Abstract
Hepatitis C virus (HCV) establishes a persistent infection and causes chronic hepatitis. Chronic hepatitis patients often develop hepatic cirrhosis and progress to liver cancer. The development of this pathological condition is linked to the persistent infection of the virus. In other words, viral replication/multiplication may contribute to disease pathology. Accumulating clinical studies suggest that HCV infection alters lipid metabolism, and thus causes fatty liver. It has been reported that this abnormal metabolism exacerbates hepatic diseases. Recently, we revealed that lipid droplets play a key role in HCV replication. Understanding the molecular mechanism of HCV replication will help elucidate the pathogenic mechanism and develop preventive measures that inhibit disease manifestation by blocking persistent infection. In this review, we outline recent findings on the function of lipid droplets in the HCV replication cycle and describe the relationship between the development of liver diseases and virus replication.
Keywords: hepatitis C virus, lipid droplet, replication, liver disease, VLDL, apolipoprotein
Introduction
Patients infected with hepatitis C virus (HCV) develop acute hepatitis. While 20% of acute hepatitis patients eliminate the virus and recover from the infection, the remaining infectants maintain subclinical state or develop chronic hepatitis as a result of persistent infection. About one-fourth of chronic hepatitis patients progress to hepatic cirrhosis, and half of hepatic cirrhosis patients develop hepatocellular carcinoma (HCC). Many patients with hepatic cirrhosis caused by HCV infection die from the exacerbation of the disease and from HCC. An estimated 15–20% of HCV-infected patients succumb to these hepatic diseases.
Interferon and ribavirin combination therapy is an effective treatment for chronic hepatitis C. Approximately half of patients who complete this therapy eliminate the virus and, therefore, will not develop hepatic disorders thereafter. Generally, it is thought to be difficult to eliminate persistently or latently infected viruses from host. However, in case of HCV, interferon and ribavirin combination therapy can eliminate HCV in half of infected patients, suggesting that HCV infection is more curable with anti-HCV drugs as compared to other persistently infecting viruses. Because persistent virus replication is one of the causes of chronic diseases, a better understanding of the mechanisms of HCV replication, and modification of host cell proliferation by viral infection, will help elucidate the molecular pathogenic mechanism of the disease.
HCV genome and viral proteins
HCV is classified in the Hepacivirus genus of the Flaviviridae family. Viruses of the Flaviviridae family have the positive-sense, single-strand RNA genome that is packaged into an enveloped viral particle.1) All the viral proteins are encoded in the largest reading frame in the genome. Translation from the open reading frame of the HCV genome starts by the mechanism using internal ribosome entry site (IRES) to produce a precursor poly-protein for viral proteins.2) The precursor poly-protein is subsequently cleaved by a cellular signal peptidase and viral peptidases to produce about 10 different viral proteins.3),4) Some proteins receive further modification such as glycosylation or phosphorylation to become functionally matured forms. From the N-terminus of the reading frame, Core, envelope1 (E1), envelope2 (E2), p7, non-structural 2 (NS2), NS3, NS4A, NS4B, NS5A and NS5B are produced in this order (Fig. 1). Viral proteins are divided into two categories: structural proteins and non-structural (NS) proteins. The structural proteins compose the virus particle, while the NS proteins function only inside the infected cell and are not packaged in the virion. The HCV structural proteins include the Core, envelope-1 (E1), and -2 (E2) proteins. The remaining NS proteins have unique functions. One important function of the NS proteins is to construct a stage for viral-genome replication and mRNA synthesis by forming a viral replication complex. This replication complex is a specialized structure protected by a cellular membrane that is induced and built by the virus infection. In addition, another type of NS protein complex is thought to contribute to virus particle assembly.
Fig. 1.
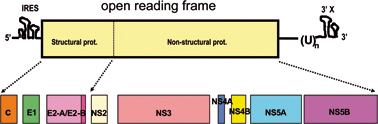
Structure of the HCV genome and the proteins produced by the genome. The HCV genome (upper panel) consists of a positive-sense RNA strand comprising about 10,000 nucleotides. Virus proteins are first produced as a precursor protein encoded in the largest open reading frame that comprises about 90% of the entire genome, then processed by cellular signal protease(s) followed by two virus proteases which are encoded by NS2–NS3, and NS3, respectively. 5′ one-thirds of the open reading frame encodes proteins for virus particle and the rest encodes non-structural virus proteins that function in virus-infected cells (lower panel).
HCV replicates in the cytoplasm of infected cells (Fig. 2). Current models propose that the virus infects a cell by binding to a cell surface receptor, after which the virus particle is endocytosed into the cytoplasm by clathrin-mediated endocytosis. To date, several candidate proteins including virus receptor(s) have been identified as host factors involved in this process. It seems that the LDL receptor and the scavenger receptor class B type I function in the initial stage of HCV infection, after which the HCV particles interact with the tetraspanin CD81 and the tight-junction protein cluadin-1 to establish clathrin dependent internalization.5)–12) However, the function of these proteins—from the moment the virus and target cell make contact until infection is established—is not fully understood.
Fig. 2.
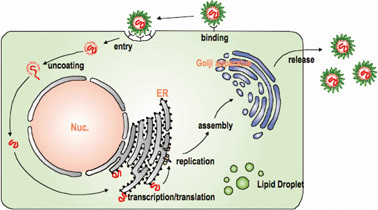
HCV replication cycle. Since the detailed molecular mechanism of HCV infection/multiplication is yet to be clarified, this figure is drawn as a general view of replication of viruses in Flaviviridae, in which HCV is classified. In cells being established virus entry, most of the events for virus replication are conducted in cytoplasm.
The viral genome replicates in a peculiar membranous structure that is sensitive to detergent around the endoplasmic reticulum13)–16)
For most RNA viruses, the viral genome replicates in the environment associating with membrane components of the host cell. It is expected that HCV genome replication also undergoes a process similar to other RNA viruses. While developing the in vitro infection and replication system of HCV, the HCV subgenome replicon, which lacks the coding region for structural proteins but carries a drug-resistant marker, is established.17) Since this replicon can replicate autonomously, cell lines bearing the subgenome replicon can be selected by selectable marker for neomycin. Electron microscopic observation revealed that such cells have an unusual membrane structure and that part of the endoplasmic reticulum (ER) membrane is notably deformed.13) This membranous structure has a complicated shape as well as a botryoidal structure in some cases and is also seen in liver specimen from a HCV infected individual. This structure is called “a membranous web” and is unique to HCV genome bearing cells. Under the labeling condition of de novo synthesized RNA, viral-RNA synthesis could be shown to occur around this membranous web.15) Biochemical analysis of HCV genome-bearing cells was carried out by treating with digitonin, which partially destroys plasma membrane of cells and thus makes cells accessible by exogenous nucleases or proteases present in certain buffer. Through this work the following results were obtained:14) (1) Surfactants including NP-40 destroyed the replication function of the membranous web; (2) Micrococcal nuclease treatment, under a condition that digests ribosomal RNA completely, did not destroy HCV RNA. However, HCV RNA was immediately destroyed when nuclease treatment was performed in the presence of NP-40, suggesting that viral RNA is protected by membrane components; (3) When digitonin-treated cells were treated with protease K, most viral proteins were hydrolyzed, but about one-tenth or less of the viral proteins remained without being degraded. However, when the protease digestion was performed in the presence of NP-40, the HCV proteins were no longer resistant to protease digestion. This observation indicates that a small portion of the viral proteins is also protected by membrane structures; (4) When digitonin-treated cells were digested with a protease without surfactants, viral RNA synthesis was not affected even when most viral proteins had disappeared. These results suggest that HCV RNA (both positive and negative strands) is synthesized in a membrane-protected complex where only a small portion of the total HCV proteins in cells is present (Fig. 3).14) Furthermore, the analyses indicate the presence of a protease-sensitive viral protein complex that is not protected by the membranous component in the outer side of the replication complex. It is presumed that the protease-sensitive complex is directly exposed to the cytoplasm without the protection of the membranous component. Hereinafter, the complex involving viral-genome replication is referred to as the “replication complex” (RC). The complexes consist of HCV RNA and about 10% of the total viral NS proteins in the host cell (data not shown). The other complex that is retained on the membrane structure but directly exposed to the cytoplasmic environment is referred to as the “non-structural protein complex” (NPC). The function of the NPC is not fully understood. Although detailed analyses are still in progress, it is presumed that NPC plays a role in the assembly process of virus particles (see below). A previous study has reported that one of these viral proteins, NS4B, is capable of changing the membrane structure by acting on the ER membrane.13)
Fig. 3.
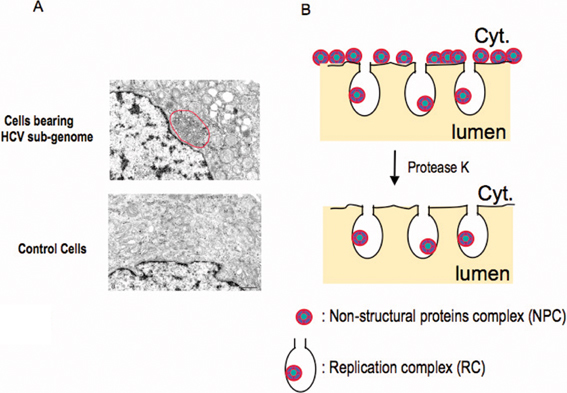
Altered membranous structure observed in HCV genome replicating cells, in which synthesis of the positive as well as the negative HCV genome is conducted. (A) Electron micrographs of the generation of complex membranous structures (called membranous web) around the ER in HCV genome-replicating cells. The web structure is highlighted by the red circle. (The web image was from Gosert, R et al. (2003) J. Virol. 77, 5487 with permission). (B) Diagram of the distribution of NPC (non-structural proteins complex) and RC (replication complex) around the ER membrane. HCV-genome replicating cells treated with digitonin, a detergent that permeabilizes the plasma membrane but does not affect other membranes (e.g., ER-lumen and nuclear membrane)—were analyzed for virus proteins as well as HCV RNA synthesizing activity after incubation of exogenously added protease K. The majority of NS proteins were found to be sensitive to the hydrolysis; only a small portion of NS proteins were found to be resistant to the digestion because of the membrane’s protection. Further, the NS protein complex protected by membranous fraction is fully active to synthesize HCV RNA. From this result the NS protein complex exposed to outside of cytoplasm (NPC) and the NS protein complex surrounded with membrane structure (RC) are distinguished and found to have different functions (see text for details).
Cytoplasmic lipid droplets play a key role in the production of infectious HCV particles18)
As mentioned earlier, most HCV proteins localize around the ER membrane in cells that autonomously replicate the HCV genome. In addition, it has been shown that the Core, a structural HCV protein, associates with lipid droplets when it is singularly expressed.19) It has also been shown that a small portion of NS5A localizes to lipid droplets when NS5A is singularly expressed.20) What is the relationship between the intracellular location of viral proteins, particularly the association of Core and NS5A with the lipid droplet, and viral replication? To understand this phenomenon, we also analyzed the role of Core association with the lipid droplet in virus reproduction using a cell-culture system that produces an infectious virus.18)
When infectious clone JFH1 of HCV genomic RNA is introduced into HuH7 cells (a human liver cancer-derived cell line), genome replication begins and virus particles are produced.21) Analysis of the buoyant density of virus particles determined by a sucrose density-gradient centrifugation method indicates the presence of two types of HCV. Most viral particles were detected in a fraction of 1.15 g/ml density, but these particles were not infectious to HuH7 cells. On the other hand, a small amount of infectious virus particles was detected in a 1.12 g/ml density fraction (Fig. 4). In the HCV-replicating cells, the core protein was found to localize around lipid droplets similar to a previous report that identified it via an experiment on the solitary expression of Core proteins in cells.19) In addition, many of the NS proteins (NS3–NS5B) were co-localized to the ER as previously reported. However, detailed examination revealed that the NS proteins localized around the lipid droplets as well as ER.18) HCV RNA could also be detected around the lipid droplets. The fraction containing lipid droplets, partially purified by floating centrifugation method, was capable of supporting HCV RNA synthesis. Furthermore, the surrounding environment of the lipid droplets in HCV replicating cells is rich in membrane-like structure (see below). This accumulation of membranous structures around the lipid droplet is unique to the infected cells and is not observed in uninfected control cells.
Fig. 4.
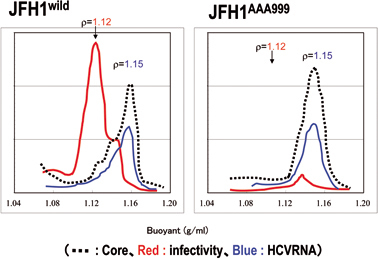
Characteristics and infectivity of virus particles produced into the culture supernatant. The characteristics of virus particles released from HCV(JFH1wild)-replicating cells (left) or cells replicating a mutated virus, JFH1AAA999, encoding proteins that cannot associate with lipid droplets (right) were analyzed by the sucrose density gradient centrifugation method. Dotted, red, and blue lines indicate the amount of virus particles measured by the amount of viral core protein, infectivity and viral RNA, respectively. Buoyant density of sucrose is indicated.
Core proteins promote the localization of NS proteins to lipid droplets18)
In cells bearing the defective HCV genome not expressing the core protein, the NS proteins were mainly localized on the ER and were not seen around the lipid droplet. However, in cells producing the infectious virus, the NS proteins were localized to both lipid droplets and the ER membrane. The Core and other structural proteins (E2), co-localized to lipid droplets. It is worth noting that the Core promotes the localization of the NS proteins to lipid droplets. This was found using the defective HCV genome that does not express the Core. When this mutant genome was expressed, NS5A localized to the ER but not to the lipid droplet. However, when the Core was provided exogenously into the cells, both NS5A and the Core were co-localized to the lipid droplets. Association of other HCV NS proteins with the lipid droplets was also observed by Core dependent manner. This suggests that the Core recruits NS proteins to the lipid droplets, although the mechanism for this association in the presence of Core is unclear.
HCV proteins are arranged in order around lipid droplets in laminae according to protein type
When HCV Core-dependent association of viral NS proteins to the lipid droplets was examined by confocal light and electron microscopy, the Core was shown to be directly localized on the surface of the lipid droplets. However, NS5A was mainly observed in the membranous structure that surrounds the Core-coated lipid droplets.18) This membrane structure is not of the lipid droplet. More specifically, the Core could be seen as a lamina surrounding the lipid droplet, and the membranous components that are rich in NS proteins surrounded the Core-coated lipid droplets. While examining intracellular localization and behavior of the Core-coated lipid droplets, majority of Core coated-lipid droplets were found to reside near the ER around the nuclear membrane (data not shown).
Abnormal membrane structures are enriched around lipid droplets in HCV-producing cells
Regarding generation of lipid droplet, several mechanisms have been proposed.22),23) One mechanism is that ER accumulates neutral fat, mainly tri-glycerides and cholesterol ester, in the space of the membrane bilayer. Increasing the lipid contents increases the mass of the body and the lipid droplet covered with ER-derived membrane buds from the ER.22) The membrane structure of the lipid droplet is, thus, monolayer. Lipid droplet-specific proteins (such as TAP: TIP47, ADRP and Perilipin) associate with the monolayer membrane of the lipid droplet. This typical structure of the lipid droplet can be seen in human liver cancer-derived cell line, HuH7 (Fig. 5). However, in HuH7 cells where the infectious HCV genome could replicate and produce virus particles, a part of surface of the lipid droplets were often seen to be covered with several layers of membrane (Fig. 5). Moreover, complicated membrane structures accumulated around the lipid droplets. Although the origin of these membrane structures is unknown, it is assumed that they are derived from ER membranes because the Core-coated lipid droplets are frequently observed around the ER. The ER membranes are also rich in NPC and RC of HCV as shown in the graphic of Fig. 6(A).
Fig. 5.
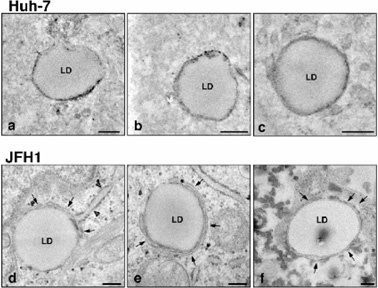
Membranous structures are frequently observed around the lipid droplets in JFH1-bearing cells. Electron micrographs of the lipid droplets and the surrounding area are shown. These show different images of lipid droplets in cells; a to c, lipid droplets in control HuH-7 cells, and d to f, those in JFH1 bearing HuH7 cells. Arrows indicate enriched membrane structure around the lipid droplet. Arrowhead shows rough ER membrane-like structure attached to the lipid droplet (The data was from Miyanari et al. (2007) Nat. Cell Biol. 9, 1089–1097).
Fig. 6.
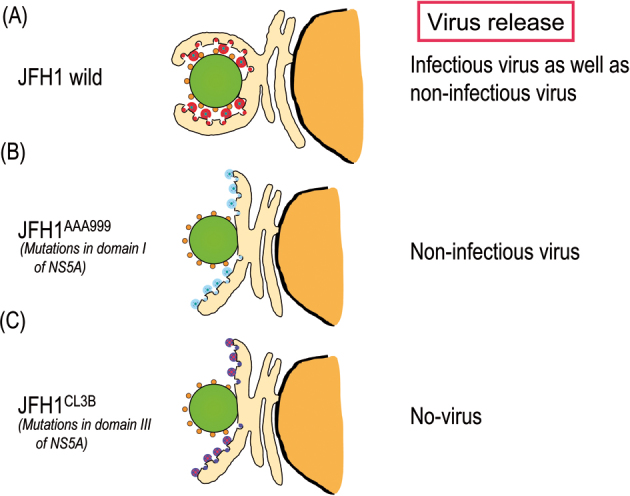
Model of the association of the Core-coated lipid droplets with NPC- and RC-rich endoplasmic reticulum. Association of Core-coated lipid droplets with RC- and NPC-rich ER in HuH7 cells bearing the infectious HCV genome, JFH1 (A). Association of Core coated-lipid droplets with the lipid droplet was not observed in cells bearing the mutant JFH1, JFH1AAA999, expressing NS5A with mutations in domain-I (B),18) and in cells bearing the mutant JFH1, JFH1CL3B encoding NS5A with mutations in domain-III (C).28) Green circles represent lipid droplets. The small orange circles around the lipid droplets are the Core. The largest and the smallest circles with a mosaic represent NPC and RC, respectively. The color of NPC and RC indicates the noted mutations in NS5A in these complexes. Note that the NPC- and RC-rich ER lobes with a mutant NS5A (B and C) do not associate with the core-coated lipid droplet. However, as shown in (B) noninfectious virus particles are released into the culture medium, while NPC and RC with mutations in domain-III of NS5A do not produce virus particles, indicating the lack of virus assembly with this mutant. For NPC and RC localization, see the graphic in Fig. 3.
Functional analysis of lipid droplets in virus production using the mutated viral genomes
How does the association of viral proteins around the lipid droplets affect virus production? Previously, the domain of the Core to interact with the lipid droplet is disclosed.24),25) Mutations of two proline residues to alanine in that region of Core, CorePP/AA, no longer associate with the lipid droplet.25) When the mutated viral genome that produces CorePP/AA is introduced into cells, the Core as well as HCV NS proteins no longer localize around the lipid droplets.18) This result indicates that the Core, which is able to associate with the lipid droplet, functions to recruit other viral proteins to the lipid droplets. The cells expressing this mutant genome do not produce virus particles in the culture medium. Under these conditions, virus production may be inhibited due to lack of association of viral proteins with the lipid droplet that is probably a prerequisite for virus production or virus assembly, or, the mutated Core itself is no more able to execute morphogenesis of virus particle.
An experiment was performed to examine whether or not the association of Core and HCV non-structural proteins to the lipid droplet is essential to virus particle formation.18) For this purpose the HCV genome with mutation in an NS protein was constructed. NS5A was selected for introducing the mutation among other non-structural proteins as NS5A associates with the lipid droplets more frequently than other NS proteins and, thus, is thought to play a proactive role in lipid droplet association with other NS proteins. As shown in Fig. 7, NS5A of HCV-1b genotype is a protein with 466-aminoacid residues that can be divided into three domains (domain-I, -II, and -III from the N-terminus). Mutations were introduced into domain-I using alaninescanning mutagenesis. NS5A mutants that are not severely interfered with genome replication, but fail to associate with lipid droplet were selected among the mutants. By doing so, NS5A mutant in which amino acid residue, APK, of 99–101th or, PPT, of 102–104th, was converted to 3 consecutive alanine residues. When the viral RNA genomes encoding one of these NS5A mutants were introduced into cells and virus production to culture medium was examined, only noninfectious virus particles with 1.15 g/ml buoyant density were produced (Fig. 4).18)
Fig. 7.
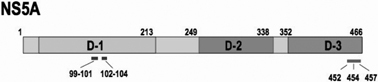
Structure of the HCV NS5A protein and mutant NS5A used for the analysis described in this paper.18),28) D-1- D-3 indicates domain-I - domain-III.
This result confirmed the importance of the lipid droplet for production of “infectious” virus particle. It is likely that infectious virus production requires aggregation of NPC- and RC-rich structures derived from the ER membrane around the lipid droplets. Cells with these structures produced both infectious and noninfectious particles. However, cells bearing the viral genome with mutations in NS5A that failed to associate with the Core-coated lipid droplet produced virus particles that lacked infectivity. Thus, it is likely that lipid droplets play a key role in generating infectious viral particles. Meanwhile, mutants with deletions or point mutations in domain-III in the C-terminus of NS5A had no defect in genome replication but were unable to associate with Core-coated lipid. Neither infectious nor noninfectious viral particles were produced from these cells bearing the genome having these mutations.26)–28)
This observation suggests that NS proteins, which constitute NPC around the Core-coated lipid droplet, are involved in the assembly of infectious virus particles.18),26)–28)
Function of lipid droplets in the production of infectious particles
As mentioned earlier, lipid droplet plays an important role in the production of infectious HCV particles. For this role, one could think the following possibilities: (1) the lipid droplet generates an aggregate together with NPC and RC for the production of infectious virus particles, (2) it provides a factor required for infectivity to the virus particles, and (3) it incorporates virus particle into a vesicle transport system of lipid or lipid related materials to export infectious virus outside the cells. The lipid droplets and their surrounding environment including NPC may be utilized for particle formation because virus-like particles are observed around the lipid droplets by electron microscopy.18) However, as shown in the model (Fig. 6), non-infectious virus particles can be produced even when the HCV NS proteins are not aggregated with lipid droplets. Therefore, lipid droplets seem to be non-essential organelles involved in the assembly of viral particle. Regarding the second possibility, the buoyant density of infectious virus particles was lower than those of noninfectious virus particles by 0.03 g/ml. This suggests that virus particles produced around the Core-coated lipid droplets have a composition of viral protein and nucleic acid different from that of non-infectious particles which are produced from environment other than that produced with association of the Core-coated lipid droplet, to give rise to virus with lighter density. Or, viruses produced from the lipid droplet-associated environment are composed of, or have associated with components with lower density, which is essential for infectivity. The third proposed role of lipid droplets attracts attention to a new function of lipid droplets as a transporting machinery of virus embedded vesicle. It has been known that the LD is surrounded by a monolayer of phospholipids and dynamically moves through the cytoplasm interacting with other organelles, including mitochondria, peroxisomes, endosomes, and the endoplasmic reticulum (ER).29) It is possible that these interactions facilitate the transport of lipids and lipid-associated proteins to other organelles. Thus, it is also possible that these dynamic movements of the lipid droplet facilitate transport of virus particles to where secretion of virus can take place.
A model of production of infectious HCV
When HCV in the blood stream of patients is analyzed by density-gradient centrifugation, virus is observed as a heterogeneous population with different densities.30) The reasons for this heterogeneity may include the association of HCV with blood components, such as lipoprotein and others, rather than differences in the viral protein or nucleic acid composition. It has been shown that HCV associates with lipoproteins including very low-density lipoprotein (VLDL), low density lipoprotein (LDL), and high-density lipoprotein (HDL).30) In addition, the possibility that virus particles associate with antibodies against the virus itself, rheumatoid factor or cryoglobulin has also been reported. However, the significance of these associations with blood components in the HCV replication cycle is still unknown and many questions remain unanswered. It has been suggested that endocytosis of the Flaviviridae viruses such as HCV and bovine viral diarrheal virus was mediated by low density lipoprotein (LDL) receptors on cultured cells.31) Recently, it has been shown that the association with VLDL has an important role in the secretion of virus particles as well as in the acquisition of infectivity.32)–34)
VLDL is a particle that consists of a hydrophobic core of neutral lipid surrounded by a monolayer of amphipathic hospholipids and free cholesterol in which apolipoproteins reside.35) VLDL synthesis is mediated by the function of microsomal triglyceride transfer protein (MTP), which regulates the association of triglyceride with apolipoproteins B-100 to synthesize VLDL precursor. Cholesterol ester and additional triglyceride complexed with apolipoproteins E are incorporated while the VLDL precursor is simultaneously transformed to the matured form of VLDL. In HCV-producing cells, it is possible that VLDL precursor accumulates in the microenvironment of the lumen of ER where Core-coated lipid droplets associate with ER membranes rich in RC and NPC, which may increase the amount of the precursor VLDL and allow to associate with HCV which is also budded into ER lumen. It is presumed that HCV/VLDL complexes are secreted to outside of the cell via the Golgi pathway. Importance of lipoproteins to infectivity of HCV is suggested by the fact that an inhibitor of MTP suppresses release of infectious virus particle.32),33) In cells treated with MTP, it was observed that despite the suppression of infectious viruses released into culture medium, intracellular infectious virus particles have a slightly higher density than those released into culture medium. Another possibility for the result obtained by MTP inhibitor experiment is that MTP inhibitor may affect production or function of other cellular factor(s) that associates with HCV or regulates production of infectious HCV.
Recently, we found that Apo-E interactd with HCV NS5A (Hishiki et al., unpublished data). Moreover, Apo-E knockdown cells suppressed the production of infectious virus in a culture medium.36) while Apo-A1 knockdown cells did not (Hishiki et al. unpublished data). Taking this data together with others, it is suggested that Apo-E as well as Apo-B are important host factors, at least in part, involved in endowing infectivity to HCV.
A model of infectious virus particle production that reflects our experimental results, which indicate that the association of Core-coated lipid droplets with the ER membrane is important for infectious virus particle production, is shown in Fig. 8. In this model, we hypothesize that the association of lipid droplets with the ER membrane increases the production efficiency of VLDL precursor in micro environmental space of the lumen. This higher concentration of VLDL precursor may facilitate the association with HCV which is also budded into ER lumen. But it is unclear whether the formation of this complex occurs actively or passively. Since NS5A binds with Apo-B20) as well as Apo-E, it is possible that the NS proteins promote interaction of VLDL with HCV. Alternatively, integration of Apo-E and/or Apo-B to micro-environmental membranous structure where HCV particle buds may also be likely. In this case Apo-B and/or Apo-E should be integrated as a membranous component of HCV membranous structure. Although involvement of VLDL for infectious HCV production was suggested, it remains to be clarified whether or not VLDL itself associates with HCV before virus release to culture medium. There is no direct evidence to show this association in an in vitro experiment.
Fig. 8.
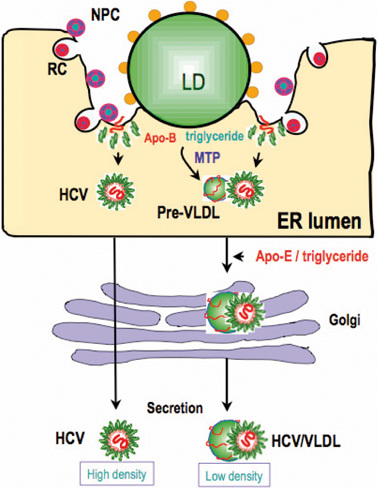
Model of the predicted role of the Core-coated lipid droplet in the production of infectious HCV. The association of the Core-coated lipid droplet with NPC-and RC-rich ER may enhance the interaction of HCV and VLDL. VLDL is generated frequently and enhanced in the micro-environment where lipid droplets associate with the ER. This increased concentration may increase the frequency of the association of HCV and VLDL. HCV/VLDL is released as an infectious particle with a low density, whereas HCV particles that are not associated with VLDL are secreted into the culture medium as noninfectious, dense particles. However, this model does not discriminate the possibility that noninfectious virus particle also associates with or is integrated with some lipoprotein like structure.
It has been reported that HCV infection is mediated by various cellular receptors and the LDL receptor (LDLR) is one of them.31),34) VLDL or its derivative(s) in the VLDL/HCV complex, or Apo-B and -E in HCV may act as a ligand of LDLR on surface protein of hepatocyte.
Increase in lipid droplets in HCV-infected cells
It has been shown that the lipid droplet is an important cellular organelle that produces infectious HCV.18) Then, how does HCV infection affect lipid droplets in the cell? As described above, the structure of the membrane monolayer of the lipid droplet is modified in HCV-replicating cells; multilayered membrane structure on the surface of the lipid droplet are observed often and, moreover, lipid droplets are surrounded by an excess membranous structure. In addition, the amount of lipid droplets increases in HCV-replicating cells.18) Enhanced production of the lipid droplet seems to be mediated by the expression of Core, which is suggested by the following experiment. When autonomous HCV replicon lacking Core production was expressed in HuH7 cells, no enhanced expression of lipid droplet was observed by confocal microscopy analysis of stained cells with BODIPY 493/503 (Invitrogen), a marker for the lipid droplet. In contrast, cells expressing the full HCV replicon showed substantial enhanced production of lipid droplet. Enhancement of lipid droplet production was also observed by ectopic expression of Core in cells bearing HCV replicon lacking expression of the Core. Therefore, it was concluded that HCV infection increases the amount of lipid droplets and that the Core is required for this increase. The amount of intercellular lipids depends on a balance between lipid synthesis, degradation, and lipid transport to the outside of the cell. The Core enhances lipid synthesis by activating SREBP, a transcription factor necessary for lipid synthesis, as well as by suppressing MTP activity slightly. NS5A is also reported to suppress MTP activity some extent. These roles of Core and NS5A are probably significant for balancing intracellular lipid level in HCV infected cells.
Relationship between abnormal cellular metabolism of lipid in HCV-infected cells and HCV related hepatic disease
Many questions about the molecular basis between infection/replication of HCV and the development of chronic hepatitis, hepatic cirrhosis, and liver cancer remain unsolved. It is believed that chronic hepatitis results from the continuous destruction and regeneration of hepatic cells. It has been suggested that continuous immuno-mediated destruction of liver plays a central role in liver pathogenesis by HCV infection. On the other hand, many HCV-infected patients can experience complicated diabetes due to inhibition of insulin signal transduction and can also experience abnormal metabolism, including increased liver lipid content, because of abnormal lipid accumulation. In addition, these disorders are thought to exacerbate liver diseases.37)–39)
Transgenic mice producing HCV core protein develop steatosis and subsequent liver cancer.40) This suggests that abnormal lipid metabolism caused by the HCV core protein can lead to the development of liver cancer at least in mouse model system. The physiological significance of the mechanism by which the HCV core protein controls lipid metabolism became clear when lipid droplets were shown to be essential in the production of infectious virus particles in infected cells. In other words, HCV requires activation of lipid metabolism in the host cell to produce its progeny efficiently, which is, at least in part, a requisite of abnormal metabolism of lipid that may be linked to subsequent liver diseases in the host.
Conclusions
HCV establishes persistent infection and causes chronic hepatitis, which causes the development of hepatic cirrhosis and liver cancer in some patients. Therefore, it is very important to prevent infection and disease progression. A better understanding of the molecular mechanisms of virus replication would help to establish preventative measures. Recent studies have uncovered a mechanism of HCV replication in which lipid droplets are used to produce infectious virus. This finding may contribute to uncovering the link between HCV infection/replication and liver pathogenesis. A detailed analysis of the mechanism of HCV replication will further increase our understanding of the interaction between the virus and host cell, to explain disease development.
Acknowledgments
This work was supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology; by grants-in-aid for the Research for the Future Program from the Japanese Society for the Promotion of Science; and by grants-in-aid for the Program for Promotion of Fundamental Studies in Health Science from the Organization for Pharmaceutical Safety.
Profile

Kunitada Shimotohno was born in 1944. After he graduated from the Graduate School at Hokkaido University in 1972, he investigated the mechanisms of transcriptional regulation of a double stranded RNA virus (silkworm cytoplasmic polyhedrosis virus) at the National Institute of Genetics. He then studied the molecular mechanisms of avian retrovirus replication as a post-doctoral fellow in the laboratory of Dr. Howard M. Temin at the McArdle Laboratory for Cancer Research, at the University of Wisconsin. During his very productive post-doctoral studies, he elucidated the structural similarities of the avian retrovirus proviruses to bacterial transposable elements. He also determined that there was no apparent sequence specificity at the proviral integration sites in the chromosomes of virus-infected cells, which was also known to be a characteristic feature of transposable elements. This leads to the concept of insertional mutagenesis as when proviruses integrate, they can disrupt the normal functions of a gene by insertion (mutagenesis) into the host chromosomal DNA. It turns out that insertional mutagenesis by endogenous retroviruses and retroviral-like sequences, is now a well documented mechanism of carcinogenesis. Insertional mutagenesis occurs in human cancers as well as other diseases. He also demonstrated, for the first time, that retroviruses have the ability to tranduce foreign genes into cells irrespective of the gene being transduced by the retrovirus. By taking advantage of this intrinsic feature, retroviruses are now widely used as genetic agents to transduce foreign genes into cells, allowing gene transfer and therapy.
Returning to Japan, he initiated studies on the genomes and biological functions of human T cell leukemia virus (HTLV) and hepatitis C virus (HCV) to elucidate their roles in human tumorigenesis as well as to help establish preventive measures. He started this work at National Cancer Center Research Institute in 1983. There he decoded the entire structure of HTLV-2 genome and the immortalizing activity of HTLV-1-encoded Tax gene in primary human lymphocytes. He also discovered nucleotide sequence diversity in the HCV genome and decoded the nearly entire HCV genome that originated from Japanese patients with chronic hepatitis C, which is now classified into a subtype of HCV-1 genotype.
In 1996, he relocated to the Institute for Virus Research at Kyoto University as a professor and served as a director from 2002 to 2006. Dr. Shimotohno extended his work on HCV and determined that cyclosporin A and its derivatives could exert anti-HCV activities without inducing immunosuppressive effects. He also discovered that lipid droplets played important roles in HCV proliferation.
After retirement from Kyoto University in 2007, Dr. Shimotohno has continued his research at the Research Institute at Chiba Institute of Technology as a professor. He is continuing to focus his research on HCV in order to elucidate the role of HCV in human hepatocellular carcinoma (HCC). Dr. Shimotohno continues to unravel the mysteries of HCV replication and transformation with the ultimate goal to aid in the prevention of HCV infection and to ameliorate the treatment of HCC patients.
Dr. Shimotohno received Noguchi Hideyo-Memorial Award for Medicine in 1997 and Tomizo Yoshida Award by Japanese Cancer Association in 2006.
References
- 1).Lindenbach, B.D., Thiel, H.J. and Rice, C. (2007) The viruses and their replication. InField’s Virology (eds. Knipe, D.M. and Howley, P.M.). Lippincott-Raven, Philadelphia, pp. 1101–1152 [Google Scholar]
- 2).Tsukiyama-Kohara, K., Iizuka, N., Kohara, M. and Nomoto, A. (1992) Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66, 1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Grakoui, A., Wychowski, C., Lin, C., Feinstone, S.M. and Rice, C.M. (1993) Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67, 1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Tanji, Y., Hijikata, M., Hirowatari, Y. and Shimotohno, K. (1994) Hepatitis C virus polyprotein processing: kinetics and mutagenic analysis of serine proteinase-dependent cleavage. J. Virol. 68, 8418–8422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Pileri, P., Uematsu, Y., Campagnoli, S., Galli, G., Falugi, F., Petracca, R.et al. (1998) Binding of hepatitis C virus to CD81. Science 282, 938–941 [DOI] [PubMed] [Google Scholar]
- 6).Agnello, V., Abel, G., Elfahal, M., Knight, G.B. and Zhang, Q.X. (1999) Hepatitis C virus and other Flaviviridae viruses enter cell via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96, 12766–12771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Germi, R., Crance, J.M., Garin, D., Guimet, J., Lortat-Jacob, H., Ruigrok, R.W.et al. (2002) Cellular gylcosaminoglycans and low density lipoproteins receptor are involved in hepatitis C virus adsorption. J. Med. Virol. 68, 206–215 [DOI] [PubMed] [Google Scholar]
- 8).Scarselli, E., Ansuini, H., Cerino, R., Roccasecca, R.M., Acali, S., Filocamo, G.et al. (2002) The human scavenger receptor class B type 1 is a novel candidate receptor for the hepatitis C virus. EMBO J. 21, 5017–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Lozach, P.Y., Lortat-Jacob, H., de Lacroix de Lavalette, A., Staropoli, I., Foung, S., Amara, A.et al. (2003) DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biological Chem. 279, 20358–20366 [DOI] [PubMed] [Google Scholar]
- 10).Evans, M.J., von Hahn, T., Tscheme, D.M., Syder, A.J., Panis, M., Wolk, B.et al. (2007) Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446, 801–805 [DOI] [PubMed] [Google Scholar]
- 11).Helle, F. and Dubuisson, J. (2008) Hepatitis C virus entry into host cells. Cell Mol. Life Sci. 65, 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Cocquerel, L., Voisset, C. and Dubuisson, J. (2006) Hepatitis C virus entry: potential receptors and their biological functions. J. Gen. Virol. 87, 1075–1084 [DOI] [PubMed] [Google Scholar]
- 13).Egger, D., Wölk, B., Gosert, R., Bianchi, L., Blum, H.E., Moradpour, D.et al. (2002) Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76, 5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Miyanari, Y., Hijikata, M., Yamaji, M., Hosaka, M., Takahashi, H. and Shimotohno, K. (2003) Hepatitis C Virus Non-structural proteins in the probable membranous compartment function in viral genome replication. J. Biol. Chem. 278, 50301–50308 [DOI] [PubMed] [Google Scholar]
- 15).Gosert, R., Egger, D., Lohmann, V., Bartenschlager, R., Blum, H.E., Bienz, K.et al. (2003) Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77, 5487–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Aizaki, H., Lee, K.J., Sung, V.M., Ishiko, H. and Lai, M.M. (2004) Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324, 450–461 [DOI] [PubMed] [Google Scholar]
- 17).Lohman, V., Korner, F., Koch, J., Herian, U., Theilann, L. and Bartenchalger, R. (1999) Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285, 110–113 [DOI] [PubMed] [Google Scholar]
- 18).Miyanari, Y., Atsuzawa, K., Usuda, N., Watashi, K., Hishiki, T., Zayas, M.et al. (2007) The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9, 1089–1097 [DOI] [PubMed] [Google Scholar]
- 19).Barba, G., Harper, F., Harada, T., Kohara, M., Goulinet, S., Matsuura, Y.et al. (1997) Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl. Acad. Sci. USA 94, 1200–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Shi, S.T., Polyak, S.J., Tu, H., Taylor, D.R., Gretch, D.R and Lai, M.M. (2002) Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology 292, 198–210 [DOI] [PubMed] [Google Scholar]
- 21).Wakita, T., Pietschmann, T., Kato, T., Data, T., Miyamaoto, M., Zhao, Z.et al. (2005) Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nature Med. 11, 791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Martin, S. and Parton, R.G. (2006) Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7, 373–378 [DOI] [PubMed] [Google Scholar]
- 23).Olofsson, S.O., Boström, P., Andersson, L., Rutberg, M., Perman, J and Borén, J. (2008) Lipid droplets as dynamic organelles connecting storage and efflux of lipids. Biochim. Biophys. Acta, 10.1016/j.bbalip.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 24).Hope, R.G. and McLauchlan, J. (2000) Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 81, 1913–1925 [DOI] [PubMed] [Google Scholar]
- 25).Hope, R.G., Murphy, D.J. and McLauchlan, J. (2002) The domains required to direct core proteins of hepatitis C virus and GB virus-B to lipid droplets share common features with plant oleosin proteins J. Biol. Chem. 277, 4261–4270 [DOI] [PubMed] [Google Scholar]
- 26).Appel, N., Zayas, M., Miller, S., Krijnse-Locker, J., Schaller, T., Friebe, P.et al. (2008) Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4(3), e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Tellinghuisen, T.L., Foss, K.L. and Treadaway, J. (2008) Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4(3), e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Masaki, T., Suzuki, R., Murakami, K., Aizaki, H., Ishii, K., Murayama, A.et al. (2008) Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82, 7964–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Murphy, S., Martin, S and Parton, R.G. (2008) Lipid droplet-organelle interactions; sharing the fats, Biochim. Biophys. Acta, 10.1016/j.bbalip.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 30).Nielsen, S.U., Bassendine, M.F., Burt, A.D., Martin, C., Pumeechockchai, W. and Toms, G.L. (2006) Association between hepatitis C virus and very-low-density lipoprotein (CLDL)/LDL analyzed in iodixanol density gradients. J. Virology 80, 2418–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Agnello, V., Ábel, G., Elfahal, M., Knight, G.B. and Zhang, Q.X. (1999) Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96, 12766–127671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Huang, H., Sun, H., Owen, F., Li, D.M., Chen, W., Gale, Y.et al. (2007) Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. USA 104, 5848–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Gastaminza, P., Cheng, G., Wieland, S., Zhong, J., Liao, W. and Chisari, F.V. (2008) Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J. Virol. 82, 2120–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Molina, S., Castet, V., Fournier-Wirth, C., Pichard-Garcia, L., Avner, R., Harats, D.et al. (2007) The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J. Hepatol. 46, 411–419 [DOI] [PubMed] [Google Scholar]
- 35).Fisher, E.A. and Ginsberg, H.N. (2002) Complexity in the secretary pathway: the assembly and secretion of apolipoproteins B-containing lipoproteins. J. Biol. Chem. 277, 17377–17380 [DOI] [PubMed] [Google Scholar]
- 36).Chang, K.S., Jiang J., Cai, Z. and Luo, G. (2007) Human apolipoprotein E is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 81, 13783–13793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Negro, F. (2006) Mechanisms and significance of liver steatosis in hepatitis C virus infection. World J. Gastroentero. 12, 6756–6765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Powell, E.E., Jonsson, J.R. and Clouston, A.D. (2005) Steatosis: co-factor in other liver diseases. Hepatology 42, 5–13 [DOI] [PubMed] [Google Scholar]
- 39).Pekow, J.R., Bhan, A.K., Zheng, H. and Chung, R.T. (2007) Hepatic steatosis is associated with increased frequency of hepatocellular carcinoma in patients with hepatitis C-related cirrhosis. Cancer 109, 2490–2496 [DOI] [PubMed] [Google Scholar]
- 40).Moriya, K., Fujie, H., Shintani, Y., Yotsuyanagi, H., Tsutsumi, T., Ishibashi, K.et al. (1998) The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 4, 1065–1067 [DOI] [PubMed] [Google Scholar]


