Abstract
Background
Using the genome-wide association approach in individuals of European ancestry, we and others recently identified single nucleotide polymorphisms (SNPs) at 19 loci as associated with blood lipids; eight of these loci were novel. Whether these same SNPs associate with lipids in a broader range of ethnicities is unknown.
Methods and Results
We genotyped index SNPs at 19 loci in the Third United States National Health and Nutrition Examination Survey (n=7159), a population-based probability sample of the U.S. comprised primarily of non-Hispanic blacks, Mexican Americans, and non-Hispanic whites. We constructed ethnic-specific residual blood lipid levels after adjusting for age and gender. Ethnic-specific linear regression was used to test the association of genotype with blood lipids. To summarize the statistical evidence across three racial groups, we conducted a fixed-effects variance-weighted meta-analysis.
After exclusions, there were 1627 non-Hispanic blacks, 1659 Mexican Americans, and 2230 non-Hispanic whites. At five loci (1p13 near CELSR2/PSRC1/SORT1, HMGCR, CETP, LPL, and APOA5), the index SNP was associated with low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, or triglycerides in all three ethnic groups. At the remaining loci, there was mixed evidence by ethnic group. In meta-analysis, we found that, at 14 of the 19 loci, SNPs exceeded a nominal P < 0.05.
Conclusions
At five loci including the recently-discovered region on 1p13 near CELSR2/PSRC1/SORT1, the same SNP discovered in whites associates with blood lipids in non-Hispanic blacks and Mexican Americans. For the remaining loci, fine-mapping and resequencing will be required to definitively evaluate the relevance of each locus in individuals of African and Hispanic ancestries.
Keywords: lipids, genetics, epidemiology, risk factors
Introduction
Genome-wide association (GWA) studies in subjects of European ancestry have successfully identified genetic loci associated with plasma level of low-density lipoprotein (LDL) cholesterol, high-density lipoprotein (HDL) cholesterol, and/or triglycerides. We and others recently identified single nucleotide polymorphisms (SNPs) at 19 loci as associated with blood lipids; eight of these loci were novel.1–6 However, the relevance of these variants in the diverse U.S. population is unclear.
When investigating these loci in diverse ethnic populations such as African-American and Hispanic groups, three potential outcomes are possible. First, association of the same polymorphism at the same locus may be confirmed. Second, a different polymorphism at the same locus may be associated in differing ethnicities. Finally, loci identified in cohorts of European ancestry may not replicate in individuals of non-European ancestry, suggesting that unique loci in the genome are responsible for blood lipid traits in groups of differing ancestries.
Here, we test the first of these possibilities - the hypothesis that the index SNP at each of 19 loci validated in whites will also be associated with blood lipid traits in individuals of African-American and Hispanic ancestry. We address this hypothesis in the United States Third National Health and Nutrition Examination Survey (NHANES III), a multiethnic representative sample of non-institutionalized persons in the U.S.
Methods
Participants
The NHANES III cohort and DNA sample collection have been previously described.7 Briefly, NHANES III is a representative, cross-sectional sample of the non-institutionalized U.S. population surveyed between 1988 and 1994 by the National Center for Health Statistics (NCHS) at the Center for Disease Control and Prevention (CDC). Minority groups (non-Hispanic blacks and Mexican Americans), children, and the elderly were over sampled. The survey consisted of a household interview, including a detailed dietary questionnaire. In addition, a physical examination was performed, and blood samples were collected. For the genetic component of NHANES III, DNA was isolated from cell lines created from human blood samples from participants of NHANES III phase 2 (1991–1994). The total number of participants in NHANES III phase 2 was 16,530. DNA was available from 7157 of these individuals aged 12 and older.7 The present study was approved by the CDC Ethics Review Board and the institutional review board at the Massachusetts Institutes of Technology, and all subjects provided written informed consent.
Laboratory Measures
HDL cholesterol and triglycerides were measured using standard enzymatic methods. LDL cholesterol was calculated using the Friedewald formula with assignment of a missing value for participants with triglycerides exceeding 400 mg/dL. Fasting blood samples were available in 41% of individuals (n=2916), and the remainder were non-fasting. We included both fasting and nonfasting samples in our analyses to increase statistical power.
SNP Selection and Genotyping
We selected 21 SNPs from 19 loci that exceeded genome-wide statistical significance (P < 5 × 10−8) in two recently published GWA studies for LDL cholesterol, HDL cholesterol, and triglycerides.2, 3 Eight of these loci had been newly identified. SNPs at the new loci included the following: rs3794991 (near CILP2/PBX4, LDL cholesterol and triglycerides); rs646776 (near CELSR2/PSRC1/SORT1, LDL cholesterol); rs4846914 (in GALNT2, HDL cholesterol and triglycerides); rs10774708 (in MMAB/MVK, HDL cholesterol); rs17145738 (near BCL7B/TBL2/MLXIPL, triglycerides); rs17321515 (near TRIB1, triglycerides); rs1748195 (ANGPTL3/DOCK7/ATG4C, triglycerides); and rs1260326 (GCKR, triglycerides). SNPs at the 11 additional loci included the following: rs6511720 and rs1529729 (LDLR, LDL cholesterol); rs11591147 (PCSK9, LDL cholesterol); rs12654264 (HMGCR, LDL cholesterol); rs4420638 (APOE-C1-C4-C2 cluster, LDL cholesterol); rs693 (APOB, LDL cholesterol); rs2156552 (LIPG, HDL cholesterol); rs1800588 (LIPC, HDL cholesterol); rs1800775 (CETP, HDL cholesterol); rs328 (LPL, HDL cholesterol, triglycerides); rs3890182 (ABCA1, HDL cholesterol); and rs3135506 and rs662799 (APOA5, HDL cholesterol and triglycerides).8–16 rs6511720 and rs1529729 have a low degree of linkage disequilibrium within the LDLR gene in populations of European (r2=0.085) and African (r2=0.167) ancestry. rs3135506 and rs662799 tag two known haplotypes within the APOA5 gene. 17
Genotyping was performed on the Sequenom platform, which utilizes matrix-assisted laser-desorption ionization time-of-flight mass spectroscopy as described previously.18 Using 356 samples in duplicate, we found the genotyping error to be 0.34%. SNP rs693 in the APOB gene was excluded from analysis due to a low genotyping call rate (88%). SNP rs10774708 near the MMAB/MVK genes was excluded from analysis due to a low genotyping call rate (70%). In sum, 19 SNPs from 17 loci were successfully genotyped and the genotyping success rate for each of these 19 SNPs exceeded 92%.
Statistical analysis
All statistical analyses were performed at the NCHS at the CDC and conducted using SAS software version 9.0 (Cary, North Carolina). Participants in NHANES III who were missing phenotype data, those under 18 years of age, and those on lipid lowering therapy were excluded from analysis. LDL cholesterol, HDL cholesterol, and triglyceride values were log transformed.
Ethnic-specific residual lipid levels were generated after adjusting for age and gender. Linear regression stratified by self-reported race/ethnicity was used to test the association of residual values with genotype, assuming an additive model of inheritance. To summarize the statistical evidence across the three race/ethnic groups, we conducted a fixed-effects variance-weighted meta-analysis. We computed a weighted average of the beta-coefficient estimates and standard errors (from the linear regression models above) for each genotype, using the inverse of the variance in each group as weights.
We denote significance to be an association P ≤ 0.05 for the same allele at the same SNP for the same trait as reported in the original discovery GWA studies.1–3 The authors had full access to the data and take responsibility for its integrity. All authors have read and agree to the manuscript as written.
Results
Participant characteristics are summarized in Table 1. After exclusions, there were 1692 non-Hispanic blacks, 1715 Mexican Americans, and 2295 non-Hispanic whites with genotype and phenotype data available for analysis.
Table 1.
Participant Characteristics
| Characteristic | NHANES: Non-Hispanic Blacks N=1692 |
NHANES: Mexican Americans N=1715 |
NHANES: Non-Hispanic Whites N=2295 |
|---|---|---|---|
| Mean age, yrs | 40.44 (0.72) | 40.70 (1.21) | 52.84 (1.45) |
| Female sex, % | 58 | 50 | 60 |
| Body mass index, kg/m2 | 28.17 (0.24) | 27.68 (0.10) | 26.58 (0.15) |
| Total cholesterol, mg/dL | 196.68 (0.96) | 199.01 (1.60) | 207.43 (1.65) |
| Low-density lipoprotein cholesterol, mg/dL | 120.56 (0.78) | 119.74 (0.85) | 127.94 (1.16) |
| High-density lipoprotein cholesterol, mg/dL | 54.09 (0.39) | 47.79 (0.45) | 50.62 (0.58) |
| Triglycerides, mg/dL | 114.41 (1.57) | 163.54 (5.53) | 148.22 (2.76) |
| Systolic blood pressure, mmHg | 124.12 (0.63) | 121.89 (1.13) | 126.45 (0.96) |
| Diabetes mellitus, % | 5.3 | 5.3 | 4.1 |
Data are expressed as mean (standard deviation).
LDL Cholesterol
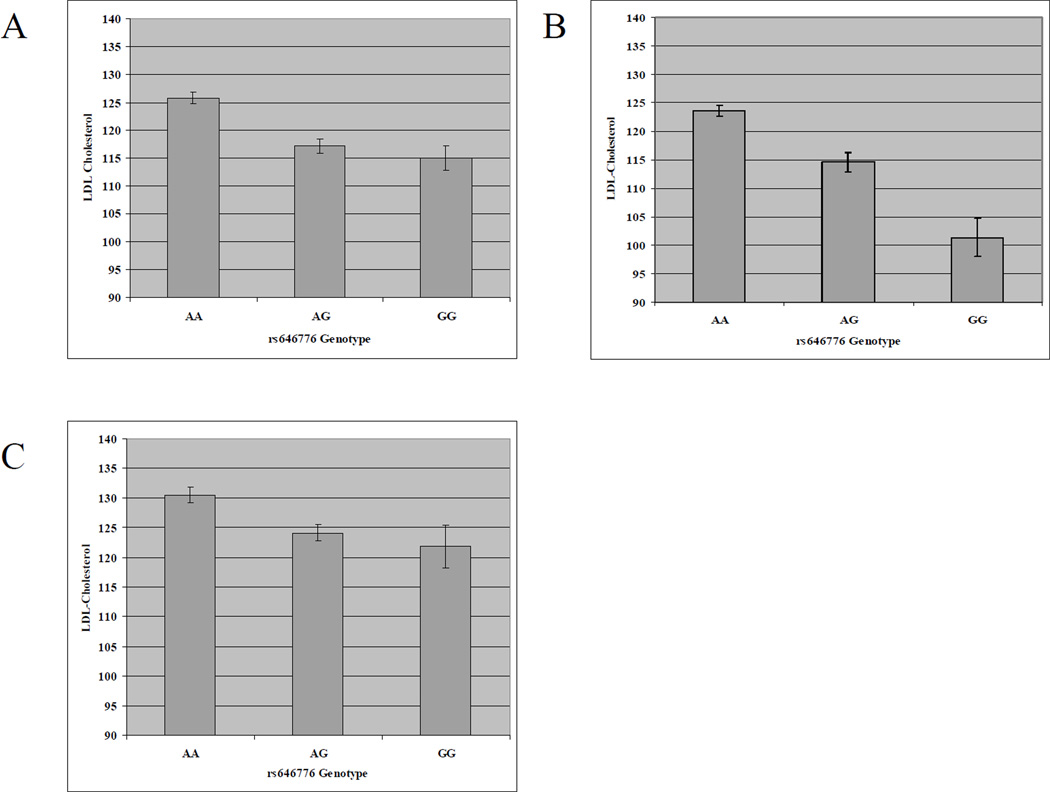
Associations of genotypes with LDL cholesterol concentrations are detailed in Table 2. The association between rs646776 (at 1p13 near CELSR2/PSRC1/SORT1) and LDL cholesterol was highly significant in all three ethnic groups (P = 0.0005 in non-Hispanic blacks, P = 1.8 × 10−6 in Mexican Americans, and P = 0.002 in non-Hispanic whites). The effects of rs646776 on LDL cholesterol are summarized in the Figure. In each ethnic group, individuals with at least one copy of the minor G allele had lower LDL cholesterol. Each copy of the minor allele lowered LDL cholesterol by ~5mg/dL in non-Hispanic blacks, ~11 mg/dL in Mexican Americans, and ~4 mg/dL in non-Hispanic whites.
Table 2.
Associations of genotypes with low-density lipoprotein cholesterol
| Index SNP | Locus | Alleles | Modeled allele (Frequency in Non-Hispanic Blacks) (Frequency in Non-Hispanic Whites) (Frequency in Mexican Americans) |
Non-Hispanic Blacks P |
Mexican Americans P |
Non-Hispanic Whites P |
Combined evidence in all three ethnic groups Beta (SE) P |
|---|---|---|---|---|---|---|---|
| rs646776 | 1p13 near PSRC1/CESLR2/ SORT1 |
A/G | G (0.36) (0.22) (0.20) |
0.0004 | 1.8×10−6 | 0.002 | −0.06 (0.01) 2.0 × 10−15 |
| rs17321515 | 8q24 3’ downstream of TRIB1 |
A/G | G (0.53) (0.46) (0.40) |
0.92 | 0.0004 | 0.17 | −0.02 (0.006) 9.2 × 10−5 |
| rs3794991 | 19p13 near CILP2/PBX4 |
C/T | T (0.06) (0.08) (0.05) |
0.04 | 0.16 | 0.03 | −0.04 (0.01) 0.00052 |
| rs6511720 | LDLR | G/T | T (0.13) (0.12) (0.10) |
8.6 × 10−5 | 0.10 | 2.2 × 10−5 | −0.06 (0.01) 2.2 × 10−11 |
| rs1529729 | LDLR | A/G | G (0.30) (0.46) (0.51) |
0.97 | 0.02 | 0.03 | 0.00061 (0.006) 0.91 |
| rs12654264 | HMGCR | A/T | T (0.32) (0.38) (0.40) |
0.006 | 0.01 | 0.05 | 0.02 (0.005) 2.3 × 10−5 |
| rs11591147 | PCSK9 | G/T | T (0.004) (0.03) (0.02) |
0.62 | 0.08 | 0.0002 | −0.16 (0.03) 7.3 × 10−6 |
Figure. LDL cholesterol by rs646776 genotype at 1p13.
LDL cholesterol concentration decreases as the modeled allele copy number increases in non-Hispanic blacks (panel A), non-Hispanic whites (panel B), and Mexican Americans (panel C).
In addition to 1p13, HMGCR rs12654264 was associated with LDL cholesterol in non-Hispanic blacks (P = 0.006), Mexican Americans (P = 0.01), and non-Hispanic whites (P = 0.05). As in the original discovery studies, the minor T allele was associated with higher LDL cholesterol in each of the ethnic groups from NHANES III.2, 18
SNPs at three loci (TRIB1, LDLR, and CILP2/PBX4) were significantly associated with LDL cholesterol in at least one non-white ethnic group. TRIB1 rs17321515 and LDLR rs1529729 were significantly associated with LDL cholesterol (P = 0.0004 and P = 0.02, respectively) in Mexican Americans. CILP2/PBX4 rs3794991 and LDLR rs6511720 were associated with LDL cholesterol in non-Hispanic blacks (P = 0.04 and P = 8.6 × 10−5, respectively).
At PCSK9, rs11591147 (or R46L) failed to reach statistical significance in both non-Hispanic blacks or Mexican Americans but was highly significant in non-Hispanic whites (P = 0.0002).
HDL-Cholesterol
The results for HDL cholesterol are summarized in Table 3. SNPs at CETP and LPL were strongly associated with HDL cholesterol in all three ethnic groups. Each increase in minor C allele copy number in CETP polymorphism rs1800775 resulted in a 0.05 standard deviation (SD) unit decrease in HDL cholesterol across all ethnicities (P=5.0 × 10−20). LPL rs328 was associated an increase of 0.04 SD unit in HDL cholesterol per allele (P = 5 × 10−8).
Table 3.
Associations of genotypes with high-density lipoprotein cholesterol
| Index SNP | Locus | Alleles | Modeled allele (Frequency in Non-Hispanic Blacks) (Frequency in Non-Hispanic Whites) (Frequency in Mexican Americans) |
Non-Hispanic Blacks P |
Mexican Americans P |
Non-Hispanic Whites P |
Combined evidence in all three ethnic groups Beta (SE) P |
|---|---|---|---|---|---|---|---|
| rs17145738 | 7q11 near BCL7B/ TBL2/ MLXIPL |
C/T | T (0.09) (0.11) (0.07) |
0.11 | 0.19 | 0.004 | 0.01 (0.01) 0.04 |
| rs4846914 | 1q42 in GALNT2 | G/A | A (0.15) (0.60) (0.58) |
0.94 | 0.02 | 0.39 | 0.006 (0.005) 0.23 |
| rs17321515 | 8q24 3’ downstream of TRIB1 |
A/G | G (0.53) (0.46) (0.40) |
0.31 | 0.59 | 0.16 | 0.003 (0.004) 0.53 |
| rs1800775 | CETP | A/C | C (0.43) (0.53) (0.46) |
0.004 | 5.7 × 10−7 | 1.9 × 10−5 | −0.05 (0.006) 5.0 × 10−20 |
| rs328 | LPL | C/G | G (0.07) (0.10) (0.07) |
0.04 | 0.006 | 0.0003 | 0.04 (0.008) 4.8 × 10−8 |
| rs1800588 | LIPC | C/T | T (0.51) (0.23) (0.54) |
0.10 | 0.15 | 0.19 | 0.01 (0.006) 0.01 |
| rs3890182 | ABCA1 | G/A | A (0.12) (0.12) (0.08) |
0.68 | 0.27 | 0.94 | −0.007 (0.009) 0.35 |
| rs2156552 | LIPG | A/T | T (0.04) (0.16) (0.07) |
0.58 | 0.21 | 0.09 | −0.02 (0.01) 0.03 |
At one locus - GALNT2 - the same SNP discovered in whites was significantly associated with HDL cholesterol in at least one non-white ethnic group. SNP rs4846914 at GALNT2 was associated with HDL cholesterol in Mexican Americans (P = 0.02) but not in non-Hispanic blacks. SNPs at the remaining loci failed to replicate in either non-white ethnic group.
Triglycerides
The results for triglycerides are summarized in Table 4. At APOA5, rs3135506 was associated with triglyceride concentrations across all ethnicities. In each ethnicity, increase in minor allele copy number resulted in a ~0.12 SD increase in triglyceride level (P = 4.3 × 10−11).
Table 4.
Associations of genotypes with triglycerides
| Index SNP | Locus | Alleles | Modeled allele (Frequency in Non-Hispanic Blacks) (Frequency in Non-Hispanic Whites) (Frequency in Mexican Americans) |
Non-Hispanic Blacks P |
Mexican Americans P |
Non-Hispanic Whites P |
Combined evidence in all three ethnic groups Beta (SE) P |
|---|---|---|---|---|---|---|---|
| rs17321515 | 8q24 3’ downstream of TRIB1 |
A/G | G (0.53) (0.46) (0.40) |
0.55 | 7.1 × 10−5 | 0.02 | −0.03 (0.009) 0.0005 |
| rs17145738 | 7q11 near BCL7B/TBL2/ MLXIPL |
C/T | T (0.09) (0.11) (0.07) |
0.94 | 0.01 | 0.01 | −0.05 (0.01) 0.002 |
| rs1748195 | 1p31 near ANGPTL3/ DOCK7/ATG4C |
C/G | G (0.64) (0.33) (0.40) |
0.43 | 0.07 | 0.16 | −0.02 (0.001) 0.01 |
| rs3794991 | 19p13 near CILP2/PBX4 |
C/T | T (0.06) (0.08) (0.05) |
0.01 | 0.37 | 0.78 | −0.04 (0.02) 0.01 |
| rs1260326 | 2p23 in GCKR | C/T | T (0.16) (0.43) (0.34) |
0.75 | 0.17 | 0.06 | 0.02 (0.01) 0.04 |
| rs4846914 | in GALNT2 | G/A | A (0.15) (0.60) (0.58) |
0.06 | 0.25 | 0.46 | −0.02 (0.01) 0.06 |
| rs3135506 | APOA5 | G/C | C (0.06) (0.06) (0.14) |
8.1 × 10−5 | 0.001 | 0.01 | 0.12 (0.02) 4.3 × 10−11 |
| rs662799 | APOA5 | A/G | G (0.13) (0.06) (0.15) |
0.29 | 0.0001 | 0.05 | 0.09 (0.02) 7.2 × 10−6 |
| rs328 | LPL | C/G | G (0.07) (0.10) (0.07) |
0.10 | 0.64 | 0.0002 | −0.08 (0.02) 1.3 × 10−5 |
SNPs at three loci (MLXIPL, TRIB1, and CILP2/PBX4) were significantly associated with triglycerides in at least one non-white ethnic group. SNPs rs17321515 near MLXIPL and rs17145738 near TRIB1 replicated in Mexican Americans (P = 0.01 and 7.1 × 10−5, respectively). In non-Hispanic blacks, we observed an association between rs3734991 near CILP2/PBX4 and triglycerides (P = 0.01). LPL rs328 was strongly associated with triglycerides in non-Hispanic whites (P = 0.0002). SNPs at the remaining four loci failed to replicate in either non-white ethnic group.
As non-fasting status may affect triglyceride levels, we conducted secondary analyses with the phenotype of race-specific triglyceride residuals adjusted for age, gender, and fasting status and the results were not materially different (data not shown).
Discussion
In a multiethnic sample representative of the U.S. population, we tested DNA sequence variants at 19 loci that had been discovered in individuals of European descent. We addressed the hypothesis that these same variants would relate to blood lipid traits in other ethnic groups. At five loci (CELSR2/PSRC1/SORT1, HMGCR, CETP, LPL, and APOA5), we found that the same variant identified in whites was nominally associated in both non-Hispanic blacks and Mexican Americans. At other loci, there was mixed evidence for SNPs by ethnic group.
For SNPs at five loci discovered in whites, the same polymorphism was confirmed across all three NHANES III ethnic groups. This suggests that these loci impact blood lipid traits across multiple ethnic groups. These results set the stage for fine-mapping and resequencing of these gene regions in all three ethnic groups.
For the remaining loci, variable replication among ethnicities likely occurred for one or more of the following reasons. First, individual genetic variants contributing to complex traits have very modest effect sizes. Therefore, each individual ethnic group may have been underpowered to detect an association. This problem could be addressed by genotyping the polymorphisms in additional individuals of African-American or Hispanic ancestry. Second, the locus discovered in whites may be relevant in other ethnicities but the specific polymorphisms associated with lipids may differ across ethnicities. Older populations such as those of African descent have a more complex linkage disequilibrium (LD) structure.19 Therefore, if the causal variant in African Americans is poorly correlated with the index SNP discovered from whites, no association will be detected. In this case, testing a comprehensive set of variants that tag the entire locus (i.e. fine-mapping the region in other ethnicities) would be required to detect the relevant polymorphism(s). Finally, genetic variants in different ethnicities may lie in regions of the genome distinct from those observed in whites. New variant discovery through additional GWA studies or resequencing efforts in other ethnicities would be necessary to locate these variants. Such efforts are currently underway.
In prior literature, several of the loci have been studied in African-Americans and/or Hispanic-Americans including LDLR, PCSK9, CETP, LPL, and APOA5. Our study supports and builds upon these prior reports. While other variants have been reported in multi-ethnic groups20, we are the first to describe associations of the LDLR intronic SNPs rs6511720 and rs1529729 with LDL cholesterol in a sample including African-Americans and Mexican-Americans. We confirm that rs11591147 (R46L) in the PCSK9 gene is common and decreases LDL cholesterol in whites (minor allele frequency of 3%) but is rare in African-Americans (minor allele frequency of 0.4%).9, 21 As in our study, associations between polymorphisms in the CETP and LPL genes and HDL cholesterol have been consistently reported in cohorts including African American and/or Hispanic subjects.22–24 Finally, we confirm the robust association between APOA5 and triglycerides in all three ethnic groups.25, 26 By reproducing previously reported findings, we demonstrate that our patient sample is representative and that our methods are valid.
Our study has several strengths and limitations. To our knowledge, this is the first study to assess loci recently discovered through GWA studies in a multiethnic cohort. Next, compared to prior studies assessing few variants in multiethnic cohorts, our study queried a larger set of lipid loci. Finally, because the NHANES III cohort over-sampled minority groups in the U.S. population, our sample includes a large number of African-Americans and Mexican Americans. That being said, due to the modest contribution of each polymorphism to trait variation, low statistical power is a key limitation. Lipids in NHANES III were measured in both fasting and non-fasting samples and our use of both may have further reduced statistical power, particularly for lipid variables sensitive to fasting status such as triglycerides. In addition, in NHANES III, ethnic groups are designated by self-reported ethnicity and this assessment is not always accurate. For a more precise assessment of ethnicity using genetic data, ancestry-informative markers are necessary. Lastly, we did not evaluate a comprehensive set of 'tag' SNPs across each of the 19 loci. Such an effort would be required to definitively study each locus in African-Americans and Mexican-Americans.
In conclusion, we evaluated 21 SNPs in 19 loci from recent GWA studies for blood lipids in a U.S. population-based multi-ethnic sample. We found that at five loci, the exact same SNP identified in whites was also associated with lipids in both African-Americans and Mexican-Americans. At 1p13 near CELSR2/PSRC1/SORT1, the strongest new locus identified in GWA studies of LDL cholesterol, the same index SNP was associated with LDL cholesterol in all three ethnic groups, suggesting that this locus is of broad relevance. Our findings set the stage for comprehensive fine-mapping and sequencing efforts across these loci in multiple ethnic groups.
A principal goal of genetic association studies has been to augment current disease prediction algorithms by identifying genetic variants associated with common diseases. Genome-wide association (GWA) studies along with candidate gene studies have identified many variants associated with plasma lipid traits. However, most GWA studies published to date and the majority of candidate gene studies have been conducted exclusively in samples of European ancestry. Therefore, it is unclear whether these variants are relevant in a broader range of ethnic groups such as African Americans and Hispanic Americans. In the current study, we tested polymorphisms from candidate gene loci and loci identified through GWA in the Third United States National Health and Nutritional Examination Survey (NHANES III), a population-based probability sample consisting of non-Hispanic black, Mexican American, and non-Hispanic white participants. Our study takes the first step toward addressing whether the same genetic variants identified in populations of European ancestry will be associated with blood lipids and, therefore, possibly predictive in other ethnic groups. While further work is needed, our findings suggest that for many loci, the same variant identified in whites will also be relevant in other ethnic groups.
Acknowledgements
None
Funding Sources: M.K. was supported by the Charles A. King Trust, Bank of America, Co-Trustee. S.K. is supported by a Doris Duke Charitable Foundation Clinical Scientist Development Award, a charitable gift from the Fannie E. Rippel Foundation, the Donovan Family Foundation, a career development award from the United States National Institutes of Health (NIH), and institutional support from the Department of Medicine and Cardiovascular Research Center at Massachusetts General Hospital.
Footnotes
Disclosures: The authors have no disclosures to report.
Contributor Information
Mary E. Keebler, Massachusetts General Hosp, Boston, MA.
Christopher L. Sanders, Ctrs for Disease Control, Atlanta, GA.
Aarti Surti, Broad Inst of MIT/Harvard, Cambridge, MA.
Candace Guiducci, Broad Inst of MIT/Harvard, Cambridge, MA.
Noel P Burtt, Broad Inst of MIT/Harvard, Cambridge, MA.
Sekar Kathiresan, Massachusetts General Hosp, Boston, MA.
References
- 1.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 2.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M, Ahmadi K, Dobson RJ, Marcano AC, Hajat C, Burton P, Deloukas P, Brown M, Connell JM, Dominiczak A, Lathrop GM, Webster J, Farrall M, Spector T, Samani NJ, Caulfield MJ, Munroe PB. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kooner JS, Chambers JC, Aguilar-Salinas CA, Hinds DA, Hyde CL, Warnes GR, Gomez Perez FJ, Frazer KA, Elliott P, Scott J, Milos PM, Cox DR, Thompson JF. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat Genet. 2008;40:149–151. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- 6.Sandhu MS, Waterworth DM, Debenham SL, Wheeler E, Papadakis K, Zhao JH, Song K, Yuan X, Johnson T, Ashford S, Inouye M, Luben R, Sims M, Hadley D, McArdle W, Barter P, Kesaniemi YA, Mahley RW, McPherson R, Grundy SM, Bingham SA, Khaw KT, Loos RJ, Waeber G, Barroso I, Strachan DP, Deloukas P, Vollenweider P, Wareham NJ, Mooser V. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371:483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford DC, Sanders CL, Qin X, Smith JD, Shephard C, Wong M, Witrak L, Rieder MJ, Nickerson DA. Genetic variation is associated with C-reactive protein levels in the Third National Health and Nutrition Examination Survey. Circulation. 2006;114:2458–2465. doi: 10.1161/CIRCULATIONAHA.106.615740. [DOI] [PubMed] [Google Scholar]
- 8.Knoblauch H, Bauerfeind A, Toliat MR, Becker C, Luganskaja T, Gunther UP, Rohde K, Schuster H, Junghans C, Luft FC, Nurnberg P, Reich JG. Haplotypes and SNPs in 13 lipid-relevant genes explain most of the genetic variance in high-density lipoprotein and low-density lipoprotein cholesterol. Hum Mol Genet. 2004;13:993–1004. doi: 10.1093/hmg/ddh119. [DOI] [PubMed] [Google Scholar]
- 9.Kotowski IK, Pertsemlidis A, Luke A, Cooper RS, Vega GL, Cohen JC, Hobbs HH. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am J Hum Genet. 2006;78:410–422. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sing CF, Davignon J. Role of the apolipoprotein E polymorphism in determining normal plasma lipid and lipoprotein variation. Am J Hum Genet. 1985;37:268–285. [PMC free article] [PubMed] [Google Scholar]
- 11.Benn M, Nordestgaard BG, Jensen JS, Grande P, Sillesen H, Tybjaerg-Hansen A. Polymorphism in APOB associated with increased low-density lipoprotein levels in both genders in the general population. J Clin Endocrinol Metab. 2005;90:5797–5803. doi: 10.1210/jc.2005-0974. [DOI] [PubMed] [Google Scholar]
- 12.Guerra R, Wang J, Grundy SM, Cohen JC. A hepatic lipase (LIPC) allele associated with high plasma concentrations of high density lipoprotein cholesterol. Proc Natl Acad Sci U S A. 1997;94:4532–4537. doi: 10.1073/pnas.94.9.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boekholdt SM, Sacks FM, Jukema JW, Shepherd J, Freeman DJ, McMahon AD, Cambien F, Nicaud V, de Grooth GJ, Talmud PJ, Humphries SE, Miller GJ, Eiriksdottir G, Gudnason V, Kauma H, Kakko S, Savolainen MJ, Arca M, Montali A, Liu S, Lanz HJ, Zwinderman AH, Kuivenhoven JA, Kastelein JJ. Cholesteryl ester transfer protein TaqIB variant, high-density lipoprotein cholesterol levels, cardiovascular risk, and efficacy of pravastatin treatment: individual patient meta-analysis of 13,677 subjects. Circulation. 2005;111:278–287. doi: 10.1161/01.CIR.0000153341.46271.40. [DOI] [PubMed] [Google Scholar]
- 14.Rip J, Nierman MC, Ross CJ, Jukema JW, Hayden MR, Kastelein JJ, Stroes ES, Kuivenhoven JA. Lipoprotein lipase S447X: a naturally occurring gain-of-function mutation. Arterioscler Thromb Vasc Biol. 2006;26:1236–1245. doi: 10.1161/01.ATV.0000219283.10832.43. [DOI] [PubMed] [Google Scholar]
- 15.Frikke-Schmidt R, Nordestgaard BG, Jensen GB, Tybjaerg-Hansen A. Genetic variation in ABC transporter A1 contributes to HDL cholesterol in the general population. J Clin Invest. 2004;114:1343–1353. doi: 10.1172/JCI20361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pennacchio LA, Olivier M, Hubacek JA, Cohen JC, Cox DR, Fruchart JC, Krauss RM, Rubin EM. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 2001;294:169–173. doi: 10.1126/science.1064852. [DOI] [PubMed] [Google Scholar]
- 17.Lai CQ, Demissie S, Cupples LA, Zhu Y, Adiconis X, Parnell LD, Corella D, Ordovas JM. Influence of the APOA5 locus on plasma triglyceride, lipoprotein subclasses, and CVD risk in the Framingham Heart Study. J Lipid Res. 2004;45:2096–2105. doi: 10.1194/jlr.M400192-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, Altshuler DM, Newton-Cheh C, Orho-Melander M. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 19.A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muallem H, North KE, Kakoki M, Wojczynski MK, Li X, Grove M, Boerwinkle E, Wilhelmsen KC, Heiss G, Maeda N. Quantitative effects of common genetic variations in the 3'UTR of the human LDL-receptor gene and their associations with plasma lipid levels in the Atherosclerosis Risk in Communities study. Hum Genet. 2007;121:421–431. doi: 10.1007/s00439-007-0327-1. [DOI] [PubMed] [Google Scholar]
- 21.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 22.Tsai MY, Johnson C, Kao WH, Sharrett AR, Arends VL, Kronmal R, Jenny NS, Jacobs DR, Jr, Arnett D, O'Leary D, Post W. Cholesteryl ester transfer protein genetic polymorphisms, HDL cholesterol, and subclinical cardiovascular disease in the Multi- Ethnic Study of Atherosclerosis. Atherosclerosis. 2008 doi: 10.1016/j.atherosclerosis.2007.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chamberlain AM, Folsom AR, Schreiner PJ, Boerwinkle E, Ballantyne CM. Low-density lipoprotein and high-density lipoprotein cholesterol levels in relation to genetic polymorphisms and menopausal status: The Atherosclerosis Risk in Communities (ARIC) Study. Atherosclerosis. 2008 doi: 10.1016/j.atherosclerosis.2007.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humphries SE, Berglund L, Isasi CR, Otvos JD, Kaluski D, Deckelbaum RJ, Shea S, Talmud PJ. Loci for CETP, LPL, LIPC, and APOC3 affect plasma lipoprotein size and sub-population distribution in Hispanic and non-Hispanic white subjects: the Columbia University BioMarkers Study. Nutr Metab Cardiovasc Dis. 2002;12:163–172. [PubMed] [Google Scholar]
- 25.Hallman DM, Srinivasan SR, Chen W, Boerwinkle E, Berenson GS. Longitudinal analysis of haplotypes and polymorphisms of the APOA5 and APOC3 genes associated with variation in serum triglyceride levels: the Bogalusa Heart Study. Metabolism. 2006;55:1574–1581. doi: 10.1016/j.metabol.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 26.Pennacchio LA, Olivier M, Hubacek JA, Krauss RM, Rubin EM, Cohen JC. Two independent apolipoprotein A5 haplotypes influence human plasma triglyceride levels. Hum Mol Genet. 2002;11:3031–3038. doi: 10.1093/hmg/11.24.3031. [DOI] [PubMed] [Google Scholar]