Abstract
Mammalian sperm are differentiated germ cells that transfer genetic material from the male to the female. Owing to this essential role in the reproductive process, an understanding of the complex mechanisms that underlie sperm function has implications ranging from the development of novel contraceptives to the treatment of male infertility. While the importance of phosphorylation in sperm differentiation, maturation and fertilization has been well established, the ability to directly determine the sites of phosphorylation within sperm proteins and to quantitate the extent of phosphorylation at these sites is a recent development that has relied almost exclusively on advances in the field of proteomics. This review will summarize the work that has been carried out to date on sperm phosphoproteomics and discuss how the resulting qualitative and quantitative information has been used to provide insight into the manner in which protein phosphorylation events modulate sperm function. The authors also present the proteomics process as it is most often utilized for the elucidation of protein expression, with a particular emphasis on the way in which the process has been modified for the analysis of protein phosphorylation in sperm.
Keywords: peptide quantitation, phosphoproteomics, phosphorylation, proteomics, sperm
Sperm maturation & function
As a result of their high level of specialization, sperm have abandoned much of the cellular machinery that is found in other cell types. During spermiogenesis, postmeiotic spermatids discard several organelles and retain only the minimal protein complement necessary to deliver the male genetic information to the ovum [1]. At the same time, the DNA condenses tightly around sperm-specific proteins called protamines, making it unavailable for transcription [2]. Upon leaving the testes, sperm may be morphologically mature, but they are immotile and have not yet gained the ability to fertilize. Sperm acquire progressive motility during their transit through the epididymis in a process known as epididymal maturation. After ejaculation, mammalian sperm move actively, but they need to reside in the female reproductive tract before acquiring fertilization potential through a series of events collectively referred to as capacitation.
Although capacitation and epididymal maturation are poorly understood at the molecular level, the belief is that sperm are both transcriptionally and translationally silent after leaving the testes [3,4]. This lack of protein synthesis in mature sperm supports the current view that regulation of post-testicular development is controlled almost exclusively by the addition of exogenous proteins (e.g., during epididymal transit) [5,6] or by the post-translational modification (PTM) of their intrinsic protein complement [7,8]. An understanding of sperm function is therefore dependent upon the ability to unequivocally identify proteins, quantitate their relative abundances and determine the exact sites of protein PTM. While traditional genomic approaches are of limited utility in this regard, the field of proteomics is uniquely suited to address these challenges [9].
Previous phosphoproteomic studies in sperm
Although the word ‘proteome’, defined as the entire set of proteins encoded by a genome, was not coined until 1996 [10], attempts to identify the protein complement of sperm began in the early 1950s [11]. In fact, the identification of specific sites of protein phosphorylation in sperm, an area of study currently referred to as ‘sperm phosphoproteomics’, began several decades ago. In 1967, Ingles and Dixon isolated protamines from trout spermatids undergoing differentiation and, in a remarkably advanced series of experiments for the time, determined seven distinct sites of serine phosphorylation [12]. Notably these sites appeared to be completely dephosphorylated in mature spermatozoa, leading the authors to hypothesize that phosphorylation was being used as a means to prevent nucleic acids from binding to the newly synthesized, highly basic protamines until they were properly localized to the nucleus. Not long after this ground-breaking research, phosphorylation was implicated in sperm movement when it was discovered that cAMP signaling is essential for the regulation of motility [13] and that cAMP-dependent protein kinases, such as protein kinase A (PKA), are highly active in mammalian spermatozoa [14]. The role of phosphorylation in sperm motility was confirmed in 1980 when it was shown that partially-purified sperm protein fractions incorporated 32P after cAMP treatment [15]. While these early studies were essential in establishing a link between protein phosphorylation and sperm function, the identification of novel phosphoproteins and their specific sites of modification remained elusive. Unfortunately, the specificity of the protein enrichment methods and the sensitivity of the protein sequencing used at the time prevented all but the most abundant proteins, such as the protamines, from being successfully analyzed.
The difficulties associated with the isolation and identification of novel phosphoproteins were partially addressed with the development of antiphosphotyrosine (anti-pY) antibodies in the early 1980s [16]. Leyton and Saling first used monoclonal antibodies against phosphotyrosine while attempting to elucidate the mechanism by which the acrosome reaction is initiated in mouse sperm [17]. They used radioactively labeled ZP3, a known acrosome reaction-stimulating ligand, and an anti-pY antibody to probe sperm western blots. Although a single 95 kDa band was visualized using both methods, identification of the purified protein was not accomplished until 1994 when Kalab et al. utilized partial ingel tryptic digestion, HPLC-based peptide separation and Edman sequencing of three specific peptides to unambiguously identify the 95 kDa protein as a testis-specific form of hexokinase [18]. Not long after, anti-pY antibodies were used to investigate the role of phosphorylation in the capacitation of mouse sperm [19]. Using gel electrophoresis, western blotting and anti-pY labeling, it was observed that a global increase in protein tyrosine phosphorylation is temporally associated with progression of the capacitation process in mouse and that these changes correlate with an increase in cAMP production and concomitant activation of PKA [20–23]. Additional phosphotyrosine-containing proteins were seen when a similar approach was used to study the phosphorylation of human sperm surface proteins. Naaby-Hansen et al. determined the approximate molecular weight (MW) and isoelectric point (pI) for 22 tyrosine phosphorylated human proteins, but were unable to unequivocally identify any of these proteins as the sequencing techniques used at the time required relatively large amounts of highly purified material [24].
While Naaby-Hansen et al. did not sequence the proteins recognized by anti-pY antibodies, they suggested a technique for phosphoprotein microsequencing utilizing mass spectrometry (MS) [24]. Their approach, wherein a protein spot on a gel, known to be phosphorylated through alignment with a corresponding western blot, is cored, in-gel digested and identified using tandem MS (MS/MS), was first applied to sperm in 1999. Mandal et al. sequenced 18 peptides from a 95 kDa human sperm protein (FSP95) that is tyrosine phosphorylated during capacitation and which shows reactivity to antibodies present in the sera of infertile men [25]. Not only did this information lead to the identification of the antigen as a novel A-kinase anchor protein (AKAP), but it also allowed primers to be generated, cDNA for FSP95 to be isolated, and protein expression to be carried out in E. coli. In 2001, the same group used MS/MS-based peptide sequencing on an ion trap mass spectrometer to identify eight sperm surface proteins cored from two-dimensional (2D) gels [26]. In the same year, Bohring et al. used an alternative MS-based protein identification approach, peptide mass fingerprinting (PMF), to identify six autoantigenic sperm surface proteins [27]. While these studies were not specifically focused on phosphorylation, this same approach is still being used today for the identification of phosphoproteins involved in the capacitation process [28–30]. However, these studies have been largely limited to the identification of pY-containing proteins as the lower inherent immunogenicity of phosphoserine (pS) and phosphothreonine (pT) has prevented the production of anti-pS/pT antibodies that demonstrate specificities equivalent to those seen with anti-pY antibodies [31].
In an effort to address the poor specificity associated with anti-pS/pT antibodies and to more closely track changes in phosphorylation within a given signaling pathway, antibodies recognizing specific phosphorylated epitopes have been developed. Not only can these antibodies be used to monitor and quantitate the level of phosphorylation at particular sites, but they can also be coupled with immunofluorescence to detect phosphorylation-induced changes in cellular localization. As an example, antiphospho-ERK1/2 antibodies were used to determine that the MAPK pathway is activated during sperm capacitation [32,33]. However, antibodies of this type are often too specific to be used for the identification of novel kinase substrates. Zhang et al. addressed this issue by creating a broad range, phospho-specific substrate antiserum by raising antibodies against a degenerate mix of phosphopeptides [34]. Using this same approach, Harrison took advantage of antibodies developed against a phosphopeptide library containing the consensus sequence RXXp(S/T) in an effort to identify the PKA substrates involved in capacitation [35]. However, as this consensus sequence is known to be phosphorylated by other kinases, such as Protein Kinase B (AKT), a control experiment using a known PKA inhibitor was required in order to exclude non-PKA substrates. This work identified two outer dense fiber proteins as components of the cAMP/PKA-dependent pathway in boar sperm and similar strategies have since been used to identify both PKA dependent [36–40] and proline-directed [41] phosphorylation in sperm from other species. As useful as these studies have been, experiments relying on antibody binding can only provide indirect evidence of protein phosphorylation at known sites of modification. In order to truly understand the role which phosphorylation plays in the regulation of sperm function, direct localization of the specific sites of phosphorylation in unknown proteins is essential.
In 2003, an attempt to identify the specific sites of phosphorylation on all of the proteins present in capacitated human sperm was carried out by Ficarro et al. [28]. Recognizing the limitations of antibody-based phosphoprotein visualization, the authors used immobilized metal ion affinity chromatography (IMAC) for unbiased phosphopeptide enrichment. Combining this methodology with MS/MS-based sequencing resulted in the identification of five tyrosine, 56 serine and two threonine phosphorylation sites. Notably, this research was the first to identify specific sites of tyrosine phosphorylation in AKAPs and provided direct evidence connecting this protein family to the capacitation process. An alternative method for phosphopeptide enrichment, based upon the affinity of phosphate for titanium dioxide (TiO2), was first applied to sperm by Baker et al. in 2010 [42]. In this study, 288 distinct sites of phosphorylation were identified from 120 proteins in capacitated rat sperm. Among the reported phosphoproteins, many are known to be involved in sperm–egg binding, suggesting a linkage between capacitation-induced phosphorylation and fertilization capability. Importantly, the results of these global phosphoproteomic studies have the potential to be used for the future generation of novel, site-specific antibodies designed to monitor phosphorylation changes associated with sperm function.
Recent developments in sperm phosphoproteomics obviate the need for antibody-based quantitation and permit the relative extent of phosphorylation at a specific site to be determined from MS data acquired during MS/MS-based phosphopeptide sequencing. In 2009, Platt et al. utilized a differential isotopic labeling scheme based on the Fischer esterification [43] to label peptides derived from both capacitated and noncapacitated mouse sperm proteins [44]. Of the 55 phosphorylation sites reported in this study, relative quantitation was achieved for 53 of these sites by comparing the differentially labeled MS peak areas of 42 unique phosphopeptides. Consistent with previously published reports, a general increase in phosphorylation was seen following sperm capacitation. Of note, these experiments also unambiguously identified the site of tyrosine phosphorylation within hexokinase which was originally visualized by Kalab et al. in 1994 using anti-pY antibodies [18]. Relative quantitation of phosphorylation levels as a consequence of the capacitation process in rat sperm was subsequently carried out by Baker et al. in 2010 [42]. In this study a label-free approach, relying on the ratio of MS peak areas measured in multiple analyses, was used to identify 15 proteins whose phosphorylation status changed as a result of capacitation. The following year, Baker et al. used the same approach on rat sperm extracted from the caput and caudal regions of the epididymis to quantitate the extent of modification at 77 specific phosphorylation sites during epididymal maturation [45]. Importantly, these studies also represent the first use of electron transfer dissociation (ETD) for the sequencing of phosphopeptides derived from sperm proteins. Although only three phosphopeptides were successfully sequenced using ETD in these two studies, this method overcomes difficulties associated with traditional phosphopeptide fragmentation [46] and promises to play an integral role in future studies designed to answer particular biological questions in sperm phosphoproteomics.
The proteomics process for current investigations into sperm phosphorylation
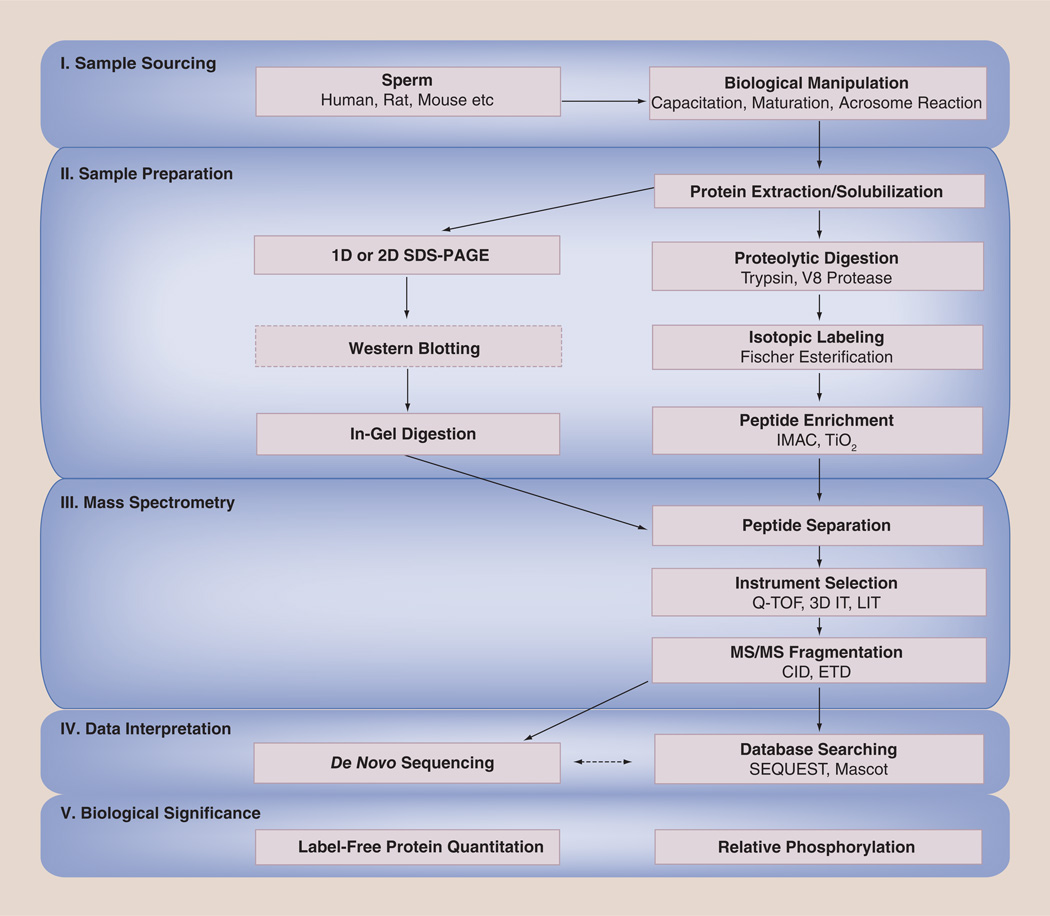
The fact that the proteomics workflow is largely a linear process is apparent in Figure 1. Every sperm phosphoproteomics experiment begins with the selection of a particular organism for analysis. Although this may seem trivial, the source of spermatozoa determines protein concentration and directly impacts downstream data analysis. Once an organism is selected for study, the overall goal of the project (i.e., quantitation, phosphorylation site determination or phosphoprotein identification) is the primary consideration which drives sample preparation. The appropriate type of mass spectrometer can be chosen only after the sample is prepared properly, as each instrument has its own capabilities and limitations. Ultimately, the approaches taken to analyze the resulting data are perhaps the most critical as a compromise must be made between speed and accuracy. Only when all steps in the process have been considered and integrated into an overall experimental workflow can valid biological conclusions be drawn from the acquired data.
Figure 1. Potential phosphoproteomic workflows.
Diagram illustrating current approaches used to investigate the role of phosphorylation in sperm function.
3D IT: Three-dimensional ion trap mass spectrometer; CID: Collisionally induced dissociation; ETD: Electron transfer dissociation; IMAC: Immobilized metal ion affinity chromatography; LIT: Linear ion trap mass spectrometer; MS: Mass spectrometry; SDS-PAGE: Sodium dodecyl sulfate polyacrylamide gel electrophoresis; Q-TOF: Quadrupole time-of-flight mass spectrometer.
Sample sourcing
The quality of the information obtained in any phosphoproteomics experiment is dependent upon the concentration, complexity and contaminating proteins present in the sample selected for analysis. High concentrations of protein can be obtained from mammalian sperm because they are readily accessible and protein dense. They are also uniform in their starting state and a number of techniques are available which can induce them to undergo in vitro maturational changes as an entire population, thereby limiting their proteomic complexity. The major difficulty in the analysis of sperm is the presence of highly abundant species such as protamines, tubulins and outer dense fiber proteins that result in a wide range of phosphoprotein concentrations [47]. This complicates the MS-based sequencing approaches used in current phosphoproteomic workflows because the 103 dynamic range, achievable in a single MS analysis on even the most advanced mass spectrometer, prevents detection of any analyte whose concentration is 1000-times lower than that of the most abundant species [48].
Sample preparation
Some sample preparation protocols used in sperm phospho-proteomics involve heating the samples in the presence of solubilization reagents in an effort to extract the greatest number of proteins [28,42,44]. While the majority of these compounds, such as salts and detergents, demonstrate minimal interference with traditional gel-based protein separation methods, their presence can be detrimental to modern MS-based phosphoproteomic workflows. For example, heating sperm samples in the presence of urea may aid in membrane protein solubilization [41], but the cyanic acid found in urea solutions [49] can carbamylate the primary amines found on lysine residues and protein N-termini [50]. This chemically-induced modification has a significant impact on all subsequent steps in the proteomics process: it can suppress ionization, it can inhibit digestion and the altered MW can confound automated database searching algorithms. It is important to note that modifications such as this do not necessarily preclude MS-based sequencing, but they do add variables that must be taken into account in order to avoid peptide sequence misassignment and protein misidentification.
Denaturing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), which separates proteins on the basis of their length (which closely correlates with protein MW), was relied upon heavily in early phosphoproteomics experiments [12] because the Edman sequencing used at the time required concentrated protein samples relatively free of contamination [51]. Even today, SDS-PAGE may still be used before MS if salts and detergents are used for certain experimental strategies [52,53] or if western blotting using a specific antibody is the basis for phosphoprotein ‘identification’ [17]. The moderate resolution afforded by SDS-PAGE is also exploited in some proteomic workflows as a means to reduce sample complexity prior to MS [54]. An orthogonal method of separation, in which zwitterionic molecules align themselves at their pIs in an immobilized pH gradient, is known as isoelectric focusing (IEF) [55]. This technique was used successfully to separate peptides derived from mouse sperm proteins but, in this case, rapid processing of the IEF-embedded peptides was necessary to minimize diffusion and sample loss [56]. When used for the resolution of intact proteins, the problem of diffusion is greatly reduced and IEF can be coupled with SDS-PAGE to separate a complex protein mixture into thousands of well-resolved spots [57]. As mentioned earlier, this combination, referred to as 2D gel electrophoresis (2DE) has been adopted extensively for the identification of phosphorylated proteins in sperm and is still used today [24–28,30,58]. Despite this widespread acceptance, there are several caveats to keep in mind when incorporating 2DE into an overall phosphoproteomics workflow. To begin with, protein identification from gels requires visualization, and the sensitivity of the reagents used for staining is often incompatible with the sensitivity exhibited by current MS-based analytical approaches. For example, Coomassie Blue, the most commonly used protein stain, is at least an order of magnitude less sensitive than the lower limit of detection achieved using most modern mass spectrometers [25–27], meaning that it is not possible to visualize with Coomassie all of the proteins which could potentially be identified using MS. At the other extreme, fluorescent protein stains, such as Sypro® Ruby, exhibit greater sensitivity than most MS instrumentation, meaning that although a protein may be visualized, it may not be present in sufficient quantity for subsequent in-gel digestion and MS-based identification [58]. In our experience, a staining approach relying on the reduction of ionic silver [59] demonstrates a limit of detection most compatible with downstream MS analysis [28]. However, once visualization is achieved, identification of the exact site(s) of phosphorylation in a protein cored from a gel may still remain elusive due to the low stoichiometric level of phosphorylation and the relatively high concentration of enzymatic autolysis products present following in-gel digestion [60].
As the extensive handling associated with gel-based protein separation can lead to significant sample loss [61], solution-based digestion procedures are often a better choice. The enzyme most commonly used for this purpose is trypsin, largely because of its specificity and tendency to generate peptides which are particularly amenable to sequencing when using positive ionization MS [62]. Trypsin cleaves the peptide bond C-terminal to the basic amino acids lysine and arginine, such that most peptides generated from trypsin digestion carry two charges: one at the C-terminal basic residue and another on the primary amine at the peptide N-terminus. Importantly, the spatial separation of these charges increases the amount of structural information obtained following peptide fragmentation [63] and the relative abundances of lysine and arginine mean that most tryptic peptides fall within a narrow mass-to-charge (m/z) range of 400–800. Of course, as useful as trypsin is in preliminary studies, it may not be ideal if the protein or proteins of interest do not contain lysine or arginine, or if cleavage at these residues generates peptides which are too long or too short for subsequent analysis. In those situations, chemical digestion [64] or an alternative enzyme, such as V8 protease, may be required [28].
Regardless of the procedure used for proteolysis, the presence of more abundant nonphosphorylated peptides in the digestion mixture requires the selective enrichment of phosphorylated species. Although attempts have been made to use anti-pY antibodies to enrich for phosphorylated sperm proteins prior to digestion, these approaches still suffer from the presence of significant numbers of nonphosphorylated peptides after digestion [28]. As a result, several methods for the enrichment of all phosphate-containing species have been developed and applied at the peptide level [65–69]. One of the most widely used techniques for global phosphopeptide enrichment, IMAC, is based upon the affinity which phosphate exhibits towards immobilized metal ions, such as iron(III) [70–72]. A drawback to the application of this technology, which was recognized early on, is that the affinity material also binds carboxylate-rich peptides (e.g., those containing aspartic and/or glutamic acid residues), unnecessarily complicating the sample for analysis [73]. To prevent the binding of nonphosphorylated peptides to the IMAC resin, a modification of this technique was developed wherein acidic residues are converted to their corresponding methyl esters using the Fischer esterification [28,74]. Our group has successfully used this reaction in sperm phosphoproteomic studies to minimize the binding of nonphosphorylated peptides to an alternative phosphate-binding material composed of TiO2 beads. However, other groups [42,45] have relied on the inclusion of a ‘displacer’, such as dihydroxybenzoic acid (DHB), into the phosphopeptide loading buffer to compete with carboxylic acids for binding sites on the TiO2 [75]. While this competitive approach has proven to be successful, the bridging interaction that characterizes the binding of phosphate to TiO2 makes the elution of multiply phosphorylated species more difficult [76]. This suggests that it may be best to view TiO2 not as an alternative to IMAC, but rather as a complementary method: TiO2 for the enrichment of singly phosphorylated peptides and IMAC for the enrichment of multiply phosphorylated species [74].
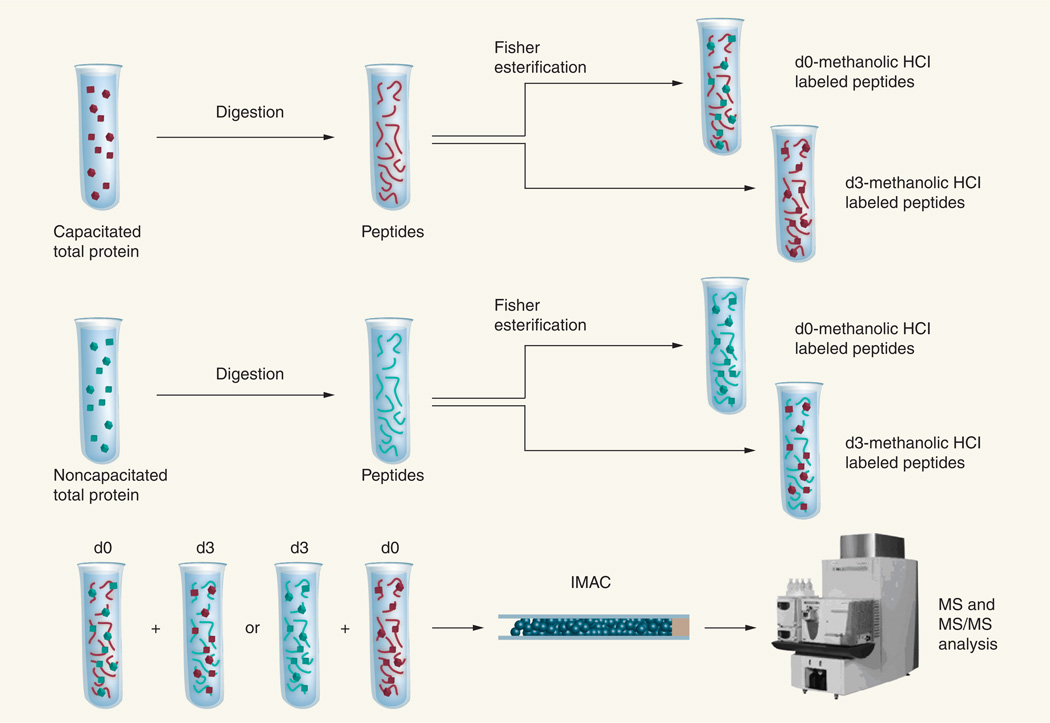
The lists of phosphoproteins and phosphopeptides obtained in studies attempting to link sperm protein PTM with function take on greater biological significance when associated with measurements permitting their relative quantitation. Difference in-gel electrophoresis (DIGE) has been used in conjunction with anti-pS antibodies for the relative quantitation of a protein believed to be phosphorylated to a greater extent in caput versus caudal spermatozoa [58]. This approach, which relies upon protein labeling using fluorescent dyes with unique excitation wavelengths, permits the visualization and volume measurement of the same protein spot from two differentially labeled samples on a single 2DE gel, thereby eliminating the problems associated with gel to gel variability [77]. However, this technique does not allow for the localization of specific phosphorylation sites and, as mentioned earlier, the sensitivity of fluorescent labeling is often incompatible with downstream protein identification. A more suitable approach for the relative quantitation of phosphorylation changes occurring on a particular residue is the differential chemical labeling of peptides following protein digestion [78–80]. To this end, our group has again taken advantage of the Fischer esterification, which is required for the enrichment of phosphorylated species using IMAC as explained earlier, and carried out the reaction on capacitated and noncapacitated sperm protein digests using methanol (d0) and deuterated methanol (d3) as reagents [44]. In the simplest experiment, the two isotopically labeled peptide samples are combined and examined together in a single MS analysis for the relative quantitation of protein expression. Alternatively, as shown in Figure 2, the pooled sample can be enriched for phosphopeptides and a single MS experiment can be used to simultaneously identify the phosphoproteins, pinpoint the specific sites of phosphorylation and quantitate the relative extent of modification at these sites as a consequence of the biological changes occurring within sperm.
Figure 2. Differential isotopic labeling strategy for mass spectrometry-based identification and quantitation of phosphorylation changes associated with capacitation.
Proteins extracted from both capacitated and noncapacitated sperm samples are first digested and split into two aliquots. The Fischer esterification is carried out on one aliquot from each sample using anhydrous methanol and on another aliquot using anhydrous, deuterated methanol to convert the peptides to their corresponding methyl esters. Differentially labeled peptide samples (e.g., d0-labeled capacitated peptides and d3-labeled noncapacitated peptides) are combined and subjected to IMAC-based phosphopeptide enrichment and MS analysis. Carrying out the same analysis in the ‘reverse’ direction (e.g., d3-labeled capacitated peptides and d0-labeled noncapacitated peptides) permits the calculation of an odds ratio for quantitation.
IMAC: Immobilized metal ion affinity chromatography; MS: Mass spectrometry; MS/MS: Tandem mass spectrometry.
Adapted with permission from [44]. © 2009 American Chemical Society.
Mass spectrometry
Although it is by no means the only factor to be considered in the proteomics process, MS represents the major enabling technology for the identification of sperm phosphoproteins and the localization of phosphorylation sites. Following sample preparation, consideration must be given to the residual protein complexity, concentration and potential contamination in order to select the most appropriate MS instrumentation for downstream analysis. As each type of mass spectrometer uses a specific method of ionization, mass analysis, fragmentation and detection, each has a unique set of capabilities making it particularly well suited for a given phosphoproteomic study.
For the rapid identification of sperm phosphoproteins resolved on 2DE gels, several studies have avoided amino acid sequencing of the peptides resulting from in-gel digestion, and have instead used the determination of several intact peptide m/z values as a means to identify the protein from which they are derived [27,58,81–83]. The success of this approach, known as PMF, requires that multiple peptide m/z values be matched to a single known protein sequence. For this reason, an instrument coupling time-of-flight (TOF) detection with matrix-assisted laser desorption/ionization (MALDI) is often used, as this combination exhibits a high resolving power stemming from the method of ion generation. MALDI uses a brief laser pulse applied to a dried matrix-sample droplet in a vacuum chamber to generate predominantly singly charged gas-phase ions in a discrete location and with a narrow distribution of kinetic energies [84]. As a consequence, the initial spread of velocities for ions with the same m/z is minimized, ensuring that their arrival time at the TOF detector can be measured accurately [85]. This permits the peptide m/z values to be determined with a high degree of precision, minimizing the number of potential amino acid sequences associated with each and significantly improving the likelihood of correctly identifying the corresponding protein of interest.
When irrefutable phosphoprotein identification is required however, a partial determination of the primary amino acid sequence of the protein must be obtained. MS/MS of peptides involves the gas-phase isolation of species with a given m/z, their fragmentation, and the detection of the resulting product ions. Carrying out this process on a MALDI-TOF/TOF instrument involves an initial round of TOF to isolate ions of a specific m/z in a trapping cell so that they may undergo a process known as collisionally induced dissociation (CID). The peptide ions are confined for a finite amount of time, during which they undergo multiple collisions with an inert gas held at relatively high pressure within the cell. The energy imparted to the ions through each impact gradually raises their internal vibrational energies in an ergodic process that eventually induces them to fragment [86]. Following fragmentation, the product ions are accelerated out of the cell and into a second TOF region, providing a high mass accuracy determination of the fragment m/z values and permitting both peptide sequencing and phosphoprotein identification [30].
As useful as it is, a MALDI-dependent approach becomes impractical when the peptide mixture is of even moderate complexity (i.e., derived from more than two proteins) as MALDI ionizes all species in a sample simultaneously and may suffer from reproducibility issues associated with the nonhomogeneity of matrix crystallization and the localization of peptides to particular regions (‘hot spots’) within the matrix-sample droplet [87]. As an alternative, electrospray ionization (ESI) involves the application of high potential (±1–5 kV) on a capillary emitter tip to produce multiply charged, gas-phase peptide ions from an aerosol of small, highly charged liquid droplets [88,89]. This method is particularly well suited for the generation of ions from a complex mixture because it directly transfers solvated species into the gas phase and, as it occurs at atmospheric pressure, it can easily be interfaced with upfront liquid chromatographic (LC) peptide separation [54,90,91].
Although ESI has been successfully used in conjunction with TOF detection [42,45,82], it has most often been coupled with ion trapping instrumentation. In such mass spectrometers, the kinetic energies of the species generated by ESI are reduced through low energy collisions with an inert bath gas present in the ion trap (i.e., ‘collisional cooling’) and are then sequentially ejected to an electron multiplier in order to produce a mass spectrum. MS/MS is accomplished by first isolating ions of a given m/z value within the same trapping region, forcing these ions to undergo CID through more energetic collisions with the bath gas and then scanning out the fragments [29,40,56,90–92]. While this tandem-in-time approach to MS/MS allows successive rounds of dissociation to be carried out on the resulting fragments in order to obtain additional structural information (MSn), mass spectrometers of this type have a low duty cycle due to the use of the ion trap for both MS and MS/MS data acquisition. In addition, the effective resolution of these instruments is limited by the rate at which ions can be scanned out of the trap [93,94]. In order to address these issues, ESI sources have been coupled with Fourier transform (FT)-based detectors that exhibit greatly improved duty cycle (i.e., the number of ions detected in a given time) and resolving power relative to their scanning, ion trapping counterparts [28,85]. The major drawback of these instruments is the extremely low vacuum required for FT-based detection that prevents the concomitant use of a CID bath gas in the trapping cell for controlled peptide fragmentation. Hybrid mass spectrometers work around this issue by incorporating both ion trapping and FT-based detectors into a single instrument: while the resolving power and associated mass accuracy of FT-based detection is being exploited for the determination of m/z, CID can be carried out simultaneously in the ion trap to generate a linked MS/MS spectrum for a given precursor [44].
Although advances in peptide ionization and MS instrumentation have resulted in the unequivocal identification of many sperm phosphoproteins, a molecular-level understanding of sperm function requires determination of the specific sites of phosphorylation within these proteins. The electronegativity and unique chemical characteristics of the phosphate moiety, which make it useful in biological signaling, lead to a number of issues that must be addressed in order to successfully identify specific sites of protein phosphorylation in sperm. For example, it has been reported that phosphorylated peptides ionize poorly when traditional MALDI matrices are used [95]. To overcome this issue, different matrices [96] and acidic additives [97,98] have been utilized to increase ionization yields and improve phosphopeptide identification. The complications associated with MALDI-based sequencing of phosphopeptide mixtures have been circumvented by using reversed phase LC coupled with ESI MS/MS, but this approach is not without its own inherent difficulties, even when upstream phosphopeptide enrichment is used. To begin with, phosphorylated species exhibit decreased hydrophobicity when compared with their unmodified counterparts, so shallow reversed phase LC elution gradients and minimal column rinsing need to be used to ensure phosphopeptide retention and chromatographic resolution. In addition, the phosphodiester bond that links the phosphate moiety to serine and threonine residues is labile, making it the bond most readily broken when CID is used for phosphopeptide fragmentation [56,75–78]. This produces MS/MS spectra in which the majority of the detected ion current represents species that have undergone the neutral loss of phosphoric acid and in which there are few, if any, sequence-informative peaks [28,45].
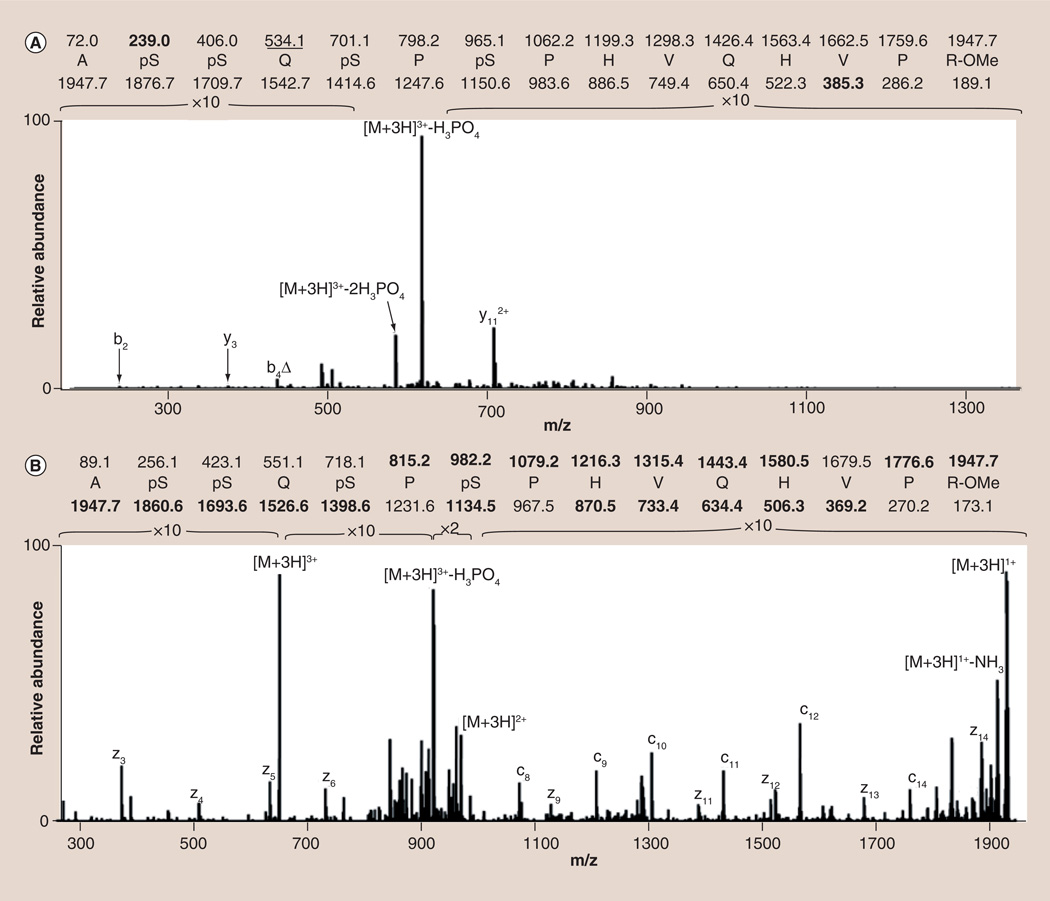
The creation of a linear ion trap (LIT) capable of isolating significantly more precursor ions [99] partially addresses this problem by trapping more fragments and increasing, above the noise threshold, the signals resulting from peptide backbone cleavage [42,44,45]. As shown in Figure 3A, while there are a few detectable sequence-informative peaks in the MS/MS spectrum acquired on a LIT mass spectrometer, they are low level and, alone, they are insufficient to allow for phosphopeptide sequencing. In this instance, it may have been beneficial to conduct the analysis on a hybrid instrument in which the high mass accuracy afforded by FT-based detection could have been used in combination with the limited sequence information to successfully identify the phosphorylation sites [44,99]. Ultimately, however, the limitation in sperm phosphopeptide sequencing using this approach stems from the ergodic nature of the CID process itself. This means that the energy deposited gradually becomes randomized throughout the peptide and leads to fragmentation occurring primarily at the labile phosphodiester bond [100]. While identification of the phosphorylated peptide can be improved by using alkaline phosphatase to remove the phosphate moiety prior to CID, this procedure alone cannot be used to unequivocally identify the site(s) of phosphorylation [28]. As an alternative, a novel, nonergodic technique referred to as ETD has been developed wherein the transfer of a low energy (<10 eV) electron from a radical anionic gas leads to fragmentation of the phosphopeptide without inducing the neutral loss of phosphoric acid [101]. As illustrated in Figure 3B, the greater extent of backbone cleavage shown in the ETD MS/MS spectrum permits unambiguous assignment of the amino acid sequence and localization of the sites of phosphorylation within the large, multiply phosphorylated peptide [42,45].
Figure 3. Comparison between the (A) CID and (B) ETD spectra of a +3, quadruply phosphorylated peptide from testis-specific serine/proline-rich protein.
While the CID spectrum is dominated by the neutral loss of phosphoric acid from the intact precursor, the ETD spectrum shows significant, sequence-informative peptide backbone fragmentation. Calculated m/z values found in the spectra are shown in bold while those m/z values exhibiting the neutral loss of phosphoric acid are underlined. Δ denotes assigned sequence ions which have undergone the neutral loss of phosphoric acid.
CID: Collisionally induced dissociation; ETD: Electron transfer dissociation.
Mass spectral data interpretation
The ability to draw meaningful biological conclusions from any sperm phosphoproteomics experiment depends entirely upon an unbiased interpretation of the data. However, the complexity of the proteomics process means that it is possible to unintentionally neglect a critical consideration, which, in turn, severely compromises the quality of the peptide and protein assignments. In addition, the large number of spectra obtained in a given MS experiment is often used to justify complete reliance on software for sequence assignments, which can further compound the problem. For example, the database searching tool Mascot [102] has been utilized extensively for the identification of sperm phosphoproteins in PMF experiments [58,83,103]. The software works by extracting the most abundant peptide m/z values from an experimental MS spectrum, comparing them with values generated from the in silico enzymatic digestion of proteins contained within a user-selected database and then assigning a protein or proteins to each spectrum on the basis of the matched peptide m/z values. Although the manner in which Mascot carries out these steps is proprietary, it uses the probability-based MOWSE algorithm [104] to associate a significance threshold with each protein assignment such that a sequence with a score above this threshold has a low probability of being assigned by chance. Unfortunately, the significance of this score is easily altered when an incomplete set of PTMs are considered or when fewer proteins are present in the interrogated dataset [105], as is the case when a species-specific subdatabase is used to expedite the data analysis process.
If the goal of the project is not simply the identification of sperm phosphoproteins but rather the visualization of specific phosphorylation sites, determination of the primary amino acid sequence is required. The Mascot algorithm can be used for this purpose by matching the m/z values in an acquired CID MS/MS spectrum with the m/z values generated from in silico peptide backbone fragmentation [106]. However, even though Mascot is the most widely used software for phosphopeptide sequencing [30,42,45], the proprietary nature of the software severely limits user understanding and control of the process [107]. By contrast, the operation of the second most commonly used database searching algorithm in sperm phosphoproteomic studies [28,40,42,45,108], SEQUEST, is well documented [109,201]. SEQUEST consists of two rounds of scoring: the first round prioritizes potential peptide assignments by matching the m/z values for fragment peaks in the MS/MS spectrum with fragment m/z values determined through in silico peptide fragmentation. The second round involves the creation of idealized MS/MS spectra for the highest scoring peptide sequences from the first round and then their translation across the experimental spectrum using the Fast Fourier Transform (FFT). Unlike the probability-based scoring used in Mascot, this process results in the calculation of a cross-correlation (XCorr) score that is a measure of how well a given peptide assignment matches the distribution of peaks within the experimental spectrum.
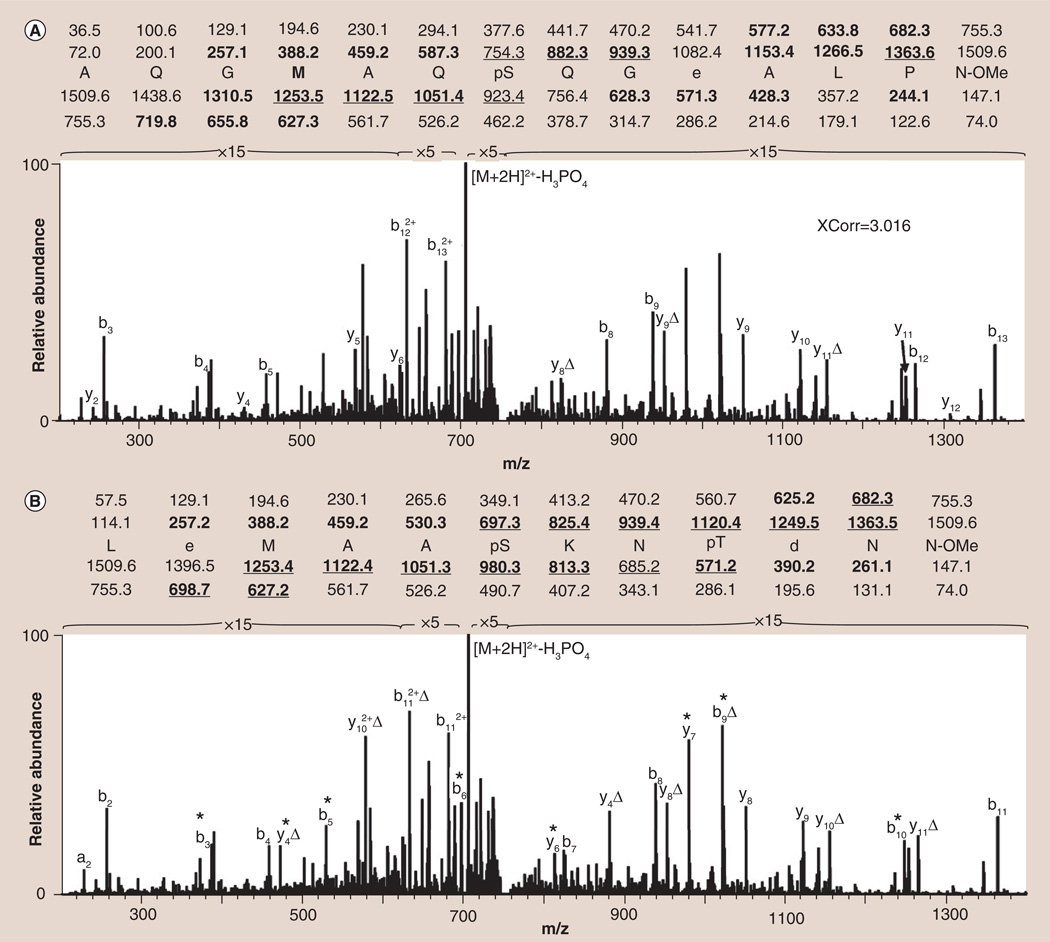
Regardless of the software tool, successful use of automated searching algorithms requires that the correct peptide be present in the database and that the sequence fit the constraints programmed by the user. As shown in Figure 4A, the best SEQUEST assignment for the given MS/MS spectrum is the singly phosphorylated peptide AQGMAQpSQGEALPN. Although this peptide assignment has a relatively high XCorr of 3.016, it actually represents a false-positive identification: a problem frequently encountered in phosphopeptide sequencing due to the large number of peaks corresponding to the neutral loss of phosphoric acid. As shown in Figure 4B, a greater number of fragment ions within the spectrum are actually explained by the doubly phosphorylated peptide LEMAApSKNpTDNN. The sequence was misassigned by SEQUEST because the correct sequence contains an unanticipated modification, deamidation of an asparagine residue to aspartic acid, and therefore does not exist in any protein database. It is important to note that even if this modification had been considered, the correct sequence would still not have been determined using SEQUEST as the aspartic acid, which was generated following deamidation, was subsequently methylated using the Fischer esterification.
Figure 4. Different sequence assignments for a single tandem mass spectrum.
(A) Most probable (XCorr 3.016) sequence assignment when the spectrum is searched against a human, rat and mouse protein database using SEQUEST. (B) The amino acid sequence as determined through manual de novo peptide sequencing. The sequence shown in (B) is more probable due to the presence of additional fragment ions (marked with *) in the spectrum. Calculated m/z values found in the spectra are shown in bold while those m/z values exhibiting the neutral loss of phosphoric acid are underlined. D denotes assigned sequence ions that have undergone the neutral loss of phosphoric acid.
The correct phosphopeptide sequence (Figure 4B) was only assigned to the MS/MS spectrum shown in Figure 4 through de novo peptide sequencing, an unbiased, but more technically challenging, alternative to automated database searching [301]. In its simplest form, de novo sequencing involves the preliminary identification of sequence informative peaks within the spectrum by finding complementary pairs of ions whose m/z values sum to the approximate mass of the intact peptide. Notably, this process can be carried out using the c- and z-ion pairs that are commonly encountered in ETD or with the b- and y-ion pairs that are found in CID spectra [110]. Once identified, the spacing between all of the peaks is examined to find m/z differences that correspond to mass shifts characteristic of particular amino acids. In this manner, a string of MS/MS peaks can be found which explain the entire peptide sequence or which explain enough of the sequence (a sequence ‘tag’) to permit a subsequent database search, such as the Basic Local Alignment Search Tool (BLAST) at the National Center for Biotechnology Information (NCBI), to be used for peptide/protein identification [111].
Although a more detailed description of de novo sequencing is beyond the scope of this review, successful use of the process frequently involves the consideration of several disparate lines of evidence, especially when it is applied to phosphopeptides. For instance, our group has used de novo sequencing to assign phosphopeptide sequences to CID MS/MS spectra derived from isotopically labeled, IMAC-enriched, capacitated and noncapacitated mouse sperm digests [44]. This is often extremely challenging due to the aforementioned neutral loss of phosphoric acid, but in this case the ability to compare a given MS/MS spectrum with its differentially labeled counterpart greatly improved our de novo sequencing capability. To begin with, calculation of the difference between the ‘heavy’ and ‘light’ precursor masses allowed the number of glutamic and/or aspartic acid residues in the phosphopeptide to be determined prior to sequencing. Likewise, inspection of the MS/MS spectrum of the deuterated version of the peptide for fragment peaks whose masses shifted as a consequence of the ‘heavy’ label simplified the discovery of complementary ion pairs, permitted the rapid determination of the ion series corresponding to the peptide C-terminus and made the localization of acidic residues within the sequence remarkably straightforward. As an example of the utility of this approach, 11 phosphopeptides whose amino acid sequences were not present in the NCBI non-redundant (nr) protein database were sequenced in this study. A subsequent search of these sequences against the mouse genome revealed an unknown gene in a region which had previously been considered noncoding. The identification of the protein encoded by this gene, which the authors have named testis-specific serine/proline-rich protein [112], represents an exception to the currently accepted paradigm: rather than relying on translated genetic information for protein sequencing it is possible to identify a target gene directly through de novo peptide sequencing.
Quantitation of site-specific phosphorylation
Metabolic labeling strategies (e.g., SILAC) commonly used for relative quantitation in other cell types are of limited utility in sperm because, as mentioned earlier, sperm are believed to be both transcriptionally and translationally silent after leaving the testes [3,4]. Nonetheless, alternative strategies have been used in sperm phosphoproteomic studies to not only quantitate phosphoprotein expression [56] but to quantify the extent of modification at a specific phosphorylation site [42,44,45]. One of these methods, label-free quantitation, uses the ratio of chromatographic peak areas obtained in separate LC-MS analyses to quantify differences between the samples being compared [113]. Although this method avoids the additional sample handling associated with chemical labeling, quantitation using this approach is difficult due to the influence which the sample medium and chromatographic conditions have on ionization efficiency and, ultimately, on MS-based detection. These inherent variations require the analysis of a significant number of biological replicates. For example, Baker et al. carried out eight replicate MS analyses of both caput and caudal spermatozoa and used specialized software (DeCyder™ MS) to average out the variability among these 16 analyses in order to quantitate phosphorylation changes associated with epididymal maturation [45]. In an alternative strategy, our group circumvented the issue of run-to-run variability by analyzing the heavy and light labeled versions of capacitated and noncapacitated sperm digests simultaneously in a single MS experiment [44]. We also carried out the analysis with the labeling reversed in order to correct for any variation in chromatographic retention, ionization efficiency or MS detection caused by the presence of the isotopic label. Within each individual analysis, the chromatographic peak area of a phosphopeptide derived from the capacitated sample was divided by the peak area of the identical, differentially labeled phosphopeptide derived from the noncapacitated sample. These two ratios were then used to calculate an odds ratio [114], which more accurately reflects the relative change in phosphorylation occurring on a particular phosphopeptide as a result of the capacitation process.
Expert commentary
The importance of phosphorylation in sperm has been well documented, but only recently, phosphoproteomic workflows have been developed that can concomitantly identify phosphoproteins, localize specific sites of phosphorylation and quantitate the extent of modification at a particular site. In our experience, a workflow incorporating both IMAC and TiO2 phosphopeptide enrichment is necessary to address the low stoichiometric level of phosphorylation and to obtain a more comprehensive view of phosphorylation events in sperm. However, chemical modification should be used to minimize the binding of nonphosphorylated peptides to the enrichment resins. For this purpose, we use the Fischer esterification as it has a number of additional advantages: the reagents are relatively inexpensive, the reaction goes to completion under anhydrous conditions and every peptide in the mixture contains at least one label (at the peptide C-terminus). In addition, the Fischer esterification can be carried out using heavy and light methanol if the phosphoproteomic analysis involves a comparison of dissimilar samples. As mentioned earlier, this allows the enriched and differentially labeled peptides to be mixed and analyzed in a single LC-MS experiment. We recommend carrying out this experiment on a hybrid mass spectrometer, as the high resolving power can be used to more accurately measure chromatographic peak areas for purposes of quantitation, while the associated increase in accurate mass determination can be used to improve phosphopeptide sequencing. Furthermore, some of these instruments are capable of utilizing ETD as an alternative phosphopeptide fragmentation method if the CID MS/MS spectra exhibit the dominant neutral loss of phosphoric acid [115]. However, ETD is a relatively recent development and, despite its commercial availability, optimization of the process is an active area of research [116]. In addition, ETD involves a different fragmentation mechanism than CID and, as a result, current database searching algorithms have difficulty assigning amino acid sequences to ETD spectra. In our laboratory, we capitalize upon the differential isotopic labeling strategy to aid in the de novo sequencing of both CID and ETD phosphopeptide MS/MS spectra, but we recognize that this approach is technically challenging and impractical for extremely large datasets. As an alternative, we suggest that most laboratories use either SEQUEST or Andromeda [117], a well-documented probability-based scoring algorithm similar to Mascot, for automated phosphopeptide sequence assignment. We also recommend that validation of the sequence involve more than a ‘manual check’ to identify the presence of several fragment m/z values within the spectrum: this only serves to confirm the operation of the software and says nothing about the quality of the peptide assignment. In our opinion, no phosphopeptide sequence should be accepted if the associated MS/MS spectrum contains peak pairs that are unaccounted for, or if a significant portion of the ion current (~20%) remains unexplained. Under these circumstances, the probability that this assignment is a false-positive identification is simply too high and, with more and more laboratories relying on proteomic data, the potential for this information to be used as the basis for unwarranted research effort is simply too great.
Five-year view
Our current understanding of the role which phosphorylation plays in the regulation of sperm function has relied almost entirely on recent advances in the field of proteomics. As methodologies for the global enrichment of phosphorylated species and MS-based sequencing techniques are adopted by more and more laboratories involved in sperm research, it is likely that the number of identified protein phosphorylation sites will increase dramatically in the next 5 years. In addition, the ability to quantitate changes in the extent of modification at particular amino acids, coupled with advances in germ cell transplantation and transgenic rescue of knock-out models will begin to reveal the role of these specific sites in sperm function. However, the utility of this information is absolutely dependent upon the quality of the experimental design and the resultant data. As we move forward, the greatest challenge in the field of sperm phosphoproteomics will be in ensuring that non-proteomic specialists fully understand the complexity of the overall proteomics process and that they adequately validate and correctly interpret their results.
Key issues.
As sperm are transcriptionally and translationally silent after leaving the testes and because their post-testicular development is controlled almost exclusively by the addition of exogenous proteins or by the post-translational modification of their intrinsic protein complement, proteomics is required to investigate sperm function.
Phosphorylation has been shown to be one of the major post-translational modifications controlling sperm function, both during epididymal maturation, development, and later on in the female reproductive tract during capacitation. Although more is known about the regulation of capacitation-associated changes in tyrosine phosphorylation, other pathways still need to be elucidated.
Drawing meaningful conclusions from a proteomics experiment requires the successful integration of the diverse areas of sample sourcing/preparation, mass spectrometric analysis, data interpretation and determination of biological significance.
The development of immobilized metal affinity chromatography and titanium dioxide for phosphopeptide enrichment expands on antibody affinity-based methods and permits phosphorylation analysis on a global scale.
The dominant neutral loss of phosphoric acid from phosphopeptides fragmented using collisionally induced dissociation has recently been addressed by the development of more sensitive mass spectrometers capable of high mass accuracy measurements and electron transfer dissociation-based fragmentation.
Manual validation of phosphopeptide sequence assignments and the quantitation of relative changes in phosphorylation are absolutely critical as more laboratories adopt proteomic technologies for the investigation of sperm phosphorylation and as they carry out additional experiments based on this data.
Acknowledgments
This work was supported by Rensselaer Polytechnic Institute start-up funds (to MD Platt) and grants (HD38082 and HD44044) from the NIH (to PE Visconti).
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.De Jonge CJ, Barratt CLR, editors. The Sperm Cell Production, Maturation, Fertilization, Regeneration. NY, USA: Cambridge University Press; 2006. [Google Scholar]
- 2.Bloch D. A catalog of sperm histones. Genetics. 1969;61(1):93–111. [PubMed] [Google Scholar]
- 3.Díez-Sánchez C, Ruiz-Pesini E, Montoya J, Pérez-Martos A, Enríquez JA, López-Pérez MJ. Mitochondria from ejaculated human spermatozoa do not synthesize proteins. FEBS Lett. 2003;553(1–2):205–208. doi: 10.1016/s0014-5793(03)01013-5. [DOI] [PubMed] [Google Scholar]
- 4.Aitken RJ, Nixon B, Lin M, Koppers AJ, Lee YH, Baker MA. Proteomic changes in mammalian spermatozoa during epididymal maturation. Asian J. Androl. 2007;9(4):554–564. doi: 10.1111/j.1745-7262.2007.00280.x. [DOI] [PubMed] [Google Scholar]
- 5.Roberts KP, Ensrud KM, Wooters JL, Nolan MA, Johnston DS, Hamilton DW. Epididymal secreted protein Crisp-1 and sperm function. Mol. Cell. Endocrinol. 2006;250(1– 2):122–127. doi: 10.1016/j.mce.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 6.O’Rand MG, Widgren EE, Hamil KG, Silva EJ, Richardson RT. Epididymal protein targets: a brief history of the development of EPPIN as a contraceptive target. J. Androl. 2011;32(6):698–704. doi: 10.2164/jandrol.110.012781. [DOI] [PubMed] [Google Scholar]
- 7.Vijayaraghavan S, Stephens DT, Trautman K, et al. Sperm motility development in the epididymis is associated with decreased glycogen synthase kinase-3 and protein phosphatase 1 activity. Biol. Reprod. 1996;54(3):709–718. doi: 10.1095/biolreprod54.3.709. [DOI] [PubMed] [Google Scholar]
- 8.Visconti PE, Krapf D, de la Vega-Beltrán JL, Acevedo JJ, Darszon A. Ion channels, phosphorylation and mammalian sperm capacitation. Asian J. Androl. 2011;13(3):395–405. doi: 10.1038/aja.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cox J, Mann M. Is proteomics the new genomics? Cell. 2007;130(3):395–398. doi: 10.1016/j.cell.2007.07.032. • Introduces current proteomic capabilities and highlights the application of these technologies to comprehensive protein identification and quantitation.
- 10.Wilkins MR, Sanchez JC, Gooley AA, et al. Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol. Genet. Eng. Rev. 1996;13:19–50. doi: 10.1080/02648725.1996.10647923. [DOI] [PubMed] [Google Scholar]
- 11.Felix K, Krekels A. Die endaminosauren einiger protamine. Hoppe-Seylers Zeitschrift Fur Physiologische Chemie. 1953;295:107–109. doi: 10.1515/bchm2.1953.295.1.107. [DOI] [PubMed] [Google Scholar]
- 12. Ingles CJ, Dixon GH. Phosphorylation of protamine during spermatogenesis in trout testis. Proc. Natl Acad. Sci. USA. 1967;58(3):1011–1018. doi: 10.1073/pnas.58.3.1011. • This is the first paper to detail procedures, some of which are used even today, for the direct identification of specific sites of phosphorylation in sperm protamines.
- 13.Garbers D, Lust W, First N, Lardy H. Effects of phosphodiesterase inhibitors and cyclic nucleotides on sperm respiration and motility. Biochemistry. 1971;10(10):1825–1831. [Google Scholar]
- 14.Hoskins DD, Casillas ER, Stephens DT. Cyclic AMP-dependent protein kinases of bovine epididymal spermatozoa. Biochem. Biophys. Res. Commun. 1972;48(6):1131–1138. doi: 10.1016/0006-291x(72)90858-3. [DOI] [PubMed] [Google Scholar]
- 15.Brandt H, Hoskins DD. A cAMP-dependent phosphorylated motility protein in bovine epididymal sperm. J. Biol. Chem. 1980;255(3):982–987. [PubMed] [Google Scholar]
- 16.Frackelton AR, Jr, Ross AH, Eisen HN. Characterization and use of monoclonal antibodies for isolation of phosphotyrosyl proteins from retrovirus-transformed cells and growth factor-stimulated cells. Mol. Cell. Biol. 1983;3(8):1343–1352. doi: 10.1128/mcb.3.8.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leyton L, Saling P. 95 kd sperm proteins bind ZP3 and serve as tyrosine kinase substrates in response to zona binding. Cell. 1989;57(7):1123–1130. doi: 10.1016/0092-8674(89)90049-4. [DOI] [PubMed] [Google Scholar]
- 18.Kalab P, Visconti P, Leclerc P, Kopf GS. p95, the major phosphotyrosine-containing protein in mouse spermatozoa, is a hexokinase with unique properties. J. Biol. Chem. 1994;269(5):3810–3817. [PubMed] [Google Scholar]
- 19. Visconti PE, Bailey JL, Moore GD, Pan D, Olds-Clarke P, Kopf GS. Capacitation of mouse spermatozoa. I. Correlation between the capacitation state and protein tyrosine phosphorylation. Development. 1995;121(4):1129–1137. doi: 10.1242/dev.121.4.1129. • First paper to establish an unequivocal link between protein phosphorylation and the critical cellular process of sperm capacitation.
- 20.Visconti PE, Moore GD, Bailey JL, et al. Capacitation of mouse spermatozoa. II. Protein tyrosine phosphorylation and capacitation are regulated by a cAMP-dependent pathway. Development. 1995;121(4):1139–1150. doi: 10.1242/dev.121.4.1139. [DOI] [PubMed] [Google Scholar]
- 21.Galantino-Homer HL, Visconti PE, Kopf GS. Regulation of protein tyrosine phosphorylation during bovine sperm capacitation by a cyclic adenosine 3´5´-monophosphate-dependent pathway. Biol. Reprod. 1997;56(3):707–719. doi: 10.1095/biolreprod56.3.707. [DOI] [PubMed] [Google Scholar]
- 22.Leclerc P, de Lamirande E, Gagnon C. Cyclic adenosine 3´,5´monophosphate-dependent regulation of protein tyrosine phosphorylation in relation to human sperm capacitation and motility. Biol. Reprod. 1996;55(3):684–692. doi: 10.1095/biolreprod55.3.684. [DOI] [PubMed] [Google Scholar]
- 23.Kalab P, Peknicová J, Geussová G, Moos J. Regulation of protein tyrosine phosphorylation in boar sperm through a cAMP-dependent pathway. Mol. Reprod. Dev. 1998;51(3):304–314. doi: 10.1002/(SICI)1098-2795(199811)51:3<304::AID-MRD10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Naaby-Hansen S, Flickinger CJ, Herr JC. Two-dimensional gel electrophoretic analysis of vectorially labeled surface proteins of human spermatozoa. Biol. Reprod. 1997;56(3):771–787. doi: 10.1095/biolreprod56.3.771. [DOI] [PubMed] [Google Scholar]
- 25.Mandal A, Naaby-Hansen S, Wolkowicz MJ, et al. FSP95, a testis-specific 95-kilodalton fibrous sheath antigen that undergoes tyrosine phosphorylation in capacitated human spermatozoa. Biol. Reprod. 1999;61(5):1184–1197. doi: 10.1095/biolreprod61.5.1184. [DOI] [PubMed] [Google Scholar]
- 26. Shetty J, Diekman AB, Jayes FC, et al. Differential extraction and enrichment of human sperm surface proteins in a proteome: identification of immunocontraceptive candidates. Electrophoresis. 2001;22(14):3053–3066. doi: 10.1002/1522-2683(200108)22:14<3053::AID-ELPS3053>3.0.CO;2-K. •• First paper in which sperm proteins were identified using MS/MS-based peptide sequencing. Identification of the same protein in several distinct 2D gel spots suggested that these proteins exist in multiple, post-translationally modified forms.
- 27.Bohring C, Krause E, Habermann B, Krause W. Isolation and identification of sperm membrane antigens recognized by antisperm antibodies, and their possible role in immunological infertility disease. Mol. Hum. Reprod. 2001;7(2):113–118. doi: 10.1093/molehr/7.2.113. [DOI] [PubMed] [Google Scholar]
- 28. Ficarro S, Chertihin O, Westbrook VA, et al. Phosphoproteome analysis of capacitated human sperm. Evidence of tyrosine phosphorylation of a kinase-anchoring protein 3 and valosin-containing protein/p97 during capacitation. J. Biol. Chem. 2003;278(13):11579–11589. doi: 10.1074/jbc.M202325200. •• Utilized a 2D gel approach to identify tyrosine phosphorylated sperm proteins and was the first to determine the exact sites of protein phosphorylation in sperm using a combination of Fisher esterification and immobilized metal affinity chromatography.
- 29.Arcelay E, Salicioni AM, Wertheimer E, Visconti PE. Identification of proteins undergoing tyrosine phosphorylation during mouse sperm capacitation. Int. J. Dev. Biol. 2008;52(5–6):463–472. doi: 10.1387/ijdb.072555ea. [DOI] [PubMed] [Google Scholar]
- 30.Kota V, Dhople VM, Shivaji S. Tyrosine phosphoproteome of hamster spermatozoa: role of glycerol-3-phosphate dehydrogenase 2 in sperm capacitation. Proteomics. 2009;9(7):1809–1826. doi: 10.1002/pmic.200800519. [DOI] [PubMed] [Google Scholar]
- 31.Johnson SA, Hunter T. Kinomics: methods for deciphering the kinome. Nat. Methods. 2005;2(1):17–25. doi: 10.1038/nmeth731. [DOI] [PubMed] [Google Scholar]
- 32.de Lamirande E, Gagnon C. The extracellular signal-regulated kinase (ERK) pathway is involved in human sperm function and modulated by the superoxide anion. Mol. Hum. Reprod. 2002;8(2):124–135. doi: 10.1093/molehr/8.2.124. [DOI] [PubMed] [Google Scholar]
- 33.Luconi M, Barni T, Vannelli GB, et al. Extracellular signal-regulated kinases modulate capacitation of human spermatozoa. Biol. Reprod. 1998;58(6):1476–1489. doi: 10.1095/biolreprod58.6.1476. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Zha X, Tan Y, et al. Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J. Biol. Chem. 2002;277(42):39379–39387. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- 35.Harrison RA. Rapid PKA-catalysed phosphorylation of boar sperm proteins induced by the capacitating agent bicarbonate. Mol. Reprod. Dev. 2004;67(3):337–352. doi: 10.1002/mrd.20028. [DOI] [PubMed] [Google Scholar]
- 36.Krapf D, Arcelay E, Wertheimer EV, et al. Inhibition of Ser/Thr phosphatases induces capacitation-associated signaling in the presence of Src kinase inhibitors. J. Biol Chem. 2010;285(11):7977–7985. doi: 10.1074/jbc.M109.085845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rotman T, Etkovitz N, Spiegel A, Rubinstein S, Breitbart H. Protein kinase A and protein kinase C(alpha)/PPP1CC2 play opposing roles in the regulation of phosphatidylinositol 3-kinase activation in bovine sperm. Reproduction. 2010;140(1):43–56. doi: 10.1530/REP-09-0314. [DOI] [PubMed] [Google Scholar]
- 38.Morgan DJ, Weisenhaus M, Shum S, et al. Tissue-specific PKA inhibition using a chemical genetic approach and its application to studies on sperm capacitation. Proc Natl Acad. Sci USA. 2008;105(52):20740–20745. doi: 10.1073/pnas.0810971105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Brien ED, Krapf D, Cabada MO, Visconti PE, Arranz SE. Transmembrane adenylyl cyclase regulates amphibian sperm motility through protein kinase A activation. Dev. Biol. 2011;350(1):80–88. doi: 10.1016/j.ydbio.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su YH, Chen SH, Zhou H, Vacquier VD. Tandem mass spectrometry identifies proteins phosphorylated by cyclic AMP-dependent protein kinase when sea urchin sperm undergo the acrosome reaction. Dev. Biol. 2005;285(1):116–125. doi: 10.1016/j.ydbio.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 41.Jha KN, Salicioni AM, Arcelay E, et al. Evidence for the involvement of proline-directed serine/threonine phosphorylation in sperm capacitation. Mol. Hum. Reprod. 2006;12(12):781–789. doi: 10.1093/molehr/gal085. [DOI] [PubMed] [Google Scholar]
- 42.Baker MA, Smith ND, Hetherington L, et al. Label-free quantitation of phosphopeptide changes during rat sperm capacitation. J. Proteome Res. 2010;9(2):718–729. doi: 10.1021/pr900513d. [DOI] [PubMed] [Google Scholar]
- 43.Fischer E, Speier A. Darstellung der Ester. Berichte der deutschen chemischen Gesellschaft. 1895;28(3):3252–3258. [Google Scholar]
- 44. Platt MD, Salicioni AM, Hunt DF, Visconti PE. Use of differential isotopic labeling and mass spectrometry to analyze capacitation-associated changes in the phosphorylation status of mouse sperm proteins. J. Proteome Res. 2009;8(3):1431–1440. doi: 10.1021/pr800796j. •• Details the use of immobilized metal affinity chromatography for global phosphopeptide enrichment and differential isotopic labeling to quantitate the extent of modification occurring at individual phosphorylation sites as a result of the capacitation process.
- 45. Baker MA, Smith ND, Hetherington L, Pelzing M, Condina MR, Aitken RJ. Use of titanium dioxide to find phosphopeptide and total protein changes during epididymal sperm maturation. J. Proteome Res. 2011;10(3):1004–1017. doi: 10.1021/pr1007224. •• Details the use of titanium dioxide for global phosphopeptide enrichment and label-free quantitation to determine the protein phosphorylation changes occurring in sperm during epididymal maturation. It also is the first to use electron transfer dissociation for the sequencing of phosphopeptides from sperm proteins.
- 46.Coon JJ. Collisions or electrons? Protein sequence analysis in the 21st century. Anal. Chem. 2009;81(9):3208–3215. doi: 10.1021/ac802330b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li LW, Fan LQ, Zhu WB, et al. Establishment of a high-resolution 2-D reference map of human spermatozoal proteins from 12 fertile sperm-bank donors. Asian J. Androl. 2007;9(3):321–329. doi: 10.1111/j.1745-7262.2007.00261.x. [DOI] [PubMed] [Google Scholar]
- 48.Hu Q, Noll RJ, Li H, Makarov A, Hardman M, Graham Cooks R. The Orbitrap: a new mass spectrometer. J. Mass Spectrom. 2005;40(4):430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 49.Hagel P, Gerding JJ, Fieggen W, Bloemendal H. Cyanate formation in solutions of urea. I. Calculation of cyanate concentrations at different temperature and pH. Biochim. Biophys. Acta. 1971;243(3):366–373. doi: 10.1016/0005-2795(71)90003-1. [DOI] [PubMed] [Google Scholar]
- 50.Gerding JJ, Koppers A, Hagel P, Bloemendal H. Cyanate formation in solutions of urea. II. Effect of urea on the eye lens protein -crystallin. Biochim. Biophys. Acta. 1971;243(3):375–379. [PubMed] [Google Scholar]
- 51.Edman P, Begg G. A protein sequenator. Eur. J. Biochem. 1967;1(1):80–91. doi: 10.1007/978-3-662-25813-2_14. [DOI] [PubMed] [Google Scholar]
- 52.Sleight SB, Miranda PV, Plaskett NW, et al. Isolation and proteomic analysis of mouse sperm detergent-resistant membrane fractions: evidence for dissociation of lipid rafts during capacitation. Biol. Reprod. 2005;73(4):721–729. doi: 10.1095/biolreprod.105.041533. [DOI] [PubMed] [Google Scholar]
- 53.Krisfalusi M, Miki K, Magyar PL, O’Brien DA. Multiple glycolytic enzymes are tightly bound to the fibrous sheath of mouse spermatozoa. Biol. Reprod. 2006;75(2):270–278. doi: 10.1095/biolreprod.105.049684. [DOI] [PubMed] [Google Scholar]
- 54.Johnston DS, Wooters J, Kopf GS, Qiu Y, Roberts KP. Analysis of the human sperm proteome. Ann. NY Acad. Sci. 2005;1061:190–202. doi: 10.1196/annals.1336.021. [DOI] [PubMed] [Google Scholar]
- 55.Haglund H. Isoelectric focusing in pH gradients – a technique for fractionation and characterization of ampholytes. Methods Biochem. Anal. 1971;19:1–104. doi: 10.1002/9780470110386.ch1. [DOI] [PubMed] [Google Scholar]
- 56.Baker MA, Reeves G, Hetherington L, Aitken RJ. Analysis of proteomic changes associated with sperm capacitation through the combined use of IPG-strip pre-fractionation followed by RP chromatography LC-MS/MS analysis. Proteomics. 2010;10(3):482–495. doi: 10.1002/pmic.200900574. [DOI] [PubMed] [Google Scholar]
- 57.O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 58.Baker MA, Witherdin R, Hetherington L, Cunningham-Smith K, Aitken RJ. Identification of post-translational modifications that occur during sperm maturation using difference in two-dimensional gel electrophoresis. Proteomics. 2005;5(4):1003–1012. doi: 10.1002/pmic.200401100. [DOI] [PubMed] [Google Scholar]
- 59.Wray W, Boulikas T, Wray VP, Hancock R. Silver staining of proteins in polyacrylamide gels. Anal. Biochem. 1981;118(1):197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- 60.Fujinoki M, Kawamura T, Toda T, et al. Identification of 36-kDa flagellar phosphoproteins associated with hamster sperm motility. J. Biochem. 2003;133(3):361–369. doi: 10.1093/jb/mvg048. [DOI] [PubMed] [Google Scholar]
- 61.Staudenmann W, Hatt PD, Hoving S, Lehmann A, Kertesz M, James P. Sample handling for proteome analysis. Electrophoresis. 1998;19(6):901–908. doi: 10.1002/elps.1150190605. [DOI] [PubMed] [Google Scholar]
- 62.Tabb DL, Smith LL, Breci LA, Wysocki VH, Lin D, Yates JR., 3rd Statistical characterization of ion trap tandem mass spectra from doubly charged tryptic peptides. Anal. Chem. 2003;75(5):1155–1163. doi: 10.1021/ac026122m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCormack AL, Somogyi A, Dongré AR, Wysocki VH. Fragmentation of protonated peptides: surface-induced dissociation in conjunction with a quantum mechanical approach. Anal. Chem. 1993;65(20):2859–2872. doi: 10.1021/ac00068a024. [DOI] [PubMed] [Google Scholar]
- 64.Quach TT, Li N, Richards DP, Zheng J, Keller BO, Li L. Development and applications of in-gel CNBr/tryptic digestion combined with mass spectrometry for the analysis of membrane proteins. J. Proteome Res. 2003;2(5):543–552. doi: 10.1021/pr0340126. [DOI] [PubMed] [Google Scholar]
- 65.Porath J. Immobilized metal ion affinity chromatography. Protein Expr. Purif. 1992;3(4):263–281. doi: 10.1016/1046-5928(92)90001-d. [DOI] [PubMed] [Google Scholar]
- 66.Posewitz MC, Tempst P. Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal. Chem. 1999;71(14):2883–2892. doi: 10.1021/ac981409y. [DOI] [PubMed] [Google Scholar]
- 67.Annan RS, Huddleston MJ, Verma R, Deshaies RJ, Carr SA. A multidimensional electrospray MS-based approach to phosphopeptide mapping. Anal. Chem. 2001;73(3):393–404. doi: 10.1021/ac001130t. [DOI] [PubMed] [Google Scholar]
- 68.Cao P, Stults JT. Phosphopeptide analysis by on-line immobilized metal-ion affinity chromatography-capillary electrophoresis-electrospray ionization mass spectrometry. J. Chromatogr. A. 1999;853(1–2):225–235. doi: 10.1016/s0021-9673(99)00481-1. [DOI] [PubMed] [Google Scholar]
- 69.Cao P, Stults JT. Mapping the phosphorylation sites of proteins using on-line immobilized metal affinity chromatography/capillary electrophoresis/electrospray ionization multiple stage tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2000;14(17):1600–1606. doi: 10.1002/1097-0231(20000915)14:17<1600::AID-RCM68>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 70.Porath J, Carlsson J, Olsson I, Belfrage G. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature. 1975;258(5536):598–599. doi: 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- 71.Porath J, Olin B. Immobilized metal ion affinity adsorption and immobilized metal ion affinity chromatography of biomaterials. Serum protein affinities for gel-immobilized iron and nickel ions. Biochemistry. 1983;22(7):1621–1630. doi: 10.1021/bi00276a015. [DOI] [PubMed] [Google Scholar]
- 72.Muszynska G, Andersson L, Porath J. Selective adsorption of phosphoproteins on gel-immobilized ferric chelate. Biochemistry. 1986;25(22):6850–6853. doi: 10.1021/bi00370a018. [DOI] [PubMed] [Google Scholar]
- 73.Muszynska G, Dobrowolska G, Medin A, Ekman P, Porath JO. Model studies on iron(III) ion affinity chromatography. II. Interaction of immobilized iron(III) ions with phosphorylated amino acids, peptides and proteins. J. Chromatogr. 1992;604(1):19–28. doi: 10.1016/0021-9673(92)85524-w. [DOI] [PubMed] [Google Scholar]
- 74.Ficarro SB, McCleland ML, Stukenberg PT, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 2002;20(3):301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 75.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jørgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell Proteomics. 2005;4(7):873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 76.Thingholm TE, Jensen ON, Robinson PJ, Larsen MR. SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol. Cell Proteomics. 2008;7(4):661–671. doi: 10.1074/mcp.M700362-MCP200. [DOI] [PubMed] [Google Scholar]
- 77.Marouga R, David S, Hawkins E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal. Bioanal. Chem. 2005;382(3):669–678. doi: 10.1007/s00216-005-3126-3. [DOI] [PubMed] [Google Scholar]
- 78.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 1999;17(10):994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 79.Thompson A, Schäfer J, Kuhn K, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003;75(8):1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 80.Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics. 2004;3(12):1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 81.Fujinoki M, Kawamura T, Toda T, et al. Identification of 36 kDa phosphoprotein in fibrous sheath of hamster spermatozoa. Comp. Biochem. Physiol. B, Biochem. Mol. Biol. 2004;137(4):509–520. doi: 10.1016/j.cbpc.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 82.Lalancette C, Faure RL, Leclerc P. Identification of the proteins present in the bull sperm cytosolic fraction enriched in tyrosine kinase activity: a proteomic approach. Proteomics. 2006;6(16):4523–4540. doi: 10.1002/pmic.200500578. [DOI] [PubMed] [Google Scholar]
- 83.Baker MA, Aitken RJ, Hetherington L. Identification of SRC as a key PKA-stimulated tyrosine kinase involved in the capacitation-associated hyperactivation of murine spermatozoa. J Cell Sci. 2006;119(15):3182–3192. doi: 10.1242/jcs.03055. [DOI] [PubMed] [Google Scholar]
- 84.Hillenkamp F, Karas M, Beavis RC, Chait BT. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal. Chem. 1991;63(24):1193A–1203A. doi: 10.1021/ac00024a002. [DOI] [PubMed] [Google Scholar]
- 85.Marshall AG, Hendrickson CL. High-resolution mass spectrometers. Annu. Rev. Anal. Chem. (Palo Alto. Calif.) 2008;1:579–599. doi: 10.1146/annurev.anchem.1.031207.112945. [DOI] [PubMed] [Google Scholar]
- 86.McLuckey S, Goeringer D. Slow heating methods in tandem mass spectrometry. J. Mass Spectrometry. 1997;32(5):461–474. [Google Scholar]
- 87.Zheng J, Li N, Ridyard M, Dai H, Robbins SM, Li L. Simple and robust two-layer matrix/sample preparation method for MALDI MS/MS analysis of peptides. J. Proteome Res. 2005;4(5):1709–1716. doi: 10.1021/pr050157w. [DOI] [PubMed] [Google Scholar]
- 88.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246(4926):64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 89.Emmett M, Caprioli R. Micro-electrospray mass-spectrometry – ultra-high-sensitivity analysis of peptides and proteins. J. Am. Soc. Mass Spectrom. 1994;5(7):605–613. doi: 10.1016/1044-0305(94)85001-1. [DOI] [PubMed] [Google Scholar]
- 90.Baker MA, Hetherington L, Reeves GM, Aitken RJ. The mouse sperm proteome characterized via IPG strip prefractionation and LC-MS/MS identification. Proteomics. 2008;8(8):1720–1730. doi: 10.1002/pmic.200701020. [DOI] [PubMed] [Google Scholar]
- 91.Baker MA, Hetherington L, Reeves G, Müller J, Aitken RJ. The rat sperm proteome characterized via IPG strip prefractionation and LC-MS/MS identification. Proteomics. 2008;8(11):2312–2321. doi: 10.1002/pmic.200700876. [DOI] [PubMed] [Google Scholar]
- 92.Fujinoki M, Kawamura T, Toda T, et al. Identification of the 58-kDa phosphoprotein associated with motility initiation of hamster spermatozoa. J. Biochem. 2003;134(4):559–565. doi: 10.1093/jb/mvg174. [DOI] [PubMed] [Google Scholar]
- 93.March R. An introduction to quadrupole ion trap mass spectrometry. J. Mass Spectrom. 1997;32(4):351–369. [Google Scholar]
- 94.Brancia FL. Recent developments in ion-trap mass spectrometry and related technologies. Expert Rev. Proteomics. 2006;3(1):143–151. doi: 10.1586/14789450.3.1.143. [DOI] [PubMed] [Google Scholar]
- 95.Craig AG, Hoeger CA, Miller CL, Goedken T, Rivier JE, Fischer WH. Monitoring protein kinase and phosphatase reactions with matrix-assisted laser desorption/ionization mass spectrometry and capillary zone electrophoresis: comparison of the detection efficiency of peptide-phosphopeptide mixtures. Biol. Mass Spectrom. 1994;23(8):519–528. doi: 10.1002/bms.1200230810. [DOI] [PubMed] [Google Scholar]
- 96.Yang X, Wu H, Kobayashi T, Solaro RJ, van Breemen RB. Enhanced ionization of phosphorylated peptides during MALDI TOF mass spectrometry. Anal. Chem. 2004;76(5):1532–1536. doi: 10.1021/ac035203v. [DOI] [PubMed] [Google Scholar]
- 97.Asara JM, Allison J. Enhanced detection of phosphopeptides in matrix-assisted laser desorption/ionization mass spectrometry using ammonium salts. J. Am. Soc. Mass Spectrom. 1999;10(1):35–44. doi: 10.1016/S1044-0305(98)00129-9. [DOI] [PubMed] [Google Scholar]
- 98.Stensballe A, Jensen ON. Phosphoric acid enhances the performance of Fe(III) affinity chromatography and matrix-assisted laser desorption/ionization tandem mass spectrometry for recovery, detection and sequencing of phosphopeptides. Rapid Commun. Mass Spectrom. 2004;18(15):1721–1730. doi: 10.1002/rcm.1542. [DOI] [PubMed] [Google Scholar]
- 99.Schwartz JC, Senko MW, Syka JE. A two-dimensional quadrupole ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 2002;13(6):659–669. doi: 10.1016/S1044-0305(02)00384-7. [DOI] [PubMed] [Google Scholar]
- 100.DeGnore JP, Qin J. Fragmentation of phosphopeptides in an ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 1998;9(11):1175–1188. doi: 10.1016/S1044-0305(98)00088-9. [DOI] [PubMed] [Google Scholar]
- 101. Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl Acad. Sci. USA. 2004;101(26):9528–9533. doi: 10.1073/pnas.0402700101. • First to describe the electron transfer dissociation process and to demonstrate its utility in phosphorylation site determination as a consequence of improved phosphopeptide MS/MS spectral quality.
- 102.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 103.Fardilha M, Esteves SL, Korrodi-Gregório L, et al. Identification of the human testis protein phosphatase 1 interactome. Biochem. Pharmacol. 2011;82(10):1403–1415. doi: 10.1016/j.bcp.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 104.Pappin DJ, Hojrup P, Bleasby AJ. Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol. 1993;3(6):327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- 105.Chamrad DC, Körting G, Stühler K, Meyer HE, Klose J, Blüggel M. Evaluation of algorithms for protein identification from sequence databases using mass spectrometry data. Proteomics. 2004;4(3):619–628. doi: 10.1002/pmic.200300612. [DOI] [PubMed] [Google Scholar]
- 106.Johnson RS, Martin SA, Biemann K, Stults JT, Watson JT. Novel fragmentation process of peptides by collision-induced decomposition in a tandem mass spectrometer: differentiation of leucine and isoleucine. Anal. Chem. 1987;59(21):2621–2625. doi: 10.1021/ac00148a019. [DOI] [PubMed] [Google Scholar]
- 107.Cottrell JS, Creasy DM. Response to: the problem with peptide presumption and low Mascot scoring. J. Proteome Res. 2011;10(11):5272–5273. doi: 10.1021/pr200726c. [DOI] [PubMed] [Google Scholar]
- 108.Li YF, He W, Kim YH, et al. CABYR isoforms expressed in late steps of spermiogenesis bind with AKAPs and ropporin in mouse sperm fibrous sheath. Reprod. Biol. Endocrinol. 2010;8:101. doi: 10.1186/1477-7827-8-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 110.Biemann K. Contributions of mass spectrometry to peptide and protein structure. Biomed. Environ. Mass Spectrom. 1988;16(1–12):99–111. doi: 10.1002/bms.1200160119. [DOI] [PubMed] [Google Scholar]
- 111.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 112.Salicioni A, Platt M, Visconti P. A novel testis-specific serine-proline rich protein is present in mammalian sperm and becomes phosphorylated during capacitation. Presented at: Society for the Study of Reproduction 41st Annual Meeting; 27–30 May 2008; Kailua-Kona, HI, USA. [Google Scholar]
- 113.Neilson KA, Ali NA, Muralidharan S, et al. Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics. 2011;11(4):535–553. doi: 10.1002/pmic.201000553. [DOI] [PubMed] [Google Scholar]
- 114.Bland JM, Altman DG. Statistics notes. The odds ratio. BMJ. 2000;320(7247):1468. doi: 10.1136/bmj.320.7247.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Swaney DL, McAlister GC, Coon JJ. Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat. Methods. 2008;5(11):959–964. doi: 10.1038/nmeth.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Compton PD, Strukl JV, Bai DL, Shabanowitz J, Hunt DF. Optimization of electron transfer dissociation via informed selection of reagents and operating parameters. Anal. Chem. 2012;84(3):1781–1785. doi: 10.1021/ac202807h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
Patent
- 201.Yates Jr. US5538897. Eng J. 1994
Website
- 301.Ion source: online tutorial for peptide de novo sequencing from tandem MS/MS spectra. www.ionsource.com/tutorial/DeNovo/DeNovoTOC.htm.