Abstract
Phage display is commonly used to isolate peptides that bind to a desired cell type. While chemical synthesis of selected peptides often results in ligands with low affinity, a multivalent tetrameric presentation of the peptides dramatically improves affinity. One of the primary uses of these peptides is conjugation to nanoparticle-based therapeutics for specific delivery to target cell types. We set out to optimize the path from phage display peptide selection to peptide presentation on a nanoparticle surface for targeted delivery. Here, we examine the effects of peptide valency, density, and affinity on nanoparticle delivery and therapeutic efficacy, using the αvβ6-specific H2009.1 peptide as a model phage-selected peptide and liposomal doxorubicin as a model therapeutic nanoparticle. Liposomes displaying the higher affinity multivalent H2009.1 tetrameric peptide demonstrate 5-10 fold higher drug delivery than liposomes displaying the lower affinity monomeric H2009.1 peptide, even when the same number of peptide subunits are displayed on the liposome. Importantly, a 6-fold greater toxicity is observed towards αvβ6-expressing cells for liposomes displaying tetrameric verses monomeric H2009.1 peptides. Additionally, liposomal targeting and toxicity increase with increasing concentrations of H2009.1 tetrameric peptide on the liposome surface. Thus, both the multivalent peptide and the multivalent liposome scaffold work together to increase targeting to αvβ6-expressing cells. This multi-layered approach to developing high affinity targeted nanoparticles may improve the utility of moderate affinity peptides. As tetramerization is known to increase affinity for a variety of phage-selected peptides, it is anticipated that the tetrameric scaffold may act as a general method for taking peptides from phage display to nanoparticle display.
INTRODUCTION
Targeting ligands that specifically recognize certain cell types or biological structures are emerging as important tools for cell-specific delivery of therapeutics and imaging agents. While antibodies have traditionally been the gold standard for cell-targeting,1, 2 peptides are an attractive alternative. Unlike antibodies, peptides are easy to synthesize in large quantities3 and their smaller size improves tissue penetration while preventing nonspecific uptake by the reticuloendoethial system. Additionally, peptides can be chemically modified to alter affinity, charge, hydrophobicity, stability, and solubility. In this manner, peptides can be optimized for in vivo use through reiterative modifications.
Phage display4-6 is a powerful method for screening large peptide libraries for specific binding to a desired target,7, 8 including proteins,5 whole cells9 and tissues.10 However, chemical synthesis of peptides identified by phage display often results in ligands with significantly lower affinity than the corresponding phage, likely due to a loss of multivalency. The widely used and commercially available M13 phage libraries present peptides at the N-terminus of the phage pIII coat protein, which is displayed at one end of the filamentous phage in 3-5 copies. Thus, target-specific binding of the phage is likely driven by both the peptide sequence and the multimeric presentation of peptides.
Many naturally occurring ligands bind via multivalent interactions,11 and multivalency is a proven synthetic approach to improving affinity of moderate binders.11, 12 We previously demonstrated that peptide affinity is increased by mimicking the multimeric presentation of the phage.13 Employing a trilysine dendrimeric core, we synthesized tetrameric peptides that display four peptide copies in the same valency and orientation as the phage. In a similar fashion, dendrimeric wedges displaying 2-5 peptide copies have also been used to emulate phage displayed peptides for increased affinity.14, 15 Our tetrameric peptide presentation works as a general construct for peptides selected by phage display against a variety of cell types, allowing the peptides to maintain their cell specificity while increasing affinity relative to the monovalent peptides.13, 16-19 Importantly, tetramerization increases affinity of the peptides for their target cells by >45-fold compared to the corresponding monomeric peptides. The non-additive increase in affinity likely results from multivalent binding. While dimer and trimer peptides displaying either two or three peptide copies also increase affinity compared to the corresponding monomer, the tetrameric peptide construct displays the highest affinity.20 Importantly, the tetrameric peptides rival the affinity of antibodies.20 We recently developed a convergent method for the synthesis of tetrameric peptides with high yield and purity, expanding the utility of these tetrameric peptides.20
One of the downstream applications of phage display isolated peptides is incorporation into drug delivery or molecular imaging systems. As nanoparticles are able to encapsulate a variety of imaging agents or therapeutics, they are particularly attractive for ligand-guided delivery to cells or organs. It has been assumed that conjugation of multiple copies of a ligand to the surface of a nanoparticle will impart multivalent binding and improve affinity of the ligand for its target.11 An assortment of monomeric peptides selected from phage display libraries have been attached to nanoparticle platforms for imaging or therapy applications in animals.21-23 However, it is unclear whether these platforms display the peptides in an ideal multimeric conformation. Additionally, increasing the copy number of the ligand on the nanoparticle to improve the effects of multivalent binding can result in increased non-specific binding. Nanoparticle display of higher affinity, multimeric peptides may further increase targeting, resulting in optimized therapeutic or imaging outcomes.
Multivalent sugar ligands have been used for targeting of liposomes24-27 and the dendritic display of mannose, a known multivalent ligand, on the surface of nanoparticles has been shown to increase affinity for a Concanavalin A target protein.28 However, there are no reports of multimeric peptide ligands conjugated to nanoparticles. It is unknown whether the multivalency of the nanoparticle is sufficient to provide selective targeting or whether a multivalent phage-mimicking peptide could further enhance nanoparticle targeting. The goal of this work is to examine the effects of peptide concentration, affinity, and valency on nanoparticle delivery.
For this study, we chose the H2009.1 peptide as a model ligand for studying the effects of peptide affinity and valency with the goal of translating phage display selected peptides to nanoparticle-targeting peptides. The H2009.1 peptide was originally selected by biopanning a phage display library against the non0small cell lung cancer (NSCLC) cell line H2009.13.The peptide was subsequently discovered to bind the restrictively expressed integrin αvβ6 on the cell surface.29 αvβ6 is emerging as a viable target for many epithelial-derived cancers,30-40 including NSCLC; it is expressed in 56% of NSCLC patient tumor samples and only rarely expressed in normal tissue.41 Importantly, αvβ6 is “turned on” early in the disease progression of NSCLC indicating that it may be a good biomarker for early cancer detection and treatment.41 While a monomeric version of the H2009.1 peptide binds αvβ6-expressing H2009 cells with a half-maximal binding affinity of 9.2 nM, displaying the peptide on a tetrameric scaffold increases affinity three orders of magnitude, down to 11 pM. The H2009.1 peptide specifically binds and internalizes into αvβ6-expressing cells and targets αvβ6-positive tumors in mice, making this peptide a good model ligand with clinical relevance. In this study, we conjugated either monomeric or tetrameric H2009.1 peptides to liposomal doxorubicin for a systematic comparison of the effects of peptide concentration, affinity, and valency in targeting αvβ6-positive NSCLC cells (Figure 1), with the goal of translating peptides isolated from phage displayed libraries to nanoparticle display.
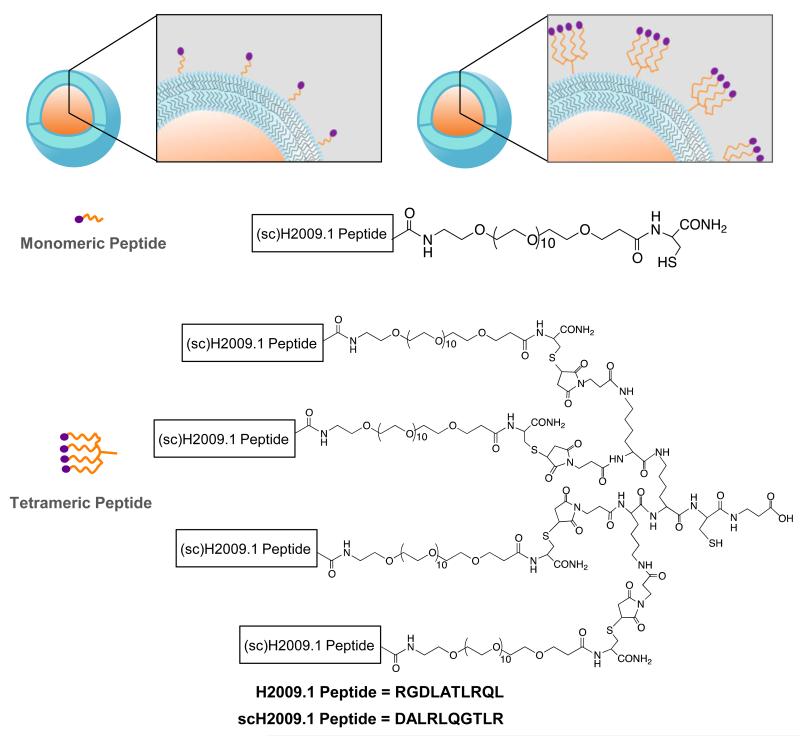
Figure 1. Liposomes displaying either monomeric or tetrameric H2009.1 or scH2009.1 peptides.
Liposomes were prepared according to standard procedure and contained 0.64-2.0 molar percent DSPE-PEG2000-maleimide. The peptides were conjugated to the liposome via reaction of DSPE-PEG2000-maleimide with a unique thiol on the peptide.
EXPERIMENTAL METHODS
Materials
Fmoc amino acids were purchased from EMD Millipore, lipids from Avanti Polar Lipids, Inc., and doxorubicin HCl from Bedford Laboratories. For cell culture, RPMI 1640 was purchased from Mediatech, Inc., FBS from Gemini Bio Products, and Trypsin/EDTA from Fisher Scientific. Sepharose CL-4B and Sephadex G-50 were purchased from Sigma-Aldrich.
Cell Culture
Human NSCLC lines were provided by the UT Southwestern Hamon Center for Therapeutic Oncology Research and maintained according to published protocols.42 Cells were grown at 37°C and 5% CO2 in RPMI 1640 supplemented with 5% fetal bovine serum.
Peptide Synthesis
Monomeric and tetrameric H2009.1 peptides were synthesized as previously described.20 Briefly, the monomeric peptides and tetrameric core were synthesized on a Symphony Synthesizer (Rainin Instruments, Protein Technologies, Inc.) using standard Fmoc solid-phase synthesis. The tetrameric peptides were synthesized by reaction of 5-fold excess monomeric peptide with the maleimide activated tetrameric core in PBS + 10mM EDTA by shaking at RT for 2h. All peptides were purified by reverse phase HPLC on a Vydac PR-C18 column using previously published HPLC elution conditions.20 Matrix-assisted laser desorption/ionization time of flight mass spectrometry was used to confirm peptide mass. H2009.1 monomeric peptide mass (average mass calculated/found:1843.02/1844.18) was determined in reflective mode using α-cyano-4-hydroxycinnamic acid as a matrix and H2009.1 tetrameric peptide mass (average mass calculated/found: 8557.94/8558.38) was determined in linear mode using sinapinic acid as a matrix.
Liposome Preparation and Characterization
Liposomes were prepared from solutions of the lipids HSPC, cholesterol, DSPE-PEG20000, and DSPE-PEG2000-maelimide in 2:1 chloroform:methanol (Table S1). Solvent was removed under a slow stream of nitrogen at 45°C and the lipid film was left under vacuum overnight. The dried lipid film was hydrated with 155mM (NH4)2SO4 buffer, pH 5.5, by intermittently heating at 65°C and vortexing. Liposomes were subsequently extruded 20 times through double-stacked 100nm membranes and PD-10 columns were used to change the outer liposomal buffer to 123mM sodium citrate, pH 5.5. Doxorubicin was loaded remotely (post-liposome formation) by incubation of liposomes and doxorubicin at 65°C for 1h. Free doxorubicin was removed using a Sephadex G-50 column equilibrated with HEPES buffered saline. Peptides were conjugated to liposomes by reaction for 24h at a ratio of 2:1 peptide: DSPE-PEG2000-maelimide in a solution of HEPES buffered saline and excess peptide was removed using Sepharose CL-4B columns. Liposome size was determined by dynamic light scattering using a Viskotec instrument and phospholipid content was determined using the Modified Bartlett Procedure.43 Doxorubicin concentration was determined by absorbance at 480nm after the addition of 1% Triton X-100 and heating at 95°C as detailed elsewhere.44 The amounts of peptide conjugated to the liposomes were determined using the CBQCA Protein Quantitation Kit (Life Technologies) according to manufacturer’s protocol. In brief, peptide quantities are measured by the reaction of (3-(4-caroxybenzoyl)quinolone-2-carboxaldehyde and KCN with the modified liposomes. Fluorescence was measured using excitation/emission wavelengths of 465/550 nm. A stock solution of H2009.1 peptide with a known concentration was used to generate a standard curve. Peptide number/liposome was calculated by assuming 80,000 phospholipid molecules/liposome for a liposome size of 100nm.45
Doxorubicin Fluorescence in Cells
H2009 cells were plated in 12 well plates and 48h later, once the cells reached 90% confluency, incubated with doxorubicin liposomes for 1h. After washing the cells twice with PBS, PBS + 5% Triton was added and the plates were shaken for 15 minutes to lyse both the cells and liposomes. Doxorubicin concentration was determined by absorbance at 590nm compared to doxorubicin standards in untreated cells.
Liposome Accumulation in Cells as Monitored by Flow Cytometry
H2009 or H1299 cells were plated in 12 well plates and 48h later, once the cells reached 90% confluency, incubated with 8-20 NM (based on doxorubicin concentration) doxorubicin liposomes for 1h. All wells were then washed (with gentle shaking) with 4 × 1mL of PBS + 0.1% BSA and cells were removed from the plates after incubation on ice for 30 min. in 1mL/well of Enzyme-free Cell Dissociation Buffer (Life Technologies). A Cell Lab Quanta™ SC flow cytometer (Beckman Coulter) was used to measure doxorubicin fluorescence for 10,000 cells per treatment group in channel 2 (excitation 488nm, emission 550-600nm). Data were analyzed by WinMDI 2.9 software. No significant fluorescence quenching by the membranes of the intact cells and liposomes was observed in these assays.
Cell Viability
H2009 or H1299 cells were seeded at 2,000 cells per well in 96 well plates in a volume of 50 NL of R5 media. After 24h, cells were treated with doxorubicin liposomes at varying concentrations (10-10240 nM based on total doxorubicin concentration). After incubation with the liposomes for 1h, all wells were washed twice with 100 NL of R5 before the addition of 100 NL of fresh R5. The cells were maintained at 37°C and 5% CO2 for 96h longer, with one media change at 72h. At 120h, cell viability was determined using the ATP-based CellTiter-Glo® Luminescent Cell Viability Assay (Promega). IC50 values were calculated using DIVISA software.
Confocal Microscopy
H2009 cells were plated at 1,000 cells/plate in poly-d-lysine coated glass bottom culture dishes (MatTek Corporation). 24h later, the cells were incubated with 4 μM (based on total doxorubicin concentration) of either free doxorubicin or 1.3% H2009.1 tetrameric liposomes. After 1h, the cells were washed twice with R5 before the addition of fresh R5. The cells were then examined using a Nikon TE2000-E microscope, observing doxorubicin fluorescence (excitation 485 nm, emission 595 nm).
RESULTS
Preparation of Liposomes Functionalized with either Monomeric or Tetrameric H2009.1 Peptides
Liposomal doxorubicin was chosen as a model nanoparticle system due to its widespread clinical use. As liposomes are small vesicles formed from lipid bilayers, their aqueous interior allows encapsulation of a variety of hydrophilic or amphipathic drugs or imaging agents. Additionally, their ~100nm size is ideal for tumor targeting, falling within the desired range for extravasation from the blood vessels into the tumor.44 The stealth formulation which contains surface polyethylene glycol was employed. Polyethylene glycol (PEG) lipids prolong liposome in vivo circulation times by reducing interaction of the liposomes with serum proteins and delaying clearance of the nanoparticles by the reticuloendoethial system.46 PEG2000 is typically employed as it is long enough to impart these favorable characteristics while keeping the liposome within the size range required for tumor penetration. While there are a growing number of nanomedicine platforms which we could explore, using a well characterized nanoparticle drug delivery system for our study will speed advances from chemical optimization to clinical use.
Liposomes were prepared by mixing hydrogenated soy phosphatidylcholine (HSPC), cholesterol, DSPE-PEG2000 (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[carbonyl-methoxypolyethyelene glycol-2000]), and DSPE(PEG2000-maelimide (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000]) at a molar ratio of 2:1:0.08:0.02, resulting in liposomes with 0.64% of the total lipid displaying maleimide groups. This formulation has been used for antibody conjugation to liposomes and serves as a reasonable starting point. The maleimide-activated lipid allows for conjugation of cysteine containing peptides to the liposome, displaying the peptides outside the PEG brush layer coating the liposome. To examine the effect of peptide density on liposome targeting ability, additional liposome formulations were prepared with increasing amounts of maleimide-activated lipid, keeping the total amount of pegylated lipid constant. All the liposome formulations are named according to the percentage of lipid bearing maleimide functionality – 0.64%, 1.3%, or 2.0% liposomes (Table S1). Replacing all pegylated lipid with maleimide-pegylated lipid, to make 3.2% maleimide liposomes, resulted in liposomes with high nonspecific cellular binding (data not shown).
To investigate whether peptide valency affects liposomal targeting, we synthesized four different peptides for conjugation to the doxorubicin liposomes – the targeting H2009.1 monomeric and tetrameric peptides and sequence scrambled control versions of these peptides, denoted as scH2009.1 peptides (Figure 1). The scH2009.1 peptides serve as important controls as they contain the same amino acids as the H2009.1 peptides but display the amino acids in a scrambled order that renders the peptides unable to bind αvβ6. Carboxy-terminal PEG groups were included in the monomeric peptides to increase solubility and to provide space between the peptide sequence responsible for binding and the point of attachment to either the nanoparticle or tetrameric core. The tetrameric peptides were synthesized from the monomeric peptides and a maleimide-activated tetrameric core using a convergent synthesis previously described.20 All monomeric and tetrameric peptides display a unique C-terminal cysteine for reaction with the maleimide-activated lipid on the liposome surface.
Peptide functionalized liposomes were synthesized using thiol-maleimide chemistry from the reaction of cysteine-bearing peptides with maleimide-bearing liposomes. β-Mercaptoethanol was used to quench any unreacted maleimide groups. This provides a more closely related control than liposomes that no not contain any DSPE-PEG2000-maelimide. The peptide functionalized liposomes ranged in size from 110-130nm as determined by dynamic light scattering (Figure S1); addition of peptide or increasing peptide density did not significantly affect the liposome size. Liposomes reached optimal doxorubicin loading levels of 166-186 Ngrams doxorubicin/Nmoles phospholipid. As shown in Table 1, peptide coupling efficiency to the liposomes was typically >90% and correlated with the amount of maleimide-activated lipid. Based on a liposome size of 100nm, the peptide density ranges from approximately 740 peptides/liposome for the 0.64% liposomes to approximately 2,400 peptides/liposome for the 2.0% liposomes. Due to anticipated lipid partitioning between the exterior and interior lipid layers, it was expected that only half of the maleimide-lipid would be available for coupling. Accordingly, peptide coupling efficiency was predicted to be 50% based on the total amount of DSPE-PEG2000-maelimide. However, > 90% peptide coupling is consistently observed, indicative of maleimide-lipid partitioning to the outer lipid layer. Protein quantification is performed on intact liposomes; it is unlikely that the CBCQA peptide quantitation assay, which employs (3-(4-caroxybenzoyl)quinolone-2-carboxaldehyde and KCN, would detect peptides in the internal compartment of the liposome, further suggesting that the maleimide-lipid-peptide conjugate is partitioning to the outside face of the liposome. To ensure that the peptides were not inserting into the lipid bilayer, we added cysteine-bearing peptides to liposomes in which the maleimide groups had already been quenched with β-mercaptoethanol. After following our normal 24 hour reaction time period and removing excess peptide, we were unable to detect any peptide associated with the liposomes. This result indicates that the peptide is not directly inserting into the lipid bilayer. It is also highly unlikely that the positively charged peptides would traverse the membrane to become entrapped in the core of the liposome. Thus, our peptide coupling values appear to reflect the actual numbers of peptides displayed on the exterior of the liposomes due to partitioning of maleimide-lipid to the outer lipid layer.
Table 1. Characterization of Peptide Coupling to Liposomes.
| liposome maleimide percentage |
peptide | nmole peptide/μmole phospholipid |
peptides/ liposomea |
|---|---|---|---|
| 0.64 | H2009.1 tetramer | 9.1 | 720 |
| scH2009.1 tetramer | 9.4 | 760 | |
|
| |||
| 1.3 | H2009.1 monomer | 15 | 1200 |
| H2009.1 tetramer | 18 | 1400 | |
| scH2009.1 tetramer | 19 | 1500 | |
|
| |||
| 2.0 | H2009.1 monomer | 30 | 2400 |
| H2009.1 tetramer | 30 | 2400 | |
| scH2009.1 tetramer | 32 | 2600 | |
Peptide number/liposome was calculated by assuming 80,000 phospholipid molecules/liposome for a liposome size of 100nm (reference 26).
Effects of H2009.1 Peptide Valency and Density on Liposome Targeting to αvβ6-Expressing Cells
The ability of the monomeric and tetrameric peptide bearing liposomes to bind and target their receptor, αvβ6, was examined by measuring the amount of liposomal doxorubicin internalized into αvβ6-expressing cells. After incubating liposomes with cells and subsequently washing to remove any unbound nanoparticles, both the cells and liposomes were lysed with detergent and the amount of cell-associated doxorubicin quantified by measuring total doxorubicin fluorescence. Based on a standard curve of free doxorubicin, this assay was able to detect doxorubicin amounts above ~0.4 pmole. Cells were counted prior to fluorescence measurements so that the doxorubicin concentration could be normalized on a per cell basis. Our results are calculated as picomoles of doxorubicin per 10,000 cells. As a note of reference, delivery of a single 100 nm liposome with a loading of 180 Ngrams of doxorubicin/Nmoles phospholipid should deliver ~2.6 × 104 molecules (4.3 × 10−8 pmoles) of doxorubicin.
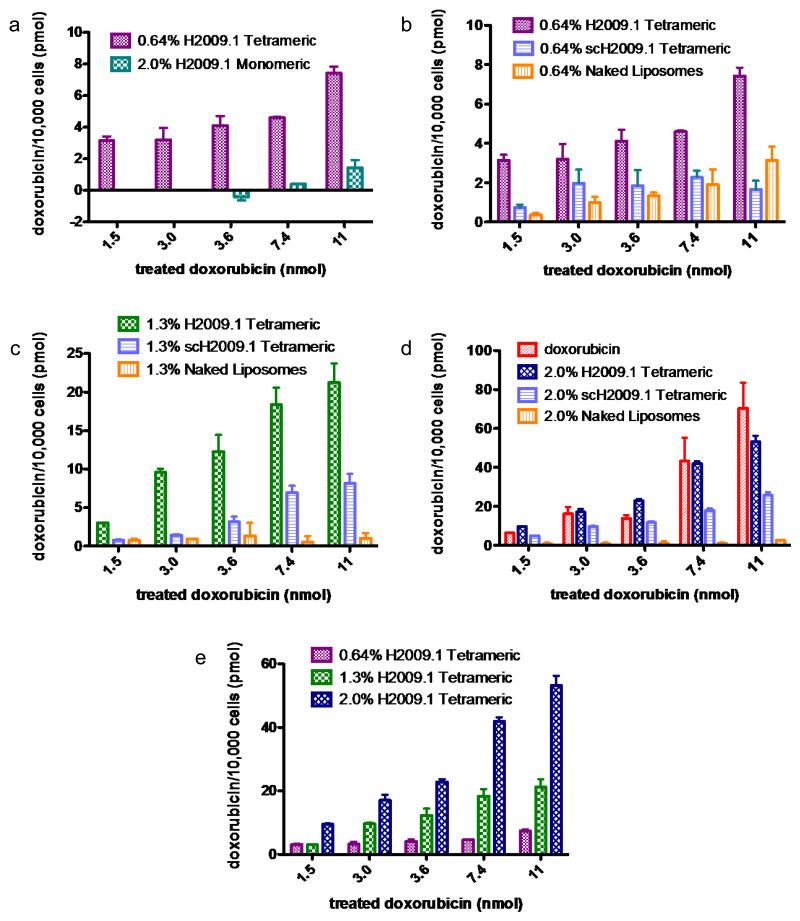
To determine whether peptide valency plays a role in targeting αvβ6, αvβ6-positive H2009 cells were incubated with either 2.0% H2009.1 monomeric or 0.64% H2009.1 tetrameric liposomes, which bear similar numbers of individual peptide units. Cell associated doxorubicin was not measurable for the 2.0% monomeric liposomes until the two highest drug treatment concentrations. However, all treatment concentrations of the 0.64% H2009.1 tetrameric liposomes resulted in detectable cellular doxorubicin levels. Significantly, the 0.64% H2009.1 tetrameric liposomes demonstrated 5-10 fold more drug uptake than the 2.0% H2009.1 monomeric liposomes (Figure 2a). Thus, although the 0.64% H2009.1 tetrameric and 2.0% H2009.1 monomeric liposomes display similar numbers of monomeric H2009.1 peptide units, the liposomes displaying tetrameric peptides receive a synergistic boost in targeting αvβ6-positive H2009 cells. The multivalency inherently given to the monomeric peptides by conjugation to the liposome surface is not as effective for targeting as the multimeric tetrameric peptides displayed in a multivalent fashion on the liposomal surface. This is the first demonstration that a multimeric peptide functions better than a monomeric peptide on a nanoparticle platform.
Figure 2. H2009.1 tetrameric liposomes target αvβ6-expressing cells better than monomeric liposomes and targeting increases with increasing peptide density.
αvβ6-expressing H2009 cells were incubated with increasing concentrations of different liposome formulations. Cellular doxorubicin concentration was determined by absorbance at 590nm compared to doxorubicin standards mixed with untreated cells. Accumulation of drug is represented as pmol doxorubicin/10,000 cells.
To determine the effect of peptide density on liposomal targeting, we compared the amount of doxorubicin internalized into cells incubated with the 0.64%, 1.3%, and 2.0% H2009.1 tetrameric liposomes. Increasing liposomal peptide density increased targeting to αvβ6-expressing H2009 cells (Figure 2e). While the 1.3% H2009.1 tetrameric liposomes display twice as much peptide as the 0.64% H2009.1 liposomes, they target H2009 cells 3-4 fold better at all concentrations except for the lowest treatment concentration. The discrepancy at the lower concentration is most likely due to the sensitivity of the assay. Similarly, the 2.0% H2009.1 tetrameric liposomes display 1.5 times more peptide than the 1.3% H2009.1 tetrameric liposomes yet target H2009 cells 2-3-fold better than the 1.3% liposomes. The nonlinear increase in targeting with increasing peptide density indicates that the peptide amount alone is not sufficient for the improved specificity of the 1.3% and 2.0% liposomes. The extra layer of multivalency provided by the liposomes also plays a role in targeting.
To ensure that the H2009.1 tetrameric peptide mediates αvβ6-receptor binding, control scH2009.1 tetrameric liposomes and control “naked”, no peptide, liposomes were synthesized and tested for binding to H2009 cells. At all treatment concentrations, the 0.64% H2009.1 tetrameric liposomes delivered 2-4 fold more doxorubicin than the scH2009.1 tetrameric liposomes and 2-8 fold more doxorubicin than the naked liposomes (Figure 2b). Similar trends emerged for the 1.3% and 2.0% liposomes functionalized with higher concentrations of peptide (Figure 2c-d). Thus, the specific sequence of the H2009.1 tetrameric peptide, and not just the presence of a peptide, mediates receptor specific binding, resulting in enhanced delivery of doxorubicin to H2009 cells.
One concern with increasing liposomal peptide number is that higher peptide concentrations could lead to higher nonspecific uptake merely due to charge effects. Positively charged lipids are commonly used to courier cargo across the negatively charged cell membrane. The H2009.1 monomeric peptide has a +2 charge at neutral pH and the tetrameric peptide has a +8 charge. Thus, the 2.0% liposomes, with approximately 2400 tetrameric peptides per liposome, bear a +19E3 charge. As the scH2009.1 peptide bears the same charge as the H2009.1 peptide without αvβ6 specificity, any affinity of the scH2009.1 peptide modified liposomes for αvβ6-expressing cells is the result of nonspecific accumulation. Accordingly, the scH2009.1 functionalized liposomes exhibit an increased cell binding at the 2.0% density. While the 0.64% and 1.3% scH2009.1 tetrameric liposomes generally accumulate in H2009 cells at similar levels, as expected for liposomes displaying a control peptide, the 2.0% scH2009.1 tetrameric liposomes consistently target the H2009 cells 3-7 fold better than the other scH2009.1 peptide liposome formulations (Figure 2 b-d). Binding of the 2.0% scH2009.1 tetramer liposomes is driven entirely by the presence of the peptide; the naked, no peptide, 2.0% liposomes accumulate in cells to the same extent as the 0.64% and 1.3% naked liposomes. Thus, the increased affinity of the 2.0% scH2009.1 tetrameric liposomes is likely due to charge effects. Despite nonspecific cellular accumulation, the 2.0% liposomes functionalized with the H2009.1 tetrameric peptide still target αvβ6 2-fold better than 2.0% liposomes functionalized with the scH2009.1 tetrameric peptide (Figure 2d). Thus, a combination of receptor-specific binding and nonspecific cellular interactions contribute to cellular binding when the peptide density is increased to 2.0%.
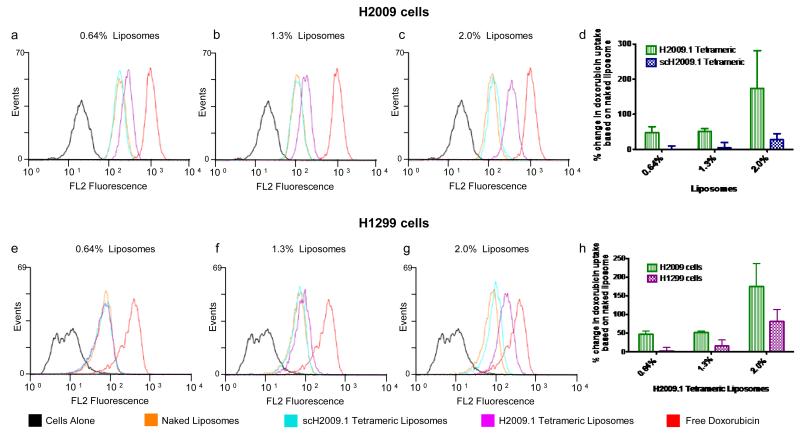
Targeting of the H2009.1 tetrameric liposomes was verified using flow cytometry to determine the relative amount of cell associated doxorubicin. Although this assay cannot provide absolute quantification of doxorubicin uptake, flow cytometry can provide relative ratios between liposomal formulations and is expected to be more sensitive at lower doxorubicin concentrations. Additionally, it can determine if uptake is homogeneous throughout the cell population or if a subset of non-binding cells exists. Consistent with the previous data, the 0.64%, 1.3%, and 2.0% H2009.1 tetrameric liposomes exhibit increased binding to H2009 cells compared to corresponding scH2009.1 tetrameric and naked liposomes (Figure 3a-c). In three independent flow assays, we determined the doxorubicin uptake for either the H2009.1 tetrameric or scH2009.1 tetrameric liposomes as compared to the corresponding naked liposomes. Modification of liposomes with the H2009.1 tetrameric peptide dramatically improved targeting compared to the naked liposomes, with values ranging from a 47% increase for the 0.64% liposomes to a 170% increase for the 2.0% liposomes (Figure 3d). Additionally, the uniform shift in the fluorescence channel suggests that the bulk of the cell population is binding the H2009.1 liposomes (Figure 3).
Figure 3. H2009.1 tetrameric peptide liposomes display specificity for αvβ6-expressing H2009 cells compared to αvβ6-low-expressing H1299 cells.
Cells were incubated with different liposome formulations for 1h and liposome accumulation in cells measured using flow cytometry. A total of 10,000 cells were counted per treatment group and doxorubicin fluorescence was measured (excitation 488nm, emission 550-600nm). Representative flow cytometry analyses for H2009 cells are shown in panels a-d and for H1299 cells are shown in panels e-h. Panels d & h show the averages from 3 independent flow assays in which the mean fluorescence intensities of the H2009.1 tetrameric and scH2009.1 tetrameric liposomes were normalized to the mean fluorescence intensity of the naked liposomes.
As previously observed, increasing H2009.1 peptide density on the liposome surface also increases targeting, with the highest peptide concentration 2.0% liposomes targeting 4-fold better than the lowest peptide concentration 0.64% liposomes (Figure 3d). Additionally, while each H2009.1 tetramer liposome targets better than its corresponding scH2009.1 tetramer liposome, this targeting differential decreases as the amount of peptide increases. The lowest peptide concentration 0.64% H2009.1 tetrameric liposomes bind 190-fold better than the corresponding scH2009.1 tetrameric liposomes, while the highest peptide concentration 2.0% H2009.1 tetrameric liposomes only bind 6-fold better than the corresponding scH2009.1 tetrameric liposomes. Despite this decrease in the targeting difference between the H2009.1 tetrameric liposomes and the scH2009.1 tetrameric liposomes, it is still clear that the specific sequence of the H2009.1 tetrameric peptide, and not just the presence of a peptide, directs liposome targeting.
These results demonstrate that multimeric display of a phage-selected peptide on a nanoparticle increases cell binding compared to monomeric presentation, even at high densities of the monomeric peptide. While the liposome itself is inherently a multivalent platform, functionalizing the nanoparticle with multiple copies of the monomeric peptide is not as effective as attaching multiple copies of the phage-mimicking tetrameric peptide. Yet increasing the number of multimeric peptides on the liposome also increases cell binding, suggesting the liposome platform provides a second level of multivalent binding. Thus, both the multivalent peptide and the multivalent liposome scaffold work together to increase targeting to αvβ6-expressing cells.
Specificity of H2009.1 Tetrameric Peptide Liposomes for αvβ6
To further verify the specificity of H2009.1 peptide liposomes for αvβ6, we compared liposomal targeting to αvβ6-positive verses αvβ6-negative cells. Flow cytometry was used to follow binding of the peptide-modified liposomes to an αvβ6-expressing cell line (H2009) verses an αvβ6-negative NSCLC cell line (H1299). The 0.64%, 1.3%, and 2.0% H2009.1 tetrameric liposomes exhibit increased cellular association to the H2009 cells compared to corresponding scH2009.1 tetrameric and naked liposomes (Figure 3a-d). However, these same 0.64% and 1.3% H2009.1 tetrameric liposomes display no specific binding to H1299 cells, accumulating to the same extent as the scH2009.1 tetrameric and naked liposomes in the non-targeted cells (Figure 3e,f, & h). At higher peptide concentrations, the αvβ6-specificity is dampened; the 2.0% H2009.1 tetrameric liposomes display a 2-fold preference for H1299 cells over the control scH2009.1 liposomes (Figure 3h). While we observe some nonspecific uptake of the 2.0% liposomes into H2009 cells, peptide charge is unlikely the only driving force behind the accumulation of 2.0% H2009.1 tetrameric liposomes into H1299 cells. If the effect were exclusively charge dependent, the 2.0% scH2009.1 tetrameric liposomes would bind to the same extent as the 2.0% H2009.1 tetrameric liposomes, since the H2009.1 and scH2009.1 peptides contain the same amino acids and same charge. Therefore, while some of the cellular association of the 2.0% H2009.1 tetrameric liposomes with H1299 cells may be the result of nonspecific binding, the H2009.1 tetrameric peptide liposomes may also be binding to low levels of αvβ6 on the H1299 cells that are detected as the affinity of the liposomes increases. Consistent with this observation, the H2009.1 peptide exhibits a low level of binding to H1299 cells, albeit at levels at least 60-fold lower than the H2009 cells, and low levels of αvβ6 have been detected in H1299 cells.29 This highlights a challenge in cell-specific targeting, especially in regards to targeting tumor cells. It is unlikely that a cell is completely devoid of a cellular receptor; instead, there is differential receptor expression between the cancer cell and its normal counterpart. As the affinity of a cell-targeting ligand increases, so does the likelihood of its binding the low levels of its receptor found on normal cells. However, the 2.0% H2009.1 tetrameric liposomes still demonstrated 2-fold greater affinity for the αvβ6-positive H2009 cells over the αvβ6-negative H1299 cells (Figure 3h). In summary, the higher density of the tetrameric peptide on the liposome results in increased cellular targeting yet this increase comes at the cost of higher nonspecific binding and decreased cell specificity.
Drug Release
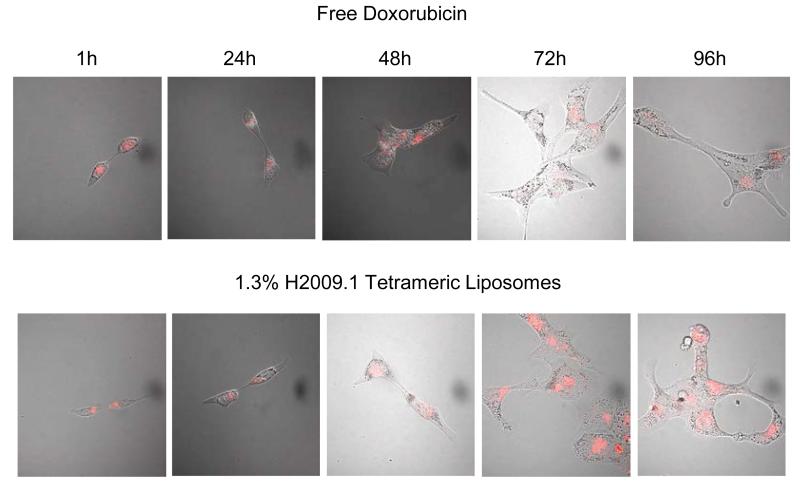
Doxorubicin must enter the cell nucleus in order to exert its effects. To visualize the subcellular localization and entry of H2009.1 liposomes into the nucleus, H2009 cells were incubated with either free doxorubicin or the 1.3% H2009.1 tetrameric liposomes and doxorubicin fluorescence was examined by confocal microscopy (Figure 4). Free doxorubicin was visible in the cell nucleus as early as 1 hour after incubation and began to visibly affect cell structure 48 hours after treatment. By 72 hours, cells treated with free doxorubicin began to bleb, indicative of apoptosis. In contrast, the 1.3% H2009.1 tetrameric liposomes initially appeared perinuclear and doxorubicin did not localize to the nucleus until 48 hours after treatment. By 72 hours, the liposome-treated cells were bloated and abnormal and at 96 hours they began to bleb. Thus, it requires approximately 48 hours for the internalized liposomes to release their cargo allowing for observable nuclear accumulation of doxorubicin. While liposomal drug release is delayed, alterations in cellular structure occur only 24 hours later than the free drug. These results demonstrate that the H2009.1 tetrameric peptide does not prevent liposomal drug release. Additionally, the H2009.1 tetrameric liposomes remain intact extracellularly, releasing doxorubicin only after internalization into cells. Importantly, even though the liposomes are initially trapped within a perinuclear compartment, they degrade and release free doxorubicin to its site of action in the nucleus.
Figure 4. Time course for release of doxorubicin from 1.3% H2009.1 Tetrameric Liposomes.
Cells were treated with either free doxorubicin or 1.3% H2009.1 tetrameric liposomal doxorubicin for 1 hour before the drug was removed. Doxorubicin fluorescence was observed on a Nikon TE2000-E microscope at 60X magnification at the indicated time after initial treatment. Bright field and fluorescence images are overlaid to show the overall cell morphology.
Cytotoxicity of H2009.1 Monomeric and Tetrameric Peptide Liposomes
In order to serve as viable treatment options, the H2009.1 peptide liposomes must not only accumulate in αvβ6-positive cells, but also exert specific cytotoxic effects to these cells. We anticipated that increased cellular binding would improve the cytotoxicity of liposomal doxorubicin. To examine the ability of the H2009.1 liposomes to induce cell death, we compared the cytotoxic effects of the various peptide liposome constructs. As expected from the ability of the H2009.1 peptide to specifically direct liposomes to these cells, all of the H2009.1 tetrameric liposomes were more cytotoxic than the corresponding scH2009.1 tetrameric and naked liposomes. The IC50 values were determined for each liposome formulation and are detailed in Table 2. The IC50 for each H2009.1 peptide liposome was approximately 2-fold lower than the corresponding scH2009.1 peptide liposome. Additionally, the IC50 decreased with increasing peptide concentration. Importantly, the IC50 values correlate with the amount of internalized doxorubicin measured in Figure 2. Significantly, the 2.0% H2009.1 tetrameric liposomes (360 ± 35 nM) gave an IC50 value similar to that of free, non-liposome encapsulated, doxorubicin (430 ± 75 nM) as expected from their similar levels of accumulation quantified by the doxorubicin uptake studies. Due to its ability to easily cross the cell membrane, free doxorubicin accumulates quickly in the cell nucleus and often demonstrates better in vitro cytotoxicity than liposome-encapsulated doxorubicin.
Table 2.
IC50 Values of Different Liposome Formulations on αvβ6-Positive H2009 Cellsa
| Drug Formulation | IC50 on H2009 cells (nM) |
|---|---|
| 0.6% H2009.1 Tetrameric | 1500 ± 200 |
| 0.6% scH2009.1 Tetrameric | 2400 ± 120 |
| 0.6% Naked | 7400 ± 1400 |
|
| |
| 1.3% H2009.1 Monomeric | 5700 ± 120 |
| 1.3% H2009.1 Tetrameric | 970 ± 130 |
| 1.3% scH2009.1 Tetrameric | 2000 ± 330 |
| 1.3% Naked | Not reachedb |
|
| |
| 2.0% H2009.1 Tetrameric | 360 ± 35 |
| 2.0% scH2009.1 Tetrameric | 610 ± 90 |
| 2.0% Naked | 6900 ± 960 |
|
| |
| Free Doxorubicin | 430 ± 75 |
Cells were incubated with liposomes for 1 hour followed by a 120 hour recovery in media.
Not reached at the highest treatment concentration of 10.2 μM.
While the 2.0% H2009.1 tetrameric liposomes gave the lowest IC50 value (360 ± 35 nM), the 1.3% H2009.1 tetrameric liposomes (970 ± 130 nM) have the best αvβ6-specific cytotoxicity based on their differential cytotoxicity compared to both the scH2009.1 and naked liposomes. The IC50 value of the 1.3% scH2009.1 tetrameric liposomes (2000 ± 330 nM) is 2-fold lower than the value for the 1.3% H2009.1 tetrameric liposomes, and the 1.3% naked liposomes have the highest IC50 value of the liposomes tested (IC50 not reached in our assays); thus, the 1.3% liposomes inherently have low background uptake into the H2009 cells. The 1.3% H2009.1 tetrameric liposomes also demonstrate αvβ6-specific cytotoxicity, selectively killing H2009 cells, and not the H1299 cells when compared to the control scH2009.1 tetrameric liposomes (Table S2). The 2.0% scH2009.1 tetrameric liposomes display increased cytotoxicity compared to the other scH2009.1 liposome formulations, consistent with the doxorubicin uptake data demonstrating increased nonspecific accumulation of these liposomes in cells. While the 0.64% scH2009.1 tetrameric (2400 ± 115 nM) and 1.3% scH2009.1 tetrameric liposomes (2030 ± 328 nM) have similar IC50 values, the 2.0% scH2009.1 tetrameric liposomes have a much lower IC50 value (608 ± 89.7 nM). Therefore, although the higher peptide density displayed on the 2.0% liposomes increases cell cytotoxicity, it reduces αvβ6-specificity. Thus, the 1.3% liposomes represent the best specific targeting formulation. The efficacy and specificity of peptide-targeted liposomes ride a fine balance and the liposome with the lowest IC50 may not be the best for targeted therapy.
We also examined the effects of peptide valency on the cytotoxicity of the ideal 1.3% liposome platform, comparing liposomes functionalized with H2009.1 monomeric or tetrameric peptides. Consistent with the liposomal targeting data that demonstrated more than additive targeting for the tetrameric verses monomeric liposomes, the monomeric liposomes (5700 ± 120) were 6-fold less cytotoxic than the tetrameric liposomes (970 ± 130). These data further support the benefit of the layered valency of multivalent tetrameric peptides displayed in a multivalent fashion on the liposomal surface.
DISCUSSION
Phage display is a powerful method for screening a library of peptides for specific binding to a desired cell type. Peptides with specificity for a particular cell type can be identified after 3-5 rounds of screening. However, removal of peptides from the phage context through chemical synthesis often results in ligands with significantly lower affinity, likely due to loss of multivalency. As the phage displays 3-5 copies of a given peptide, multivalent interactions are expected to contribute to phage peptide binding. Additionally, when whole cells serve as the bait for the selection, peptides that bind clustered cellular receptors are likely to dominate because of the multivalent presentation of the peptide ligand on the phage. The synthesis of only one peptide copy, therefore, fails to reconstitute this ideal structure. To address the loss in peptide affinity upon synthesis, we developed a higher affinity tetrameric peptide construct that mimics the original peptide presentation of the phage13 and demonstrated the utility of this construct for peptides selected against a variety of cell types.13, 16-19
One of the primary uses of peptides selected by phage display is conjugation of the peptides to therapeutics or imaging agents for specific delivery to target cell types. The peptides can either be linked directly to the desired cargo or to a cargo-carrier such as a nanoparticle. Nanoparticles are receiving considerable attention for targeted delivery applications due to their ability to encapsulate a variety of imaging agents or therapeutics. With the goal of optimizing the path from peptides selected by phage display to presentation of these peptides on a nanoparticle surface for targeted delivery, we set out to examine the effects of peptide concentration, affinity, and valency on nanoparticle delivery and therapeutic efficacy. As a nanoparticle is considered a multivalent platform for ligand display, we were particularly interested in the effects of multivalent peptide presentation on the multivalent nanoparticle.
Here we report the first demonstration that a multivalent peptide functions better than a monomeric peptide on a nanoparticle platform, using the H2009.1 peptide as a model. The αvβ6-specific H2009.1 peptide was originally selected from a pIII phage displayed peptide library using the NSCLC cell line H2009 as bait. The H2009.1 peptide retains its binding specificity when displayed on a liposome, delivering liposomes to αvβ6-expressing cells. Significantly, liposomes displaying the higher affinity multivalent H2009.1 tetrameric peptide demonstrate higher specificity and greater toxicity towards αvβ6-expressing cells than liposomes displaying the lower affinity monomeric H2009.1 peptide. Additionally, we demonstrate that tetrameric peptide concentration plays a role in liposomal targeting. Liposomes displaying approximately 1400 tetrameric peptides per liposome (1.3% formulation) target better than liposomes displaying approximately 700 tetrameric peptides per liposome (0.64% formulation) and have less nonspecific toxicity than liposomes displaying higher concentrations of peptide (2.0% formulation). Our results suggest that the layered valency effect given by the multivalent liposomal presentation of the already multivalent H2009.1 tetrameric peptide renders more efficient cell targeting.
The enhancement in binding and targeting imparted by liposomal display of the tetrameric verses monomeric H2009.1 peptide is likely due to differences in their ability to engage in multivalent binding with αvβ6 on the surface of the cell. Even when the total number of individual peptide units are the same, the tetrameric H2009.1 liposome binds to H2009 cells 5-10 fold better than the monomeric peptide formulation. The tetrameric peptide has been optimized for optimal multivalent binding outside the context of the liposome. However, the monomeric peptide is dependent on the liposome to provide the multivalent scaffolding. For a 1.3% liposome with a diameter of 100 nm, there is approximately 1 peptide per 22 nm2 on the liposome surface and increasing the density of peptides to 2.0% provides 1 peptide for every 13 nm2. Accordingly, the monomeric peptides may be less likely to either achieve cooperative binding or to influence the binding of other monomeric peptides. Furthermore, binding multiple separate monomers is entropically unfavorable compared to binding multiple peptides from the tetrameric. We cannot rule out the possibility that there is some multivalent binding involved with the monomeric peptides, however, our results clearly demonstrate that tetrameric display of the peptide improves liposome binding and drug delivery.
In contrast to the monomeric peptides, the tetrameric peptide scaffold places the peptide branches in close proximity, leaving them primed for multivalent interactions with αvβ6. However, increasing the density of multimeric peptides on the liposome also increases cell binding indicating that the liposome platform does provide a second level of multivalent binding. Thus, both the multivalent peptide and the multivalent liposome scaffold work together to increase targeting to αvβ6-expressing cells. This multi-layered approach to developing high affinity targeted nanoparticles may improve the utility of moderate affinity peptides.
It is important to note that the curvature of the liposome limits the initial contact interface between the cell and liposome. The surface area of contact is dependent on the radius of the liposomal sphere as well as the extended length of the PEG linker. As a rough estimate, a 100 nm liposome with 1400 peptides will only display ~200 of these peptides to the cell surface upon initial interaction with the cell.47 Thus, while liposomes displaying monomeric peptides will only present 200 peptide units to the cell, liposomes displaying tetrameric peptides will present 800 peptide units, increasing the local concentration of ligand. Clearly this is an oversimplification and cell wrapping around the nanoparticle during endocytosis will increase ligand-receptor interactions. However, the tetrameric display will present a greater number of ligands in the correct orientation for receptor binding.
There exist several different models for multivalent ligand binding that are applicable in this context.12 In the “chelate effect,” multimeric ligands bind to clustered receptors. In a second model, after one arm of a multivalent ligand binds to its receptor, other receptors are recruited to the first receptor and subsequently bind additional arms of the multivalent ligand. A third model suggests that the higher local concentration of peptide units displayed by a multimeric ligand lends higher affinity for binding to a single receptor. Although we speculate that in our system receptor clustering leads to a chelate effect, each of these models requires a multimeric ligand with appropriate spacing and conformation. The distance between monomeric peptides on the liposome surface in conjunction with the curvature of the liposome membrane likely keeps the individual monomeric chains too far apart for simultaneous binding to clustered receptors or for achieving a high local concentration of peptide. By contrast, the tetrameric presentation of peptides keeps the local concentration of monomeric chains high while displaying each chain in an orientation suitable for binding to multiple receptors.
Creating different types of nanoparticles that display the monomeric peptides in closer contact may also recapitulate multimeric binding. However, our studies suggest that high concentrations of peptide on the liposomal surface increase nonspecific binding and this observation is likely to hold true for other nanoparticle platforms. In order to achieve the same absolute number of peptide branches as the ideal 1.3% tetrameric liposome, a 100 nm nanoparticle would have to bear 5600 monomeric peptides. This would require an increase of DSPE-PEG-Maleimide to 5.2% of the total lipid content and may destabilize the liposome. Additionally, the coupling efficiency is likely to decrease as the ratio of DSPE-PEG-maleimide increases. Finally, we have observed that an increase of the DSPE-PEG-Maleimide past 3.0% increases nonspecific binding even in the absence of peptide. Smaller sized nanoparticles could display monomeric peptides in closer proximity for multivalent interactions, but care must be taken to ensure that the nanoparticle size is large enough to avoid rapid renal clearance for in vivo applications. Additionally, peptides displayed on smaller sized nanoparticles will suffer more from curvature effects of the nanoparticle; even though the peptides may display in closer proximity, the curvature of the nanoparticle surface may prevent ideal multimeric interactions.
These studies were performed using whole cells instead of a target receptor immobilized on a solid surface. As receptor number, orientation, and localization could greatly affect ligand binding, it is important to study receptor-ligand interactions in their native context. Meijer et al. previously demonstrated that the binding of multivalent peptide dendrimers is affected by receptor density.15 By using cells that express αvβ6 as our target, we maintain a more relevant receptor display and optimize the liposome system within a biological context. While the tetrameric peptide delivers nanoparticles more effectively to these physiologically relevant cells in vitro, it remains to be seen whether this will hold true in vivo. Biodistribution for tumor targeting in vivo depends on many aspects not present in the in vitro context such as nanoparticle half-life, tumor vasculature leakiness and size, and receptor levels and availability. Nevertheless, our in vitro studies are promising and the tetrameric H2009.1 peptide liposomes merit further in vivo evaluation.
Peptides selected using whole cells as the phage library target are especially likely to benefit from tetrameric nanoparticle display as the receptor is in its native, multimeric-peptide binding context; the receptor may reside in clusters or diffuse through the cell membrane to bind different arms of the multivalent phage peptide. Of particular advantage, high affinity peptide-targeted nanoparticles can be designed without knowledge of the amount of receptor, arrangement of the receptor on the cell surface, or even the identity of the receptor. Biopanning pIII phage displayed peptide libraries on whole cells does not require A priori knowledge of the receptor and the selection process identifies peptides that bind to cell surface receptors in their native context. Assuming the peptides can transition from the phage particle to the trilysine core without loss of affinity or specificity, it is likely that the tetrameric peptide will be functional on the liposome. This alleviates the need for rational design to achieve a targeted liposome with the optimized ligand spacing. The same cannot be said for the monomeric peptide as the liposome scaffold does not recapitulate the peptide display found on the phage.
Peptide tetramerization for nanoparticle delivery is expected to apply to the majority of peptides selected by phage display. We have tested an assortment of peptides selected from different phage libraries against a variety of targets; typically a synergetic increase of >25-fold in affinity is observed upon tetramerization. Preliminary experiments in our lab with another cancer cell-targeting peptide isolated by phage display suggest that a tetrameric version of this peptide can also be used for effective liposomal delivery to cells (BG and KCB unpublished data). Thus, the tetrameric scaffold may act as a general method for taking peptides from phage
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the Welch Foundation (I1622 to KCB), the NIH (1RO1CA106646 to KCB). BPG is supported by a fellowship from the Cancer Research and Prevention Institute of Texas (RP 101496). We thank Dr. Michael J McGuire for insightful comments and discussions, and Ms. Dorothy Cupka for assistance in preparing figures.
ABBREVIATIONS
- NSCLC
non-small cell lung cancer
- PEG
polyethylene glycol
- CBQCA
3-(4-caroxybenzoyl)quinolone-2-carboxaldehyde
- HSPC
hydrogenated soy phosphatidylcholine cholesterol
- DSPE-PEG2000
1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[carbonyl-methoxypolyethyelene glycol-2000]
- DSPE-PEG2000-maelimide
1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000]
Footnotes
Supporting Information Available: Liposome formulations (Table S1), DLS characterization of the liposomes (Figure S1), detailed cell cytotoxicity data (Figure S2) and IC50 data on the control H1299 cells (Table S2). This material is available free of charge via the Intranet at http://pubs.acs.org
The authors declare no competing financial interest.
REFERENCES
- 1.Alley SC, Okeley NM, Senter PD. Antibody-drug conjugates: targeted drug delivery for cancer. Curr. Opin. Chem. Biol. 2010;14:529–537. doi: 10.1016/j.cbpa.2010.06.170. [DOI] [PubMed] [Google Scholar]
- 2.Reichert JM, Valge-Archer VE. Development trends for monoclonal antibody cancer therapeutics. Nat. Rev. Drug Discovery. 2007;6:349–356. doi: 10.1038/nrd2241. [DOI] [PubMed] [Google Scholar]
- 3.Bray BL. Innovation: Large-scale manufacture of peptide therapeutics by chemical synthesis. Nat. Rev. Drug Discovery. 2003;2:587–593. doi: 10.1038/nrd1133. [DOI] [PubMed] [Google Scholar]
- 4.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 5.Smith GP, Petrenko VA. Phage Display. Chem. Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 6.Kehoe JW, Kay BK. Filamentous phage display in the new millennium. Chem. Rev. 2005;105:4056–4072. doi: 10.1021/cr000261r. [DOI] [PubMed] [Google Scholar]
- 7.Brown KC. Peptidic Tumor Targeting Agents: The Road from Phage Display Peptide Selections to Clinical Applications. Curr. Pharm. Des. 2010;16:1040–1054. doi: 10.2174/138161210790963788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aina OH, Liu R, Sutcliffe JL, Marik J, Pan CX, Lam KS. From combinatorial chemistry to cancer-targeting peptides. Mol. Pharm. 2007;4:631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- 9.Barry MA, Dower WJ, Johnston SA. Toward cell-targeting gene therapy vectors: Selection of cell-binding peptides from random peptide-presenting phage libraries. Nat. Med. 1996;2:299–305. doi: 10.1038/nm0396-299. [DOI] [PubMed] [Google Scholar]
- 10.Pasqualini R, Ruoslahti E. Organ targeting In vivo using phage display peptide libraries. Nature. 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 11.Mammen M, Choi SK, Whitesides GM. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 12.Kiessling LL, Gestwicki JE, Strong LE. Synthetic multivalent ligands in the exploration of cell-surface interactions. Curr. Opin. Chem. Biol. 2000;4:696–703. doi: 10.1016/s1367-5931(00)00153-8. [DOI] [PubMed] [Google Scholar]
- 13.Oyama T, Sykes KF, Samli KN, Minna JD, Johnston SA, Brown KC. Isolation of lung tumor specific peptides from a random peptide library: generation of diagnostic and cell-targeting reagents. Cancer Lett. 2003;202:219–230. doi: 10.1016/j.canlet.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Helms BA, Reulen SWA, Nijhuis S, Graaf-Heuvelmans P. T. H. M. d., Merkx M, Meijer EW. High-Affinity Peptide-Based Collagen Targeting Using Synthetic Phage Mimics: From Phage Display to Dendrimer Display. J. Am. Chem. Soc. 2009;131:11683–11685. doi: 10.1021/ja902285m. [DOI] [PubMed] [Google Scholar]
- 15.Bastings MMC, Helms BA, van Baal I, Hackeng TM, Merkx M, Meijer EW. From Phage Display to Dendrimer Display: Insights into Multivalent Binding. J. Am. Chem. Soc. 2011;133:6636–6641. doi: 10.1021/ja110700x. [DOI] [PubMed] [Google Scholar]
- 16.De J, Chang Y-C, Samli KN, Schisler JC, Newgard CB, Johnston SA, Brown KC. Isolation of a mycoplasma-specific binding peptide from an unbiased phage-displayed peptide library. Mol. Biosyst. 2005;1:149–157. doi: 10.1039/b504572j. [DOI] [PubMed] [Google Scholar]
- 17.McGuire MJ, Sykes KF, Samli KN, Timares L, Barry MA, Stemke-Hale K, Tagliaferri F, Logan M, Jansa K, Takashima A. A library-selected, Langerhans cell-targeting peptide enhances an immune response. DNA Cell Biol. 2004;23:742–752. doi: 10.1089/dna.2004.23.742. [DOI] [PubMed] [Google Scholar]
- 18.McGuire MJ, Samli KN, Chang Y-C, Brown KC. Novel ligands for cancer diagnosis: selection of peptide ligands for identification and isolation of B-cell lymphomas. Exp. Hematol. 2006;34:443–452. doi: 10.1016/j.exphem.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 19.Oyama T, Rombel IT, Samli KN, Zhou X, Brown KC. Isolation of multiple cell-binding ligands from different phage displayed-peptide libraries. Biosens. Bioelectron. 2006;21:1867–1875. doi: 10.1016/j.bios.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 20.Li S, McGuire MJ, Lin M, Liu Y-H, Oyama T, Sun X, Brown KC. Synthesis and characterization of a high-affinity {alpha}v{beta}6-specific ligand for in vitro and in vivo applications. Mol. Cancer Ther. 2009;8:1239–1249. doi: 10.1158/1535-7163.MCT-08-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loi M, Marchio S, Becherini P, Di Paolo D, Soster M, Curnis F, Brignole C, Pagnan G, Perri P, Caffa I, Longhi R, Nico B, Bussolino F, Gambini C, Ribatti D, Cilli M, Arap W, Pasqualini R, Allen TM, Corti A, Ponzoni M, Pastorino F. Combined targeting of perivascular and endothelial tumor cells enhances anti-tumor efficacy of liposomal chemotherapy in neuroblastoma. J. Controlled Release. 2010;145:66–73. doi: 10.1016/j.jconrel.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 22.Lowery A, Onishko H, Hallahan DE, Han Z. Tumor-targeted delivery of liposome-encapsulated doxorubicin by use of a peptide that selectively binds to irradiated tumors. J. Controlled Release. 2011;150:117–124. doi: 10.1016/j.jconrel.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park JH, von Maltzahn G, Xu MJ, Fogal V, Kotamraju VR, Ruoslahti E, Bhatia SN, Sailor MJ. Cooperative nanomaterial system to sensitize, target, and treat tumors. Proc. Natl. Acad. Sci. U. S. A. 2010;107:981–986. doi: 10.1073/pnas.0909565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN, Jayaraman M, Rajeev KG, Cantley WL, Dorkin JR, Butler JS, Qin L, Racie T, Sprague A, Fava E, Zeigerer A, Hope MJ, Zerial M, Sah DW, Fitzgerald K, Tracy MA, Manoharan M, Koteliansky V, Fougerolles A. d., Maier MA. Targeted Delivery of RNAi Therapeutics With Endogenous and Exogenous Ligand-Based Mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Copland MJ, Baird MA, Rades T, McKenzie JL, Becker B, Reck F, Tyler PC, Davies NM. Liposomal delivery of antigen to human dendritic cells. Vaccine. 2003;21:883–890. doi: 10.1016/s0264-410x(02)00536-4. [DOI] [PubMed] [Google Scholar]
- 26.Espuelas S, Haller P, Schuber F, Frisch B. Synthesis of an Amphiphilic Tetraantennary Mannosyl Conjugate and Incorporation into Liposome Carriers. Bioorg. Med. Chem. Lett. 2003;13:2557–2560. doi: 10.1016/s0960-894x(03)00472-4. [DOI] [PubMed] [Google Scholar]
- 27.Heurtault B, Gentine P, Thomann J-S, Baehr C, Frisch B, Pons F. Design of a Liposomal Candidate Vaccine Against Pseudomonas aeruginosa and its Evaluation in Triggering Systemic and Lung Mucosal Immunity. Pharm. Res. 2008;26:276–285. doi: 10.1007/s11095-008-9724-y. [DOI] [PubMed] [Google Scholar]
- 28.Martin AL, Li B, Gillies ER. Surface Functionalization of Nanomaterials with Dendritic Groups: Toward Enhanced Binding to Biological Targets. J. Am. Chem. Soc. 2009;131:734–741. doi: 10.1021/ja807220u. [DOI] [PubMed] [Google Scholar]
- 29.Elayadi AN, Samli KN, Prudkin L, Liu Y-H, Bian A, Xie X-J, Wistuba II, Roth JA, McGuire MJ, Brown KC. A peptide selected by biopanning identifies the integrin alphavbeta6 as a prognostic biomarker for nonsmall cell lung cancer. Cancer Res. 2007;67:5889–5895. doi: 10.1158/0008-5472.CAN-07-0245. [DOI] [PubMed] [Google Scholar]
- 30.Ahmed N, Riley C, Rice GE, Quinn MA, Baker MS. αvβ6 integrin-A marker for the malignant potential of epithelial ovarian cancer. J. Histochem. Cytochem. 2002;50:1371. doi: 10.1177/002215540205001010. [DOI] [PubMed] [Google Scholar]
- 31.Arihiro K, Kaneko M, Fujii S, Inai K, Yokosaki Y. Significance of α9β1 and αvβ6 integrin expression in breast carcinoma. Breast Cancer. 2000;7:19–26. doi: 10.1007/BF02967183. [DOI] [PubMed] [Google Scholar]
- 32.Bates RC, Bellovin DI, Brown C, Maynard E, Wu B, Kawakatsu H, Sheppard D, Oettgen P, Mercurio AM. Transcriptional activation of integrin β6 during the epithelial-mesenchymal transition defines a novel prognostic indicator of aggressive colon carcinoma. J. Clin. Invest. 2005;115:339–347. doi: 10.1172/JCI23183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamidi S, Salo T, Kainulainen T, Epstein J, Lerner K, Larjava H. Expression of αvβ6 integrin in oral leukoplakia. Br. J. Cancer. 2000;82:1433–1440. doi: 10.1054/bjoc.1999.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breuss J, Gallo J, DeLisser H, Klimanskaya I, Folkesson H, Pittet J, Nishimura S, Aldape K, Landers D, Carpenter W, Gillett N, Sheppard D, Matthay M, Albelda S, Kramer R, Pytela R. Expression of the beta 6 integrin subunit in development, neoplasia and tissue repair suggests a role in epithelial remodeling. J. Cell Sci. 1995;108:2241–2251. doi: 10.1242/jcs.108.6.2241. [DOI] [PubMed] [Google Scholar]
- 35.Kawashima A, Tsugawa S, Boku A, Kobayashi M, Minamoto T, Nakanishi I, Oda Y. Expression of [alpha] v Integrin Family in Gastric Carcinomas: Increased [alpha] v [beta] 6 is Associated with Lymph Node Metastasis. Pathol. Res. Pract. 2003;199:57–64. doi: 10.1078/0344-0338-00355. [DOI] [PubMed] [Google Scholar]
- 36.Hazelbag S, Kenter G, Gorter A, Dreef E, Koopman L, Violette S, Weinreb P, Fleuren G. Overexpression of the αvβ6 integrin in cervical squamous cell carcinoma is a prognostic factor for decreased survival. J. Pathol. 2007;212:316–324. doi: 10.1002/path.2168. [DOI] [PubMed] [Google Scholar]
- 37.Hecht JL, Dolinski BM, Gardner HA, Violette SM, Weinreb PH. Overexpression of the αvβ6 Integrin in Endometrial Cancer. Appl. Immunohistochem. Mol. Morphol. 2008;16:543–547. doi: 10.1097/PAI.0b013e31816bc5ee. [DOI] [PubMed] [Google Scholar]
- 38.Jones J, Watt FM, Speight PM. Changes in the expression of αv integrins in oral squamous cell carcinomas. J. Oral Pathol. Med. 1997;26:63–68. doi: 10.1111/j.1600-0714.1997.tb00023.x. [DOI] [PubMed] [Google Scholar]
- 39.Regezi JA, Ramos DM, Pytela R, Dekker NP, Jordan RCK. Tenascin and β6 integrin are overexpressed in floor of mouth in situ carcinomas and invasive squamous cell carcinomas. Oral Oncol. 2002;38:332–336. doi: 10.1016/s1368-8375(01)00062-8. [DOI] [PubMed] [Google Scholar]
- 40.Thomas GJ, Nystrom ML, Marshall JF. Alphavbeta6 integrin in wound healing and cancer of the oral cavity. J. Oral Pathol. Med. 2006;35:1–10. doi: 10.1111/j.1600-0714.2005.00374.x. [DOI] [PubMed] [Google Scholar]
- 41.Prudkin L, Liu DD, Ozburn NC, Sun M, Behrens C, Tang X, Brown KC, Bekele BN, Moran C, Wistuba II. Epithelial-to-mesenchymal transition in the development and progression of adenocarcinoma and squamous cell carcinoma of the lung. Modern Pathol. 2009;22:668–678. doi: 10.1038/modpathol.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phelps RM, Johnson BE, Ihde DC, Gazdar AF, Carbone DP, McClintock PR, Linnoila RI, Matthews MJ, Bunn PA, Carney D, Minna JD, Mulshine JL. NCI-navy medical oncology branch cell line data base. J. Cell. Biochem. 1996;63:32–91. doi: 10.1002/jcb.240630505. [DOI] [PubMed] [Google Scholar]
- 43.Shmeeda H, Even-Chen S, Honen R, Cohen R, Weintraub C, Barenholz Y. Methods Enzymol. Elsevier; 2003. Enzymatic Assays for Quality Control and Pharmacokinetics of Liposome Formulations: Comparison with Nonenzymatic Conventional Methodologies. [DOI] [PubMed] [Google Scholar]
- 44.Abraham SA, Waterhouse DN, Mayer LD, Cullis PR, Madden TD, Bally MB. The Liposomal Formulation of Doxorubicin. Vol. 391. Elsevier; 2005. [DOI] [PubMed] [Google Scholar]
- 45.Dubey PK, Mishra V, Jain S, Mahor S, Vyas S. Liposomes modified with cyclic RGD peptide for tumor targeting. J. Drug Targeting. 2004;12:257–264. doi: 10.1080/10611860410001728040. [DOI] [PubMed] [Google Scholar]
- 46.Owens DE, 3rd, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int. J. Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 47.Moore NW, Kuhl TL. The role of flexible tethers in multiple ligand-receptor bond formation between curved surfaces. Biophys. J. 2006;91:1675–1687. doi: 10.1529/biophysj.105.079871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.