Summary
Histone H3.3 is a constitutively expressed H3 variant implicated in the epigenetic inheritance of chromatin structures. Recently, the PML-nuclear body (PML-NB)/Nuclear Domain 10 (ND10) proteins, Daxx and ATRX, were found to regulate replication-independent histone H3.3 chromatin assembly at telomeres and pericentric heterochromatin. As it is not completely understood how PML-NBs/ND10s regulate transcription and resistance to viral infection, we have used a CMV-promoter-regulated inducible transgene array, at which Daxx and ATRX are enriched, to delineate the mechanisms through which they regulate transcription. When integrated into HeLa cells, which express both Daxx and ATRX, the array is refractory to activation. However, transcription can be induced when ICP0, the HSV-1 E3 ubiquitin ligase required to reverse latency, is expressed. As ATRX and Daxx are depleted from the activated array in ICP0-expressing HeLa cells, this suggests that they are required to maintain a repressed chromatin environment. As histone H3.3 is strongly recruited to the ICP0-activated array but does not co-localize with the DNA, this also suggests that chromatin assembly is blocked during activation. The conclusion that the Daxx and ATRX pathway is required for transcriptional repression and chromatin assembly at this site is further supported by the finding that an array integrated into the ATRX-negative U2OS cell line can be robustly activated and that histone H3.3 is similarly recruited and unincorporated into the chromatin. Therefore, this study has important implications for understanding gene silencing, viral latency and PML-NB/ND10 function.
Key words: Daxx, ATRX, Histone H3.3, ICP0, Transcription, Chromatin
Introduction
For a gene to be silenced or expressed, the functions of multiple factors must be coordinated at the chromatin/DNA interface. Since many of these factors were discovered using techniques, which disrupt cellular architecture and average effects in cell populations, it is not known how or when critical regulators are selectively recruited to specific genomic sites in intact cells. One of the pivotal advances to imaging chromatin in single cells has been the development of reporter plasmids carrying sequence elements from bacteria and bacteriophage that allow DNA and RNA to be visualized in living cells (Rafalska-Metcalf and Janicki, 2007). The genomic sites into which these plasmids integrate can be visualized when their binding proteins, fused to auto-fluorescent proteins are expressed. As such, these types of systems provide an unparalleled opportunity to observe the temporal and spatial organization of gene regulatory events directly at chromatin (Darzacq et al., 2007; Shanbhag et al., 2010; Shav-Tal et al., 2004; Zhao et al., 2011).
Histone H3.3 is a constitutively expressed variant of histone H3, which is linked to the epigenetic inheritance of chromatin states (Elsaesser et al., 2010). Originally associated with sites of active transcription (Ahmad and Henikoff, 2002), it is now known to also be incorporated into heterochromatin with assembly at these distinct sites regulated by different chaperones (Drané et al., 2010; Goldberg et al., 2010; Hake et al., 2005; Wong et al., 2009). HIRA is an H3.3-specific chaperone, which functions at single copy genes (Goldberg et al., 2010; Ray-Gallet et al., 2002; Tagami et al., 2004), and Daxx, together with the Snf2-type chromatin-remodeling factor, ATRX, is required for H3.3 incorporation into telomeres and pericentric heterochromatin (Drané et al., 2010; Goldberg et al., 2010; Lewis et al., 2010). The finding that Daxx/ATRX and HIRA regulate mutually exclusive sites suggests that their targeting signals are critical regulators of replication independent histone H3.3 chromatin assembly.
As histone H3.3 is incorporated into both transcriptionally active and inactive chromatin, it is not known how these sites are functionally linked by its deposition. Interestingly, disruption of the HIRA and Daxx/ATRX pathways has markedly different effects on transcription and chromatin. Although histone H3.3 is not incorporated into single-copy genes in HIRA−/− embryonic stem cells (ESCs), large-scale changes in gene expression are not detected (Goldberg et al., 2010). However, knockdown of either histone H3.3 or ATRX in ESCs induces a telomere dysfunction phenotype, which indicates that this pathway is critical for maintaining telomeric chromatin organization (Wong et al., 2010; Wong et al., 2009).
Before the mechanistic role of Daxx and ATRX in histone H3.3 chromatin assembly was identified, both of these factors were known to be components of PML nuclear bodies (PML-NBs)/nuclear domain 10s (ND10s) and to function in the transcriptional repression of genes and viruses. In fact, many viruses encode proteins that disorganize and/or degrade PML-NB components, including Daxx, ATRX and PML, in the effort to evade cellular resistance to infection (Geoffroy and Chelbi-Alix, 2011). Specifically, the degradation of Daxx by the CMV viral tegument protein, pp71, is important for the initiation of efficient HCMV gene expression (Cantrell and Bresnahan, 2006; Hofmann et al., 2002; Hwang and Kalejta, 2007; Ishov et al., 2002; Preston and Nicholl, 2006). Additionally, ICP0, the HSV-1 E3 ubiquitin ligase required for viral replication and activation of the latent genome, promotes the disaggregation of PML-NBs/ND10s and degradation of their components including PML (Everett et al., 1998).
In addition to viral infections, Daxx and ATRX are significantly linked to several other diseases. ATRX is mutated in the intellectual disability syndrome, α-thalassemia/mental retardation, X-linked (Picketts et al., 1996). Daxx and ATRX are mutated in pancreatic neuroendocrine tumors (PanNETs) (Jiao et al., 2011), ATRX is mutated in oligodendrogliomas and medulloblastomas (Heaphy et al., 2011; Molenaar et al., 2012), and Daxx, ATRX and H3.3 are all mutated in pediatric glioblastoma (Schwartzentruber et al., 2012; Wu et al., 2012). As all PanNETs with inactivating mutations in Daxx or ATRX are positive for the telomerase-independent telomere maintenance mechanism termed ALT (alternative lengthening of telomeres) (Heaphy et al., 2011), this further confirms the importance of these factors in telomere regulation. As ICP0-deficient HSV-1 replicates as well as wild-type virus in the ATRX-null and ALT-positive human osteosarcoma cell line, U2OS (Heaphy et al., 2011; Lukashchuk and Everett, 2010; Yao and Schaffer, 1995), this also highlights the critical position of this pathway at the interface between gene silencing and intrinsic cellular defense.
We previously reported that an array of an inducible CMV-promoter regulated transgene stably integrated into U2OS cells can be robustly activated and that histone H3.3 is strongly recruited to the active site (Janicki et al., 2004). Here, we report that Daxx, ATRX and PML are enriched at the CMV promoter of this transgene and that when it is integrated into HeLa cells, which express both Daxx and ATRX, the array is refractory to activation. As ICP0 depletes Daxx, ATRX and PML from the array, permits its activation, and prevents histone H3.3 from being incorporated into its chromatin, this suggests that histone H3.3 chromatin assembly is blocked during activation.
Results
The histone H3.3 chaperone, Daxx, but not HIRA, is recruited to the transgene array in the U2OS cell line
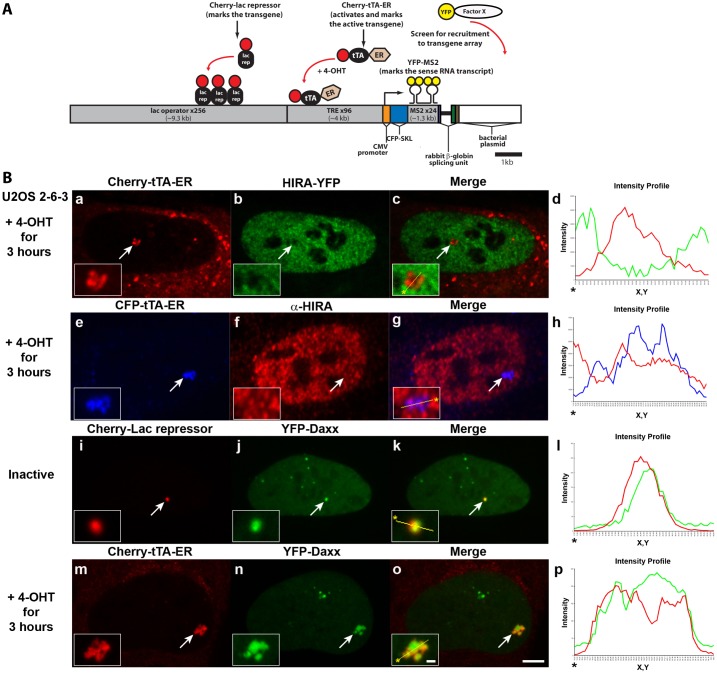
To dissect the mechanisms through which replication-independent histone H3.3 chromatin assembly factors regulate transcription, we used an engineered transgene system, which allows DNA, RNA and proteins to be visualized in single living cells (Janicki et al., 2004). Cell lines with stably integrated multi-copy arrays of this transgene can be isolated and the inactive array visualized by expressing lac repressor fused to an auto-fluorescent protein (Fig. 1A). Transcription is induced from the CMV minimal promoter using tetracycline-regulated activators. When the tetracycline-regulated transcriptional activator (tTA) tagged with an auto-fluorescent protein is fused to the estrogen receptor hormone binding domain (Cherry/CFP/YFP-tTA-ER), it is retained in the cytoplasm until the addition of 4-hydroxytamoxifen (4-OHT) to the media induces its entry into the nucleus where it binds to the TRE repeats and activates transcription (Fig. 1A) (Rafalska-Metcalf et al., 2010). Newly synthesized RNA can be monitored through the interaction of the MS2 coat protein with the stem loops in the transcript, and translation can be confirmed by the appearance of the CFP-SKL protein in the peroxisomes in the cytoplasm. Therefore, this ‘synthetic’ gene contains all of the elements required for RNA pol II transcription, mRNA processing, and protein translation. As such, it is a powerful tool for defining the temporal and spatial organization of gene regulatory events directly at chromatin (Janicki et al., 2004).
Fig. 1.
The histone H3.3 chaperone, Daxx, is recruited to the transgene array in U2OS cells. (A) Diagram of the inducible transgene drawn to scale. Expression of Cherry-lac repressor allows the inactive transgene array to be visualized. Transcription is induced by the activator, Cherry-tTA-ER, in the presence of 4-hydroxytamoxifen (4-OHT). The transcribed RNA encodes CFP fused to a peroxisomal targeting signal (SKL). The RNA is visualized by YFP-MS2, which binds to the stem loops in the transcript. The 3′ end of the transcription unit is the intron 2 splicing unit of the rabbit β-globin gene. The recruitment of YFP-tagged factors to the array can be monitored by co-expression with Cherry-lac repressor or Cherry-tTA-ER. (B) Localization of HIRA-YFP (a–d) and endogenous HIRA labeled with α-HIRA antibody (e–h) in relation to Cherry- or CFP-tTA-ER, at the activated transgene array in U2OS 2-6-3 cells. YFP-Daxx is enriched at the inactive array, marked by Cherry-lac repressor (i–l), and the activated array, marked by Cherry-tTA-ER (m–p). Yellow lines in enlarged merge insets show the path through which the red, green and blue intensities were measured in the intensity profiles (d, h, l and p). Asterisks mark the start of the line. Scale bar: 5 µm; inset: 1 µm.
To investigate replication independent histone H3.3 chromatin assembly using this system, we evaluated the localization of HIRA and Daxx at a transgene array stably integrated into U2OS cells (cell line, 2-6-3) (Janicki et al., 2004). Neither YFP-tagged nor antibody labeled endogenous HIRA (Fig. 1B, panels a−h) accumulated at the activated array marked by the activator, Cherry/CFP-tTA-ER. In contrast, both YFP-tagged and endogenous Daxx were enriched at the inactive and activated arrays, marked by Cherry-lac repressor and Cherry-tTA-ER, respectively (Fig. 1B, panels i–p; supplementary material Fig. S1). The enrichment of Daxx, but not HIRA, suggests that it, together with ATRX, regulates histone H3.3 chromatin assembly at the array and is consistent with reports that Daxx and HIRA function at mutually exclusive sites (Elsaesser and Allis, 2010; Goldberg et al., 2010).
ATRX represses activation of the transgene array in U2OS cells
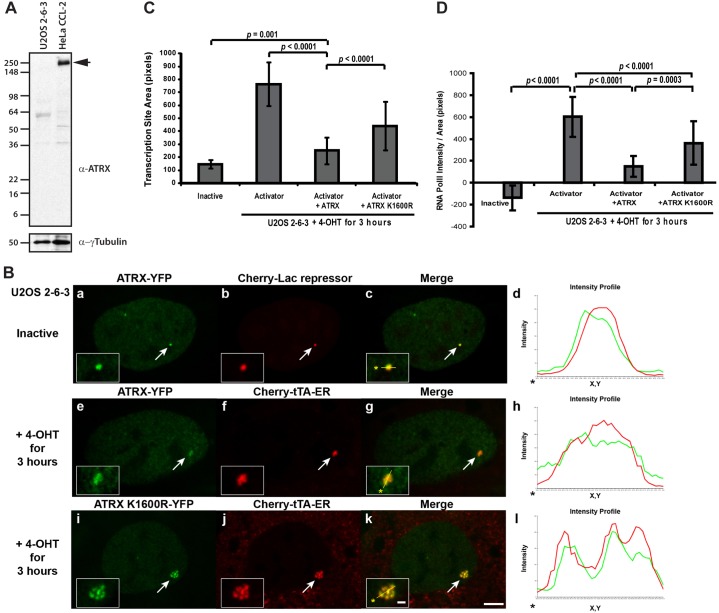
As U2OS cells do not express ATRX (Heaphy et al., 2011; Lukashchuk and Everett, 2010) (Fig. 2A), this suggests that the Daxx and ATRX pathway is not functional in this cell line. To determine whether exogenously expressed ATRX is recruited to the array, we co-expressed ATRX-YFP and Cherry-lac repressor. As they co-localized (Fig. 2B, panels a–d), this suggested that ATRX functions at the site.
Fig. 2.
ATRX represses transcriptional activation of the transgene array in U2OS cells. (A) Western blot of endogenous ATRX in U2OS 2-6-3 and HeLa cells. γ-Tubulin is used as a loading control. (B) Enrichment of ATRX-YFP at the inactive array, marked by Cherry-lac repressor (a–d). Enrichment of ATRX-YFP (e–h) and ATRX(K1600R)-YFP (i–l) at the activated array, marked by Cherry-tTA-ER. Yellow lines in enlarged merge insets show the path through which the red and green intensities were measured in the intensity profiles (d, h and l). Asterisks mark the start of the line. Scale bar: 5 µm; inset: 1 µm. (C) Pixel areas of the inactive array, marked by Cherry-lac repressor (n = 16), and the Cherry-tTA-ER activated arrays in control (n = 16), ATRX-YFP (n = 82) and ATRX(K1600R)-YFP (n = 68) in U2OS 2-6-3 cells. (D) Mean intensity levels of RNA pol II (4H8 Ab staining) at the inactive (n = 16) and CFP-tTA-ER activated arrays in control (n = 12), ATRX-YFP (n = 14) and ATRX(K1600R)-YFP (n = 18) expressing U2OS 2-6-3 cells. Standard deviations, in the form of error bars, are shown in the graph. P values were calculated using unpaired t-test.
Previously, we reported that the transgene array in this cell line significantly decondenses during activation (Janicki et al., 2004; Rafalska-Metcalf et al., 2010). To evaluate the effects of ATRX on array decondensation, we measured the pixel areas of the regions bound by Cherry-lac repressor (inactive array) and Cherry-tTA-ER (activated arrays). In activated control cells, the array was ∼5× larger than the inactive site (Fig. 2C). However, in ATRX-YFP-expressing cells, it was only ∼1.7× larger indicating that ATRX represses decondensation (Fig. 2B, panels e–h, Fig. 2C). To evaluate the function of the ATRX helicase domain, we expressed a construct containing the mutation, K1600R, which is commonly used to abolish ATPase activity in SWI/SNF proteins (Mitson et al., 2011). In ATRX (K1600R)-YFP expressing cells, the area of the activated array increased ∼3 fold (Fig. 2B, panels i–l, Fig. 2C) indicating that the helicase activity is required to maintain the condensed state.
We also previously reported that transcription is required for chromatin decondensation because in α-amanatin-treated cells, the array remains condensed despite activator and transcription factor recruitment (Rafalska-Metcalf et al., 2010). To determine whether ATRX inhibits decondensation by preventing transcription, we measured RNA pol II levels at the array. At the inactive array RNA pol II levels are below background, which indicates that the activator is required for its recruitment (Fig. 2D). As the RNA pol II at the activated array in ATRX-YFP expressing cells is ∼4-fold lower compared to control cells, this indicates that it prevents RNA pol II accumulation (Fig. 2D; supplementary material Fig. S2). Consistent with this result, RNA levels at the transcription site in ATRX-YFP expressing cells, measured by Cherry-MS2 intensity, are ∼7-fold lower than in control cells (Fig. 3C). In ATRX(K1600R)-YFP expressing cells, the RNA pol II (Fig. 2D; supplementary material Fig. S2) and RNA levels (Fig. 3C) at the activated array are ∼2.5 and 4× higher, respectively, compared to ATRX-YFP expressing cells indicating that the helicase activity is required to repress transcription.
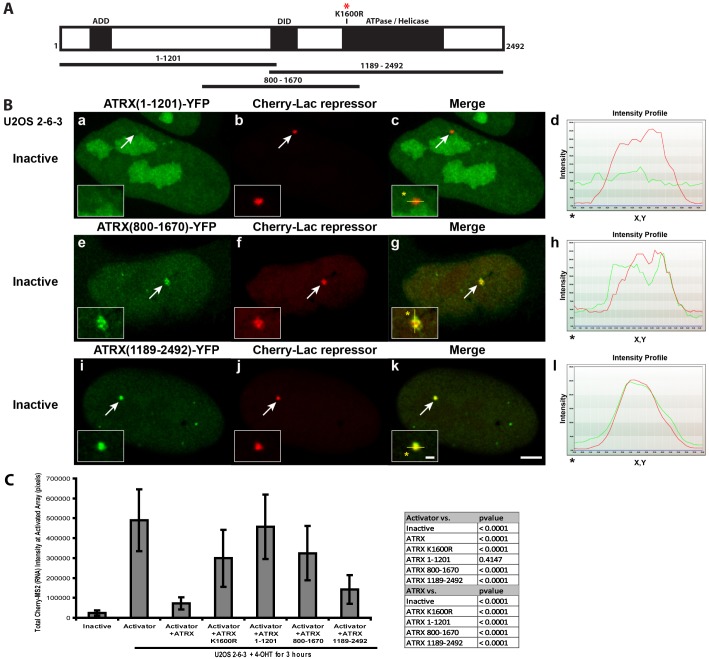
Fig. 3.
Analysis of the ATRX domains required for recruitment and repression of the transgene array in U2OS cells. (A) Diagram of ATRX showing the location of the ADD domain, the Daxx interaction domain (DID), the ATPase/helicase domain, the K1600R catalytic mutation and the amino acid composition of the YFP-tagged deletion constructs. (B) Localization of the ATRX deletion constructs at the inactive array in U2OS 2-6-3 cells marked by Cherry-lac repressor. Yellow lines in enlarged merge insets show the path through which the red and green intensities were measured in the intensity profiles (d, h and l). Asterisks mark the start of the line. Scale bar: 5 µm; inset: 1 µm. (C) Integrated intensity measurements of the accumulation of Cherry-MS2 (RNA) at the inactive array (n = 20) and CFP-tTA-ER activated arrays in control (n = 34), ATRX-YFP (n = 41), ATRX(K1600R)-YFP (n = 57), ATRX(1–1201)-YFP (n = 28), ATRX(800–1670)-YFP (n = 37) and ATRX(1189–2492)-YFP (n = 38) expressing U2OS 2-6-3 cells. Standard deviations, in the form of error bars, are shown in the graph. P values were calculated using unpaired t-test and are shown in the table.
The ATRX helicase domain represses transcription
As Daxx is enriched at the transgene array in U2OS cells, this indicates that it is not required for its recruitment ATRX. To identify the region within ATRX required for its recruitment, we examined the localization of a series of YFP-tagged ATRX deletion constructs at the inactive array, marked by Cherry-lac repressor (Fig. 3A,B). ATRX(1–1201), which includes the ADD domain, an atypical histone H3-lysine 9 trimethylation binding module required for ATRX recruitment to pericentric heterochromatin (Eustermann et al., 2011; Iwase et al., 2011), was not enriched suggesting that histone modifications do not recruit ATRX to the array (Fig. 3A,B, panels a–d). However both ATRX(800–1670) and ATRX(1189–2492), which had previously been shown to be recruited to PML-NBs/ND10s (Bérubé et al., 2008; Tang et al., 2004), were enriched at the array suggesting that the recruitment signal is within the 1189–1670 amino acid region (Fig. 3A,B, panels e–l). As it contains a previously identified Daxx interaction domain (DID) (Tang et al., 2004), this suggests that ATRX is recruited through its interaction with Daxx.
To evaluate the effects of these deletion constructs on transcription, we used Cherry-MS2 to measure RNA accumulation at the activated array (Fig. 3C). Consistent with its lack of recruitment to the array, ATRX(1–1201) did not have a repressive effect. Among the deletion constructs, RNA levels were the lowest in ATRX(1189–2492) expressing cells indicating that the helicase domain has the most significant effect on repression.
Daxx, ATRX and PML are enriched at the CMV promoter of the transgene
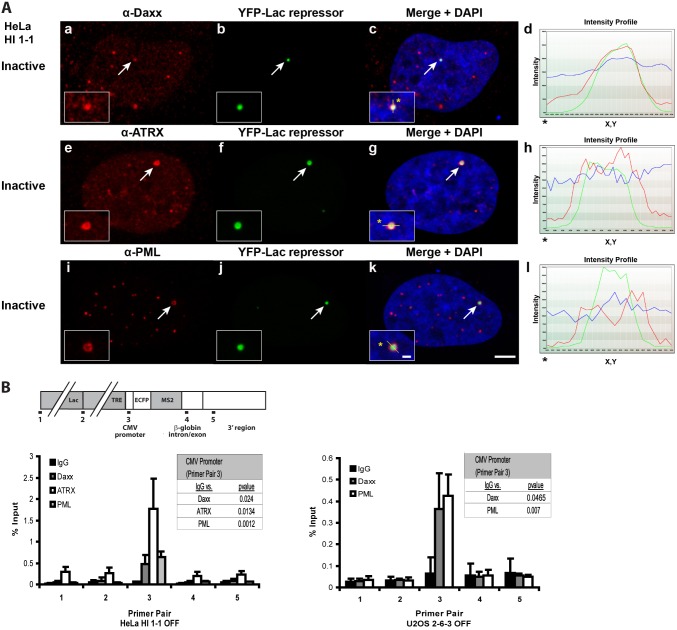
To evaluate transcription in a cell line, which expresses both Daxx and ATRX, we stably integrated the inducible transgene (Fig. 1A) into HeLa cells (cell line, HI 1-1). By immunofluorescence, Daxx and ATRX both co-localized with YFP-lac repressor at the multi-copy array (Fig. 4A, panels a–h). Daxx and ATRX are also found in PML-NBs/ND10s, therefore, we evaluated the localization of PML protein, which is the defining component of these structures (Geoffroy and Chelbi-Alix, 2011). As PML also co-localized with YFP-lac repressor (Fig. 4A, panels i–l), this suggests that the transgene array is a PML-NB/ND10. PML is also enriched at both the inactive and activated array in the U2OS cell line, which indicates that ATRX is not required for its recruitment and that it is not displaced during activation (supplementary material Fig. S3). To identify the transgene sequence to which Daxx, ATRX and PML are recruited, we used chromatin immunoprecipitation (ChIP). All three are enriched at the CMV promoter in the HeLa cell line; in U2OS cells, Daxx and PML are similarly enriched at the promoter (Fig. 4B). These results, therefore, indicate that the promoter sequence regulates the recruitment of Daxx, ATRX and PML to these transgene arrays.
Fig. 4.
Daxx, ATRX and PML are enriched at the CMV promoter of the transgene in HeLa and U2OS cells. (A) Immunofluorescence localization of Daxx (a–d), ATRX (e–h) and PML (i–l) at the inactive array marked YFP-lac repressor in HeLa, HI 1-1 cells. Yellow lines in enlarged merge insets show the path through which the red, green and DAPI intensities were measured in the intensity profiles (d, h and l). Asterisks mark the start of the line. Scale bar: 5 µm; inset: 1 µm. (B) Diagram of the transgene showing the location of primers used for real-time PCR in the ChIP assays. Daxx, ATRX, and PML were evaluated in chromatin lysates prepared from inactive HeLa, HI 1-1 and U2OS 2-6-3 cells. Results are the average of at least three independent experiments. Standard deviations, in the form of error bars, are presented in the graphs. P values were calculated using unpaired t-test and are shown in the tables.
Histone H3.3 is specifically enriched at the transgene array in HeLa cells
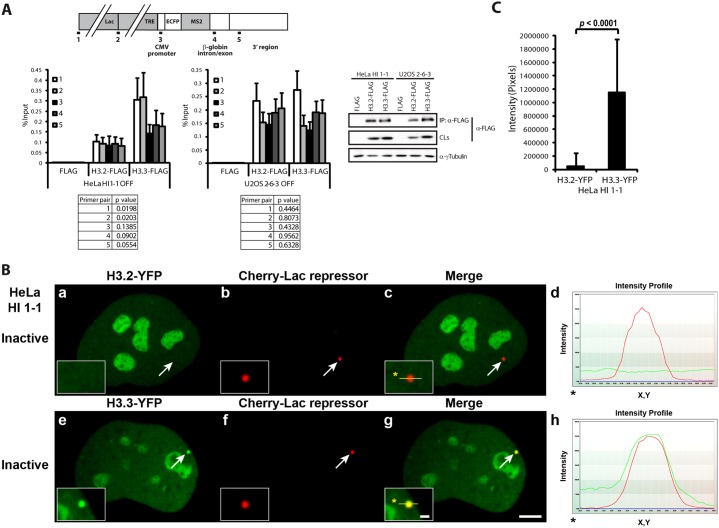
To evaluate the incorporation of histone H3.3 into the chromatin of the transgene arrays in the HeLa and U2OS cell lines, we used flag-tagged constructs in a native ChIP assay (Fig. 5A). In HeLa cells, histone H3.3 is significantly more enriched in the lac operator repeats compared to the replication-dependent variant, H3.2 (Fig. 5A). In contrast, they were comparably incorporated into the transgene chromatin in U2OS cells (Fig. 5A). The increased incorporation of histone H3.3 into the transgene array in HeLa cells could also be detected visually when YFP-tagged constructs were transiently expressed (Fig. 5B; note overlap of red and green signals in intensity profile, panel h). Intensity measurements indicated that H3.3-YFP is ∼24× more enriched at the site compared to H3.2-YFP (Fig. 5C). ChIP analysis of histone H3.3-YFP and H3.2-YFP stably expressed in HeLa cells confirmed that histone H3.3 is more enriched in the lac operator repeats (supplementary material Fig. S4). These result, therefore, indicate that in HeLa cells, which express both Daxx and ATRX, histone H3.3 is preferentially incorporated into the chromatin of the lac operator repeats in the transgene.
Fig. 5.
Histone H3.3 is specifically incorporated into the transgene array chromatin in HeLa cells. (A) Diagram of the transgene showing the location of primers used for real-time PCR in the ChIP assay. Flag-tagged histone H3.2 and H3.3 were evaluated by native ChIP in inactive HeLa, HI 1-1 and U2OS, 2-6-3 cells. Results are the average of at least three independent experiments. Standard deviations, in the form of error bars, are presented in the graphs. P values for comparison of H3.2 and H3.3 levels using each primer pair are presented in the tables and were calculated using unpaired t-test. Western blot shows histone H3.2 and H3.3 immunoprecipitated with flag antibody and in crude lysates (CLs). γ-Tubulin is used as a loading control. (B) Enrichment of transiently expressed histone H3.2-YFP (a–d) and H3.3-YFP (e–h) at the inactive transgene array, marked by Cherry-lac repressor, in HeLa HI 1-1 cells. Yellow lines in enlarged merge insets show the path through which the red and green intensities were measured in the intensity profiles (d and h). Asterisks mark the start of the line. Scale bar: 5 µm; inset: 1 µm. (C) Intensity measurements of transiently expressed H3.2-YFP (n = 25) and H3.3-YFP (n = 23) at the inactive array, marked by Cherry-lac repressor in HeLa HI 1-1 cells. Example images are shown in B. Standard deviations, in the form of error bars, are presented in the graphs. P values were calculated using unpaired t-test.
Daxx is not required for transgene array activation in U2OS cells
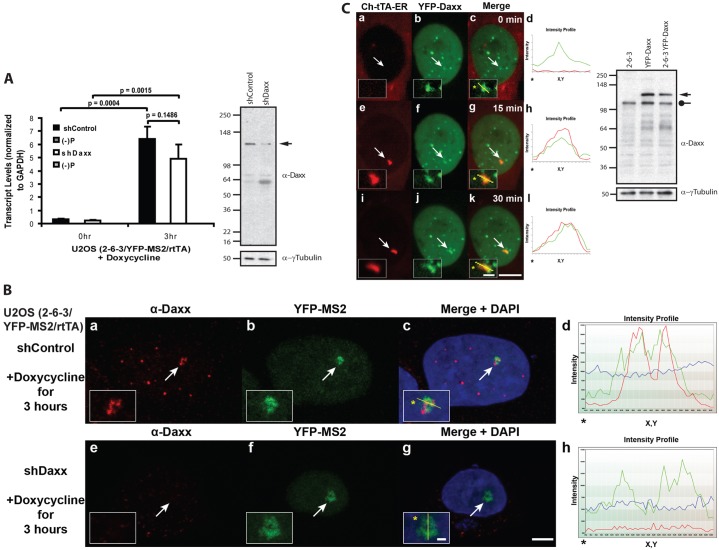
As the transgene array in U2OS cells can be rapidly induced, we wanted to determine whether Daxx is required for activation. To evaluate the effects of depleting Daxx on transcription, we knocked it down using a validated shRNA construct (Lukashchuk and Everett, 2010). Quantitative RT-PCR (qRT-PCR) analysis indicated that transcription is induced to comparable levels in shControl and shDaxx-expressing cells (Fig. 6A). However, as knocking down Daxx induced high levels of cell death, the incomplete depletion of the protein seen in the western blot (Fig. 6A) likely reflects the selective advantage of cells, which still express it. Therefore, we used YFP-MS2 to evaluate transcription in single cells (Fig. 6B). As YFP-MS2 was present at the activated array in Daxx-depleted cells, this confirmed that it is not required for activation.
Fig. 6.
Daxx is not required for transgene array activation in U2OS cells. (A) Quantitative RT-PCR analysis of RNA collected from U2OS (2-6-3/YFP-MS2/rtTA) cells expressing control and Daxx shRNAs. Results are the average of three independent experiments. Standard deviations, in the form of error bars, are presented in the graphs. P values were calculated using unpaired t-test. Western blot shows Daxx protein levels in knockdown cells. γ-Tubulin is used as a loading control. (B) Single-cell analysis of YFP-MS2 accumulation at the activated transcription site in U2OS (2-6-3/YFP-MS2/rtTA) cells expressing control and Daxx shRNAs. The same exposure settings were used to image Daxx levels in these cells. Scale bar: 5 µm; inset: 1 µm. (C) Single-cell time lapse imaging of YFP-Daxx at the transgene array, marked by Cherry-tTA-ER, in U2OS 2-6-3 stably expressing YFP-Daxx. Western blot shows the levels of endogenous (circle) and stably expressed YFP-Daxx (arrow) in the U2OS 2-6-3 cell line. Images were taken at 0 min and 15 and 30 min after the addition of 4-OHT. Yellow lines in enlarged merge insets show the path through which the red and green intensities were measured in the intensity profiles (d, h and l). Asterisks mark the start of the line. Scale bar:10 µm; inset: 2 µm.
To evaluate the interaction of Daxx with the transgene array in single cells, we expressed the activator, Cherry-tTA-ER, in a U2OS 2-6-3 cell line stably expressing YFP-Daxx and imaged cells 0, 15 and 30 min after activation (Fig. 6C). By comparing the region of the nucleus in which Cherry-tTA-ER accumulates at 15 min to the 0 min picture (Fig. 6C, compare panels c and g), we determined that Daxx is enriched at the array prior to activator binding. This indicates that Daxx recruitment is not due to the presence of proteins which bind to the repeat sequences in the transgene (Tsukamoto et al., 2000).
ICP0 permits transcriptional activation of the transgene array in HeLa cells
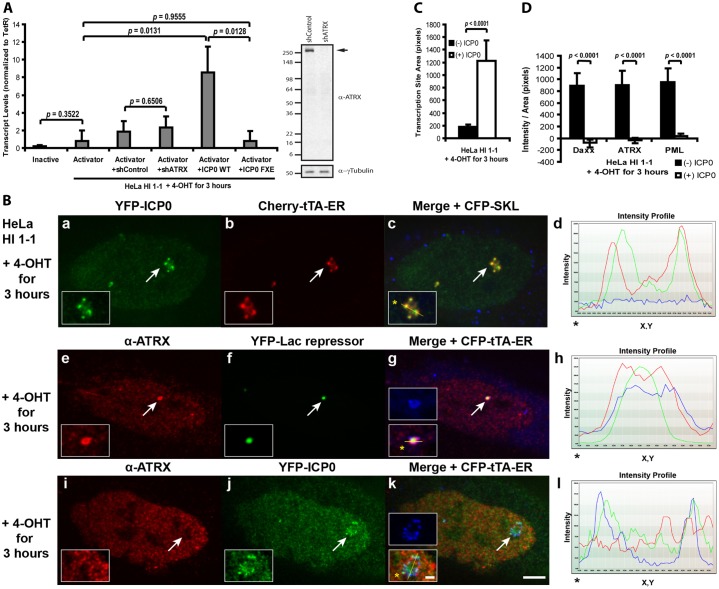
To further delineate the mechanism through which ATRX represses transcription of the CMV-promoter regulated transgene, we evaluated transcriptional activation in the HeLa cell line. As RNA was not induced in activator expressing cells, this indicates that the array is refractory to activation (Fig. 7A). To determine whether depleting ATRX is sufficient to permit activation, we knocked it down using a validated shRNA construct (Lukashchuk and Everett, 2010) (Fig. 7A). As transcription was not significantly induced in ATRX knock down cells, this suggests that either low levels of ATRX, sufficient to maintain repression, are still present or that in this cell type, the ATRX/Daxx pathway establishes a repressive chromatin environment that cannot be efficiently reversed by depleting ATRX alone.
Fig. 7.
ICP0 makes the transgene array in HeLa cells permissible to activation. (A) Quantitative RT-PCR analysis of RNA collected from HeLa HI 1-1 cells. Inactive and Cherry-tTA-ER expressing control cells are compared to those co-expressing control and ATRX shRNAs, ICP0 and ICP0-FxE, in which the zinc-binding motif in the RING finger domain is deleted. Results are an average of at least three independent experiments. Western blot shows ATRX protein levels in knockdown cells. γ-Tubulin is used as a loading control. (B) Recruitment of YFP-ICP0 to the activated array, marked by Cherry-tTA-ER, in HeLa HI 1-1 cells (a–d). Immunofluorescence staining of ATRX at the CFP-tTA-ER activated transgene array in control (e–h) and ICP0-expressing (i–l) HeLa HI 1-1 cells. Yellow lines in enlarged merge insets show the path through which the red, green and blue intensities were measured in the intensity profiles (d, h, and l). Asterisks mark the start of the line. Scale bar: 5 µm; inset: 1 µm. (C) Measurement of the area of the activated transgene array in HeLa HI 1-1cells, (−)ICP0 (n = 43) and (+)ICP0 (n = 38). (D) Measurement of mean intensity of Daxx [(−)ICP0, n = 25; (+)ICP0, n = 25], ATRX [(−)ICP0, n = 18; (+)ICP0, n = 21] and PML [(−)ICP0, n = 25; (+)ICP0, n = 24] at the activated array in HeLa, HI 1-1, cells. Standard deviations, in the form of error bars, are presented in the graphs. P value was calculated using unpaired t-test.
However, viral proteins, including the HSV-1 E3 ubiquitin ligase, ICP0, which is required to promote lytic replication and to reverse latency, are able to rapidly convert repressed chromatin to an active state (Hagglund and Roizman, 2004). Therefore, to determine whether ICP0 expression would permit activation of the transgene array in the HeLa cell line, we co-expressed YFP-ICP0 and the activator, Cherry-tTA-ER; indeed, activation was robust. Cherry-tTA-ER accumulated at the array, which decondensed ∼7-fold compared to the inactive site, and the CFP-SKL protein accumulated in the peroxisomes in the cytoplasm (Fig. 7B, panels a–d; Fig. 7C). As YFP-ICP0 co-localizes with Cherry-tTA-ER, this suggests that its effects on the array chromatin are direct. qRT-PCR analysis indicates that RNA levels are ∼11-fold higher in ICP0-expressing cells compared to controls (Fig. 7A). As RNA levels did not increase in cells expressing ICP0-FxE, a mutant deleted of the RING finger zinc-binding motif, which is required for PML degradation and viral genome reactivation (Everett et al., 1998; Parkinson and Everett, 2000; Smith et al., 2011), this indicates that the ICP0 RING finger E3 ubiquitin ligase activity is required for activation (Fig. 7A).
To identify the mechanism through which ICP0 creates a transcriptionally permissive chromatin environment, we evaluated ATRX, Daxx and PML levels at the activated array. In control ICP0(-) cells, ATRX remained enriched and CFP-tTA-ER formed only a faint ring around the array, suggesting that it is unable to access its binding sites (Fig. 7B, panels e–h; Fig. 7D). In fact, co-expression of YFP-lac repressor was needed to identify the site. In contrast, ATRX is depleted and the activator becomes strongly enriched at the array in ICP0-expressing cells (Fig. 7B, panels i–l; Fig. 7D). As Daxx and PML levels were similarly reduced by ICP0 expression, this suggests that ICP0 permits activation by altering the association of multiple regulatory factors, including but likely not limited to Daxx, ATRX and PML, with the CMV-promoter-regulated array (Fig. 7D).
Histone H3.3 is strongly recruited to the activated transgene array but is not incorporated into the chromatin
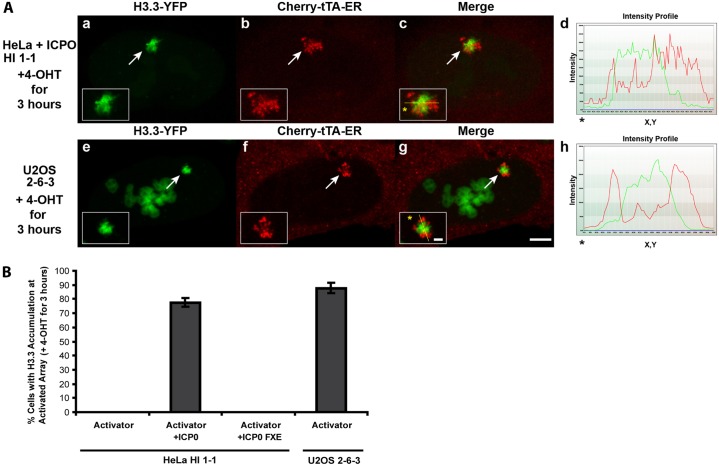
As transgene array activation is robustly induced in the ATRX-null-U2OS and ICP0-expressing HeLa cell lines, this suggests that repression is relieved when Daxx and ATRX-mediated chromatin assembly is blocked. To investigate the effect on histone H3.3 recruitment, we examined H3.3-YFP localization in relation to Cherry-tTA-ER. In both the U2OS and HeLa cell lines, H3.3-YFP is strongly recruited to the majority of the activated sites (Fig. 8A,B). However, in contrast to it localization pattern at the inactive array in HeLa cells where it completely co-localizes with Cherry-lac repressor (Fig. 5B, panels e–h), H3.3-YFP accumulated in regions of the activated array which were adjacent but non-overlapping with Cherry-tTA-ER (Fig. 8A). As the activator binds directly to the TRE repeats (∼4 kb) in the transgene, this suggests that H3.3-YFP is not incorporated into the nucleosomes in this region. As ATRX and Daxx in ICP0-expressing cells are not present at the activated arrays, this also indicates that they are not required for histone H3.3 recruitment.
Fig. 8.
Histone H3.3 is recruited to the activated transgene array in U2OS and ICP0-expressing HeLa cells. (A) H3.3-YFP enrichment at the activated transgene array, marked by Cherry-tTA-ER, in ICP0-expressing HeLa, HI 1-1 cells (a–d) and U2OS, 2-6-3 cells (e–h). Yellow lines in enlarged merge insets show the path through which the red and green intensities were measured in the intensity profiles (d and h). Asterisks mark the start of the line. Scale bar: 5 µm; inset: 1 µm. (B) Percentage of cells in which H3.3-YFP accumulated at the activated transgene array in control, ICP0 and ICP0-FxE expressing HeLa, HI 1-1 cells and U2OS 2-6-3 cells. 100 cells were counted in three independent transfections.
To evaluate histone H3.3-YFP in relation to the lac operator repeats (∼10 kb) localization at the activated array, we co-expressed Cherry-lac repressor in the U2OS line. Again, H3.3-YFP did not co-localize with the DNA binding protein (supplementary material Fig. S5, panels a–d). To evaluate its localization in relation to factors that accumulate on the transcription unit (∼1.7 kb), we labeled with RNA pol II. H3.3-YFP and RNA pol II localized in adjacent but non-overlapping regions suggesting that histone H3.3 is also not incorporated into the chromatin in this region (supplementary material Fig. S5, panels e–h). The co-localization of RNA pol II with the activator, YFP-tTA-ER, however, confirms that factors, which bind directly to the transgene DNA and/or function on chromatin, occupy the same regions of the activated array (supplementary material Fig. S5, panels i–l).
To determine whether the H3.3-YFP accumulation at the activated array is due to the lac operator and/or MS2 stem loop repeats, we evaluated its recruitment to an activated array composed of a transgene without these elements, which is stably integrated into U2OS cells (cell line, 60-1). The enrichment of histone H3.3-YFP at the activated array in this cell line indicates that its recruitment is not due to these sequence elements, the absence of which was confirmed by the lack of Cherry-lac repressor and Cherry-MS2 accumulation at the site (supplementary material Fig. S6). These results, therefore, further strengthen the conclusion that histone H3.3 chromatin assembly is blocked at the transgene array when the Daxx and ATRX pathway is disrupted and suggest that the recruitment of histone H3.3 to the activated array is a result of transcription.
Discussion
In this study, we show that Daxx and ATRX regulate transcriptional repression at a multi-copy array of a transgene regulated by the CMV promoter. Transcription can be robustly induced, however, when this pathway is disrupted by either ATRX-deletion or ICP0 expression, which depletes Daxx and ATRX from the chromatin. Additionally, activation-induced chromatin decondensation, mRNA expression and RNA pol II accumulation at the array are significantly reduced when ATRX is expressed in the ATRX-null U2OS cell line indicating that the repression defect can be rescued. Taken together, these results indicate that the Daxx and ATRX pathway regulates transcriptional repression at the viral CMV promoter.
Before the functions of Daxx and ATRX in replication-independent histone H3.3 chromatin assembly were elucidated (Drané et al., 2010; Goldberg et al., 2010; Lewis et al., 2010), both were known to localize to PML-NBs/ND10s, which are defined by the presence of PML protein (Geoffroy and Chelbi-Alix, 2011). As Daxx, ATRX and PML are all enriched at the transgene arrays in both HeLa and U2OS cells, this suggests that the array can be classified as a PML-NB/ND10, and conversely, that at least some PML-NBs/ND10s are transcription sites. Using similar transgene engineering methodology, two other groups demonstrated that transcription and RNA seed the formation of nuclear bodies, further supporting the link between nuclear organization and molecular function (Mao et al., 2011; Shevtsov and Dundr, 2011).
We also show by ChIP, that ATRX, Daxx and PML are enriched at the CMV promoter of the transgene, which suggests that the promoter sequence regulates the recruitment of histone H3.3 chromatin assembly factors. Daxx has previously been reported to bind to and/or transcriptionally repress cellular and viral promoters, including c-met, RelB target genes and CMV (Croxton et al., 2006; Morozov et al., 2008; Puto and Reed, 2008; Reeves et al., 2010; Zhang et al., 2010). It is not known whether ATRX is required for the Daxx-mediated repression of these genes, however, the Drosophila ATRX homolog was recently shown to repress the pannier gene through a promoter-mediated mechanism (Valadez-Graham et al., 2012). Additionally, the mutually exclusive enrichment of active factors, including RNA pol II, and repressive factors, including Daxx, at the promoters of the murine CMV immediate early (IE) genes during the lytic and latent phases of infection, respectively (Liu et al., 2010), is reminiscent of our result showing that RNA pol II accumulation at the activated array is significantly reduced by ATRX expression in the U2OS cell line. This suggests that an important step in the establishment of latency is the creation of a chromatin environment that excludes transcription factors.
Our demonstration that ICP0 depletes Daxx and ATRX from the array in HeLa cells and permits transcriptional activation further supports the conclusion that chromatin organization is a critical regulator of repression and viral latency. Although the role of replication independent histone H3.3 chromatin assembly in this process is not completely clear, ChIP analyses indicate that histone H3.3 is specifically incorporated into the lac operator repeats of the transgene in HeLa cells, which express both Daxx and ATRX. Its incorporation into this repetitive region is consistent with reports that histone H3.3 is enriched in repetitive regions of the genome including telomeres in ESCs (Goldberg et al., 2010) and satellite DNA in Drosophila (Schneiderman et al., 2009).
Although loss of chromatin assembly provides a satisfying explanation for how disrupting the Daxx and ATRX pathway relieves transcriptional repression and is consistent with reports that the levels of histones associated with the HSV-1 genome are lower during lytic infection (Cliffe and Knipe, 2008), the surprising finding in this study is that histone H3.3 is strongly recruited to the activated array. Additionally, its enrichment at the site in the absence of ATRX (U2OS cells) as well as Daxx in ICP0-expressing HeLa cells indicates that Daxx and ATRX are not required for its recruitment. This result, therefore, suggests that histone H3.3 is recruited to the activated array by an as yet unidentified chaperone or through a chaperone-independent mechanism. The dramatic effects that ICP0 has on the localization of regulatory factors at the array provides visible proof of how viruses are able to harness cellular regulatory mechanisms to promote their own replication. The recruitment of histone H3.3 to the activated array in the absence of chromatin assembly suggests that it may have a non-nucleosomal function, which has been unmasked by these conditions. The recent discovery that histone H3.3 acquires gain of function driver mutations in pediatric glioblastoma (Schwartzentruber et al., 2012; Wu et al., 2012) also supports the idea that it may be an active participant in cellular signaling events and not merely delivered passively to the nucleosome.
Materials and Methods
Cell culture
The U2OS cell line, 2-6-3, and its growth conditions were previously described (Janicki et al., 2004; Rafalska-Metcalf et al., 2010). The HeLa cell line (HI 1-1) containing the stably integrated array of p3216PECMS2β was made as previously described (Janicki et al., 2004; Tsukamoto et al., 2000) with selection in 150 µg/ml of Hygromycin B (Sigma, St. Louis, MO, USA). HI 1-1 cells are grown in Eagle's minimal essential medium (MEM) supplemented with 10% Tet-System Approved FBS (Clontech, Mountain View, CA, USA), Penicillin-streptomycin (1:100), sodium pyruvate (1 mM) and L-glutamine (2 mM) (Life Technologies, Grand Island, NY, USA). The HI 1-1 cell lines stably expressing H3.3-YFP (HI 1-1/H3.3-YFP) and H3.2-YFP (HI 1-1/H3.2-YFP) were made by selecting in 700 µg/ml of G418 (Life Technologies, Grand Island, NY, USA). The U2OS 2-6-3 cell line stably expressing YFP-MS2 and rtTA (U2OS/2-6-3/YFP-MS2/rtTA) and YFP-Daxx (2-6-3/YFP-Daxx) were made by selecting in 400 µg/ml of G418 (Life Technologies, Grand Island, NY, USA). The U2OS cell line, 60-1, was made by stably integrating a multi-copy array of the p16PEChβglobin transgene, which contains the TRE repeats, the CMV minimal promoter, CFP-SKL coding sequence and the 3′ end of the human β-globin gene as previously described (Janicki et al., 2004) by selection in 100 µg/ml of Hygromycin B (Sigma, St Louis, MO, USA).
Plasmids
SV2-Cherry/CFP-lac repressor, Cherry/YFP-MS2, Cherry/YFP/CFP-tTA-ER, pLU-YFP/Cherry-tTA-ER, and the pLU lentiviral plasmid (Rafalska-Metcalf et al., 2010), H3.3-YFP (Ahmad and Henikoff, 2002; Janicki et al., 2004) (gift of S. Henikoff and K. Ahmad) were previously described. H3.2-YFP was made by overlapping PCR to include the same 7 aa linker (Ser-Arg-Pro-Pro-Val-Ala-Thr) in the H3.3-YFP construct. To make pLU-H3.3-3XFLAG, an H3.3 PCR product was cloned into p3XFLAG-CMV-14 (Sigma, St Louis, MO, USA) (NotI/BamHI) then moved with the FLAG tag to pLU (XbaI/SalI). To make pLU-H3.2-3XFLAG, an H3.2 PCR product was cloned into p3XFLAG-CMV-14 (NotI/BamHI) then moved with the FLAG tag (XbaI/XhoI) to pLU (XbaI/SalI). YFP-Daxx-C1 was made by moving the Daxx cDNA (BamHI) from GFP-DAXX-C1 (Ishov et al., 1999) (gift of Gerd Maul). HIRA-YFP-N1 was made by cloning a PCR product of HIRA (gift of Genevieve Almouzni), which included a 23-aa linker (XhoI/SacII). ATRX-YFP-N1 was made by cloning a PCR product of full length ATRX (gift of David Picketts) (Bérubé et al., 2008) into pLU (XbaI/SalI) then replacing the 3′ end, which had been fused in frame to YFP YFP-N1 at the AgeI site (BamHI/SalI). pLU-ATRX(K1600R)-YFP (Mitson et al., 2011) was made by introducing a point mutation using QuikChange kit (Agilent Technologies, Santa Clara, CA, USA). ATRX(1–1201) was made by cloning PCR product (XhoI/SacII) into YFP-N1 (For: ccgctcgagATGACCGCTGAGCCCATGAGT; Rev: gctctagaTCATGTAATGTCAGCTTGCTTCCTTTTAGT) ATRX(800–1670)(For: ccgctcgagATGAGCTCTATAATTTCTAAAAAGAAACGACAAAC; Rev: gctctagaTCACTGCAGCATGTAGCTTCTCTCCTG) and ATRX(1189–2492)(For: ccgctcgagATGCTAAGAACAAGCACTAAAAGGAAGCAAG; Rev: gctctagaTCACATTGATTTCCCTTGGGAAG) were made by cloning PCR products (XhoI/XbaI) into YFP-C3. ICP0 and ICP0-FxE cDNAs (gift of Roger Everett) were cloned (blunt NcoI/EcoRI) into YFP-C3 (blunt Xho/EcoRI). For lentiviral expression, YFP tagged ICP0s (NheI/BamHI) were moved into pLU (XbaI/BamHI).
RT-PCR and qPCR analysis
1 µg of TRIzol (Life Technologies, Grand Island, NY, USA) isolated RNA was treated with DNase I (Promega, Madison, WI, USA), heated to 65°C (15 min), cooled on ice (1 hr), heated again 65°C (15 min), cooled on ice (5 min) and purified with the RNeasy kit (Qiagen, Valencia, CA, USA). Strand-specific RT reactions were done using OmniScript (Qiagen, Valencia, CA, USA) followed by qPCR with SYBR Green (Sigma, St Louis, MO, USA) using the 7500 Fast Real-Time PCR system (Applied Biosystems, Life Technologies, Carlsbad, CA, USA). qPCR data was analyzed using ddCt method according to AB guidance using GAPDH in U2OS cells and the tetracycline activator in HeLa cells for normalization. The following primer pairs were used: 5′-GGTGCAGGCTGCCTATCAG-3′ and 5′-TTTGTGAGCCAGGGCATTG-3′ (rabbit β-globin exon 3); 5′-ATGGAAATCCCATCACCATCTT-3′ and 5′-CGCCCCACTTGATTTTGG-3′ (hGAPDH); 5′-ACAGCGCATTAGAGCTGCTTAAT-3′ and 5′-GGCGAGTTTACGGGTTGTTAAA-3′ (tetR of tTA activator).
Cross-linked chromatin immunoprecipitation
Cross-linked ChIP was done as described (Zeng et al., 2006). Cells grown to ∼80% confluency were trypsinized, resuspended in 1× PBS and incubated with freshly prepared 1.5 mM EGS (ethylene glycolbis[succinimidyl succinate])(Pierce Biotechnology, Rockford, IL, USA) dissolved in DMSO (Sigma, St. Louis, MO, USA) for 25 min at room temperature with gentle shaking. Formaldehyde (1%) (Sigma, St. Louis, MO, USA) was added for another 10 min. Cross-linking was quenched with glycine-PBS (50 mM) for 10 min. Cells were centrifuged (1500 rpm; 5 min; 4°C), washed twice with 1× PBS, resuspended in 1 mL SDS Lysis Buffer (1% SDS, 10 mM EDTA, 50 mM Tris pH 8.0, freshly added protease inhibitors) and incubated on ice for 20 min. Immunoprecipitation was done as previously described (Rafalska-Metcalf et al., 2010) using 2 µg of rabbit α-DAXX (D7810) (Sigma, St Louis, MO, USA), 4 µg of rabbit α-ATRX (H300) (Santa Cruz Biotechnology, Santa Cruz, CA, USA) 4 µg of mouse α-PML (PG-M3) (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and 2 µg rabbit IgG (Abcam, Cambridge, MA, USA).
Native chromatin immunoprecipitation
Cells grown to ∼80% confluency were scraped from dishes in hypotonic TMS buffer (20 mM HEPES pH 7.5; 2.5 mM MgCl2; 250 mM Sucrose; and 0.1 mM PMSF), incubated on ice for 25 min, then spun at 3,600 rpm (10 min) to collect nuclei. For micrococcal nuclease digestion, nuclei were resuspended in 1 ml of MNase digestion buffer (20 mM HEPES, pH 7.5, 10 mM NaCl, 3 mM MgCl2, 3 mM CaCl2, and fresh protease inhibitors) and incubated for 10 min at 37°C with 200 U/ml of MNase (N-5386) (Sigma, St Louis, MO, USA). 50 mM EDTA was added to stop the reaction and nuclei were pelleted by centrifugation at 8000 g (5 min). For Flag and YFP-tagged histones, the chromatin supernatants were incubated overnight at 4°C with 20 µl of Anti-flag M2 agarose (A-2220) (Sigma, St Louis, MO, USA) and 2 µg of ChIP-grade α-GFP antibody (Ab290) (Abcam, Cambridge, MA, USA), respectively. GFP antibody was bound with protein A agarose/salmon sperm DNA (Millipore, Temecula, CA, USA) for 1 hr at 4°C. Antibody-bound beads were washed 3× with BC50 buffer (20 mM Tris-Cl pH 7.5; 0.2 mM EDTA; 50 mM NaCl; 5% glycerol; 0.1 mM PMSF). Flag IPed chromatin was eluted with 3× Flag peptide (F4799) (Sigma, St Louis, MO, USA) and GFP IPed chromatin with elution buffer (10 mM Tris pH 8.0, 5 mM EDTA, 1% SDS). DNA was isolated using QIAquick PCR purification kit (Qiagen, Valencia, CA, USA). Samples were analyzed by qPCR as previously described (Rafalska-Metcalf et al., 2010).
Imaging
Imaging methodologies were described previously (Rafalska-Metcalf et al., 2010). Intensity profiles for merged pictures were generated using SimplePCI software (Hamamatsu Corporation, Sewickley, PA, USA). Image analysis was done using ImageJ software (National Institutes of Health, USA). The averaged value of three background regions was used for normalization. Confocal images with DAPI staining were taken using the Leica TCD SP5 II confocal microscope (Leica Microsystems, Wetzler, Germany).
Knock down analysis
Depletion of endogenous ATRX and Daxx in HeLa and U2OS cells, respectively, was achieved using lentiviral vectors pLKO-shATRX90 and pLKO-shDaxx2, (gifts of Roger Everett) as previously described (Lukashchuk and Everett, 2010). Activation was induced 96–120 hrs after shRNA infection.
Immunofluorescence and western blotting
For immunofluorescence, staining was done as previously described (Rafalska-Metcalf et al., 2010). The following antibodies were used: RNA pol II (1∶500; 4H8; Covance, Emeryville, CA, USA); DAXX (1∶4000; D7810; Sigma, St Louis, MO, USA); ATRX (1∶250; H300; Santa Cruz Biotechnology, Santa Cruz, CA, USA); PML (1∶250; PG-M3; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and HIRA (Zhang et al., 2007) (gift of Peter Adams). Western blotting was done with the following antibodies: ATRX (1∶1000; α-ATRX; H300; Santa Cruz Biotechnology, Santa Cruz, CA, USA), Daxx (1∶1000; 07-471; EMD Millipore, Billerica, MA, USA), γ-tubulin (1∶1000; Sigma, St Louis, MO, USA), and GFP (1∶2000; Roche, Indianapolis, IN, USA); and Flag (1∶10,000; M2; Sigma, St Louis, MO, USA).
Supplementary Material
Acknowledgments
We thank Peter Adams, Genevieve Almouzni, David Picketts and Roger Everett for reagents, Gary LeRoy for assistance with the native ChIP, Laurence Bell and Elizabeth Strumfels for technical assistance, David White for contributions to ChIP primer design, Nadia Dahmane and Gerd Maul for critical discussions and Ellen Puré, Gerd Blobel and Dario Altieri for critical comments on the manuscript.
Footnotes
Funding
This work was funded by start-up funding from The Wistar Institute, PA Department of Health (CURE) funding; the Beckman Young Investigator Award; The Mallinckrodt Foundation; The Emerald Foundation; The March of Dimes Basil O'Connor Award, National Institutes of Health [grant number R01 GM 093000-02]; The Wistar Cancer Center Core Grant [grant number P30 CA10815; and the Imaging and Genomics/Sequencing Facilities. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.110148/-/DC1
References
- Ahmad K., Henikoff S. (2002). The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9, 1191–1200 10.1016/S1097-2765(02)00542-7 [DOI] [PubMed] [Google Scholar]
- Bérubé N. G., Healy J., Medina C. F., Wu S., Hodgson T., Jagla M., Picketts D. J. (2008). Patient mutations alter ATRX targeting to PML nuclear bodies. Eur. J. Hum. Genet. 16, 192–201 10.1038/sj.ejhg.5201943 [DOI] [PubMed] [Google Scholar]
- Cantrell S. R., Bresnahan W. A. (2006). Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J. Virol. 80, 6188–6191 10.1128/JVI.02676-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe A. R., Knipe D. M. (2008). Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J. Virol. 82, 12030–12038 10.1128/JVI.01575-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxton R., Puto L. A., de Belle I., Thomas M., Torii S., Hanaii F., Cuddy M., Reed J. C. (2006). Daxx represses expression of a subset of antiapoptotic genes regulated by nuclear factor-kappaB. Cancer Res. 66, 9026–9035 10.1158/0008-5472.CAN-06-1047 [DOI] [PubMed] [Google Scholar]
- Darzacq X., Shav–Tal Y., de Turris V., Brody Y., Shenoy S. M., Phair R. D., Singer R. H. (2007). In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 14, 796–806 10.1038/nsmb1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drané P., Ouararhni K., Depaux A., Shuaib M., Hamiche A. (2010). The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24, 1253–1265 10.1101/gad.566910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsaesser S. J., Allis C. D. (2010). HIRA and Daxx constitute two independent Histone H3.3-Containing predeposition complexes. Cold Spring Harb. Symp. Quant. Biol. 2010, 27–34 [DOI] [PubMed] [Google Scholar]
- Elsaesser S. J., Goldberg A. D., Allis C. D. (2010). New functions for an old variant: no substitute for histone H3.3. Curr. Opin. Genet. Dev. 20, 110–117 10.1016/j.gde.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S., Yang J. C., Law M. J., Amos R., Chapman L. M., Jelinska C., Garrick D., Clynes D., Gibbons R. J., Rhodes D.et al. (2011). Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat. Struct. Mol. Biol. 18, 777–782 10.1038/nsmb.2070 [DOI] [PubMed] [Google Scholar]
- Everett R. D., Freemont P., Saitoh H., Dasso M., Orr A., Kathoria M., Parkinson J. (1998). The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72, 6581–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy M. C., Chelbi–Alix M. K. (2011). Role of promyelocytic leukemia protein in host antiviral defense. J. Interferon Cytokine Res. 31, 145–158 10.1089/jir.2010.0111 [DOI] [PubMed] [Google Scholar]
- Goldberg A. D., Banaszynski L. A., Noh K. M., Lewis P. W., Elsaesser S. J., Stadler S., Dewell S., Law M., Guo X., Li X.et al. (2010). Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 10.1016/j.cell.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagglund R., Roizman B. (2004). Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78, 2169–2178 10.1128/JVI.78.5.2169-2178.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S. B., Garcia B. A., Kauer M., Baker S. P., Shabanowitz J., Hunt D. F., Allis C. D. (2005). Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc. Natl. Acad. Sci. USA 102, 6344–6349 10.1073/pnas.0502413102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy C. M., de Wilde R. F., Jiao Y., Klein A. P., Edil B. H., Shi C., Bettegowda C., Rodriguez F. J., Eberhart C. G., Hebbar S.et al. (2011). Altered telomeres in tumors with ATRX and DAXX mutations. Science 333, 425 10.1126/science.1207313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Sindre H., Stamminger T. (2002). Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J. Virol. 76, 5769–5783 10.1128/JVI.76.11.5769-5783.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J., Kalejta R. F. (2007). Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology 367, 334–338 10.1016/j.virol.2007.05.037 [DOI] [PubMed] [Google Scholar]
- Ishov A. M., Sotnikov A. G., Negorev D., Vladimirova O. V., Neff N., Kamitani T., Yeh E. T., Strauss J. F., 3rd, Maul G. G. (1999). PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147, 221–234 10.1083/jcb.147.2.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov A. M., Vladimirova O. V., Maul G. G. (2002). Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains. J. Virol. 76, 7705–7712 10.1128/JVI.76.15.7705-7712.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S., Xiang B., Ghosh S., Ren T., Lewis P. W., Cochrane J. C., Allis C. D., Picketts D. J., Patel D. J., Li H.et al. (2011). ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat. Struct. Mol. Biol. 18, 769–776 10.1038/nsmb.2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki S. M., Tsukamoto T., Salghetti S. E., Tansey W. P., Sachidanandam R., Prasanth K. V., Ried T., Shav–Tal Y., Bertrand E., Singer R. H.et al. (2004). From silencing to gene expression: real-time analysis in single cells. Cell 116, 683–698 10.1016/S0092-8674(04)00171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Shi C., Edil B. H., de Wilde R. F., Klimstra D. S., Maitra A., Schulick R. D., Tang L. H., Wolfgang C. L., Choti M. A.et al. (2011). DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331, 1199–1203 10.1126/science.1200609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P. W., Elsaesser S. J., Noh K. M., Stadler S. C., Allis C. D. (2010). Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. USA 107, 14075–14080 10.1073/pnas.1008850107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. F., Yan S., Abecassis M., Hummel M. (2010). Biphasic recruitment of transcriptional repressors to the murine cytomegalovirus major immediate-early promoter during the course of infection in vivo. J. Virol. 84, 3631–3643 10.1128/JVI.02380-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashchuk V., Everett R. D. (2010). Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J. Virol. 84, 4026–4040 10.1128/JVI.02597-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y. S., Sunwoo H., Zhang B., Spector D. L. (2011). Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 13, 95–101 10.1038/ncb2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitson M., Kelley L. A., Sternberg M. J., Higgs D. R., Gibbons R. J. (2011). Functional significance of mutations in the Snf2 domain of ATRX. Hum. Mol. Genet. 20, 2603–2610 10.1093/hmg/ddr163 [DOI] [PubMed] [Google Scholar]
- Molenaar J. J., Koster J., Zwijnenburg D. A., van Sluis P., Valentijn L. J., van der Ploeg I., Hamdi M., van Nes J., Westerman B. A., van Arkel J.et al. (2012). Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483, 589–593 10.1038/nature10910 [DOI] [PubMed] [Google Scholar]
- Morozov V. M., Massoll N. A., Vladimirova O. V., Maul G. G., Ishov A. M. (2008). Regulation of c-met expression by transcription repressor Daxx. Oncogene 27, 2177–2186 10.1038/sj.onc.1210865 [DOI] [PubMed] [Google Scholar]
- Parkinson J., Everett R. D. (2000). Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74, 10006–10017 10.1128/JVI.74.21.10006-10017.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picketts D. J., Higgs D. R., Bachoo S., Blake D. J., Quarrell O. W., Gibbons R. J. (1996). ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum. Mol. Genet. 5, 1899–1907 10.1093/hmg/5.12.1899 [DOI] [PubMed] [Google Scholar]
- Preston C. M., Nicholl M. J. (2006). Role of the cellular protein hDaxx in human cytomegalovirus immediate-early gene expression. J. Gen. Virol. 87, 1113–1121 10.1099/vir.0.81566-0 [DOI] [PubMed] [Google Scholar]
- Puto L. A., Reed J. C. (2008). Daxx represses RelB target promoters via DNA methyltransferase recruitment and DNA hypermethylation. Genes Dev. 22, 998–1010 10.1101/gad.1632208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalska–Metcalf I. U., Janicki S. M. (2007). Show and tell: visualizing gene expression in living cells. J. Cell Sci. 120, 2301–2307 10.1242/jcs.008664 [DOI] [PubMed] [Google Scholar]
- Rafalska–Metcalf I. U., Powers S. L., Joo L. M., LeRoy G., Janicki S. M. (2010). Single cell analysis of transcriptional activation dynamics. PLoS ONE 5, e10272 10.1371/journal.pone.0010272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray–Gallet D., Quivy J. P., Scamps C., Martini E. M., Lipinski M., Almouzni G. (2002). HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9, 1091–1100 10.1016/S1097-2765(02)00526-9 [DOI] [PubMed] [Google Scholar]
- Reeves M., Woodhall D., Compton T., Sinclair J. (2010). Human cytomegalovirus IE72 protein interacts with the transcriptional repressor hDaxx to regulate LUNA gene expression during lytic infection. J. Virol. 84, 7185–7194 10.1128/JVI.02231-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiderman J. I., Sakai A., Goldstein S., Ahmad K. (2009). The XNP remodeler targets dynamic chromatin in Drosophila. Proc. Natl. Acad. Sci. USA 106, 14472–14477 10.1073/pnas.0905816106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J., Korshunov A., Liu X. Y., Jones D. T., Pfaff E., Jacob K., Sturm D., Fontebasso A. M., Quang D. A., Tönjes M.et al. (2012). Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 10.1038/nature10833 [DOI] [PubMed] [Google Scholar]
- Shanbhag N. M., Rafalska–Metcalf I. U., Balane–Bolivar C., Janicki S. M., Greenberg R. A. (2010). ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141, 970–981 10.1016/j.cell.2010.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav–Tal Y., Darzacq X., Shenoy S. M., Fusco D., Janicki S. M., Spector D. L., Singer R. H. (2004). Dynamics of single mRNPs in nuclei of living cells. Science 304, 1797–1800 10.1126/science.1099754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov S. P., Dundr M. (2011). Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 13, 167–173 10.1038/ncb2157 [DOI] [PubMed] [Google Scholar]
- Smith M. C., Boutell C., Davido D. J. (2011). HSV-1 ICP0: paving the way for viral replication. Future Virol. 6, 421–429 10.2217/fvl.11.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H., Ray–Gallet D., Almouzni G., Nakatani Y. (2004). Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 10.1016/S0092-8674(03)01064-X [DOI] [PubMed] [Google Scholar]
- Tang J., Wu S., Liu H., Stratt R., Barak O. G., Shiekhattar R., Picketts D. J., Yang X. (2004). A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J. Biol. Chem. 279, 20369–20377 10.1074/jbc.M401321200 [DOI] [PubMed] [Google Scholar]
- Tsukamoto T., Hashiguchi N., Janicki S. M., Tumbar T., Belmont A. S., Spector D. L. (2000). Visualization of gene activity in living cells. Nat. Cell Biol. 2, 871–878 10.1038/35046510 [DOI] [PubMed] [Google Scholar]
- Valadez–Graham V., Yoshioka Y., Velazquez O., Kawamori A., Vázquez M., Neumann A., Yamaguchi M., Zurita M. (2012). XNP/dATRX interacts with DREF in the chromatin to regulate gene expression. Nucleic Acids Res. 40, 1460–1474 10.1093/nar/gkr865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L. H., Ren H., Williams E., McGhie J., Ahn S., Sim M., Tam A., Earle E., Anderson M. A., Mann J.et al. (2009). Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 19, 404–414 10.1101/gr.084947.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L. H., McGhie J. D., Sim M., Anderson M. A., Ahn S., Hannan R. D., George A. J., Morgan K. A., Mann J. R., Choo K. H. (2010). ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 20, 351–360 10.1101/gr.101477.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Broniscer A., McEachron T. A., Lu C., Paugh B. S., Becksfort J., Qu C., Ding L., Huether R., Parker M.et al. St. Jude Children's Research Hospital–Washington University Pediatric Cancer Genome Project (2012). Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 44, 251–253 10.1038/ng.1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao F., Schaffer P. A. (1995). An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J. Virol. 69, 6249–6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng P. Y., Vakoc C. R., Chen Z. C., Blobel G. A., Berger S. L. (2006). In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques 41, 694, 696, 698 10.2144/000112297 [DOI] [PubMed] [Google Scholar]
- Zhang R., Chen W., Adams P. D. (2007). Molecular dissection of formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 27, 2343–2358 10.1128/MCB.02019-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Hu Y. Z., Xueli L., Li S., Wang M., Kong X., Li T., Shen P., Ma Y. (2010). The inhibition of CMV promoter by heat shock factor 4b is regulated by Daxx. Int. J. Biochem. Cell Biol. 42, 1698–1707 10.1016/j.biocel.2010.06.024 [DOI] [PubMed] [Google Scholar]
- Zhao R., Nakamura T., Fu Y., Lazar Z., Spector D. L. (2011). Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell Biol. 13, 1295–1304 10.1038/ncb2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.