Abstract
Acrolein (Acr) is a ubiquitous environmental pollutant found in cigarette smoke and automobile exhaust. It can also be produced endogenously by oxidation of polyunsaturated fatty acids. The Acr-derived 1,N2-propanodeoxyguanosine (Acr-dG) adducts in DNA are mutagenic lesions that are potentially involved in human cancers. In this study, monoclonal antibodies were raised against Acr-dG adducts and characterized using ELISA. They showed strong reactivity and specificity towards Acr-dG, weaker reactivity towards crotonaldehyde- and trans-4-hydroxy-2-nonenal-derived 1,N2-propanodeoxyguanosines, and weak or no reactivity towards 1,N6-ethenodeoxyadenosine and 8-oxo-deoxyguanosine. Using these novel antibodies, we developed assays to detect Acr-dG in vivo: First, a simple and quick FACS-based assay for detecting these adducts directly in cells; Second, a highly sensitive direct ELISA assay for measuring Acr-dG in DNA of cells and tissues using only one μg DNA without DNA digestion and sample enrichment; And third, a competitive ELISA for better quantitative measurement of Acr-dG levels in DNA samples. The assays were validated using Acr-treated HT29 cell DNA samples or calf thymus DNA and the results were confirmed by LC-MS/MS-MRM. An immunohistochemical assay was also developed to detect and visualize Acr-dG in HT29 cells as well as in human oral cells. These antibody-based methods provide useful tools for the studies of Acr-dG as a cancer biomarker and of the molecular mechanisms by which cells respond to Acr-dG as a ubiquitous DNA lesion.
Introduction
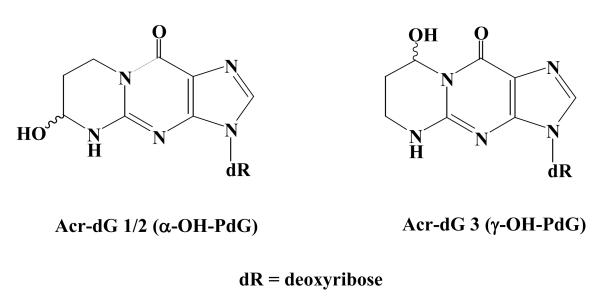
Acrolein (Acr), the simplest α,β-unsaturated aldehyde, or enal, is a major component of cigarette smoke. It can also be generated through the combustion of fossil fuel and high temperature cooking oil. 1,2 More importantly, Acr is formed endogenously via oxidation of polyunsaturated fatty acids (PUFAs). Acr is a major lipid peroxidation product that can arise from both ω-3 and ω-6 PUFAs, although ω-3 fatty acids represent a main source of endogenous Acr. 3-6 Perhaps the most ubiquitous and abundant enal, Acr is a highly reactive electophile towards DNA and proteins and readily forms adducts via Michael addition, thereby rendering the enal highly genotoxic and cytotoxic. 1,4,7,8 Cyclic 1,N2-propanodeoxyguanosines (PdG) are the major products formed by enals in DNA. 7,8 Earlier studies observed that Acr-derived PdG (Acr-dG) adducts, designated Acr-dG1/2 (α-OHPdG) and Acr-dG3 (γ-OHPdG), are formed as a pair of regio-isomers via opposite ring closure of dG with Acr (Figure 1). 8 Using a 32P-postlabeling method, it was found Acr-dG3 is the predominant isomeric lesion present in rodent and human tissue DNA. 9 These findings have now been confirmed by LC-MS/MS-MRM. 10,11
Figure 1. Structures of 1,N2-propanodeoxyguanosine adducts of Acr as regio-isomers.
Acr-dG1/2 (α-OHPdG) and Acr-dG3 (γ-OHPdG).
Ever since the discovery of the cyclic Acr-dG in vivo, their mutagenicity, repair and potential roles in cancer have been extensively studied in cultured cells and animals. 7,12-19 Acr itself is mutagenic in Salmonella typhimurium tester strains TA 100 and 104 20 and Acr-dG adducts were detected in DNA from these strains under the conditions that mutations occurred. 21 They were also detected in DNA isolated from Chinese hamster ovary cells incubated with Acr. 22 Site-specific mutagenesis studies have shown that Acr-dG adducts are pro-mutagenic lesions that can lead to base substitution and frame-shift mutations. 13,17 Results of these studies are, however, inconclusive. For example, the mutagenicity of the major adduct in vivo, Acr-dG 3 (γ-isomer) varies based on the nucleotide sequence context, single-vs, double-stranded DNA, and the host system. The adduct was not found to be mutagenic in E. coli, while it is mutagenic in mammalian cells when present in single-stranded DNA, not in double-stranded DNA. 13,15,17 Recent studies on repair of Acr-dG shows it is repaired by the nucleotide excision repair pathway. 23 The finding that the binding sites of Acr in the human p53 gene, particularly to guanines, coincide with p53 mutational hotspots supports the role of Acr and Acr-dG in human lung cancer caused by cigarette smoking. 24 A recent study showed, upon reaching a threshold level, Acr-dG may serve as a trigger of apoptosis in human colon cancer HT29 cells as the formation of Acr-dG correlates with apoptosis induced by Acr and docosahexaenoic acid, an ω-3 PUFA. 25 Despite ample studies on Acr-dG, to date there has been no antibody raised specifically against these adducts. The development of antibodies against Acr-dG is of critical importance, not only that Acr-dG may serve as a cancer biomarker, but also because the antibodies can be used as tools to study the formation and repair of Acr-dG in cells and tissues as well as how it interacts with proteins involved in cell cycle and apoptosis signaling.
In this study, we report the characterization of the monoclonal antibodies raised specifically against Acr-dG. The specificity and reactivity of the antibodies were determined. These antibodies were used to detect adducts in intact cells using a flow cytometry-based FACS method and to measure the adduct levels in DNA isolated from cells using highly sensitive ELISA assays. The levels of Acr-dG in the cells measured by using the antibodies were then confirmed by LC-MS/MS-MRM. An immunohistochemical assay was also developed using the antibodies to detect and visualize Acr-dG in human colon cancer HT29 cells and human oral cells.
Experimental Procedures
Chemicals
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Fair Lawn, NJ) unless otherwise stated. Acr-dG adducts were synthesized by a previously published method. 8 Calf thymus DNA (CTDNA) was modified with Acr as previously described. 26 Briefly, DNA was incubated with Acr at 37°C with shaking for 4 h. At the end of the reaction, excess Acr was removed by extraction with chloroform and the DNA was precipitated with cold ethanol and stored at −20°C. Other adduct standards were either synthesized 10 or purchased from Sigma-Aldrich.
Synthesis of immunogens
The guanosine adducts of Acr (Acr-Guo) were synthesized under conditions similar to those of Acr-dG. 8 The structures were confirmed by their characteristic UV spectra. Acr-Guo 3 and Acr-Guo 1 and 2 were collected separately using reverse-phase HPLC and were further confirmed by ring-opening and ring-reduction reactions with NaBH4, respectively, as previously described for Acr-dG. 6,27
The Acr-Guo adducts were then individually coupled to bovine serum albumin (BSA) through the periodate oxidation method 28 with some modifications. The ribose moiety was opened by mixing 14 mg of Acr-G with 300 μL of ice cold 0.1 M NaIO4, and the pH was adjusted to 5.0 with 10% (v/v) acetic acid. The mixture was stirred on ice for 20 min and added to ice cold 2.5 mg/ml BSA in H2O. The pH was adjusted to 8.5 with 5% (w/v) NaHCO3 and the mixture was again stirred on ice for 20 min. Freshly prepared 200 μL of 0.25% (w/v) sodium cyanoborohydride, NaBH3(CN), was then added to the solution. The pH of the mixture was quickly adjusted to 6.5~7.0 with 10% (v/v) acetic acid and stirred for 40 min on ice. At the end of the reaction, Zeba desalting spin columns (Promega, Madison WI) were used to remove the salt and the collected Acr-G conjugated BSA fraction was then measured by spectrophotometry to determine concentration. The identities of the conjugated BSA were confirmed with SDS gel electrophoresis and mass spectrometry. One μl of Acr-Guo-conjugated BSA samples was mixed with 1 μl of 10 mg/ml sinapinic acid in 30% (v/v) acrylonitrile containing 0.3% (v/v) trifluoroacidic acid in water and then spotted on a MALDI plate. Mass spectra were acquired in linear high mass mode using a 4800 MALDI-TOF-TOF Analyzer (Applied Biosystems, CA).
Immunization
Immunization was performed as previously described. 29 Briefly, BALB/C mice were immunized with 100 μg of either Acr-Guo 1/2- or Acr-Guo 3-cojugated BSA in 0.1 ml of saline emulsified with an equal amount of Freund’s complete adjuvant, given in a split dose, i.p. and s.c. A second immunization was given 2 weeks after the first one in incomplete Freund’s adjuvant. Mice were boosted with 100 μg of the conjugate in saline given i.p. on days 1, 2, 3 and 4 on fourth week after the second injection. On day 5, mice were sacrificed, spleens were removed for fusion. Test bleeds were taken to check the antibody reactivity towards immunogens using ELISA.
Cell fusion and screening of hybridomas
Satisfactory immune responses were seen in all mice, four of which were used for cell fusion and hybridoma production. A total of 14 hybridoma cell lines derived from seven parental clones were produced. Splenocytes and myelomas were fused, plated into 96-well culture plates and screened by ELISA to detect the positive clones. The selected clones were then subcloned by limiting dilution until they were monoclonal and stable hybridomas. Two subclones from each parental clone were expanded into culture flasks, and 4-6 vials of cells for each subclonal cell line were cryopreserved.
Characterization of monoclonal antibodies with ELISA
The ELISA plate wells (Immulon 4 HBX flat bottom 1×12 strips from Thermo, part number 6404) were coated with 100 μl of 1 μg/ml antigens of Acr-Guo 1/2- or Acr-Guo 3-cojugated BSA in PBS at 37°C for 2 h and washed twice with PBS containing 0.1% (v/v) Tween 20 (PBST). The wells were then blocked with 100 μl of 1 μg/ml BSA in PBS at room temperature with moderate shaking for 1 h and washed twice with PBST. For ELISA, 100 μl of antibody solutions with different dilutions of antibodies or test bleeds in PBS containing 0.1% (w/v) BSA were added. For competitive ELISA, the tested competing reagents were first mixed with antibody solution then added into the wells. The plates were incubated at 37°C for 1 h and washed twice with PBST. Horseradish peroxidase conjugated goat anti-mouse IgG secondary antibodies in 100 μl PBS containing 0.1% (w/v) BSA were added and the plates were incubated at room temperature with moderate shaking for 30 min. The wells were washed and developed with 100 μl of 3,3′,5,5′-tetramethylbenzidine for 30 min, and the reaction was stopped with 100 μl of 2 M H2SO4/1 M HCl. The absorbance was measured at 450 nm.
Slot-blot assay
Plasmid pSP189 DNA samples were modified with Acr (0.5, 1, 2, 5 mM) at 37 °C for 24 h, and purified with repeated phenol/ether extraction. After purification, modified DNA was precipitated with ethanol and dissolved in TE (pH 8.0). Malondialdehyde (MDA)-modified CTDNA samples were kindly provided by Dr. Lawrence J. Marnett at Vanderbilt University. DNA was modified with benzo(a)pyrene diol epoxide (BPDE, 2 μM) or H2O2 (10 mM) the same as previously described. 24 Modified DNA samples (2 μg) were denatured at 95 °C for 10 min, loaded onto nylon membrane using Bio-Dot SF microfiltration apparatus (Bio-Rad, Hercules, CA) and the membrane was air-dried for 30 min. After blocking in PBST with 5% (w/v) non-fat milk for 1h at room temperature, the membrane was probed with anti-Acr-dG mouse monoclonal antibody for overnight at 4 °C. HRP-conjugated secondary antibodies (1:5000) (Santa Cruz Biotechnology, Santa Cruz, CA) were then added in PBST with 5% (w/v) non-fat milk for 2 hr at room temperature. DNA adducts were detected using the ECL plus chemiluminescence kit (PerkinElmer, Boston, MA) and films (Fuji medical X-ray film, Düsserldorf, Germany) were scanned with a Laser Scanning Densitometer (CanoScan 8800F). After antibody detection, the same membrane was stained with methylene blue (Molecular Research Center, Cincinnati, OH) to indicate the amount of DNA.
Cell culture and treatment
Human colonic carcinoma HT29 cells were routinely cultured at 37°C with 5% (v/v) CO2 in Dulbecco’s modified Eagle’s medium (DMEM) with 10% (v/v) fetal bovine serum (FBS), 50 IU/ml penicillin, and 50 g/ml streptomycin (all from Mediatech Inc., Herndon, VA). For immunohistochemical studies, cells were grown on cover glasses placed into the wells of 12-well plate and treated with 0, 100 and 200 M of Acr for 20 h. For FACS and ELISA assays, cells in regular dishes were treated with 0, 50, 100, 150 and 200 M of Acr dissolved in 50 μl of PBS.
Measuring Acr-dG in HT29 cells with FACS analysis
HT29 cells treated with Acr were harvested with trypsin and washed with PBS, followed by fixation in 70% (v/v) ethanol for at least 1 h. Cells were then permeabilized with PBS containing 0.5% (v/v) Triton X-100 and 2 M HCl for 5 min. From this point on, cells were washed twice with 0.5% (v/v) Triton X-100 in PBS after each incubation step. Cells were incubated in 10% (v/v) FBS, 5% (w/v) BSA, and 0.5% (v/v) Triton X-100 in PBS for 1 h to block non-specific bindings, and incubated with anti Acr-dG monoclonal antibody for 1 h. After incubation with the Alexa 488 labeled goat anti-mouse IgG secondary antibody for 1 h, cells were washed and finally suspended in PBS for FACS flow cytometry analysis. The FACS assay was done on a Becton Dickinson FACSort system and the data was analyzed with FCS Express Version 4. All incubations were carried out at room temperature.
Measuring Acr-dG in HT29 cells with direct or competitive ELISA assays
For direct ELISA, DNA was extracted from HT29 cells treated with different concentrations of Acr as previously described 25 and dissolved in PBS at 1 μg/ μl. The samples were further diluted with PBS containing 0.05 mM GSH to 0.02 μg/ μl for a direct ELISA assay. 30 Briefly, 50 μl of DNA (1 μg) solution heat-denatured at 100°C for 10 min was chilled on ice for at least 15 min and then added to the ELISA plates (FluoroNunc/LumiNunc black Maxisorp 96-well plates, Thermo Scientific) pre-coated with 0.003% (w/v) protamine sulfate. The DNA was coated over night at 37°C, blocked with 200 μl 2% (v/v) FBS in PBS at 37°C for 1 h, followed by sequential incubation with anti-Acr-dG antibodies and the horseradish peroxidase conjugated goat anti-mouse IgG secondary antibodies at 37°C for 1 h. To improve the sensitivity, a highly sensitive chemiluminescent HRP substrate (SuperSignal ELISA Femto Maximum Sensitivity Substrate, Thermo Scientific) for ELISA was used. The controls included a blank control and background controls with the same DNA sample subjected to all steps excluding the addition of anti-Acr-dG antibodies. For competitive ELISA, 1 μg of DNA was coated overnight and blocked in the same way as in direct ELISA. After adding the anti-Acr-dG antibodies, either different amount of Acr-dG standards or DNA samples to be detected were also added to compete with Acr-dG adduct in coated DNA samples. The data from the standards were used to construct a standard curve and the signals from the unknown DNA samples were compared with the standard curve to calculate the amount of Acr-dG.
Detection and quantification of Acr-dG in cell DNA by LC-MS/MS-MRM
To confirm the adduct levels, the same HT29 cell DNA samples used for ELISA assay were then subjected to an LC-MS/MS-MRM assay 11 with some modifications. Briefly, approximately 50 μg of DNA dissolved in PBS and mixed with [13C10,15N5]-Acr-dG as internal standards were hydrolyzed enzymatically with DNase I, alkaline phosphatase and phosphodiesterase I. The hydrolysate was then purified by solid phase extraction (SPE). The fraction containing Acr-dG adducts was collected, dried and injected on an LC-MS/MS-MRM column. Detection and quantification of Acr-dG adducts was carried out with an ACQUITY UPLC liquid chromatography system (Waters Corporation, Milford, MA) equipped with 50 × 2.1 mm, 1.7 μm particle size C18 column (Waters Acquity UPLC BEH C18) and coupled Applied Biosystems/MDS SCIEX 4000 Q TRAP (Life Technologies Corporation, Carlsbad, CA) triple quadrupole mass spectrometer. The separation of Acr-dG adducts was performed by isocratical eluting with 3% acetonitrile (v/v) in 1 mM ammonium formate buffer over 3.5 min at 0.5 ml/min at 40°C, followed by 100% (v/v) acetonitrile wash. The ESI source operated in positive mode. The MRM experiment was performed using ion transitions of 324.2→208.1 m/z (Acr-dG) and 339.2→218.1 m/z ([13C10, 15N5]-Acr-dG) for quantitation, and those of 324.2→190.1 m/z (Acr-dG) and 339.2→200.1 m/z ([13C10, 15N5]-Acr-dG) for structural confirmation. All other parameters were optimized to achieve maximum signal intensity. Calibration curves were constructed for all three Acr-dG regio-isomers before each analysis using standard solutions of Acr-dG and [13C10, 15N5]-Acr-dG. A constant concentration of [13C10, 15N5]-Acr-dG (1 fmol/μl) was used with different concentrations of Acr-dG (1.68 amol/μl – 220 fmol/μl) and analyzed using 37 μl injections by LC-MS/MS-MRM.
Immunohistochemical assay
Acr treated and control HT29 cells on cover glass in the 12-well plate were washed with PBS and fixed for 20 min with 2% (v/v) paraformaldehyde in PBS pre-warmed to room temperature, followed by permeabilization with 0.5% Triton X-100/ 2 M HCl in PBS for 5 min. From this point onwards, cells were washed twice with 0.5% (v/v) Triton X-100 in PBS after each incubation step. After blocking in 10% (v/v) FBS, 5% (w/v) BSA, and 0.5% (v/v) Triton X-100 in PBS at room temperature for 1 h, cells were incubated with anti Acr-dG antibody at room temperature for 1 h, followed by incubation of Alexa 488 labeled goat anti-mouse IgG secondary antibody with 80 ng/ml DAPI at room temperature for 1 h. Lastly, the glass covers were washed with distilled water, dried and mounted to slides with Fluoro-Gel (Electron Microscope Sciences, Hatfield, PA), and stored at 4°C until microscopic observation.
Oral keratinocytes were harvested from the oral cavity from smokers and non-smokers by cytobrushing. Cells from identical sites in the right and left buccal mucosa were brushed at least 10 times, and the left lateral borders of the tongue were brushed. Cells were washed and fixed with 1% (v/v) paraformaldehyde for 24 h. Immunofluorescent detection of Acr-dG in paraffin embedded (FFPE) oral cells was performed using a similar method for other adducts described in a previous study 31 with some modifications. Tissue sections were de-paraffinized and incubated with 1% H2O2 for 20 min at room temperature. Sections were subjected to heat-induced antigen retrieval using sodium citrate and blocked with 10% (v/v) normal goat serum in PBS, incubated overnight at 4°C in a humidified chamber with anti-Acr antibody (1:4000). Sections were washed in PBS-Brij and incubated with biotinylated anti-mouse secondary antibody (1:100; Vector Laboratories) for 45 min at room temperature. Samples were washed again and incubated with streptavidin Cy-3 tertiary antibody (1:100; Invitrogen) for 45 min at room temperature in the dark. Following a final wash, coverslips were applied to the sections using Prolong Gold Antifade Reagent with DAPI (Molecular Probes) and allowed to cure for 24 h at room temperature in the dark. All cells were visualized using an Olympus Fluoview-FV300 laser scanning confocal microscope.
Results and Discussion
Developing monoclonal antibodies against Acr-dG
To synthesize the immunogens, the periodate oxidation reaction was performed to couple the Acr-Guo with BSA. Because the propano ring is liable to ring-opening under basic conditions to yield an aldehyde, sodium borohydride can not be used as reducing agent for generating the conjugated immunogens as described in the earlier method. 28 After different concentrations of milder reducing reagents were used, 0.25% of sodium cyanoborohydride, NaBH3(CN), was chosen for this reaction. The SDS gel showed that the conjugated products have higher molecular weights than BSA, and mass spectrometry further confirmed that the difference in molecular weight represents the modification level of 3.8 and 6.7 adducts per BSA for Acr-Guo1/2 and Acr-Guo3, respectively. Considering the fact that Acr-dG3 is the predominant isomer detected in vivo and that Acr-dG isomers may possess different mutagenic potencies, 13,16-18,32 we raised antibodies against each of the two Acr-dG regio-isomers in hopes that they would display a certain degree of specificity towards each isomer.
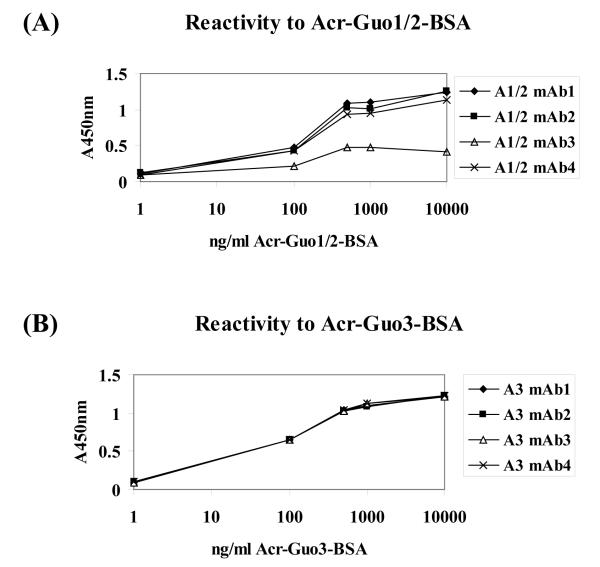
The supernatants of selected fused cells were screened by ELISA for their reactivity towards the conjugates. Eight positive clones, four from each immunogen, were chosen, subcloned and again tested for their antibody activity and specificity. Lyophilized supernatants from the selected cell lines were used as monoclonal antibodies (mAbs) without further purification. They were incubated with different concentrations of Acr-Guo1/2-BSA or Acr-Guo3-BSA. Figure 2 shows the reactivity of mAbs towards the antigens measured by ELISA. All four anti Acr-dG3 mAbs (A3-mAbs 1, 2, 3 and 4) displayed binding activities, however, only three of the four anti Acr-dG1/2 mAbs (A1/2-mAbs 1, 2 and 3) showed activity.
Figure 2. Reactivity of mAbs against immunogens by ELISA.
The mAbs were incubated with varying concentrations of immunogens, Acr-Guo1/2- or Acr-Guo3-conjugated BSA. All four A3-mAbs and three A1/2-mAbs showed strong binding to their respective immunogens, while A1/2-mAb3 displayed only low reactivity.
Determining reactivity and specificity of antibodies
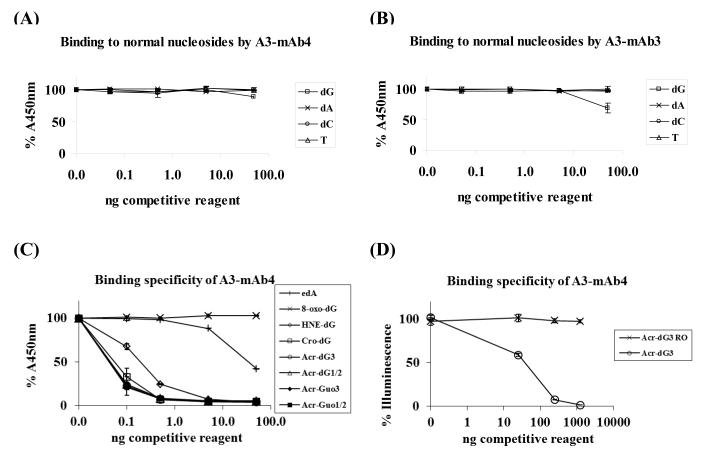
To examine the specificity of the mAbs, we first carried out competitive ELISA to determine their reactivity towards immunogens in the presence of dC, dG, T and dA. Among the mAbs for Acr-dG3, A3-mAb4 and A3-mAb1 yielded the best results; both showed little or no reactivity towards the normal nucleosides including dG (Figure 3A shows the results of A3-mAb4. Similar results were obtained for A3-mAb1). Other A3-mAbs and all A1/2-mAbs showed no reactivity for dA, T or dC, but some reactivity towards dG at the highest concentration (Figure 3B shows the results of A3-mAb3 and similar results were obtained for A3-mAb2 and A1/2-mAbs 1, 2, 3 and 4). The cross-reactivity towards dG by the mAbs is probably due to the relatively small structural differences between dG and Acr-dG. Based on these results, A3-mAb4 and A3-mAb1 were chosen for FACS, ELISA and immunohistochemical assays.
Figure 3. Specificity of mAbs by competitive ELISA and Slot-Blotting.
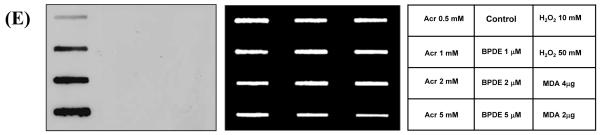
(A) Different amounts of dG, dA, dC and T were used to determine their effects on the binding to mAbs to immunogens. Of all the mAbs examined, A3-mAb4 did not bind to normal nucleosides, as did A3-mAb1 (not shown). (B) All other mAbs, represented by A3-mAb3, showed cross-reactivity to dG at the highest concentration. (C) The A3-mAb4 and A3-mAb1 were further studied in a competitive ELISA assay coated with Acr-Guo3-conjugated BSA for their reactivity towards Acr-dG1/2, Acr-dG3, Acr-Guo1/2, Acr-Guo3, Cro-dG, HNE-dG, 8-oxo-dG, edA. No stereospecificity was observed; A3-mAb4 bound equally well to Acr-dG3 and Acr-dG1/2 as to Acr-Guo1/2 and Acr-Guo3. A3-mAb4 displayed significant cross-reactivity towards Cro-dG, but less for HNE-dG. It did not at all recognize 8-oxo-dG and showed minimal reactivity towards edA only at the highest concentration. (D) Acr modified CTDNA was coated on plates and Acr-dG3 and ring-opened form of AcrdG3 (Acr-dG3 RO) were added in competitive ELISA. A3-mAb4 showed binding to Acr-dG3 and no reactivity towards ring-opened form of Acr-dG3. The coated antigens were different in (C) and (D), so the competitive binding curves for Acr-dG3 were different. Data were obtained from duplicate experiments. (E) Slot-blot assay further demonstrated the specificity of these antibodies towards Acr modified plasmid DNA with no reactivity towards BPDE, H2O2, MDA modified DNA. The left panel of blot image shows the binding of antibodies to the modified DNA samples and binding only occurred with Acr-Modified DNA in a dose-dependent manner, the middle panel shows the DNA loading markers and the right panel is the information for the corresponding modified DNA samples.
To further examine the specificity, we used a competitive ELISA to determine the reactivity of A3-mAb4 and A3-mAb1 towards Acr-dG1/2 and Acr-dG3 as well as other structurally-related in vivo DNA adducts. As expected, both A3-mAbs bound strongly to Acr-dG1/2 and Acr-dG3. Figure 3C shows the results of A3-mAb4; similar results were obtained with A3-mAb1. However, they displayed no preferential binding to the regio-isomers of Acr-dG as Acr-dG1/2 and Acr-dG3 both competitively bound in a similar manner to A3-mAbs 1 or 4 when Acr-Guo3-BSA was used as the coating antigen. Similar results were observed using Acr-Guo1/2-BSA as the coating antigen. Therefore, it can be concluded that the mAbs raised against Acr-Guo1/2 and Acr-Guo3 do not differentiate between Acr-dG1/2 and Acr-dG3 as we had hoped.
The antibodies also recognized crotonaldehyde-derived PdG (Cro-dG) and 4-hydroxy-2-nonenal-derived PdG (HNE-dG). However, the binding appeared to decrease as the carbon chain length increased in the order of Acr-dG≥Cro-dG>HNE-dG. In sharp contrast to the 1,N2-propanodG adducts, A3-mAbs showed no reactivity towards 8-hydroxydeoxyguanosine (8-oxo-dG) and showed minimal binding, only at the highest concentration, to 1,N6-ethenodeoxyadenosine (edA). The results clearly demonstrate that the antibodies strongly recognize the PdG ring moiety as an epitope with considerably less affinity towards the etheno ring. Since the cyclic propano ring of Acr-dG3 can be opened under certain conditions, we determined if A3-mAb1 and A3-mAb4 recognize the ring-opened form by coating the ELISA plate with Acr-modified CTDNA and determined the competitive binding of Acr-dG3 or ring-opened Acr-dG3 to the antibodies using a competitive ELISA assay. Figure 3D shows that, as expected, AdG3-mAb4 strongly binds to Acr-dG3 with no binding activity towards its ring-opened form.
The antibodies specificities were further evaluated against other commonly studied in vivo DNA adducts with slot-blot experiments. As shown in Figure 3E, plasmid DNA was modified with different amount of Acr, or BPDE, H2O2 and MDA. The antibodies showed only reactivity towards Acr modified DNA, but no binding activity towards the other modified DNA adducts. An earlier study reported a monoclonal antibody for Cro-dG and it was used in an HPLC-based fluorescence immunoassay to detect Cro-dG as well as Acr-dG in the enzyme-digested DNA. 21, 22, 29 However, this antibody has never been used in immunohistochemical assays to detect these adducts in tissues or intact cells. Despite the antibodies show a similar affinity for Cro-dG and a minimal reactivity towards HNE-dG, the low abundance of in vivo Cro- and HNE-dG, at least 10 to 100 times lower than Acr-dG 5,10 is unlikely to significantly compromise their usefulness in detecting Acr-dG in cells and tissues. A monoclonal antibody against the Acr-modified DNA was previously described and used to detect the Acr-derived dA adducts. Interestingly, this antibody did not display any reactivity towards Acr-dG. 33
Detection of Acr-dG in HT29 cells by a FACS assay using A3-mAb4
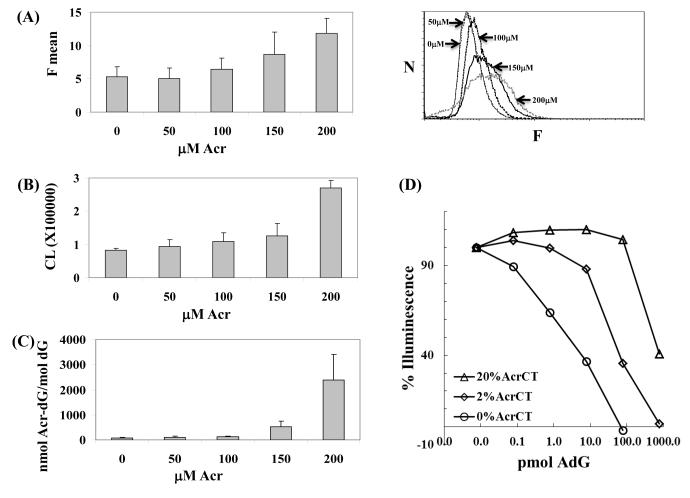
Using a highly sensitive HPLC-based 32P-postlabeling method, we previously demonstrated that the levels of Acr-dG were increased in the DNA of HT29 cells treated with Acr. 25 Here, we used the antibodies to detect Acr-dG in HT29 cells incubated with Acr using a FACS assay. Cells were fixed with ethanol, permeabilized, labeled with A3-mAb4, and incubated with an Alexa-488 labeled secondary antibody. As shown in Figure 4A, the fluorescence intensity increased in a concentration-dependent manner in HT29 cells treated with Acr from 0 to 200 μM, indicating increased levels of Acr-dG. Because these antibodies also recognize Acr-Guo (Figure 3C), the increased fluorescence signal may also reflect the Acr adduct levels in RNA. The background fluorescence in untreated cells may arise from the following sources: the actual background levels of Acr-dG in cells, auto-fluorescence, and non-specific binding. The FACS assay offers a simple and rapid way to detect and quantify Acr-derived adducts in intact cells.
Figure 4. Acr-dG levels in HT29 cells measured with A3-mAb4 by FACS analysis, sensitive ELISA assay and LC-MS/MS-MRM.
(A) In the FACS assay, the left panel shows that the average fluorescence intensity (F mean) indicates the antibodies bound in a concentration-dependent manner to the cellular DNA containing Acr-dG. The right panel shows the original histograms with X-axis as the fluorescence (F) and Y axis as the number of cells (N). The p values from t-test for 0 vs 50, 50 vs 100, 100 vs 150 and 150 vs 200 μM comparisons are 0.4, 0.14, 0.14 and 0.08, respectively. (B) A highly sensitive ELISA assay shows the Acr-dG levels in DNA extracted from HT29 cells. Only one μg DNA from each sample was used, and the chemiluminescence signal (CL), representing the binding of antibodies to DNA, showed that the Acr-dG levels increased in a concentration-dependent manner. The corresponding p values as (A) are 0.26, 0.3, 0.3 and 0.02, respectively. (C) To validate this, Acr-dG levels in the same DNA samples were quantified with by LC-MS/MS-MRM. The corresponding p values as (A) are 0.2, 0.3, 0.08 and 0.06, respectively. All results were obtained from duplicate experiments. (D) Quantification of Acr-dG by a competitive ELISA assay. To the ELISA plates were coated with different levels of modified CTDNA, different amount of Acr-dG standards were added together with the anti-Acr-dG antibodies. The approximate detection ranges that can be used to quantify the Acr-dG adducts are: 100pmol to more than 1000pmol for the 20% Acr-CTDNA coated plates, 10pmol to 1000pmol for the 2% Acr-CTDNA coated plates and <100fmol to 100pmol for and the un-modified CTDNA coated plates.
Quantification of Acr-dG in HT29 cells by an ELISA assay using A3-mAb4
In these experiments, a sensitive ELISA assay was developed using the antibodies to directly measure Acr-dG in DNA. The DNA samples extracted from tissue or cells were heat-denatured and coated on ELISA plates. The plates were then incubated with A3-mAb4 and HRP conjugated secondary antibodies. An ultra sensitive chemiluminescent HRP substrate was added to enhance the sensitivity. Figure 4B shows that the chemiluminescence increased gradually in DNA obtained from HT29 cells treated with Acr from 0 to 150 μM, and that a steep increase was observed in cells treated with 200 μM Acr. To confirm these results the same DNA samples were analyzed by LC-MS/MS-MRM after enzymatic digestion to nucleosides (Figure 4C). Both assays showed an incremental increase of Acr-dG levels at lower concentrations of Acr (0 to 150 μM), reflecting the persistent low background levels due to efficient repair, followed by a sharp increase at 200 μM Acr, possibly due to a combined effect of increased formation and impaired repair activity. 24
It is useful to compare the pros and cons of these antibody-based detection methods. While the increase of Acr-dG in cells treated with Acr is seen using all three methods, the magnitude of this increase was noticeably different among methods. For example, a four-fold increase of Acr-dG levels was observed between cells treated with 150 μM Acr and cells treated with 200 μM Acr when using the LC-MS/MS-MRM method. In contrast, the ELISA detected only a two-fold increase in the same samples. The LC-MS/MS-MRM method with the isotope-labeled internal standards detects Acr-dG adducts in DNA at the nucleoside level in a highly specific and quantitative manner with minimal background interference, whereas the ELISA assay relies on mAb binding to the lesions in denatured DNA. Because the binding can occur with other structurally related adducts, the specificity of ELISA is likely to be somewhat compromised. It is also possible that not all Acr-dG in DNA can be accessible to mAbs due to factors such as sequence and structural folding. The background auto-chemiluminescence in ELISA may also affect sensitivity. Despite the limitations, the ELISA method has a few obvious advantages. First, only one μg of DNA is needed, whereas the 32P-postlabeling and LC-MS/MS-MRM methods normally require 10 to 100 μg or more DNA. Second, unlike the other DNA adduct detection methods, no DNA digestion or sample enrichment procedure is involved, thereby minimizing the variations inherited via lengthy procedures, making this assay more reproducible and efficient. Among these methods, the FACS assay is perhaps the most convenient, but it is the least quantitative. In this method, mAbs are required to be transported into the nuclei of cells and bind to the Acr-dG lesions in the chromatin. Furthermore, they may also recognize Acr adducts in RNA.
The direct ELISA will be useful to detect Acr-dG with relatively small amounts of DNA and is suitable for comparing DNA samples with significantly differences in adducts levels. However, it could present challenges to measure small changes in DNA samples. A competitive ELISA based on previously published method 34 was also tested aiming to improve the quantification of the assay. The ELISA plates were coated with 1 μg of different percentage of Acr modified CTDNA and then different amount of Acr-dG standards were added together with the anti-Acr-dG antibodies. By measuring the signal decrease, standard curves can be constructed as shown in Figure 4D. The results showed that the linear response ranges of the assay are dependent on the total amount of Acr-dG in the coating DNA samples: For the 20%, 2% Acr-modified and un-modified CTDNA coated plates, the assay may be used to detect DNA samlpes containing Acr-dG from ~100pmol to more than 1000pmol, ~10pmol to 1000pmol and sub 100fmol to 100pmol, respectively. Because a standard curve can be easily obtained and up to 100 μg of DNA can be used as compared to 1 μg in the direct ELISA, the competitive ELISA is expected to have considerably better quantitative consistency and accuracy.
Immunohistochemical detection of Acr-dG in HT29 and human oral cells
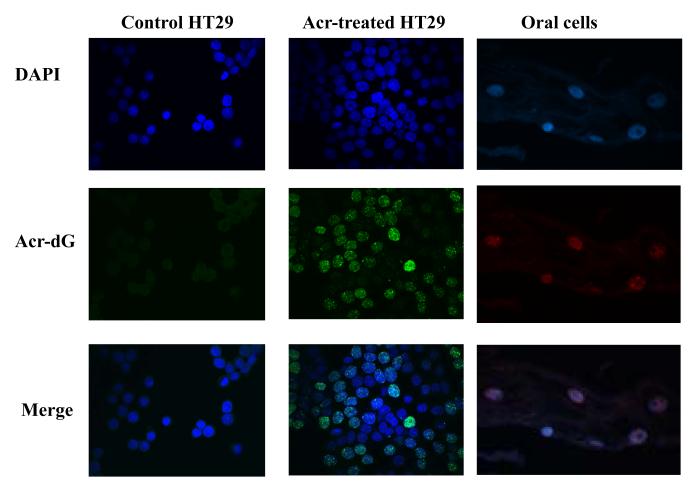
In these experiments we used the new mAbs to develop immunohistochemical staining-based assays to detect and visualize Acr-dG in intact cells. Protease K was used to increase the accessibility of mAb by removing histone and non-histone proteins, 0.5% (v/v) of Triton X-100 was used in each step after permeation, and 2 M HCl treatment was included to denature DNA. Figure 5 shows weak green fluorescence of A3-mAb4 staining in control HT29 cells, and the fluorescent intensity increased significantly upon treatment with 200 μM Acr. A3-mAb4 staining was observed primarily in nuclei and superimposed with DAPI staining. It is important to point out that not all cells stained positively for A3-mAb4, indicating the heterogeneity of Acr-dG formation within the cell population. We also applied the mAbs to the formalin fixed paraffin embedded oral cells and detected different Acr-dG levels between smokers and non-smokers (results to be published separately). Figure 5 shows a typical staining of an oral cell sample.
Figure 5. Immunohistochemical staining of Acr-dG in HT29 cells and human oral cells using A3-mAb4.
HT29 cells treated with 200 μM Acr showed increased fluorescent intensity in nuclei (Alexa 488 labeled green) as compared to untreated controls. Human oral cells were subjected to FFPE-based staining with A3-mAb4. The positive cells show red fluorescence in nuclei stained with Cy3.
In this study, monoclonal antibodies with relatively high reactivity and specificity for Acr-dG were raised. The antibodies were used to develop flow cytometry-based FACS, ELISA and immunohistochemical staining assays. Whereas these antibodies could not distinguish isomeric Acr-dG, the assays developed clearly demonstrate that the antibodies usefulness in detecting and visualizing Acr-dG in human cells. The sensitive direct ELISA assay enables an efficient detection of Acr-dG in only one μg DNA and a more quantitative competitive ELISA can be potentially developed for a faster and more accurate Acr-dG assay kit. In addition to measuring Acr-dG as a biomarker of environmental and endogenous exposure to Acr, these antibodies will be invaluable tools in the molecular mechanism studies of cellular responses to this ubiquitous DNA lesion.
Acknowledgements
The authors thank the Shared Resources of Histopathology and Tissue, Microscopy and Imaging, Flow Cytometry and Cell Sorting, Proteomics and Metabolomics, and Tissue Culture at the Lombardi Comprehensive Cancer Center of Georgetown University for their technical support and assistance.
Funding Support This work was supported by National Cancer Institute grant CA043159.
Abbreviations
- Acr
acrolein
- PUFA
polyunsaturated fatty acid
- Acr-dG
acrolein-derived 1,N2-propanodeoxyguanosine
- PdG
1,N2-propanodeoxyguanosine
- Acr-Guo
acrolein-derived 1,N2-propanoguanosine
- MDA
malondialdehyde
- BPDE
benzo(a)pyrene diol epoxide
- mAb
monoclonal antibody
- Cro-dG
crotonaldehyde-derived PdG
- HNE-dG
4-hydroxy-2-nonenal-derived PdG
- 8-oxo-dG
8-hydroxydeoxyguanosine
- edA
1,N6-ethenodeoxyadenosine
- CTDNA
calf thymus DNA
- BSA
bovine serum album
References
- (1).Stevens JF, Maier CS. Acrolein: Sources, metabolism, and biomolecular interactions relevant to human health and disease. Mol. Nutr. Food Res. 2008;52:7–25. doi: 10.1002/mnfr.200700412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Evaluation of the carcinogenic risk of chemicals to humans, dry cleaning, some chlorinated solvents and other industrial chemicals. Volume 63. International Agency for Research on Cancer; Lyon: 1995. pp. 337–372. IARC Monograph. [PMC free article] [PubMed] [Google Scholar]
- (3).Nair U, Bartsch H, Nair J. Lipid peroxidation-induced DNA damage in cancer-prone inflammatory diseases: A review of published adduct types and levels in humans. Free Radical Biol. Med. 2007;43:1109–1120. doi: 10.1016/j.freeradbiomed.2007.07.012. [DOI] [PubMed] [Google Scholar]
- (4).Uchida K, Kanematsu M, Sakai K, Matsuda T, Hattori N, Mizuno Y, Suzuki D, Miyata T, Noguchi N, Niki E, Osawa T. Protein-bound acrolein: Potential markers for oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 1998;95:4882–4887. doi: 10.1073/pnas.95.9.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Chung F-L, Chen H-J, Nath RG. Lipid peroxidation as a potential endogenous source for the formation of exocyclic DNA adducts. Carcinogenesis. 1996;17:2105–2111. doi: 10.1093/carcin/17.10.2105. [DOI] [PubMed] [Google Scholar]
- (6).Pan J, Chung F-L. Formation of cyclic deoxyguanosine adducts from ω-3 and ω-6 polyunsaturated fatty acids under oxidative conditions. Chem. Res. Toxicol. 2002;15:367–372. doi: 10.1021/tx010136q. [DOI] [PubMed] [Google Scholar]
- (7).Minko IG, Kozekov ID, Harris TM, Rizzo CJ, Lloyd RS, Stone MP. Chemistry and biology of DNA containing 1,N2-deoxyguanosine adducts of the α,β-unsaturated aldehydes acrolein, crotonaldehyde, and 4-hydroxynonenal. Chem. Res. Toxicol. 2009;22:759–778. doi: 10.1021/tx9000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chung F-L, Young R, Hecht SS. Formation of cyclic 1,N2-propano-deoxyguanosine adducts in DNA upon reaction with acrolein or crotonaldehyde. Cancer Res. 1984;44:990–995. [PubMed] [Google Scholar]
- (9).Nath RG, Chung F-L. Detection of exocyclic 1,N2-propanodeoxyguanosine adducts as common DNA lesions in rodents and humans. Proc. Natl. Acad. Sci. U.S.A. 1994;91:7491–7495. doi: 10.1073/pnas.91.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Zhang S, Villalta PW, Wang M, Hecht SS. Detection and quantitation of acrolein-derived 1,N2-propanodeoxyguanosine adducts in human lung by liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem. Res. Toxicol. 2007;20:565–571. doi: 10.1021/tx700023z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Chung F-L, Wu MY, Basudan A, Dyba M, Nath RG. Regioselective formation of acrolein-derived cyclic 1,N2-propanodeoxyguanosine adducts mediated by amino acids, proteins, and cell lysates. Chem. Res. Toxicol. 2012;25:1921–1928. doi: 10.1021/tx3002252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Exocyclic DNA Adducts in Mutagenesis and Carcinogenesis. Proceedings of the 2nd international conference; Heidelberg, Germany. September 1998; Lyon: International Agency for Research on Cancer; 1999. pp. 1–361. (IARC Scientific Publication No. 150) [PubMed] [Google Scholar]
- (13).VanderVeen LA, Hashim MF, Nechev LV, Harris TM, Harris CM, Marnett LJ. Evaluation of the mutagenic potential of the principal DNA adduct of acrolein. J. Biol. Chem. 2001;276:9066–9070. doi: 10.1074/jbc.M008900200. [DOI] [PubMed] [Google Scholar]
- (14).Benamira M, Singh U, Marnett LJ. Site-specific frameshift mutagenesis by a propanodeoxyguanosine adduct positioned in the (CpG)4 hot-spot of Salmonella typhimurium his D3052 carried on an M13 vector. J. Biol. Chem. 1992;267:22392–22400. [PubMed] [Google Scholar]
- (15).Moriya M, Zhang W, Johnson F, Grollman AP. Mutagenic potency of exocyclic DNA adducts: marked differences between Escherichia coli and simian kidney cells. Proc. Natl. Acad. Sci. U.S.A. 1994;91:11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Yang I,Y, Hossain M, Miller H, Khullar S, Johnson F, Grollman A, Moriya M. Responses to the major acrolein-derived deoxyguanosine adduct in Escherichia coli. J. Biol. Chem. 2001;276:9071–9076. doi: 10.1074/jbc.M008918200. [DOI] [PubMed] [Google Scholar]
- (17).Yang IY, Chan G, Miller H, Huang Y, Torres MC, Johnson F, Moriya M. Mutagenesis by acrolein-derived propanodeoxyguanosine adducts in human cells. Biochemistry. 2002;41:13826–13832. doi: 10.1021/bi0264723. [DOI] [PubMed] [Google Scholar]
- (18).Kanuri M, Minko IG, Nechev LV, Harris TM, Harris CM, Lloyd RS. Error prone translesion synthesis past γ-hydroxypropano deoxyguanosine, the primary acrolein-derived adduct in mammalian cells. J. Biol. Chem. 2002;277:18257–18265. doi: 10.1074/jbc.M112419200. [DOI] [PubMed] [Google Scholar]
- (19).Wang HT, Zhang S, Hu Y, Tang MS. Mutagenicity and sequence specificity of acrolein-DNA adducts. Chem. Res. Toxicol. 2009;22:511–517. doi: 10.1021/tx800369y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Marnett LJ, Hurd HK, Hollstein MC, Levin DE, Esterbauer H, Ames BN. Naturally occurring carbonyl compounds are mutagens in Salmonella tester strain TA104. Mutat. Res. 1985;148:25–34. doi: 10.1016/0027-5107(85)90204-0. [DOI] [PubMed] [Google Scholar]
- (21).Foiles PG, Akerkar SA, Chung F-L. Application of an immunoassay for cyclic acrolein deoxyguanosine adducts to assess their formation in DNA of Salmonella typhimurium under conditions of mutation induction by acrolein. Carcinogenesis. 1989;10:87–90. doi: 10.1093/carcin/10.1.87. [DOI] [PubMed] [Google Scholar]
- (22).Foiles PG, Akerkar SA, Miglietta LM, Chung F-L. Formation of cyclic deoxyguanosine adducts in Chinese hamster ovary cells by acrolein and crotonaldehyde. Carcinogenesis. 1990;11:2059–2061. doi: 10.1093/carcin/11.11.2059. [DOI] [PubMed] [Google Scholar]
- (23).Wang HT, Hu Y, Tong D, Huang J, Gu L, Wu XR, Chung F-L, Li GM, Tang MS. Effect of carcinogenic acrolein on DNA repair and mutagenic susceptibility. J. Biol. Chem. 2012;287:12379–12386. doi: 10.1074/jbc.M111.329623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Feng Z, Hu W, Hu Y, Tang MS. Acrolein is a major cigarette-related lung cancer agent: preferential binding at p53 mutational hotspots and inhibition of DNA repair. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15404–15409. doi: 10.1073/pnas.0607031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Pan J, Keffer J, Emami A, Ma X, Lan R, Goldman R, Chung F-L. Acrolein-derived DNA adduct formation in human colon cancer cells: its role in apoptosis induction by docosahexaenoic acid. Chem. Res. Toxicol. 2009;22:798–806. doi: 10.1021/tx800355k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Pan J, Davis W, Trushin N, Amin S, Nath RG, Salem N, Jr., Chung F-L. A solid-phase extraction/high-performance liquid chromatography-based 32P-postlabeling method for detection of cyclic 1,N2-propanodeoxyguanosine adducts derived from enals. Anal. Biochem. 2006;348:15–23. doi: 10.1016/j.ab.2005.10.011. [DOI] [PubMed] [Google Scholar]
- (27).Emami A, Dyba M, Cheema AK, Pan J, Nath RG, Chung F-L. Detection of the acrolein-derived cyclic DNA adduct by a quantitative 32P - postlabeling/solid-phase extraction/HPLC method: Blocking its artifact formation with glutathione. Anal. Biochem. 2008;374:163–172. doi: 10.1016/j.ab.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Erlanger BF, Berser SM. Antibodies specific for ribonucleosides and ribonucleotides and their reaction with DNA. Proc. Natl. Acad. Sci. U.S.A. 1964;52:68–74. doi: 10.1073/pnas.52.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Foiles PG, Chung F-L, Hecht SS. Development of a monoclonal antibody-based immunoassay for cyclic DNA adducts resulting from exposure to crotonaldehyde. Cancer Res. 1987;47:360–363. [PubMed] [Google Scholar]
- (30).Okahashi Y, Iwamoto T, Suzuki N, Shibutani S, Sugiura S, Itoh S, Nishiwaki T, Ueno S, Mori T. Quantitative detection of 4-hydroxyequilenin-DNA adducts in mammalian cells using an immunoassay with a novel monoclonal antibody. Nucleic Acids Res. 2010;38:e133. doi: 10.1093/nar/gkq233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Schwartz JL, Muscat JE, Baker V, Larios E, Stephenson GD, Guo W, Xie T, Gu X, Chung F-L. Oral cytology assessment by flow cytometry of DNA adducts, aneuploidy, proliferation and apoptosis shows differences between smokers and non-smokers. Oral Oncol. 2003;39:842–854. doi: 10.1016/s1368-8375(03)00107-6. [DOI] [PubMed] [Google Scholar]
- (32).Sanchez AM, Minko IG, Kurtz AJ, Kanuri M, Moriya M, Lloyd RS. Comparative evaluation of the bioreactivity and mutagenic spectra of acrolein-derived α-HOPdG and γ-HOPdG regioisomeric deoxyguanosine adducts. Chem. Res. Toxicol. 2003;16:1019–1028. doi: 10.1021/tx034066u. [DOI] [PubMed] [Google Scholar]
- (33).Kawai Y, Furuhata A, Toyokuni S, Aratani Y, Uchida K. Formation of acrolein-derived 2′-deoxyadenosine adduct in an iron-induced carcinogenesis model. J. Biol. Chem. 2003;278:50346–50354. doi: 10.1074/jbc.M309057200. [DOI] [PubMed] [Google Scholar]
- (34).Santella RM. Immunological methods for detection of carcinogen-DNA damage in humans. Cancer Epidemiol. Biomarkers Prev. 1999;8:733–739. [PubMed] [Google Scholar]