Abstract
In metazoans, the nuclear lamina is thought to play an important role in the spatial organization of interphase chromosomes, by providing anchoring sites for large genomic segments named lamina-associated domains (LADs). Some of these LADs are cell-type specific, while many others appear constitutively associated with the lamina. Constitutive LADs (cLADs) may contribute to a basal chromosome architecture. By comparison of mouse and human lamina interaction maps, we find that the sizes and genomic positions of cLADs are strongly conserved. Moreover, cLADs are depleted of synteny breakpoints, pointing to evolutionary selective pressure to keep cLADs intact. Paradoxically, the overall sequence conservation is low for cLADs. Instead, cLADs are universally characterized by long stretches of DNA of high A/T content. Cell-type specific LADs also tend to adhere to this “A/T rule” in embryonic stem cells, but not in differentiated cells. This suggests that the A/T rule represents a default positioning mechanism that is locally overruled during lineage commitment. Analysis of paralogs suggests that during evolution changes in A/T content have driven the relocation of genes to and from the nuclear lamina, in tight association with changes in expression level. Taken together, these results reveal that the spatial organization of mammalian genomes is highly conserved and tightly linked to local nucleotide composition.
The spatial architecture of interphase chromosomes is thought to be important for gene regulation and genome maintenance (Misteli and Soutoglou 2009; Kind and van Steensel 2010). However, the organization of chromosomes inside the nucleus is still poorly understood. While folding of the chromatin fiber is to some degree stochastic, most genomic loci are nonrandomly positioned with respect to each other and relative to fixed landmarks in the nucleus. Two classes of biochemical mechanisms are thought to contribute to this nonrandom positioning (van Steensel and Dekker 2010). First, a variety of protein complexes mediate specific physical associations between linearly distant loci. Second, specific loci may be anchored to large nuclear structures that serve as scaffolds. One of the main candidates for such a scaffold function is the nuclear lamina (NL).
The NL is a filamentous structure of proteins lining the inner nuclear membrane of metazoans. Lamins are intermediate filament proteins that form the major component of the NL. In mammals, these are represented by lamin A/C (A-type) and lamin B1 and B2 (B-type). By DamID of B-type lamins, we have previously shown that genomes of organisms evolutionarily as distant as fruit fly, mouse, and man have large nuclear lamina–associated domains (LADs) (Pickersgill et al. 2006; Guelen et al. 2008; Peric-Hupkes et al. 2010; van Bemmel et al. 2010). LADs are generally very large regions (typically hundreds of kilobases) and collectively cover ∼35% of the genome. Genes within LADs are generally transcriptionally inactive. Comparison of mouse embryonic stem (ES) cells and differentiated cell types revealed that hundreds of genes interact with the NL in a cell-type specific (facultative) manner. These genes lose NL association upon or prior to their activation during differentiation, or gain NL association if they are no longer expressed (Peric-Hupkes et al. 2010).
Despite these dynamics, there appear to be many regions in the genome that interact with the NL in a cell type independent manner. Such constitutive LADs (cLADs) may provide chromosomes with a basic “backbone” structure that is shared among most or all cell types. Insight into the nature of cLADs is therefore of importance to our understanding of the mechanisms that determine the spatial architecture of chromosomes. Here, we report a detailed analysis of cLADs. We find that they are highly conserved between mouse and human, indicating that they are functionally important. Sequence analysis reveals that cLADs can be predicted based on their high A/T content. Furthermore, we demonstrate that divergence of spatial positioning of paralogous genes strongly correlates with a divergence in their overall A/T content. We propose that A/T-rich stretches in mammalian genomes serve as NL-anchoring sequences that form a structural backbone of interphase chromosomes.
Results
Genomic regions of constitutive NL interactions have distinctive properties
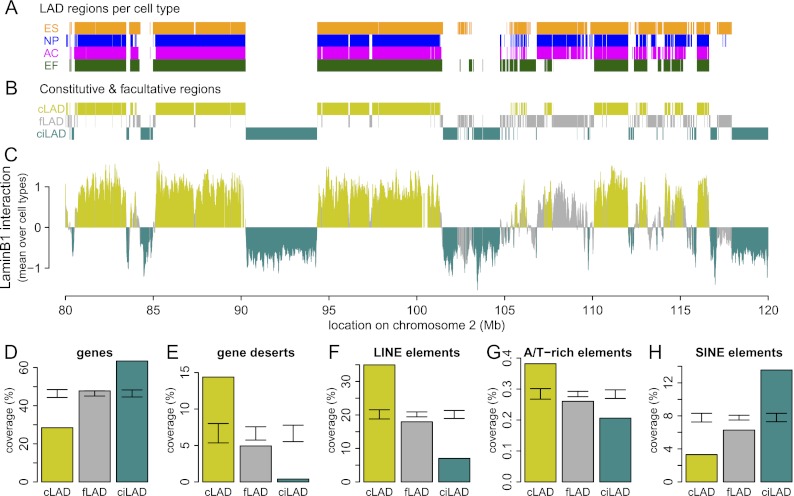
To compare regions that exhibit constitutive and facultative NL interactions, we used previously reported genome-wide DamID lamin B1 interaction data from four different mouse cell types: embryonic stem cells (ESCs), neural precursor cells (NPCs), astrocytes (ACs), and embryonic fibroblasts (MEFs) (Peric-Hupkes et al. 2010). The DamID data were obtained using genomic tiling arrays with a median probe spacing of ∼1.2 kb. To compare NL interactions among the four cell types, we first used a hidden Markov model (HMM) to classify all microarray probes in each of the cell types as either LAD or inter-LAD (Fig. 1A). Next, we defined constitutive LADs (cLADs) as regions that are LAD in all four cell types; constitutive inter-LADs (ciLADs) as regions that are inter-LAD in all four cell types, and facultative LADs (fLADs) as regions for which LAD-status is cell type dependent (Fig. 1B). Furthermore, we refer to cLADs and ciLADs collectively as “constitutive regions” because they are both invariant between cell types. We find that ∼71% of the genome is organized in a constitutive manner, consisting of roughly equal parts of cLADs (33%) and ciLADs (38%) (Fig. 1C). We note that these definitions are operational; a comprehensive definition would require maps of genome–NL interactions in all possible cell types. Nevertheless, as shown below, the operational definitions used here provide useful insights.
Figure 1.
A core architecture of genome–nuclear lamina interactions. (A) Regions classified by HMM as lamin B1 interacting in mouse ESC (orange), NPC (blue), astrocyte (magenta), and MEF (dark green) cell cultures, shown for a 40-Mb region of chromosome 2. (B) Regions common to all cell types are termed cLADs (mustard) and ciLADs (cyan), with dynamic regions termed fLADs (gray). (C) Mean profile of lamin B1 association in the cell types indicated in A. Colors as in B. The core architecture of constitutive regions makes up ∼71% of the genome, with 33% in cLAD and 38% in ciLAD regions. (D–H) Percent coverage of cLAD (mustard), fLAD (gray), and ciLAD regions for genes (D), gene deserts (E), LINE elements (F), simple A/T-rich elements (G), and SINE elements (H). Error bars indicate two standard deviations around mean coverage values for random regions obtained through circular permutations.
We have previously shown that LADs are relatively gene poor in multiple species (Pickersgill et al. 2006; Guelen et al. 2008; Peric-Hupkes et al. 2010; van Bemmel et al. 2010). cLADs are also very gene poor, even more so than fLADs (Fig. 1D). Consistent with this, gene deserts are substantially enriched in cLADs (Fig. 1E). Furthermore, LINE elements (Fig. 1F), of which most are L1 elements, and simple A/T-rich elements (Fig. 1G) are enriched, while SINE elements are depleted in cLADs (Fig. 1H). Importantly, cLADs and fLADs differ for all five genomic features (Fig. 1D–H), indicating that these two classes of LADs are distinct even though they were defined based on four cell types only.
cLADs are conserved across species
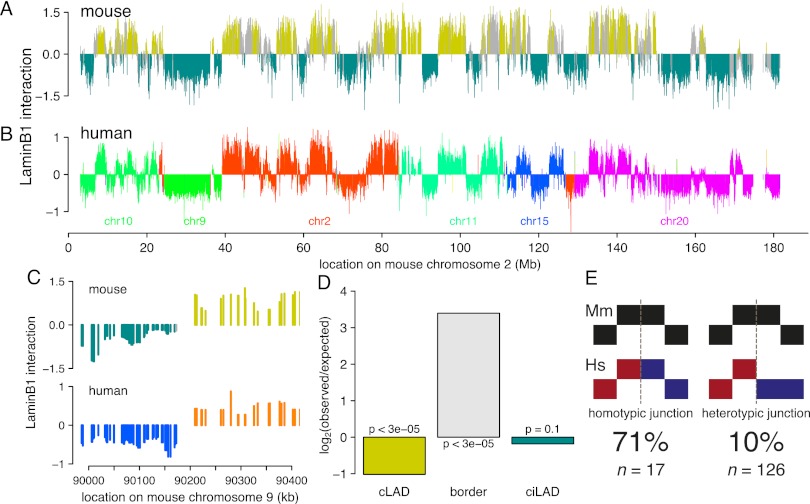
We reasoned that if cLADs are important, then they are likely to be conserved across species. To test this, we generated a genome-wide map of NL interactions in human ESCs and compared it with the previously derived map from mouse ESCs (Peric-Hupkes et al. 2010). After correction for divergence in synteny, we observed a remarkable similarity of the NL interactions between mouse and human ESCs (Fig. 2A,B; Supplemental Fig. 1), with an overall concordance of 83%. Remarkably, this concordance is even higher in constitutive regions (91%) and relatively low in fLADs (67%, which is only modestly higher than the 52% concordance expected by random chance, although this is also statistically significant) (Table 1). The evolutionary conservation of NL interactions is not restricted to ESCs, because a separate comparison between fibroblasts from both species yielded similar results (Supplemental Fig. 2; Table 1). To further investigate conservation of constitutive regions between mouse and human, we also generated DamID lamin B1 maps for the human HT1080 fibrosarcoma cell line and found that as much as 71% of mouse constitutive regions are also constitutive across human ESCs, fibroblasts, and HT1080 cells. Taken together, the pattern of cLADs and ciLADs is highly conserved between mouse and human, which are separated by ∼75 million years in the evolutionary tree.
Figure 2.
Core architecture is highly conserved between mouse and human. (A) Map of NL interactions in mouse ESCs for chromosome 2 ([mustard] cLADs; [cyan] ciLADs; [gray] fLADs), and (B) in human ESCs for the corresponding syntenic regions (each color denotes a different human chromosome). (C) Example of co-occurrence of synteny breakpoint and LAD border. Color scheme as in A and B. (D) Ratio of observed and expected occurrence of breakpoints in cLADs, ciLADs, or their border regions. (E) Cartoon representations of homotypic and heterotypic junctions; breakpoints indicated by dashed gray lines. All breakpoints in constitutive regions (n = 143) were scored based on whether they coincide with an LAD in mouse or human.
Table 1.
Concordance scores between mouse and human cells
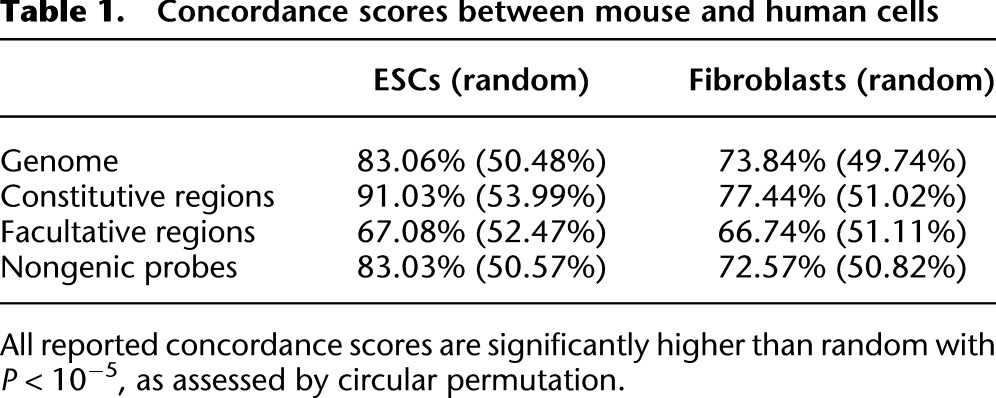
The cell types that we studied may have similar gene expression repertoires between mouse and human. Because gene expression is overall inversely correlated to NL association (Peric-Hupkes et al. 2010), it is possible that the observed evolutionary conservation of NL interaction patterns is merely a result of conserved gene expression patterns. To rule out this possibility, we investigated whether the conservation of NL association is linked to the presence of genes. This revealed that this is not the case: The interspecies concordance of NL interactions does not decrease when genic regions are removed from the analysis (Table 1). Thus, conserved gene expression patterns cannot account for the remarkable conservation of the pattern of cLADs and ciLADs.
The high overall conservation of cLADs suggests that chromosomal rearrangements during evolution that disrupt this organization would be deleterious. Such deleterious effects would be minimized if rearrangement breakpoints are restricted to cLAD/ciLAD borders, thereby keeping both cLADs and ciLADs mostly intact. To test this prediction, we studied 143 loci representing chromosome rearrangements between human and mouse, as evidenced by transitions between mapped chromosomes (i.e., change of colors in Fig. 2B) in constitutive regions. Strikingly, these breakpoints occur preferentially close to cLAD/ciLAD borders and are significantly depleted from cLADs (Fig. 2C,D). This suggests an overall importance of cLAD integrity, even though cLADs have relatively low gene densities and thus may be expected to be under low selective pressure. Although this analysis is restricted to constitutive regions only, the same analysis for the full genome yields very similar results (Supplemental Fig. 3A).
Although synteny breakpoints are significantly depleted from cLADs (24 out of 143, P < 3 × 10−5), they are not completely absent. We asked whether these tolerated junctions are homotypic (joining two LAD segments) rather than heterotypic (joining a LAD segment to a non-LAD segment). Indeed, we found that breaks that do occur within cLADs are not likely to cause heterotypic rearrangement junctions, compared with those outside LADs (P = 1.126 × 10−7) (Fig. 2E). When also incorporating facultative regions in the analysis, the same conclusion holds (Supplemental Fig. 3B). Because homotypic rather than heterotypic junctions would prevent fragmentation into increasingly smaller LADs and thereby loss of the overall architecture, these observations suggest that disruptions of the cLAD core architecture are under negative selection pressure.
cLAD sequences are characterized by high A/T content
We next asked whether this strong evolutionary conservation of constitutive NL interactions is also reflected in conservation of the underlying genomic sequence. To address this, we compared average basewise conservation scores (derived from sequence alignments of 28 placental mammals) (Pollard et al. 2010) in cLADs, ciLADs, and fLADs. We restricted this analysis to intergenic and intronic regions because coding sequences are subject to a variety of selective pressures that could confound this analysis. Surprisingly, the results show that both intergenic and intronic sequences in cLADs are generally less well conserved than in ciLADs and fLADs (Supplemental Fig. 4). Thus, the strong overall conservation of cLAD positions is not accompanied by strong conservation of cLAD nucleotide sequence.
We reasoned that, despite the poor overall sequence conservation, there may still be specific sequence motifs dispersed throughout cLADs that could mediate NL interactions. To investigate this, we systemically searched for sequence motifs of length 1–5 bp that could discriminate cLADs from ciLADs. For this purpose, we selected 1000 cLAD and 1000 ciLAD regions of 1 kb from the mouse genome that show the most extreme lamin B1 association across all four mouse cell types. Furthermore, we excluded all genic regions, because those may introduce a bias in the analysis. Strikingly, the result of this motif search revealed that A/T content alone is sufficient to discriminate cLADs from ciLADs with 94% accuracy (10-fold cross-validation accuracy). cLADs and ciLADs have a mean A/T content of ∼63% and 54%, respectively, with very little overlap in the distributions (Fig. 3A).
Figure 3.
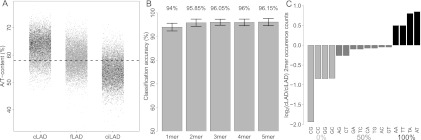
A/T content is a strong predictor of constitutive lamina association. (A) A/T content versus mean mouse lamin B1 association for a random sample of 10,000 defined cLAD, fLAD, and ciLAD regions (gray dots). (Black dots) The regions used for classification (1000 per group). (Dashed line) Genome-wide average (58%). (B) Tenfold cross-validation classification accuracies of classifiers using k-mers as predictive features, with k = 1 … 5. Bars show mean accuracies, with error bars indicating one standard deviation. (C) Log2 ratios of occurrence counts in cLAD versus ciLAD regions for each dinucleotide. Bar shading indicates the A/T content of dinucleotides.
The predictive accuracy of the cLAD versus ciLAD classifier increases slightly, albeit nonsignificantly, when the occurrence of dinucleotides is used as a predictive feature (95.85%) but does not significantly increase further when longer k-mers are used (Fig. 3B). It is therefore unlikely that recurrent sequence motifs larger than 1–2 bp explain NL interactions of constitutive regions. In fact, a survey of the enrichment of individual dinucleotides in cLADs versus ciLADs shows that the main driving force behind the classification accuracies is simple A/T content (Fig. 3C).
We considered two potential confounding factors in these analyses. First, we ruled out that the high abundance of AT-rich LINE-1 elements in cLADs explained the classification result. Removal of all LINE-1 elements did not affect the classification accuracy (data not shown). Second, we asked whether A/T content might simply correlate with gene density, which is low in cLADs and high in ciLADs. Within ciLADs (which have the broadest diversity of gene density), we found no significant correlation between A/T content and gene density (see Methods), indicating that this is not a major confounding factor. In summary, intergenic regions in cLADs almost invariably have a high A/T content, while those in ciLADs have a consistently low A/T content.
The observed segmentation of the genome into ciLADs and cLADs of low and high A/T content, respectively, reminded us of the long-known partitioning of mammalian genomes into isochores, which are long DNA segments of relatively homogenous base composition (Bernardi et al. 1985; Eyre-Walker and Hurst 2001). Indeed, comparison of the mouse cLAD/ciLAD pattern to that of isochores (Costantini et al. 2009) revealed a strikingly high concordance of 93% (Supplemental Fig. 5).
A/T content predicts fLADs only in ESCs
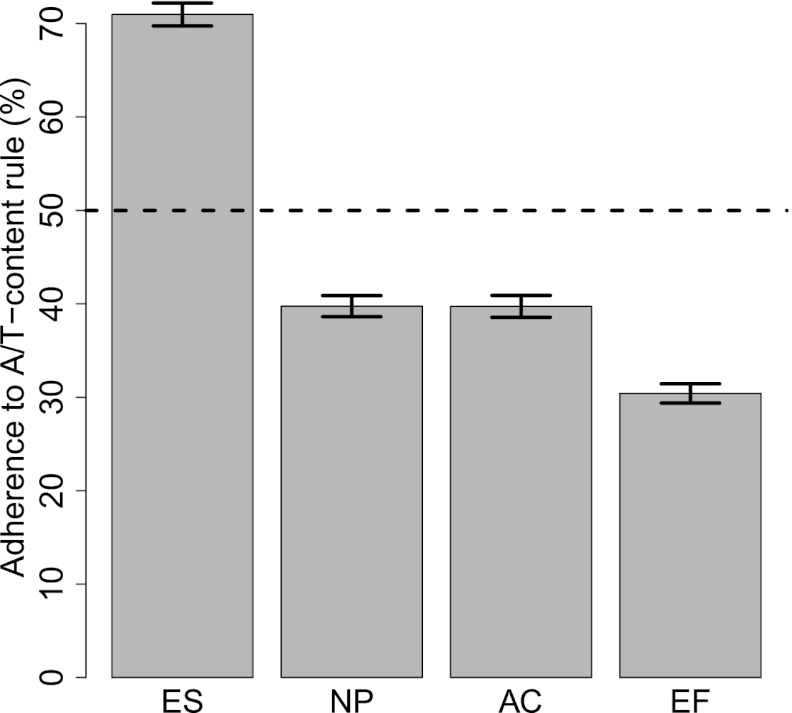
Because of the unique pluripotent nature of ESCs, it has been suggested that these cells contain a set of default NL interactions, together forming a “basal” state, that are progressively modified during subsequent differentiation steps (Peric-Hupkes et al. 2010). In other words, NL interactions in ESCs may be driven by a default mechanism; upon differentiation, this default mechanism could be partially overruled by cell-type specific mechanisms that relocate fLADs to or from the NL. We wondered whether the A/T rule that we identified for cLADs could constitute the default mechanism in this model. If so, then fLADs should adhere to the A/T rule in ESCs, but not in differentiated cells. Indeed, the A/T rule predicts the NL interaction status of fLADs in ESCs with an accuracy of 71% (Fig. 4). In contrast, in the three differentiated cell types, this accuracy drops to 30%–40%. The latter numbers are less than expected by chance, suggesting that differentiated cells have one or more mechanisms that overrule A/T-driven interactions of fLADs. These results are consistent with a model in which A/T-driven NL interactions of the genome constitute a default principle that is locally overruled by cell-type specific mechanisms.
Figure 4.
A/T-content rule during differentiation. Adherence to A/T-content rule by regions that show cell-type specific lamin interactions, as measured by classification accuracy using the classifier (k = 1) trained on cLAD and ciLAD regions. Bars show mean bootstrapped classification accuracies, with error bars indicating standard deviation. (Dotted horizontal line) Random performance.
A/T-content divergence of paralogous genes is linked to differential NL interaction
If A/T content is a determinant of NL interactions, then during evolution some genes may have acquired a high A/T content to enhance their NL associations, while other genes may have become C/G rich in order to keep them in the nuclear interior. Because paralogous genes are derived from a common ancestor, they are particularly useful to investigate this model. We therefore compared differences in A/T content of paralogous genes with differences in NL association. Remarkably, paralogous genes with a clear difference in A/T content generally differ strongly in their NL interactions, in a manner consistent with the A/T-content rule (Fig. 5A; Supplemental Fig. 6A,C,E, P < 1 × 10−4). Also, differential NL interaction levels of paralogous genes correlate with differences in expression status (Fig. 5B; Supplemental Fig. 6B,D,F, P < 1 × 10−4). More specifically, a gene that underwent an increase in A/T content, relative to its non-lamina-associated paralogous gene, typically shows enhanced association with the NL, as well as a strongly decreased level of expression. These results suggest that the A/T-content rule may be an important evolutionary tool to modify gene localization and activity. However, it cannot be ruled out that genes that move to LADs become more A/T rich once they reside there, due to regional biases in DNA replication or repair (Eyre-Walker and Hurst 2001). Regardless of the underlying evolutionary mechanism, this analysis of paralogs further underscores the overall relationship between NL interactions and A/T content.
Figure 5.
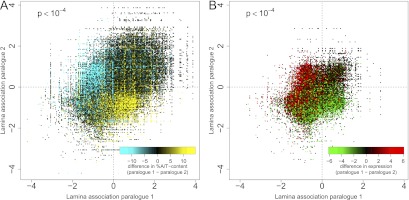
Correlation of A/T content with behavior of paralogous gene pairs. Panels show mean NL interaction scores in mouse ES cells for all known paralog pairs. Each dot represents a paralog pair, colored by percent difference in A/T content (A) and log2 difference in expression level (B). The assignment of paralog 1 or 2 is redundant, hence the symmetry across the diagonal. Expression data for mouse ES cells are taken from Mikkelsen et al. (2007), based on Affymetrix arrays, which is why for a number of genes no value is reported in B.
A/T content also correlates with NL interactions in other metazoans
We wondered to what extent NL interactions are also correlated with A/T content in other species. We investigated this for Caenorhabditis elegans and Drosophila melanogaster, for which NL interactions were mapped in embryos and cultured embryonic cells, respectively (Ikegami et al. 2010; van Bemmel et al. 2010). In both species, LADs have significantly higher A/T content than inter-LAD regions, although the magnitudes of the differences are relatively small (Supplemental Fig. 7). Not enough data are available to discriminate cLADs from fLADs in these species.
No detectable role for POU2F1 in genome–NL anchoring
The presence of a clear sequence signature (i.e., high A/T content) in cLADs suggested that one or more proteins in the NL may bind directly to A/T-rich sequences and thereby tether cLADs to the NL. Previously, we reported that human LADs are enriched for binding motifs of POU2F1 (Guelen et al. 2008), which is a ubiquitous DNA-binding factor (also known as Oct1) that is localized at the nuclear periphery and interacts with lamin B1 (Malhas et al. 2009). Since all published POU2F1-binding motifs are relatively A/T rich (consensus sequence: TATGCAAAT), it is possible that this protein mediates NL association by binding both its genomic targets and lamins simultaneously. To test this, we generated genome-wide maps of lamin B1 interactions in wild-type and POU2F1−/− MEFs (Wang et al. 2004). The results indicate that genome–NL interactions are largely independent of POU2F1, with up to 96% concordance for constitutive regions between wild-type and POU2F1-null cells (Supplemental Fig. 8A–C). Furthermore, plotting of changes in lamin B1 DamID signals around POU2F1-binding motifs failed to detect any local effect of POU2F1 (Supplemental Fig. 8D). These results indicate that POU2F1 does not play an essential role in genome–NL interactions, at least not in MEFs.
Lamin B1 and lamin A interact with largely the same genomic regions
Most cells express both A- and B-type lamins. Several lines of evidence indicate that these have in part distinct functions (for review, see Dechat et al. 2010). For example, A-type lamins are essentially absent in ESCs; mutations in lamin A/C can cause a broad range of human disorders not associated with mutations in B-type lamins; A- and B-type lamins have been reported to form spatially distinct meshworks inside the NL; and finally, while B-type lamins are primarily restricted to the NL, A-type lamins are typically also present in the nuclear interior. These differences raised the possibility that A- and B-type lamins interact with distinct parts of the genome. Because in vivo interactions so far have only been mapped for lamin B1 (Guelen et al. 2008; Peric-Hupkes et al. 2010; Handoko et al. 2011), we used DamID to generate genome-wide maps of lamin A interactions. We focused on NPCs and ACs, which express lamin A endogenously.
The resulting lamin A binding profiles are very similar to those of lamin B1, with an overall concordance of 94%–95% between lamins A and B1 in both cell types (Fig. 6). In particular, the interactions for constitutive regions are virtually identical at 99% and 98% concordance in NPCs and ACs, respectively (Fig. 6C,F). Within fLADs the concordance between lamin A and lamin B1 binding is somewhat lower but still substantial (83%–86%). To assess whether the strong overall agreement in binding between lamin A and lamin B1 is restricted to the tested mouse cell types, we generated and compared genome-wide DamID maps for both proteins in human HT1080 fibrosarcoma cells. This again yielded a very high concordance between the two lamins (∼97%) (Supplemental Fig. 9). Taken together, these findings demonstrate that cLADs interact robustly with both lamin A and lamin B1 in mouse and human, while fLADs may have slight preferences for one lamin type over the other.
Figure 6.
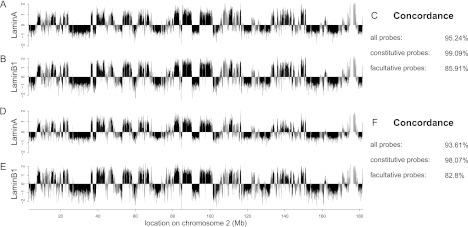
The core architecture is shared among lamin A and lamin B1. Genome–NL interaction profiles for mouse chromosome 2 as assayed by DamID of lamin A (A,D) and lamin B1 (B,E) in NPCs (A,B) and astrocytes (D,E). Data in B and E are from Peric-Hupkes et al. (2010). (Black) Constitutive regions; (gray) facultative regions. (C,F) Concordance scores are shown for all, constitutive (cLAD and ciLAD) and facultative regions.
Discussion
Using maps of genome–NL interactions in various mouse and human cell types, we conducted a detailed analysis of cell type invariant (constitutive) NL interactions. We find that the positions of cLADs along the genome are highly conserved between mouse and human. This contrasts with the overall poor conservation of transcription factor binding sites, which has been reported to be <15% between mouse and human (Schmidt et al. 2010; Soccio et al. 2011), and the moderate conservation (∼30%) of CTCF-binding sites (Schmidt et al. 2012). Interestingly, recent systematic mapping of chromatin contacts in human and mouse cells identified hundreds of megabase-sized domains that are conserved by >50%. These “topological domains” overlap in part with LADs (Dixon et al. 2012; Nora et al. 2012), but limited genome coverage or a >10-fold difference in the mapping resolution precludes more detailed comparisons.
Our analyses indicate that synteny breaks are depleted from cLADs, despite the fact that cLADs are gene poor. We cannot entirely rule out that LAD border regions are somehow intrinsically fragile and thus have accumulated synteny breakpoints, or that LADs are somehow less susceptible to DNA breaks. Furthermore, those synteny breaks that do occur in LADs tend to favor homotypic junctions, which leads to preservation of LADs as large units. Possibly, peripheral positioning of cLADs requires cooperative interactions of long A/T-rich stretches of DNA with NL components. Accordingly, fragmentation of such stretches by genomic rearrangements might cause weakening of NL association and thereby loss of spatial organization. Clustering of cLADs in the peripheral nuclear compartment may promote homotypic rearrangements and thereby preserve the large sizes of cLADs. It is also possible that heterotypic junctions lead to repression of genes in ciLADs near the junction borders as a result of the heterochromatic nature of cLADs. Regardless of the underlying mechanisms, our results indicate that the positions and size of cLADs have been remarkably conserved.
Despite this conservation of size and positions, noncoding DNA sequence in cLADs is relatively poorly conserved. This appears to match well with an earlier observation that DNA sequences in compacted chromatin are less conserved than in decondensed chromatin (Prendergast et al. 2007), assuming that the transcriptionally inactive cLADs harbor primarily compacted chromatin.
A defining characteristic of cLADs is their high A/T content. Likewise, paralogous genes that differ in their NL associations show a concomitant difference in A/T content. These observations reveal a tight link between spatial organization and simple nucleotide composition of genomic regions. The molecular mechanisms that might be responsible for the peripheral positioning of A/T-rich DNA are presently unknown. Even though POU2F1 was a likely candidate because it binds an A/T-rich motif and is known to associate with the NL, we found that MEFs lacking POU2F1 exhibit no detectable changes in NL–genome interactions. Lamins themselves are prime candidates to mediate the anchoring of cLADs, because they have been reported to interact with A/T-rich sequences in vitro (Luderus et al. 1994). However, a recent study of mouse ES cells lacking all lamins found few changes in the expression of genes in LADs (Kim et al. 2011). It is possible that other NL-associated proteins provide redundancy for adherence of A/T-rich stretches. Furthermore, epigenetic mechanisms, e.g., involving certain histone modifications, may also play a role. Finally, nuclear organization is likely also dependent on other inter- and intrachromosomal contacts (e.g., transcription factories, nucleolar interactions, etc.). We emphasize that a potential causal relationship between A/T richness and peripheral positioning needs to be addressed further.
We found that the cLAD/ciLAD segmentation coincides largely with isochore distribution. Several explanations have been suggested for the isochore organization of vertebrate genomes (Eyre-Walker and Hurst 2001), but a role for A/T-rich isochores as NL-anchoring sites has not been considered so far. Our results raise the interesting possibility that the evolution of isochores is linked to the spatial organization of chromosomes in the nucleus.
Centromeric satellite repeats have A/T contents ∼63%–66%, which is in the range of cLADs. Indeed, in most mouse and human cell types, centromeres show a preferential localization near the NL (Weierich et al. 2003; Wiblin et al. 2005). An exception to this is human ES cells (Wiblin et al. 2005; Bartova et al. 2008). Centromeres have a unique chromatin composition (Bergmann et al. 2012), which may modulate NL interactions. Human telomeres are not typically found at the periphery (Luderus et al. 1996; Weierich et al. 2003; Ramirez and Surralles 2008), consistent with their relatively low (50%) A/T content. Mouse telomeres, which have the same sequence, can be peripheral to some degree (Vourc'h et al. 1993; Weierich et al. 2003), but much of this is accounted for by the telocentric nature of mouse chromosomes (i.e., half of the telomeres are linked to pericentric regions, which tend to be peripheral). Telomeric repeats and centromeric satellite repeats lack GATC sequence motifs, and therefore their NL interactions cannot be probed by DamID.
Several of the findings we describe here are reminiscent of what has been found for replication timing (RT) of DNA in S phase. Analogous to LADs, late-replicating domains are generally gene poor, A/T rich, and often located at the nuclear periphery (Hiratani et al. 2009). Recently it has been shown that there are cell-type invariant (constitutive) and cell-type specific (facultative) RT domains (Hiratani et al. 2008, 2010) and that RT patterns are conserved for syntenic regions between mouse and human (Ryba et al. 2010; Yaffe et al. 2010). However, LADs and late-replicating domains overlap only partially (Peric-Hupkes et al. 2010), and there are also a few other notable differences. First, we observed an enrichment of synteny breakpoints on cLAD–ciLAD borders, whereas RT domains seem to shift slightly across synteny breaks (Yaffe et al. 2010). Second, RT becomes more tightly correlated to A/T content as cells differentiate (Hiratani et al. 2010), which contrasts our observation that fLADs adhere to the A/T-content rule in ESCs but not in differentiated cell types. These discrepancies underscore that RT organization and LAD organization are only partially linked.
Our results indicate that lamins A and B1 have largely overlapping binding patterns along the genome, although some subtle differences may exist in fLADs. Our data therefore imply that most loci do not differentiate between microdomains in the NL consisting of only one type of lamin (Shimi et al. 2008). Moreover, it appears that the pool of lamin A present in the nuclear interior (Broers et al. 1999; Dechat et al. 2000) does not bind to a distinct set of genomic loci. Possibly, internal lamin A does not interact with the genome at all, which is consistent with the high mobility of this pool (Broers et al. 1999). Alternatively, LADs may detach from the NL in a stochastic manner, and this internally located set of LADs may still interact with lamin A. Both scenarios would yield the high similarity of DamID maps for lamin A and lamin B1 as we observed here. Future studies should be aimed at elucidating the molecular mechanisms that drive the interactions of specific genomic regions with NL components.
Methods
Data sets
DamID maps of lamin B1 in mouse ESCs, ACs, NPCs, and MEFs were taken from Peric-Hupkes et al. (2010), and of lamin B1 in human Tig3 fibroblasts from Guelen et al. (2008). Coordinates of LADs in fly and worm were from van Bemmel et al. (2010) and Ikegami et al. (2010), respectively. For this study, we generated new DamID maps of lamin B1 in human ESCs and HT1080 cells and in mouse POU2F1−/− and matching wild-type MEFs; and of lamin A in human HT1080 cells and in mouse NPCs and ACs.
Cell culture and DamID
Human embryonic stem cell line SHEF2 was cultured under feeder-free conditions as described (Braam et al. 2008), with the modification that they were maintained in mTeSR medium (Stem Cell Technologies). Viral transduction with either Dam-lamin B1 or Dam-only virus was performed as follows. Almost confluent cells in a six-well plate were washed twice with DMEM-F12 (GIBCO BRL), trypsinized for 120 sec at 37°C; following two additional washes, cells were resuspended thoroughly to single cells in mTeSR medium. Cells were then collected by centrifugation for 5 min at 350g and directly resuspended in a mixture containing 400 μL of virus (in mTeSR), 1.6 mL of mTeSR, and 8 μg/mL Polybrene. Cells were seeded at high density (typically 1:1, never exceeding 1:2) onto a six-well plate. Medium was changed every day, and genomic DNA was harvested 72 h after plating. A lentiviral Dam-lamin A-expressing construct was made through Gateway cloning as described in Vogel et al. (2006), and correct subnuclear localization to the NL of the fusion protein was confirmed by immunofluorescence microscopy using an antibody against the v5 epitope-tag (data not shown). POU2F1−/− MEFs and matching isogenic wild-type MEFs were obtained from D. Tantin and cultured as described (Kang et al. 2009). All subsequent DamID steps were performed as described before (Peric-Hupkes et al. 2010).
NimbleGen microarray design
Microarrays were custom-designed for DamID purposes. A selection of NimbleGen whole-genome, high-density, ChIP-on-chip probe sequences was made to attain the desired probe spacing and exclude probes containing GATC motifs. The tiling arrays cover the entire nonrepetitive genome, with a median probe spacing of 1 kb (human) and 1.2 kb (mouse).
Interspecies syntenic region mapping
To construct full-chromosome maps of syntenic regions between two species, we mapped microarray probe coordinates from a master species to a slave species genome coordinates. In Figure 2, for example, the master species is mouse (Fig. 2A) and the slave species is human (Fig. 2B). To perform the actual mapping, we used the UCSC liftOver tool (http://hgdownload.cse.ucsc.edu/admin/exe/) together with liftOver-compatible chain files. These files were obtained from UCSC (http://hgdownload.cse.ucsc.edu/goldenPath/mm9/liftOver/) and converted to reciprocal best chain files using code based on Jim Kent's source tree (http://hgdownload.cse.ucsc.edu/admin/jksrc.zip, doRecipBest.pl). The liftOver procedure was initiated from every probe in the master species. For each match, we tried to pair the original probe in the master species to a representative probe in the slave species. This was achieved by searching for the probe nearest to the matched region in the slave species, within a fixed-width window. The width of this window was determined by the median spacing between matched regions in the slave species, which was 1905 bp in the case of the mouse-to-human mapping. During a final step, results were curated to remove ambiguous probe matches so as to retain only one-to-one matches. This yielded final species-to-species maps with a resolution of ∼2.5 kb, derived from original maps with a resolution of ∼1 kb.
Calling of NL interaction status
We fitted a two-state hidden Markov model (HMM) whereby emissions are distributed as Student's t variables. The mean and variance of DamID signals differ between states, but the degree of freedom (nu) is the same. Gaps in the probe coverage were filled by evenly spaced null probe values. The parameters were estimated by an adaptation of the ECME algorithm to the HMM framework, showing faster convergence than regular EM when nu is unknown (Filion et al. 2010). State calls were derived through the Viterbi algorithm. This process was repeated separately for each cell type and species, yielding per-probe calls. Probes in the “bound” state are indicated as LAD-probes, probes in the “unbound” state as inter-LAD-probes. Note that in order to obtain a probe-by-probe readout of lamina status, we did not use the algorithm as used before (Guelen et al. 2008; Peric-Hupkes et al. 2010).
Concordance scoring
The HMM calls are the basis for the concordance score, which is defined as the percentage of all calls that is in agreement (LAD vs. LAD or inter-LAD vs. inter-LAD) between two data sets (e.g., two different cell types or species). Statistical significance of concordance scores was assessed by comparing scores against a null distribution constructed of concordance scores for 100,000 random circular permutations of calls.
Definition of constitutive and facultative regions
The HMM procedure described above was performed on data for all four cell types described in Peric-Hupkes et al. (2010). Regions that were in perfect agreement were termed “constitutive” (70.76%) and the remainder “facultative” (29.24%). For the subset of mappable regions between mouse and human, these numbers are 66.70% and 33.30%, respectively. We chose to use an HMM-based method to identify constitutive regions rather than the method described in Peric-Hupkes et al. (2010) because the latter has the opposite purpose of identifying differentially bound regions and was used to call these differences in a gene-centric fashion between pairs of cell types. Here, we were interested in regions showing qualitatively identical binding across multiple cell types.
Gene annotations
A compendium of gene annotations was constructed using annotations downloaded from the UCSC Table Browser. The annotations used are UCSC genes (table “knownGene”), Consensus Coding Sequence (table “ccdsGene”), RefSeq (table “refGene”), Vega (table “vegaGene”), Vega pseudogenes (table “vegaPseudoGene”), and Ensembl (table “ensGene”). Merged together, the compendium describes a total of 34,129 mouse genes, collectively covering 45.62% of the mouse genome.
Coverage of genes, gene deserts, and LINE, A/T-rich, and SINE elements
Gene deserts were defined as regions >500 kb containing no genes. LINE, simple A/T-rich, and SINE elements were obtained from the UCSC Table Browser. Coverage of cLAD, facultative, and ciLAD regions was obtained by calculating the overlap of these regions with genes, gene deserts, LINEs, A/T-rich elements, and SINEs. Error bars were obtained by performing 1000 random circular permutations of cLAD, ciLAD, and fLAD regions, calculating the overlap for these random regions and reporting two standard deviations around mean random overlap numbers.
Concordance between species in absence of genes
The interspecies concordance of nongenic regions was assessed by removing mouse probes, and their representative human probes, that overlap at least 50% with any of the 34,129 genes in the mouse gene compendium described above.
Calling of chromosome breaks
Breaks in synteny, i.e., chromosomal rearrangements, were called by finding transitions in reconstituted slave chromosomes. This is illustrated by transitions in color in Figure 2B and Supplemental Figure 1. We used a sliding probe–based window corresponding on average to 50 kb to identify transitions. Majority voting was used as a pre-processing step to remove spurious transitions. This resulted in a set of 152 major breaks for the full genome and 143 breaks for constitutive regions only.
Chromosome breaks versus lamina status
For each of the rearrangement loci within constitutive regions, we calculated the average mouse NL interaction status in a window corresponding to on average 50 kb, centered around the break. The binary LAD/inter-LAD calls yielded by the HMM were used as indicators for lamina status. The result could naturally be clustered into three groups: LADs (24), LAD-borders (39), and inter-LADs (80), where the size of borders has an expected upper bound of ∼17 kb. Distributions of random overlap for the three groups were obtained by circularly permuting the positions of the 143 chromosome breaks, for all possible positions, and recalculating overlap. P-values were obtained by comparing the real overlap against these distributions. Note that this approach retains the relative genomic distribution of cLADs, ciLADs, and LAD borders, thereby correcting for differences in genomic coverage of these three categories. The same analysis was performed for the full genome using ESC data, over 152 identified chromosomal breaks, with similar results (see text).
Homo/heterotypic analysis
One-tailed Fisher's exact tests were performed to test whether breakpoints inside mouse cLADs are enriched for coinciding with human LADs. For the 143 breakpoints in constitutive regions, this is, indeed, the case (P = 1.126 × 10−7). For the 152 breakpoints in the full genome, using data for mouse and human ESCs, this also holds true (P = 3.56 × 10−7).
Definition of regions used for classification
The regions selected for classification consisted of regions that share a common lamina status in mouse (i.e., either LAD or inter-LAD in all assayed cell types). These are obtained by extending microarray probe positions to 1-kb regions. Regions annotated in the mouse gene compendium are removed, including 2 kb upstream and downstream of genes, to disregard any gene-specific patterns that may be present in these regions. Regions smaller than 1 kb are also disregarded. The remainder of the regions are scored based on their mean ranked lamina association across the assayed mouse cell types. For facultative regions, the standard deviation of the ranked lamina association is used as the score. Scores are then used to order regions, i.e., high and low lamina associations for cLAD and ciLAD regions and high variation in lamina association for facultative regions. The top 1000 cLAD/ciLAD/facultative regions are used for subsequent analyses. Facultative LAD and inter-LAD regions specific to a particular cell type (Fig. 4) were selected in a similar manner, with HMM state calls being specific to the cell type of interest and scores based on the level of lamina association of the same cell type only. For the assessment of phyloP conservation scores in cLAD, ciLAD, and facultative regions (Supplemental Fig. 3), we used 1000 1-kb regions for each group. Intronic regions were selected by size matching introns positioned in cLAD, ciLAD, and facultative regions, giving preference to large regions. The center 1 kb of each region was subsequently selected for assessing conservation levels.
Construction of classifiers
Genomic sequences were obtained for all selected regions, and the number of occurrences of k-mers (k = 1 … 5) was scored for each region. Support vector machine classifiers were trained using the e1071 R-package with default parameters, using a radial basis kernel. Their performance was assessed using 10-fold cross-validation. Cross-validation accuracy [i.e., (TN + TP)/(TN + TP + FN + FP)] scores are the result of averaging accuracies over folds.
Isochore analysis
Mouse isochore definitions were obtained from Costantini et al. (2009), who report five types of genomic regions based on their G/C content (L1, L2, H1, H2, and H3). For Supplemental Figure 5, we have grouped L1 and L2 in one high A/T-content class, and H1, H2, and H3 in one low A/T-content class.
Analysis of A/T content versus gene density in ciLAD regions
We calculated gene density in windows of 99 microarray probes, roughly corresponding to 100 kb, as the number of probes that overlap with an annotated gene. Per-probe A/T content was calculated within windows of 1 kb, centered on the genomic probe position. We then scored intergenic iLAD regions based on their gene density and/or A/T content being below or above the genome-wide median, resulting in an odds ratio. Ten thousand random circular permutations of A/T-content scores were performed to derive a null distribution of odds ratios. Comparing the original odds ratio to this null yielded a P-value of 0.0986, indicating that there is not a significant difference in A/T content between gene-rich and gene-poor intergenic ciLAD regions.
Adherence to A/T-content rule of cell-type specific LADs
The classifier constructed based on A/T content for discriminating between cLAD and ciLAD regions was applied to regions that show a cell-type specific lamina association. In other words, these regions are LAD or inter-LAD exclusively for one particular cell type. The adherence to the A/T-content rule is reported by way of the classification accuracy, of which the variance is estimated using a bootstrapping procedure. The number of cell-type specific LAD/inter-LAD regions was set to 1000, as in the cLAD/ciLAD classification procedure.
Paralog analysis
Within-species paralog information was obtained from BioMart (http://www.biomart.org; obtained on Oct 10, 2011). We included one-to-one as well as many-to-many paralogs. The mean DamID log2-ratio score was calculated for each gene and plotted against (each of) its paralog partners. For many-to-many paralogs, all gene pairs are separately represented in Figure 5 and Supplemental Figure 6. A/T-content information for entire gene bodies was also retrieved from BioMart, and mouse gene expression data were obtained from Mikkelsen et al. (2007). To assess the statistical significance of the patterns observed in Figure 5 and Supplemental Figure 6, we used the mean difference in A/T content or expression for the upper-left quadrant as a test statistic. The null distribution was estimated by repeatedly (10,000×) randomly shuffling A/T content and gene expression values of genes, while keeping paralog pairings intact, and calculating the mean upper-left quadrant value for each iteration. In all cases, the observed means were more extreme than those expected under the null (P < 1 × 10−4).
POU2F1 motif analysis
We scanned the mouse genome for occurrences of the TRANSFAC (Matys et al. 2003) OCT1_01 motif, at a precision of 90% of the information content. We then aligned the differential signal obtained from subtracting DamID lamin B1 scores of POU2F1+/+ cells from those of POU2F1−/− cells around the identified motif sites (Supplemental Fig. 8D). As a control, we performed the same alignment for control sites positioned exactly halfway in between OCT1_01 motif sites.
Data access
Human lamin A (HT1080) and lamin B1 (ESC and HT1080) data are available from the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE22428. Mouse lamin A (NPC and AC) and lamin B1 (POU2F1−/− and POU2F1+/+ MEFs) data are available from GEO under accession number GSE36132.
Acknowledgments
We thank Gaetano Gargulio for help with human ESC culture and transduction, Martijn Huynen and Berend Snel for suggesting the paralog analysis, and Maarten van Lohuizen for support. D. Tantin is acknowledged for providing the POU2F1 mutant and matching control cells. This study was funded by NWO-ALW VICI and a European Young Investigator Award to B.v.S., an ESTOOLS FP7 EU grant to J-B.B., and an ALW-VENI Fellowship to J.K.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.141028.112.
References
- Bartova E, Galiova G, Krejci J, Harnicarova A, Strasak L, Kozubek S 2008. Epigenome and chromatin structure in human embryonic stem cells undergoing differentiation. Dev Dyn 237: 3690–3702 [DOI] [PubMed] [Google Scholar]
- Bergmann JH, Martins NM, Larionov V, Masumoto H, Earnshaw WC 2012. HACking the centromere chromatin code: Insights from human artificial chromosomes. Chromosome Res 20: 505–519 [DOI] [PubMed] [Google Scholar]
- Bernardi G, Olofsson B, Filipski J, Zerial M, Salinas J, Cuny G, Meunier-Rotival M, Rodier F 1985. The mosaic genome of warm-blooded vertebrates. Science 228: 953–958 [DOI] [PubMed] [Google Scholar]
- Braam SR, Denning C, Matsa E, Young LE, Passier R, Mummery CL 2008. Feeder-free culture of human embryonic stem cells in conditioned medium for efficient genetic modification. Nat Protoc 3: 1435–1443 [DOI] [PubMed] [Google Scholar]
- Broers JL, Machiels BM, van Eys GJ, Kuijpers HJ, Manders EM, van Driel R, Ramaekers FC 1999. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J Cell Sci 112: 3463–3475 [DOI] [PubMed] [Google Scholar]
- Costantini M, Cammarano R, Bernardi G 2009. The evolution of isochore patterns in vertebrate genomes. BMC Genomics 10: 146 doi: 10.1186/1471-2164-10-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Korbei B, Vaughan OA, Vlcek S, Hutchison CJ, Foisner R 2000. Lamina-associated polypeptide 2α binds intranuclear A-type lamins. J Cell Sci 113: 3473–3484 [DOI] [PubMed] [Google Scholar]
- Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD 2010. Nuclear lamins. Cold Spring Harb Perspect Biol 2: a000547 doi: 10.1101/cshperspect.a000547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Hurst LD 2001. The evolution of isochores. Nat Rev Genet 2: 549–555 [DOI] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951 [DOI] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, et al. 2011. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet 43: 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM 2008. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6: e245 doi: 10.1371/journal.pbio.0060245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Takebayashi S, Lu J, Gilbert DM 2009. Replication timing and transcriptional control: Beyond cause and effect—Part II. Curr Opin Genet Dev 19: 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. 2010. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res 20: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Egelhofer TA, Strome S, Lieb JD 2010. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol 11: R120 doi: 10.1186/gb-2010-11-12-r120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Gemberling M, Nakamura M, Whitby FG, Handa H, Fairbrother WG, Tantin D 2009. A general mechanism for transcription regulation by Oct1 and Oct4 in response to genotoxic and oxidative stress. Genes Dev 23: 208–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan CM, Gaiano N, Ko MS, Zheng Y 2011. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science 334: 1706–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, van Steensel B 2010. Genome-nuclear lamina interactions and gene regulation. Curr Opin Cell Biol 22: 320–325 [DOI] [PubMed] [Google Scholar]
- Luderus ME, den Blaauwen JL, de Smit OJ, Compton DA, van Driel R 1994. Binding of matrix attachment regions to lamin polymers involves single-stranded regions and the minor groove. Mol Cell Biol 14: 6297–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luderus ME, van Steensel B, Chong L, Sibon OC, Cremers FF, de Lange T 1996. Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J Cell Biol 135: 867–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhas AN, Lee CF, Vaux DJ 2009. Lamin B1 controls oxidative stress responses via Oct-1. J Cell Biol 184: 45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. 2003. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Soutoglou E 2009. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 10: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38: 603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B 2006. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet 38: 1005–1014 [DOI] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20: 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast JG, Campbell H, Gilbert N, Dunlop MG, Bickmore WA, Semple CA 2007. Chromatin structure and evolution in the human genome. BMC Evol Biol 7: 72 doi: 10.1186/1471-2148-7-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez MJ, Surralles J 2008. Laser confocal microscopy analysis of human interphase nuclei by three-dimensional FISH reveals dynamic perinucleolar clustering of telomeres. Cytogenet Genome Res 122: 237–242 [DOI] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Wilson MD, Ballester B, Goncalves A, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT 2012. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 148: 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, et al. 2008. The A- and B-type nuclear lamin networks: Microdomains involved in chromatin organization and transcription. Genes Dev 22: 3409–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soccio RE, Tuteja G, Everett LJ, Li Z, Lazar MA, Kaestner KH 2011. Species-specific strategies underlying conserved functions of metabolic transcription factors. Mol Endocrinol 25: 694–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bemmel JG, Pagie L, Braunschweig U, Brugman W, Meuleman W, Kerkhoven RM, van Steensel B 2010. The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PLoS ONE 5: e15013 doi: 10.1371/journal.pone.0015013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Dekker J 2010. Genomics tools for unraveling chromosome architecture. Nat Biotechnol 28: 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel MJ, Guelen L, de Wit E, Peric-Hupkes D, Loden M, Talhout W, Feenstra M, Abbas B, Classen AK, van Steensel B 2006. Human heterochromatin proteins form large domains containing KRAB-ZNF genes. Genome Res 16: 1493–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourc'h C, Taruscio D, Boyle AL, Ward DC 1993. Cell cycle-dependent distribution of telomeres, centromeres, and chromosome-specific subsatellite domains in the interphase nucleus of mouse lymphocytes. Exp Cell Res 205: 142–151 [DOI] [PubMed] [Google Scholar]
- Wang VE, Schmidt T, Chen J, Sharp PA, Tantin D 2004. Embryonic lethality, decreased erythropoiesis, and defective octamer-dependent promoter activation in Oct-1-deficient mice. Mol Cell Biol 24: 1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weierich C, Brero A, Stein S, von Hase J, Cremer C, Cremer T, Solovei I 2003. Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes. Chromosome Res 11: 485–502 [DOI] [PubMed] [Google Scholar]
- Wiblin AE, Cui W, Clark AJ, Bickmore WA 2005. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J Cell Sci 118: 3861–3868 [DOI] [PubMed] [Google Scholar]
- Yaffe E, Farkash-Amar S, Polten A, Yakhini Z, Tanay A, Simon I 2010. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet 6: e1001011 doi: 10.1371/journal.pgen.1001011 [DOI] [PMC free article] [PubMed] [Google Scholar]