SUMMARY
T-bet and Bcl-6 are required to establish TH1 or TFH gene expression profiles, respectively. Here, we demonstrated that high interleukin 2 (IL-2) concentrations inhibited Bcl-6 expression in polarized TH1 cells. Mechanistically, the low amounts of Bcl-6 normally found in effector TH1 cells could not repress its target genes because a T-bet-Bcl-6 complex masked the Bcl-6 DNA-binding domain. TH1 cells increased their Bcl-6/T-bet ratio in response to limiting IL-2 conditions, allowing excess Bcl-6 to repress its direct target Prdm1 (which encodes Blimp-1). The Bcl-6-dependent repression of Blimp-1 effectively induced a partial TFH-profile because Blimp-1 directly repressed a subset of TFH-signature genes, including Cxcr5. Taken together, IL-2-signaling regulates the Bcl-6-Blimp-1 axis in TH1 cells to maintain flexibility with a TFH-like gene profile.
INTRODUCTION
CD4+ T helper cells can develop into a variety of functionally distinct subtypes after their initial encounter with foreign antigen. These subtypes include T helper 1 (TH1), TH2, TH17, and T follicular helper (TFH) cells1–6. The proper development and maintenance of T helper cell subsets is required to clear specific pathogens without causing self-damage. For example, TH1 cells coordinate the immune response to intracellular pathogens, but inappropriate activation results in autoimmunity7, 8.
Unique, signature gene expression programs define each specialized T helper cell subtype. T helper cell specific gene profiles are created by the induction of key lineage-defining transcription factors in response to cytokine signaling events at the time of initial antigen encounter9–14. For example, naïve helper T cells exposed to interleukin 12 (IL-12) and/or interferon-γ (IFNγ) upregulate the T-box transcription factor T-bet, which is required to establish the TH1 gene expression profile12. In contrast, IL-4 induces GATA3 to create a TH2 gene program, while a combination of IL-6 and transforming growth factor-β (TGFβ) upregulate RORγt to activate the TH17 profile9, 14. Additionally, IL-6 and IL-21 can induce the transcriptional repressor Bcl-6 to functionally regulate the TFH-gene program10, 11, 13. To date, the signaling pathways that initially induce T helper cell lineage-defining transcription factors have been well characterized, but it is currently unclear whether changing environmental conditions can alter their composition after the primary commitment decision.
Much research has been performed to examine the mechanisms by which T-bet activates TH1-signature genes15–20. Recently, several studies have uncovered diverse ways in which T-bet antagonizes alternative T helper cell fates16, 21–23. One mechanism T-bet utilizes to directly repress transcription is by physically recruiting the transcriptional repressor Bcl-6 to a subset of target genes in committed TH1 cells24. Thus, low amounts of Bcl-6 are necessary for T-bet to effectively repress alternative T helper cell gene programs24. These findings raised the question of how are Bcl-6 activity and expression tightly controlled in TH1 cells to prevent it from tipping the balance towards a TFH-gene profile.
Bcl-6 is a member of the BTB/POZ transcriptional repressor family. This family represses transcription by directly binding to specific DNA sequences through its zinc finger DNA binding domain, with the BTB/POZ domain mediating transcriptional repression25. It is currently unclear how Bcl-6 functionally activates TFH-signature genes. In TFH cells, Bcl-6 represses a miRNA gene cluster to effectively stabilize the expression of several TFH-signature genes13, but this does not explain the initial activation of TFH-signature gene transcription in response to Bcl-6.
In this study, we found that a T-bet-Bcl-6 complex masked the Bcl-6 DNA binding domain, which blocked Bcl-6 from repressing its target genes. This finding raised the possibility that there may be flexibility between TH1 and TFH-like gene expression patterns if there are environmental conditions that change the ratio of Bcl-6 to T-bet in TH1 cells. We demonstrated that strong IL-2-signaling, acting through STAT5, inhibited Bcl6 expression in effector TH1 cells, but when IL-2 was limiting, Foxo factors were able to activate Bcl6 transcription. Enhanced Bcl-6 expression in polarized TH1 cells resulted in the induction of Cxcr5 and a subset of TFH-genes. Mechanistically, altering the Bcl-6 to T-bet ratio in TH1 cells allowed Bcl-6 to repress its direct target gene Prdm1 (the gene that encodes Blimp-1). Notably, Blimp-1 was directly responsible for the repression of a subset of TFH-signature genes in effector TH1 cells. Therefore, the Bcl-6-dependent repression of Blimp-1 translated the repressive activity of Bcl-6 into the induction potential for a subset of TFH-genes.
RESULTS
T-bet interacts with the Bcl-6 DNA binding domain
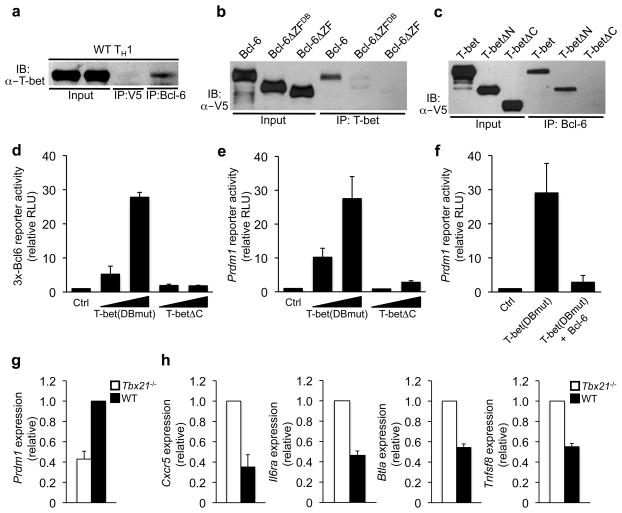
We have previously shown that T-bet physically interacts with Bcl-6 in TH1 cells (Fig. 1a and 24), which targets T-bet-Bcl-6 complexes to a subset of T-bet DNA binding elements24. These findings raised the question of why are T-bet-Bcl-6 complexes preferentially targeted to the T-bet, rather than Bcl-6, DNA binding elements. To begin to address this question, we performed co-immunoprecipitation (co-IP) experiments to define the domains within Bcl-6 and T-bet that were required for their interaction. A Bcl-6 truncation construct deleting its entire C-terminal zinc finger domain (Bcl-6ΔZF) failed to associate with T-bet (Fig. 1b, Supplementary Fig. 1a and 24). This domain contains six zinc fingers, with the four most C-terminal zinc fingers required for DNA binding26. A more detailed Bcl-6 truncation analysis demonstrated that the zinc fingers known to mediate Bcl-6 DNA-binding activity were also the ones required for its interaction with T-bet (Fig. 1b; Bcl-6ΔZFDB).
Figure 1. A T-bet-Bcl-6 complex inhibits Bcl-6-dependent repression.
(a) A co-IP was performed from wild-type TH1 cells with either a control or Bcl-6-specific antibody, followed by immunoblot analysis with a T-bet-specific antibody. (b) EL4 T cells were transfected with an untagged T-bet expression construct in combination with V5-epitope tagged wild-type Bcl-6, Bcl-6ΔZFDB or Bcl-6ΔZF as indicated. (c) EL4 T cells were transfected with an untagged Bcl-6 expression construct in combination with V5-epitope tagged wild-type T-bet, T-betΔN or T-betΔC as indicated. Lysates were immunoprecipitated with an antibody to (b) T-bet or (c) Bcl-6. (b, c) Immunoblots were probed with a V5-epitope specific antibody. (d, e) EL4 T cells were co-transfected with (d) 3x-Bcl6-promoter reporter or (e) Prdm1-promoter reporter constructs and either a control empty expression plasmid or increasing concentrations of a T-bet DNA-binding mutant construct [T-bet(DBmut)] or T-betΔC. (f) EL4 T cells were co-transfected with a Prdm1-promoter reporter and either an empty vector control or T-bet(DBmut) alone or in combination with Bcl-6. (d–f) Luciferase values were normalized to a renilla control and are represented relative to the control sample (relative RLU). (g, h) RNA was isolated from wild-type (black bars) or Tbx21−/− (white bars) primary CD4+ T cells polarized in TH1 conditions. Quantitative RT-PCR results for the genes indicated on the y-axis were first normalized to an Rps18 control, then represented relative to the (g) WT TH1 or (h) Tbx21−/− sample. Data in (a–c) are representative of at least three independent experiments. Data in (d–h) represent the mean of at least three (d–f, h) or five (g) independent experiments (error bars, s.e.m.).
Next, we localized the domain within T-bet that was required for its association with Bcl-6. T-bet is composed of a central T-box DNA binding domain as well as N- and C-terminal domains that mediate protein-protein interactions and transactivation events (Supplementary Fig. 1a). Truncating the N-terminal domain of T-bet (T-betΔN) did not impair its ability to interact with Bcl-6, while a T-bet C-terminal truncation construct (T-betΔC) failed to associate with Bcl-6 in co-IP experiments (Fig. 1c). Collectively, these data suggest that the interaction between T-bet and Bcl-6 has the potential to inhibit Bcl-6 DNA binding activity, while leaving the T-box DNA binding domain exposed.
A T-bet-Bcl-6 complex inhibits Bcl-6-dependent repression
To begin to address whether the interaction between T-bet and Bcl-6 interferes with Bcl-6 DNA binding activity, we performed transfection experiments with luciferase-reporter constructs containing either the Prdm1-promoter alone or multimerized Bcl-6 DNA binding elements upstream of the minimal SV40 promoter (3x-Bcl6-promoter reporter). The 3x-Bcl6-promoter reporter construct represents a simplified scenario where the repression of a minimal promoter is solely dependent upon Bcl-6 DNA binding elements. The Prdm1-promoter reporter represents a direct Bcl-6 target gene in the context of a physiologically relevant, and thus more complex, promoter setting. We utilized EL4 T cells for these experiments because they express endogenous Bcl-6, but do not express T-bet15, 24. As a control, we first confirmed that Bcl-6 repressed the activity of the 3x-Bcl6- and Prdm1-promoter reporters (Supplementary Fig. 1b, c).
If the interaction between T-bet and Bcl-6 inhibits Bcl-6 DNA binding, then a T-bet-Bcl-6 complex would prevent Bcl-6 from targeting to its own binding sites. In this scenario, increasing T-bet expression would enhance the formation of T-bet-Bcl-6 complexes and effectively block Bcl-6 from repressing its own target genes. To test this possibility, we examined whether increasing T-bet expression inhibited Bcl-6 from repressing the 3x-Bcl6-and Prdm1-promoter reporter constructs. For these experiments, we utilized a T-bet DNA binding mutant construct [T-bet(DBmut)] to exclude the possibility that T-bet may directly bind to and activate the 3x-Bcl6- or Prdm1-promoter reporters. Importantly, overexpression of T-bet(DBmut) alone substantially enhanced 3x-Bcl6- and Prdm1-promoter reporter activity, but did not activate an Ifng-promoter reporter as a control (Fig. 1d, e, Supplementary Fig. 1d, e). We also performed the promoter reporter experiments with a T-bet construct that cannot interact with Bcl-6 (T-betΔC; see Fig. 1c). T-betΔC did not inhibit Bcl-6-dependent repression, suggesting that the interaction between T-bet and Bcl-6 was required for T-bet’s ability to alleviate the Bcl-6-dependent repression of the 3x-Bcl6- and Prdm1-promoter reporters (Fig. 1d, e, Supplementary Fig. 1d).
We next hypothesized that the relative expression between T-bet and Bcl-6 defines the functional capability of Bcl-6 when both are expressed in the same cell. That is, if there is excess T-bet, T-bet-Bcl-6 complex formation will inhibit the majority of Bcl-6 from localizing to its own DNA binding elements (Fig. 1d, e). However, increasing Bcl-6 abundance in the presence of constant T-bet expression would allow excess Bcl-6 to interact with its own DNA binding elements. To test this hypothesis, we overexpressed Bcl-6 in conjunction with T-bet(DBmut) and examined the functional consequence on Bcl-6-dependent repression. Bcl-6 overexpression rescued the repression of the 3x-Bcl6- and Prdm1-promoter reporters despite the presence of T-bet(DBmut) (Fig. 1f, Supplementary Fig. 1f, g). These data suggest that the relative ratio of T-bet to Bcl-6 determines whether Bcl-6 can repress its direct target genes.
Enhanced TFH-signature gene expression in Tbx21−/− cells
Our previous study demonstrating that a T-bet-Bcl-6 complex is functionally important for repressing a subset of T-bet target genes did not address whether the T-bet-Bcl-6 complex prevents Bcl-6 from repressing its direct target genes in TH1 cells24. To begin to explore this question, we examined endogenous Prdm1 gene expression in primary CD4+ T cells isolated from either wild-type or Tbx21 (the gene that encodes T-bet)−/− mice polarized in TH1 conditions. In this experimental setting, Bcl-6 is expressed at a constant, low amount in both wild-type and Tbx21−/− cells24, but because T-bet is not present to form a complex with Bcl-6, this may allow the “free” Bcl-6 in the Tbx21−/− cells to repress its direct target genes. Consistent with this hypothesis, Prdm1 expression was reduced in Tbx21−/− TH1 cells (Fig. 1g).
We next wanted to determine whether changes in the functional activity of Bcl-6 in the T-bet-deficient setting would induce a TFH-like gene expression profile. A subset of TFH-signature genes1, 27, including Cxcr5, Il6ra, Btla, and Tnfsf8 (the gene that encodes CD30L) had increased expression in Tbx21−/− relative to wild-type TH1 polarized cells (Fig. 1h). To further explore these results, we also performed Tbx21 siRNA knockdown experiments in the context of wild-type TH1 polarized cells. In this experimental strategy, CD4+ T cells commit to the TH1 pathway in the presence of T-bet, which allowed us to examine the functional consequence of reducing T-bet expression in a natural TH1 setting. Similar to the data from the T-bet-deficient cells, the knockdown of Tbx21 in wildtype TH1 cells resulted in the induction of a subset of TFH-signature genes (Supplementary Fig. 2). Collectively, these experiments suggest that the interaction between T-bet and Bcl-6 functionally regulates the activities of both T-bet and Bcl-6 in TH1 cells (24, Fig. 1 and Supplementary Fig. 1, 2).
IL-2R-signaling inhibits Bcl-6 expression in TH1 cells
The mechanistic findings presented thus far suggest that there may be flexibility between the TH1 and TFH gene programs if environmental signaling events can regulate Bcl-6 expression in TH1 cells. Therefore, we wanted to determine whether there are signaling pathways in developing TH1 cells that modulate Bcl-6 expression. Previous research has suggested that IL-2-signaling regulates Bcl-6 expression in some circumstances. Specifically, Bcl-6 is repressed when CD8+ T cells are exposed to high concentrations of IL-2, whereas Bcl-6 expression is upregulated in limiting IL-2 conditions28. Also, an inverse correlation exists between IL-2Rα and Bcl-6 expression in CD4+ TFH cells29. Thus, we hypothesized that IL-2-receptor (IL-2R)-signaling may regulate Bcl-6 expression in TH1 cells.
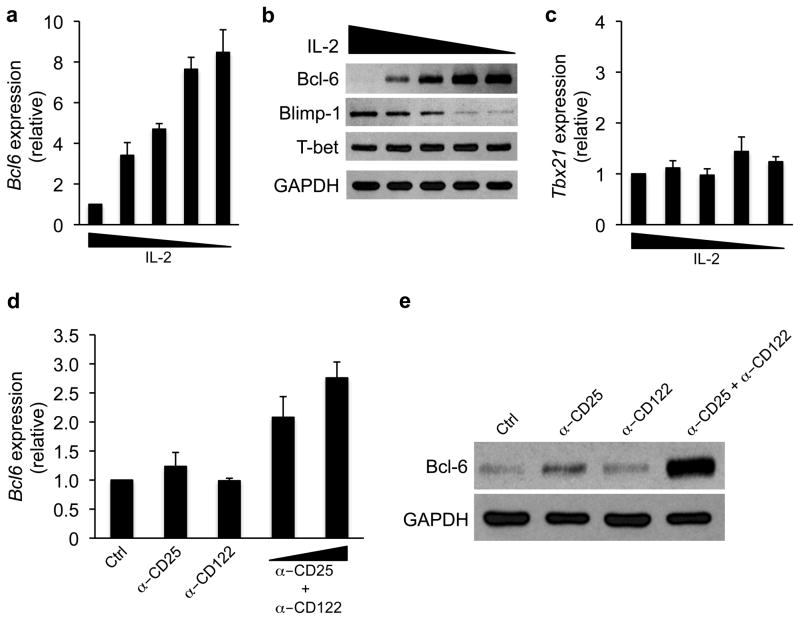
To test this possibility, we monitored Bcl-6 expression in CD4+ T cells cultured in TH1 polarizing conditions with a range of IL-2 concentrations. For these experiments, CD4+ T cells were stimulated with plate bound αCD3 and αCD28 for three days in the presence of TH1 polarizing conditions and IL-2. The cells were then split and maintained in TH1 polarizing conditions in the presence of variable IL-2 concentrations for an additional three days. In developing TH1 cells, Bcl-6 transcript and protein expression inversely correlated with the concentration of IL-2 (Fig. 2a, b, Supplementary Fig. 3a; and see Supplementary Fig. 3b to compare expression in high versus low IL-2). In contrast, T-bet was similarly expressed in all IL-2 conditions, indicating that TH1 polarizing conditions are dominant over IL-2 concentrations for regulating T-bet expression (Fig. 2b, c, and Supplementary Fig. 3b, c). Together these data suggest that the environmental concentration of IL-2 regulates Bcl-6, but not T-bet, expression in developing TH1 cells.
Figure 2. IL-2-signaling inhibits Bcl-6 expression in TH1 cells.
(a–c) Primary CD4+ T cells were continuously cultured in TH1 polarizing conditions for six days. All cells were initially stimulated with plate bound αCD3 and αCD28 and IL-2. At day three, the cells were split and cultured in the presence of decreasing concentrations of IL-2 for an additional 3 days. (a, c) RNA was then isolated and transcripts for (a) Bcl6 or (c) Tbx21 were determined by quantitative RT-PCR. Samples were first normalized to the Rps18 control followed by their graphical representation as a ratio relative to the most concentrated IL-2 condition. (b) An immunoblot analysis was performed with antibodies to the indicated proteins and GAPDH as a control for equal protein loading. (d, e) Primary wild-type CD4+ T cells were cultured in TH1 conditions and incubated with antibodies to CD25, CD122, or both in combination. The cells were harvested after 36 hours and (d) Bcl6 transcripts were assessed as in (a), or (e) Bcl-6 protein expression was determined by immunoblot analysis. Data represent the mean of either four (a, c) or three (d) independent experiments (error bars, s.e.m.). Data in (b, e) are representative of at least (b) two or (e) three independent experiments.
To further explore whether signaling through the IL-2R inhibits Bcl-6 expression in TH1 cells, we incubated developing TH1 cells with blocking antibodies to the CD25 (IL-2Rα) and/or CD122 (IL-2Rβ) subunits of the IL-2R complex. We found that blocking both CD25 and CD122 in combination enhanced Bcl-6 transcript and protein expression (Fig. 2d, e). These data are consistent with the findings from the IL-2 titration experiments and together provide evidence that strong IL-2R-signaling inhibits Bcl-6 expression in TH1 cells.
IL-2-signaling regulates STAT binding to the Bcl6 promoter
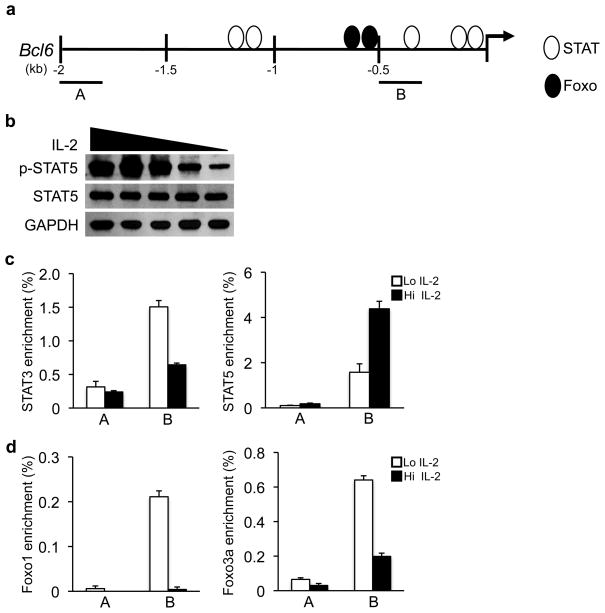
We next wanted to determine the mechanism by which IL-2R-signaling regulates Bcl6 expression in TH1 cells. A scan of the transcription factor binding elements within the Bcl6 promoter identified DNA binding sites for two transcription factor families, STAT and Foxo (Fig. 3a), whose activities are responsive to IL-2R-signaling in T cells30–32. STAT5 has been suggested to inhibit a subset of genes by either displacing activating STAT3 complexes or recruiting repressive chromatin modifying complexes to the promoter33, 34. Therefore, we wanted to test whether strong IL-2-signaling inhibits Bcl6 expression by enhancing STAT5 binding and/or the relative STAT5/STAT3 ratio at the Bcl6 promoter in TH1 cells.
Figure 3. STAT and Foxo transcription factors regulate Bcl6.
(a) Schematic representing STAT and Foxo DNA-binding element locations in the Bcl6 promoter. Regions monitored in ChIP analysis (A or B) are shown. (b) Immunoblot analysis of p-STAT5 and total STAT5 protein in wild-type TH1 cells exposed to variable IL-2 concentrations. (c, d) ChIP experiments were performed with wild-type TH1 polarized cells maintained in either high (Hi; black bars) or low (Lo; white bars) IL-2 conditions. Chromatin samples were immunoprecipitated with either an antibody to (c) STAT3, STAT5 or (d) Foxo1, Foxo3a, or (c, d) an IgG antibody control and quantitated by qPCR to monitor the identified binding elements (B) or a negative control region (A) in the Bcl6 promoter. To calculate the percent input, the values for the precipitated samples were first normalized to a standardized aliquot of the input chromatin for each condition, followed by subtraction of the IgG antibody control as the nonspecific background of the experiment. Data in (b) are representative of three independent experiments. Data in (c, d) represent the mean of three independent experiments (error bars, s.e.m.).
We first confirmed that STAT5 phosphorylation (p-STAT5) was enhanced with increasing IL-2 concentrations in TH1 cells (Fig. 3b). Next, we performed a chromatin immunoprecipitation (ChIP) analysis examining STAT3 and STAT5 binding to the Bcl6 promoter in TH1 cells cultured in either high or low IL-2 conditions (Fig. 3c). The ChIP experiments demonstrated that the ratio of STAT3 versus STAT5 bound to the Bcl6 promoter varied with the IL-2 concentration. Specifically, STAT5 binding to the Bcl6 promoter increased, while STAT3 binding decreased, in TH1 cells under high IL-2 conditions (Fig. 3c). Enhanced STAT5 binding correlated with the inhibition of Bcl6 expression in TH1 cells exposed to increasing concentrations of IL-2 (Fig. 2a, b). These data are consistent with a repressive role for STAT5 in the IL-2-dependent regulation of Bcl6 expression in TH1 cells.
Foxo factors regulate Bcl6 expression in TH1 cells
Recent studies suggest that strong IL-2R-signaling inhibits Foxo transcription factor activity in T cells. IL-2 induces a miRNA that inhibits Foxo1 expression and it also prevents the nuclear translocation of Foxo family members32, 35. Notably, there are Foxo binding elements in the Bcl6 promoter and Foxo transcription factors were able to activate Bcl6 promoter activity (Fig. 3a, Supplementary Fig. 4a, b and 36). Therefore, we wanted to determine whether Bcl6 is a direct, IL-2-responsive Foxo transcription factor target gene in TH1 cells.
Consistent with previous findings in non-polarized CD4+ T cells32, Foxo1 expression was reduced when TH1 cells were cultured in high IL-2 conditions (Supplementary Fig. 4c). We next performed ChIP experiments to assess Foxo1 and Foxo3a binding to the Bcl6 promoter in TH1 cells maintained in either high or low IL-2 (Fig. 3d). Both Foxo1 and Foxo3a bound to the Bcl6 promoter when TH1 cells were maintained in low IL-2, correlating with Bcl6 expression in these conditions. In contrast, Foxo1 and Foxo3a binding to the Bcl6 promoter were substantially reduced when TH1 cells were cultured in high IL-2 (Fig. 3d). Collectively, these data suggest that IL-2 regulates the binding of the transcriptional activators Foxo1 and Foxo3a to the Bcl6 promoter in primary TH1 cells.
TH1 cells upregulate a TFH-like profile in low IL-2
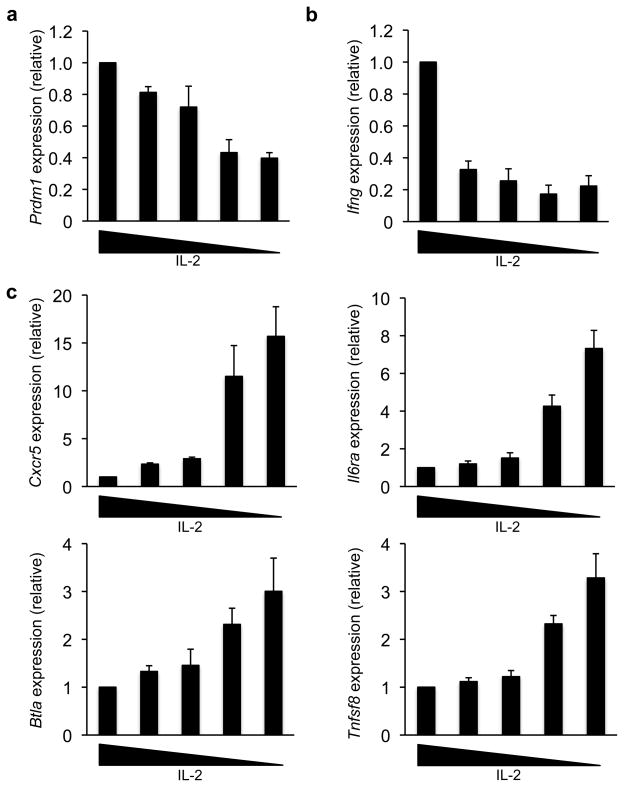
We next wanted to determine whether enhanced Bcl-6 abundance in TH1 cells could alter the gene expression profile of the cell (Fig. 4a–c). We first examined the expression of the direct Bcl-6 target gene Prdm1 in TH1 cells cultured in variable IL-2 conditions. Prdm1 transcript and Blimp-1 protein expression were substantially reduced in TH1 cells maintained in limiting IL-2 (Fig. 2b, 4a and Supplementary Fig. 3b). It is interesting to note that there appeared to be a threshold for the amount of Bcl-6 that was needed to effectively repress Blimp-1 expression. These data highly suggest that similar to the findings in Fig. 1, naturally increasing the Bcl-6 to T-bet ratio past a threshold point in primary TH1 cells results in the functional repression of a prototypic Bcl-6 target gene.
Figure 4. IL-2 regulates Prdm1 and TFH-gene expression in TH1 cells.
(a–c) Primary wild-type CD4+ T cells were continuously cultured in TH1 polarizing conditions for six days, with variable IL-2 concentrations from days 3–6 as described in Fig. 2a. Transcript amounts for the indicated gene were determined by quantitative RT-PCR. Values were normalized and expressed as described in Fig. 2a. Data in (a–c) represent the mean of four independent experiments (error bars, s.e.m.).
We then assessed whether enhanced Bcl-6 expression in TH1 cells was sufficient to upregulate TFH-genes. Cxcr5 was induced more than 15-fold in developing TH1 cells cultured in low IL-2 (Fig. 4c). Three other TFH-associated genes, Il6ra, Btla and Tnfsf8, were also upregulated in this setting (Fig. 4c). In contrast, the TH1 cytokine gene, Ifng, was reduced when TH1 cells were maintained in limiting IL-2, potentially indicating a shift in the balance of the T helper cell program (Fig. 4b). It is important to note that not all TFH-signature genes were induced in TH1 cells coincident with Bcl-6 upregulation. In particular, Pdcd1 (the gene that encodes PD-1) and Icos expression were unchanged (Supplementary Fig. 5). These data suggest that enhanced Bcl-6 expression in TH1 cells induces a partial TFH-profile, but additional events are required to establish the complete program.
IL-2 regulates Bcl6 expression in polarized TH1 cells
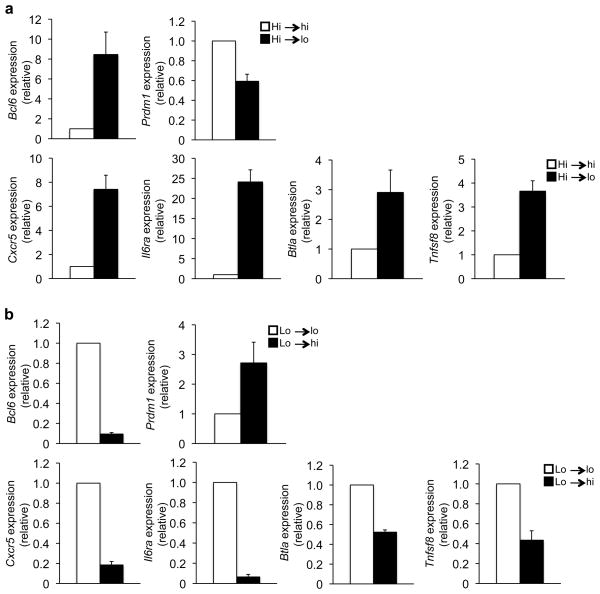
We next wanted to determine whether fully polarized TH1 cells retain the flexibility to modulate Bcl-6 expression in response to IL-2-signaling. To examine this question, CD4+ T cells were continuously cultured in TH1 polarizing conditions for nine days, with the IL-2 concentration either held constant or altered after six days of polarization. Bcl6 expression was induced when fully polarized TH1 cells maintained in high IL-2 were switched to low IL-2 (Fig. 5a). Notably, the upregulation of Bcl6 in fully polarized TH1 cells correlated with the repression of Prdm1 and the functional induction of TFH-signature genes including Cxcr5 (Fig. 5a). In contrast, the enhanced expression of Bcl6 and TFH-genes observed in low IL-2 conditions was substantially downregulated when TH1 polarized cells were exposed to high IL-2 conditions (Fig. 5b). Collectively, these data indicate that IL-2 regulates Bcl-6 expression in both developing and fully polarized TH1 cells.
Figure 5. TH1 cells maintain IL-2-sensitive Bcl-6 and TFH-gene regulation.
(a) Primary wild-type CD4+ T cells were cultured in TH1 conditions and high IL-2 for six days. On day 6, the polarized TH1 cells were split and maintained in TH1 conditions and either high IL-2 (Hi→hi; white bars) or low IL-2 (Hi→lo; black bars) for an additional 3 days. (b) Primary wild-type CD4+ T cells were cultured in TH1 conditions with the cells maintained in low IL-2 after the day 3 split. On day 6, the TH1 polarized cells were split and maintained for an additional three days in TH1 polarizing conditions with either low IL-2 (Lo→lo; white bars) or high IL-2 (Lo→hi; black bars). (a, b) RNA was isolated and transcript amounts for the genes indicated on the y-axis were determined by quantitative RT-PCR. Results were first normalized to the Rps18 control and are graphically represented as the ratio relative to the sample maintained in (a) high IL-2 (Hi→hi) or (b) low IL-2 (Lo→lo). Data in (a–b) represent the mean of at least three independent experiments (error bars, s.e.m.).
Blimp-1 directly represses TFH-genes in TH1 cells
Bcl-6 is a transcriptional repressor and thus it is unlikely to directly activate TFH-genes. Notably, the Bcl-6-dependent repression of Blimp-1 correlated with the induction of TFH-signature genes in decreasing IL-2 concentrations (Fig. 2b, 4a, c). Like Bcl-6, Blimp-1 is a transcriptional repressor37, 38. Mechanistically, if Blimp-1 directly represses TFH-signature genes in effector TH1 cells, then increasing the activity of Bcl-6, which directly represses Blimp-1, would effectively reduce a “Blimp-1-brake” in place on the TFH-associated genes. Therefore, Prdm1 is a good candidate for the direct Bcl-6 target gene that translates Bcl-6-mediated repression into the downstream activation potential for a subset of TFH-genes.
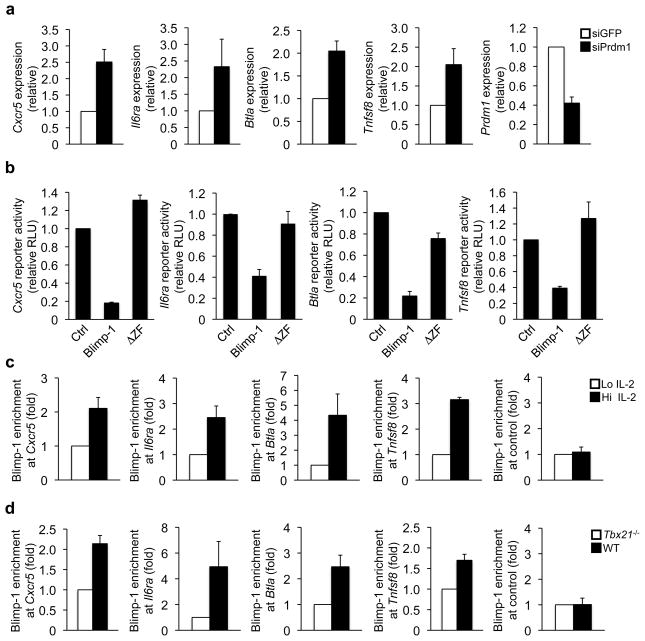
We first examined whether Blimp-1 represses endogenous TFH-signature gene expression in primary TH1 cells. We transfected wild-type TH1 cells with either a control or Prdm1 siRNA and analyzed the consequences on endogenous gene expression. Prdm1 expression was reduced in TH1 cells transfected with Prdm1-specific versus control siRNA (Fig. 6a). Consistent with a role for Blimp-1 in repressing TFH-gene expression in TH1 cells, Cxcr5, Il6ra, Btla, and Tnfsf8 expression were substantially enhanced when Blimp-1 expression was reduced (Fig. 6a). These data indicate that Blimp-1 functionally represses a subset of TFH-associated genes in primary effector TH1 cells.
Figure 6. Blimp-1 directly represses TFH-genes in effector TH1 cells.
(a) Wild-type TH1 cells were transfected with either a control (siGFP; white bars) or Prdm1 (siPrdm1; black bars) siRNA. The genes indicated on the y-axis were analyzed as in Fig. 1g and are represented relative to the siGFP sample. (b) EL4 T cells were transfected with either a Cxcr5, Il6ra, Btla, or Tnfsf8 promoter reporter and either an empty expression vector control, wild-type Blimp-1, or Blimp-1ΔZF. Normalized luciferase values are expressed relative to the vector control. (c) ChIP experiments were performed with wild-type TH1 polarized cells maintained in either high (Hi; black bars) or low (Lo; white bars) IL-2 conditions. Chromatin samples were immunoprecipitated with either an antibody to Blimp-1 or an IgG control. Blimp-1-precipitated samples were normalized as in Fig. 3c to obtain the percent input value (Supplementary Fig. 6b). The data are graphically represented as fold enrichment, which is the ratio of the percent input for each sample relative to the low IL-2 sample from the same experiment (Hi; black bars or Lo; white bars). (d) Chromatin was isolated from either wild-type (black bars) or Tbx21−/− (white bars) CD4+ T cells polarized in TH1 conditions. The ChIP experiments were performed and normalized as in (c) with the exception that the fold enrichment is relative to the Tbx21−/− percent input values (Supplementary Fig. 6c). Data in (a–d) represent the mean of three independent experiments (error bars, s.e.m.).
To start to address whether Blimp-1 plays a direct role in repressing these genes, we cloned the promoters for Cxcr5, Il6ra, Btla, and Tnfsf8 into luciferase reporter vectors to determine whether they are responsive to Blimp-1-mediated repression. Each of these promoters was repressed by the overexpression of wild-type Blimp-1, but not a DNA binding mutant Blimp-1 construct (Blimp-1ΔZF) (Fig. 6b and Supplementary Fig. 6a). Thus, the promoter reporter data support the hypothesis that Blimp-1 directly represses Cxcr5, Il6ra, Btla, and Tnfsf8 transcription.
Blimp-1 binding inversely correlates with TFH-expression
If Blimp-1 directly represses the IL-2-sensitive TFH-associated genes in effector TH1 cells, then Blimp-1 binding would inversely correlate with their expression. In ChIP experiments, Blimp-1 was associated with the Cxcr5, Il6ra, Btla, and Tnfsf8 promoters in TH1 cells maintained in high IL-2, coinciding with the repression of these genes (Fig. 6c and Supplementary Fig. 6b). In contrast, Blimp-1 binding was substantially reduced at the Cxcr5, Il6ra, Btla and Tnfsf8 promoters in response to limiting IL-2 (Fig. 6c and Supplementary Fig. 6b). The reduction in Blimp-1 binding correlated with the induction of these genes (Fig. 4c and 6c). Collectively, these data suggest that Blimp-1 directly binds to and represses the IL-2-sensitive TFH-signature genes in effector TH1 cells.
Finally, we wanted to explore whether Blimp-1 is a key regulatory factor that translates an increased Bcl-6/T-bet ratio in TH1 cells into the downstream activation potential for TFH-associated genes. To test this question, we examined Blimp-1 binding to the Cxcr5, Il6ra, Btla, and Tnfsf8 promoters in the setting of Tbx21−/− cells. Blimp-1 binding was diminished at these promoters in Tbx21−/− in comparison to wild-type TH1 polarized cells (Fig. 6d and Supplementary Fig. 6c), coinciding with enhanced gene expression (Fig. 1h). Taken together, Blimp-1 binding inversely correlates with increasing the Bcl-6 to T-bet ratio in TH1 cells by either natural environmental cues (Fig. 6c) or genetic manipulation (Fig. 6d). Therefore, the IL-2-sensitive regulation of Bcl-6 expression in TH1 cells determines the downstream potential of a subset of TFH-signature genes by controlling the “Blimp-1-brake” (Supplementary Fig. 7).
DISCUSSION
This study demonstrates that variable IL-2-signaling regulates Bcl-6 expression in polarized TH1 cells. In effector TH1 cells, a high T-bet to Bcl-6 ratio promoted T-bet-Bcl-6 complex formation, which masked the Bcl-6 DNA binding domain. As Bcl-6 expression was enhanced in TH1 cells maintained in low IL-2 conditions, excess Bcl-6 repressed its target gene Prdm1. Blimp-1 directly repressed a subset of TFH-signature genes in effector TH1 cells. Therefore, the Bcl-6-dependent repression of Blimp-1 was responsible for regulating TFH-like gene expression activation potential in CD4+ T cells. Collectively, these data suggest that TH1 cells retain flexibility with a TFH-like gene profile by maintaining their ability to regulate the Bcl-6-Blimp-1 axis in response to IL-2.
A long-held view in the field has been that opposing T helper cell lineage-defining transcription factors are expressed in a mutually exclusive pattern, but recent research has questioned this simplistic paradigm. There is now increasing awareness that opposing T helper cell lineage-defining transcription factors are co-expressed in many circumstances, and their co-expression is functionally important for regulating the gene expression profile of the cell21, 24, 39–41. This concept raises the question of how are the expression and functional activities of these factors precisely regulated during an immune response. Our study demonstrates that T-bet can dominantly control Bcl-6 activity because a T-bet-Bcl-6 complex masks the Bcl-6 DNA-binding domain, but leaves the T-bet DNA-binding domain available. This effectively lets T-bet keep Bcl-6 in check in effector TH1 cells. However, if Bcl-6 expression increases past the threshold of T-bet control, the balance of the cell can shift towards a TFH-like gene profile.
The environmental concentration of IL-2, translated through STAT and Foxo transcription factor activity, regulates Bcl-6 expression in polarized TH1 cells. This means that TH1 cells retain flexibility with a TFH-like gene profile because they have the capacity to alter Bcl-6 expression in response to IL-2-signaling. A recent study found that IL-2-signaling is critical for the formation of multiple T helper cell lineages42. Our findings add to this concept and suggest that IL-2-signaling can change the phenotype of polarized TH1 cells. Intriguingly, new research suggests that IL-2Rα expression inversely correlates with Bcl-6 expression to create a continuum of TCM, TFH, or effector TH1 characteristics 29, 43. It is possible that the IL-2R subunit expression pattern on a CD4+ T cell will allow individual cells in a population to respond differently to the same environmental IL-2 conditions.
Our study provides new insight into how the transcriptional repressor Bcl-6 serves to promote TFH-signature gene expression. We found that the Bcl-6-dependent repression of Prdm1 was directly responsible for regulating the transcriptional potential for some TFH-genes. Interestingly, Blimp-1 is expressed in multiple effector T helper cell subtypes, but is repressed during TFH cell differentiation in vivo10, 37. It is possible that Blimp-1 commonly represses TFH-signature genes in other effector T helper cell subtypes as well.
We need to envision the activation of the TFH-gene program as a multi-step process, with the Bcl-6-dependent removal of the “Blimp-1-brake” representing the first step. Recent studies demonstrate that Icos-IcosL interactions in the follicle are required for the full induction of a TFH-gene profile29. Additionally, transcriptional regulators such as Batf and c-Maf play a role in TFH-differentiation44, 45. It will be important to determine the complete series of molecular events that occur downstream of removing the “Blimp-1-brake” needed to fully activate TFH-genes and whether polarized TH1 cells can initiate all of these events. Of note, not all TFH-signature genes, such as PD-1 and Icos, were upregulated in low IL-2 indicating that distinct classes of TFH-target genes exist that either require additional, or completely independent, events. Interestingly, Cxcr5high/PD-1low T helper cells exist outside of the germinal center, representing a TFH-like cell prior to homing within the follicle for full differentiation46. We hypothesize that the Cxcr5+ TH1 cells will need to home to the follicle where the next events required in the TFH gene program occur. Importantly, the overexpression of Bcl-6 alone is sufficient for complete TFH cell differentiation in vivo10, 11, 13. Therefore, because Bcl-6 expression is regulated by IL-2-signaling in TH1 cells, given the right circumstances in vivo, TH1 cells may retain complete flexibility towards the TFH-gene program.
METHODS
Cell Culture and Transfection
Primary T Cells
Primary CD4+ T cells were isolated from the spleen and lymph nodes of either wild-type C57BL/6 or Tbx21−/− mice using the Mag Cellect kit (R&D) as previously described15. Following isolation, cells were grown on plate-bound αCD3/αCD28 in the presence of IL-2 and TH1 polarizing cytokines [αIL-4 (10 μg/mL) and IL-12 (5 ng/mL)]. On day 3, cells were split and maintained in TH1 conditions for an additional three days. During this time, the cells were cultured in a range of IL-2 concentrations (500, 100, 50, 10, or 1 IU/mL). When indicated, high IL-2 is 500 IU/mL and low IL-2 is 10 IU/mL. For the IL-2R blocking experiments, antibodies to CD25 (PC61, BD Biosciences) and CD122 (TM-β1, BD Biosciences) were used. Primary CD4+ T cell transfections with the Lonza nucleofection system using mouse primary T cell solutions and program X-01 were performed as previously described18, 19. The siRNA transfections were performed as previously described with an siRNA smartpool to either Tbx21 or Prdm1 (Dharmacon) or GFP as a control (Ambion)18, 19. All experiments involving mice were conducted with IACUC approval.
EL4 T Cells
Murine EL4 T cells transfections using the Lonza nucleofection system program 0–17 and solution V were performed as previously described15. Immunoblot analysis was conducted to determine the expression of the transfected proteins.
Promoter Reporter Assay
The 3x-Bcl6-promoter reporter construct was made by cloning three Bcl-6 DNA binding elements upstream of the minimal SV40 promoter in the pGL3-promoter reporter vector (Promega). Bcl6 (+1998 to +1 bp), Prdm1 (+1991 to −222 bp), Cxcr5 (+1841 to −12 bp), Btla (+913 to −16 bp), Tnfsf8 (+1458 to −226 bp), and Il6ra (+1209 to −123 bp) promoter reporter constructs were prepared by cloning each of the promoters into the pGL3-basic luciferase reporter construct (Promega). EL4 cells were co-transfected with the promoter reporter constructs in combination with either T-bet(DBmut), T-betΔC, Bcl-6, Blimp-1, or Blimp-1ΔZF V5-tagged expression vectors, or an empty vector control as indicated. The TK-renilla control plasmid was also co-transfected and used to normalize for transfection efficiency. Transfections were harvested after 16–24 hours and samples were analyzed with the Dual-Luciferase Reporter system (Promega). Expression amounts for transfected expression constructs were monitored by immunoblot analysis (Supplementary Fig. 1d, f, 6a)
Co-Immunoprecipitation (co-IP)
The co-IP assay was performed as described previously18, 19, 24. A T-bet specific antibody (H-210; Santa Cruz Biotechnologies) or a Bcl-6 specific antibody (C-19; Santa Cruz Biotechnologies) was used for immunoprecipitation. The co-immunoprecipitated proteins were detected using a V5-epitope tag specific antibody (R960-25; Invitrogen). For the endogenous co-IP studies in primary wild-type TH1 cells, the samples were immunoprecipitated with either an antibody specific for Bcl-6 or a negative control V5 antibody. The immunoblots were then probed with a T-bet specific antibody (4B10; Santa Cruz).
RNA and qRT-PCR
RNA was obtained by Nucleospin RNA purification (Machery-Nagel) and cDNA was prepared with the First Strand Superscript II Synthesis System (Invitrogen). qPCR reactions using 20 nanograms of the cDNA template were performed with gene specific primers and the qPCR Sybr Green Mix (Biorad). All samples were first normalized to the Rps18 (ribosomal protein S18) control. Graphs represent the normalized expression for the sample as a ratio to relative to the indicated comparison condition in each Figure panel.
Immunoblot Analysis
An equal number of primary wild-type TH1 cells were harvested for each IL-2 treatment condition and subjected to an immunoblot analysis to determine protein expression. Antibodies for T-bet (4B10), STAT5 (C-17), and GAPDH were from Santa Cruz, while the antibodies to Bcl-6 (561520) and p-STAT5 (611964) were from BD Pharmingen. The antibody for Blimp-1 (A01647) was from Genscript.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed as described previously15, 17, 18, 24. The STAT3 (C-20), STAT5 (C-17), Foxo1 (H-128), and Foxo3a (H-144) antibodies were from Santa Cruz Biotechnology, while the Blimp-1-specific antibody was from Genscript (A01647). Chromatin was harvested from either primary polarized TH1 wild-type cells maintained in variable IL-2 conditions (high or low IL-2) or from Tbx21−/− CD4+ T cells as indicated. The precipitated DNA was analyzed by qPCR with promoter-specific primers (for primer sequences see Supplementary Table 1). Samples were normalized to a standardized total input DNA control followed by subtraction of the IgG antibody control as the nonspecific background for the precipitation. This value represented the percent input for each sample. Fold enrichment values were calculated by dividing the percent input of the samples by that of either the low IL-2 or Tbx21−/− samples from the same experiment.
STATISTICS
The error bars for all graphs were calculated as the standard error of the mean (s.e.m).
Supplementary Material
Acknowledgments
We would like to thank members of the Weinmann lab for helpful discussions and Michael Wijaranakula for technical assistance. The project described was supported by grants from the NIAID (AI061061 and AI07272) and the American Cancer Society (RSG-09-045-01-DDC) to A.S.W. We also would like to thank the NCI preclinical repository for IL-2 and anti-IL-4.
Footnotes
AUTHOR CONTRIBUTIONS
K.J.O and A.S.W. designed, performed experiments, analyzed data and wrote the manuscript; S.E.M contributed to performing the experiments for Fig. 1c, 2b, e, Supplementary Fig. 4a, b.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Crotty S. Follicular Helper CD4 T Cells (T(FH)) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 2.Murphy KM, Stockinger B. Effector T cell plasticity: flexibility in the face of changing circumstances. Nat Immunol. 2010;11:674–680. doi: 10.1038/ni.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reinhardt RL, Kang SJ, Liang HE, Locksley RM. T helper cell effector fates--who, how and where? Curr Opin Immunol. 2006;18:271–277. doi: 10.1016/j.coi.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng SL. The T-box transcription factor T-bet in immunity and autoimmunity. Cell Mol Immunol. 2006;3:87–95. [PubMed] [Google Scholar]
- 8.Lazarevic V, Glimcher LH. T-bet in disease. Nat Immunol. 2011;12:597–606. doi: 10.1038/ni.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 10.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szabo SJ, et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 13.Yu D, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 15.Beima KM, et al. T-bet binding to newly identified target gene promoters is cell type-independent but results in variable context-dependent functional effects. J Biol Chem. 2006;281:11992–12000. doi: 10.1074/jbc.M513613200. [DOI] [PubMed] [Google Scholar]
- 16.Djuretic IM, et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol. 2007;8:145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 17.Lewis MD, Miller SA, Miazgowicz MM, Beima KM, Weinmann AS. T-bet’s ability to regulate individual target genes requires the conserved T-box domain to recruit histone methyltransferase activity and a separate family member-specific transactivation domain. Mol Cell Biol. 2007;27:8510–8521. doi: 10.1128/MCB.01615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22:2980–2993. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szabo SJ, et al. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 21.Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005;307:430–433. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- 22.Lazarevic V, et al. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol. 2011;12:96–104. doi: 10.1038/ni.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mehta DS, Wurster AL, Weinmann AS, Grusby MJ. NFATc2 and T-bet contribute to T-helper-cell-subset-specific regulation of IL-21 expression. Proc Natl Acad Sci U S A. 2005;102:2016–2021. doi: 10.1073/pnas.0409512102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oestreich KJ, Huang AC, Weinmann AS. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J Exp Med. 2011;208:1001–1013. doi: 10.1084/jem.20102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basso K, Dalla-Favera R. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv Immunol. 2010;105:193–210. doi: 10.1016/S0065-2776(10)05007-8. [DOI] [PubMed] [Google Scholar]
- 26.Mascle X, Albagli O, Lemercier C. Point mutations in BCL6 DNA-binding domain reveal distinct roles for the six zinc fingers. Biochem Biophys Res Commun. 2003;300:391–396. doi: 10.1016/s0006-291x(02)02873-5. [DOI] [PubMed] [Google Scholar]
- 27.Vinuesa CG, Tangye SG, Moser B, Mackay CR. Follicular B helper T cells in antibody responses and autoimmunity. Nat Rev Immunol. 2005;5:853–865. doi: 10.1038/nri1714. [DOI] [PubMed] [Google Scholar]
- 28.Pipkin ME, et al. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi YS, et al. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 2011;34:932–946. doi: 10.1016/j.immuni.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao W, Lin JX, Leonard WJ. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr Opin Immunol. 2011;23:598–604. doi: 10.1016/j.coi.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merkenschlager M, von Boehmer H. PI3 kinase signalling blocks Foxp3 expression by sequestering Foxo factors. J Exp Med. 2010;207:1347–1350. doi: 10.1084/jem.20101156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stittrich AB, et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. 2011;11:1057–1062. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- 33.Mandal M, et al. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat Immunol. 2011;12:1212–1220. doi: 10.1038/ni.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang XP, et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011;12:247–254. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riou C, et al. Convergence of TCR and cytokine signaling leads to FOXO3a phosphorylation and drives the survival of CD4+ central memory T cells. J Exp Med. 2007;204:79–91. doi: 10.1084/jem.20061681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurtz C, et al. BCL6-mediated repression of p53 is critical for leukemia stem cell survival in chronic myeloid leukemia. J Exp Med. 2011;208:2163–2174. doi: 10.1084/jem.20110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol. 2010;11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 39.Hegazy AN, et al. Interferons direct Th2 cell reprogramming to generate a stable GATA-3(+)T-bet(+) cell subset with combined Th2 and Th1 cell functions. Immunity. 2010;32:116–128. doi: 10.1016/j.immuni.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 40.Koch MA, et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011;35:337–348. doi: 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao W, Lin JX, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol. 2011;12:551–559. doi: 10.1038/ni.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, Jenkins MK. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583–595. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bauquet AT, et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat Immunol. 2009;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ise W, et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol. 2011;12:536–543. doi: 10.1038/ni.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee SK, et al. B cell priming for extrafollicular antibody responses requires Bcl-6 expression by T cells. J Exp Med. 2011;208:1377–1388. doi: 10.1084/jem.20102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.