Summary
LSH, a protein related to the SNF2 family of chromatin-remodelling ATPases, is essential for the correct establishment of DNA methylation levels and patterns in plants and mammalian cells. However, some of the phenotypes resulting from LSH deficiency cannot be explained easily by defects in DNA methylation. Here we show that LSH-deficient mouse and human fibroblasts show reduced viability after exposure to ionizing radiation and repair DNA double-strand breaks less efficiently than wild-type cells. A more detailed characterisation of this phenotype revealed that, in the absence of LSH, the histone variant H2AX is not efficiently phosphorylated in response to DNA damage. This results in impaired recruitment of MDC1 and 53BP1 proteins to DNA double-strand breaks and compromises phosphorylation of checkpoint kinase CHK2. Furthermore, we demonstrate that the ability of LSH to hydrolyse ATP is necessary for efficient phosphorylation of H2AX at DNA double-strand breaks and successful repair of DNA damage. Taken together, our data reveal a previously unsuspected role of LSH ATPase in the maintenance of genome stability in mammalian somatic cells, which is independent of its function in de novo DNA methylation during development.
Key words: DNA damage, DNA methylation, LSH, Chromatin, Chromatin remodelling
Introduction
Genetic information within the eukaryotic nucleus is organised into a highly conserved structural polymer, chromatin, which supports and controls crucial functions of the genome. The organisation of DNA into chromatin is inhibitory to most biological processes that utilise DNA as a template, such as transcription, replication, recombination and repair. There are several mechanisms by which chromatin structure and composition can be altered. One of the most fundamental of these is ATP-dependent chromatin-remodelling carried out by specialised proteins (Becker and Hörz, 2002; Lusser and Kadonaga, 2003). A number of chromatin remodelling proteins, often organised into large multi-subunit complexes, have been identified in mammalian cells (Lusser and Kadonaga, 2003; Narlikar et al., 2002). The catalytic subunit in these complexes, the ATPase, uses the energy from ATP hydrolysis in order to reposition, reorganize, modify or evict nucleosomes from DNA. Several chromatin-remodelling complexes including INO80, SWI/SNF, CHD4 and ISWI have been implicated in DNA damage repair in yeast, plants and mammalian cells (Polo and Jackson, 2011).
The Lymphoid-Specific Helicase LSH, also known as HELLS or PASG, is a ubiquitously-expressed 100 kDa protein related to the SNF2 family of chromatin-remodelling ATPases. Knockout of Lsh (Hells) gene in mice leads to postnatal lethality and 50–70% reduction in the global levels of DNA methylation, including repetitive sequences and large chromosomal domains throughout the genome (Dennis et al., 2001; Myant et al., 2011; Tao et al., 2011). In addition, Lsh−/− embryos exhibit defects in male and female meiosis, which are manifested by incomplete chromosome synapses and failure to load crossover-associated foci (De La Fuente et al., 2006; Zeng et al., 2011). It has been hypothesised that LSH remodels chromatin to render DNA accessible to DNA methyltransferase enzymes and therefore it supports de novo DNA methylation and stable gene silencing (Zhu et al., 2006). In agreement with this, it has been reported that LSH is required for developmentally programmed DNA methylation during embryogenesis (Myant et al., 2011). Nevertheless, ATP-dependent chromatin remodelling activity of LSH has never been demonstrated in vitro. In vivo, deletion of exons 10–12 was shown to truncate the catalytic SNF2 domain generating a hypomorph Lsh allele. Mice homozygous for the truncated Lsh allele also display DNA methylation defects, but they survive longer after birth, exhibit signs of premature ageing and upregulated expression of senescence-associated markers (Sun et al., 2004). It is yet unclear whether the loss of DNA methylation and cellular senescence are distinct or related to each other phenotypes resulting from impaired function of LSH in chromatin remodelling.
Here, we investigate the response of LSH-deficient mouse and human cells to DNA damage induced by ionizing radiation (IR). We find that LSH-deficient cells display reduced survival and inefficient repair of DNA double-strand breaks (DSBs) compared to cells with wild-type levels of LSH. Our characterisation of this phenotype reveals that the LSH-deficient cells show normal activation of DNA damage-responsive kinase ATM, but weaker and more transient phosphorylation of ATM substrate, the variant histone H2AX (γH2AX). This results in inefficient recruitment and retention of DNA damage response mediator proteins MDC1 and 53BP1 at DSBs and leads to compromised ATM-dependent phosphorylation of checkpoint kinase CHK2. We also show that the DSB repair defects in LSH-deficient cells are independent of changes in DNA methylation and can be reversed by re-expression of wild-type, but not a catalytically inactive, LSH protein in Lsh−/− mouse embryonic fibroblasts (MEFs). Taken together our data imply that, in addition to promoting DNA methylation during development, LSH has a conserved, DNA methylation-independent function in DNA DSB repair.
Results
Inefficient repair of ionizing radiation-induced DNA damage in LSH-deficient cells
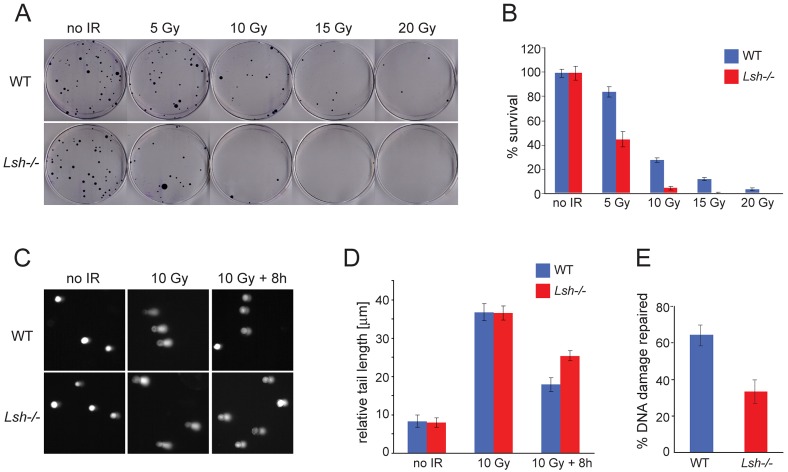
In order to investigate whether LSH is required for repair of DSBs in mammalian cells, we plated wild-type and Lsh−/− MEFs at clonal density and subjected them to increasing doses of ionizing radiation (IR). We monitored the survival of non-irradiated and irradiated cells by colony formation assay. Interestingly, the viability of Lsh−/− MEFs after IR was significantly impaired compared to wild-type MEFs, indicating that LSH-deficient cells are more sensitive to DNA damage induced by IR (Fig. 1A,B).
Fig. 1.
Inefficient repair of DNA damage induced by IR in Lsh−/− MEFs. (A) Wild-type (WT) and Lsh−/− MEFs were plated at clonal density and subjected to increasing doses of IR. The number of colonies appearing after ten days on the irradiated plates indicate the reduced viability of Lsh−/− MEFs after exposure to IR. (B) Quantification of experiments (as shown in A) performed in triplicate. ‘% survival’ represents the number of colonies appearing on irradiated plates relative to the number of colonies on non-irradiated plates seeded in parallel. (C) Comet assays show DNA fragmentation in wild-type and Lsh−/− MEFs without IR, immediately after treatment with 10 Gy of IR and 8 hours of recovery post-IR. (D) Quantification of the relative tail length from the experiments shown in C. A total of 50 cells were examined at each time point for each of the two genotypes. (E) The % repaired DNA damage was calculated from the values shown in D for 10 Gy and 10 Gy+8 hours time points. All error bars represent standard deviation (S.D.).
As LSH is involved in the formation of heterochromatin (Myant and Stancheva, 2008; Myant et al., 2011; Yan et al., 2003), it is possible that the same dose of IR generates a greater number of breaks in Lsh−/− fibroblasts than in wild-type MEFs due to general chromatin relaxation. To examine this, we employed single cell electrophoresis (comet assay) to measure the relative extent of DNA damage induced by IR in wild-type and LSH-deficient MEFs. We also examined whether the IR-induced DSBs are repaired equally well in both cell types. As is apparent from the comparable comet tail length after exposure to 10 Gy of IR, the wild-type and the Lsh−/− cells acquired a similar number of DNA breaks (Fig. 1C,D). However, the Lsh−/− MEFs failed to repair ∼50% of these lesions compared to wild-type cells when investigated 8 hours after irradiation (Fig. 1C–E). Taken together, these data suggest that reduced viability of Lsh−/− MEFs after IR exposure reflects inefficient repair of DSBs rather than acquisition of greater DNA damage.
LSH-deficient MEFs display normal activation of DNA damage signalling
In mammalian cells, IR-induced DSBs lead to activation of a DNA damage response (DDR) signalling which promotes the recruitment of mediator and repair proteins to the sites of DNA damage and simultaneously orchestrates cell cycle arrest while the breaks are repaired (Polo and Jackson, 2011). DDR signalling cascade in G1 and G2 of the cell cycle is initiated by the ATM (Ataxia telangiectasia mutated) kinase, which undergoes auto-phosphorylation at Serine 329 in response to DNA damage (Bakkenist and Kastan, 2003). ATM subsequently binds to the MRN (MRE11, RAD50, NBS) complex that assembles at DNA breaks (Lee and Paull, 2004; Lee and Paull, 2005). The MRN-bound ATM phosphorylates multiple effectors proteins, including Serine 139 of the histone variant H2AX which is present in approximately every fifth nucleosome throughout chromatin (Burma et al., 2001). Phosphorylated H2AX (γH2AX) appears within minutes of DNA damage, spreads over large megabase-long domains flanking the DSB and serves as a platform for the recruitment of DNA damage repair machinery (Iacovoni et al., 2010; Rogakou et al., 1999; Rogakou et al., 1998). Accumulation of γH2AX and repair proteins at the sites of the DNA damage leads to the formation of discrete cytologically detectable foci which disperse as DNA damage is repaired (Polo and Jackson, 2011).
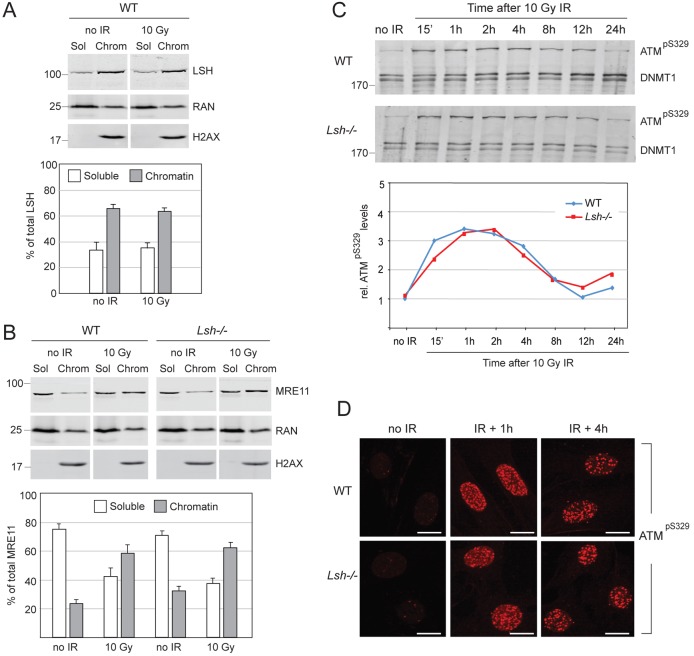
To investigate whether DDR initiates correctly in the Lsh−/− MEFs, we monitored the association of MRE11 with chromatin and the phosphorylation and localisation of ATM before and after IR. Chromatin retention assays showed that, unlike LSH, which stably associates with chromatin independently of DNA damage (Fig. 2A), MRE11 was enriched on DNA after exposure to IR and this occurred at similar levels in wild-type and Lsh−/− cells (Fig. 2B). Moreover, we detected no difference between wild-type and Lsh−/− MEFs in the kinetics of ATM phosphorylation after IR (Fig. 2C), the localisation of ATM to DSBs (Fig. 2D) and the number of ATM foci at the onset of DNA repair (supplementary material Fig. S1A). Collectively, our data suggest that the activation of ATM, detection of DSBs and initiation of DNA damage response are not impaired in the Lsh−/− MEFs.
Fig. 2.
Normal activation of DNA damage response in wild-type (WT) and Lsh−/− MEFs. (A) Western blot detecting LSH before and after IR in the soluble (Sol) and chromatin-bound (Chrom) fractions of nuclear proteins. Quantification of western blots from several independent experiments indicates that before and after IR, ∼60% of LSH is constitutively bound to DNA/chromatin and ∼40% is in the soluble nucleoplasm fraction. RAN and H2AX serve as loading controls for the soluble and chromatin fractions, respectively. (B) MRE11 accumulates equally well on chromatin after IR in wild-type and Lsh−/− MEFs and is partly depleted from the soluble nucleoplasm fraction. The bar graph shows quantification of MRE11 in the soluble and chromatin fractions from three independent experiments. The error bars in A and B represent S.D. (C) The activating phosphorylation of ATM at Serine 329 (pS329) displays similar kinetics after irradiation of the wild-type and Lsh−/− MEFs. DNMT1 is a loading control. The numbers on the left indicate molecular weight in kDa. The graph shows quantification of phosphorylated ATM relative to DNMT1 during the time course. (D) Phosphorylated ATM is recruited to DNA damage foci after irradiation of the wild-type and Lsh−/− MEFs. The scale bars represent 10 µm.
Reduced phosphorylation of H2AX in LSH-deficient cells
To examine the events that take place downstream of ATM activation, we followed the kinetics of γH2AX accumulation after IR in wild-type and Lsh−/− MEFs by indirect immunofluorescence and western blots (Fig. 3A–C). In wild-type cells, γH2AX was detectable 15 minutes after the exposure of cells to IR, peaked 1 hour post-IR, persisted at lower, but significant levels at 2- and 4-hour time points, and gradually declined to basal levels by the 24-hour time point (Fig. 3A,C, top panels). In contrast, the accumulation and the persistence of γH2AX in the Lsh−/− MEFs were significantly reduced (Fig. 3A,C, bottom panels; see also the graph in Fig. 3B). In the irradiated LSH-deficient cells, γH2AX never reached the same levels as in the wild-type cells at the 1-hour time point and decreased dramatically 4 hours post-IR (Fig. 3A,C, bottom panels). Quantification of γH2AX in immunofluorescence experiments and western blots showed that the phosphorylation of the variant histone is reduced by ∼50% in the Lsh−/− MEFs compared to their wild-type counterparts (Fig. 3B; supplementary material Fig. S2). The inefficient phosphorylation of H2AX was neither due to a reduced total amount of the variant histone in chromatin of LSH-deficient cells (Fig. 2B, Fig. 3C; supplementary material Fig. S2), nor caused by the overexpression of a known phosphatase that may dephosphorylate γH2AX prematurely (supplementary material Table S1). Thus, despite the normal activation and localisation of ATM kinase to IR-induced DSBs, the phosphorylation of H2AX is significantly impaired in the absence of LSH.
Fig. 3.
Reduced phosphorylation of H2AX in Lsh−/− MEFs subjected to IR. (A) Immunostaining of wild-type (WT) and Lsh−/− MEFs with antibodies against γH2AX (red) before and after 10 Gy of IR. The nuclei were counterstained with DAPI (blue). The analysed time points are shown above the panels. The scale bars represent 10 µm. Note that Lsh−/− MEFs display significantly weaker staining for γH2AX. (B) Quantification of relative intensity of γH2AX staining in the immunofluorescence experiments shown in A. A total of 100 cells of each genotype were analysed at every time point. The error bars represent S.D. (C) Western blots showing the kinetics of γH2AX relative to H2AX and histone H4 in wild-type and Lsh−/− MEFs. All samples were run on the same gel and detected simultaneously. The numbers on the left indicate molecular weight in kDa.
Potentially, the lack of LSH could affect γH2AX either globally, at all DSBs, or locally – only at a subset of DNA lesions. To investigate this, we counted the γH2AX foci in wild-type and Lsh−/− MEFs exposed to IR and found that the number of detectable γH2AX foci in Lsh−/− MEFs is reduced compared to their wild-type counterparts (supplementary material Fig. S1B). This indicates that LSH may not be required for high levels of γH2AX at all DSBs.
Reduction of γH2AX is independent of DNA methylation levels and is conserved between mouse and human LSH-deficient fibroblasts
In the Lsh−/− MEFs, the global levels of DNA methylation are reduced by ∼50%, affecting many gene promoters and large chromosomal domains (Myant et al., 2011; Tao et al., 2011). Potentially, DNA hypomethylation may compromise the efficiency of DSBs repair either directly, by altering chromatin structure, or indirectly, by changing the expression of DNA repair genes. In order to investigate whether lack of DNA methylation affects the efficiency of H2AX phosphorylation, we irradiated MEFs genetically null for maintenance DNA methyltransferase DNMT1 and p53 (Dnmt1−/−; p53−/−) as well as p53-null MEFs (p53−/−) (Lande-Diner et al., 2007) as controls and followed the γH2AX levels during DNA damage repair. Despite the almost complete lack of DNA methylation (<10%), Dnmt1−/−; p53−/− MEFs displayed normal kinetics of γH2AX compared to wild-type and p53−/− control MEFs (Fig. 4A). This indicates that DNA methylation is not essential for efficient phosphorylation of H2AX near the sites of DNA damage.
Fig. 4.
Inefficient phosphorylation of H2AX is independent of DNA methylation levels in mouse and human fibroblasts. (A) Western blots probed with anti-γH2AX and anti-H2AX antibodies before and after irradiation of wild-type (WT), Lsh−/−, p53−/− and DNA methylation-deficient Dnmt1−/−; p53−/− MEFs. Note that Dnmt1−/−; p53−/− MEFs show normal levels of γH2AX when compared to controls. (B) Western blot detecting the successful stable KD of LSH (LSH KD) in hTERT-immortalized MRC5 human lung fibroblasts in comparison to non-infected and control cells infected with non-silencing shRNA lentiviruses (Con KD). (C) Quantitative RT-PCR detecting LSH mRNA levels relative to GAPDH in control and LSH KD MRC5 cells. (D) The levels of 5-methyl cytosine (5-meC) in LSH KD and control cells were detected by ELISA and quantified as % 5-meC detected in non-infected MRC5 cells. (E) MRC5 cells in which LSH was knocked down show reduced γH2AX after irradiation when compared to control cells. All samples were run on the same gel and detected simultaneously. H2AX and H4 serve as loading controls. (F) Quantification of γH2AX signal relative to H2AX from three independent experiments. (G) LSH-deficient MRC5 fibroblasts show reduced survival in colony formation assays in comparison to control cells expressing non-silencing shRNA. The error bars in C, D, F and G represent S.D.
In order to determine whether reduced γH2AX in response to IR is specific to LSH-deficient mouse fibroblasts or can be observed in other mammalian cell types, we stably knocked down LSH by small hairpin RNA (shRNA) in primary human lung fibroblasts, MRC5, immortalized by expression of the catalytic subunit of human telomerase (hTERT) (Fig. 4B,C). Consistent with previous reports (Suzuki et al., 2008), the LSH knockdown (KD) human fibroblasts maintained initially normal DNA methylation levels (Fig. 4D). However, similar to LSH-deficient MEFs, LSH KD MRC5 cells displayed reduced phosphorylation of H2AX and impaired survival after exposure to IR (Fig. 4E–G) and other damaging agents inducing DSBs (supplementary material Fig. S3). Taken together, these experiments strongly suggest that the efficiency of H2AX phosphorylation and DSB repair is impaired by the absence of LSH, but not by the loss of DNA methylation. This effect is conserved between mouse and human fibroblasts.
Compromised recruitment of mediators of DDR signalling at DSBs in Lsh−/− MEFs
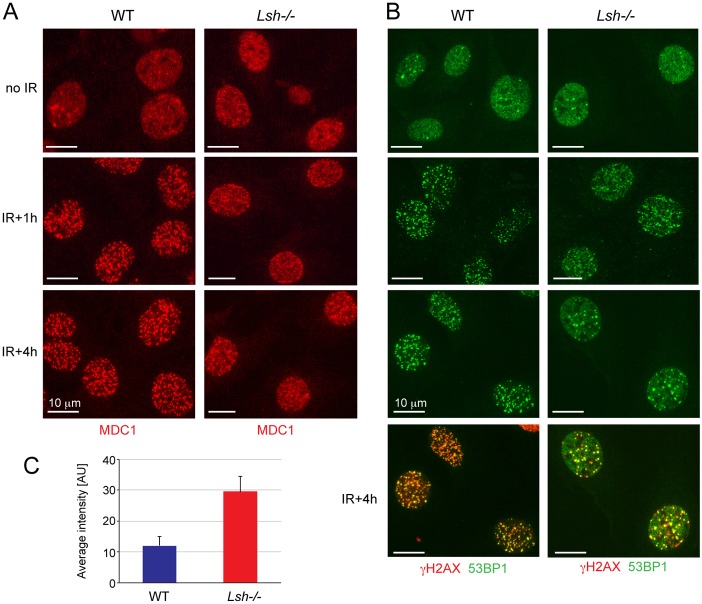
Given that Lsh−/− MEFs exposed to IR inefficiently phosphorylate H2AX, we expected that either the recruitment or the long-term retention of proteins that bind to chromatin in a γH2AX-dependent manner at DSBs might be compromised in these cells. To investigate this, we stained non-irradiated and irradiated wild-type and LSH-null cells with antibodies against MDC1 (mediator of DNA damage checkpoint 1), which directly binds to γH2AX via its BRCT domain (Glover et al., 2004), and antibodies against 53BP1, a protein that requires MDC1 and exposed modified histone H3 tails for binding at IR-induced DSBs (Eliezer et al., 2009; Huyen et al., 2004; Sanders et al., 2004). As expected, MDC1 and 53BP1 were diffusely distributed throughout the nucleus in non-irradiated cells and began to accumulate at IR-induced foci (IRIF) after irradiation in wild-type MEFs (Fig. 5A,B). However, MDC1 and 53BP1 localised to IRIF less efficiently in the Lsh−/− MEFs at the 1-hour time point, as evident by the high proportion of cells (50%) showing a more diffuse nuclear staining (Fig. 5A,B, IR+1h; supplementary material Fig. S4). Also, while large and robust foci of MDC1 and 53BP1 were observed in the nuclei of wild-type cells 4 hours after irradiation, such foci were fewer in the Lsh−/− MEFs and diffuse nucleoplasm staining for MDC1 and 53BP1 was readily detectable (Fig. 5A–C, IR+4h; supplementary material Fig. S4). In all cases the MDC1 and 53BP1 foci co-localised with γH2AX (Fig. 5B; data not shown). These observations are in agreement with the kinetics of γH2AX at DSBs observed by immunofluorescence and western blots in the Lsh−/− MEFs (Figs 3, 4) and indicate that mediator proteins of DDR signalling accumulate less efficiently and dissociate prematurely from DSBs in the mutant cells. This could, potentially, result in inefficient DNA repair and cell cycle arrest in cells lacking LSH.
Fig. 5.
Impaired recruitment of MDC1 and 53BP1 to the sites of DNA damage in Lsh−/− MEFs. (A) Immunostaining of non-irradiated and irradiated wild-type (WT) and Lsh−/− MEFs with antibodies detecting γH2AX binding protein MDC1. Note that diffuse staining for MDC1 can be seen in Lsh−/− MEFs 1 and 4 hours after exposure to IR. (B) Immunostaining of non-irradiated and irradiated wild-type and Lsh−/− MEFs with antibodies against 53BP1. Similar to MDC1, there is more diffuse staining for 53BP1 in Lsh−/− MEFs at 1- and 4-hour time points. The co-localisation of 53BP1 with γH2AX shown in the bottom two panels clearly shows diffuse 53BP1 staining in Lsh−/− MEFs that does not overlap with γH2AX. Note the reduced number of γH2AX foci in Lsh−/− MEFs at this time point in comparison to the wild-type MEFs. The scale bars represent 10 µm. (C) Quantification of 53BP1 staining in the nucleoplasm in wild-type and Lsh−/− MEFs 4 hours post-IR. The error bars represent S.D.
Impaired activation of CHK2 in LSH-deficient cells
It was reported that in response to IR-induced DNA damage the DDR checkpoint kinase CHK2 is transiently recruited to DSBs via MDC1 where it becomes activated by ATM-dependent phosphorylation at threonine 68 (T68) (Ahn et al., 2000; Lukas et al., 2003). Phosphorylated CHK2 dissociates rapidly from DSBs to induce cell cycle arrest by inhibitory phosphorylation of CDC25 phosphatase (Lukas et al., 2003; Matsuoka et al., 1998). On the other hand, 53BP1 is required for recruitment to DSBs of other ATM substrates such as BRCA1 and transcription factor p53 (Abraham, 2002). In response to IR, p53 undergoes stabilisation and ATM-dependent phosphorylation at Serine 15 (S15) (Canman et al., 1998). S15-phosphorylated p53 can either induce cell cycle arrest or promote apoptosis in cells that fail to repair DNA damage (Dumaz and Meek, 1999; Lambert et al., 1998).
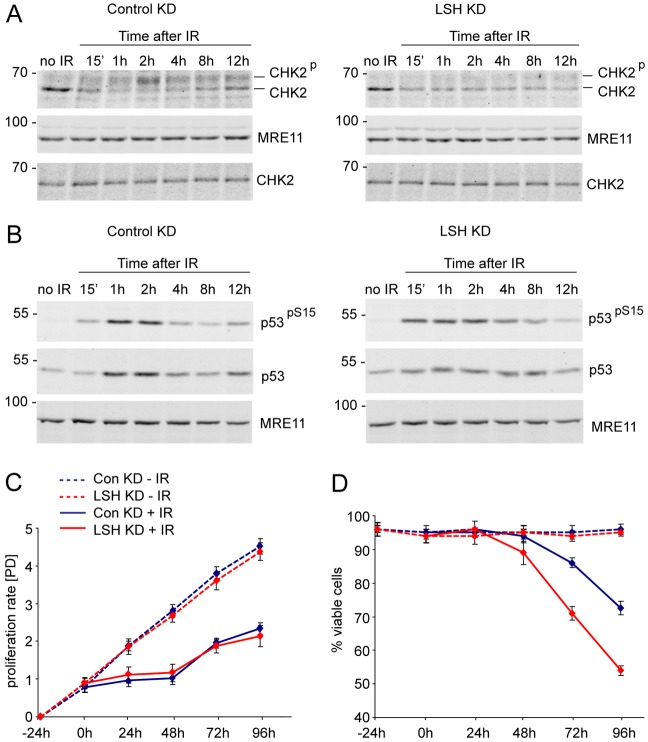
In order to investigate whether reduced γH2AX and premature dissociation from DSBs of mediator proteins MDC1 and 53BP1 compromise DNA damage checkpoints, we examined the kinetics of CHK2 and p53 phosphorylation after IR in wild-type and LSH-deficient cells. As MEFs often spontaneously immortalize and lose p53 expression, we performed these analyses in human MRC5 fibroblasts infected with either the control non-silencing shRNA vector or MRC5 cells with stable KD of LSH (Fig. 4B). Consistent with γH2AX kinetics and MDC1 recruitment to IRIF, the CHK2 phosphorylation increased gradually after IR and peaked at the 2-hour time point in the control cells (Fig. 6A, Control KD). In contrast, the phosphorylated CHK2 was barely detectable in LSH-deficient cells at all time points after irradiation (Fig. 6A, LSH KD). However, LSH-deficient human fibroblasts displayed relatively normal ATM-dependent phosphorylation of p53 at Serine 15 (pS15) in response to IR. Although in the LSH KD cells the p53 protein levels did not peak as dramatically at 1- and 2-hour time points as in the control cells, we observed earlier and more persistent phosphorylation of p53 at S15 after DNA damage (Fig. 6B). These observations suggest that despite the reduced phosphorylation of CHK2 the LSH-deficient cells either undergo p53-dependent cell cycle arrest or apoptosis.
Fig. 6.
Reduced phosphorylation of CHK2 in LSH-deficient cells. (A) Western blots detecting CHK2, phosphorylated CHK2 and MRE11 in nuclear extracts of non-irradiated and irradiated control KD and LSH KD MRC5 cells. Note that the slower migrating, phosphorylated form of CHK2 is barely detectable throughout the time course of recovery after IR in the LSH KD cells. Extracts prepared without phosphatase inhibitors (bottom panels) do not show the slower migrating phosphorylated form of CHK2. (B) The control KD and LSH-deficient cells display stabilisation of p53 and ATM-dependent phosphorylation of p53 at Serine 15 (pS15) after IR. MRE11 serves as a loading control. The numbers on the left indicate molecular weight markers in kDa. (C) Proliferation of non-irradiated (−IR) and 2.5 Gy irradiated (+IR) control and LSH KD MRC5 cells at indicated time points. ‘[PD]’ indicates population doublings. (D) Viability of non-irradiated and irradiated control and LSH KD cells as detected by trypan blue staining at indicated time points. The error bars represent standard error of the mean.
To investigate whether the control and LSH KD cells can arrest in response to DNA damage we followed their proliferation rate and viability after exposure to IR. Both cell lines showed robust exponential growth and no apparent cell death when grown under normal conditions (Fig. 6C,D, −IR). In response to radiation, both cell lines underwent cell cycle arrest, but resumed proliferation after 48 hours (Fig. 6C, +IR). However, the viability of LSH-deficient cells rapidly decreased after resuming proliferation. 42% of LSH KD cells compared to 24% of controls stained positive with trypan blue 96 hours post-irradiation (Fig. 6D, +IR). This indicates that despite the reduced phosphorylation of CHK2, LSH-deficient cells can undergo transient cell cycle arrest in response to DNA damage, but as they repair DSBs less efficiently, they exit the cell cycle arrest with unrepaired DNA damage which results in accelerated cell death.
The ATPase activity of LSH is required for efficient phosphorylation of H2AX
Despite having a conserved SNF2 domain, the ability of mammalian LSH to hydrolyse ATP and remodel chromatin remains largely unknown. In our hands, recombinant full-length mouse LSH did not reposition nucleosomes in vitro, but had a detectable, albeit weak, DNA-dependent ATPase activity (supplementary material Fig. S5). ATP hydrolysis by recombinant LSH in vitro was completely abolished by substitution of a highly conserved Lysine to Glutamine (K237Q) in the ATP-binding site (supplementary material Fig. S5C).
Considering this, we asked whether the wild-type and mutant LSH can rescue the kinetics of γH2AX after exposure of Lsh−/− MEFs to DNA damage. To do so, we infected the Lsh−/− MEFs with lentiviral vectors expressing FLAG-tagged either wild-type or K237Q mutant form of LSH at levels close to endogenous (Fig. 7A). Lsh−/− MEFs infected with empty vector (MSCV) served as a control. After exposure of these cell lines to radiation, we found that Lsh−/− MEFs expressing wild-type LSH survived IR as well as the wild-type MEFs, while cells expressing ATPase-deficient LSH K237Q had reduced survival, similar to Lsh−/− MEFs infected with empty vector (Fig. 7B). In addition, the expression of wild-type LSH in Lsh−/− MEFs restored to normal levels the phosphorylation of H2AX after IR (Fig. 7C) and the recruitment of 53BP1 to IRIF (Fig. 7D). In contrast, the Lsh−/− MEFs expressing the mutant LSH K237Q retained reduced phosphorylation of H2AX post-IR, diffuse localisation of 53BP1 to IRIF and were undistinguishable from Lsh−/− MEFs carrying the empty vector (Fig. 7C,D). From these experiments we conclude that the ability of LSH to hydrolyse ATP is required for efficient H2AX phosphorylation and repair of DNA damage.
Fig. 7.
Wild-type LSH, but not the ATP-binding-deficient LSH K237Q, can rescue the kinetics of γH2AX in irradiated Lsh−/− MEFs. (A) Western blots with antibodies against LSH and FLAG show the levels of LSH protein in wild-type (WT) MEFs, Lsh−/− MEFs infected with empty MSCV lentivirus and cells infected with lentivirus expressing either wild-type or mutant 3× FLAG-tagged LSH protein. MRE11 serves as a loading control. (B) % survival after IR of Lsh−/− MEFs carrying empty MSCV vector, wild-type LSH and mutant LSH K237Q. The error bars indicate S.D. (C) Lsh−/− MEFs expressing wild-type LSH, but not cells expressing the K237Q mutant form of LSH, display normal kinetics of γH2AX after IR when compared to control cells. H2AX serves as a loading control. All samples were run on the same gel and detected simultaneously. The numbers on the left indicate molecular weight markers in kDa. (D) Immunostaining with anti-FLAG and anti-53BP1 antibodies detect normal localisation of 53BP1 to DNA damage foci in Lsh−/− MEFs expressing wild-type LSH. Irradiated Lsh−/− MEFs infected either with empty MSCV virus or expressing LSH K237Q show diffuse staining for 53BP1. The scale bars represent 10 µm.
Discussion
Several chromatin-remodelling proteins are implicated in the efficient repair of DNA damage in mammalian cells, including BRG1, CHD1L/ALC1, CHD4, INO80 and ISWI proteins ACF1 and SNF2H (Ahel et al., 2009; Kashiwaba et al., 2010; Lan et al., 2010; Larsen et al., 2010; Lee et al., 2010; Nakamura et al., 2011; Park et al., 2009; Park et al., 2006; Polo et al., 2010; Sánchez-Molina et al., 2011; Smeenk et al., 2010). Most of the chromatin-remodelling proteins are recruited to DNA breaks in a γH2AX-dependent manner and function to modulate chromatin structure and modifications in order to facilitate the access to either DNA or chromatin of factors involved in DNA damage signalling and repair. The function of chromatin remodellers in supporting DNA repair is often conserved from yeast to mammalian cells, indicating that chromatin reorganization during DNA repair is vital for maintenance of genome stability.
LSH has been extensively studied as a protein that promotes DNA methylation and silencing of retrotransposons and genes in plants and mammalian cells (Dennis et al., 2001; Lippman et al., 2004; Myant et al., 2011; Tao et al., 2011; Teixeira et al., 2009). However, genetics screens in budding yeast (Alvaro et al., 2007; Costanzo et al., 2010), which carry an LSH homologue YFR038W/IRC5 (Flaus et al., 2006), but entirely lack DNA methylation, and experiments in Arabidopsis thaliana (Costanzo et al., 2010; Shaked et al., 2006) have suggested that LSH may have additional functions that are independent of its role in DNA methylation and regulation of gene expression. Here, we provide experimental evidence that LSH is essential for efficient repair of DNA DSBs in human and mouse fibroblasts. This function of LSH is neither related to alterations in DNA methylation nor caused by mis-expression of known genes involved in DNA repair (supplementary material Table S1). Thus Lsh−/− MEFs, but not Dnmt1−/− MEFs lacking DNA methylation, exhibit reduced levels of γH2AX in response to IR. This effect is conserved in human cells as stable KD of LSH in hTERT-immortalized lung fibroblasts compromised the phosphorylation of H2AX in response to DNA damage without detectable change in DNA methylation levels. Our data are consistent with earlier observations in plants (Shaked et al., 2006), and strongly suggest that SNF2 family ATPase LSH has at least two distinct functions in vivo, namely, to promote de novo DNA methylation during development and to facilitate repair of DSBs in somatic cells. Our KD experiments in human fibroblasts demonstrate that these two functions can be successfully uncoupled from each other.
Whether deficient DNA repair can explain the meiotic defects observed in Lsh−/− germ cells (De La Fuente et al., 2006) and premature ageing phenotype in mice expressing hypomorph alleles of LSH (Sun et al., 2004) is yet to be investigated in detail. The formation of synaptonemal complexes during meiosis requires DNA recombination initiated at SPO11-generated DSBs. If such breaks are formed, but not correctly marked by γH2AX, this may compromise recombination between homologous chromosomes and thus impair meiosis (Celeste et al., 2002; Xu et al., 1996). However, γH2AX and RAD51 staining seem to persist longer in pachytene stage Lsh−/− oocytes than in their wild-type counterparts (De La Fuente et al., 2006). This may indicate that the lack of LSH confers additional defects in processing of meiotic DSBs during oogenesis and spermatogenesis. On the other hand, accumulation of endogenous unrepaired DNA damage in mice expressing hypomorphic Lsh alleles may lead to cellular senescence and premature ageing. Thus, some of the phenotypes resulting from LSH deficiency can be, at least in part, a direct consequence of the defective repair of DNA DSBs.
Although the detection of DNA damage, as evident by ATM activation and binding of MRE11 and ATM to DSBs, are not affected by LSH deficiency, reduced phosphorylation of H2AX at DNA repair sites in LSH-deficient cells indicates that LSH is involved in the early steps of DSB repair. The inefficient recruitment to DSBs of DNA damage signalling mediators MDC1 and 53BP1, compromised phosphorylation of CHK2 and reduced viability are expected outcomes of altered γH2AX levels and patterns in irradiated LSH-deficient cells. However, p53 stabilisation and phosphorylation by ATM remain unaffected by the lack of LSH and may contribute to cell cycle arrest and apoptosis of LSH-deficient cells with unrepaired DNA damage.
How LSH promotes γH2AX accumulation in response to DNA damage is currently unclear. As is evident from our rescue experiments in the Lsh−/− MEFs, the ability of LSH to hydrolyse ATP is essential for normal levels of γH2AX and successful DNA repair. Unlike BRG1, which also supports efficient accumulation of γH2AX (Lee et al., 2010; Park et al., 2006), and other chromatin-remodelling proteins implicated in DNA damage repair (Ahel et al., 2009; Larsen et al., 2010; Polo et al., 2010; Smeenk et al., 2010; van Attikum et al., 2004), LSH neither accumulates at repair foci (Fig. 7D), nor binds more tightly to chromatin specifically after the induction of DNA damage (Fig. 2A). In fact, chromatin retention assays indicate that a significant proportion of LSH is constitutively bound to DNA or chromatin in mammalian cells. Therefore, it is possible that LSH renders the C-terminus of H2AX more receptive to ATM-mediated phosphorylation by acting locally, at DSBs that occur in the vicinity of DNA/chromatin-bound LSH. Alternatively, LSH may not function specifically during DSB repair, but instead facilitate the even distribution of variant histone H2AX in chromatin throughout the genome. It is currently accepted that in mammalian cells on average every fifth nucleosome contains H2AX (Rogakou et al., 1999). However, it is yet unclear when during the cell cycle and how H2AX is incorporated into chromatin. Uneven distribution of H2AX throughout the genome of LSH-deficient cells would result in areas that lack H2AX and repair DSBs less efficiently. Either one of these two hypotheses could explain the reduced levels of γH2AX and number of detectable γH2AX foci in the Lsh−/− MEFs (supplementary material Fig. S1B). Future experiments will aim to investigate the accumulation and spreading of γH2AX at defined DNA lesions (Iacovoni et al., 2010) as well as the distribution of H2AX in the genome of LSH-deficient cells. It will also be essential to determine whether or not LSH and BRG1 have redundant function in promoting γH2AX accumulation in response to IR-induced DNA damage.
Collectively, the data we provide in this report identify LSH as an important and functionally conserved, from plants to mammals, component of DSB repair process. Frequent deletions within the SNF2 domain of LSH have been detected in human lymphoid malignancies and may contribute to stress-induced genomic instability in cancers carrying LSH mutations (Lee et al., 2000). Compared to quiescent cells, LSH is highly expressed in all rapidly proliferating cell types, including most immortalized and cancer-derived cell lines (Lee et al., 2000). Therefore, it is conceivable that development of small molecules that inhibit the ATPase activity of LSH would be beneficial for applications aiming to sensitise cancer cells to DNA damage in order to aid successful radiotherapy.
Materials and Methods
Cell lines and viral infections
Wild-type and Lsh−/− MEFs were cultured in DMEM supplemented with 10% FCS, penicillin, streptomycin and L-glutamine. Vectors expressing 3×FLAG C-terminally-tagged LSH and K237Q mutant LSH were generated by cloning synthetic cDNAs (Life Technologies) into pMSCV-puro vector (Clontech). The vectors were packaged into lentiviral particles in amphotropic Phoenix cell line and used for infection of Lsh−/− MEFs at multiplicity of infection (MOI) = 1. The cells were selected with 2.5 µg/ml of puromycin for two weeks and individual stable colonies were expanded and tested for LSH expression. Human foetal lung fibroblast MRC5 (ATCC number: CRL-171) were cultured in MEM supplemented with 10% FCS, non-essential amino acids, sodium pyruvate, penicillin, streptomycin and L-glutamine. MRC5 cells were immortalized by transduction with pBabe-hTERT-Neo plasmid packaged into retroviral particles in Phoenix cells. The infected cells were selected with 400 µg/ml G418 for 10 days. LSH was stably knocked down in MRC5hTERT cells by introducing at MOI = 4 two independent pGIPZ-shRNAmir plasmids V2LHS_155499 and V2LHS_155497 packaged into lentiviral particles (Open Biosystems). Control cells were infected under the same conditions with non-silencing pGIPZ-shRNAmir particles RHS4348 (Open Biosystems). The infected cells were selected with 2.5 µg/ml of puromycin for 7 days and the LSH knockdown assessed by western blots and quantitative RT-PCR.
Irradiation experiments
Wild-type and LSH-deficient either mouse or human fibroblasts were plated on 10 cm dishes in triplicate at a density of 200 cells/plate and irradiated with indicated doses of IR in a Torrex X-ray cabinet (Faxitron Ltd). In survival assays, the irradiated cells were left to recover and form colonies for 10–14 days, fixed, stained with Giemsa and counted. For time course experiments, the cells were plated on 20 cm dishes at ∼60% confluency, subjected to indicated dose of IR (usually 10 Gy) and collected 15 minutes, 1, 2, 4, 8, 12 and 24 hours after irradiation. Nuclear extracts were prepared as described previously (Myant et al., 2011) in buffers supplemented with phosphatase (Pierce) and protease (Sigma Aldrich) inhibitors. For short-term analyses of cell proliferation and viability, human fibroblasts were plated on 6-well plates (2×105 cells/well) irradiated with 2.5 Gy and counted in trypan blue solution by Countess Cell Counter (Life Technologies).
Comet assays
Cells were plated on 10 cm dishes at ∼60% confluency, subjected to 10 Gy of IR and either collected immediately or left to recover for 8 hours. Cells were diluted to 6×105 cells/ml with PBS, mixed 1∶10 with 0.6% low melting point agarose and placed onto glass slides pre-coated with 1% agarose. Slides were placed in lysis buffer (2.5 M NaCl, 10 mM Tris, 1% Triton-X, pH 10) overnight at 4°C. Lysis buffer was removed, the slides washed twice with dH2O and electrophoresis was carried out in a cold alkaline buffer (300 mM NaCl, 1 mM EDTA, pH 13) at 100 A, 12 V for 20 minutes. Slides were neutralised with 3×5 minute washes with cold 0.4 M Tris, pH 7.5 followed by 10 minute wash with cold dH2O. Staining was carried out with a 20 µg/ml propidium iodide solution followed by 2×5 minute washes in cold dH2O. Images were collected by an Olympus BX61 microscope equipped with 20× objectives and AnalySIS software (Soft Imaging Systems). Comet tail lengths were quantified using customized macro in ImagePro 9.0 software (Media Cybernetics).
Chromatin retention assays
Cells were plated on 20 cm dishes at ∼60% confluency, subjected to the indicated dose of IR and collected at the indicated time point after irradiation. Nuclei were prepared by disrupting the cells in hypotonic buffer (NE1) as previously described (Myant et al., 2011). Soluble nuclear proteins and those loosely associated with chromatin were extracted using NE1 supplemented with 100 mM NaCl. After centrifugation the pellets containing chromatin bound proteins were resuspended in NE1 buffer supplemented with 25 U of Benzonase nuclease (Merck). Following 1 hour incubation on ice, NaCl was added to a final concentration of 500 mM and incubated at 4°C for 1 hour. After centrifugation the supernatant containing the chromatin associated proteins was collected. All buffers were supplemented with phosphatase (Pierce) and protease inhibitors (Sigma Aldrich).
Western blots
50 µg of each nuclear extract was resolved on either 8.5% or 15% SDS-PAGE, transferred to nitrocellulose membrane (BioRad) and detected by anti-LSH (Santa Cruz; sc-46665), anti-γH2AX (Millipore; 05-636), anti-H2AX (Active Motif), anti-H4 (Millipore; 07-108), anti-phospho-ATM (Millipore; 05-740), anti-MRE11 (Calbiochem; PC388), anti-CHK2 (Cell Signaling; 2662), anti-p53 (Santa Cruz, sc126 for human cells and Cell Signaling 2524 for mouse cells) and anti-p53 pS15 (Cell Signaling; 9284) antibodies followed by secondary either anti-mouse IR800 or anti-rabbit IR680 (LiCOR Biosciences). The western blots were imaged on Odyssey Imager (LiCOR Biosciences) and quantified where indicated with the aid of Odyssey V3.0 software.
Immunofluorescence
Cells were fixed either with 4% paraformaldehyde or with cold 100% methanol for 5 minutes, permeabilized by incubation in PBS with 0.1% Tween for 10 minutes, blocked with 4% BSA in PBS, incubated with diluted primary antibodies for 2 hours and after washes with secondary anti-rabbit Alexa 488 or anti-mouse Alexa 565 labelled antibodies. The slides were washed with PBS, counterstained with DAPI and mounted in Vectashield (Vector Laboratories). Images were collected by Olympus BX61 microscope equipped with 20× and 40× PlanApo objectives (Olympus), ColourViewII camera and AnalySIS software (Soft Imaging Systems). Where indicated, fluorescent images were quantified using a customized macro in ImagePro 9.0 software (Media Cybernetics). Antibodies used for immunofluorescence were: anti-pATM (Millipore; 05-740) 1∶2000 dilution; anti-γH2AX (Millipore; 05-636) 1∶4000 dilution; anti-MDC1 (Abcam; ab11171) 1∶1000 dilution; anti-53BP1 (Cell Signaling; 4397) 1∶1000; and anti-FLAG M2 (Sigma Aldrich) 1∶2000.
DNA methylation assays
The global levels of 5-methyl cytosine in the genome of MRC5hTERT, LSH KD and control KD cells were investigated by ELISA assays using an ImprintTM kit (Sigma Aldrich) according to manufacturer’s instructions.
Quantitative RT-PCR
RNA was extracted from cells with Trizol reagent (Life Technologies). 4 µg of each RNA were treated with RNase-free DNase (Fermentas) and reverse transcribed using poly-dT primer and SuperscriptIII reverse transcriptase (Life Technologies). Q-PCRs were carried out in triplicate with SYBR Green Master Mix (Roche) and primers detecting LSH and GAPDH mRNA on a Lightcycler 480 instrument (Roche). Relative levels of LSH mRNA were quantified relative to GAPDH using the standard methods (Pfaffl, 2001).
Chromatin remodelling and ATPase assays
Full-length cDNA of wild-type and K237Q mutant mouse LSH were cloned into a shuttle pFastBac vector (Invitrogen) in frame with a N-terminal 6× Histidine tag and introduced into DH10Bac E. coli (Life Technologies) to obtain recombinant Bacmids. Expression in SF2 cells and purification by nickel affinity and ion exchange chromatography were carried out using standard protocols. Chromatin remodelling assays were performed as described (Stockdale et al., 2006) with nucleosomes assembled by salt dialysis (Narlikar et al., 2002) on Cy5 labelled 601 nucleosome positioning sequence (Lowary and Widom, 1998) carrying either 54 bp of linker DNA on either site (54A54) or 601 with 54 bp linker DNA on one side and 0 on the other side (54A0) in the presence of either recombinant wild-type LSH or S. cerevisiae RSC protein as control. Remodelling reactions were resolved on 5% acrylamide gel in 0.5× TBE buffer and scanned on ProXPRESS proteomics imaging system (Perkin Elmer Life Sciences, UK). ATPase activity assays were carried out with either wild-type or mutant LSH in the presence of 32P-γATP and indicated substrates for 30 minutes at 37°C. The reactions were stopped on ice and spotted on PEI Cellulose TLC plates (Merck). Separation was achieved in 0.5 M LiCl/1 M formic acid buffer. The plates were dried, exposed and scanned on Storm 860 Phosphorimager (Molecular Dynamics). ATP hydrolysis was calculated from the signal intensity of the released free phosphate (32Pi) using ImageQuant software. Detailed protocols of these methods are available upon request.
Supplementary Material
Acknowledgments
We thank Kathrin Muegge (National Cancer Institute, Frederick, MD, USA) for kindly providing the wild-type and Lsh−/− MEFs, Eric So (King’s College, London, UK) for MSCV vectors, David Kelly (Wellcome Trust Centre for Cell Biology, COIL facility) for help with imaging and image quantification and the members of Stancheva laboratory for reading the manuscript and helpful suggestions.
Footnotes
Funding
This research was supported by Cancer Research UK Senior Fellowship [grant number C7215/A8983] and Wellcome Trust Centre for Cell Biology Core grant [grant number 077707]; J.B. is a Biotechnology and Biological Sciences Research Council CASE PhD student supported by CellCentric Ltd.; and A.T. is a PhD student funded by Cancer Research UK [grant number C7215/A9218]. Deposited in PMC for release after 6 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.111252/-/DC1
References
- Abraham R. T. (2002). Checkpoint signalling: focusing on 53BP1. Nat. Cell Biol. 4, E277–E279 10.1038/ncb1202-e277 [DOI] [PubMed] [Google Scholar]
- Ahel D., Horejsí Z., Wiechens N., Polo S. E., Garcia–Wilson E., Ahel I., Flynn H., Skehel M., West S. C., Jackson S. P.et al. (2009). Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325, 1240–1243 10.1126/science.1177321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J. Y., Schwarz J. K., Piwnica–Worms H., Canman C. E. (2000). Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 60, 5934–5936 [PubMed] [Google Scholar]
- Alvaro D., Lisby M., Rothstein R. (2007). Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 3, e228 10.1371/journal.pgen.0030228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist C. J., Kastan M. B. (2003). DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421, 499–506 10.1038/nature01368 [DOI] [PubMed] [Google Scholar]
- Becker P. B., Hörz W. (2002). ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71, 247–273 10.1146/annurev.biochem.71.110601.135400 [DOI] [PubMed] [Google Scholar]
- Burma S., Chen B. P., Murphy M., Kurimasa A., Chen D. J. (2001). ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276, 42462–42467 10.1074/jbc.C100466200 [DOI] [PubMed] [Google Scholar]
- Canman C. E., Lim D. S., Cimprich K. A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M. B., Siliciano J. D. (1998). Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281, 1677–1679 10.1126/science.281.5383.1677 [DOI] [PubMed] [Google Scholar]
- Celeste A., Petersen S., Romanienko P. J., Fernandez–Capetillo O., Chen H. T., Sedelnikova O. A., Reina–San–Martin B., Coppola V., Meffre E., Difilippantonio M. J.et al. (2002). Genomic instability in mice lacking histone H2AX. Science 296, 922–927 10.1126/science.1069398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S.et al. (2010). The genetic landscape of a cell. Science 327, 425–431 10.1126/science.1180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fuente R., Baumann C., Fan T., Schmidtmann A., Dobrinski I., Muegge K. (2006). Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nat. Cell Biol. 8, 1448–1454 10.1038/ncb1513 [DOI] [PubMed] [Google Scholar]
- Dennis K., Fan T., Geiman T., Yan Q., Muegge K. (2001). Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 15, 2940–2944 10.1101/gad.929101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumaz N., Meek D. W. (1999). Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 18, 7002–7010 10.1093/emboj/18.24.7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliezer Y., Argaman L., Rhie A., Doherty A. J., Goldberg M. (2009). The direct interaction between 53BP1 and MDC1 is required for the recruitment of 53BP1 to sites of damage. J. Biol. Chem. 284, 426–435 10.1074/jbc.M807375200 [DOI] [PubMed] [Google Scholar]
- Flaus A., Martin D. M., Barton G. J., Owen–Hughes T. (2006). Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34, 2887–2905 10.1093/nar/gkl295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J. N., Williams R. S., Lee M. S. (2004). Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem. Sci. 29, 579–585 10.1016/j.tibs.2004.09.010 [DOI] [PubMed] [Google Scholar]
- Huyen Y., Zgheib O., Ditullio R. A., Jr, Gorgoulis V. G., Zacharatos P., Petty T. J., Sheston E. A., Mellert H. S., Stavridi E. S., Halazonetis T. D. (2004). Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432, 406–411 10.1038/nature03114 [DOI] [PubMed] [Google Scholar]
- Iacovoni J. S., Caron P., Lassadi I., Nicolas E., Massip L., Trouche D., Legube G. (2010). High-resolution profiling of γH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 29, 1446–1457 10.1038/emboj.2010.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwaba S., Kitahashi K., Watanabe T., Onoda F., Ohtsu M., Murakami Y. (2010). The mammalian INO80 complex is recruited to DNA damage sites in an ARP8 dependent manner. Biochem. Biophys. Res. Commun. 402, 619–625 10.1016/j.bbrc.2010.10.066 [DOI] [PubMed] [Google Scholar]
- Lambert P. F., Kashanchi F., Radonovich M. F., Shiekhattar R., Brady J. N. (1998). Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 273, 33048–33053 10.1074/jbc.273.49.33048 [DOI] [PubMed] [Google Scholar]
- Lan L., Ui A., Nakajima S., Hatakeyama K., Hoshi M., Watanabe R., Janicki S. M., Ogiwara H., Kohno T., Kanno S.et al. (2010). The ACF1 complex is required for DNA double-strand break repair in human cells. Mol. Cell 40, 976–987 10.1016/j.molcel.2010.12.003 [DOI] [PubMed] [Google Scholar]
- Lande–Diner L., Zhang J., Ben–Porath I., Amariglio N., Keshet I., Hecht M., Azuara V., Fisher A. G., Rechavi G., Cedar H. (2007). Role of DNA methylation in stable gene repression. J. Biol. Chem. 282, 12194–12200 10.1074/jbc.M607838200 [DOI] [PubMed] [Google Scholar]
- Larsen D. H., Poinsignon C., Gudjonsson T., Dinant C., Payne M. R., Hari F. J., Rendtlew Danielsen J. M., Menard P., Sand J. C., Stucki M.et al. (2010). The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J. Cell Biol. 190, 731–740 10.1083/jcb.200912135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Paull T. T. (2004). Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304, 93–96 10.1126/science.1091496 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Paull T. T. (2005). ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308, 551–554 10.1126/science.1108297 [DOI] [PubMed] [Google Scholar]
- Lee D. W., Zhang K., Ning Z. Q., Raabe E. H., Tintner S., Wieland R., Wilkins B. J., Kim J. M., Blough R. I., Arceci R. J. (2000). Proliferation-associated SNF2-like gene (PASG): a SNF2 family member altered in leukemia. Cancer Res. 60, 3612–3622 [PubMed] [Google Scholar]
- Lee H. S., Park J. H., Kim S. J., Kwon S. J., Kwon J. (2010). A cooperative activation loop among SWI/SNF, γ-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 29, 1434–1445 10.1038/emboj.2010.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z., Gendrel A. V., Black M., Vaughn M. W., Dedhia N., McCombie W. R., Lavine K., Mittal V., May B., Kasschau K. D.et al. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430, 471–476 10.1038/nature02651 [DOI] [PubMed] [Google Scholar]
- Lowary P. T., Widom J. (1998). New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 10.1006/jmbi.1997.1494 [DOI] [PubMed] [Google Scholar]
- Lukas C., Falck J., Bartkova J., Bartek J., Lukas J. (2003). Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat. Cell Biol. 5, 255–260 10.1038/ncb945 [DOI] [PubMed] [Google Scholar]
- Lusser A., Kadonaga J. T. (2003). Chromatin remodeling by ATP-dependent molecular machines. Bioessays 25, 1192–1200 10.1002/bies.10359 [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Huang M., Elledge S. J. (1998). Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282, 1893–1897 10.1126/science.282.5395.1893 [DOI] [PubMed] [Google Scholar]
- Myant K., Stancheva I. (2008). LSH cooperates with DNA methyltransferases to repress transcription. Mol. Cell. Biol. 28, 215–226 10.1128/MCB.01073-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myant K., Termanis A., Sundaram A. Y., Boe T., Li C., Merusi C., Burrage J., de Las Heras J. I., Stancheva I. (2011). LSH and G9a/GLP complex are required for developmentally programmed DNA methylation. Genome Res. 21, 83–94 10.1101/gr.108498.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Kato A., Kobayashi J., Yanagihara H., Sakamoto S., Oliveira D. V., Shimada M., Tauchi H., Suzuki H., Tashiro S.et al. (2011). Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell 41, 515–528 10.1016/j.molcel.2011.02.002 [DOI] [PubMed] [Google Scholar]
- Narlikar G. J., Fan H. Y., Kingston R. E. (2002). Cooperation between complexes that regulate chromatin structure and transcription. Cell 108, 475–487 10.1016/S0092-8674(02)00654-2 [DOI] [PubMed] [Google Scholar]
- Park J. H., Park E. J., Lee H. S., Kim S. J., Hur S. K., Imbalzano A. N., Kwon J. (2006). Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting γ-H2AX induction. EMBO J. 25, 3986–3997 10.1038/sj.emboj.7601291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Park E. J., Hur S. K., Kim S., Kwon J. (2009). Mammalian SWI/SNF chromatin remodeling complexes are required to prevent apoptosis after DNA damage. DNA Repair (Amst.) 8, 29–39 10.1016/j.dnarep.2008.08.011 [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S. E., Jackson S. P. (2011). Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 25, 409–433 10.1101/gad.2021311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S. E., Kaidi A., Baskcomb L., Galanty Y., Jackson S. P. (2010). Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 29, 3130–3139 10.1038/emboj.2010.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. (1998). DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Rogakou E. P., Boon C., Redon C., Bonner W. M. (1999). Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146, 905–916 10.1083/jcb.146.5.905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez–Molina S., Mortusewicz O., Bieber B., Auer S., Eckey M., Leonhardt H., Friedl A. A., Becker P. B. (2011). Role for hACF1 in the G2/M damage checkpoint. Nucleic Acids Res. 39, 8445–8456 10.1093/nar/gkr435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S. L., Portoso M., Mata J., Bähler J., Allshire R. C., Kouzarides T. (2004). Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119, 603–614 10.1016/j.cell.2004.11.009 [DOI] [PubMed] [Google Scholar]
- Shaked H., Avivi–Ragolsky N., Levy A. A. (2006). Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics 173, 985–994 10.1534/genetics.105.051664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk G., Wiegant W. W., Vrolijk H., Solari A. P., Pastink A., van Attikum H. (2010). The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J. Cell Biol. 190, 741–749 10.1083/jcb.201001048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdale C., Flaus A., Ferreira H., Owen–Hughes T. (2006). Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J. Biol. Chem. 281, 16279–16288 10.1074/jbc.M600682200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. Q., Lee D. W., Zhang Q., Xiao W., Raabe E. H., Meeker A., Miao D., Huso D. L., Arceci R. J. (2004). Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev. 18, 1035–1046 10.1101/gad.1176104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Farrar J. E., Yegnasubramanian S., Zahed M., Suzuki N., Arceci R. J. (2008). Stable knockdown of PASG enhances DNA demethylation but does not accelerate cellular senescence in TIG-7 human fibroblasts. Epigenetics 3, 281–291 10.4161/epi.3.5.6914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Xi S., Shan J., Maunakea A., Che A., Briones V., Lee E. Y., Geiman T., Huang J., Stephens R.et al. (2011). Lsh, chromatin remodeling family member, modulates genome-wide cytosine methylation patterns at nonrepeat sequences. Proc. Natl. Acad. Sci. USA 108, 5626–5631 10.1073/pnas.1017000108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira F. K., Heredia F., Sarazin A., Roudier F., Boccara M., Ciaudo C., Cruaud C., Poulain J., Berdasco M., Fraga M. F.et al. (2009). A role for RNAi in the selective correction of DNA methylation defects. Science 323, 1600–1604 10.1126/science.1165313 [DOI] [PubMed] [Google Scholar]
- van Attikum H., Fritsch O., Hohn B., Gasser S. M. (2004). Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119, 777–788 10.1016/j.cell.2004.11.033 [DOI] [PubMed] [Google Scholar]
- Xu Y., Ashley T., Brainerd E. E., Bronson R. T., Meyn M. S., Baltimore D. (1996). Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 10, 2411–2422 10.1101/gad.10.19.2411 [DOI] [PubMed] [Google Scholar]
- Yan Q., Huang J., Fan T., Zhu H., Muegge K. (2003). Lsh, a modulator of CpG methylation, is crucial for normal histone methylation. EMBO J. 22, 5154–5162 10.1093/emboj/cdg493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., Baumann C., Schmidtmann A., Honaramooz A., Tang L., Bondareva A., Dores C., Fan T., Xi S., Geiman T.et al. (2011). Lymphoid-specific helicase (HELLS) is essential for meiotic progression in mouse spermatocytes. Biol. Reprod. 84, 1235–1241 10.1095/biolreprod.110.085720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Geiman T. M., Xi S., Jiang Q., Schmidtmann A., Chen T., Li E., Muegge K. (2006). Lsh is involved in de novo methylation of DNA. EMBO J. 25, 335–345 10.1038/sj.emboj.7600925 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.