Abstract
Clear cell sarcoma (CCS) is an aggressive soft tissue malignant tumor characterized by a unique t(12;22) translocation that leads to the expression of a chimeric EWS/ATF1 fusion gene. However, little is known about the mechanisms underlying the involvement of EWS/ATF1 in CCS development. In addition, the cellular origins of CCS have not been determined. Here, we generated EWS/ATF1-inducible mice and examined the effects of EWS/ATF1 expression in adult somatic cells. We found that forced expression of EWS/ATF1 resulted in the development of EWS/ATF1-dependent sarcomas in mice. The histology of EWS/ATF1-induced sarcomas resembled that of CCS, and EWS/ATF1-induced tumor cells expressed CCS markers, including S100, SOX10, and MITF. Lineage-tracing experiments indicated that neural crest–derived cells were subject to EWS/ATF1-driven transformation. EWS/ATF1 directly induced Fos in an ERK-independent manner. Treatment of human and EWS/ATF1-induced CCS tumor cells with FOS-targeted siRNA attenuated proliferation. These findings demonstrated that FOS mediates the growth of EWS/ATF1-associated sarcomas and suggest that FOS is a potential therapeutic target in human CCS.
Introduction
Clear cell sarcoma (CCS) is an aggressive malignancy of adolescents and young adults that was first described by Enzinger (1). It typically arises in the deep soft tissues of the lower extremities closed to tendon, fascia, and aponeurosis (2). Chemotherapy and radiotherapy are not of any benefit (3–5), and a high rate of local and distant recurrence results in poor survival rates (3, 6, 7). CCSs harbor the potential for melanocytic differentiation and melanin synthesis (8). Gene expression profiles support the classification of CCS as a distinct genomic subtype of melanomas (9). These melanocytic features often make the distinction from malignant melanoma (MM) difficult. However, in contrast to MM, CCS is characterized by a chromosomal translocation, t(12;22)(q13;q12), that leads to the fusion of activating transcription factor 1 (ATF1) gene localized to 12q13 to Ewing’s sarcoma oncogene (EWS) gene at 22q12 in up to 90% of cases, resulting in expression of the EWS/ATF1 fusion gene (10–12). Given that CCS and MM have such similar characteristics, it has been proposed that CCSs may arise from a neural crest progenitor. However, the exact origin of CCS still remains to be determined.
The biological role of the EWS/ATF1 fusion protein is still unclear. EWS contains a transcriptional activation domain in the N-terminal region (13–15) and several conserved RNA binding motifs in the C-terminal region (16). Binding of the N-terminal region of EWS to the RNA polymerase II subunit hsRPB7 has been proposed to be important for transactivation of the target genes (17). In contrast, ATF1 is a member of the CREB transcription factor family, whose activity is regulated through phosphorylation of its kinase inducible domain (KID) by protein kinase A (18). ATF1 mediates the activation of cAMP-responsive genes through binding to a conserved cAMP-responsive element (CRE) as a dimmer (19, 20). However, the N-terminal activation domain of EWS replaces the KID in the EWS/ATF1 fusion protein, rendering it unable to support a typical inductive signal (21). Therefore, EWS/ATF1 can act as constitutive transcriptional activator in a cAMP-independent fashion with normal CRE DNA binding activity (14, 22, 23).
Previous studies have revealed some target genes of EWS/ATF1, but their true function in tumorigenesis is still not well understood (24). Expression of MITF is constitutively activated by EWS/ATF1 in CCS in vitro (25). Consistent with this finding, several studies have identified the expression of MITF protein or mRNA in CCS (26–28). MITF is a master regulator of melanocyte development and plays a role in melanoma development (29, 30). Importantly, activation of MITF by EWS/ATF1 is required for CCS proliferation as well as for melanocytic differentiation of CCS in vitro (25).
Although previous studies have demonstrated that EWS/ATF1 is associated with oncogenic potential in CCS, the effect of in vivo expression of EWS/ATF1 on sarcoma formation is still not known. In the present study, we established EWS/ATF1 transgenic mice using a doxycycline-dependent expression system in order to investigate the role of EWS/ATF1 on CCS development in vivo. Our results showed that forced expression of EWS/ATF1 induced CCS-like sarcoma in the transgenic mice. This mouse model was used to identify the origin of EWS/ATF1-induced sarcomas as well as the direct target of EWS/ATF1 in these sarcomas.
Results
Inducible expression of EWS/ATF1 in mice.
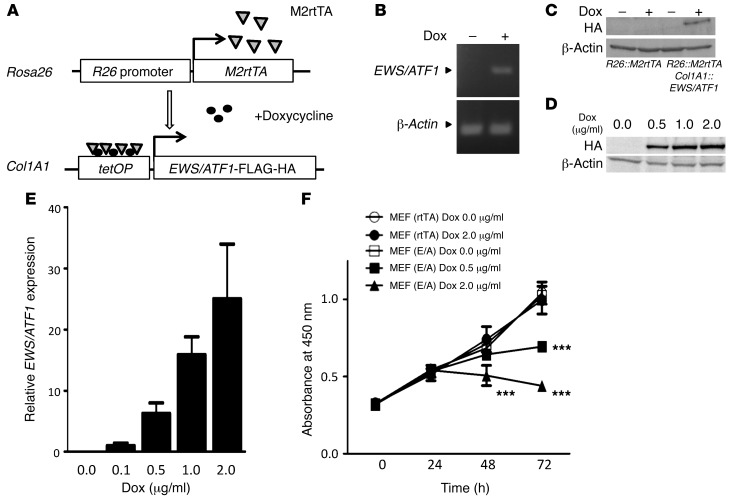
We first generated doxycycline-inducible EWS/ATF1 ES cells, in which the human EWS/ATF1 type 2 fusion gene (26, 31) can be induced under the control of a tetracycline-responsive regulatory element (Figure 1A). Upon treatment of these ES cells with doxycycline, expression of the EWS/ATF1 fusion transcript was detected by RT-PCR (Figure 1B). We also confirmed the expression of EWS/ATF1 protein upon doxycycline treatment (Figure 1C), which was regulated in a dose-dependent manner (up to 2 μg/ml; Figure 1D).
Figure 1. Inducible expression of EWS/ATF1.
(A) Schematic of the doxycycline-inducible EWS/ATF1 alleles. (B) EWS/ATF1 expression in ES cells, detected by RT-PCR, after exposure to doxycycline for 12 hours. (C) EWS/ATF1 expression in ES cells, detected by Western blot, after exposure to doxycycline for 24 hours. (D) Dose-dependent induction of EWS/ATF1 protein in EWS/ATF1-inducible ES cells by doxycycline. ES cells were exposed to doxycycline concentrations up to 2 μg/ml for 24 hours. Western blot analysis was performed using an anti-HA antibody. (E) Dose-dependent doxycycline induction of EWS/ATF1 mRNA in EWS/ATF1-inducible MEFs. MEFs were exposed to different concentrations of doxycycline for 24 hours. Transcript levels were normalized to β-actin. Data are mean ± SD (n = 3). (F) EWS/ATF1 expression suppressed MEF growth. Cell viability was determined by WST-8 assay. Data are mean ± SD (n = 4). Control MEFs (rtTA) and EWS/ATF1-inducible MEFs (E/A) were derived from heterozygous Rosa26::M2rtTA and Col1A1::tetO-EWS/ATF1 mice, respectively. ***P < 0.001 vs. MEF (rtTA) Dox 0.0 μg/ml, MEF (rtTA) Dox 2.0 μg/ml, and MEF (E/A) Dox 0.0 μg/ml.
Heterozygous Rosa26::M2rtTA mice with heterozygous tetO-EWS/ATF1 allele were used to induce the EWS/ATF1 fusion gene. Cultured murine embryonic fibroblasts (MEFs) derived from EWS/ATF1-inducible mice were first exposed to doxycycline to test the effect of EWS/ATF1 expression on somatic cells. EWS/ATF1 expression at the mRNA level was confirmed 24 hours after exposure (Figure 1E). Unexpectedly, the cell proliferation rate of MEFs decreased after EWS/ATF1 induction in a doxycycline dose–dependent manner (Figure 1F).
EWS/ATF1 induces sarcoma formation in mice.
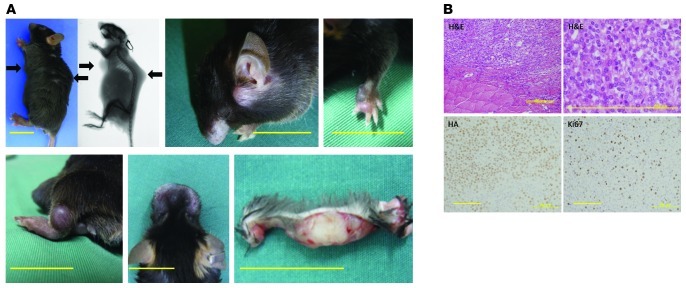
To investigate the effect of EWS/ATF1 expression in vivo, we treated EWS/ATF1-inducible mice at 6 weeks of age with doxycycline in the drinking water (50 μg/ml). The EWS/ATF1-inducible mice given doxycycline started to develop multiple macroscopic soft tissue tumors after 4 weeks. After doxycycline treatment, EWS/ATF1 protein was detected in a variety of tissues, including the intestine, liver, epidermis, and deep soft tissue (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI63572DS1). Doxycycline treatment for 3 months resulted in tumor formation in the deep soft tissues of all mice (n = 39), whereas control mice without doxycycline treatment developed no detectable tumors. EWS/ATF1-induced tumors typically arose in the trunks, heads, limbs, and whisker pads (Figure 2A). Macroscopically, tumors consisted of circumscribed and lobulated gray-white mass (Figure 2A). In most cases, the tumors were attached to fascia or aponeuroses (Figure 2, A and B), which indicates that the tumors specifically arose from the deep soft tissues. Importantly, 36 of 39 mice (92%) developed tumors in the trunk, which suggests that cells located in the trunk are particularly permissive for tumorigenesis by EWS/ATF1 expression. Despite expression of EWS/ATF1 protein, no tumor formation was observed in other tissues, such as the epidermis and intestine, even in mice given doxycycline for 3 months.
Figure 2. EWS/ATF1-induced tumors resemble human CCS.
EWS/ATF1 transgenic mice were administered 50 μg/ml doxycycline in their drinking water for 3 months. (A) EWS/ATF1 expression caused tumor formation (arrows) in various locations: trunk, head, limbs, and whisker pads. X-ray examination revealed multiple tumors in deep soft tissue. The cut surface of a large tumor on the ventral trunk of an EWS/ATF1-inducible mouse revealed a lobulated gray-white mass in the deep soft tissue. Scale bars: 20 mm. (B) Histological analysis of EWS/ATF1-induced tumors. Tumors were composed of round or fusiform cells with prominent basophilic nuclei and clear cytoplasm, which were surrounded by fibrous fascicles. HA immunostaining confirmed EWS/ATF1 expression in the tumor cells. Frequent Ki67-positive cells were present throughout the lesions. Scale bars: 200 μm (H&E, left); 50 μm (H&E, right); 100 μm (HA and Ki67).
Microscopic examination of these tumors revealed striking similarities to human CCSs. The tumors showed a rather uniform pattern of compact nests or fascicles of rounded or fusiform cells, which were divided by a framework of fibrocollagenous tissue (Figure 2B). The individual tumor cells had a homogeneous appearance. They had round to ovoid vesicular nuclei with prominent basophilic nucleoli and clear or pale-staining cytoplasm (Figure 2B). The majority of the tumor cells expressed EWS/ATF1 fusion protein in nuclei (Figure 2B and Supplemental Figure 2A). Ki67-positive proliferating cells were observed in about 30%–40% of tumor cells (Figure 2B), indicative of active proliferative activity. The survival curves of EWS/ATF1-induced mice were analyzed to evaluate the overall effect of EWS/ATF1 expression on life span. The transgenic mice treated with doxycycline became moribund within 3–10 months, suggestive of multiple tumor formation in the deep soft tissue, whereas mice without doxycycline treatment survived much longer, and no tumor formation was observed. The median survival time of EWS/ATF1-inducible mice treated with doxycycline was 20 weeks (Supplemental Figure 2B).
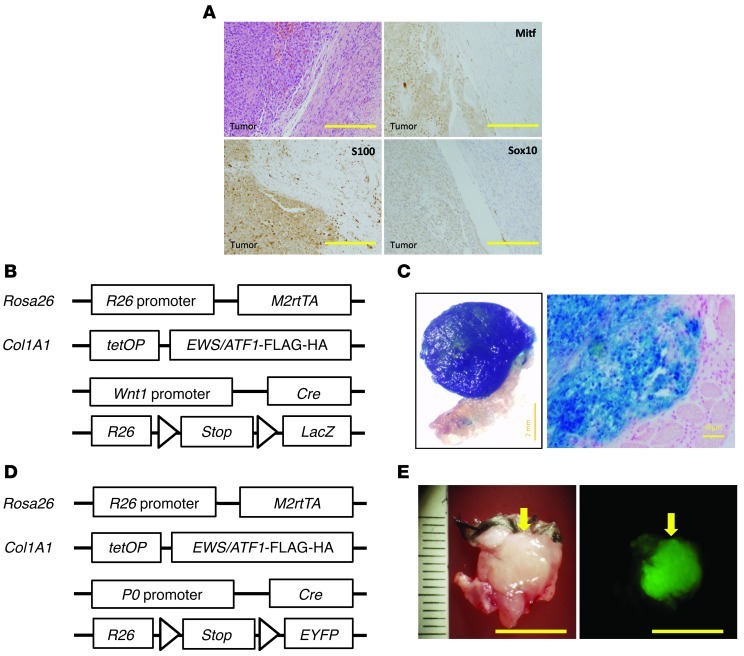
Previous studies demonstrated that human CCSs express markers for neural crest lineage as well as melanocytic differentiation (8, 9). Therefore, to examine the similarity of mouse EWS/ATF1-induced tumors with human CCSs, we performed immunohistochemical analysis for CCS-expressing markers; EWS/ATF1-induced tumor cells showed the expression such markers, including S100, Sox10, and Mitf (Figure 3A).
Figure 3. EWS/ATF1-induced tumors arise from neural crest–lineage cells.
(A) Immunohistochemical analysis for CCS markers. Nuclear staining for S100, Sox10, and Mitf was observed in tumor cells. Sections were counterstained with hematoxylin. Scale bars: 100 μm. (B) Schematic representation of reporter alleles for the lineage-tracing experiment using Wnt1-Cre allele. Doxycycline-inducible EWS/ATF1 alleles were introduced into reporter mice containing the Wnt1-Cre and floxed LacZ reporter alleles. (C) X-gal staining for EWS/ATF1-induced tumors with Wnt1-Cre and floxed LacZ reporter alleles. Positive staining for X-gal indicated that the tumor arose from a neural crest–lineage cell. Histological analysis revealed that neoplastic cells were stained with X-gal. Counterstaining was performed with fast red. Scale bars: 2 mm (left); 50 μm (right). (D) Schematic representation of reporter alleles for the lineage-tracing experiment using P0-Cre allele. Doxycycline-inducible EWS/ATF1 alleles were introduced into reporter mice containing the P0-Cre and floxed EYFP reporter alleles. (E) Representative image of a tumor (arrow) in the trunk of an EWS/ATF1-induced mouse with P0-Cre and floxed EYFP reporter alleles. Fluorescent signals for EYFP expression were detected in the. Scale bars: 10 mm.
Neural crest–lineage cells are permissive to EWS/ATF1-driven sarcoma development.
The cell of origin for CCS remains to be determined. Based on the potential of CCSs for melanocytic differentiation and melanin synthesis, previous studies proposed that CCS may arise from a neural crest progenitor. To determine whether EWS/ATF1-induced sarcomas actually arise from neural crest–derived cells, we performed a lineage-tracing experiment in which neural crest–derived cells were tagged by reporter in vivo (32). To label neural crest–derived cells in vivo, we first used transgenic mice containing Wnt1-Cre and floxed LacZ reporter alleles. We further introduced doxycycline-inducible EWS/ATF1 alleles into the reporter mice to generate compound transgenic mice (Figure 3B). We confirmed that EWS/ATF1-induced tumor cells did not express Wnt1 (Supplemental Figure 3A). Transgenic mice were treated with doxycycline in the drinking water to induce subcutaneous tumors and the developed tumors were then analyzed for the expression of the reporter gene. Importantly, all 14 EWS/ATF1-induced tumors were ubiquitously positive for X-gal staining (Figure 3C and Supplemental Figure 3D), which suggests that neural crest–lineage cells are a cell of origin for EWS/ATF1-associated sarcomas. We further performed another lineage-tracing experiment using transgenic mice containing P0-Cre and floxed EYFP reporter alleles (Figure 3D), which have been also widely used to label neural crest–derived cells. Again, we found that all 6 EWS/ATF1-induced tumors were positive for EYFP (Figure 3E and Supplemental Figure 4, C and D).
Establishment of tumor cell lines.
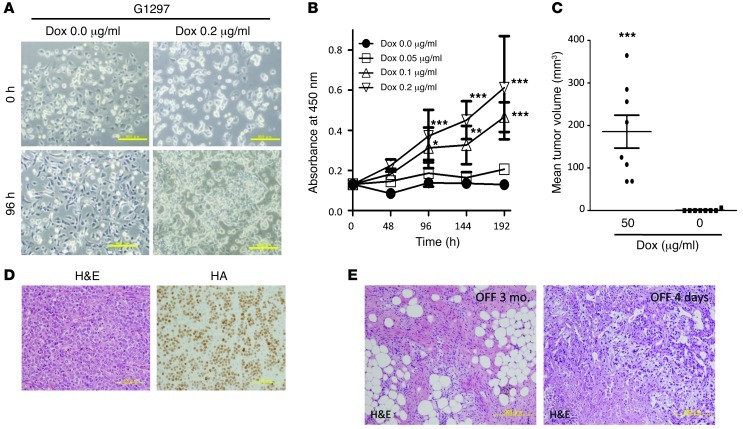
Tumor samples were obtained from primary tumors of EWS/ATF1-induced mice to establish cell lines from EWS/ATF1-induced tumors. We established 2 tumor cell lines, G1297 and G1169, from 2 independent mice. These cells grew in the form of an adherent monolayer in the presence of doxycycline (0.2 μg/ml). We cultured the cells up to the fourth passage in medium containing 0.2 μg/ml doxycycline in order to avoid contamination by fibroblasts. We examined the effect of different concentrations of doxycycline on the growth and morphology of the established cell lines. We confirmed that the expression of EWS/ATF1 transcript and protein increased in response to doxycycline in a dose-dependent manner in both established cell lines (Supplemental Figure 5, A–C). The growth and morphology of the tumor cells varied in a doxycycline dose–dependent manner: small, round tumor cells grew rapidly at concentrations above 0.1 μg/ml, whereas dendritic fibroblast–like spindle cells were observed below 0.05 μg/ml (Figure 4A). Notably, doxycycline withdrawal caused rapid morphological changes, into a fibroblast-like shape, and these tumor cells did not proliferate up to the next passage (Figure 4A). Consistent with these findings, cell viability assay revealed that the number of cells was increased by doxycycline treatment in a dose-dependent manner (Figure 4B). We next examined the effect of EWS/ATF1 expression on tumorigenesis ability in the subcutaneous tissue of immunocompromised mice. The established cell line G1297 was cultured in medium containing 0.2 μg/ml doxycycline, and 5.0 × 106 cells were transplanted into the subcutaneous tissue of nude mice. It is important to note that all mice treated with 50 μg/ml doxycycline in the drinking water developed tumors within 3 weeks, whereas no tumor formation was observed in mice without doxycycline treatment (Figure 4C). Histological analysis revealed that the subcutaneous tumors in nude mice consisted of neoplastic cells that resembled the primary tumor cells in EWS/ATF1-induced transgenic mice (Figure 4D). Positive immunoreactivity for HA-Tag was observed in all tumor cells (Figure 4D).
Figure 4. Establishment and analysis of tumor cell lines.
G1297 and G1169 cell lines were established from 2 independent EWS/ATF1-induced tumors. (A) Morphology of the G1297 line after treatment without or with doxycycline (0 and 0.2 μg/ml, respectively). At concentrations above 0.1 μg/ml, small, round tumor cells grew rapidly, while dendritic fibroblast-like spindle cells were observed; tumor cell growth almost stopped at concentrations less than 0.05 μg/ml. Scale bars: 50 μm. (B) Effect of different levels of EWS/ATF1 on tumor cell growth. G1297 cells were cultured in different concentrations of doxycycline (0, 0.05, 0.1, and 0.2 μg/ml), and cell viability was determined by WST-8 assay. Data are mean ± SD (n = 8). *P < 0.05, **P < 0.01, ***P < 0.001 vs. Dox 0.0 μg/ml. (C) Subcutaneous transplantation of 5.0 × 106 G1297 cells in immunocompromised mice resulted in tumor formation in mice treated with 50 μg/ml doxycycline (n = 8). Mean tumor volumes ± SEM are shown. ***P < 0.005. (D) Representative histology and HA immunostaining of tumors in nude mice. The tumor resembled the original sarcoma with EWS/ATF1 expression. Scale bars: 100 μm. (E) Doxycycline withdrawal led to rapid tumor regression. At 3 months after doxycycline withdrawal, no viable tumor cells were observed, and tumors were replaced by fibrous tissue. Widespread cell death was observed 4 days after doxycycline withdrawal. Scale bars: 200 μm.
Continuous expression of EWS/ATF1 is required for tumor growth maintenance.
To further examine whether continuous expression of EWS/ATF1 is necessary for the growth of EWS/ATF1-induced tumors, we withdrew doxycycline in tumor-bearing EWS/ATF1 transgenic mice that had been given doxycycline for 3 months. Importantly, doxycycline withdrawal resulted in a rapid reduction of tumor mass in 4 independent mice (7 tumors total). The regressed tumors contained fibrous tissue, but no viable neoplastic cells were observed 3 months after doxycycline withdrawal (Figure 4E), which suggests that EWS/ATF1-induced tumor growth depends on continuous EWS/ATF1 expression. We next examined the histological changes shortly after doxycycline withdrawal in order to investigate the mechanisms of tumor regression. We found widespread cell death within the tumor mass, accompanied by massive infiltration of inflammatory cells, at 4 days after doxycycline withdrawal (Figure 4E), which indicates that neoplastic cells cannot survive in vivo without EWS/ATF1 expression. Taken together, these results clearly indicate that EWS/ATF1 plays a pivotal role in the proliferation and maintenance of EWS/ATF1-induced tumor cells in vivo.
Fos is a direct target of EWS/ATF1.
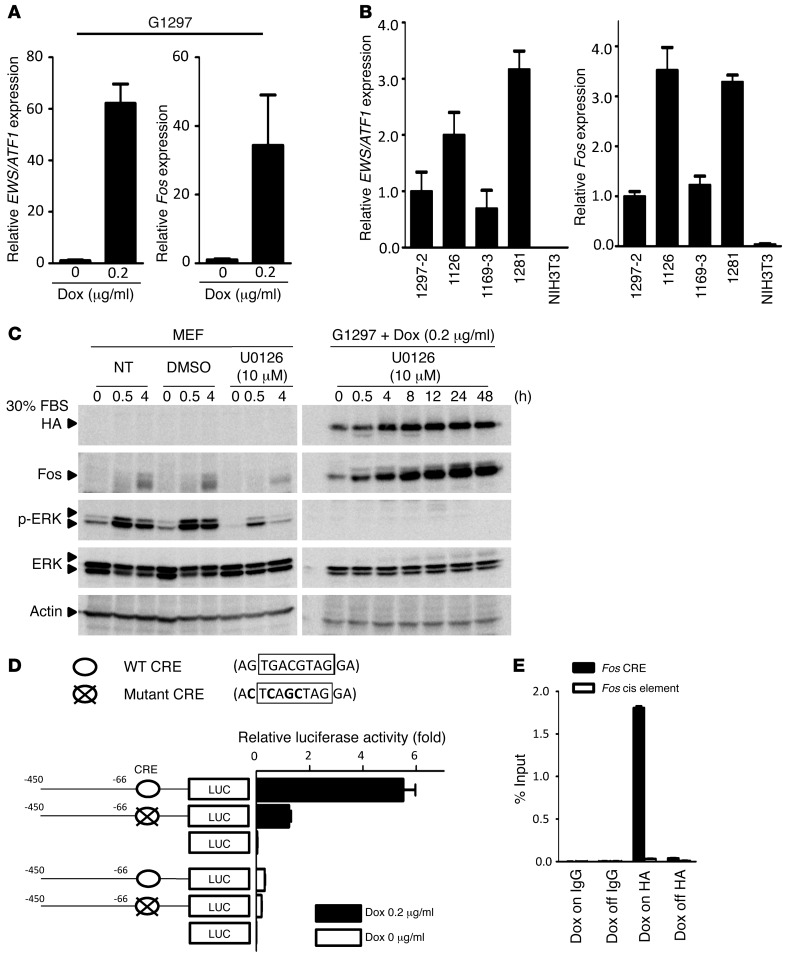
To determine the downstream targets regulated by EWS/ATF1, we next performed gene expression analysis using G1297 cells. First, we confirmed that withdrawal of doxycycline for 96 hours resulted in no detectable expression of EWS/ATF1 RNA or protein in cultured tumor cells. Next, the tumor cells were exposed again to doxycycline at a concentration of 0.05 or 0.2 μg/ml, and microarray analysis was performed at 3 and 48 hours after doxycycline exposure. Induction of EWS/ATF1 resulted in altered expression of a number of genes associated with cell growth, such as growth factor genes (Areg and Ereg), cell cycle regulators (Cenpa, Ccna2, Ccnb2, Cdkn1b, Plk1, and Aurka), and a proto-oncogene (Fos) at either time point (Supplemental Figure 6A). Although a previous study demonstrated that MITF-M is a direct target of EWS/ATF1 in human CCS cell lines (25), we failed to detect its expression in our EWS/ATF1-induced tumor cell lines and primary tumor samples (Supplemental Figure 7, A and B). Among the transcripts upregulated by EWS/ATF1, we focused on the proto-oncogene Fos, because this was one of the most highly upregulated genes by EWS/ATF1 after doxycycline exposure in the microarray analysis (Supplemental Figure 6A). Quantitative real-time RT-PCR (qRT-PCR) confirmed upregulation of both Fos and EWS/ATF1 transgenes in 2 independent tumor cell lines as early as 3 hours after doxycycline exposure (Figure 5A and Supplemental Figure 7C). We also found that the EWS/ATF1-induced tumor specimens expressed higher levels of both Fos and EWS/ATF1 transgenes (Figure 5B). Expression of Fos is induced by numerous stimuli, which are transmitted through the RAS/Raf/MAP kinase or cAMP-dependent protein kinase pathway (33). In order to investigate the mechanism of Fos induction by EWS/ATF1, we next examined whether the RAS/Raf/MAP kinase pathway is involved in EWS/ATF1-mediated Fos activation. In contrast to the rapid and transient induction of Fos in MEFs after serum stimulation (Supplemental Figure 7D), expression of Fos in the EWS/ATF1-expressing tumor cell line was detected even under serum-free conditions, and it gradually increased after serum stimulation (Supplemental Figure 7E). Interestingly, whereas serum-stimulated MEFs revealed immediate phosphorylation of ERK1 and ERK2 (Supplemental Figure 7D), phosphorylation of ERK1/2 was not observed in the EWS/ATF1-induced tumor cell line, even after serum stimulation (Supplemental Figure 7E), which suggests that continuous upregulation of Fos in EWS/ATF1-induced tumor cells is independent of the RAS/Raf/ERK signaling pathway. We treated EWS/ATF1-induced tumor cells with the MEK inhibitor U0126 to block activation of ERK1/2 in order to further confirm the ERK-independent activation of Fos. Although inhibition of ERK1/2 resulted in a substantial decrease of Fos in MEFs, U0126 failed to suppress Fos expression in EWS/ATF1-induced tumor cells (Figure 5C). These data indicate that constitutive overexpression of Fos in EWS/ATF1-induced tumor cells was mediated by an ERK-independent mechanism.
Figure 5. Fos is a direct target of EWS/ATF1.
(A) Real-time RT-PCR analysis of G1297 cells revealed significant upregulation of both EWS/ATF1 and Fos 3 hours after doxycycline exposure. (B) Relative expression of EWS/ATF1 and Fos in 4 EWS/ATF1-induced tumors from 4 independent mice. NIH3T3 cells served as a control. Transcript levels were normalized to β-actin. Data are mean ± SD (n = 3). (C) Fos induction by EWS/ATF1 was independent of the ERK pathway. Serum-starved MEFs and G1297 cells were stimulated with 30% FBS for the indicated times. Cells were also treated with 10 μM of the MEK inhibitor U0126. Whereas ERK1/2 inhibition by U0126 decreased Fos in MEFs, U0126 failed to suppress Fos expression in G1297 cells. NT, not treated. (D) Mouse Fos promoter–luciferase reporter constructs and pRL-SV40 vector (as an internal control) were cotransfected in G1297 cells treated with or without 0.2 μg/ml doxycycline. Luciferase activity of each construct was normalized to internal control activity. Data are mean ± SD (n = 3). (E) ChIP-PCR analysis was performed for the Fos promoter region containing CRE or the negative control cis element using HA-tag antibody or IgG as nonimmune immunoprecipitation, respectively. EWS/ATF1 was enriched at the CRE element of the Fos promoter in G1297 cells after treatment with 0.2 μg/ml doxycycline. Data (mean ± SD) were quantified by qRT-PCR and expressed as percent of input DNA.
Previous studies demonstrated an interaction of ATF1 at a CRE in the Fos promoter (34, 35), which suggests that EWS/ATF1 may induce Fos expression through interaction with the CRE. Conversely, in the present study, regulatory motif analysis of the upregulated genes by EWS/ATF1 demonstrated enrichment of CRE near the transcription start site (from –1,000 bp to +200 bp; Supplemental Figure 6B). To evaluate the functional importance of this element in EWS/ATF1-mediated activation of Fos, we constructed a reporter plasmid containing the mouse Fos promoter with wild-type and mutated CRE and examined transcriptional activity by luciferase assay (Figure 5D). We confirmed that induction of EWS/ATF1 resulted in remarkably increased Fos promoter activity with wild-type CRE in G1297 cells. Importantly, luciferase activity of the mutated promoter significantly decreased compared with that of the wild-type promoter. We further examined whether EWS/ATF1 directly binds to the CRE of the Fos promoter. ChIP-PCR analysis revealed that doxycycline-induced EWS/ATF1 was enriched at the CRE of the Fos promoter, but not at the negative control cis element (Figure 5E). Our results indicated that the CRE is crucial for EWS/ATF1-mediated transcriptional activity of Fos in EWS/ATF1-induced tumor cells.
Expression of FOS in human CCS.
To investigate whether overexpression of FOS is linked to human CCS, we analyzed FOS expression in the human CCS cell lines MP-CCS-SY and KAS and in the control lung fibroblast cell line WI38 by qRT-PCR. FOS was found to be highly expressed in both human CCS cell lines compared with WI38 (Supplemental Figure 8A). We also found that surgically resected clinical CCS specimens also expressed higher levels of FOS than did WI38 (Supplemental Figure 8B), which indicates that human CCS expresses higher levels of FOS.
We examined the effect of EWS/ATF1 knockdown on FOS expression in human CCS cell lines to further investigate the association between EWS/ATF1 expression and increased FOS expression in human CCS. Human CCS cell lines MP-CCS-SY and KAS carry the EWS/ATF1 type 1 and type 2 fusion genes, respectively (Supplemental Figure 9, A and B). We next designed a specific siRNA targeting the breakpoint of the EWS/ATF1 type 1 fusion gene, which had no effect on the expression of ATF1 or of the EWS/ATF1 type 2 fusion gene in KAS (Supplemental Figure 9, E and F). siRNA treatment targeting EWS/ATF1 type 1 in MP-CCS-SY led to significant downregulation of FOS 48 hours after treatment (Supplemental Figure 9G) as well as of EWS/ATF1 type 1 itself (Supplemental Figure 9, C and D), which indicates that FOS is a direct target of EWS/ATF1 in human CCS. In contrast to FOS, we observed a modest reduction of MITF-M expression after EWS/ATF1 knockdown in MP-CCS-SY cells (Supplemental Figure 9H).
FOS could be a promising therapeutic target for human CCS.
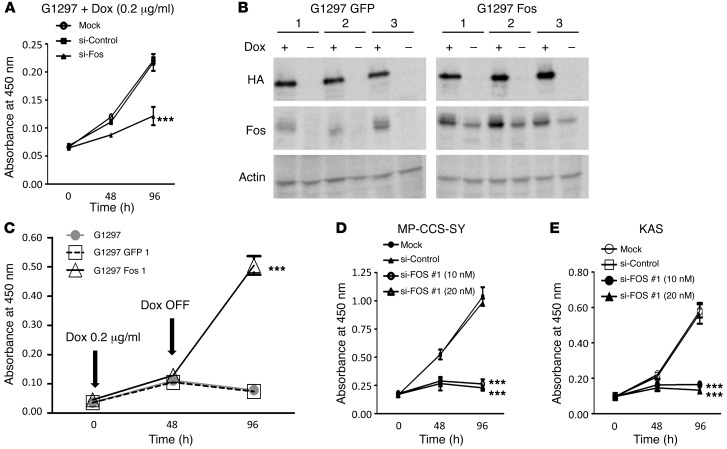
To examine whether Fos overexpression facilitates proliferation of tumor cells expressing EWS/ATF1, we knocked down Fos in EWS/ATF1-induced tumor cells using siRNA. The G1297 cell line was treated with siRNA for Fos in the presence of doxycycline. siRNA treatment (10 nM) decreased the expression of Fos at the mRNA level by 75% at 24 hours after transfection, although it had no effect on expression of the EWS/ATF1 transgene compared with the control siRNA (Supplemental Figure 10A). In addition, we confirmed that Fos protein levels were also decreased 48 hours after transfection (Supplemental Figure 10B). A WST-8 assay was performed in EWS/ATF1-induced tumor cells transfected with the nonfunctional control siRNA or with functional Fos siRNA to examine the effect of Fos knockdown on the cellular kinetics. The siRNA targeting Fos efficiently inhibited cell proliferation of EWS/ATF1-induced tumor cells, even in the presence of doxycycline (Figure 6A). In order to further confirm the importance of Fos expression for EWS/ATF1-induced tumor cell growth, we established EWS/ATF1-induced tumor cell lines in which Fos is overexpressed (Figure 6B). We found that Fos-overexpressed EWS/ATF1-inducible cells retained the ability to proliferate for at least 48 hours after doxycycline withdrawal, whereas control cells in which GFP is overexpressed stopped their proliferation soon after withdrawal (Figure 6C). We also examined the effect of FOS knockdown on cell growth of human CCS cell lines using siRNA targeting FOS. Consistent with the results in EWS/ATF1-induced tumor cells, siRNA treatment strongly suppressed the growth of CCS cell lines (Figure 6, D and E, and Supplemental Figure 11, A–F). Taken together, these data suggest that FOS, a direct target of EWS/ATF1, mediates the oncogenic growth of EWS/ATF1-related sarcomas and could be a potent therapeutic target for human CCS.
Figure 6. Fos plays a key role in EWS/ATF1-induced cell proliferation.
(A) Effect of Fos knockdown on proliferation of EWS/ATF1-induced cells. G1297 cells were treated with siRNA targeting Fos (si-Fos; 10 nM), a control siRNA (si-Control; 10 nM), or lipofectamine alone (Mock). 48 and 96 hours later, cell viability was determined by WST-8 assay. Results are mean ± SD (n = 4). ***P < 0.001 vs. si-Control and Mock. (B) EWS/ATF1-induced tumor cell lines overexpressing Fos or EGFP (G1297 Fos and G1297 GFP, respectively). pCAG-Fos-IZ vector or pCAG-EGFP-IZ vector were stably transfected in G1297 cells. Western blot analysis revealed that G1297 Fos cells stably expressed Fos protein even in the absence of doxycycline. (C) Cell proliferation assay for G1297, G1297 GFP, and G1297 Fos cells before and after doxycycline withdrawal. Doxycycline treatment (0.2 μg/ml) was withdrawn for 48 hours. Cell viability was determined by WST-8 assay. ***P < 0.001 vs. G1297 and G1297 GFP. (D and E) Effect of FOS knockdown on growth of human CCS cell lines. MP-CCS-SY and KAS cells were treated with siRNA#1 targeting FOS (si-FOS #1; 10 nM and 20 nM), control siRNA (si-Control; 20 nM), or lipofectamine alone (Mock). 48 and 96 hours later, cell viability was determined by WST-8 assay. Data are mean ± SD (n = 4). ***P < 0.001 vs. si-Control and Mock.
Discussion
The current study revealed that forced expression of the EWS/ATF1 fusion gene induced sarcoma formation in EWS/ATF1 transgenic mice. The histology of the tumors in EWS/ATF1 transgenic mice showed a striking similarity to that of human CCS. In addition, immunohistochemistry demonstrated that EWS/ATF1-induced tumor cells express neural crest–associated markers, such as S100, Mitf, and Sox10, which are also expressed in human CCS. Given that the EWS/ATF1 fusion gene is detected in CCS, our EWS/ATF1 transgenic mouse is the first mouse model for investigating CCS pathogenesis. Our present results demonstrated that continuous expression of EWS/ATF1 was required for growth and tumor formation of EWS/ATF1-induced tumor cells. These results indicate that EWS/ATF1 plays a pivotal role in both development and maintenance of EWS/ATF1-associated sarcomas, implying that CCS exhibits oncogene addiction (36) to EWS/ATF1, and provide a rationale for targeting EWS/ATF1 itself to treat CCS.
It is interesting to note that sarcoma formation was observed only in deep soft tissue, although EWS/ATF1 was induced in a variety of cell types in this experimental system (37, 38). In addition, the cell proliferation rate of MEFs in vitro was reduced by EWS/ATF1 induction. These results clearly demonstrated that the abnormal proliferation by the forced expression of EWS/ATF1 requires a specific cell type of origin, accompanied by a specific microenvironment. Consistent with these findings, recent studies of other sarcoma-related genes revealed that introduction of SYT/SSX, a synovial sarcoma-related gene, into Myf5-positive immature myoblasts specifically resulted in sarcoma formation, whereas its expression in more differentiated cells induced myopathy without tumor induction (39). In addition, introduction of EWS/FLI1, a fusion gene detectable in Ewing sarcomas, results in transformation specifically in bone marrow–derived mesenchymal progenitor cells in vitro (40). Taken together, these findings are suggestive of cell type–specific carcinogenesis by expression of sarcoma-related fusion oncogenes.
Our lineage-tracing experiments in vivo suggested that EWS/ATF1-associated tumor cells are derived from neural crest–derived cells. This result is consistent with several lines of evidence that CCS often shows melanocytic differentiation and resembles MM. However, our present results do not exclude the possibility that EWS/ATF1-induced tumors can arise from non–neural crest–derived cells. In addition, the exact cell type of origin of EWS/ATF1-induced tumors remains unclear, since neural crest–lineage progenitors can differentiate into many different cell types, such as neuronal cells, melanocytes, and Schwann cells. Recently, Schwann cell precursors along the peripheral nerve have been shown to be a cellular source of large numbers of melanocytes in the skin during development in mice and chicks (41). Moreover, Schwann cells also retain the potential to differentiate into melanocytes, resulting from a loss of nerve contact (41). Given the finding that EWS/ATF1-induced tumor cells expressed markers for melanocytic differentiation, it is possible that the neural crest–derived Schwann cells could be the origin of EWS/ATF1-associated sarcomas.
We found that Fos was one of the direct targets of EWS/ATF1 in EWS/ATF1-induced tumor cells. Fos is an immediate early gene that can be activated by a variety of mitogens and growth factors. The present study showed that Fos induction by forced expression of EWS/ATF1 was independent of the ERK signaling pathway. In contrast, we found that Fos upregulation was mediated by a CRE of the Fos promoter, accompanied by direct interaction of EWS/ATF1 with the CRE on the Fos promoter. The direct interaction of EWS/ATF1 at CRE may induce continuous transcriptional activation of Fos in EWS/ATF1-induced tumor cells. Previous studies have demonstrated a higher expression level of FOS to be involved in tumor growth in several cancers (42–44), and overexpression of Fos results in osteosarcoma formation in transgenic mice (45, 46). Here we showed that FOS was also upregulated in CCS by the EWS/ATF1 fusion transcript and that the increased FOS promoted the growth of EWS/ATF1-related sarcomas. Accordingly, blocking the FOS pathway might be a promising therapeutic strategy for treating CCS (Supplemental Figure 12).
Methods
Molecular cloning and gene targeting in ES cells.
Human EWS/ATF1-FLAG-HA was amplified by RT-PCR from the human CCS cell line KAS using primers ACATGGCGTCCACGGATTACAG and CCTAGGCGTAGTCGGGCACGTCGTAGGGGTATCCTCCAGCGGCCGACTTGTCATCGTCGTCCTTGTAGTCTCCTCCAACACTTTTATTGGAATAAAGAT and cloned into pcr2.1-TOPO. Sequence-verified EWS/ATF1-FLAG-HA cDNA was subcloned into a unique EcoRI site of pBS31 prime (37, 38). KH2 ES cells (obtained from Open Biosystems) were used to insert a single copy of EWS/ATF1-FLAG-HA by Flipase (PLP) recombination into the Col1A1 locus under the control of a minimal CMV tetracycline-inducible promoter using a previously described method (37), and ES cells were selected for hygromycin resistance.
Mouse generation.
For blastocyst injections, fertilized zygotes were isolated from the oviducts of day-0.5 pregnant B6D2F1 females and allowed to develop to the blastocyst stage in culture. 7–12 ES cells were injected per blastocyst. The injected blastocysts were transferred into day-2.5 pseudopregnant recipient females.
Doxycycline treatment.
6-week-old mice were administered 50 μg/ml doxycycline (Sigma-Aldrich) in their drinking water supplemented with 2 mg/ml sucrose. For cultured cells, doxycycline was used at a concentration of 0.05–0.2 μg/ml.
RNA preparation and RT-PCR.
Total RNA was isolated using a RNeasy mini kit (Qiagen). Total RNA was reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit with RNase inhibitor (Applied Biosystems). qRT-PCR analysis using the fluorescent SYBR green method (Bio-Rad) was performed in accordance with the manufacturer’s instructions. The data generated from each reaction were subjected to gene expression analysis using an iCycler iQ Real-Time PCR Detection System (Bio-Rad). See Supplemental Table 1 for specific primer pairs used for amplification. Microarray analysis was performed with SurePrint G3 Mouse GE 8X60K microarray (Agilent Technologies) and Mouse Gene 1.0 ST Array (Affymetrix) according to the manufacturer’s instructions. All analyses were performed by Genespring GX (version 12; Agilent Technologies).
Western blot analysis.
Western blot analyses were carried out as described previously (47, 48). The following antibodies were used: anti-HA (rabbit IgG, 1:1,000 dilution; Cell Signaling), anti-Fos (rabbit IgG, 1:1,000 dilution; Cell Signaling), anti-ERK1/2 (rabbit IgG, 1:1,000 dilution; Cell Signaling), anti–phospho-ERK1/2 (rabbit IgG, 1:1,000 dilution; Cell Signaling), anti-ATF1 (rabbit IgG, 1:5,000 dilution; EPITOMICS), and anti–β-actin (mouse IgG, 1:5,000 dilution; Calbiochem).
Cell proliferation assay.
Cell growth was determined by WST-8 assay using a Cell Counting Kit-8 (Dojindo Laboratories). Absorbance at 450 nm is indicative of the die amount of formazan, which is directly proportional to the number of living cells.
Histological analysis.
Normal and tumor tissue samples were fixed in 10% buffered formalin for 24 hours and embedded in paraffin. 4-μm sections were stained with H&E, and serial sections were used for immunohistochemical analyses. Immunostaining was performed using an avidin-biotin immunoperoxidase assay. The primary antibodies used were anti–HA-Tag (1:600 dilution; Cell Signaling), anti-Ki67 (1:250 dilution; Dako), anti-S100 (1:800 dilution; Dako), anti-SOX10 (1:200 dilution; R&D Systems), anti-MITF (1:500 dilution; Exalpha), and anti-GFP (1:1,000 dilution; Abcam).
X-gal staining.
Briefly, tumor tissue samples were embedded in OCT compound and frozen. 8-μm cryostat sections were immediately fixed in 0.2% glutalaldehyde for 10 minutes. The sections were stained overnight in an X-gal staining solution, then counterstained with fast red for 3 minutes.
Tumorigenicity studies.
4-week-old male BALB/c athymic mice were obtained from Japan SLC. A total of 5.0 × 106 G1297 cells in 0.1 ml serum-free DMEM was inoculated subcutaneously through a 26-gauge needle into the posterior flank of each mouse. 3 weeks after inoculation, the tumor diameters were measured with digital calipers, and tumor volume was calculated as (w2 × l)/2 and expressed in mm3.
siRNA transfection.
We performed transient knockdown assays with a siRNA targeting Fos (Santa Cruz), FOS (Santa Cruz and Dharmacon) or the breakpoint of EWS/ATF1 type 1 (sense, GCGGUGGAAUGGGAAAAAUTT; antisense, AUUUUUCCCAUUCCACCGCTT; KOKEN) using Lipofectamine RNAiMAX (Invitrogen). We used nontargeting siRNA (Cosmo Bio Co.) as a control.
Cell lines.
MP-CCS-SY and KAS are CCS cell lines carrying EWS/ATF1 type 1 and type 2, respectively. MP-CCS-SY was established as described previously (49), and KAS was provided by T. Nakamura (Cancer Institute, Japanese Foundation for Cancer Research, Tokyo, Japan; ref. 24). HOS (osteosarcoma), U2OS (osteosarcoma), NIH3T3 (embryonic fibroblast), and WI38 (lung fibroblast) cells were purchased from the American Type Culture Collection. B16-F1 (mouse melanoma) and A375 (human MM) were purchased from the European Collection of Cell Cultures.
Mice.
Wnt1-Cre mice were provided by S. Iseki (Tokyo Medical and Dental University, Tokyo, Japan; ref. 50), P0-Cre mice were provided by K. Yamamura (Kumamoto University, Kumamoto, Japan; ref. 51), and floxed LacZ mice were provided by M. Okabe (Osaka University, Suita, Japan; ref. 52). Floxed EYFP mice (53) were obtained from Jackson Laboratory.
Construction of the reporter plasmid.
To obtain the Fos reporter plasmid (pGL3A-1486), the genomic DNA fragment containing –450 to +0 of the 5′-flanking sequence was amplified by PCR with the primer set 5′-TCTATCGATAGGTACGAATGTTCGCTCGCCTTCTC-3′ and 5′-ACGCGTAAGAGCTCGGGAGTAGTAGGCGCCTCAGC-3′ and subcloned into the Kpnl site of the pGL3 vector (Promega). The Fos reporter plasmid with mutant CRE element was generated by PCR-targeted mutagenesis with the primer set 5′-CCAGTTCCGCCCACTCAGCTAGGAAGTCCATCC-3′ and 5′-GGATGGACTTCCTAGCTGAGTGGGCGGAACTGG-3′.
Luciferase assay.
Reporter genes were transfected into the G1297 EWS/ATF1-induced tumor cell line together with phRL-SV40 (Promega) using lipofectamine LTX (Invitrogen), and luciferase activity was measured with a luminometer (VERITAS; Promega). Firefly luciferase activities, derived from each reporter construct, were normalized to Renilla luciferase activities from phRL-SV40.
Stable transfection.
To obtain Fos expression plasmid (pCAG-Fos-IZ vector), Fos cDNA was amplified by RT-PCR from the G1297 cell line with the primer set 5′-ACATGATGTTCTCGGGTTTCAA-3′ and 5′-ACTCACAGGGCCAGCAGCGTGG-3′ and subcloned into the ECoR site of the pCAG-EGFP-IZ vector (provided by H. Niwa, RIKEN, Kobe, Japan). pCAG-EGFP-IZ vector or pCAG-Fos-IZ vector was transfected into the G1297 cell line using Lipofectamine LTX Reagent (Invitrogen) and selected for Zeocine resistance (600 μg/ml).
Patients and tumor tissue collection.
Anonymized tumor specimens were obtained by surgical resection or biopsy at Gifu University Hospital or Kyoto University Hospital in accordance with an approved protocol from the Institutional Review Board. Total RNA was isolated using a RNeasy mini kit (Qiagen).
ChIP analysis.
A total of 5.0 × 106 EWS/ATF1-inducible tumor cells was fixed in 1% formaldehyde for 10 minutes, followed by treatment with 1 ml glycine buffer for 5 minutes. Cells were pelleted, washed, and then resuspended in lysis buffer for 30 minutes. After centrifugation, the pellet was resuspended in NP40 buffer with protease inhibitors (Sigma-Aldrich). Sonication was performed using a XL-2000 (MISONIX), after which the supernatant was used as the input sample for immunoprecipitation experiments. Antibodies used were rabbit HA (Cell Signaling) and rabbit normal IgG (Abcam). Protein G–coated magnetic beads were used to purify specific antibody/DNA complexes. After washes, immunoprecipitated DNA was decrosslinked by elution buffer at 65°C for 12 hours. To remove protein and RNA, samples were incubated with RNaseA for 1 hour at 37°C and proteinase K treatment for 1 hour at 37°C. Samples were purified by PCR purification kit (Qiagen). The amount of DNA immunoprecipitated with HA-tagged protein was quantified by real-time PCR with primers flanking the Fos promoter, including the CRE element (forward, 5′-TCCTACACGCGGAAGGTCTAGG-3′; reverse, 5′-TAGAAGCGCTGTGAATGGATGG-3′). Primers 5′-GCACTAATTAGTCGCGGTGGTGG-3′ (forward) and 5′-CAGGGTCTTAGTGGGATCAAGG-3′ (reverse) were used as a negative control.
Accession number.
The profiling data cited in Supplemental Figure 6 are available at GEO (accession no. GSE41123).
Statistics.
Statistical analyses were carried out using GraphPad Prism (version 5.01; GraphPad Software). Data were analyzed using ANOVA, and P values less than 0.05 were considered statistically significant.
Study approval.
Animal experiments were approved by the Gifu University Animal Experiment Committee, and the care of the animals was in accordance with institutional guidelines. All clinical samples were approved for analysis by the Ethics Committee at Kyoto University Graduate School and Faculty of Medicine (Kyoto, Japan). Written informed consent was obtained from all patients with cancers analyzed in this study.
Supplementary Material
Acknowledgments
The authors thank K. Woltjen and T. Yamamoto (CiRA, Kyoto University) for careful reading of the manuscript and helpful comments. We also thank T. Motohashi (Tissue and Organ Development Regeneration and Advanced Medical Science) for helpful discussion and the members of the Department of Orthopedic Surgery and Department of Tumor Pathology, Gifu University Graduate School of Medicine; the Department of Orthopaedic Surgery, Graduate School of Medicine, Kyoto University; and the Toguchida and Yamada laboratories for their valuable technical assistance. This study was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and from the Ministry of Health, Labor, and Welfare of Japan.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(2):600–610. doi:10.1172/JCI63572.
References
- 1.Enzinger FM. Clear-cell sarcoma of tendons and aponeuroses. An analysis of 21 cases. Cancer. 1965;18:1163–1174. doi: 10.1002/1097-0142(196509)18:9<1163::AID-CNCR2820180916>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 2.Covinsky M, Gong S, Rajaram V, Perry A, Pfeifer J. EWS-ATF1 fusion transcripts in gastrointestinal tumors previously diagnosed as malignant melanoma. Hum Pathol. 2005;36(1):74–81. doi: 10.1016/j.humpath.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 3.Deenik W, Mooi WJ, Rutgers EJ, Peterse JL, Hart AA, Kroon BB. Clear cell sarcoma (malignant melanoma) of soft parts: A clinicopathologic study of 30 cases. Cancer. 1999;86(6):969–975. doi: 10.1002/(SICI)1097-0142(19990915)86:6<969::AID-CNCR11>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 4.Ferrari A, et al. Clear cell sarcoma of tendons and aponeuroses in pediatric patients: a report from the Italian and German Soft Tissue Sarcoma Cooperative Group. Cancer. 2002;94(12):3269–3276. doi: 10.1002/cncr.10597. [DOI] [PubMed] [Google Scholar]
- 5.Finley JW, Hanypsiak B, McGrath B, Kraybill W, Gibbs JF. Clear cell sarcoma: the Roswell Park experience. J Surg Oncol. 2001;77(1):16–20. doi: 10.1002/jso.1057. [DOI] [PubMed] [Google Scholar]
- 6.Eckardt JJ, Pritchard DJ, Soule EH. Clear cell sarcoma. A clinicopathologic study of 27 cases. Cancer. 1983;52(8):1482–1488. doi: 10.1002/1097-0142(19831015)52:8<1482::AID-CNCR2820520825>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 7.Kawai A, et al. Clear cell sarcoma of tendons and aponeuroses: a study of 75 patients. Cancer. 2007;109(1):109–116. doi: 10.1002/cncr.22380. [DOI] [PubMed] [Google Scholar]
- 8.Kindblom LG, Lodding P, Angervall L. Clear-cell sarcoma of tendons and aponeuroses. An immunohistochemical and electron microscopic analysis indicating neural crest origin. Virchows Arch A Pathol Anat Histopathol. 1983;401(1):109–128. doi: 10.1007/BF00644794. [DOI] [PubMed] [Google Scholar]
- 9.Segal NH, et al. Classification of clear-cell sarcoma as a subtype of melanoma by genomic profiling. J Clin Oncol. 2003;21(9):1775–1781. doi: 10.1200/JCO.2003.10.108. [DOI] [PubMed] [Google Scholar]
- 10.Bridge JA, Borek DA, Neff JR, Huntrakoon M. Chromosomal abnormalities in clear cell sarcoma. Implications for histogenesis. Am J Clin Pathol. 1990;93(1):26–31. doi: 10.1093/ajcp/93.1.26. [DOI] [PubMed] [Google Scholar]
- 11.Bridge JA, Sreekantaiah C, Neff JR, Sandberg AA. Cytogenetic findings in clear cell sarcoma of tendons and aponeuroses. Malignant melanoma of soft parts. Cancer Genet Cytogenet. 1991;52(1):101–106. doi: 10.1016/0165-4608(91)90059-4. [DOI] [PubMed] [Google Scholar]
- 12.Sandberg AA, Bridge JA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: clear cell sarcoma (malignant melanoma of soft parts). Cancer Genet Cytogenet. 2001;130(1):1–7. doi: 10.1016/S0165-4608(01)00462-9. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, Lee K, Pelletier J. The DNA binding domains of the WT1 tumor suppressor gene product and chimeric EWS/WT1 oncoprotein are functionally distinct. Oncogene. 1998;16(8):1021–1030. doi: 10.1038/sj.onc.1201616. [DOI] [PubMed] [Google Scholar]
- 14.Lessnick SL, Braun BS, Denny CT, May WA. Multiple domains mediate transformation by the Ewing’s sarcoma EWS/FLI-1 fusion gene. Oncogene. 1995;10(3):423–431. [PubMed] [Google Scholar]
- 15.Pan S, Ming KY, Dunn TA, Li KK, Lee KA. The EWS/ATF1 fusion protein contains a dispersed activation domain that functions directly. Oncogene. 1998;16(12):1625–1631. doi: 10.1038/sj.onc.1201671. [DOI] [PubMed] [Google Scholar]
- 16.Ohno T, Ouchida M, Lee L, Gatalica Z, Rao VN, Reddy ES. The EWS gene, involved in Ewing family of tumors, malignant melanoma of soft parts and desmoplastic small round cell tumors, codes for an RNA binding protein with novel regulatory domains. Oncogene. 1994;9(10):3087–3097. [PubMed] [Google Scholar]
- 17.Petermann R, Mossier BM, Aryee DN, Khazak V, Golemis EA, Kovar H. Oncogenic EWS-Fli1 interacts with hsRPB7, a subunit of human RNA polymerase II. Oncogene. 1998;17(5):603–610. doi: 10.1038/sj.onc.1201964. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59(4):675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 19.Comb M, Birnberg NC, Seasholtz A, Herbert E, Goodman HM. A cyclic AMP- and phorbol ester-inducible DNA element. Nature. 1986;323(6086):353–356. doi: 10.1038/323353a0. [DOI] [PubMed] [Google Scholar]
- 20.Montminy MR, Sevarino KA, Wagner JA, Mandel G, Goodman RH. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc Natl Acad Sci U S A. 1986;83(18):6682–6686. doi: 10.1073/pnas.83.18.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li KK, Lee KA. MMSP tumor cells expressing the EWS/ATF1 oncogene do not support cAMP-inducible transcription. Oncogene. 1998;16(10):1325–1331. doi: 10.1038/sj.onc.1201649. [DOI] [PubMed] [Google Scholar]
- 22.Brown AD, Lopez-Terrada D, Denny C, Lee KA. Promoters containing ATF-binding sites are de-regulated in cells that express the EWS/ATF1 oncogene. Oncogene. 1995;10(9):1749–1756. [PubMed] [Google Scholar]
- 23.Fujimura Y, Ohno T, Siddique H, Lee L, Rao VN, Reddy ES. The EWS-ATF-1 gene involved in malignant melanoma of soft parts with t(12;22) chromosome translocation, encodes a constitutive transcriptional activator. Oncogene. 1996;12(1):159–167. [PubMed] [Google Scholar]
- 24.Jishage M, Fujino T, Yamazaki Y, Kuroda H, Nakamura T. Identification of target genes for EWS/ATF-1 chimeric transcription factor. Oncogene. 2003;22(1):41–49. doi: 10.1038/sj.onc.1206074. [DOI] [PubMed] [Google Scholar]
- 25.Davis IJ, et al. Oncogenic MITF dysregulation in clear cell sarcoma: defining the MiT family of human cancers. Cancer Cell. 2006;9(6):473–484. doi: 10.1016/j.ccr.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 26.Antonescu CR, Tschernyavsky SJ, Woodruff JM, Jungbluth AA, Brennan MF, Ladanyi M. Molecular diagnosis of clear cell sarcoma: detection of EWS-ATF1 and MITF-M transcripts and histopathological and ultrastructural analysis of 12 cases. J Mol Diagn. 2002;4(1):44–52. doi: 10.1016/S1525-1578(10)60679-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Granter SR, Weilbaecher KN, Quigley C, Fletcher CD, Fisher DE. Clear cell sarcoma shows immunoreactivity for microphthalmia transcription factor: further evidence for melanocytic differentiation. Mod Pathol. 2001;14(1):6–9. doi: 10.1038/modpathol.3880249. [DOI] [PubMed] [Google Scholar]
- 28.Li KK, et al. The melanocyte inducing factor MITF is stably expressed in cell lines from human clear cell sarcoma. Br J Cancer. 2003;89(6):1072–1078. doi: 10.1038/sj.bjc.6601212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12(9):406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 30.Garraway LA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436(7047):117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 31.Speleman F, Delattre O, Peter M, Hauben E, Van Roy N, Van Marck E. Malignant melanoma of the soft parts (clear-cell sarcoma): confirmation of EWS and ATF-1 gene fusion caused by a t(12;22) translocation. Mod Pathol. 1997;10(5):496–499. [PubMed] [Google Scholar]
- 32.Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127(8):1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- 33.Seternes OM, Sorensen R, Johansen B, Loennechen T, Aarbakke J, Moens U. Synergistic increase in c-fos expression by simultaneous activation of the ras/raf/map kinase- and protein kinase A signaling pathways is mediated by the c-fos AP-1 and SRE sites. Biochim Biophys Acta. 1998;1395(3):345–360. doi: 10.1016/s0167-4781(97)00189-9. [DOI] [PubMed] [Google Scholar]
- 34.Ginty DD, Bonni A, Greenberg ME. Nerve growth factor activates a Ras-dependent protein kinase that stimulates c-fos transcription via phosphorylation of CREB. Cell. 1994;77(5):713–725. doi: 10.1016/0092-8674(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 35.Sassone-Corsi P, Visvader J, Ferland L, Mellon PL, Verma IM. Induction of proto-oncogene fos transcription through the adenylate cyclase pathway: characterization of a cAMP-responsive element. Genes Dev. 1988;2(12A):1529–1538. doi: 10.1101/gad.2.12a.1529. [DOI] [PubMed] [Google Scholar]
- 36.Weinstein IB. Cancer. Addiction to oncogenes — the Achilles heal of cancer. Science. 2002;297(5578):63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 37.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44(1):23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 38.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121(3):465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 39.Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 2007;11(4):375–388. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 40.Riggi N, et al. Development of Ewing’s sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Res. 2005;65(24):11459–11468. doi: 10.1158/0008-5472.CAN-05-1696. [DOI] [PubMed] [Google Scholar]
- 41.Adameyko I, et al. Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell. 2009;139(2):366–379. doi: 10.1016/j.cell.2009.07.049. [DOI] [PubMed] [Google Scholar]
- 42.Guller M, et al. c-Fos overexpression increases the proliferation of human hepatocytes by stabilizing nuclear Cyclin D1. World J Gastroenterol. 2008;14(41):6339–6346. doi: 10.3748/wjg.14.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandey MK, Liu G, Cooper TK, Mulder KM. Knockdown of c-Fos suppresses the growth of human colon carcinoma cells in athymic mice. Int J Cancer. 2012;130(1):213–222. doi: 10.1002/ijc.25997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saez E, et al. c-fos is required for malignant progression of skin tumors. Cell. 1995;82(5):721–732. doi: 10.1016/0092-8674(95)90469-7. [DOI] [PubMed] [Google Scholar]
- 45.Grigoriadis AE, Schellander K, Wang ZQ, Wagner EF. Osteoblasts are target cells for transformation in c-fos transgenic mice. J Cell Biol. 1993;122(3):685–701. doi: 10.1083/jcb.122.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang ZQ, Grigoriadis AE, Mohle-Steinlein U, Wagner EF. A novel target cell for c-fos-induced oncogenesis: development of chondrogenic tumours in embryonic stem cell chimeras. EMBO J. 1991;10(9):2437–2450. doi: 10.1002/j.1460-2075.1991.tb07783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nozawa S, et al. Inhibition of platelet-derived growth factor-induced cell growth signaling by a short interfering RNA for EWS-Fli1 via down-regulation of phospholipase D2 in Ewing sarcoma cells. J Biol Chem. 2005;280(30):27544–27551. doi: 10.1074/jbc.M411626200. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto T, et al. Simultaneous inhibition of mitogen-activated protein kinase and phosphatidylinositol 3-kinase pathways augment the sensitivity to actinomycin D in Ewing sarcoma. J Cancer Res Clin Oncol. 2009;135(8):1125–1136. doi: 10.1007/s00432-009-0554-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moritake H, et al. Newly established clear cell sarcoma (malignant melanoma of soft parts) cell line expressing melanoma-associated Melan-A antigen and overexpressing C-MYC oncogene. Cancer Genet Cytogenet. 2002;135(1):48–56. doi: 10.1016/S0165-4608(01)00641-0. [DOI] [PubMed] [Google Scholar]
- 50.Chai Y, et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127(8):1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- 51.Yamauchi Y, et al. A novel transgenic technique that allows specific marking of the neural crest cell lineage in mice. Dev Biol. 1999;212(1):191–203. doi: 10.1006/dbio.1999.9323. [DOI] [PubMed] [Google Scholar]
- 52.Sakai K, Miyazaki J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun. 1997;237(2):318–324. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- 53.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.