array was scanned with a CCD camera on the robot. Based on the fluorescent image, microchambers containing no or more than 2 fluorescent particles were excluded from further analyses. Green circles, excluded microchambers; red circles, selected microchambers (step (c)). A histogram of fluorescent intensity from each cell was generated for permutation of cells by the order of fluorescent intensities, a list of addresses, fluorescent intensities and images, as well as a transmission image of each cell was generated (step (d)). Cells of interest were automatically retrieved with a glass capillary attached to the micromanipulator. Recovery of each cell was repeated until the fluorescence from each microchamber was absent (step (e)). Each retrieved cell was transferred to an assigned reservoir well (step (f)). Scale bars = 200 μm (b) and 30 μm (e).
Single cell-based breeding of mouse ES cells
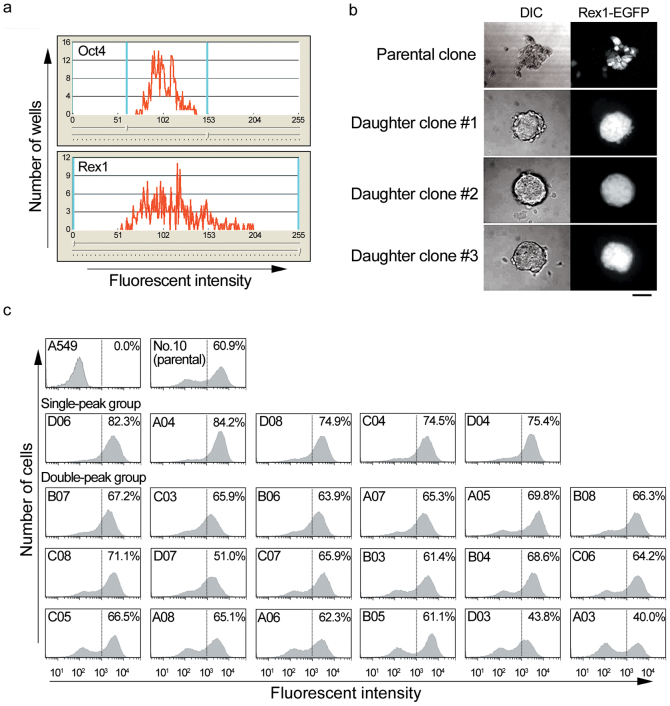
Among established ES cell lines, the expression of pluripotency markers in each cell has often been observed in a stochastic fluctuating state3,6. When ~5.0 × 104 cells of the mouse ES cell line OLG harboring the Oct4-EGFP gene were introduced to 30-μm PDMS microchambers in our system, the cells showed variety of expression level of Oct4 (Figure 3a, upper panel). The mouse ES cell line clone No. 10 harboring the Rex1-EGFP gene showed an even higher degree of variety of expression level of Rex1 (Figure 3a, lower panel), indicating that each mouse ES cell line showed a distinct distribution of stemness9. From the cell library of clone No. 10 mouse ES cells, 24 cells with the highest fluorescent intensity were transferred to culture medium and allowed to proliferate from 1 to ~1,000 cells over 7 d (Figure 3b). The daughter cells formed rounder colonies with increased homogeneous Rex1-EGFP expression, compared with that of parental cells. After 2–3 weeks, 23 clones reached ~1 × 106 cells, in which 20 clones retained a higher fluorescent intensity compared with that of the parental cell population (Figure 3c). When calculating the ratio of highest numbers of cells with higher intensity (over 103) to those with lower intensity (102 ~ 2 × 102), the daughter cells of >7.0 ratio (mean + 3SD of parental cells, n = 6) were judged as a single-peak group. Finally, we obtained 5 clones expressing higher level of Rex1, which would be suitable for further breeding process (Figure 3c). This result indicated that single cell-based breeding of cells isolated from a cell library is a powerful method to expand ES cells with the highest expression of pluripotency markers. ES and induced pluripotent stem (iPS) cells, particularly from humans, are often susceptible to mechanical and chemical stresses10. The automated single-cell isolation system is practical for isolating suitable cells under undisruptive conditions because of gentle manipulation of cells in culture medium with a glass capillary.
Figure 3. Single cell-based breeding of mouse ES cells.
(a) Oct4-EGFP and Rex1-EGFP expression in mouse ES cell lines OLG and No. 10, respectively, were analyzed by the robot. (b) Colony formation from isolated No. 10 cells (daughter cells). Scale bar = 50 μm. (c) Rex1-EGFP expression of isolated No. 10 cells. Approximately 2.0 × 104 cells were analyzed by FACS. Clone numbers are indicated in the upper-left. Contents of cells with higher fluorescent intensity (over 103) are indicated in the upper-right. A549 (an adenosquamous lung carcinoma cell line) cells were used as a negative control.
Single cell-based breeding of hybridomas
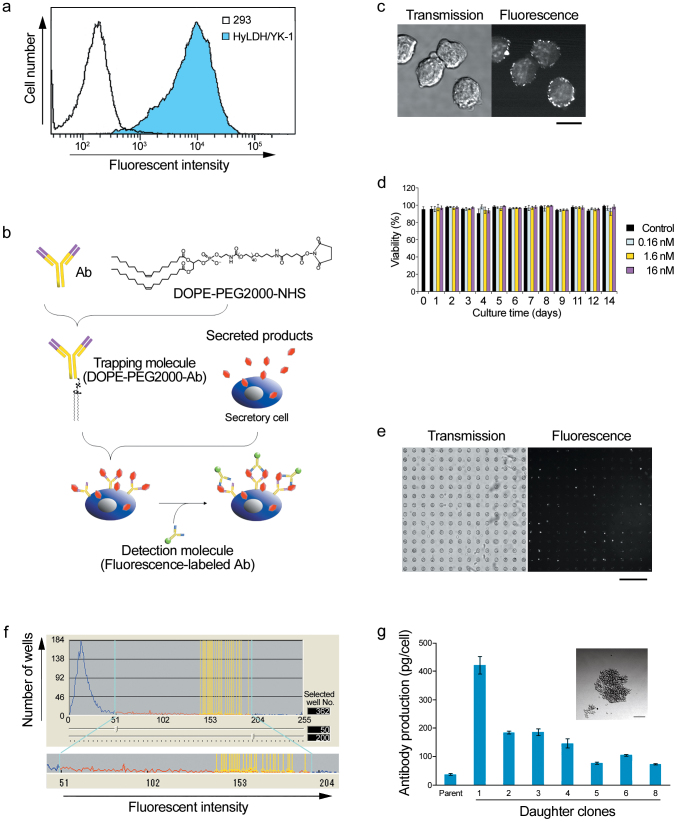
To evaluate the secretion of anti-rabbit lactate dehydrogenase IgG1 from the hybridoma cell line HyLDH/YK-1, we treated the hybridomas with brefeldin A (a protein transport inhibitor)11 to accumulate the IgG intracellularly, followed by FACS analysis. The isogenic cells were found to secrete various amounts of antibody and showed a dynamic fluorescent intensity range between ~102 and ~105 (Figure 4a). A traditional limiting dilution method to isolate mammalian cells secreting high amounts of biopharmaceuticals, deposits the cell library into 96- or 384-well plates at ~1 cell per well to form colonies. After culturing for at least 1 month, the supernatants of these cell cultures are assayed by enzyme-linked immunosorbent assay (ELISA) to detect candidate clones, followed by establishing the selected cells, which is time consuming with low scalability (up to ~100 candidate clones per screen). Recently, two automatic single-colony isolation systems became commercially available (ClonePix FL (Molecular Devices, Sunnyvale, CA, USA) and CellCelector (AVISO, Jena, Germany)). Both machines facilitate manipulation of a large number of cells, but approximately 1 month is still required for colony formation and identification of candidate cells12,13. Using a cuboid microchamber array as the single-cell container and the solid phase of the ELISA, candidate cells can be manually detected after culturing cells for several days. The selected cells are then automatically isolated by CellCelector13. However, both machines are unable to promptly and automatically isolate the most suitable cell from a cell library, and retain the issue of cell heterogeneity in the isogenic cell population. Meanwhile, an automated single cell isolation system (Cellporter system; SC World, Toyama, Japan)14 has been developed, which is likely to utilize similar picking-up system to our study. But, the machine requires additional scanning system (SC Scanner) for measurement of fluorescent intensity of each cell, indicating that Cellporter system is not a completely automated system for cell analysis and isolation. To identify hybridomas that secreted higher amounts of antibodies, we developed an undisruptive cell-surface fluorescence-linked immunosorbent assay (CS-FIA) to evaluate antibody production at a single-cell level using the robot (Figure 4b). The surfaces of secreting cells were modified with a goat anti-mouse Fc antibody conjugated to dioleoyl phosphatidylethanolamine-poly-ethylene glycol 2000 (DOPE-PEG2000) as a trapping molecule15,16. Trapping molecules spontaneously assembled around the cell surface (Figure 4c) and showed no cytotoxicity for at least 2 weeks (Figure 4d). Cells were introduced into 30-μm PDMS microchambers to allow the capture of secreted antibodies at the cell surface, followed by incubation with a fluorescein isothiocyanate (FITC)-labeled rabbit anti-mouse F(ab′)2 F(ab′)2 to establish the sandwich FIA on the cell surface (Figure 4e). After analyzing on-chip cells using the robot, 362 cells exhibited higher fluorescence among the ~5.0 × 104 cells (Figure 4f). Eight cells with the highest level of fluorescence were isolated. Seven cells formed colonies within 10 d and expanded to ~5 × 105 cells over 2 weeks (Figure 4g). When evaluating the amount of antibody secreted into the culture medium by ELISA, the daughter cells were found to secrete higher amounts of antibodies, compared with that of the parental clone. This result indicated that single cell-based analysis and isolation allowed us to obtain hybridomas with the highest antibody-secreting ability in only 1 day. Because the CS-FIA technique is applicable to other cell types and various secreted biomaterials, the combination of the automated single-cell isolation system with the CS-FIA would be more effective to improve the productivity of biomaterials secreted from various cells.
Figure 4. Single cell-based breeding of hybridomas.
(a) Antibody production by mouse hybridoma HyLDH/YK-1 cells was analyzed by FACS. HEK293 cells were used as a negative control. (b) Outline of the cell-surface FIA. Cells displaying trapping molecules, goat anti-mouse Fc antibody conjugated to DOPE-PEG2000, were allowed to capture secreted antibodies and then treated with a FITC-labeled rabbit anti-mouse F(ab′)2 antibody for detection. (c) Alexa 488-labeled trapping molecules on the surface of HyLDH/YK-1 cells. Scale bar = 20 μm. (d) Cytotoxicity of the trapping molecule on HyLDH/YK-1 cells. Cell viabilities were measured by trypan blue staining; error bars, p < 0.01, n = 12 from three independent experiments. (e) On-chip cell-surface FIA visualized by the robot. Scale bar = 240 μm. (f) Histogram of antibody secretion determined by the cell-surface FIA. Black vertical lines indicate fluorescence thresholds (range 50–200). White vertical lines indicate cells with high fluorescent intensities. (g) Antibody secretion determined by a conventional ELISA. Seven cells formed colonies at ~10 d after single-cell isolation (inset, scale bar = 100 μm) and were subjected to ELISA at ~20 d. Error bars, p < 0.05, n = 6 from three independent experiments.
Single cell isolation of adhesive cells
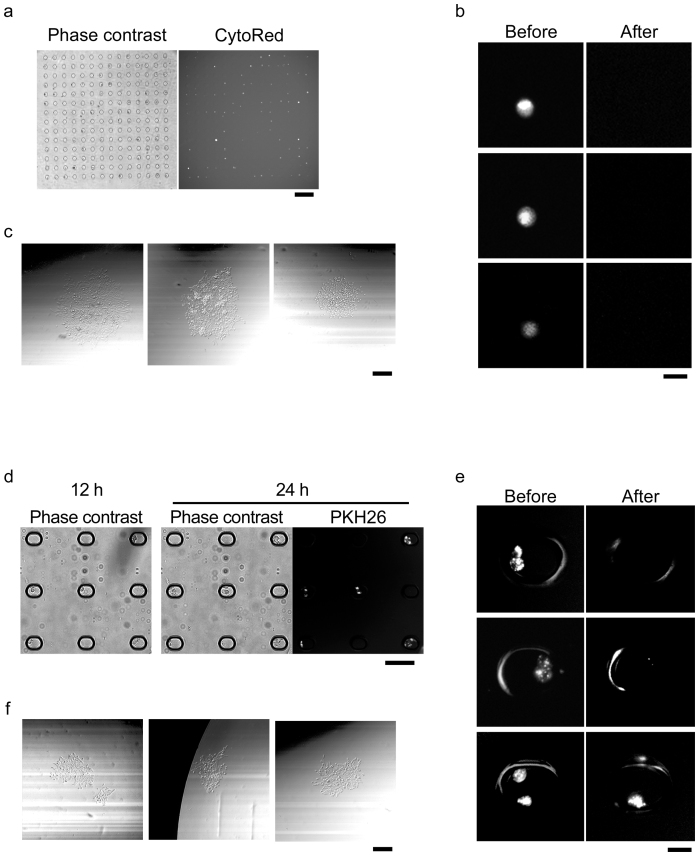
Since ES cells and hybridomas are less adhesive and non-adhesive cells, respectively, we herein examined if the robot is applicable to adhesive cells, such as CHO cells. CHO cells (~1.0 × 104 cells) were trypsinized, stained with CytoRed (fluorescent dye for viable cells; Dojin, Kumamoto, Japan) and then introduced into 30-μm PDMS microchambers at comparable efficiency of ES cells and hybridomas (Figure 5a). Within 3 h, single CHO cells could be retrieved according to their fluorescent intensities by the robot (Figure 5b), and proliferate to ~4 × 102 cells in 6 days (Figure 5c), of which the viability was more than 80% of retrieved single cells (20/24 clones). Furthermore, single CHO cells transferred into 50-μm ellipsoidal PS microchambers could propagate to several cells in microchambers (Figure 5d). After 24 h, the cells were stained with PKH26 (fluorescent dye for membrane staining; Sigma-Aldrich, St Louis, MO, USA), trypsinized briefly, and then retrieved as a cell cluster by the robot (Figure 5e). These cell clusters were allowed to grow up to ~2 × 102 cells in 5 days (Figure 5f), of which the viability was nearly 100% of retrieved cell clusters (10/10 cell clusters). The automated retrieval of cell clusters might be useful for the single cell-based breeding of vulnerable adhesive cells.
Figure 5. Single cell isolation of CHO cells.
(a) Cell array for CHO cells. Cells were stained with CytoRed solution. Scale bar = 160 μm. (b) Retrieval of single CHO cell. Scale bar = 25 μm. (c) Colony formation from isolated CHO cells. Scale bar = 200 μm. (d) On-chip culture of CHO cells. Cells were cultured on microchamber array chip and stained with PKH26. Scale bar = 100 μm. (e) Retrieval of cell cluster of CHO cells. CHO cells stained with PKH26 were trypsinized and retrieved with the robot. Scale bar = 25 μm. (f) Colony formation from isolated cell cluster of CHO cells. Scale bar = 200 μm.
Discussion
To manufacture biopharmaceuticals, each cell used for production (e.g., hybridomas and CHO cells) should express biomaterials stably and efficiently. In regenerative therapies, each stem cell should express pluripotency markers homogeneously to obtain differentiated cells with less tumorigenicity. It is an important step to isolate the most suitable cell from a candidate cell library and expand the cell population for further optimization. However, both isolation and expansion have been traditionally performed with a cell colony, which impairs isolation of suitable cells because of potential difficulties in excluding contaminating unsuitable cells. Furthermore, long-term culture necessary for colony formation often impairs the favorable properties of isolated cells7. For example, the productivity of established hybridomas is sometimes decreased by long-term cell culture17. In established ES, iPS and mesenchymal stem cells, a trace number of cells in colonies cannot fully differentiate, which sometimes leads to severe tumorigenesis18,19. These cellular instabilities are mainly caused by stochastic fluctuation of gene expression among individual cells3,4,5,6,7, which is considered a general phenomenon in various cell types. Although cellular instabilities have been conventionally circumvented by contingency-based cell screenings (i.e., random screening of naturally occurring suitable cells) to establish cells with low stochastic fluctuation, the molecular mechanisms underlying stochastic fluctuation should be delineated. For example, after purifying two types of cells with high and low stochastic fluctuation by single-cell isolation, a comparison of gene expression profiles might delineate the mechanisms. Taken together, cells used for producing biopharmaceuticals and regenerative therapies should be established by single cell-based screening to maximize favorable properties and minimize the occurrence of stochastic fluctuation.
Here, we demonstrated that an automated single-cell isolation system facilitates prompt and effective establishment of the most suitable single cells from a large number of candidate cells to avoid the occurrence of stochastic fluctuation. Mouse ES cells with the highest expression level of the pluripotency marker Rex1 were successfully isolated from a heterogeneous cell population. Although 3 isolated ES cells (clones D07, A03, D03) showed lower expression level of Rex1 at 2–3 weeks after single-cell isolation, 20 cells retained higher Rex1 expression (see Figure 3c). Particularly, 5 isolated ES cells (clones D06, A04, D08, C04, D04) were found to show homogeneous profile of Rex1 expression. In addition, hybridomas secreting the highest amount of antibodies were isolated by the robot in combination with a CS-FIA technique. All isolated hybridomas showed a higher expression level of antibody than that of parental hybridomas at ~21 days after single-cell isolation (see Figure 4g). These results indicate that single cell-based breeding is a practical way to obtain suitable cells with fewer effects from stochastic fluctuation. Moreover, these established cells would be useful to decipher the molecular basis of stochastic fluctuation.
Micromanipulation under a microscope, limiting dilution and FACS have been widely used to isolate single cells. Micromanipulation is undisruptive and reliable for manipulating single cells, but extremely laborious. Limiting dilution is also undisruptive, but not reliable because of possibly excluding suitable cells during serial dilutions. As described in the introduction, FACS is a high-throughput and reliable system for isolating cell populations, but cannot isolate a single positive cell from less than 1 × 105 cells under undisruptive conditions. In particular, the chemical and mechanical stresses intrinsically associated with FACS are likely to alter the gene expression profiles of isolated single cells, thus single-cell manipulation should be carried out under undisruptive conditions. The issues of these techniques for isolating single cells have led us to develop an automated single-cell isolation system. The robot is designed to isolate a single positive cell from less than 1 × 105 cells in about 1 ml of culture medium. Therefore, the automated single-cell isolation system is considered suitable for high-throughput single cell-based analysis and isolation.
In high-throughput identification of hybridomas that secrete high amounts of antibodies, candidate hybridomas can be selected by FACS, followed by colony formation and evaluation with a conventional ELISA. Cell-surface markers (e.g., CD19, CD20, CD38, CD138)20,21,22,23,24 expressed in hybridomas are indirectly labeled with fluorescence, and then populations of positive cells are collected as candidate hybridomas by FACS. This method cannot evaluate the antibody-secreting ability of each hybridoma directly, and only concentrates candidate hybridomas. Another method is treatment of cells with brefeldin A to accumulate antibodies intracellularly. This method can evaluate the antibody-secreting ability, but also severely affects cell viability. Thus far, no reported method can evaluate the antibody-secreting ability of hybridomas directly in an undisruptive manner and without the requirement of long-term culture for colony formation. Here, we developed a CS-FIA that fulfills the above criteria. Combination of the CS-FIA with the robot successfully shortened the time to identify positive hybridomas from ~1 month (FACS) or ~2 months (limiting dilution) to 1 day. To further optimize the CS-FIA, the use of fluorescence-labeled antigens as detection molecules could improve the signal to noise ratio by reducing cross reactions among displayed antibodies.
In addition to single cell-based high-throughput analysis and isolation, our robot can be used for the following applications. Because the robot can evaluate protein-protein interactions on the cell surface, orphan ligand screenings using cells displaying receptor libraries, and orphan receptor screenings using cells expressing cell surface-anchored forms of ligand libraries are applicable, which may lead to drug discovery. Moreover, because of the undisruptive condition during cell manipulation, the robot would be suitable for isolation of rare cells in limited clinical samples (e.g., circulating tumor cells25 and cancer stem cells26) to develop an innovative diagnosis system. Thus, our automated single-cell isolation system may offer a new technology for single cell-based engineering and bring significant progress to cell-based research and industries.
Methods
The automated single-cell analysis and isolation system
The robot consisted of a micromanipulator (<1 μm movement accuracy) equipped with a pencil pump (DENSO, Aichi, Japan) and glass capillary (40.5 μm inside diameter, 58 μm outside diameter for mammalian cell; 14.5 μm inside diameter, 21 μm outside diameter for yeast cell), a CCD camera (Nikon, Tokyo, Japan), an objective lens (Plan Fluor 10 ×/0.3, Nikon), an excitation light source (Nikon) using a super high pressure mercury lamp and power supply, filter units (for FITC, 465–495-nm excitation, 515–555-nm emission, 505 nm dichroic mirror; for Cy3, 510–560-nm excitation, 572.5–647.5-nm emission, 565 nm dichroic mirror, Nikon), a 96- or 384-well reservoir plate, a microchamber array chip and a control PC (CPU, Pentium M 1.8 GHz; RAM 980 MB). Microfabrication on the polymethylmethacrylate (PMMA) plate was performed by X-ray mask and lithography technology combined with synchrotron radiation8,27. Using the patterned PMMA plate as a master plate, a nickel (Ni) mold part was produced by an electro-forming process resulting in effective production of the Ni mold part with replicated micro-patterns. Then, the mold part was inserted into a mold base and injected to produce a microchamber array chip with polystyrene or PDMS. The robot identified a minimum 1 μm single particle at <1800 MESF from the fluorescence of GFP, EGFP, phycoerythrin and propidium iodide. The control PC regulated the vertical movement of the micromanipulator, the horizontal movement of the microchamber array chip, fluorescence acquisition and transmission of images to indentify cells of interest, followed by cell recovery using the pencil pump.
Cell culture
Mouse ES cell lines, OLG and clone No. 10, were cultured in Glasgow Minimum Essential Medium (Sigma-Aldrich, St Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS; StemCell Technologies, Vancouver, BC, Canada), 1 mM sodium pyruvate, 0.1 mM non-essential amino acids (NEAA; Invitrogen Life Technology, Carlsbad, CA, USA), 32.7 mM sodium bicarbonate, 0.1 mM 2-mercaptoethanol and 1,000 U ml−1 leukemia inhibitory factor (Invitrogen). The mouse hybridoma cell line HyLDH/YK-1 secreting anti-rabbit lactate dehydrogenase IgG1 was cultured in RPMI-1640 (Nacalai, Kyoto, Japan) supplemented with 10% FBS (Thermo, Waltham, MA, USA), 0.1 mM NEAA and 0.05 mM 2-mercaptoethanol. Human embryonic kidney (HEK) 293 and lung adenocarcinoma A549 cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS (PAA Laboratories, Pasching, Austria). Dihydrofolate reductase-deficient Chinese hamster ovary (CHO) cells (CHO-DG44 cells) were maintained in RPMI-1640 supplemented with 10% FBS (PAA Laboratories).
Flow cytometric analysis
Daughter mouse ES cells isolated by the robot were cultured for 2–3 weeks to reach ~1 × 106 cells, and then harvested by trypsinization. The fluorescent intensities of ~2.0 × 104 cells from each clone were measured with a BD FACSCanto II flow cytometer (BD, Franklin Lakes, NJ, USA). Parental mouse ES and A549 cells were used as positive and negative controls, respectively. HyLDH/YK-1 cells treated with 1 μg ml−1 brefeldin A (BD) for 6 h were fixed in PBS containing 4% paraformaldehyde (PFA) at room temperature for 15 min, permeabilized with PBS containing 0.03% Triton X-100 at room temperature for 5 min and then collected by centrifugation (200 × g, 5 min). Cells were then incubated with 2 μg ml−1 FITC-conjugated anti-mouse F(ab′)2 rabbit F(ab′)2 (Rockland, Gilbertsville, PA, USA) in PBS at room temperature for 20 min, followed by FACS analysis. HEK 293 cells were used as a negative control.
Preparation of trapping molecules
For the cell-surface FIA, 1.65 nM Immunopure anti-mouse Fc goat polyclonal antibody (Thermo) was reacted with 66 μM DOPE-PEG2000-N-hydroxysuccinimide (NHS; NOF, Tokyo, Japan) in 100 μl sodium phosphate (10 mM, pH 7.6) and 250 mM NaCl at room temperature for 10 min. Free DOPE-PEG2000-NHS molecules were removed by ultrafiltration through 50 kDa cut-off membranes (Millipore, Billerica, MA, USA). Coupling of DOPE-PEG2000 to the antibody was confirmed by SDS-PAGE and Coomassie Brilliant Blue R-250 staining. To observe trapping molecules on the HyLHD/YK-1 cell surface, ~2.0 × 104 cells were combined with 100 μl serum-free RPMI-1640 medium containing 16 nM trapping molecules, incubated at 37°C for 10 min and then fixed with 4% PFA at room temperature for 20 min. Cells were washed with PBS twice, incubated in 100 μl PBS containing 400 ng Alexa 488-conjugated anti-goat IgG antibody (Invitrogen) at room temperature for 20 min, washed with PBS twice and then observed under a confocal laser-scanning microscope (FV-1000D; Olympus, Tokyo, Japan). To evaluate the cytotoxicity of the trapping molecule (DOPE-PEG2000-antibody) in HyLDH/YK-1 cells, ~1.25 × 105 cells in 2.5 ml culture medium were treated with the trapping molecule at 0.16–16 nM final concentration (as a protein concentration) and then maintained at 37°C with 5% CO2 for 2 weeks. Cell viabilities were measured by trypan blue staining.
Cell-surface FIA
Approximately 5.0 × 104 HyLDH/YK-1 cells were combined with 100 μl serum-free RPMI-1640 medium containing 16 nM trapping molecules, incubated at 37°C for 10 min, mixed with 900 μl serum-free RPMI-1640 medium and then placed into microchambers by brief centrifugation. After addition of 100 μl FBS, cells on the microchamber array chip were incubated for 30 min in a CO2 incubator to allow antibody secretion and then treated with 2 μg ml−1 FITC-labeled anti-mouse F(ab′)2 F(ab′)2 antibody to establish the cell-surface FIA. After incubation at room temperature for 10 min, cells were briefly washed twice with medium and then subjected to the automated single-cell analysis and isolation system.
Mouse IgG ELISA
Each well of a MaxiSorp 96-well plate (Thermo) was treated with 30 μl anti-mouse Fc antibody (10 ng μl−1) at 4°C for 24 h, washed with 200 μl PBS containing 0.1% (vol/vol) Tween 20 (PBS-T) five times and then treated with 200 μl bovine serum albumin (5 μg μl−1) in PBS at room temperature for 1 h. To measure the amount of secreted antibody from HyLDH/YK-1 cells isolated by the robot, each sample was added to an anti-mouse Fc-immobilized well, incubated at room temperature for 1 h and then washed with 200 μl PBS-T five times, followed by addition of 200 μl PBS-T containing 0.2 μl horse radish peroxidase-conjugated anti-mouse Ig (GE healthcare, Little Chalfont, Buckinghamshire). Plates were incubated at room temperature for 1 h and then washed with PBS-T five times, followed by addition of 100 μl 3,3′,5,5′-tetramethylbenzidine substrate (Thermo). The coloration reaction was performed at room temperature for 30 min, stopped by 100 μl H2SO4 (2 M) and then the absorbance at 450 nm was measured.
Author Contributions
N.Y. and S.K. conceived the project, designed and performed experiments, processed and analyzed data and wrote the manuscript. A. Kida, M.I., T.N., A.D.M. and K. Tatematsu processed and analyzed data. X.J. and M.K. developed the robot and chips. I.N. and H.R.U. prepared mouse ES cell samples. A. Kondo, I.F., K. Tanizawa and S.K. coordinated the project.
Supplementary Material
Supplementary Information
Supplementary video 1
Supplementary video 2
Acknowledgments
This study was supported in part by The Japan Science Society (H22 to N.Y.), Adaptable & Seamless Technology Transfer Program through Target-driven R&D (A-STEP) by the Japan Science and Technology Agency (JST) (AS231Z04687F to N.Y., AS2311699F to S.K.), Regional New Consortium Projects (H15 to S.K. and H17 to I.F., METI, Japan), the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (H22-7, BRAIN to S.K., K. Tanizawa), the Health Labor Sciences Research Grant from the Ministry of Health Labor and Welfare (to SK), a Grant-in-Aid for Scientific Research (A) (21240052 to S.K.), the As One Corporation (to S.K., A. Kondo, I.F.), the Suzuken Memorial Foundation (10-013 to S.K.), the Nagase Science and Technology Foundation (H22-1 to S.K.) and the Canon Foundation (H22-1-7 to S.K.). We thank Satoshi Sugiyama and Ken-ichi Kimura (Furukawa) for technical support, and Masahiro Matsushita and Kenji Uemukai (As One) for advice.
References
- Caron A. W. et al. Fluorescent labeling in semi-solid medium for selection of mammalian cells secreting high-levels of recombinant proteins. BMC Biotechnol. 9, 42 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D. et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 26, 795–797 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K., Yabuta Y., Ohinata Y. & Saitou M. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat. Protoc. 2, 739–752 (2007). [DOI] [PubMed] [Google Scholar]
- Wang D. & Bodovitz S. Single cell analysis: the new frontier in ‘omics’. Trends Biotechnol. 28, 281–290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsinh K. H. et al. Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells. J. Clin. Invest. 121, 1217–1221 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y., Shimosato D., Murakami K., Takahashi K. & Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development 135, 909–918 (2008). [DOI] [PubMed] [Google Scholar]
- Pilbrough W., Munro T. P. & Gray P. Intraclonal protein expression heterogeneity in recombinant CHO cells. PLoS One 4, e8432 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura S. et al. Single-cell microarray for analyzing cellular response. Anal. Chem. 77, 8050–8056 (2005). [DOI] [PubMed] [Google Scholar]
- Casanova J. Stemness as a cell default state. EMBO Rep. 13, 396–397 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgushi M. & Sasai Y. Lonely death dance of human pluripotent stem cells: ROCKing between metastable cell states. Trends Cell Biol. 21, 274–282 (2011). [DOI] [PubMed] [Google Scholar]
- Mendez A. J. Monensin and brefeldin A inhibit high density lipoprotein-mediated cholesterol efflux from cholesterol-enriched cells. Implications for intracellular cholesterol transport. J. Biol. Chem. 270, 5891–5900 (1995). [DOI] [PubMed] [Google Scholar]
- Serpieri F. et al. Comparison of humanized IgG and FvFc anti-CD3 monoclonal antibodies expressed in CHO cells. Mol. Biotechnol. 45, 218–225 (2010). [DOI] [PubMed] [Google Scholar]
- Choi J. H. et al. Development and optimization of a process for automated recovery of single cells identified by microengraving. Biotechnol. Prog. 26, 888–895 (2010). [DOI] [PubMed] [Google Scholar]
- Suzuki M., Tanaka H. & Iribe Y. Detection and Collection System of Target Single Cell Based on pH and Oxygen Sensing. J. Rob. Mechatron. 22, 639–643 (2010). [Google Scholar]
- Kato K. et al. Immobilized culture of nonadherent cells on an oleyl poly(ethylene glycol) ether-modified surface. Biotechniques 35, 1014–1018, 1020–1021 (2003). [DOI] [PubMed] [Google Scholar]
- Kato K., Itoh C., Yasukouchi T. & Nagamune T. Rapid protein anchoring into the membranes of Mammalian cells using oleyl chain and poly(ethylene glycol) derivatives. Biotechnol. Prog. 20, 897–904 (2004). [DOI] [PubMed] [Google Scholar]
- Frame K. K. & Hu W. S. The loss of antibody productivity in continuous culture of hybridoma cells. Biotechnol. Bioeng. 35, 469–476 (1990). [DOI] [PubMed] [Google Scholar]
- Ben-David U. & Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat. Rev. Cancer 11, 268–277 (2011). [DOI] [PubMed] [Google Scholar]
- Fong C. Y., Peh G. S., Gauthaman K. & Bongso A. Separation of SSEA-4 and TRA-1-60 labelled undifferentiated human embryonic stem cells from a heterogeneous cell population using magnetic-activated cell sorting (MACS) and fluorescence-activated cell sorting (FACS). Stem Cell Rev. 5, 72–80 (2009). [DOI] [PubMed] [Google Scholar]
- Tedder T. F. & Isaacs C. M. Isolation of cDNAs encoding the CD19 antigen of human and mouse B lymphocytes. A new member of the immunoglobulin superfamily. J. Immunol. 143, 712–717 (1989). [PubMed] [Google Scholar]
- Chilosi M. et al. CD138/syndecan-1: a useful immunohistochemical marker of normal and neoplastic plasma cells on routine trephine bone marrow biopsies. Mod. Pathol. 12, 1101–1106 (1999). [PubMed] [Google Scholar]
- Harper D. et al. In vitro and in vivo investigation of a novel monoclonal antibody to plasma cells (W5 mAb). Xenotransplantation 11, 78–90 (2004). [DOI] [PubMed] [Google Scholar]
- Polson A. G. et al. Expression pattern of the human FcRH/IRTA receptors in normal tissue and in B-chronic lymphocytic leukemia. Int. Immunol. 18, 1363–1373 (2006). [DOI] [PubMed] [Google Scholar]
- Rosa E. A., Lanza S. R., Zanetti C. R. & Pinto A. R. Immunophenotyping of classic murine myeloma cell lines used for monoclonal antibody production. Hybridoma (Larchmt) 31, 1–6 (2012). [DOI] [PubMed] [Google Scholar]
- Pantel K. & Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol. Med. 16, 398–406 (2010). [DOI] [PubMed] [Google Scholar]
- Gupta P. B. et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 138, 645–659 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa M. Advanced Micro & Nanosystems, LIGA and Its Applications (Saile V., Wallrabe U., Tabata O., & Korvink J. G., eds.) 7, 323–335 (Wiley-VCH GmbH & Co. KGaA, 2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary video 1
Supplementary video 2