Abstract
Aberrations in epigenetic marks have been associated with aging of the brain while caloric restriction (CR) and upregulation of endogenous antioxidants have been suggested as tools to attenuate the aging process. We have recently observed age-related increases in levels of 5-methylcytidine (5-mC) and DNA methyltransferase 3a (Dnmt3a) in the mouse hippocampus. Most of those age-related changes in these epigenetic relevant markers were prevented by CR but not by transgenic overexpression of the endogenous antioxidant superoxide dismutase 1 (SOD1). As recent work has suggested a distinct role for hydroxymethylation in epigenetic regulation of gene expression in the brain, the current study investigated age-related changes of 5-hydroxymethylcytosine (5-hmC) in the mouse hippocampus, and furthermore tested whether CR and transgenic upregulation of SOD1 affected any age-related changes in 5-hmC. Immunohistochemical analyses of 5-hmC in 12- and 24-month-old wild-type and transgenic mice overexpressing SOD1, which were kept under either a control or a calorie restricted diet, revealed an increase of 5-hmC immunoreactivity occurring with aging in the hippocampal dentate gyrus, CA3 and CA1–2 regions. Moreover, CR, but not overexpression of SOD1, prevented the age-related increase in the CA3 region. These region-specific findings indicate that the aging process in mice is connected with epigenetic changes and suggest that the beneficial actions of CR may be mediated via epigenetic mechanisms such as methylation and hydroxymethylation of DNA.
Keywords: Aging, Epigenesis, Epigenetics, DNA hydroxymethylation, 5-hydroxymethylcytosine, Caloric restriction, Antioxidants, superoxide dismutase (SOD), Hippocampus
INTRODUCTION
The aging process in the brain is characterized by increased DNA damage, synaptic dysfunction, distinct structural alterations, and cognitive decline [1–4], with selective brain regions, including neocortical and hippocampal networks, showing increased susceptibility to such age–related pathology [5–7]. The aging brain is furthermore associated with region-specific alterations in gene expression, like downregulation of genes involved in synaptic plasticity and neurotrophic support and upregulation of immune-related genes [8–10]. It is likely that dysregulation of epigenetic mechanisms, such as DNA methylation and chromatin modifications, which have been known to modulate gene expression, are involved in the observed age-related alterations [11–15].
While DNA methylation is a well-described mechanism with a putative role in aging and age-related neurodegeneration [14], a new epigenetic modification has recently been identified, namely DNA hydroxymethylation [16]. Hydroxylation of methylated cytosines by the ten-eleven translocation (TET) enzymes leads to the production of 5-hydroxymethylcytosine (5-hmC) [17, 18], which seems to play a different role than 5-methylcytidine (5-mC) in the regulation of gene expression [19–21]. 5-hmC requires pre-existence of 5-mC [21], has a lower affinity to methyl-binding proteins [19] and, in contrast to 5-mC, is generally associated with chromatin opening and transcriptional activation [20, 22, 23]. The role of 5-hmC in aging of the brain is not known and it seems reasonable to speculate that aging affects 5-hmC in the brain. Recently, we reported an age-related increase in 5-mC immunoreactivity (IR) in the aging mouse hippocampus, correlating with increases in DNA methyltransferase 3a (Dnmt3a) IR, and prevented by caloric restriction (CR) [24, 25]. Attenuating the aging process has been a topic of intense research over the last decades. Dietary restriction of caloric intake has been shown to delay the aging process in many species and even to extend life span [26–32]. Besides CR, upregulation of antioxidants has also been speculated to attenuate the aging process and age-related neurodegenerative disorders [33–36]. A large aging cohort of wild-type and transgenic mice overexpressing the normal human antioxidant Cu/Zn superoxide dismutase (SOD1), either fed with a control diet or exposed to CR from weaning onwards, was generated for the present study. In the present study we investigate age-related changes in hippocampal 5-hmC IR as well as the potential effects of CR and SOD1 overexpression in this context. Using a specific antibody, 5-hmC IR was analyzed in the hippocampus of 12- and 24-month-old mice and possible correlations between 5-hmC IR and previously reported changes in 5-mC IR were investigated.
MATERIALS AND METHODS
ANIMALS
Brain sections from 48 wild-type C57Bl6J and transgenic mice overexpressing SOD1 were used, which were fed with a control or a calorie restricted diet. Descriptions of the generation of transgenic mice, the preparation and composition of the diets, as well as the weight and survival curves of the total cohort of mice (240 male mice) have been given previously [24, 37, 38]. In brief, the transgenic mice were generated on a C57Bl6 background and were carrying 7 copies of the entire human SOD1 sequence, inserted into chromosome 3, resulting to an increased enzyme activity in the brain and other tissues [39]. The control diet was approximately 85% of the ad libitum consumption, in order to fully control the caloric intake, while the calorie restricted diet entailed a 50% reduction in calories. The animals were housed individually, with ad libitum access to water, on a 12/12 hours light: dark circle, kept under standard temperature, humidity, and specified pathogen free (SPF) conditions. All experiments were approved by the Animals Ethics Board of Maastricht University.
EXPERIMENTAL DESIGN
Based on the genotype and diet of the animals, 4 groups were generated: 1) Wild-type mice on control diet (WT-CD), 2) SOD1 mice on control diet (SODCD), 3) WT mice on CR (WT-CR) and 4) SOD1 mice on CR (SOD-CR). From each group, 6 mice were euthanized at 12 and 6 mice at 24 months of age, yielding a total of 8 groups, for histological analyses.
TISSUE PROCESSING
The processing of the brains has been described previously [24, 37]. Briefly, following transcardial perfusion with tyrode solution and 2 fixative solutions (4% parafolmaldehyde, 0.9% NaCl, 1% acetic acid and 8% parafolmaldehyde, 0.9% NaCl, 1% acetic acid) the brains were removed and postfixed for an additional 24 hours in 8% parafolmaldehyde, at 4° C). Consequently, the brains were hemisected along the midsaggital line, cryoprotected in sucrose solutions (10%, 20% and finally 30% sucrose in Tris-HCl buffer, 2×12 hours per solution at 4° C) and embedded in Tissue Tek® (Sakura Finetec Europe, Zoeterwoude, The Netherlands). Then, the left brain halves were quickly frozen and stored at −80° C, until they were cut serially in 30 µm-thick free-floating coronal sections using a cryostat (type HM 500 OMV, Microm, Walldorf, Germany), yielding 10 subseries of every 10th section, and stored until further histological processing [37]. The right brain halves were not used in the present study.
IMMUNOHISTOCHEMICAL DETECTION OF 5-hmC
A series of sections were stained using standard immunohistochemical protocols that were previously described [24, 25], using a rabbit polyclonal anti 5-hmC antiserum (dilution 1:25,000, Active Motif, Rixensart, Belgium) as a primary antibody and a biotinylated donkey anti-rabbit (dilution 1:200, Jackson Westgrove, PA, USA) as secondary antibody. Previously published work, using 5-hmC capturing oligonucleotides has shown the specificity of this commercially available antibody [40].
Immunofluorescent labeling of 5-hmc, 5-mC, and DAPI were also performed for qualitative purposes. In this case, the same antibody against 5-hmC (dilution 1:1,500) and a mouse monoclonal anti-5-mC (dilution 1:500; GenWay Biotech, San Diego, CA, USA) were used as primary antibodies. A donkey anti-rabbit Alexa 594 (dilution 1:200, Invitrogen, Eugene, Oregon, USA) and a donkey anti-mouse biotin (dilution 1:200, Jackson Westgrove, PA, USA) coupled with streptavidin Alexa 488 (dilution 1:400, Invitrogen, Eugene, Oregon, USA) were used for detection. Further, the sections were counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma Aldrich, Zwijndrecht, The Netherlands).
ANALYSIS OF 5-hmC IR
Mean intensity and surface area of 5-hmC IR were analyzed. As indicated in Figure 1 and described previously [25], 2 images from the CA1–2 region, 2 images from the CA3 pyramidal layer and 4 images from the granule cell layer of the DG were taken at 4 different bregma levels (−1.58, −1.82, −3.40, −3.52), according to a mouse brain atlas [41], using a 40x objective. Thus, a total of 32 images were taken for every animal, with a digital camera (f-view; Olympus, Tokyo, Japan) connected to an Olympus AX70 brightfield microscope (analySIS; Imaging System, Münster, Germany). Mean intensities and surface area measurements of each image were obtained using the ImageJ software program (version 1.42q, Wayne Rasband, National Institutes of Health, Bestheda, Maryland, USA), after delineating the regions of interest and correcting for background variation by setting minimum thresholds [42]. In addition, the surface area measurements were corrected for previously reported volume differences [24, 37]. We previously found hippocampal volume reductions by age and caloric restriction [24, 37], and since surface are measurements might be affected by volume changes, they were corrected by multiplying the volume of each subregion of each animal with the corresponding surface area measurement. On the other hand, the intensity measurements are independent of volume or cell number differences and no adjustments were performed.
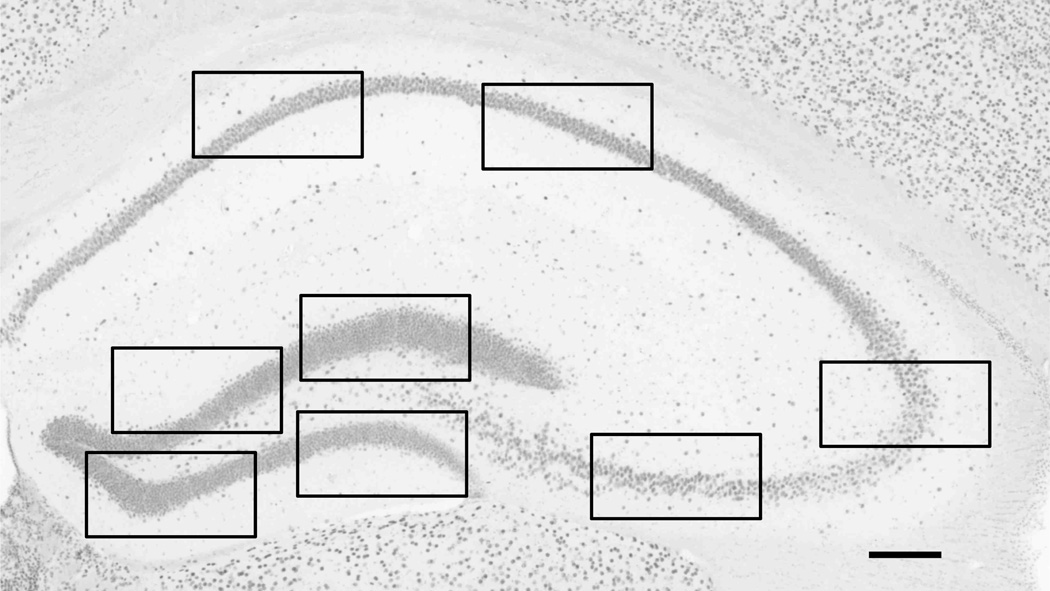
Figure 1.
Hippocampal 5-hmC immunoreactivity. Representative image of a hippocampal section stained for 5-hydroxymethylcytidine (5-hmC) (bregma level 1.58). The black boxes indicate where the high-magnification images were taken for the analysis. A total of 32 images per animal (i.e. 4 images in the dentate gyrus, 2 in the CA3 and 2 in the CA1–2 regions, at 4 different bregma levels) were taken for the analysis (see text for more details). Scale bar= 146 µm
IMAGING of 5-hmC, 5-mC AND DAPI
Imaging of the triple labeling of 5-hmC, 5-mC, and DAPI was achieved by collecting 16 µm-thick image stacks consisting of 80 confocal images (0.2 µm apart), made with a 100x objective (Olympus UPlanSApo; numerical aperture [NA] = 1.40) and the SI-SD system (MBF Bioscience, Williston, VT, USA). Projections and minor corrections in intensity and contrast were made with the Imaris software program (Bitplane AG, Zurich, Switzerland). The whole system and controlling software have been described in more detail previously [24, 43].
STATISTICAL ANALYSIS
All data are presented as mean and standard error of means. The general linear model univariate analysis of variance was used for comparisons between groups, accounting for the main and interactive effects of age, genotype and diet. Statistical significance was set at α = 0.05. Pair-wise comparisons were performed with a Bonferroni post-hoc test. In the absence of a significant interaction, main effects of age were analyzed by an additional stratified analysis per diet, in order to assess whether overall effects of age were more pronounced in a particular diet group. All statistical calculations were performed using the Statistical Package for the Social Sciences, (SPSS 16, SPSS Inc., Chicago, IL, USA). Graphs were built in GraphPad Prism (Version 4, GraphPad Software, San Diego, USA). Correlation analyses between 5-hmC and 5-mC intensity measurements, that have previously been reported [25], were carried out by calculating the Pearson’s correlation coefficient (rp).
RESULTS
QUALITATIVE ANALYSIS OF 5-hmC IR
All cells throughout the hippocampus showed nuclear 5-hmC IR (Fig. 1), appearing as a granular pattern throughout the whole nucleus of each cell. Additionally, simultaneous immunofluorescence labeling of 5-hmC, 5-mC, and DAPI confirmed previously reported colocalization of 5-mC and DAPI in heterochromatic regions of the DNA (densely DAPI-stained marks; Fig. 2). Interestingly, 5-hmC did not colocalize with 5-mC or DAPI in these heterochromatic regions. However, there were some other nuclear subregions where 5-hmC did colocalize with 5-mC (Fig. 2).
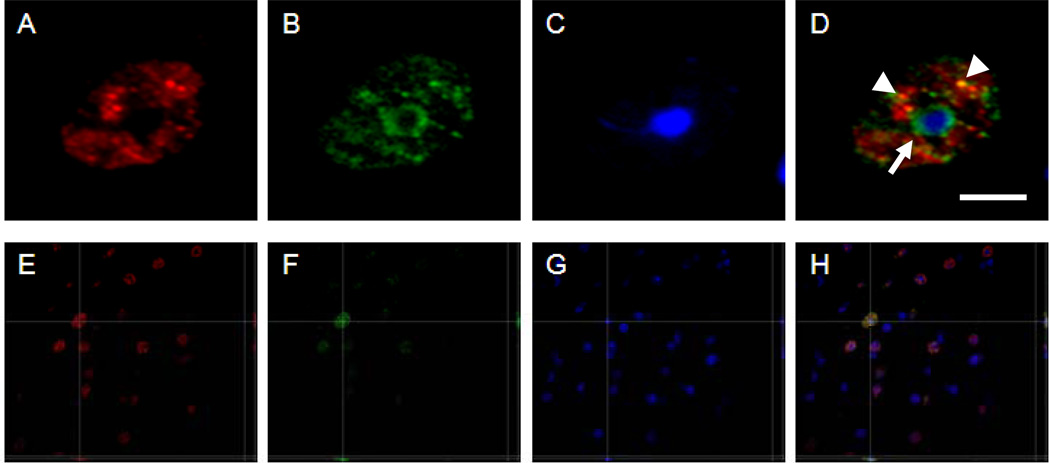
Figure 2.
5-hmC and 5-mC immunofluorescence, and DAPI labeling. Representative high magnification images of a nucleus of a cell in a 12-month-old mouse showing fluorescence labeling of 5-hmC (red) (A and E), 5-methylcytidine (5-mC) (green) in B and F, and DAPI (blue) in C and G, at the corresponding location and focal plane, with merged pictures in D and H. All images represent one stack taken with the SI-SD system (see text for more details). Note the single white arrow indicates the colocalization between 5-mC (green) and DAPI (blue) in heterochromatin, and the arrowheads indicate the colocalization between 5-hmC and 5-mC. Images E-H show a three-dimensional reconstruction of a representative image stack. The crossing lines indicate the position within the X-Y view at which the Y-Z and X-Z views were generated. Scale bar = 5 µm in A-D and 30 µm in E-H.
Semiquantitative analyses of 5-hmC IR by inspection of the sections suggested an increase of 5-hmC IR in 24-month-old mice as compared to 12-mnth-old in the three hippocampal subregions DG, CA3 and CA1–2 (see Fig 3) particularly in CD-fed, but not CR-fed, mice. Quantitative analyses were further carried on to confirm these findings.
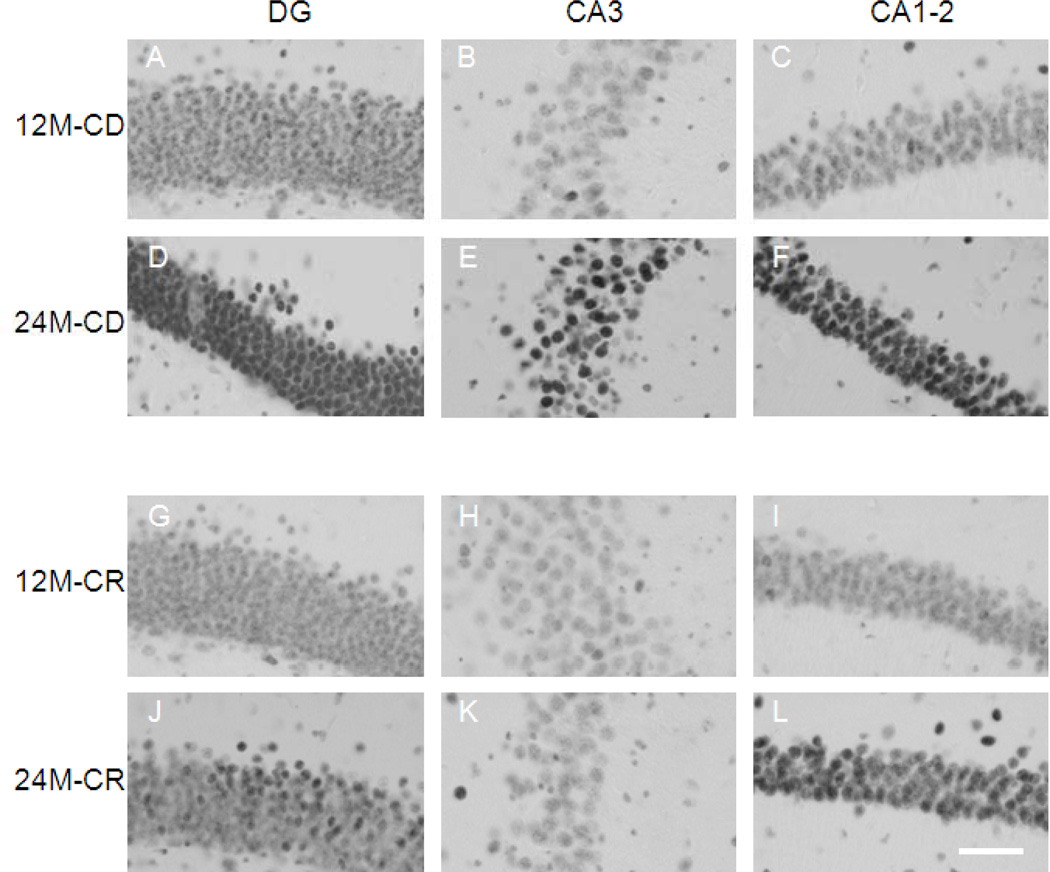
Figure 3.
Representative images of 5-hmC immunoreactivity. High magnification representative images of the hippocampal regions DG, CA3, and CA1–2. A-C represent a 12-month-old mouse on control diet (CD), D- F represent a 24-month-old CD mouse. G-I, represent a 12-month-old mouse on caloric restriction (CR), and J-L a 24-month-old CR mouse. Note: An increase in 5-hmC immunoreactivity (IR) is observed from 12 to 24 months, more pronounced in CD mice, while no differences are observed in the CA3 region of 12- and 24-monthold CR mice. The images were taken with a 40x objective. Scale bar = 45 µm.
5-hmC INTENSITY
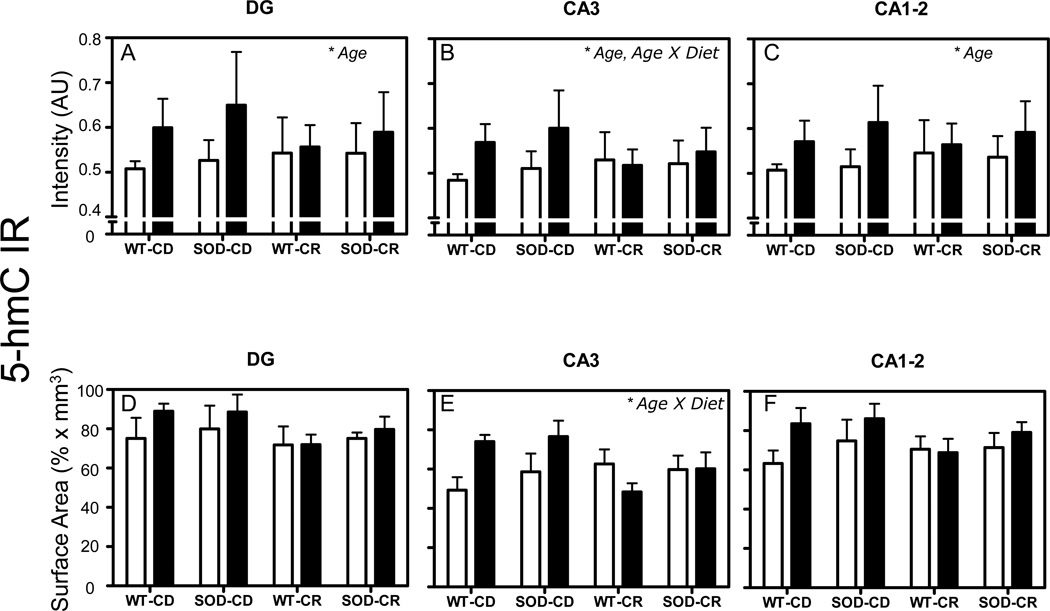
General linear model univariate analysis of variance revealed an increase of mean intensity of 5-hmC IR with aging from 12- to 24-month-old mice in all investigated regions (p = 0.002 for DG and p = 0.003 for CA3 and p = 0.001 for CA1–2) (Fig. 4A–C). A significant age×diet interaction was observed in the CA3 region only (p = 0.009) (Fig. 4B). Pair wise post-hoc comparisons using the Bonferroni correction showed that the observed changes were not specific for an individual experimental group. In addition, no significant effect of the SOD1 genotype was observed. All the p-values of the tests are given in Table 1. Additional stratified analyses per diet showed an increase of mean intensity of 5-hmC IR with aging from 12- to 24-month-old mice in the DG (p < 0.001) and CA1–2 (p < 0.001) regions in the CD animals, which was not observed in the CR animals (p = 0.367 for DG and p = 0.175 for CA1–2).
Figure 4.
5-hmC intensity and surface area. Mean and standard error of means of mean intensity value measurements of 5-hmC IR (A-F). Pooled data from the 4 groups of 12-month-old (white bars) and 24-month-old mice (black bars) are represented separately for DG (A, D), CA3 (B, E), and CA1–2 (C, F). Volume corrections were applied for the surface area occupied by 5-hmC IR measurements. Significant effects (p < 0.05 in all cases) in each analysis are indicated with an asterisk in the top right corner of each graph. AU= arbitrary units.
Table (1).
P-values of general linear model univariate analysis of variance tests of gray value measurements. Values <0.05 are indicated bold.
| P-values | DG | CA3 | CA1–2 |
|---|---|---|---|
| Age | 0.002 | 0.003 | 0.001 |
| Genotype | 0.263 | 0.181 | 0.343 |
| Diet | 0.693 | 0.563 | 0.531 |
| Age × Genotype | 0.543 | 0.549 | 0.334 |
| Age × Diet | 0.081 | 0.009 | 0.166 |
| Genotype × Diet | 0.707 | 0.540 | 0.638 |
| Age × Genotype × Diet | 0.875 | 0.461 | 0.915 |
5-hmC SURFACE AREA
General linear model univariate analysis of variance showed no significant main effect of age, genotype or diet on 5-hmC surface area in any hippocampal region (Fig. 4D–F). However, there was a significant age×diet interaction in the CA3 region (p = 0.007) (Fig. 4E). All the p-values of the tests are given in Table 2.
Table (2).
P-values of general linear model univariate analysis of variance tests of surface area measurements. Values <0.05 are indicated bold.
| P-values | DG | CA3 | CA1–2 |
|---|---|---|---|
| Age | 0.250 | 0.152 | 0.090 |
| Genotype | 0.511 | 0.297 | 0.248 |
| Diet | 0.150 | 0.172 | 0.413 |
| Age × Genotype | 0.973 | 0.694 | 0.981 |
| Age × Diet | 0.449 | 0.007 | 0.246 |
| Genotype × Diet | 0.774 | 0.887 | 0.895 |
| Age × Genotype × Diet | 0.6770 | 0.288 | 0.401 |
5-hmC AND 5-mC CORRELATION ANALYSIS
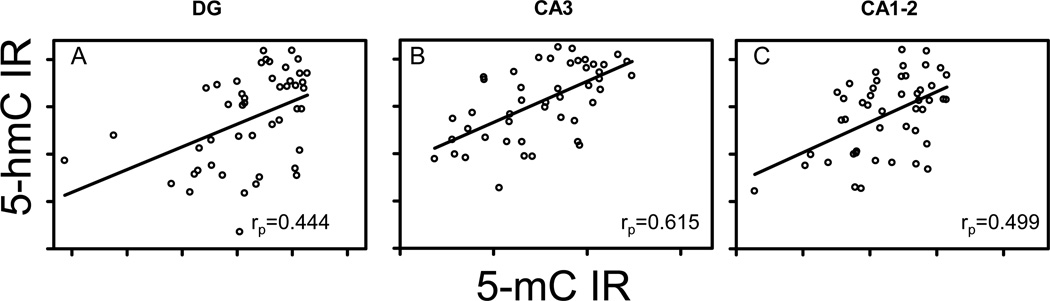
Previous work in the same cohort of mice showed age-related increases in 5-mC IR that could be prevented by CR [25]. In order to examine if the observed changes in 5-hmC IR are related to changes in 5-mC IR we performed correlation analyses between 5-hmC IR and 5-mC IR. Positive linear correlations were observed in all regions (DG: rp = 0.444, p = 0.002; CA3: rp = 0.615, p < 0.001; CA1–2: rp = 0.499, p < 0.001) (Fig. 5A–C).
Figure 5.
5-hmC and 5-mC correlation analysis. Correlation analysis of 5-hmC and 5-mC IR. Significant Spearman’s correlation coefficients are noted in the bottom right of each graph.
DISCUSSION
Immunohistochemistry and immunofluorescence of 5-hmC revealed a granular pattern of IR, observed in the nuclear area of all cells. Lack of colocalization of 5-hmC with 5-mC and DAPI in heterochromatin suggests that 5-hmC is preferentially associated with euchromatin. Quantitative assessment showed an increase of 5-hmC from 12- to 24-month-old mice in all analyzed subregions. The statistically significant age X diet interaction for the CA3 region suggested that CR delayed the age-related increase in 5-hmC particularly in that hippocampal subregion. Positive correlations between 5-hmC IR and 5-mC IR suggest that the observed changes in 5-hmC are linked in part with changes in 5-mC.
COLOCALIZATION ANALYSIS OF 5-hmC and 5-mC
While 5-hmC and 5-mC were both detected in the same cells, subcellular patterns of 5-hmC IR were different than 5-mC IR, with 5-mC preferentially detected in heterochromatin and 5-hmC in euchromatin (Fig. 2). These findings are consistent with previous findings in embryonic stem cells [22]. The different functional role of 5-hmC and 5-mC in the regulation of gene expression implies that regions high in 5-hmC are transcriptionally active (and in euchromatin status), while regions high in 5-mC are transcriptionally inactive and more likely situated in heterochromatin, which is observed using immunofluorescence. Nevertheless, there were few loci in the nucleus where 5-hmC colocalized with 5-mC (orange dots), likely indicating the dynamic process of hydroxylation of 5-mC to 5-hmC.
AGING INCREASES 5-hmC LEVELS IN THE MOUSE HIPPOCAMPUS
Qualitative and semi-quantitative assessment indicated an age-related increase in 5-hmC IR in all hippocampal subregions; more pronounced in the CD animals. Quantitative analyses further confirmed these findings. The observed age-related increase of 5-hmC IR could be associated with aberrant gene expression patterns as observed in the aging hippocampus previously [44–46]. Future studies assessing (genomic) site-specific changes and the corresponding impact on gene expression will further elucidate the role of hydroxymethylation in the aging hippocampus. The role of this recently described epigenetic modification [16, 17] is not yet fully elucidated. Interestingly, compared to 5-mC, the levels of 5-hmC are higher in the brain as compared to other tissues and 5-hmC is more specifically located at genomic regions containing genes [20]. Its presence in gene bodies (from 500 bp downstream to transcription start up to 500 bp of transcription end) and not in promoter regions has furthermore been shown to correlate positively with gene expression [16, 20, 47] and may be linked to the recent observation that DNA hydroxymethylation represents an intermediate step of active DNA demethylation [48, 49]. In support of this notion, Dnmt knock-out experiments in cell cultures have shown that 5-hmC can be produced only in the presence of 5-mC [21]. Interestingly, 5-hmC has also been shown to have lower affinity for methyl-CpG binding proteins 1,2, and 4 when compared to 5-mC [19, 50]. Altogether, these data indicate that the role of 5-hmC in transcriptional regulation might differ from, or even be opposite to, that of 5-mC.
Our findings of increased 5-hmC IR with age are compatible with previous reports of increased total levels of 5-hmC in the hippocampi of 90-day-old mice as compared to 1-day-old mice, measured by HPLC [47]. The role of 5-hmC in aging and age-related gene expression changes is not yet known. First, no correlation between DNA damage and levels of 5-hmC was observed in cerebellar cells of adult mice at 50 weeks of age, as well as in and hippocampal cells of adult mice at 12 weeks of age [16, 47]. Second, another recent investigation revealed an age-dependent increase in hydroxymethylation of genes related to neurodegeneration in the cerebellum (e.g., secretases and presenilins) of adult, 2.5-month-old mice compared to mice 21-day-old mice [23]. Furthermore, a genetic association between a single nucleotide polymorphism (SNP) of the TET1 gene, that catalyzes the conversion from 5-mC to 5-hmC, and the late-onset form of Alzheimer’s disease has been reported [51].
Aberrant epigenetic patterns have been linked with age-related neurodegeneration [14, 52–55] and the observed changes in hippocampal 5-hmC levels suggest that DNA hydroxymethylation may also contribute to age-related epigenetic dysregulation and age-related neurodegeneration. It remains unknown whether the observed changes in 5-hmC levels are connected to epigenetic mediation of environmental stimuli [56–58], to factors involved in the aging process per se, to stochastic or other unknown factors, or to a combination of these. As the environmental conditions for the present laboratory-controlled study were kept identical (except for diet) throughout life for all animals, it is unlikely that differential exposure to environmental factors (other than the diet) has had a substantial influence on the results.
Earlier studies in the same cohort of mice showed an age-related increase of 5-mC IR, concomitant with an increase in Dnmt3a IR [24, 25]. DNA methylation has been shown to be important in learning and memory, and age-related changes in the methylation of memory genes in the hippocampus, such as Arc, have been reported [13, 59, 60]. Dnmt3a has further been shown to be essential for synaptic plasticity as well as neurogenesis [61–63]. Given that 5-hmC is a product of 5-mC, the observed positive correlations suggest that the increases in 5-hmC are related to increases in 5-mC. As such, the age-related increase in DNA methylation, possibly reflecting an increased methylation potential, could thereby passively contribute to an increased hydroxymethylation potential. Whether an altered methylation-hydroxymethylation balance is responsible for functional age-related changes in gene expression remains to be elucidated.
CALORIC RESTRICTION PREVENTS AGE-RELATED INCREASE IN 5-hmC
The significant age X diet interaction indicated that the observed age-related increase of 5-hmC IR was prevented by CR particularly in the CA3 region. While 5-hmC IR increased from 12- to 24-month-old mice, this increase was not observed in the CA3 region of the 24-month-old CR mice. This region-specific finding might be associated with the selective vulnerability to aging and the differential response to CR of the CA3 subregion in rodents [26, 37, 44, 46, 64]. While the age X diet interaction was only significant for the CA3 region in the current analyses, stratified analysis indicated that the 5-hmC levels in the DG and CA1–2 regions were only higher in 24-month-old compared to 12-month-old mice fed with control diet, which is in line with qualitative inspection of the sections. The mechanism by which CR may affect 5-hmC is currently unknown, but may involve dietary impact on the one carbon metabolism pathway, which regulates the availability of methyl-groups [65–67], or may involve histone deacetylases (sirtuins) that influence life span [28, 68–72]. Thus, CR may induce a complex interplay involving chromatin remodeling as well as changes in the balance between methylation and hydroxymethylation.
SOD1 OVEREXPRESSION DID NOT AFFECT AGE-RELATED CHANGES IN 5-hmC
Overexpression of SOD1 did not affect 5-hmC IR, which was in line with previous reports of no effects of SOD1 in brain volumes, Dnmt3a IR, and 5-mC IR [24, 25, 37]. Possibly, reduction of oxidative damage by SOD1 might not involve changes in the levels of epigenetic markers, such as 5-hmC, 5-mC, and Dnmt3a. Alternatively, such effects might not be specifically observed in the hippocampus, or the upregulation of SOD1 and other antioxidants in this model were not sufficient to induce changes [39, 73].
STRENGHTS, LIMITATIONS AND FUTURE PERSPECTIVES
Major strengths and limitations of the study design have been discussed in detail previously [24, 25, 37]. A major advantage of this study is the selective detection of 5-hmC through the use of specific antibodies. The used antibody has been shown to be highly specific for visualization of 5-hmC and not 5-mC [20, 22, 50, 74]. Although numerous methods fail to detect differences between the two modifications, which have been shown to serve distinct roles in the regulation of gene expression, methods involving antibody detection have proven to be specific. Various studies have shown that widely used techniques for methylation detection, like bisulfite treatment, cannot distinguish the two modifications and that such data should therefore be treated with caution [50, 74–76]. Future studies should investigate whether the observed changes in hydroxymethylation can be attributed to specific genes. Furthermore, the possible age-dependent intricate interplay between hydroxymethylation and methylation, as well as other epigenetic processes such as histone modifications remains to be elucidated.
CONCLUSION
The present study has shown that the mouse hippocampus is subject to substantial age-related changes in levels of DNA hydroxymethylation, and that these age-related changes in levels of DNA hydroxymethylation can be prevented by CR but not by overexpression of normal SOD1. In line with previous reports using the same mouse cohort, these findings indicate that the aging process in mice is connected with profound epigenetic changes and suggest that the beneficial actions of CR may be mediated via epigenetic mechanisms such as methylation and hydroxymethylation of DNA.
ACKNOWLEDGEMENTS
Funds have been provided by the Internationale Stichting Alzheimer Onderzoek (ISAO), grant number 09552, and the Netherlands Organization for Scientific Research (NWO, Veni Award 916.11.086) to B.P.F. Rutten, by the ISAO, grant number 07551, to D.L.A. van den Hove and by a Marie Curie Host Fellowship Grant MC-EST 020589 EURON to L. Chouliaras. P.R. Hof is supported by NIH grant P50 AG05138. Figure 2 was generated with an SI-SD system (MBF Bioscience), which was obtained by NWO grant number 91106003.
REFERENCES
- 1.Morrison JH, Hof PR. Life and death of neurons in the aging brain. Science. 1997;278:412–419. doi: 10.1126/science.278.5337.412. [DOI] [PubMed] [Google Scholar]
- 2.Dickstein DL, Kabaso D, Rocher AB, Luebke JI, Wearne SL, Hof PR. Changes in the structural complexity of the aged brain. Aging Cell. 2007;6:275–284. doi: 10.1111/j.1474-9726.2007.00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rutten BP, Korr H, Steinbusch HW, Schmitz C. The aging brain: less neurons could be better. Mech Ageing Dev. 2003;124:349–355. doi: 10.1016/s0047-6374(03)00002-2. [DOI] [PubMed] [Google Scholar]
- 4.Hof PR, Morrison JH. The aging brain: morphomolecular senescence of cortical circuits. Trends Neurosci. 2004;27:607–613. doi: 10.1016/j.tins.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 5.Rutten BP, Schmitz C, Gerlach OH, et al. The aging brain: accumulation of DNA damage or neuron loss? Neurobiol Aging. 2007;28:91–98. doi: 10.1016/j.neurobiolaging.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 6.Luebke JI, Weaver CM, Rocher AB, et al. Dendritic vulnerability in neurodegenerative disease: insights from analyses of cortical pyramidal neurons in transgenic mouse models. Brain Struct Funct. 2010;214:181–199. doi: 10.1007/s00429-010-0244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu W, Brickman AM, Luchsinger J, et al. The brain in the age of old: the hippocampal formation is targeted differentially by diseases of late life. Ann Neurol. 2008;64:698–706. doi: 10.1002/ana.21557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 9.Lu T, Pan Y, Kao SY, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 10.Berchtold NC, Cribbs DH, Coleman PD, et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A. 2008;105:15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peleg S, Sananbenesi F, Zovoilis A, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 13.Penner MR, Roth TL, Barnes CA, Sweatt JD. An epigenetic hypothesis of aging-related cognitive dysfunction. Front Aging Neurosci. 2010;2:9. doi: 10.3389/fnagi.2010.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chouliaras L, Rutten BP, Kenis G, et al. Epigenetic regulation in the pathophysiology of Alzheimer's disease. Prog Neurobiol. 2010;90:498–510. doi: 10.1016/j.pneurobio.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Calvanese V, Lara E, Kahn A, Fraga MF. The role of epigenetics in aging and age-related diseases. Ageing Res Rev. 2009;8:268–276. doi: 10.1016/j.arr.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5- hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5- hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams K, Christensen J, Pedersen MT, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011 doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ficz G, Branco MR, Seisenberger S, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011 doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 23.Song CX, Szulwach KE, Fu Y, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chouliaras L, van den Hove DL, Kenis G, et al. Caloric restriction attenuates age-related changes of DNA methylltransferase 3a in mouse hippocampus. Brain Behav Immun. 2011;25:616–623. doi: 10.1016/j.bbi.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 25.Chouliaras L, van den Hove DL, Kenis G, et al. Prevention of age-related changes in hippocampal levels of 5-methylcytidine by caloric restriction. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2011.06.003. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams MM, Shi L, Linville MC, et al. Caloric restriction and age affect synaptic proteins in hippocampal CA3 and spatial learning ability. Exp Neurol. 2008;211:141–149. doi: 10.1016/j.expneurol.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson RM, Shanmuganayagam D, Weindruch R. Caloric restriction and aging: studies in mice and monkeys. Toxicol Pathol. 2009;37:47–51. doi: 10.1177/0192623308329476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 29.Colman RJ, Anderson RM, Johnson SC, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levenson CW, Rich NJ. Eat less, live longer? New insights into the role of caloric restriction in the brain. Nutr Rev. 2007;65:412–415. doi: 10.1111/j.1753-4887.2007.tb00319.x. [DOI] [PubMed] [Google Scholar]
- 31.Mattson MP, Chan SL, Duan W. Modification of brain aging and neurodegenerative disorders by genes, diet, and behavior. Physiol Rev. 2002;82:637–672. doi: 10.1152/physrev.00004.2002. [DOI] [PubMed] [Google Scholar]
- 32.Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borg J, London J. Copper/zinc superoxide dismutase overexpression promotes survival of cortical neurons exposed to neurotoxins in vitro. J Neurosci Res. 2002;70:180–189. doi: 10.1002/jnr.10404. [DOI] [PubMed] [Google Scholar]
- 34.Cadet JL, Sheng P, Ali S, Rothman R, Carlson E, Epstein C. Attenuation of methamphetamine-induced neurotoxicity in copper/zinc superoxide dismutase transgenic mice. J Neurochem. 1994;62:380–383. doi: 10.1046/j.1471-4159.1994.62010380.x. [DOI] [PubMed] [Google Scholar]
- 35.Cardozo-Pelaez F, Song S, Parthasarathy A, Epstein CJ, Sanchez-Ramos J. Attenuation of age-dependent oxidative damage to DNA and protein in brainstem of Tg Cu/Zn SOD mice. Neurobiol Aging. 1998;19:311–316. doi: 10.1016/s0197-4580(98)00067-0. [DOI] [PubMed] [Google Scholar]
- 36.Rutten BP, Steinbusch HW, Korr H, Schmitz C. Antioxidants and Alzheimer's disease: from bench to bedside (and back again) Curr Opin Clin Nutr Metab Care. 2002;5:645–651. doi: 10.1097/00075197-200211000-00006. [DOI] [PubMed] [Google Scholar]
- 37.Rutten BP, Brasnjevic I, Steinbusch HW, Schmitz C. Caloric restriction and aging but not overexpression of SOD1 affect hippocampal volumes in mice. Mech Ageing Dev. 2010;131:574–579. doi: 10.1016/j.mad.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Weindruch R, Walford RL, Fligiel S, Guthrie D. The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J Nutr. 1986;116:641–654. doi: 10.1093/jn/116.4.641. [DOI] [PubMed] [Google Scholar]
- 39.Epstein CJ, Avraham KB, Lovett M, et al. Transgenic mice with increased Cu/Zn-superoxide dismutase activity: animal model of dosage effects in Down syndrome. Proc Natl Acad Sci U S A. 1987;84:8044–8048. doi: 10.1073/pnas.84.22.8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Globisch D, Munzel M, Muller M, et al. Tissue distribution of 5- hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franklin KB, Paxinos G. The Mouse Brain in Stereotaxic Coordinates. San Diego: Academic Press; 1997. [Google Scholar]
- 42.Strackx E, van den Hove DL, Steinbusch HP, et al. Fetal asphyxia leads to a decrease in dorsal raphe serotonergic neurons. Dev Neurosci. 2008;30:358–366. doi: 10.1159/000155218. [DOI] [PubMed] [Google Scholar]
- 43.Lemmens MA, Steinbusch HW, Rutten BP, Schmitz C. Advanced microscopy techniques for quantitative analysis in neuromorphology and neuropathology research: current status and requirements for the future. J Chem Neuroanat. 2010;40:199–209. doi: 10.1016/j.jchemneu.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 44.Haberman RP, Colantuoni C, Stocker AM, Schmidt AC, Pedersen JT, Gallagher M. Prominent hippocampal CA3 gene expression profile in neurocognitive aging. Neurobiol Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weindruch R, Kayo T, Lee CK, Prolla TA. Microarray profiling of gene expression in aging and its alteration by caloric restriction in mice. J Nutr. 2001;131:918S–923S. doi: 10.1093/jn/131.3.918S. [DOI] [PubMed] [Google Scholar]
- 46.Zeier Z, Madorsky I, Xu Y, Ogle WO, Notterpek L, Foster TC. Gene expression in the hippocampus: regionally specific effects of aging and caloric restriction. Mech Ageing Dev. 2011;132:8–19. doi: 10.1016/j.mad.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munzel M, Globisch D, Bruckl T, et al. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem Int Ed Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- 48.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell. 2011 doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 50.Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morgan AR, Hamilton G, Turic D, et al. Association analysis of 528 intra-genic SNPs in a region of chromosome 10 linked to late onset Alzheimer's disease. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:727–731. doi: 10.1002/ajmg.b.30670. [DOI] [PubMed] [Google Scholar]
- 52.Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic changes in Alzheimer's disease: decrements in DNA methylation. Neurobiol Aging. 2010;31:2025–2037. doi: 10.1016/j.neurobiolaging.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mastroeni D, McKee A, Grover A, Rogers J, Coleman PD. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer's disease. PLoS One. 2009;4:e6617. doi: 10.1371/journal.pone.0006617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang SC, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer's disease. PLoS One. 2008;3:e2698. doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siegmund KD, Connor CM, Campan M, et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong CC, Caspi A, Williams B, et al. A longitudinal study of epigenetic variation in twins. Epigenetics. 2010;5:516–526. doi: 10.4161/epi.5.6.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rutten BP, Mill J. Epigenetic mediation of environmental influences in major psychotic disorders. Schizophr Bull. 2009;35:1045–1056. doi: 10.1093/schbul/sbp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chouliaras L, Sierksma AS, Kenis G, et al. Gene-environment interaction research and transgenic mouse models of Alzheimer's disease. Int J Alzheimers Dis. 2010;2010 doi: 10.4061/2010/859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murgatroyd C, Wu Y, Bockmuhl Y, Spengler D. The Janus face of DNA methylation in aging. Aging (Albany NY) 2010;2:107–110. doi: 10.18632/aging.100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Penner MR, Roth TL, Chawla MK, et al. Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol Aging. 2010 doi: 10.1016/j.neurobiolaging.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 62.Feng J, Zhou Y, Campbell SL, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu H, Coskun V, Tao J, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams MM, Donohue HS, Linville MC, Iversen EA, Newton IG, Brunso-Bechtold JK. Age-related synapse loss in hippocampal CA3 is not reversed by caloric restriction. Neuroscience. 2010;171:373–382. doi: 10.1016/j.neuroscience.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fuso A, Nicolia V, Cavallaro RA, Scarpa S. DNA methylase and demethylase activities are modulated by one-carbon metabolism in Alzheimer's disease models. J Nutr Biochem. 2011;22:242–251. doi: 10.1016/j.jnutbio.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 66.Sugden C. One-carbon metabolism in psychiatric illness. Nutr Res Rev. 2006;19:117–136. doi: 10.1079/NRR2006119. [DOI] [PubMed] [Google Scholar]
- 67.Fuso A, Seminara L, Cavallaro RA, D'Anselmi F, Scarpa S. S-adenosylmethionine/homocysteine cycle alterations modify DNA methylation status with consequent deregulation of PS1 and BACE and beta-amyloid production. Mol Cell Neurosci. 2005;28:195–204. doi: 10.1016/j.mcn.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 68.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 69.Chen D, Bruno J, Easlon E, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen D, Guarente L. SIR2: a potential target for calorie restriction mimetics. Trends Mol Med. 2007;13:64–71. doi: 10.1016/j.molmed.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 71.Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 72.Dillin A, Kelly JW. Medicine. The yin-yang of sirtuins. Science. 2007;317:461–462. doi: 10.1126/science.1146585. [DOI] [PubMed] [Google Scholar]
- 73.Przedborski S, Jackson-Lewis V, Kostic V, Carlson E, Epstein CJ, Cadet JL. Superoxide dismutase, catalase, and glutathione peroxidase activities in copper/zinc-superoxide dismutase transgenic mice. J Neurochem. 1992;58:1760–1767. doi: 10.1111/j.1471-4159.1992.tb10051.x. [DOI] [PubMed] [Google Scholar]
- 74.Nestor C, Ruzov A, Meehan R, Dunican D. Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Biotechniques. 2010;48:317–319. doi: 10.2144/000113403. [DOI] [PubMed] [Google Scholar]
- 75.Hayatsu H, Shiragami M. Reaction of bisulfite with the 5-hydroxymethyl group in pyrimidines and in phage DNAs. Biochemistry. 1979;18:632–637. doi: 10.1021/bi00571a013. [DOI] [PubMed] [Google Scholar]
- 76.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]