Abstract
Mice with a DC-specific deletion of the transcriptional repressor B lymphocyte–induced maturation protein-1 (Blimp1) exhibit a lupus-like phenotype, secondary to enhanced DC production of IL-6. Here we explored further phenotypic changes in Blimp1-deficient DCs, the molecular mechanism underlying these changes, and their relevance to human disease. Blimp1-deficient DCs exhibited elevated expression of MHC II, and exposure to TLR agonists increased secretion of proinflammatory cytokines. This phenotype reflects enhanced expression of the microRNA let-7c, which is regulated by BLIMP1. Let-7c reciprocally inhibited Blimp1 and also blocked LPS-induced suppressor of cytokine signaling-1 (SOCS1) expression, contributing to the proinflammatory phenotype of Blimp1-deficient DCs. DCs from Blimp1 SLE-risk allele carriers exhibited analogous phenotypic changes, including decreased BLIMP1 expression, increased let-7c expression, and increased expression of proinflammatory cytokines. These results suggest that let-7c regulates DC phenotype and confirm the importance of BLIMP1 in maintaining tolerogenic DCs in both mice and humans.
Introduction
DCs have long been recognized as the key APCs (1). DCs function at the interface of innate and adaptive immune responses and are distributed widely throughout the body. DC precursor cells derive from bone marrow (2), but DCs can also be differentiated from monocytes under inflammatory conditions and migrate into lymphoid tissues (3). The broad functional capabilities of DCs include the induction of antigen-specific immunity against pathogens as well as the maintenance of immune tolerance to self antigens. Depletion of the total DC population or specific alterations of DC development result in severe immunodeficiency and enhanced susceptibility to infections in humans (4), and the spontaneous development of autoimmunity in mice (5).
B lymphocyte–induced maturation protein-1 (Blimp1) is an important transcriptional repressor in B cells (6) and T cells (7). Expression of Blimp1 can be induced by various stimuli including engagement of TLRs (8, 9), B cell receptors (10), or T cell receptors (11). Its expression influences differentiation and functional homeostasis in B cells and T cells, respectively. Despite its broad range of regulatory functions, a limited number of genes directly repressed by Blimp1 have been identified. These include Ifna, c-Myc (13), Ciita (14), Pax5 (15) in B lymphocytes, Il2 in T lymphocytes (16), and Il6 in DCs (17). Polymorphisms in Blimp1 have been associated with risk for human autoimmune disorders such as SLE (18) and inflammatory bowel disease (19), although the mechanisms underlying these genetic associations have not been established.
Mice carrying a DC-specific deletion of Blimp1 develop a lupus-like phenotype with autoantibodies and glomerular inflammation (20). We now demonstrate a broadly activated phenotype in DCs from these mice, and provide evidence that a specific increase in let-7c microRNA (miRNA) is responsible for the change in DC phenotype. We show that let-7c is a target of Blimp1, and Blimp1 is reciprocally a target of let-7c in a DC-specific manner. Moreover, we found that let-7c regulates the level of suppressor of cytokine signaling-1 (SOCS1), which is induced by TLR stimulation in DCs. We propose that Blimp1 deficiency allows for increased levels of let-7c, which results in a proinflammatory DC phenotype, mediated in part through suppression of SOCS1. We also demonstrate a similar regulatory mechanism in human DCs derived from healthy donors carrying an SLE-risk allele of the Blimp1 gene. Thus, Blimp1 and let-7c reciprocally regulate DC activation, and a quantitative alteration in this relationship may be involved in autoimmune phenotypes.
Results
Activated phenotype and increased let-7c in Blimp1-deficient DCs.
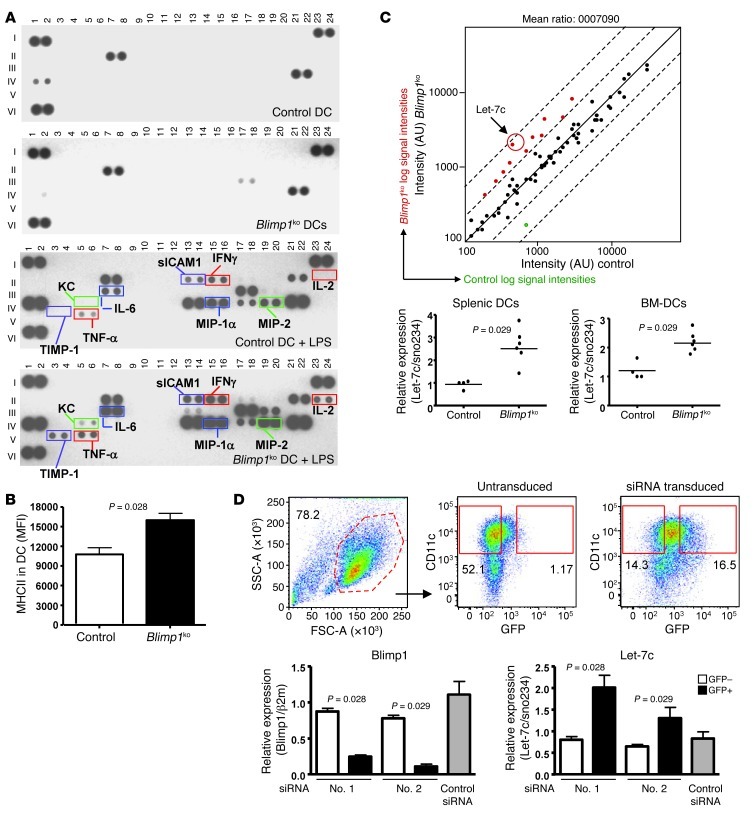
Mice with a DC-specific deletion of Blimp1 (referred to herein as DCBlimp1ko) display a lupus-like phenotype that is characterized by increased DC production of IL-6, leading to an increased number of T follicular helper cells and enhanced germinal center formation (20). The anti-DNA response present in these mice is generated within the germinal center. We asked whether the DCs exhibited additional phenotypic changes beyond increased IL-6. We performed protein arrays to characterize the phenotype of Blimp1-deficient DCs. Purified CD11chi conventional DCs (cDCs) from the spleens of control or DCBlimp1ko mice were cultured overnight in the absence or presence of LPS. Both control and Blimp1-deficient spleen cDCs secrete a very limited number of cytokines or chemokines in the absence of stimulation (Figure 1A and Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI64712DS1). LPS induced a substantial increase in both cytokine and chemokine secretion. Many cytokines were highly secreted by Blimp1-deficient DCs compared with control DCs, including IL-6, as previously shown, IFN-γ, and TNF-α. We confirmed increased secretion of these cytokines using a Meso Scale Discovery (MSD) assay (Supplemental Figure 1).
Figure 1. Proinflammatory phenotype and increased let-7c in Blimp1-deficient DCs.
(A) Cytokines and chemokines were measured by protein array in supernatant from an overnight culture of splenic DCs purified from control or DCBlimp1ko mice in the presence or absence of LPS (1 μg/ml). Legend of individual proteins is provided in Supplemental Table 1. A pool of 2 mice was used for one blot. Representative pictures of 3 independent experiments. (B) Surface expression of MHC II was measured by flow cytometry and MFI was plotted. (Mean ± SEM of 4 independent experiments) (n = 8). (C) miRNA array was performed with RNAs from sorted splenic DCs from control and DCBlimp1ko mice. A representative plot from 2 independent arrays. Pool of 6 mice were used per group and per array. Expression of let-7c was measured by qPCR and relative expression level was calculated (right panel). Relative expression was calculated based on sno234 small RNA. Each dot represents an individual mouse, and the bar represents the mean. (D) BM-DCs were infected with lentivirus encoding either anti-Blimp1 or control siRNA with a GFP reporter gene. siRNA-transduced BM-DCs were sorted by expression of GFP, and total RNA was prepared. Expression of Blimp1 and let-7c was measured by qPCR, and relative expression was normalized to indicated housekeeping genes. Flow cytometry images for sorting are representative images from 3 independent experiments (upper panel). Graph depicts the mean ± SEM of 3 independent experiments (n = 6).
Together with cytokine expression, surface expression of costimulatory molecules and MHC II are critical for the function of DCs as initiators of adaptive immune responses. We compared the expression pattern of a panel of costimulatory molecules, TLRs, CD40, CD80, CD86, CD273, and CD275 on Blimp1-deficient DCs and control DCs by flow cytometry. Expression of TLR4 and TLR7, but not TLR3 or TLR9, was higher in Blimp1-deficient DCs compared with the level observed in control DCs (Supplemental Figure 2A). There were no significant differences in either percentage of positive cells or MFI in expression of surface costimulatory molecules (Supplemental Figure 2B), even after stimulation with LPS (data not shown). However, expression of MHC II was constitutively increased in Blimp1-deficient DCs compared with control DCs (Figure 1B). This may be due to a known ability of Blimp1 to negatively regulate CIITA, a major transcription factor for MHC II gene expression in B cells (14).
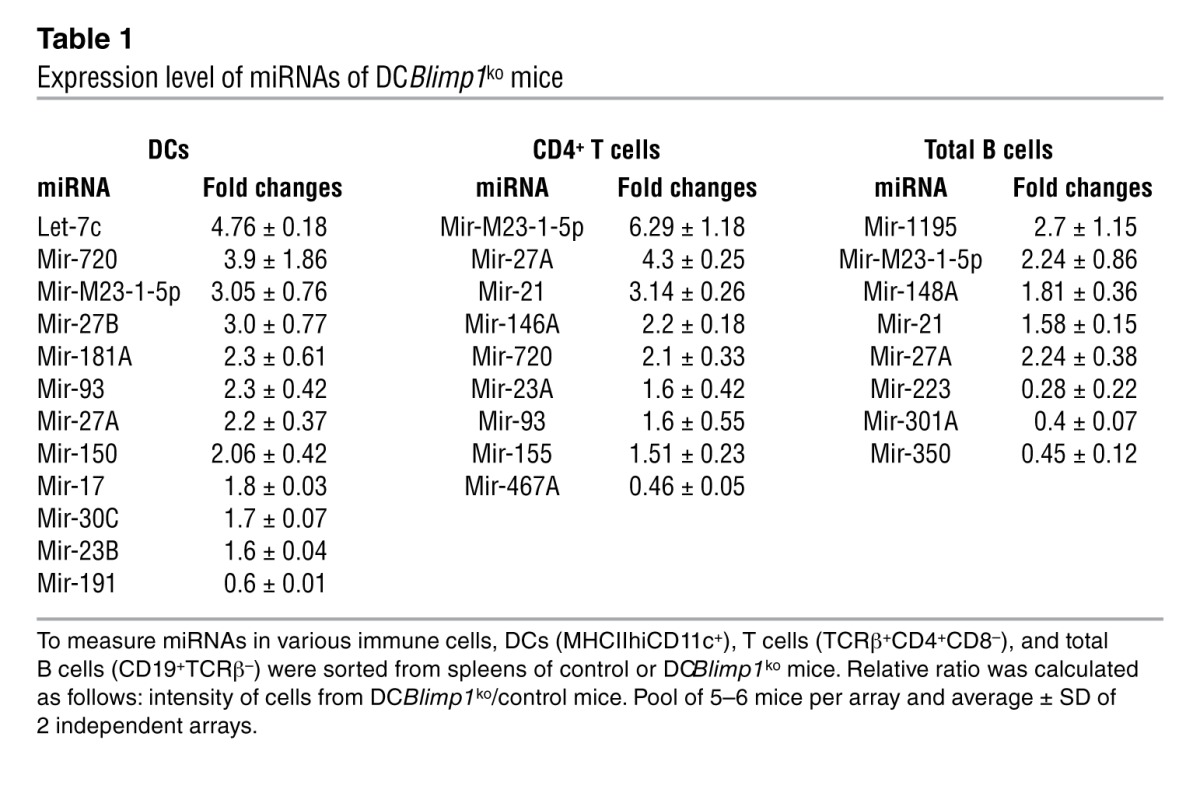
To further understand the regulatory mechanisms underlying the proinflammatory phenotype of Blimp1-deficient DCs, we compared miRNA expression in Blimp1-deficient and control DCs. A microarray of miRNA identified 71 miRNAs expressed in DCs from a total of 661 miRNAs on the chip (~10%). Of these, 11 miRNAs were increased and 1 was decreased in Blimp1-deficient DCs compared with control DCs (Figure 1C and Table 1). While some miRNAs exhibited altered expression in T and B lymphocytes as well as DCs of DCBlimp1ko mice (miR-720, MCMV-miR-M23-1-5P, miR-93, and miR-27A) (Table 1), let-7c was the most highly increased miRNA and was exclusively altered in DCs. We therefore focused on let-7c as a candidate regulatory miRNA for DC activation.
Table 1.
Expression level of miRNAs of DCBlimp1ko mice
To confirm the array results through a quantitative method, we compared the expression level of let-7c in Blimp1-deficient DCs by qPCR. There was an increased expression of let-7c in both Blimp1-deficient splenic DCs and bone marrow–derived DCs (BM-DCs) (Figure 1C). To determine whether the loss of Blimp1 was directly responsible for the increase in let-7c, we inhibited the expression of Blimp1 by siRNA in DCs. Since the reporter gene GFP is expressed concomitantly with the siRNA, we could identify cells expressing siRNA by GFP expression. Compared with the GFP– or control DCs, GFP+ DCs transduced with siRNAs #1 and #2, which target the 5′ end of the Blimp1 gene, exhibited a significantly decreased level of Blimp1 mRNA. Interestingly, we observed an increase in the level of let-7c when Blimp1 expression was reduced (Figure 1D).
Direct binding of Blimp1 to the let-7c regulatory region.
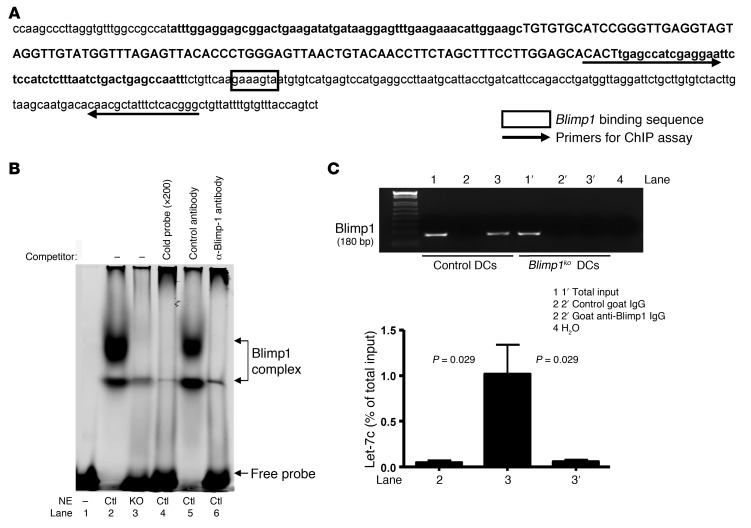
Since Blimp1 is a known transcriptional repressor, we hypothesized that it might directly downregulate expression of let-7c. To test this hypothesis, we searched the sequence around the let-7c gene for the Blimp1 consensus binding site, A/CAGT/CGAAAGT/CG/T (21). An AAGAAAGTA sequence is present immediately downstream of the 3′ terminus of the let-7c gene (Figure 2A). We performed EMSA to determine whether Blimp1 binds to this putative target sequence. Nuclear extract was prepared from LPS-stimulated BM-DCs, which exhibit high expression of Blimp1. The extract bound to an oligonucleotide probe containing the putative Blimp1 binding site, generating 2 bands (Figure 2B, lane 2). These bands were not present when the probe was incubated with nuclear extract from Blimp1-deficient DCs or when unlabeled probe was added to the extract (Figure 2B, lanes 3 and 4, respectively). Moreover, anti–Blimp1 antibodies, but not control antibodies, specifically inhibited the binding of nuclear extracts to the target oligonucleotide, confirming that Blimp1 is present in the binding complex (Figure 2B, lane 6).
Figure 2. Blimp1 binds to let-7c regulatory region in vitro and in vivo.
(A) Diagram of a mouse genome sequence encompassing let-7c. Premature let-7c gene encompassing mature miRNAs (in upper case) is in bold, and a putative Blimp1 binding sequence is boxed. Arrows indicate primers for the ChIP assay. (B) EMSA was performed with nuclear extracts (NE) prepared from BM-DCs that were stimulated with LPS for 24 hours. In vitro binding of proteins and oligos was performed under various conditions, and the reaction mixtures were separated on a 4% acrylamide gel. Images were scanned and analyzed by Odyssey (LI-COR Biosciences). (C) ChIP assay was performed with LPS-stimulated BM-DCs generated from control or DCBlimp1ko mice. Total input and elutions after immunoprecipitation with either control IgG or anti–Blimp1 IgG were diluted, and PCR was performed with primers as depicted in (A). A representative image of 4 independent experiments is shown. Quantitative PCR was performed and the amount compared with total input was calculated using mean ± SEM of 4 independent experiments.
To determine whether Blimp1 could interact with the endogenous, chromosomal let-7c gene, we performed a ChIP assay in LPS-stimulated BM-DCs using polyclonal anti–Blimp1 antibodies and PCR primers that detect a genomic sequence close to the let-7c gene, as indicated in Figure 2A. Figure 2C shows that Blimp1-specific binding was detected in control DCs (lane 3), but not in Blimp1-deficient DCs (lane 3′). Approximately 1% of let-7c genomic DNA was bound by Blimp1 (Figure 2C, graph). These in vitro and in vivo results demonstrate unequivocally that the Blimp1 transcription factor can recognize a cognate binding site on the let-7c gene.
Let-7c reciprocally downregulates Blimp1.
While these data suggest transcriptional regulation of let-7c by Blimp1 binding, we noted that the 3′ untranslated region (UTR) of Blimp1 also contains a putative target sequence of let-7c based on the database of miRNA analysis (Sanger Center miRNA Registry). We therefore sought to provide evidence for a reciprocal regulatory interaction. Let-7c miRNA or control miRNA was cotransfected with a luciferase reporter gene containing the Blimp1 3′ UTR, Luc-3′UTR/Blimp1, into the 293 cell line. There was a let-7c miRNA–mediated suppression of luciferase expression (Supplemental Figure 3), demonstrating that let-7c targets the 3′ UTR of Blimp1.
Suppression of Socs1 expression in Blimp1-deficient DCs.
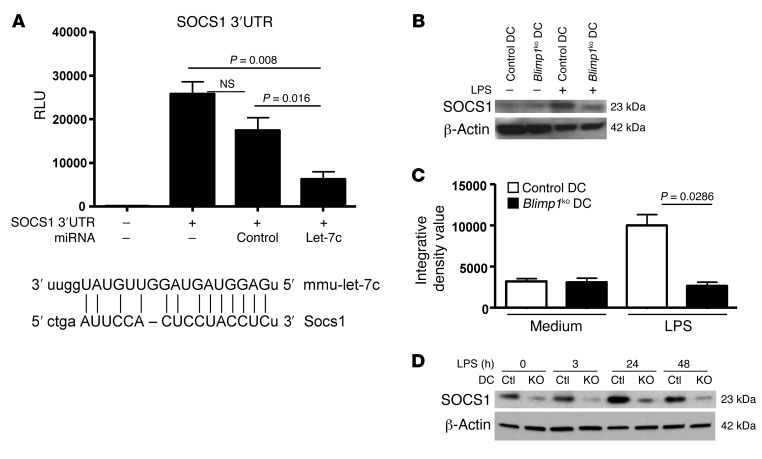
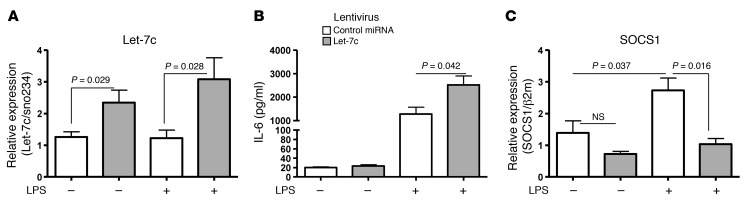
To understand the functional significance of increased let-7c in Blimp1-deficient DCs, we examined the expression of putative target genes of this miRNA. Let-7c has a large number of putative target mRNAs such as c-Myc, Fasl, Ccr7, and Socs1. We focused on Socs1, as it is a cytokine-inducible negative regulator of the Jak-Stat pathway (22), and mediates an important negative feedback loop to attenuate LPS-mediated immune cell activation (23). Socs1 has also been shown to be a key molecule for the regulation of DC maturation and activation (24). First, we examined whether Socs1 is a direct target of let-7c. Let-7c–mediated regulation of Socs1 expression was measured by a luciferase reporter assay. There was a slight, but statistically insignificant, decrease in the activity of a Socs1 luciferase vector by control miRNA transfection. Compared with control miRNA transfection, however, let-7c significantly inhibited its expression, as predicted by target sequence alignment with the 3′ UTR of Socs1 (Figure 3A). Next, we wanted to determine whether there was differential induction of Socs1 in control DCs and Blimp1-deficient DCs following stimulation with LPS. DCs purified from spleens of both control mice and DCBlimp1ko mice showed a minimal level of Socs1 in the resting state. When DCs were stimulated with LPS, Socs1 expression was significantly induced in control DCs, but not in Blimp1-deficient DCs (Figure 3, B and C). This result was confirmed in BM-DCs. Expression of Socs1 achieved a maximum level at 24 hours following LPS stimulation and was downregulated at 48 hours in control DCs. In contrast, Blimp1-deficient DCs showed only minimal induction of Socs1 by LPS at any time up to 48 hours (Figure 3D). These data demonstrate that expression of Socs1 is compromised in Blimp1-deficient DCs. Suppression of Socs1 by let-7c is the probable mechanism for the activated phenotype of Blimp1-deficient DCs, at least after LPS stimulation.
Figure 3. Regulation of SOCS1 by let-7c in DCs and defective induction in Blimp1-deficient DCs.
(A) Regulation of expression of SOCS1 mediated by let-7c was measured by luciferase assay. One microgram of SOCS1/luciferase vector alone or with 1.4 μg of either let-7c or control miRNA-expressing vectors was transfected into the 293 cell line. Luciferase activity was measure at 48 hours after transfection by luminometer. The graph represents the mean ± SEM of 3 independent experiments. Induction of SOCS1 from splenic DCs (B) or BM-DCs (D) was measured by Western blotting. (C) Quantification of SOCS1 bands from the data of splenic DCs in (B) was performed by densitometry, and the graph depicts the mean ± SEM of 3 independent experiments. DCs were purified and stimulated with LPS for 24 hours (splenic DCs) or for various durations (BM-DCs). A proteosomal inhibitor, MG-132 (25 μM), was added 3 hours before harvest to prevent degradation of SOCS1. A representative image of 3 independent experiments is shown.
Reconstitution of Blimp1 in Blimp1-deficient DCs reverses inflammatory phenotypes.
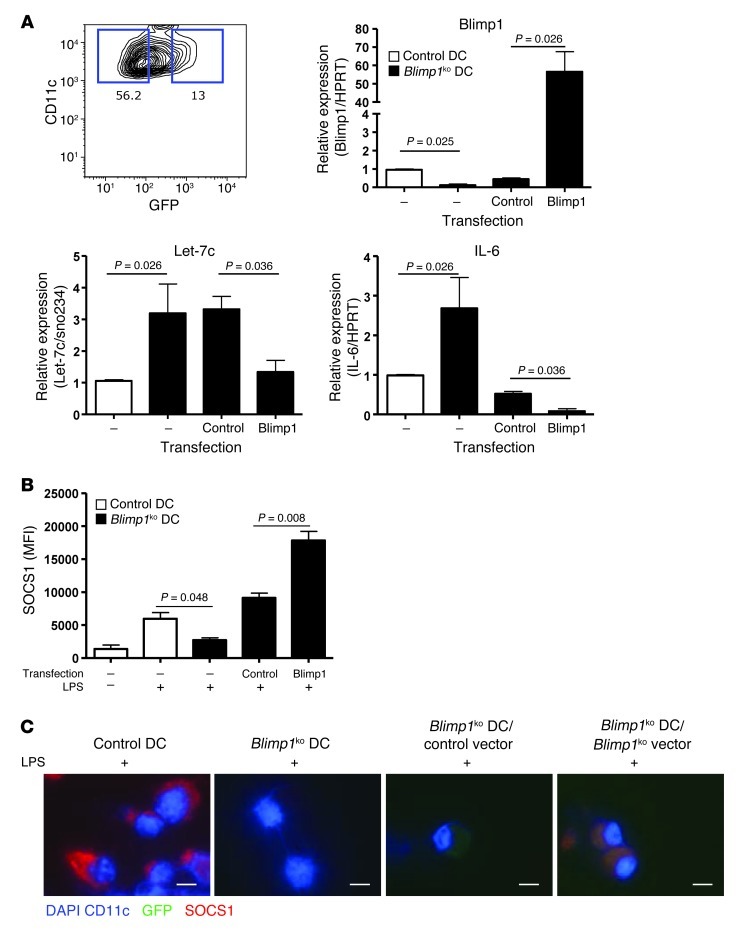
Although we demonstrated that Blimp1 deficiency is responsible for the increase in let-7c in DCs using a Blimp1-targeting siRNA, we wished to show that reconstitution of Blimp1 expression in Blimp1-deficient DCs would diminish let-7c expression and the associated proinflammatory phenotype. Since splenic DCs have a short half-life, all experiments were performed with BM-DCs. A Blimp1 open-reading frame without the 3′ UTR region was cloned into a lentiviral vector to exclude possible regulatory effects by the 3′ UTR. Lentivirus containing either Blimp1 or an irrelevant gene, firefly luciferase (control), was transduced into DC progenitor cells, and DCs were further differentiated for 4 days in vitro. CD11c+ and GFP+ cells, which express Blimp1, or control gene were isolated, and their gene expression pattern was analyzed. An increase in Blimp1 expression led to a significant downregulation of let-7c, suggesting that BLIMP1 protein is sufficient to inhibit let-7c expression in DCs, even in the absence of the 3′ UTR. As expected, IL-6, a known direct target of Blimp1, was strongly downregulated (Figure 4A).
Figure 4. Blimp1-dependent SOCS1 expression in DCs.
(A) Blimp1-deficient BM-DCs were infected with Blimp1 or control gene-expressing lentivirus as described in Methods. The GFP+ or GFP– CD11c+ population (representative flow image is in the upper left panel) was sorted, and expression of Blimp1, let-7c, and IL-6 was measured by qPCR. Data represent the mean ± SEM of 3 independent experiments, with a total of 6 per group. (B and C) LPS-mediated SOCS1 induction was measured from control DCs, Blimp1-deficient DCs, Blimp1 or control gene–transduced Blimp1-deficient DCs by flow cytometry (B) and immunofluorescence (IF) (C). Bar graph represents the mean ± SEM of 3 independent experiments. For IF, BM-DCs were cultured in a chamber slide. IF images were taken using a Zeiss microscope at original magnification, ×100 (scale bar: 15 μm). (Single color images were presented in Supplemental Figure 5.)
We also analyzed the expression of SOCS1 in Blimp1–reconstituted Blimp1–deficient DCs. There was a nonstatistically significant trend toward an increase in Socs1 mRNA in Blimp1–reconstituted DCs prior to LPS stimulation (Supplemental Figure 4A). However, when control lentivirus–transduced BM-DCs were stimulated with LPS, we observed a significant increase in SOCS1 protein in the control (Figure 4B). Blimp1–transduced Blimp1–deficient DCs exhibited higher expression of SOCS1 than hLuc-transduced Blimp1-deficient DCs after LPS stimulation (Figure 4, B and C, and Supplemental Figure 5). These differences in the level of SOCS1 were not observed in GFP– fractions (Supplemental Figure 4B), demonstrating a Blimp1-mediated effect on SOCS1 expression.
In these experiments, we reconstituted Blimp1 in Blimp1-deficient DCs. When Blimp1 expression was restored, the level of let-7c was downregulated. Decreased let-7c permitted a proper induction of SOCS1 following LPS stimulation, demonstrating that Blimp1 is a major component of the let-7c-SOCS1–regulated cytokine response in DCs. Moreover, by removing the 3′ UTR in the reconstitution construct, we demonstrated that the BLIMP1 protein, and not the 3′ UTR, is the major moiety responsible for downregulation of let-7c.
Overexpression of let-7c in control BM-DCs increases IL-6 expression.
Having demonstrated the importance of BLIMP1 expression in regulating let-7c levels in DCs, we asked whether changes in let-7c, regardless of BLIMP1, directly induce phenotypic changes in DCs. We therefore increased the level of let-7c in BM-DCs derived from control mice by transduction of let-7c–encoding lentivirus. Figure 5A shows that the level of let-7c was significantly increased in let-7c lentivirus–transduced DCs. Of note, even though expression of let-7c is controlled by a CMV promoter in the lentivirus, there was only about a 2-fold change compared with control miRNA–transduced DCs. This level of let-7c is similar to that present in Blimp1-deficient DCs.
Figure 5. Alteration of SOCS1 and IL-6 expression by overexpression of let-7c in DCs.
(A) Lentivirus carrying either control miRNA or let-7c miRNA was transduced into BM-DCs prepared from control mice. GFP+-transduced DCs were sorted, and the level of let-7c was measured by qPCR. (B) miRNA-transduced BM-DCs were cultured with or without LPS (1 μg/ml) overnight. Supernatants were harvested and IL-6 was measured by ELISA. (C) Level of SOCS1 was measured by qPCR from control miRNA or let-7c miRNA–transduced BM-DCs. Relative expression was calculated compared with β2m. Open bar represents control miRNA, and gray bar represents let-7c–transduced BM-DCs. Values are expressed as mean ± SEM, n = 4 per group.
To assess whether increased let-7c has a functional consequence, we measured IL-6 expression and SOCS1 induction in let-7c or control miRNA–transduced DCs. There was a significant increase in the level of IL-6 following LPS stimulation in let-7c–transduced DCs (Figure 5B). This probably reflects the failure of SOCS1 induction due to a high level of let-7c (Figure 5C). These data demonstrate that an increased level of let-7c might be a major mechanism in regulating SOCS1 induction, and consequently, proinflammatory cytokine production in DCs.
DCs from SLE-risk allele carriers display an activated phenotype.
Genome-wide association studies (GWAS) have shown that Blimp1 contains risk alleles for autoimmune disease, and the SNP rs548234 CC is a SLE-risk allele (18, 25). We therefore asked whether the phenotype of DCs from individuals carrying the Blimp1 SLE-risk allele was similar to that of mouse DCs with a Blimp1 deletion. To avoid the confounding issues of medication and disease activity, we obtained cells from healthy individuals with or without the SLE-risk allele and from cord blood samples.
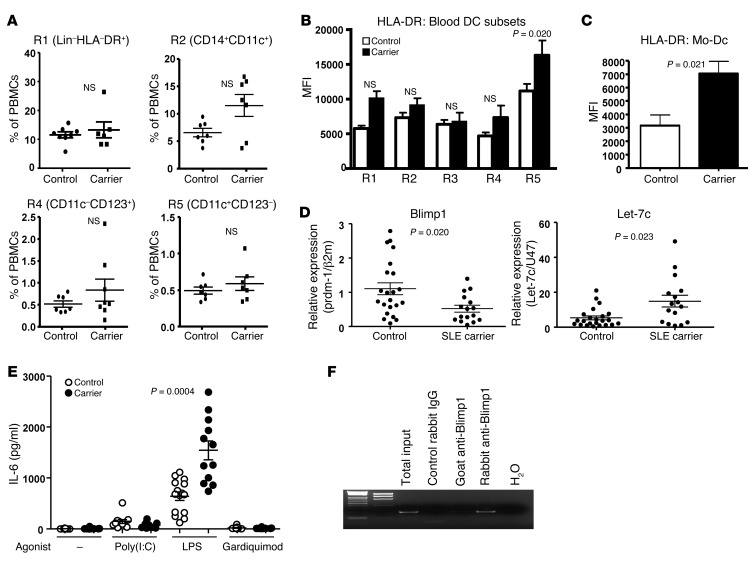
First, we compared the blood DC subsets within total PBMCs by flow cytometry. There was no significant deficit in the number of monocytes or 2 major DC populations in SLE Blimp1–risk allele carriers (Figure 6A; see Supplemental Figure 6A for the gating strategy). There was a comparable percentage of monocyte-derived DCs (Mo-DCs) differentiated with GM-CSF and IL-4 in risk allele carriers and control individuals (Supplemental Figure 6B). Consistent with the phenotype of mouse Blimp1-deficient DCs, HLA-DR expression was significantly increased in a classical DC subset (CD11c+CD123–) and in Mo-DCs of SLE-risk allele carriers (Figure 6, B and C). This increase was not observed in CD14+ or CD14–CD16+ monocytes, suggesting a DC-specific alteration. Interestingly, there was a significant decrease in Blimp1 mRNA and a 2-fold increase in let-7c miRNA in Mo-DCs from SLE-risk allele carriers compared with control Mo-DCs (Figure 6D). The level of Blimp1 and let-7c expression was lower in Mo-DCs differentiated from cells from cord blood than in those from adult blood, but the relationship between control and risk allele expression of Blimp1 and let-7c was the same in both cord and adult blood (Supplemental Figure 7). This also appears to be a DC-specific change, since purified B lymphocytes showed no statistically significant change in either Blimp1 or let-7c mRNA in risk allele carriers (Supplemental Figure 8). We also compared cytokine production in Mo-DCs from homozygous SLE-risk allele carriers compared with homozygous non-risk allele individuals. SLE-risk allele carriers secreted an increased amount of IL-6 following LPS stimulation (Figure 6E and Supplemental Figure 7 for separate analysis in cord and adult blood), but no significant increase in secretion of TNF-α and IFN-γ (Supplemental Figure 9). Interestingly, there is a putative Blimp1 binding sequence upstream of let-7c in humans. We therefore performed a ChIP assay, which showed that human Blimp1 bound to human let-7c (Figure 6F). These data suggest that there might be common pathways regulated by Blimp1 in DCs of both humans and mice.
Figure 6. Proinflammatory phenotype in DCs from Blimp1 SLE-risk allele individuals.
(A) Quantification of each blood monocyte per DC subset. Percentage of monocytes and cDCs and plasmacytoid DCs (pDCs) was analyzed and calculated based on the number of total PBMCs. Each dot represents an individual, and the bar represents the mean ± SEM. Expression of HLA-DR on each cell type (B) and Mo-DCs (C) were measured from each population (R1 = Lin–HLA+, R2 = CD14+CD11c+, R3 = CD16+CD11c+, R4 = CD14–CD16–CD11c–CD123hi, and R5 = CD14–CD16–CD11c+CD123–, and flow cytometry images are described in Supplemental Figure 6A) and compared by MFI. Values are expressed as the mean ± SEM; n = 8 for control and n = 7 for carrier (open bar represents controls and closed bar represents Blimp1 SLE carriers). (D) Blimp1 and let-7c expression was measured from Mo-DCs by qPCR. Each dot represents an individual, and the bar represents the mean ± SEM. (E) Cytokine expression was measured from Mo-DCs with or without TLR agonists (poly [I:C], 5 μg/ml; LPS, 1 μg/ml; and Gardiquimod, 2 μg/ml) overnight. IL-6 in the supernatants was measured by MSD assay. Open circle denotes a control individual, and closed circle denotes a risk carrier individual. Each dot represents an individual, and the bar represents the mean ± SEM. (F) Blimp1 binding to let-7c was assessed by ChIP assay. Nuclear extracts were prepared from 107 human Mo-DCs stimulated with LPS and incubated with control rabbit IgG, control goat IgG (not shown), Goat anti–Blimp1, or Rabbit anti–Blimp1 antibodies. PCR was performed with the primers described in Methods. Representative image is from 3 independent experiments.
Discussion
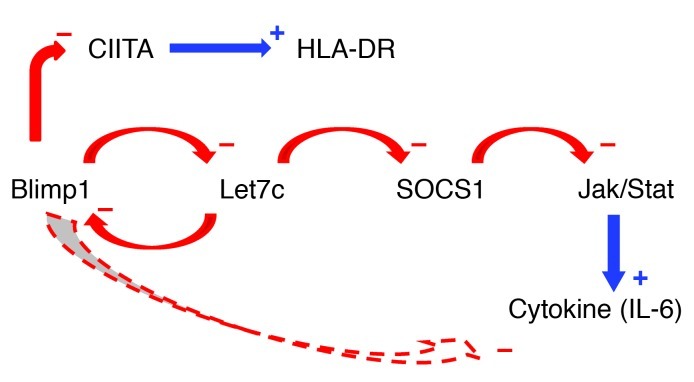
In this study, we identified a novel function of Blimp1 as a transcriptional repressor of let-7c miRNA. Blimp1 binds to the let-7c regulatory region and influences the level of let-7c in DCs. In the absence of Blimp1, an increase in let-7 miRNA results in a broad spectrum of proinflammatory features in DCs, and we provide evidence that this is mediated, in part, through suppression of SOCS1 expression. We have summarized the regulatory interactions among those molecules in Figure 7.
Figure 7. Proposed regulatory pathways for Blimp1 and let-7c in dendritic cells.
A negative feedback loop between Blimp1 and let-7c regulates levels of SOCS1 and IL-6, and may explain the function of Blimp1 risk alleles. The effects of Blimp1 KO and/or siRNA knockdown (see experiments in Figure 1) remove inhibition of let-7c, thus leading to reduced SOCS1 expression (see experiments in Figure 3). Conversely, overexpression of Blimp1 leads to reduced let-7c expression and reduced IL-6 expression, either through enhanced SOCS1 expression or through direct effects of Blimp1 on IL-6 (data in Figure 4). Finally, transfection of let-7c reduces SOCS1 expression, and leads to increased expression of IL-6 (see data in Figure 5). These relationships are consistent with data from human cells (see Figure 6), which also relate lower Blimp1 levels to increased HLA-DR expression, possibly through release of inhibition of the CIITA transcription factor.
Previously, we showed that expression of the transcriptional repressor Blimp1 in DCs is required to maintain immune tolerance (20). In this study, we show that the relative levels of the miRNA let-7c and Blimp1 determine the tolerogenic or immunogenic potential of DCs. Let-7c miRNA was significantly increased in Blimp1-deficient DCs, resulting in a proinflammatory phenotype in a cell-type specific manner. We demonstrated reciprocal regulation of Blimp1 and let-7c, similar to the reciprocal regulation of miRNA-19b-21, PTEN, and its pseudogene, PTENP1 (26). However, the coordinated regulation of Blimp1 and let-7c was primarily due to the direct binding of Blimp1 to a let-7c regulatory element.
Small RNAs are important regulators of gene expression. Among the many types of small RNAs (27, 28), miRNAs have emerged as key regulators of gene expression, acting either to facilitate translational repression by postinitiation repression (29), or to accelerate mRNA degradation (30). In particular, many recent studies have documented a critical role of miRNAs in regulating immune cell development, fine-tuning immune responses, and controlling immune homeostasis. The involvement of miRNAs in immune tolerance and autoimmunity has also been suggested by recent studies. Some miRNAs have been shown to be aberrantly expressed in the PBMCs of SLE patients (31) and in splenic lymphocytes from a mouse model of lupus (32). The importance of tight regulation of miRNAs has been demonstrated in specific cell types, such as Treg, effector T cells (Teff), and B lymphocytes (33–35). Compared with their role in adaptive immune cells, the contribution of miRNAs to DC activation has been examined in only a few studies (36).
In addition to highlighting the importance of tight regulation of let-7c in the current study, we discovered that a specific transcription factor, Blimp1, is involved in regulation of this miRNA. The let-7 family members include 9 distinguishable mature miRNAs and are widely expressed in the hematopoietic system (37). Although the mature miRNAs of individual let-7 family members have highly similar sequence homology, they are located throughout the genome and in different genomic environments. We searched for a consensus Blimp1 binding site in other family members, and none of the other let-7 genes contain the consensus sequence within 1 Kb of the coding region except let-7f-1, which is not known to be expressed in DCs. Let-7i is expressed in mature DCs and is known to regulate SOCS1, as we have now shown for let-7c; however, we did not find a putative binding site of Blimp1 near the let-7i genomic sequence. Therefore, we believe that Blimp1-dependent regulation is a unique mechanism for let-7c in DCs. To understand the consequence of Blimp1 binding to let-7c, we are currently investigating chromatin modifications mediated by Blimp1 in DCs. Previous studies have demonstrated that Blimp1 recruits the G9α histone methyl transferases to the IFN-β promoter (38). Blimp1 also has been shown to associate with the transcriptional corepressor hGroucho (39) and histone deacetylases 1 and 2 (40). We believe, therefore, that Blimp1 may lead to histone deacetylation at the let-7c gene locus.
GWAS have identified multiple loci which are implicated in immunologic disturbances in autoimmune disease. Despite a large number of SNPs that have been identified, an understanding of their functional significance has lagged. We decided to study the SNP rs548234, which is located downstream of the Blimp1 gene, and is strongly associated with SLE. We focused on the impact of the risk allele in cells of healthy donors because other studies have shown that once disease-specific pathways of inflammation are activated, phenotypic changes occur that may be independent of genotype. For example, changes in B cell signaling attributable to the protein tyrosine phosphatase nonreceptor 22 (PTPN22) risk allele can be seen in cells of non-risk allele individuals once type 1 diabetes is established (41). Mo-DCs cultured from SLE-risk allele carriers exhibit reduced Blimp1 expression and increased let-7c expression, similar to that seen in murine Blimp1-deficient DCs. This reciprocal expression of Blimp1 and let-7c is not found in purified B lymphocytes from the same donors. In fact, Zhou et al. also found that Blimp1 expression is not affected in SNP rs548234–carrying B cells from SLE patients (42), consistent with our observation in B cells. Interestingly, DCs from blood or Mo-DCs from risk allele carriers displayed increased HLA-DR on their surface, and secreted an increased amount of IL-6 after LPS stimulation. In contrast to DCs, monocytes from risk allele carriers did not overexpress HLA-DR, again attesting to the cell-specific effects of the Blimp1 polymorphism. We do not yet know why expression of Blimp1 is decreased in SLE-risk allele carriers. The SNP for SLE is located at the proximal region of the Blimp1 3′ UTR where it might have enhancer activity, act as a regulatory region for Blimp1 transcription, or be a target of multiple miRNAs, leading to increased degradation of Blimp1 mRNA. Identification of regulatory molecules or DNA modifications that are affecting the functional activity of rs548234 in DCs remains to be established.
In conclusion, we propose a novel mechanism of miRNA regulation mediated by Blimp1 in both murine and human DCs. Increased levels of let-7c that are present when Blimp1 expression is low lead to a proinflammatory phenotype in DCs, particularly after TLR stimulation. Functional changes mediated by the Blimp1/let-7c ratio may be critical in discriminating between immune tolerance and autoimmunity. Moreover, the mouse model of Blimp1 deficiency in DCs shares a strong similarity to human SLE-risk allele DCs; this similarity makes the mouse model a useful tool for studying human diseases.
Methods
Mice.
Breeder cages for DCBlimp1ko mice were maintained under specific pathogen-free conditions at the animal facility of the FIMR.
Human samples and in vitro differentiation of Mo-DCs.
Healthy Blimp1 SLE-risk allele carriers and controls (both groups were comprised of females under the age of 55 years) were identified from the (genotype and phenotype) GAP Registry at the FIMR. Fresh blood was collected, and total PBMCs were purified by Ficoll-Paque gradient centrifugation. Briefly, whole blood was diluted with PBS and gently layered on the Ficoll. Cells were centrifuged continuously at 750 g for 20 minutes. PBMCs were collected from the middle layer and washed 3 times with PBS. CD14+ monocytes were purified using EasySep Kit (StemCell Technologies Inc.), according to the manufacturer’s protocol. Purity of CD14+ cells was determined by flow cytometry; a purity of 90%–95% was consistently obtained. After purification, monocytes were cultured in RPMI 1640 with 10% FBS, penicillin-streptomycin (P/S), L-glutamine, 106 U/ml of GM-CSF (PeproTech), and 200 U/ml of IL-4 (PeproTech) for 7 days.
Flow cytometry.
For surface staining, approximately one million cells were incubated with the appropriate antibody mixture for 20 minutes on ice. Stained cells were washed with staining buffer (PBS with 2% FBS and 1 mM EDTA) 3 times, and data were acquired using an LSRII (BD) and analyzed by FlowJo software (Tree Star Inc.).
Cell lines and tissue culture.
A human embryonic kidney cell line, 293T, and a virus-producing packaging cell line, 293FT, were maintained at a concentration of 0.5 × 106 cells/ml in DMEM (Invitrogen) supplemented with 10% FBS, penicillin-streptomycin, and L-glutamine.
Antibodies.
Antibodies for flow cytometry included APC anti–mouse CD11c from eBioscience; PE anti–mouse TLR4/MD2; FITC anti–mouse IAb; FITC anti–human CD3, CD19, and CD56; PerCP anti–human HLA-DR; Pacific Blue anti–human CD14; APC anti–human CD16; PE-Cy7 anti–human CD123; PE-Cy5 anti–human CD11c; PE anti–human CD141; PE anti–human CD1a from BD Biosciences; and rabbit anti–SOCS1 (ab9870) from Abcam. Antibodies used for the ChIP assay were as follows: goat anti-mouse Blimp1 polyclonal antibodies (sc-13206X) and normal goat antibodies (sc-2028) purchased from Santa Cruz Biotechnology, and goat anti-human Blimp1 antibodies (ab106766) and rabbit anti-human Blimp1 antibodies (ab59369) purchased from Abcam.
Cytokine analysis.
The protein array was performed according to the manufacturer’s protocol. Briefly, DCs from the spleens of control mice and DCBlimp1ko mice were cultured overnight with or without 1 μg of LPS. The next day, supernatants were harvested and used for either protein array experiments or for quantitative assays. IL-1, IL-10, IL-12 (p70), IFN-γ, TNF-α, and KC were measured by proinflammatory 7-flex from MSD assays. IL-2 and TIMP-1 were measured by ELISA (R&D Systems). IL-6 was measured by both MSD and ELISA (BD Biosciences).
For human cytokine analyses, CD14+ monocyte-derived DCs were harvested at day 7, and cultured (106 cells/ml) overnight with or without various TLR agonists. Supernatants were collected and cytokines were analyzed by MSD assay (human tissue-culture proinflammatory cytokine 7-flex), according to the manufacturer’s protocol. Briefly, cytokine-precoated plates were incubated with blocking buffer (1% w/v; Blocker B) for 1 hour at room temperature. Plates were washed 3 times with washing buffer (PBS with 0.05% Tween 20), and samples were dispensed for 2 hours at room temperature. Detection antibody solution was added into each well and incubated for 2 hours. All the incubations were followed with vigorous shaking (45–130 g). After washing, Read Buffer was added, the plates were read, and data were analyzed using a SECTOR Imager (Meso Scale Discovery).
miRNA array.
Lin (CD19, CD3, CD56)– and CD11c+ MHCIIhi splenic DCs were purified by cell sorting (FACSAria) from 6- to 8-week-old DCBlimp1ko and control mice. Purified cells were spun down and immediately snap-frozen by liquid nitrogen. Total RNA was isolated and prepared by Miltenyi Biotec for miRNA array and analysis. miRNA array data have been deposited in the Gene Expression Omnibus database (GEO GSE40587).
siRNA and lentivirus transduction.
siRNAs targeting the mouse Blimp1 or luciferase (control) gene conjugated with GFP cloned in lentivirus vector were purchased from GeneCopoeia. For let-7c overexpression assays, control miRNA or let-7c miRNA cloned in GFP reporter lentivirus vectors were purchased from GeneCopoeia. To produce lentivirus, 2.5 μg of plasmid was transfected into the 293FT cell line (cells are 70%–80% confluent at the moment of transfection), together with 2.5 μg of Lenti-Pac FIV Mix by EndoFectin (GeneCopoeia). DNA-Endofectin Lenti complex was incubated with cells for 14 hours, and replaced with fresh medium supplemented with 5% FBS, penicillin-streptomycin, and 1:500 volume of the Titer Boost reagent. Virus-containing supernatant was harvested at 48 hours after transfection. Following centrifugation to remove cell debris, supernatant was filtered through 0.45 μm polyethersulfone (PES) low protein-binding filters. Virus stock was aliquoted and kept at –80°C until use.
To transduce virus into DCs, 106 day 5–cultured DCs were added to 400 μl of virus stock and 100 μl medium supplemented with 5 μg/ml of polybrene (Sigma-Aldrich). Virus and cells were spun down at 840 g for 40 minutes at 30°C. Supernatant was then removed. After a second spin, cells were incubated for 2 hours at 37°C. Medium was changed after 2 hours of incubation, and virus transduction was monitored by GFP expression at 48 hours after transduction.
ChIP assay.
The ChIP assay was performed as previously reported (43), with some modifications. Briefly, mouse CD11chi BM-DCs or CD14– human Mo-DCs were stimulated with LPS for 24 hours, and cells were fixed with 1% formaldehyde for 10 minutes at room temperature. Cells were harvested with ice-cold PBS and lysed by RIPA Buffer (Thermo Fisher Scientific Inc.). The cell suspension was sonicated (Misonix Sonicator 3000; Qsonica) 15 times for 30 seconds, allowing the suspension to cool on ice for 1 minute between pulses. Anti–Blimp1 or control goat IgG antibodies were used for overnight immunoprecipitation at 4°C with rotation. The next day, antibodies were washed with washing buffer (500 mM LiCl, 50 mM Hepes, pH 7.6, 1 mM EDTA, 0.7% DOC, and 1% NP-40), and eluted with elution buffer (50 mM Tris, pH 8.0, 10 mM EDTA and 1% SDS). PCR was performed with total input or elution. Primers used to amplify the mouse let-7c gene were: 5′-CACTTGAGCCATCG AGGAAT-3′ (forward) and 5′-CCCGTGAGAAATAGCGTTGT-3′ (reverse). PCR conditions were: 94°C for 5 minutes; 94°C for 15 seconds; 58°C for 15 seconds; 39 cycles of 72°C for 15 seconds; and 72°C for 10 minutes. Primers used for human let-7c PCR were: set#1, 5′-GGTTTGGACCAGGATCTGAA-3′ (forward) and 5′-TTGGGTTTCCAGCATAGGTC-3′ (reverse); or set#2, 5′-TGAAGCAACATTGGAAGCTG-3′ (forward) and 5′-GCCCAAATCAATGATCCAAG-3′ (reverse). PCR conditions were: 94°C for 5 minutes; 94°C for 15 seconds; 53°C for 15 seconds; 39 cycles of 72°C for 15 seconds; and 72°C for 10 minutes.
EMSA.
EMSA was performed as described previously (43), with modifications. Nuclear extract was prepared from 107 BM-DCs stimulated with LPS for 24 hours. 5′ IRDye–labeled C-terminal sequences from let-7c oligos were purchased from Integrated DNA Technologies. Oligos (100 nM) and proteins (10 μg) were incubated in binding buffer (10 mM Tris, 50 mM KCl, 3.5 mM DTT) supplemented with 2 μg poly (dI-dC) and 5 mM MgCl2 for 30 minutes at room temperature. For supershift, 1 μg of anti–Blimp1 antibodies (Santa Cruz Biotechnology Inc.) was added, and for competition, 200-fold unlabeled oligos were added to the binding mixture. Protein-DNA complexes were resolved on a prerun 5% TBE polyacrylamide gel (Bio-Rad Laboratories Inc.). The image was scanned by Odyssey (LI-COR Biosciences). Oligos used for let-7c EMSA were: 5′-ATTTCTGTTCAAGAAAGTAATGTGTCATGAG-3′ and 5′-CTCATGACACATTACTTTCTTGAACAGAAAT-3′.
Luciferase assay.
Plasmids for the luciferase and miRNA assay kits were purchased from Genecopoeia. The 293T cell line was transfected with 1 μg of luciferase vector (MmiT026645-MT01; Blimp1 3′ UTR, MmiT028883-MT01; SOCS1) with or without various amounts of miRNA plasmid (MmiR3365-MR01; mmu-let-7c; CmiR0001-MR01; control miRNA). Luciferase activity was measured at 48 hours after transfection. A firefly luciferase solution was directly added and incubated with cells for 10 minutes. Luciferase activity was measured using a Victor3 (PerkinElmer) luminometer. Substrate for Renilla luciferase was added directly to the firefly luciferase solution, and specific activity was measured by luminometer. The ratio of firefly/Renilla luciferase was calculated and compared between transfections to prevent false results due to differential transfection efficiency.
Western blotting.
5 × 106 splenic DCs or BM-DCs with or without LPS were lysed with 100 μl of RIPA buffer with complete protease inhibitor (Roche). Equal quantities of protein were loaded and separated in 4%–12% SDS-PAGE gel, separated proteins were transferred to PVDF membrane, and immunoblotting was performed. Chemiluminescent light was exposed to x-ray film, and the film developed.
Quantitative PCR.
Total RNA, including small RNA, was prepared using a mirVana miRNA Isolation Kit (Ambion; Life Technologies). An iScript cDNA Synthesis Kit (Bio-Rad Laboratories) was used for mRNA analysis, and a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems; Life Technologies) was used for miRNA assay. Gene-specific primers for qPCR and master mix were purchased from Applied Biosystems, and PCR was performed in a LightCycler 480 II (Roche Applied Science). Ct values of β2m, HPRT, or sno234 were used as housekeeping controls, and relative expression of a gene was calculated by ΔΔCt, as posted on the company’s website.
Statistics.
Statistical significance was determined with a nonparametric, Mann-Whitney U test. A P value less than or equal to 0.05 was considered significant.
Study approval.
The use of mice for this study was approved by an Association for Assessment and Accreditation of Laboratory Animal Care–accredited (AAALAC-accredited) facility under the regulation of the Institutional Animal Care and Use Committee. The protocol for the study of human cells was approved by the FIMR’s institutional review board, and prior written informed consent was obtained from all participants.
Supplementary Material
Acknowledgments
We are especially grateful to M. Keogh and M. DeFranco of the Genotype and Phenotype Registry at the Feinstein Institute for recruiting Blimp1 genotyped subjects. We also thank the Flow Cytometry Core Facility for sorting and analysis of samples, J. Goldstein for technical support, and G. Honig for valuable discussions and critical reading of the manuscript. This work was supported by a grant from the Alliance of Lupus Research (to B. Diamond); and by NIH grants RC2AR059092 (to P.K. Gregersen and B. Diamond) and K01AR059378 (to S.J. Kim).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(2):823–833. doi:10.1172/JCI64712.
References
- 1.Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J Exp Med. 1973;137(5):1142–1162. doi: 10.1084/jem.137.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Traver D, et al. Development of CD8alpha-positive dendritic cells from a common myeloid progenitor. Science. 2000;290(5499):2152–2154. doi: 10.1126/science.290.5499.2152. [DOI] [PubMed] [Google Scholar]
- 3.Randolph GJ, Inaba K, Robbiani DF, Steinman RM, Muller WA. Differentiation of phagocytic monocytes into lymph node dendritic cells in vivo. Immunity. 1999;11(6):753–761. doi: 10.1016/S1074-7613(00)80149-1. [DOI] [PubMed] [Google Scholar]
- 4.Hambleton S, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med. 2011;365(2):127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohnmacht C, et al. Constitutive ablation of dendritic cells breaks self-tolerance of CD4 T cells and results in spontaneous fatal autoimmunity. J Exp Med. 2009;206(3):549–559. doi: 10.1084/jem.20082394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19(4):607–620. doi: 10.1016/S1074-7613(03)00267-X. [DOI] [PubMed] [Google Scholar]
- 7.Martins GA, et al. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat Immunol. 2006;7(5):457–465. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- 8.Savitsky D, Calame K. B-1 B lymphocytes require Blimp-1 for immunoglobulin secretion. J Exp Med. 2006;203(10):2305–2314. doi: 10.1084/jem.20060411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fairfax KA, et al. Different kinetics of blimp-1 induction in B cell subsets revealed by reporter gene. J Immunol. 2007;178(7):4104–4111. doi: 10.4049/jimmunol.178.7.4104. [DOI] [PubMed] [Google Scholar]
- 10.Niu H, Ye BH, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 1998;12(13):1953–1961. doi: 10.1101/gad.12.13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kallies A, et al. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat Immunol. 2006;7(5):466–474. doi: 10.1038/ni1321. [DOI] [PubMed] [Google Scholar]
- 12.Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77(2):297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 13.Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276(5312):596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 14.Piskurich JF, Lin KI, Lin Y, Wang Y, Ting JP, Calame K. BLIMP-I mediates extinction of major histocompatibility class II transactivator expression in plasma cells. Nat Immunol. 2000;1(6):526–532. doi: 10.1038/82788. [DOI] [PubMed] [Google Scholar]
- 15.Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol. 2002;22(13):4771–4780. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martins GA, Cimmino L, Liao J, Magnusdottir E, Calame K. Blimp-1 directly represses Il2 and the Il2 activator Fos, attenuating T cell proliferation and survival. J Exp Med. 2008;205(9):1959–1965. doi: 10.1084/jem.20080526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan YH, et al. Absence of the transcriptional repressor Blimp-1 in hematopoietic lineages reveals its role in dendritic cell homeostatic development and function. J Immunol. 2009;183(11):7039–7046. doi: 10.4049/jimmunol.0901543. [DOI] [PubMed] [Google Scholar]
- 18.Han JW, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 19.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43(3):246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim SJ, Zou YR, Goldstein J, Reizis B, Diamond B. Tolerogenic function of Blimp-1 in dendritic cells. J Exp Med. 2011;208(11):2193–2199. doi: 10.1084/jem.20110658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo TC, Calame KL. B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites. J Immunol. 2004;173(9):5556–5563. doi: 10.4049/jimmunol.173.9.5556. [DOI] [PubMed] [Google Scholar]
- 22.Starr R, et al. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387(6636):917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa R, et al. SOCS-1 participates in negative regulation of LPS responses. Immunity. 2002;17(5):677–687. doi: 10.1016/S1074-7613(02)00449-1. [DOI] [PubMed] [Google Scholar]
- 24.Hanada T, et al. Suppressor of cytokine signaling-1 is essential for suppressing dendritic cell activation and systemic autoimmunity. Immunity. 2003;19(3):437–450. doi: 10.1016/S1074-7613(03)00240-1. [DOI] [PubMed] [Google Scholar]
- 25.Gateva V, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1, and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci U S A. 2005;102(47):16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu CY, Rana TM. Small RNAs: regulators and guardians of the genome. J Cell Physiol. 2007;213(2):412–419. doi: 10.1002/jcp.21230. [DOI] [PubMed] [Google Scholar]
- 29.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. . Dev Biol. 1999;216(2):671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 30.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;CCR4(14):1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Te JL, et al. Identification of unique microRNA signature associated with lupus nephritis. PLoS One. 2010;5(5):e10344. doi: 10.1371/journal.pone.0010344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai R, et al. Identification of a common lupus disease-associated microRNA expression pattern in three different murine models of lupus. PLoS One. 2010;5(12):e14302. doi: 10.1371/journal.pone.0014302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou X, et al. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205(9):1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9(4):405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan W, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. . J Immunol. 2010;184(12):6773–6781. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 36.Zhang M, et al. Inhibition of microRNA let-7i depresses maturation and functional state of dendritic cells in response to lipopolysaccharide stimulation via targeting suppressor of cytokine signaling 1. J Immunol. 2011;187(4):1674–1683. doi: 10.4049/jimmunol.1001937. [DOI] [PubMed] [Google Scholar]
- 37.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Gyory I, Wu J, Fejer G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol. 2004;5(3):299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- 39.Ren B, Chee KJ, Kim TH, Maniatis T. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 1999;13(1):125–137. doi: 10.1101/gad.13.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu J, Angelin-Duclos C, Greenwood J, Liao J, Calame K. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol Cell Biol. 2000;20(7):2592–2603. doi: 10.1128/MCB.20.7.2592-2603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Habib T, et al. Altered B cell homeostasis is associated with type I diabetes and carriers of the PTPN22 allelic variant. J Immunol. 2012;188(1):487–496. doi: 10.4049/jimmunol.1102176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou XJ, et al. Genetic association of PRDM1-ATG5 intergenic region and autophagy with systemic lupus erythematosus in a Chinese population. Ann Rheum Dis. 2011;70(7):1330–1337. doi: 10.1136/ard.2010.140111. [DOI] [PubMed] [Google Scholar]
- 43.Kim S, Elkon KB, Ma X. Transcriptional suppression of interleukin-12 gene expression following phagocytosis of apoptotic cells. Immunity. 2004;21(5):643–653. doi: 10.1016/j.immuni.2004.09.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.