Abstract
Pluripotent stem cells evade replicative senescence, whereas other primary cells lose their proliferation and differentiation potential after a limited number of cell divisions, and this is accompanied by specific senescence-associated DNA methylation (SA-DNAm) changes. Here, we investigate SA-DNAm changes in mesenchymal stromal cells (MSC) upon long-term culture, irradiation-induced senescence, immortalization, and reprogramming into induced pluripotent stem cells (iPSC) using high-density HumanMethylation450 BeadChips. SA-DNAm changes are highly reproducible and they are enriched in intergenic and nonpromoter regions of developmental genes. Furthermore, SA-hypomethylation in particular appears to be associated with H3K9me3, H3K27me3, and Polycomb-group 2 target genes. We demonstrate that ionizing irradiation, although associated with a senescence phenotype, does not affect SA-DNAm. Furthermore, overexpression of the catalytic subunit of the human telomerase (TERT) or conditional immortalization with a doxycycline-inducible system (TERT and SV40-TAg) result in telomere extension, but do not prevent SA-DNAm. In contrast, we demonstrate that reprogramming into iPSC prevents almost the entire set of SA-DNAm changes. Our results indicate that long-term culture is associated with an epigenetically controlled process that stalls cells in a particular functional state, whereas irradiation-induced senescence and immortalization are not causally related to this process. Absence of SA-DNAm in pluripotent cells may play a central role for their escape from cellular senescence.
Pluripotent cells have unlimited self-renewal potential. Under appropriate culture conditions, embryonic stem cells (ESC) as well as induced pluripotent stem cells (iPSC) can virtually be passaged infinitely without any signs of replicative senescence (Zeng 2007). In contrast, somatic cells enter a senescent state after a limited number of cell divisions (Hayflick and Moorhead 1961). The same holds true for the differentiated progeny of iPSC and ESC, indicating that these cells are not immortalized per se (Lapasset et al. 2011).
Senescent cells remain metabolically active, but their proliferation potential is unequivocally lost. They acquire typical morphological changes and functional deteriorations such as loss of differentiation potential. “Replicative senescence” is associated with telomere attrition during culture expansion, whereas “premature senescence” is a consequence of other stresses such as oxidative stress, DNA damaging agents, irradiation, or oncogene activation—both of these aspects may be superimposed during in vitro culture. Senescence does not occur simultaneously during culture expansion and some cells may already have entered a senescent state, whereas others are still able to proliferate (Wagner 2010). All of the above-mentioned mechanisms ultimately result in DNA damage that triggers activation of common pathways such as TP53/CDKN1A and CDKN2A/RB1 (Seluanov et al. 2001; Shay and Roninson 2004; Bazarov et al. 2012; Sperka et al. 2012). Pluripotent cells express telomerase and maintain telomere integrity over time, which may prevent replicative senescence (Marion et al. 2009b). In addition, it has been speculated that long-term culture-associated changes are related to a developmental process that might be functionally relevant for suppressing tumorigenesis in vivo (Campisi and d'Adda di 2007; Kang et al. 2011). This notion is supported by specific senescence-associated epigenetic modifications, pointing to a rather coordinated process.
DNA methylation (DNAm) predominantly occurs at cytosines in the context of CpG dinucleotides (Lister et al. 2009). It may influence gene expression by direct interference with transcription factors or with methyl-CpG-binding proteins that modify histones, and thereby inactivate promoter regions. Our group has previously demonstrated that long-term culture of mesenchymal stromal cells (MSC) and fibroblasts coincides with differential methylation at specific CpG sites (Bork et al. 2010; Koch et al. 2011; Schellenberg et al. 2011). These senescence-associated (SA) modifications are highly reproducible and continuously acquired in the course of culture expansion—they can even be used as biomarkers to account for the number of passages or the time of in vitro culture (Koch et al. 2012).
In this study we have further analyzed SA-DNAm changes. MSC were subjected to (1) long-term culture, (2) irradiation-induced senescence, (3) immortalization, and (4) reprogramming into induced pluripotent stem cells, which we refer to as iP-MSC. Subsequently, DNAm was analyzed using a novel Infinium HumanMethylation450 platform that can assay more than 480,000 CpG sites at single-base resolution (covering 99% of RefSeq genes and 96% of CpG islands) (Bibikova et al. 2011). We demonstrate that SA-DNAm changes are significantly enriched in developmental genes such as homeobox genes. Particularly, SA-hypomethylation occurs in intergenic regions or nonexpressed genes. Neither senescence induction by gamma irradiation, nor telomere extension influence SA-DNAm. In contrast, the majority of SA-DNAm is prevented by reprogramming into iP-MSC. Therefore, SA-DNAm changes do not resemble stochastic modifications that accumulate over time—they rather reflect a tightly regulated epigenetic program that is prevented by reprogramming into a pluripotent state.
Results
Long-term culture-associated DNA methylation changes
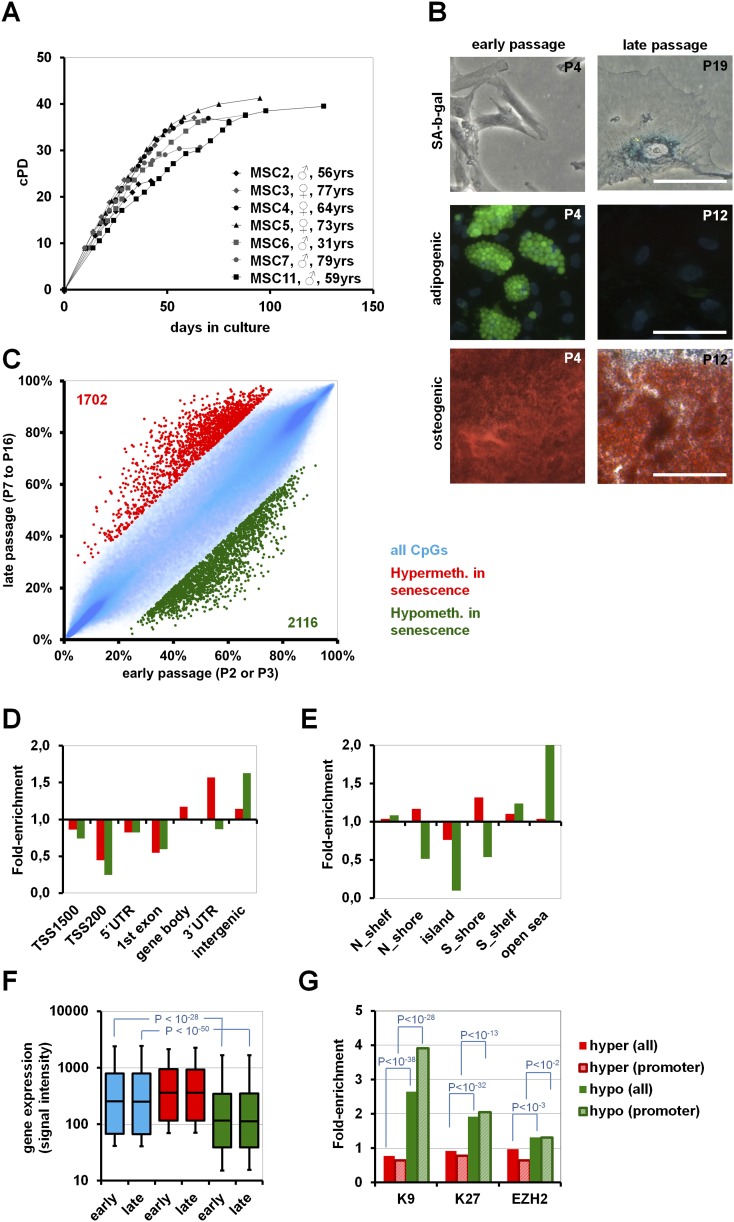
Mesenchymal stromal cells from human bone marrow were expanded until they entered growth arrest after 80 ± 25 d and 35.1 ± 5.7 cumulative population doublings (cPD) (Fig. 1A). At early passage, all cell preparations fulfilled the commonly used criteria for definition of MSC such as fibroblastoid growth, surface-marker expression (CD14−, CD29+, CD31−, CD34−, CD45−, CD73+, CD90+, and CD105+), and differentiation potential toward osteogenic, adipogenic, and chondrogenic lineages (Dominici et al. 2006). Senescent MSC acquired large and flat morphology and expressed senescence-associated beta-galactosidase (SA-β-gal). The immunophenotype was maintained throughout culture expansion, but MSC of late passages lost their adipogenic differentiation potential and revealed reduced osteogenic differentiation potential (Fig. 1B; Supplemental Fig. 1A–D). DNAm profiles were compared between MSC of early passage (P2 or P3; n = 5) and MSC of late passage just before entering senescence (P7–P16 according to inter-individual variation in long-term growth curves; n = 4). To focus on the most relevant SA-DNAm changes, we have only considered CpG sites with at least 20% differential methylation and a limma-adjusted P-value of <0.05. Under these stringent criteria 1702 CpG sites were SA-hypermethylated, whereas 2116 CpG sites were SA-hypomethylated (Fig. 1C; Supplemental Tables 1,2).
Figure 1.
Long-term culture-associated changes in MSC. (A) Long-term growth curves demonstrate that MSC reach a senescent state after 2–3 mo of culture expansion. (B) Cells at higher passage acquire morphological changes, senescence-associated beta-galactosidase staining, and loss of adipogenic differentiation potential (green: BODIPY-staining of lipid droplets; blue: DAPI) and decline in osteogenic differentiation potential (Alizarin Red staining of calcium phosphate precipitates; size bars 100 μm). (C) Comparison of DNAm between early (P2–P3) and late (P7–P16) passages. CpG sites with relevant SA-hypermethylation (red: 1702 CpG sites) and SA-hypomethylation (green: 2116 CpG sites) are depicted (adjusted P-value < 0.05 and DNAm change >20%). (D) SA-DNAm changes were classified according to gene regions, and (E) in relation to CpG islands. (F) Signal intensity of gene expression microarrays was used as an indicator for gene expression level. In particular, genes with SA-hypomethylated CpG sites were significantly less-expressed than the average. (G) Genes with SA-DNAm changes were subsequently mapped to genes with H3K9me3, H3K27me3 (Delbarre et al. 2010), and PRC2 targets (Wei et al. 2011) in human MSC. Enrichment analysis revealed that particularly genes with SA-hypomethylated CpG sites have been associated with these repressive histone marks. As an alternative, we focused on the subset of promoter-associated CpG sites (TSS1500, TSS200, or 5′UTR; striped bars), and this analysis also revealed association of genes with SA-hypomethylated CpG sites with those genes that have repressive histone marks (H3K9, P < 10−28; H3K27, P < 10−13; EZH2, P < 10−2).
DNAm changes were not simultaneously acquired within genes, but most of them were restricted to circumscribed genomic locations. CpG sites were classified according to gene regions as described before (Sandoval et al. 2011; Supplemental Fig. 2). Unexpectedly, SA-CpGs were hardly located within 200 bp upstream of the transcription start site (TSS200; P < 10−18; hypergeometric distribution) and in the first exon (P < 10−5)—those regions where differential methylation has a major impact on gene expression. In contrast, SA-hypermethylation was particularly enriched in the gene body (P < 10−7), 3′UTR (P < 10−5), and intergenic regions (P < 10−4), whereas SA-hypomethylation was highly enriched in intergenic regions (P < 10−55) (Fig. 1D). Analysis of SA-CpG sites in the context of CpG islands (CGI) revealed preferential occurrence of SA-hypermethylation in the 2-kb flanking regions of CGIs (shore-region; P < 10−5), whereas SA-hypomethylation hardly occurred in the context of CGIs at all (P < 10−264) (Fig. 1E). SA-CpG sites did not exhibit clear association with subtelomeric or centromeric regions. We searched for differentially methylated regions (DMR) where 100 adjacent CpG sites on the BeadChip are regulated accordingly—only two genomic regions revealed such a highly significant enrichment of SA-DNAm changes (P < 10−5): the HOXB locus on chromosome 17 (SA-hypermethylated) and the keratin associated protein (KRTAP) locus on chromosome 21 (SA-hypomethylated; Supplemental Fig. 3). Overall, spatio-temporal DNAm changes were highly reproducible in independent samples.
Gene expression profiles from MSC of early passage (P3–P5; n = 3) and late passage (P10; n = 3) were compared by microarray technology. Signal intensity was used as an indicator for gene expression level and correlated with DNAm profiles. As expected, higher DNAm level within 200 nucleotides upstream of the transcription start site (TSS200) and first exon coincided with markedly reduced gene expression levels. However, the specific DNAm changes upon long-term culture were hardly reflected by differential gene expression (Supplemental Fig. 4) and similar results were observed when we matched SA-DNAm with our previously published gene expression data on long-term culture of MSC (Wagner et al. 2008). Overall, SA-hypomethylated CpG sites revealed a mean signal intensity that was significantly below the average of all genes (Fig. 1F). Thus, most of these genes seem to be neither expressed at early nor at late passages. Gene ontology analysis of genes with SA-hypermethylated CpG sites or SA-hypomethylated CpG sites revealed highly significant enrichment of genes in categories associated with cellular differentiation (Supplemental Table 3). Very similar results were observed when we focused on genes with SA-CpG sites within the promoter region (TSS1500, TSS200, and 5′UTR). Using gene set enrichment analysis we found the most significant overlap of either SA-hypermethylated or SA-hypomethylated CpG sites within genes harboring trimethylated lysine 9 of histone H3 (H3K9me3; P = 2.6 × 10−13 and 8.2 × 10−13, respectively), and targets of Polycomb-group family members EED (P = 2.1 × 10−9 and 1.6 × 10−7) and SUZ12 (P = 9.3 × 10−8 and 2.6 × 10−7) in ChIP array data of human ESC (Lee et al. 2006; Ben-Porath et al. 2008). Furthermore, SA-hypomethylated CpG sites were also enriched in H3K27 targets of various cancer cells (Schlesinger et al. 2007), P = 4.0 × 10−6; (Kondo et al. 2008), P = 2.4 × 10−5; (Acevedo et al. 2008), P = 5.1 × 10−4. Enrichment of SA-hypermethylation or SA-hypomethylation was further analyzed with published ChIP array data for H3K9me3 and H3K27me3 in human MSC from adipose tissue (Delbarre et al. 2010) and for the Polycomb repressive complex 2 (PRC2) targets in human MSC from bone marrow (Wei et al. 2011); these marks were highly significantly enriched in genes with SA-hypomethylated CpG sites (Fig. 1G). Taken together, particularly SA-hypomethylation preferentially occurred in regions with repressive H3K9me3 and H3K27me3 histone marks and in PRC2 targets—this may contribute to the very low expression of the corresponding genes.
Gamma-irradiation does not affect DNAm
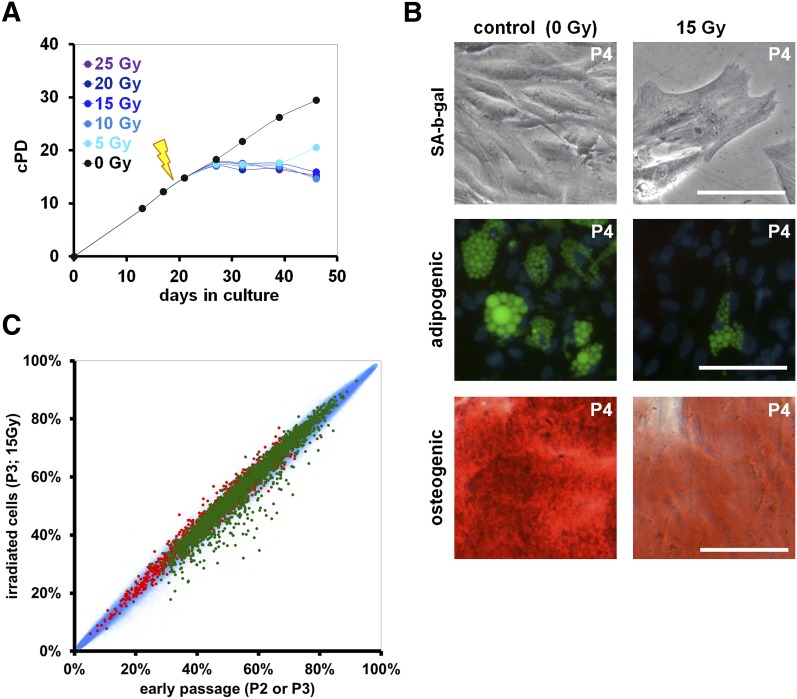
Ionizing radiation (IR) of MSC induced very similar phenotypic changes as replicative senescence. Treatment with 15 Gy completely abrogated proliferation and the majority of cells stayed metabolically active for more than one month (Fig. 2A). After 7 d the cells acquired the typical senescent morphology. Furthermore, IR impaired differentiation toward adipogenic and osteogenic lineage as described above (Fig. 2B; Supplemental Fig. 5), transiently up-regulated CDKN1A, CDKN2A, and TP53 (Supplemental Fig. 6) and other investigators demonstrated activation of the corresponding signaling cascades on a protein level (Cmielova et al. 2012). DNAm profiles were compared before and 7 d after treatment with 15 Gy (n = 4). None of the CpG sites revealed significant changes upon irradiation (limma-adjusted P < 0.05) (Fig. 2C). Accordingly, radiation-induced senescence does not seem to be associated with SA-DNAm changes. Subsequently, we measured telomere length by monochrome multiplex quantitative PCR (Cawthon 2009). As expected, telomere length decreased by long-term culture (P = 0.01), whereas irradiation did not affect telomere length. Despite phenotypic similarity of long-term culture-associated senescence and irradiation-induced senescence, the causative mechanism seems to be different.
Figure 2.
Irradiation-induced senescence does not result in SA-DNAm changes. (A) Representative growth curves of MSC after irradiation with different doses at passage 2 (0–25 Gy; black arrow). (B) Upon irradiation with 15 Gy, MSC acquire the typical senescent morphology with proliferation arrest and moderate SA-β-gal staining. Furthermore, adipogenic (green: BODIPY; blue: DAPI) and osteogenic differentiation potential (red: Alizarin Red; size bars 100 μm) is impaired after irradiation with 15 Gy. (C) Scatterplot analysis of DNAm of nonirradiated versus irradiated MSC. Long-term culture-associated hypermethylated (red) and hypomethylated CpG sites (green) are depicted. Irradiation did not result in any significant DNAm changes.
Immortalization does not reverse SA-DNAm
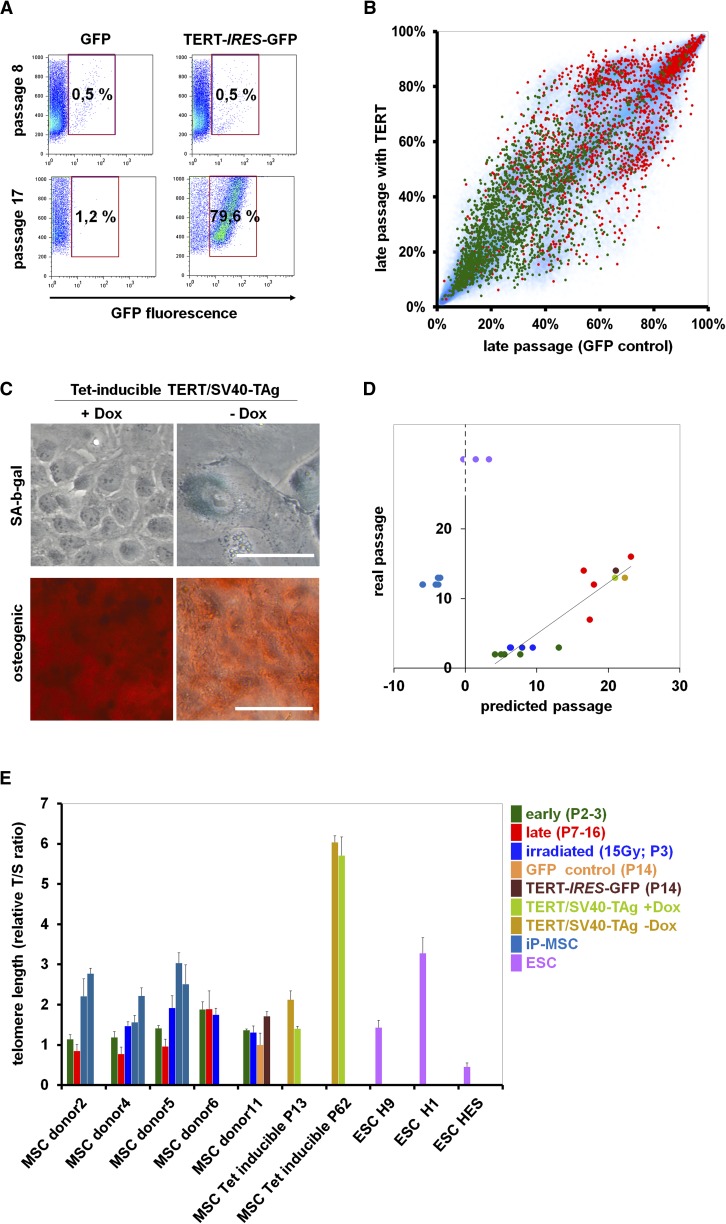
Overexpression of TERT, the catalytic subunit of human telomerase, is known to extend the lifespan of primary cells, but it is generally insufficient for immortalization (Bodnar et al. 1998; Kiyono et al. 1998). We retrovirally transfected MSC with a TERT-IRES-GFP construct in comparison to GFP-control. Constitutive overexpression resulted in growth advantage of TERT-transfected cells (Fig. 3A), but this was only observed in one of five experiments, and the cells entered a senescent state after 5 additional cPD in comparison to GFP-control (Supplemental Fig. 7). Telomere length was moderately increased by TERT overexpression, whereas SA-DNAm changes were not affected (Fig. 3B,E).
Figure 3.
Immortalization does not affect SA-DNAm. (A) MSC at passage 2 were either retrovirally transfected with an TERT-IRES-GFP construct or with IRES-GFP (control; n = 5). Flow cytometic analysis of one sample reveals a growth advantage of TERT-transduced cells at late passages. (B) The DNAm profile of this TERT-transfected sample was compared with the corresponding control (both passage 14). CpG sites that were hypermethylated (red) or hypomethylated upon long-term culture (green) are indicated. Overall, TERT expression does not affect SA-DNAm changes. (C) MSC were immortalized by tetracycline-inducible expression of TERT and SV40-TAg. Upon Dox removal from the culture medium, these cells acquire a typical senescent morphology within 10 d, express SA-β-gal, and lose osteogenic differentiation potential. (D) The Epigenetic-Senescence-Signature was used to estimate cellular aging of all samples (Koch et al. 2012). DNAm was determined at six SA-CpG sites by pyrosequencing and used for the linear regression model to predict the number of passages. The predictions correlate with the real passage number for all MSC samples, including irradiated, TERT-transfected, and TERT/SV40-TAg immortalized cells. In contrast, senescence-predictions for iP-MSC and ESC rather correspond to noncultured cells. (E) Relative telomere length was determined by the telomere to single-copy gene (T/S) ratio with quantitative multiplex PCR (each sample measured in triplicate). Telomere length is significantly reduced in MSC of later passage, whereas it is not affected by irradiation. TERT expression, TERT/SV40-TAg immortalization (particularly at passage 62), and generation of iP-MSC result in telomere extension.
Alternatively, we immortalized MSC with a lentiviral tetracycline-inducible expression system for TERT and SV40 large T antigen (SV40-TAg). In the presence of doxycycline (Dox) these cells could be passaged for more than 80 passages without any signs of senescence. However, removal of Dox from the culture medium resulted in immediate proliferation arrest, and within 10 d the cells acquired a typical senescent phenotype with SA-β-gal expression (Fig. 3C; Supplemental Fig. 8). qRT-PCR analysis revealed that TERT was highly expressed, whereas it was hardly detectable in nontransfected MSC (Supplemental Fig. 6). Telomere length was massively extended between passages 13 and 62 (Fig. 3E). We used our recently published epigenetic-senescence-signature to test this conditionally immortalized cell line for SA-DNAm changes (Koch et al. 2012). DNA was harvested at passage 13 and passage 62 (with and without Dox for 10 d) and DNAm was analyzed by pyrosequencing of the six relevant CpG sites. The predictions for passage numbers were always related to the real passage number (Fig. 3D; Supplemental Fig. 9). Taken together, neither IR nor TERT overexpression or immortalization with TERT/SV40-TAg interfered with the SA-DNAm changes.
Reprogramming into iP-MSC prevents SA-DNAm
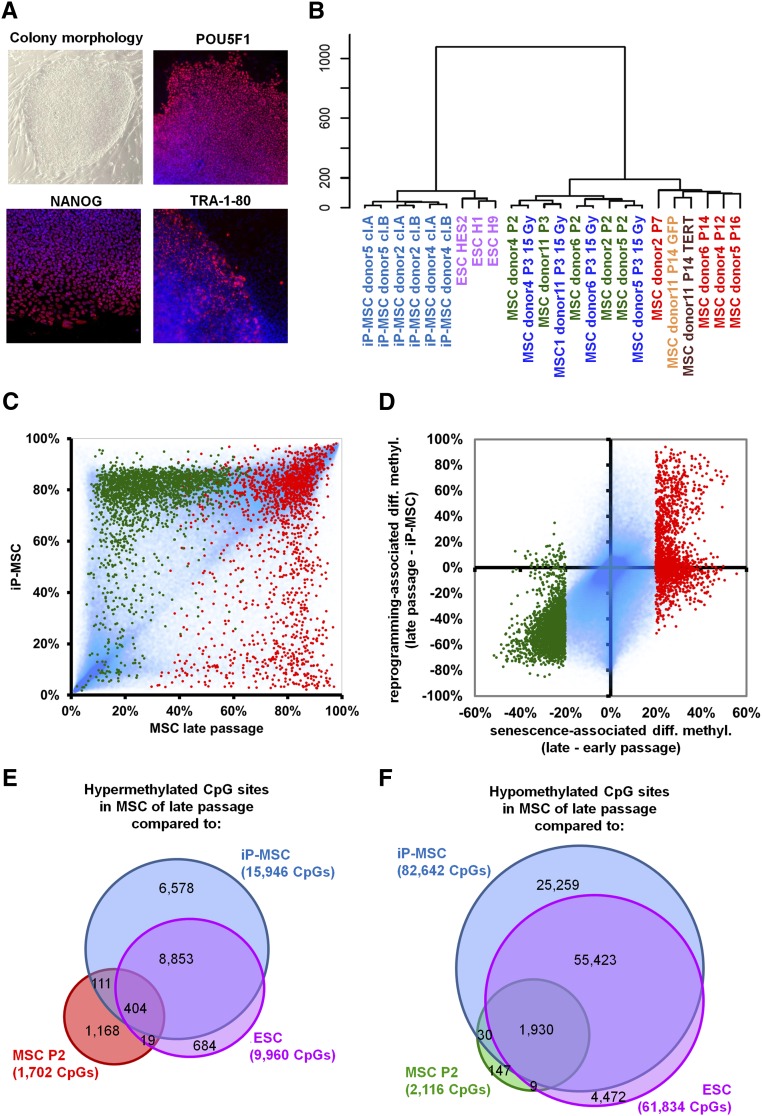
Subsequently, we analyzed SA-DNAm of induced pluripotent MSC (iP-MSC). MSC at passage 3 were retrovirally infected to express POU5F1 (OCT4), SOX2, MYC, and KLF4 (Takahashi et al. 2007). After 3–4 wk colonies with typical ESC-like morphology emerged, expressing pluripotency markers such as POU5F1, NANOG, and TRA-1-80 (Fig. 4A), whereas the expression of retrovirally encoded reprogramming factors was silenced and PluriTest analysis of global gene expression profiles supported the notion that iP-MSC were pluripotent (Muller et al. 2011; Shao et al. 2012). Furthermore, embryoid bodies derived from iP-MSC comprised derivatives of all three embryonic germ layers. For comparison we utilized three widely used human ESC lines (H1, H9, and HES2). Telomere length differed markedly between these three ESC lines, whereas iP-MSC always revealed significant telomere extension as compared with their parental MSC (P = 0.0018) (Fig. 3E).
Figure 4.
Reprogramming into iP-MSC prevents SA-DNAm changes. (A) Induced pluripotent MSC reveal typical ESC-like morphology and express the pluripotency markers POU5F1, NANOG, and TRA-1-80. (B) Unsupervised hierarchical clustering of global DNAm profiles clearly separates pluripotent cells (iP-MSC and ESC) and it discerns MSC of early and late passage. (C) Scatter plot of DNAm profiles of iP-MSC in comparison to MSC of late passage (P7–P16). In particular, CpG-sites that are hypomethylated upon replicative senescence (green) are hypermethylated in iP-MSC. In contrast, SA-hypermethylated CpG sites (red) are rather hypomethylated in iP-MSC. (D) Differential DNAm upon long-term culture is plotted against methylation changes upon reprogramming into iP-MSC. Overall, SA-DNAm changes are prevented in iP-MSC. (E,F) Venn diagrams demonstrating the overlap of SA-DNAm changes with differential methylation between MSC of late passage with either iP-MSC or ESC.
DNAm profiles were analyzed in iP-MSC that have been cultured for a similar number of passages as senescent MSC (P9–P11). Reprogramming of iP-MSC was associated with far-reaching epigenetic alterations and a global increase in DNAm level (Supplemental Fig. 10). Unsupervised hierarchical cluster analysis clearly segregated pluripotent from nonpluripotent cells and demonstrated the close relationship of iP-MSC and ESC. Notably, MSC of early and late passage were also separated (Fig. 4B). Most of the SA-DNAm changes were prevented by induced pluripotency. As expected, this association was more pronounced when we compared iP-MSC to MSC of late passage rather than MSC of early passage (Fig. 4C): 95% of the SA-hypomethylated CpG sites were significantly higher methylated in iP-MSC (2027 of 2116 CpG sites; P < 0.05). Conversely, 46% of the SA-hypermethylated CpG sites were significantly less methylated in iP-MSC in comparison to late passage MSC (796 of 1702 CpG sites) (Fig. 4D). Notably, the same tendency was also observed when we compared iP-MSC with MSC of early passage that have so far acquired relatively few SA-DNAm changes: 87% of the SA-hypomethylated CpG sites revealed a significantly higher methylation in iP-MSC (1842 of 2116 CpG sites), whereas 25% of the SA-hypermethylated CpG sites were significantly hypomethylated (419 of 1702 CpG sites; Supplemental Fig. 11). Comparison of DNAm profiles of MSC and iP-MSC may be difficult because of the stark differences between the two cell types. Yet, it is remarkable that pluripotent stem cells hardly revealed SA-DNAm changes.
We reasoned that those SA-DNAm changes particularly might be relevant, which appear to be prevented by pluripotency. SA-CpG sites were filtered to be differentially methylated in MSC of late passage in comparison to (1) MSC of early passage, (2) iP-MSC, and (3) ESC (adjusted P-value < 0.05; DNAm change >20%). A total of 404 CpG sites were overlappingly hypermethylated in MSC of late passage (Fig. 4E), which were significantly enriched in genes with sequence-specific DNA-binding transcription factor activity (GO:3700; P < 10−12), particularly those with a homeobox domain (Pfam, P < 10−6). They were also enriched in genes with promoter regions, which comprise motives for binding of POU2F1 (P = 1.1 × 10−4), growth factor independent 1 transcription repressor (GFI1; P = 1.4 × 10−4), and GATA4 (P = 3.5 × 10−4). Furthermore, they showed enrichment in targets of MIR323B (P = 2.1 × 10−5) and MIR10A/MIR10B (P = 4.6 × 10−4). In contrast, 1930 CpG sites were overlappingly hypomethylated at late passage (Fig. 4F), and these sites were significantly enriched in G-protein-coupled receptors (GO:4930; P = 1.6 × 10−8) and in genes with binding sites for POU6F1, a class 6 homeobox protein (P = 0.0025), STAT5A (P = 0.016), and GATA2 (P = 0.017).
When we used our epigenetic-senescence-signature, the predicted passage numbers for iP-MSC and ESC were close to noncultured cells, although they have been culture-expanded over many passages (Fig. 3D). Several other investigators have previously compared DNAm changes upon reprogramming (Doi et al. 2009; Lister et al. 2009). Publicly available data sets with InfiniumHumanMethylation27 BeadChip were retrieved and tested with the Epigenetic-Sensecence-Signature (Harness et al. 2011; Nishino et al. 2011; Nazor et al. 2012)—these pluripotent samples were always estimated close to noncultured cells (Supplemental Fig. 12).
Discussion
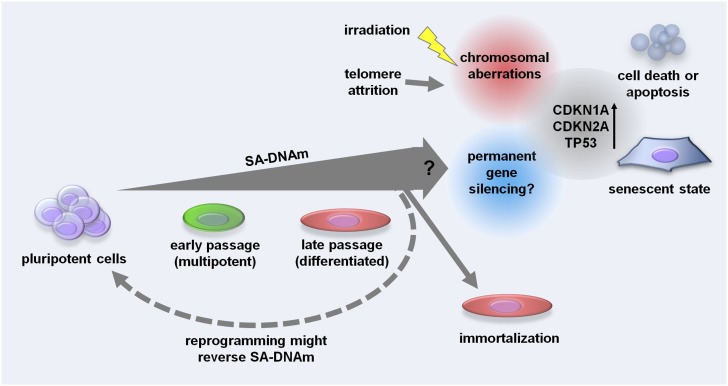
SA-DNAm changes are tightly coordinated and may entail functional consequences of cellular senescence. On the other hand, SA-DNAm does not seem to be the immediate trigger of cellular senescence. Irradiation-induced senescence, although phenotypically very similar to replicative senescence, was not associated with SA-DNAm changes. Furthermore, SA-DNAm was not influenced by TERT overexpression or immortalization. We demonstrate for the first time that most of the SA-DNAm changes do not occur in iP-MSC—and this supports the notion that these epigenetic modifications are functionally relevant. Our results indicate that long-term culture is associated with a process of cellular differentiation, which continuously locks specific genomic regions in an inactive state. Prevention of SA-DNAm upon reprogramming into iPSC may be crucial for rejuvenation and escape from senescence (Fig. 5).
Figure 5.
Schematic overview of how SA-DNAm changes may influence cellular senescence. Senescence is associated with a continuous gain of SA-DNAm changes that are enriched in genes with repressive histone marks. They may be associated with permanent gene silencing, but this link is yet unproven and therefore labeled with a question mark. SA-DNAm changes are not affected by ionizing radiation or telomere length. However, these processes evoke chromosomal aberrations and may ultimately activate the same downstream signal cascades of cellular senescence (including activation of CDKN1A, CDKN2A, and TP53). Pluripotent cells hardly reveal SA-DNAm changes; avoidance from these epigenetic modifications might contribute to the escape of pluripotent cells from cellular senescence.
MSC resemble heterogeneous cell preparations—the composition of subpopulations is greatly affected by culture conditions and changes during culture expansion (Wagner and Ho 2007; Schellenberg et al. 2012). Therefore, future work should aim for analysis of SA-DNAm changes on single cell level. On the other hand, similar SA-DNAm changes were also observed in dermal fibroblasts (Koch et al. 2011, 2012), indicating that the observations made in this study are not restricted to MSC.
We describe how SA-CpG sites are enriched in the gene body, 3′UTR, and intergenic regions. CpG-methylation in these regions is not directly related to gene expression levels (Ball et al. 2009). Particularly, SA-hypomethylated CpG sites are associated with genes showing neither expression at early, nor at late passage. In fact, none of the genes of the Epigenetic-Senescence-Signature was detected by qRT-PCR (Koch et al. 2012). This might explain why overall the SA-DNAm did not correlate with differential gene expression. Even though, SA-DNAm changes may be functionally relevant to permanently lock genes in an inactive state. DNAm directly results in tighter wrapping of DNA around the histone core (Lee and Lee 2012) and it may support reorganization of the nuclear compartment by methyl-CpG-binding proteins (MBPs) (Sasai and Defossez 2009). It has been suggested that DNAm is not the primary determinant of gene activation in MSC but rather post-translational modifications of histones (Collas 2010; Sorensen et al. 2010). DNAm may feedback on the histone code—potentially by changing the binding affinity of noncoding RNAs. PRC2 is recruited to target genes by noncoding RNAs such as HOTAIR, a transcript from the HOXC locus (Rinn et al. 2007); Xist, a noncoding RNA involved in X-chromosome inactivation (Zhao et al. 2008); and by the antisense-RNA 1 of CDKN2B (CDKN2B-AS1) (Yap et al. 2010). A recent comparison of epigenomic landscapes between human ESC and fibroblasts revealed that the repressive H3K9m3 and H3K27me3 marks expand upon differentiation into fibroblasts (Hawkins et al. 2010). In this study, we demonstrate that SA-hypomethylation, in particular, is significantly enriched in regions with H3K9me3, H3K27me3, and targets of PRC2, and is in line with previous observations (Schellenberg et al. 2011). It has been shown that aging induces DNAm in PRC2 target genes and may thereby predispose to cancer (Teschendorff et al. 2010). Many cancers are derived from epithelial cells, and our previous work indicated that similar SA-DNAm changes are also observed in other cell types including epithelial cells (Koch et al. 2012). This is of particular relevance, as SA-DNAm changes might be involved in malignant transformation. In fact, hypomethylation of CpG sites that are highly methylated in ESC has recently been shown to be a key determinant of tumor invasion and clinical outcome (Zhuang et al. 2012).
The irreversible nature of senescent cells is generally believed to be due to alterations in the chromatin structure. Upon senescence, chromatin undergoes dramatic remodeling through the formation of facultative heterochromatin domains called senescence-associated heterochromatic foci (SAHF) (Narita et al. 2003, 2006). These compacted foci of DNA disappear only 7 d after reprogramming of senescent cells, and this was even considered as a first sign of iPSC generation (Lapasset et al. 2011). Further research will be necessary to demonstrate whether the SA-DNAm reported here contributes to the formation of SAHF.
Ionizing radiation has been shown to induce a senescent phenotype in various different cell types (Di Leonardo et al. 1994; Igarashi et al. 2007); yet it is unclear whether the underlying molecular mechanisms are related to long-term culture-associated senescence (Rando and Chang 2012). We demonstrate that irradiation of MSC results in permanent proliferation arrest, typical senescent morphology, and loss of adipogenic and osteogenic differentiation potential. In contrast to replicative senescence, this stress-induced senescence was not associated with telomere attrition and did not evoke reproducible SA-DNAm changes 7 d after irradiation. It is conceivable that epigenetic alterations are initially not present at the level of DNAm, but rather on other levels of the epigenome: It has been suggested that factors involved in transcriptional repression and gene silencing are recruited to the site of DNA damage, including H3K9me3, H3K27me3, and repressive Polycomb complexes (O'Hagan et al. 2008; Chou et al. 2010; Seiler et al. 2011). This gene silencing may ultimately activate the same cell-intrinsic senescence program as described for cellular senescence—but the phenotypic and functional changes observed 7 d after irradiation are not causally related to DNAm changes at specific sites in the genome.
Immortalization is often considered a counterpart of cellular senescence, but we demonstrate that it does not reverse SA-DNAm changes. As described by many other studies, overexpression of TERT may provide a proliferative advantage, but does not assure immortalization of MSC (Bodnar et al. 1998; Kiyono et al. 1998). In contrast, combination of TERT with SV40-TAg facilitates reliable immortalization (O'Hare et al. 2001). In this study, we describe for the first time a conditionally immortalized MSC line with TERT and SV40-TAg using a tetracycline-inducible system. Despite immortalization, the in vitro differentiation potential decayed upon long-term culture—adipogenic differentiation could not be induced at passage 13 and osteogenic differentiation decreased at higher passages. TERT overexpression, as well as TERT/SV40-TAg immortalization, resulted in extended telomeres, but they did not affect SA-DNAm. Therefore, immortalization of MSC does not address the regulatory mechanisms of SA-DNAm, and these cells may still undergo functional changes throughout culture expansion.
Telomere homeostasis is also crucial for pluripotent cells. Telomere shortening is a barrier for iPSC generation (Marion et al. 2009b), whereas transfection of TERT and SV40-TAg in combination with the classical four factors (POU5F1, SOX2, MYC, and KLF4) has been shown to enhance the reprogramming efficiency (Park et al. 2008). Telomere extension upon reprogramming may be due to telomerase expression, or it may be facilitated by alternative lengthening of telomeres (ALT), which is based on homologous recombination between telomeric sequences (Dunham et al. 2000; Lapasset et al. 2011). Epigenetic modifications such as H3K9me3, H4K20me3, and histone acetylation of telomeric chromatin have been shown to be involved in the regulation of telomere length (Benetti et al. 2007; Schoeftner and Blasco 2009). Furthermore, a decrease in DNAm, particularly at subtelomeric regions, is accompanied by dramatically elongated telomeres (Gonzalo et al. 2006). SA-DNAm was not enriched in subtelomeric regions, and it is yet unclear whether or not it influences telomere length. To this end, our results demonstrate that telomere homeostasis is not the driving mechanism of SA-DNAm changes.
Replicative senescence has been described as a barrier to reprogramming of iPSC as the reprogramming efficiency is much higher in cells of early passage. A series of publications indicated that particularly up-regulation of TP53, CDKN1A, and CDKN2A may be responsible for this loss of reprogramming efficiency (Banito et al. 2009; Kawamura et al. 2009; Li et al. 2009; Marion et al. 2009a; Utikal et al. 2009). These factors are up-regulated upon senescence, and their ablation increased the reprogramming efficiency. It is also conceivable that the donor age resembles a barrier to reprogramming—other investigators demonstrated that it is possible to generate iPSC from centenarians (Lapasset et al. 2011), and it will be of interest to systematically analyze whether epigenetic rejuvenation is hampered in such samples. Overall, counteraction of senescence-associated modifications seems to play a central role for the generation of iPSC. In this regard, SA-DNAm might contribute to the reprogramming barrier at later passages.
Conclusion
All signs of cellular aging seem to be removed upon reprogramming into iPSC. It has even been demonstrated that continuous passaging of iPSC further diminishes differences in DNAm between iPSC and ESC (Nishino et al. 2011). Yet, upon redifferentiation into fibroblasts they reveal typical signs of cellular aging, and finally enter a senescent state (Lapasset et al. 2011). Our analysis reveals that SA-DNAm changes are tightly regulated. They are significantly enriched in genes with H3K9me3, H3K27me3, and PRC2-binding sites, indicating that SA-DNAm changes are associated with heterochromatin formation. Overall, our results support the notion that cellular aging is not only due to accumulation of cellular defects, but rather resembles an organized process that stalls cells in a particular functional state. It remains to be demonstrated whether SA-DNAm changes are directly related to proliferation (replicative senescence) or rather some other type of culture stress (premature senescence). Furthermore, it is still not proven whether SA-DNAm resembles the initial cause of senescence or whether it is a mere marker. MSC are heterogeneous—also with regard to cellular aging. It is therefore conceivable that our iP-MSC clones were derived from a subset with little SA-DNAm, and that this low level is then maintained throughout culture expansion. Alternatively, SA-DNAm might even be reversed in the course of reprogramming; this would suggest that pluripotency entails epigenetic rejuvenation of cellular senescence.
Methods
Expansion of mesenchymal stromal cells
MSC were isolated from tibia plateau of patients undergoing orthopaedic surgery (age range: 31–73 yr). All samples were taken after written consent using guidelines approved by the Ethic Committee on the Use of Human Subjects at the University of Aachen (Permit number: EK128/09). MSC were culture-expanded in standard medium consisting of DMEM (PAA; 1 g/L glucose) supplemented with glutamine (PAA), penicillin/streptomycin (PAA), and 10% human platelet lysate (hPL) as previously described (Horn et al. 2010). Cells grew at 37°C, in 5% CO2, and atmospheric oxygen conditions, and they were harvested by trypsinization upon 80% confluent growth, counted with a Neubauer chamber (Brand, Wertheim, Germany), and reseeded at a density of 5000 cells/cm2. cPD were calculated as described before (Cholewa et al. 2011).
Immunophenotypic analysis
Surface marker expression was analyzed on a FACS canto II (Becton Dickinson & Co.) upon staining with the following antibodies as described before (Wagner et al. 2008): CD14-allophycocyanin (APC, clone M5E2, BD), CD29-phycoerythrin (PE, clone MAR4, BD), CD31-PE (clone WM59, BD), CD34-APC (clone 8G12, BD), CD45-APC (clone HI30, BD), CD73-PE (clone AD2, BD), CD90-APC (clone 5E10, BD), CD105-fluorescein isothiocyanate (FITC, clone MEM-226, ImmunoTools).
In vitro differentiation
Adipogenic and osteogenic differentiation of MSC was induced as described before (Pittenger et al. 1999). After 18 d, osteogenic differentiation was analyzed by Alizarin Red staining. Adipogenic differentiated cells were stained with the green fluorescent dye BODIPY (4,4-difluoro-1,2,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene) and counterstained with DAPI (4′,6-Diamidin-2-phenylindol; both Molecular Probes). Quantification was performed as described in our previous work (Koch et al. 2011). Chondrogenic differentiation was induced in micromass culture for 18 d. Subsequently, the pellets were fixed with 10% formalin and first embedded in agarose and secondly in paraffin; 1 mm sections were stained as previously described (Koch et al. 2011).
Senescence-associated beta-galactosidase
Expression of pH-dependent senescence-associated beta-galactosidase (SA-β-gal) was analyzed using the SA-β-gal staining kit (Cell Signaling Technology) or by flow cytometry with the fluorogenic substrate 5-dodecanoylaminofluorescein di-beta-D-galactopyranoside (C12FDG) (Debacq-Chainiaux et al. 2009).
Senescence induction by gamma-irradiation
MSC were treated with a blood product irradiator (model IBL-437, Institute for transfusion medicine, Aachen) with 5, 10, 15, 20, or 25 Gy as indicated in the text. Proliferation rate of MSC in early and late passage as well as after irradiation was determined in triplicate using the Alamar blue assay (Life Technologies), which was measured with a Tecan infinite M200 plate reader.
TERT overexpression
The TERT sequence was excised from the pGRN145 plasmid (ATCC MBA-141) and subcloned into the retroviral expression vector pMIG (pMSCV-IRES-GFP) (Addgene plasmid 9044, Addgene) via EcoRI. MSC of five donors were transduced at passage 4 with the TERT-IRES-GFP construct, or for control with the corresponding empty pMIG vector (IRES-GFP), and the culture expanded in parallel until entering the senescent state.
Conditional immortalization with TERT/SV40-TAg
MSC were transfected with two lentiviral vectors (May et al. 2010): (1) SV40 large T antigen (SV40-TAg) under an unidirectional tetracycline-dependent promoter, which also drives a reverse transactivator through EMCV-IRES; (2) TERT under a bidirectional tetracycline-dependent promoter, additionally driving expression of a reverse transactivator and an eGFP/neomycin phosphotransferase fusion gene linked by an EMCV-IRES element. Initially, MSC were transduced on 2 consecutive d with SV40-TAg and after 2 wk with TERT. Upon neomycin selection, the resulting colonies (∼20 independent clones) were pooled and culture expanded in DMEM (PAA; 4,5 g/L glucose) supplemented with glutamine (PAA), penicillin/streptomycin (PAA), 15% FCS (PAA), and doxycycline hyclate (Dox; final conc. 2 μg/mL; Sigma, Taufkirchen). Proliferation rate with and without Dox was analyzed using the Alamar blue assay as described above.
Telomere length analysis
The average telomere length was estimated by monochrome multiplex Q-PCR as described before (Cawthon 2009; Ziegler et al. 2012). In brief, telomeres (T) and the beta-globin gene (S; single copy) were amplified on a MyIQ2 Two-color Realtime PCR Detection-System (Biorad). Analysis was performed in triplicate in parallel to serial dilutions of standard DNA. The relative telomere to single-copy gene (T/S) ratio for experimental DNA samples was calculated by dividing the copy number of the telomere template T by the copy number of the beta-globin template S. Amplification efficiency for the telomere and the beta-globin was between 90% and 100% in each run. Specificity of the amplification was confirmed by melt-curve analysis.
Generation and characterization of iP-MSC
MSC from three donors (passage 3) were infected on 2 consecutive d with equal amounts of pMXs-based retroviruses (Addgene) carrying the POU5F1, SOX2, KLF4, and MYC genes (Takahashi et al. 2007). On day 3, the medium was changed to high-glucose DMEM (Gibco), 20% FCS, 1% nonessential amino acids (NAA), 1× penicillin/streptomycin, 1× L-glutamine, 0.1 mM β-mercaptoethanol (βME), and 50 ng/mL basic fibroblast growth factor (bFGF) (Peprotech) (Cai et al. 2010). On day 6 the cells were plated on gelatin-coated dishes containing irradiated CF1 murine embryonic fibroblasts (MEFs), and on day 7 the culture medium was supplemented with 20 μg/mL vitamin C (Sigma) and 1 mM valproic acid (Sigma). Between days 21 and 30 the ESC-like colonies were observed and mechanically isolated. Established iP-MSC were maintained on MEFs in DMEM/F12 medium supplemented with Glutamax, 20% knockout serum replacer, 1% NAA, 0.1 mM βME, and 50 ng/mL bFGF. Cells were passaged by manual dissection of cell clusters every 5–6 d. Human ESC lines H1, H9, and HES-2 (WiCell Research Institute) were used as controls and cultured under the same conditions as described above. Work with human ESC has been approved by the Robert Koch Institute (permit no. Az 1710-79-1-4-57). Immunohistochemical analysis of pluripotency markers was performed with primary antibodies against POU5F1 (Santa Cruz), SOX2 (Stemgent), NANOG (Stemgent), or TRA-1-80 (Santa Cruz) by alkaline phosphatase staining (Sigma) and by flow cytometric analysis of TRA-1-60 (BD Pharmingen) and SSEA4 (Santa Cruz). Furthermore, iP-MSC differentiation was analyzed using the embryoid body differentiation system with immunostaining after 14–19 d for desmin (mesoderm), alpha-fetoprotein (endoderm), and nestin (ectoderm). Generation of iP-MSC clones and analysis of pluripotency has been described in further detail elsewhere (Shao et al. 2012).
DNA methylation analysis
Genomic DNA was isolated from 106 cells using the QIAGEN DNA Blood Midi-Kit. Bisulfite-converted DNA (EZ DNA Methylation Kit; Zymo) was prepared and then analyzed using the InfiniumHumanMethylation450 BeadChip (Illumina) according to the manufacturer's instructions. DNAm is provided as beta-values for each of the 485.577 CpG sites, ranging from 0 (nonmethylated) to 1 (100% methylation). Hybridization and initial data analysis with the BeadStudio Methylation Module was performed at the DKFZ Gene Core Facility in Heidelberg. Distribution of samples across the BeadChips is presented in Supplemental Table 4. Data have been deposited at NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/; accession no: GSE37066).
In addition, we analyzed all samples using our Epigenetic-Senescence-Signature, which has been described in detail before (Koch et al. 2012). This method is based on continuous DNAm changes at six specific CpG sites (associated with the genes GRM7, CASR, PRAMEF2, SELP, CASP14, and KRTAP13-3). DNAm at these specific CpG sites was analyzed by pyrosequencing (Varionostic GmbH) and subsequently used for linear regression models to estimate passage numbers, cPD, and days in culture. These calculations can also be performed with the online calculator, which is accessible under http://www.molcell.rwth-aachen.de/dms/.
Bioinformatic analysis of DNAm profiles
For further analysis we have only considered CpG sites on autosomes. Initially, we considered various normalization regimens, but we then decided to use raw data beta-values due to the global changes of DNAm upon reprogramming with an overall increase of DNAm level. Histograms of DNA methylation levels and principal components analysis (PCA) were calculated with R. Unsupervised hierarchical clustering according to Euclidian Distance (with Ward's Minimum Variance Method) was performed using the MultiExperiment Viewer (MeV, TM4.7.4; http://www.tm4.org/mev/) or R. SA-DNAm changes were calculated by the limma package in R with adjusted P-values <0.05 and >20% change in DNAm level between early and late passages. Affiliation of CpG sites with gene regions or CGI was used as described in detail previously (Sandoval et al. 2011). Gene Ontology analysis of genes with DNAm changes was performed using the GoMiner software (http://discover.nci.nih.gov/gominer/) and enrichment of chromosomal location was analyzed with the gene set enrichment analysis (GSEA; http://www.broadinstitute.org/gsea/index.jsp). Furthermore, we compared DNAm profiles with published chromatin immunprecipitation (ChIP) results: data for H3K9me3 and H3K27me3 in adipose tissue-derived MSC were retrieved from GSE17053 (Delbarre et al. 2010) and further processed by NimbleScan; EZH2 targets in bone marrow MSC were retrieved from the supplemental material of another manuscript (Wei et al. 2011). For comparison, DNAm and ChIP data were matched by GenBank Accession IDs and enrichment of SA-DNAm in these regions was determined by hypergeometric distribution.
Gene expression analysis
For gene expression analysis, three additional MSC samples were isolated and expanded as described above. Reseeding was performed at a density of 13,333 cells/cm2 in standard medium consisting of DMEM (PAA; 1 g/L glucose) supplemented with glutamine (PAA), penicillin/streptomycin (PAA), and 10% FSC (PAA). RNA was isolated at early passage (P3, P4, or P5) and late passage (P10) with the miRNeasy kit (Qiagen). RNA concentration and integrity were determined by an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). Gene expression analysis was performed on GeneChip Human Gene 1.0 ST Arrays (Affymetrix) at the Department of Population Genetics, Genomics, and Bioinformatics at the University of Maastricht. Raw data were normalized by RMA using Affymetrix Power Tools, and matched to DNAm data by gene name. Data are accessible at GEO Series GSE37065. To provide further evidence that iP-MSC clones are fully reprogrammed we have analyzed gene expression profiles of iP-MSC clones as described above. These hybridizations were performed at the Interdisciplinary Center for Clinical Research (IZKF) at the RWTH Aachen, and subsequently projected to PluriTest analysis as described before (Muller et al. 2011). All samples were clearly associated with pluripotent cells and data are accessible at GEO Series GSE38806.
Data access
The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE37065, GSE37066, and GSE38806.
Acknowledgments
We are grateful to Matthias Schick (gene core facility at the German Cancer Research Center, Heidelberg, Germany) for array hybridization and initial data analysis. We thank Jeannette Zauers (HZI, Braunschweig, Germany), Rebecca Dieterich, and Nadin Lange (both University of Cologne, Germany) as well as Sana Mohamad (Department of Hematology, Essen) for excellent technical assistance. This work was supported by the excellence initiative of the German federal and state governments within the START-Program of the Faculty of Medicine, RWTH Aachen (W.W.); by the state North Rhine Westphalia within the BioNRW2 project “StemCellFactory” (M.Z., W.W.), and by the Stem Cell Network NRW (T.S., W.W.); by the Köln Fortune Program (T.S.); and particularly by the Else-Kröner Fresenius Stiftung (T.S., C.M.K., W.W.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.141945.112.
References
- Acevedo LG, Bieda M, Green R, Farnham PJ 2008. Analysis of the mechanisms mediating tumor-specific changes in gene expression in human liver tumors. Cancer Res 68: 2641–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM 2009. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol 27: 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, et al. 2009. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev 23: 2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazarov AV, Jae LW, Bazarov I, Bosire M, Hines WC, Stankovich B, Chicas A, Lowe SW, Yaswen P 2012. The specific role of pRb in p16 (INK4A)-mediated arrest of normal and malignant human breast cells. Cell Cycle 11: 1008–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA 2008. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40: 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti R, Garcia-Cao M, Blasco MA 2007. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat Genet 39: 243–250 [DOI] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, et al. 2011. High density DNA methylation array with single CpG site resolution. Genomics 98: 288–295 [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279: 349–352 [DOI] [PubMed] [Google Scholar]
- Bork S, Pfister S, Witt H, Horn P, Korn B, Ho AD, Wagner W 2010. DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell 9: 54–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Li W, Su H, Qin D, Yang J, Zhu F, Xu J, He W, Guo X, Labuda K, et al. 2010. Generation of human induced pluripotent stem cells from umbilical cord matrix and amniotic membrane mesenchymal cells. J Biol Chem 285: 11227–11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, d'Adda di FF 2007. Cellular senescence: When bad things happen to good cells. Nat Rev Mol Cell Biol 8: 729–740 [DOI] [PubMed] [Google Scholar]
- Cawthon RM 2009. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res 37: e21 doi: 10/1093/nar/gkn1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholewa D, Stiehl T, Schellenberg A, Bokermann G, Joussen S, Koch C, Walenda T, Pallua N, Marciniak-Czochra A, Suschek CV, et al. 2011. Expansion of adipose mesenchymal stromal cells is affected by human platelet lysate and plating density. Cell Transplant 20: 1409–1422 [DOI] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ 2010. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci 107: 18475–18480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmielova J, Havelek R, Soukup T, Jiroutova A, Visek B, Suchanek J, Vavrova J, Mokry J, Muthna D, Bruckova L, et al. 2012. Gamma radiation induces senescence in human adult mesenchymal stem cells from bone marrow and periodontal ligaments. Int J Radiat Biol 88: 393–404 [DOI] [PubMed] [Google Scholar]
- Collas P 2010. Programming differentiation potential in mesenchymal stem cells. Epigenetics 5: 476–482 [DOI] [PubMed] [Google Scholar]
- Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O 2009. Protocols to detect senescence-associated β-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc 4: 1798–1806 [DOI] [PubMed] [Google Scholar]
- Delbarre E, Jacobsen BM, Reiner AH, Sorensen AL, Kuntziger T, Collas P 2010. Chromatin environment of histone variant H3.3 revealed by quantitative imaging and genome-scale chromatin and DNA immunoprecipitation. Mol Biol Cell 21: 1872–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leonardo A, Linke SP, Clarkin K, Wahl GM 1994. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev 8: 2540–2551 [DOI] [PubMed] [Google Scholar]
- Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. 2009. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet 41: 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E 2006. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8: 315–317 [DOI] [PubMed] [Google Scholar]
- Dunham MA, Neumann AA, Fasching CL, Reddel RR 2000. Telomere maintenance by recombination in human cells. Nat Genet 26: 447–450 [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA 2006. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol 8: 416–424 [DOI] [PubMed] [Google Scholar]
- Harness JV, Turovets NA, Seiler MJ, Nistor G, Altun G, Agapova LS, Ferguson D, Laurent LC, Loring JF, Keirstead HS 2011. Equivalence of conventionally-derived and parthenote-derived human embryonic stem cells. PLoS ONE 6: e14499 doi: 10.1371/journal.pone.0014499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. 2010. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6: 479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L, Moorhead P 1961. The serial cultivation of human diploid cell strains. Exp Cell Res 25: 585–621 [DOI] [PubMed] [Google Scholar]
- Horn P, Bokermann G, Cholewa D, Bork S, Walenda T, Koch C, Drescher W, Hutschenreuther G, Zenke M, Ho A, et al. 2010. Comparison of individual platelet lysates for isolation of human mesenchymal stromal cells. Cytotherapy 12: 888–898 [DOI] [PubMed] [Google Scholar]
- Igarashi K, Sakimoto I, Kataoka K, Ohta K, Miura M 2007. Radiation-induced senescence-like phenotype in proliferating and plateau-phase vascular endothelial cells. Exp Cell Res 313: 3326–3336 [DOI] [PubMed] [Google Scholar]
- Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, Hohmeyer A, Gereke M, Rudalska R, Potapova A, et al. 2011. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479: 547–551 [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC 2009. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460: 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396: 84–88 [DOI] [PubMed] [Google Scholar]
- Koch C, Suschek CV, Lin Q, Bork S, Goergens M, Joussen S, Pallua N, Ho AD, Zenke M, Wagner W 2011. Specific age-associated DNA methylation changes in human dermal fibroblasts. PLoS ONE 6: e16679 doi: 10.1371/journal.pone.0016679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CM, Joussen S, Schellenberg A, Lin Q, Zenke M, Wagner W 2012. Monitoring of cellular senescence by DNA-methylation at specific CpG sites. Aging Cell 11: 366–369 [DOI] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, et al. 2008. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet 40: 741–750 [DOI] [PubMed] [Google Scholar]
- Lapasset L, Milhavet O, Prieur A, Besnard E, Babled A, Ait-Hamou N, Leschik J, Pellestor F, Ramirez JM, De VJ, et al. 2011. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev 25: 2248–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Lee TH 2012. Effects of DNA methylation on the structure of nucleosomes. J Am Chem Soc 134: 173–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M 2009. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460: 1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA 2009a. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460: 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, Serrano M, Blasco MA 2009b. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell 4: 141–154 [DOI] [PubMed] [Google Scholar]
- May T, Butueva M, Bantner S, Markusic D, Seppen J, MacLeod RA, Weich H, Hauser H, Wirth D 2010. Synthetic gene regulation circuits for control of cell expansion. Tissue Eng Part A 16: 441–452 [DOI] [PubMed] [Google Scholar]
- Muller FJ, Schuldt BM, Williams R, Mason D, Altun G, Papapetrou EP, Danner S, Goldmann JE, Herbst A, Schmidt NO, et al. 2011. A bioinformatic assay for pluripotency in human cells. Nat Methods 8: 315–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113: 703–716 [DOI] [PubMed] [Google Scholar]
- Narita M, Narita M, Krizhanovsky V, Nunez S, Chicas A, Hearn SA, Myers MP, Lowe SW 2006. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 126: 503–514 [DOI] [PubMed] [Google Scholar]
- Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, Garitaonandia I, Muller FJ, Wang YC, Boscolo FS, et al. 2012. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell 10: 620–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K, Toyoda M, Yamazaki-Inoue M, Fukawatase Y, Chikazawa E, Sakaguchi H, Akutsu H, Umezawa A 2011. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet 7: e1002085 doi: 10.1371/journal.pgen.1002085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan HM, Mohammad HP, Baylin SB 2008. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet 4: e1000155 doi: 10.1371/journal.pgen.1000155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare MJ, Bond J, Clarke C, Takeuchi Y, Atherton AJ, Berry C, Moody J, Silver AR, Davies DC, Alsop AE, et al. 2001. Conditional immortalization of freshly isolated human mammary fibroblasts and endothelial cells. Proc Natl Acad Sci 98: 646–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ 2008. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451: 141–146 [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR 1999. Multilineage potential of adult human mesenchymal stem cells. Science 284: 143–147 [DOI] [PubMed] [Google Scholar]
- Rando TA, Chang HY 2012. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell 148: 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval J, Heyn HA, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M 2011. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 6: 692–702 [DOI] [PubMed] [Google Scholar]
- Sasai N, Defossez PA 2009. Many paths to one goal? The proteins that recognize methylated DNA in eukaryotes. Int J Dev Biol 53: 323–334 [DOI] [PubMed] [Google Scholar]
- Schellenberg A, Lin Q, Schüeler H, Koch C, Joussen S, Denecke B, Walenda G, Pallua N, Suschek C, Zenke M, et al. 2011. Replicative senescence of mesenchymal stem cells causes DNA-methylation changes which correlate with repressive histone marks. Aging (Albany NY) 3: 873–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg A, Stiehl T, Horn P, Joussen S, Pallua N, Ho A, Wagner W 2012. Population dynamics of mesenchymal stromal cells during culture expansion. Cytotherapy 14: 401–411 [DOI] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39: 232–236 [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA 2009. A ‘higher order’ of telomere regulation: Telomere heterochromatin and telomeric RNAs. EMBO J 28: 2323–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler DM, Rouquette J, Schmid VJ, Strickfaden H, Ottmann C, Drexler GA, Mazurek B, Greubel C, Hable V, Dollinger G, et al. 2011. Double-strand break-induced transcriptional silencing is associated with loss of tri-methylation at H3K4. Chromosome Res 19: 883–899 [DOI] [PubMed] [Google Scholar]
- Seluanov A, Gorbunova V, Falcovitz A, Sigal A, Milyavsky M, Zurer I, Shohat G, Goldfinger N, Rotter V 2001. Change of the death pathway in senescent human fibroblasts in response to DNA damage is caused by an inability to stabilize p53. Mol Cell Biol 21: 1552–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao K, Koch CM, Gupta MK, Lin Q, Lenz M, Laufs S, Denecke B, Schmidt M, Linke M, Hennies HC, et al. 2012. Induced pluripotent mesenchymal stromal cell clones retain donor-derived differences in DNA methylation profiles. Mol Ther (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Roninson IB 2004. Hallmarks of senescence in carcinogenesis and cancer therapy. Oncogene 23: 2919–2933 [DOI] [PubMed] [Google Scholar]
- Sorensen AL, Jacobsen BM, Reiner AH, Andersen IS, Collas P 2010. Promoter DNA methylation patterns of differentiated cells are largely programmed at the progenitor stage. Mol Biol Cell 21: 2066–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperka T, Song Z, Morita Y, Nalapareddy K, Guachalla LM, Lechel A, Begus-Nahrmann Y, Burkhalter MD, Mach M, Schlaudraff F, et al. 2012. Puma and p21 represent cooperating checkpoints limiting self-renewal and chromosomal instability of somatic stem cells in response to telomere dysfunction. Nat Cell Biol 14: 73–79 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, et al. 2010. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res 20: 440–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K 2009. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460: 1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner W 2010. Senescence is heterogeneous in mesenchymal stromal cells - kaleidoscopes for cellular aging. Cell Cycle 9: 2923–2924 [DOI] [PubMed] [Google Scholar]
- Wagner W, Ho AD 2007. Mesenchymal stem cell preparations-comparing apples and oranges. Stem Cell Rev 3: 239–248 [DOI] [PubMed] [Google Scholar]
- Wagner W, Horn P, Castoldi M, Diehlmann A, Bork S, Saffrich R, Benes V, Blake J, Pfister S, Eckstein V, et al. 2008. Replicative senescence of mesenchymal stem cells: A continuous and organized process. PLoS ONE 5: e2213 doi: 10.1371/journal.pone.0002213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, Yang CC, Yang JY, Lin CY, Lai CC, et al. 2011. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol 13: 87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM 2010. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X 2007. Human embryonic stem cells: Mechanisms to escape replicative senescence? Stem Cell Rev 3: 270–279 [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J, Jones A, Lee SH, Ng E, Fiegl H, Zikan M, Cibula D, Sargent A, Salvesen HB, Jacobs IJ, et al. 2012. The dynamics and prognostic potential of DNA methylation changes at stem cell gene loci in women's cancer. PLoS Genet 8: e1002517 doi: 10.1371/journal.pgen.1002517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler P, Schrezenmeier H, Akkad J, Brassat U, Vankann L, Panse J, Wilop S, Balabanov S, Schwarz K, Martens UM, et al. 2012. Telomere elongation and clinical response to androgen treatment in a patient with aplastic anemia and a heterozygous hTERT gene mutation. Ann Hematol 91: 1115–1120 [DOI] [PubMed] [Google Scholar]