Abstract
Human U1 small nuclear (sn)RNA, required for splicing of pre-mRNA, is encoded by genes on chromosome 1 (1p36). Imperfect copies of these U1 snRNA genes, also located on chromosome 1 (1q12-21), were thought to be pseudogenes. However, many of these “variant” (v)U1 snRNA genes produce fully processed transcripts. Using antisense oligonucleotides to block the activity of a specific vU1 snRNA in HeLa cells, we have identified global transcriptome changes following interrogation of the Affymetrix Human Exon ST 1.0 array. Our results indicate that this vU1 snRNA regulates expression of a subset of target genes at the level of pre-mRNA processing. This is the first indication that variant U1 snRNAs have a biological function in vivo. Furthermore, some vU1 snRNAs are packaged into unique ribonucleoproteins (RNPs), and many vU1 snRNA genes are differentially expressed in human embryonic stem cells (hESCs) and HeLa cells, suggesting developmental control of RNA processing through expression of different sets of vU1 snRNPs.
U1 small nuclear (sn)RNA, as part of a ribonucleoprotein (RNP) complex, is an essential component of the spliceosome responsible for pre-mRNA splicing. The 5′ end of the U1 snRNA base pairs with the 5′ splice site (5′ss), located at the exon/intron junction at the 5′ end of introns, and defines where the first cleavage occurs. The removal of the intron and subsequent ligation of the flanking exons requires an additional further four snRNA-containing RNP complexes, U2, U4, U5, and U6, in addition to numerous other splicing factors (Wahl et al. 2009). However, unlike the other UsnRNAs, U1 snRNA has also been shown to associate with intronless transcription units in vivo, indicating an additional biological role for this snRNA (Spiluttini et al. 2010). In agreement with this, U1 snRNA copurifies with the cyclin H component of the TFIIH complex and promotes transcription initiation by enhancing the association of the general transcription factors to the promoters of protein-coding genes (Kwek et al. 2002; O'Gorman et al. 2005; Damgaard et al. 2008). In addition, U1 snRNP can inhibit polyadenylation when bound to cryptic 5′ss located downstream from terminal exons (Gunderson et al. 1998; Fortes et al. 2003). The U1.70K component of the U1 snRNP complex has been implicated in this process, which typically leads to degradation of the transcripts. More recently it has been shown that the U1 snRNP also protects nascent transcripts from premature cleavage/polyadenylation events occurring at cryptic poly(A) sites throughout the body of pre-mRNA transcripts (Kaida et al. 2010; Vorlova et al. 2011; Berg et al. 2012). Thus, U1 snRNA can play regulatory roles in gene expression, which are independent of its constitutive role in splicing.
In humans, the U1 snRNA gene RNU1-1 (also known as RNU1) is encoded by a multigene family located on the short arm of chromosome 1, 1p36 (Naylor et al. 1984). In addition, numerous U1 snRNA pseudogenes are present throughout the genome. There are three distinct classes of the U1 snRNA pseudogenes, which have been generated by both DNA and RNA mediated mechanisms (Denison and Weiner 1982; Bernstein et al. 1985). The U1 snRNA class II and class III pseudogenes are scattered throughout the genome and have no homology in their flanking regions to each other or to the U1 snRNA gene. The U1 snRNA class I pseudogenes, in contrast, have considerable 5′ and 3′ flanking sequence homology with the U1 snRNA genes and retain good homology with the two essential promoter elements, the distal sequence element (DSE), and proximal sequence element (PSE) (Denison and Weiner 1982). The pseudogenes map to a separate location on the long arm of chromosome 1 (1q12-21) (Lindgren et al. 1985). The term “pseudogene” has been assigned to gene sequences that resemble real genes but contain base changes, deletions, and/or insertions that disrupt their ability to encode a functional protein (Balakirev and Ayala 2003; Zheng et al. 2007). Many of the U1 pseudogenes have been classified as such since they also encode imperfect or incomplete copies of the U1 snRNA (Denison et al. 1981; Denison and Weiner 1982). Since the U1 snRNA is not translated into a protein, active pseudogenes could generate variant functional snRNAs. It has been known for some time that minor U1-like snRNAs, with variation to the human U1 snRNA exist, but to date, their function remains elusive (Patton and Wieben 1987; Lund 1988; Kyriakopoulou et al. 2006). We questioned whether variant U1 snRNAs, encoded by class I pseudogenes, could play a role in regulating gene expression in vivo.
We annotated a total of 21 U1 snRNA class I pseudogenes within the 1q12-21 locus, and demonstrate that many of the pseudogene promoters have active transcription marks in human cells. The majority of these pseudogenes express an snRNA in vivo and are differentially expressed in human embryonic stem cells (hESCs) and HeLa tissue culture cells. Moreover, these variant U1 snRNAs undergo normal 3′ end processing and are packaged into ribonucleoprotein (RNP) complexes. Using antisense oligonucleotides to target the most abundant variant U1 snRNA (vU1.8 snRNA) for degradation in HeLa cells, followed by high-throughput analysis of total RNA hybridized to the Human Exon ST 1.0 array, we show that this variant U1 snRNA is required for correct expression of a subset of genes at the level of mRNA 3′ end processing. These findings indicate that this new group of snRNAs, which are encoded by sequences previously considered to be pseudogenes, play important roles in cell function.
Results
U1 snRNA class I pseudogenes are transcriptionally active in vivo
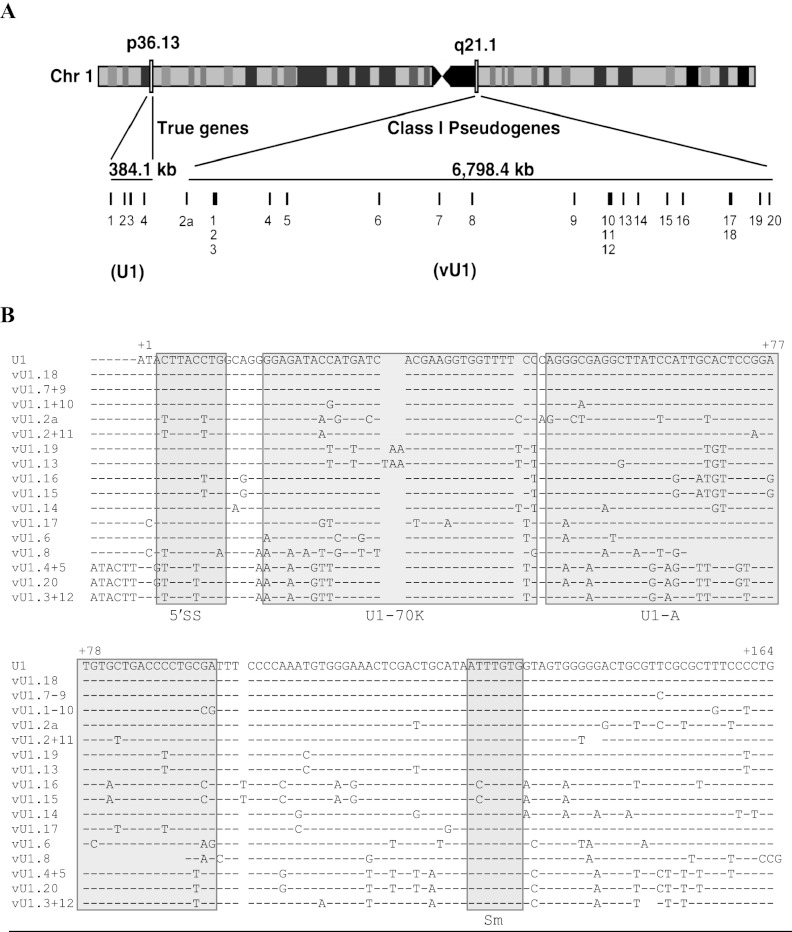
We searched the human genome for sequences similar to the prototypical human HU1.1 snRNA gene (Lund and Dahlberg 1984) using the UCSC Genome Browser BLAST-like alignment tool (BLAT) (http://genome.ucsc.edu/). A total of 21 putative and four near-perfect hits were identified within the U1 snRNA class I pseudogene locus (1q12-21) and the U1 snRNA gene locus (1p36), respectively (Fig. 1A; Supplemental Table S1). The U1 snRNA class I pseudogenes (hereafter termed variant (v)U1 snRNA genes) were further divided into nine groups based on sequence conservation (Supplemental Fig. S1) and all were aligned to the U1 snRNA gene sequence (Fig. 1B; Supplemental Figs. S2, S3). The results of our analysis demonstrate that the human U1 snRNA class I pseudogene locus spans a genomic region of ∼6.8 Mb. The 21 vU1 snRNA genes are not clustered in this region, but instead are interspersed with protein-coding genes and numerous transfer (t)RNA genes. These retain large stretches of homology with the U1 snRNA genes within the promoter and 3′ flanking sequences (Denison and Weiner 1982). While some of the vU1 snRNA-encoding regions are identical to the U1 snRNA sequence, most deviate by base changes and small deletions (Fig. 1B). Interestingly, all U1 and vU1 snRNA genes lack sequence identity immediately downstream from the snRNA gene-specific 3′ box RNA processing element, while the majority regain sequence conservation after an additional 250 bp (Supplemental Fig. S3).
Figure 1.
Annotation and sequence of the U1 snRNA class I pseudogenes. (A) Schematic of chromosome 1 illustrating the location of U1 snRNA genes (1p36) and the U1 snRNA class I pseudogenes (1q12-21). The size of each locus is shown in kilobases (kb). Each gene is represented by a vertical line and a number. Light and dark boxes on chromosome 1 refer to regions of high and low gene activity, respectively. (B) Alignment of the snRNA sequence of the U1 snRNA genes (U1.1–4) with the 21 vU1 snRNA genes. Paired vU1 snRNAs have identical sequences. Nonconserved bases are denoted with the letter code representing one of the four nucleotide bases. A dash indicates the presence of a corresponding base, and a gap indicates the absence of a base at that position. The bases are numbered, beginning with 1 for the first base of the U1 snRNA sequence. Important features of the U1 snRNA are boxed and indicated underneath the alignment: 5′ splice site recognition motif (5′ss), U1-70K protein binding region (U1-70K); U1-A protein binding region (U1-A); Sm binding site (Sm).
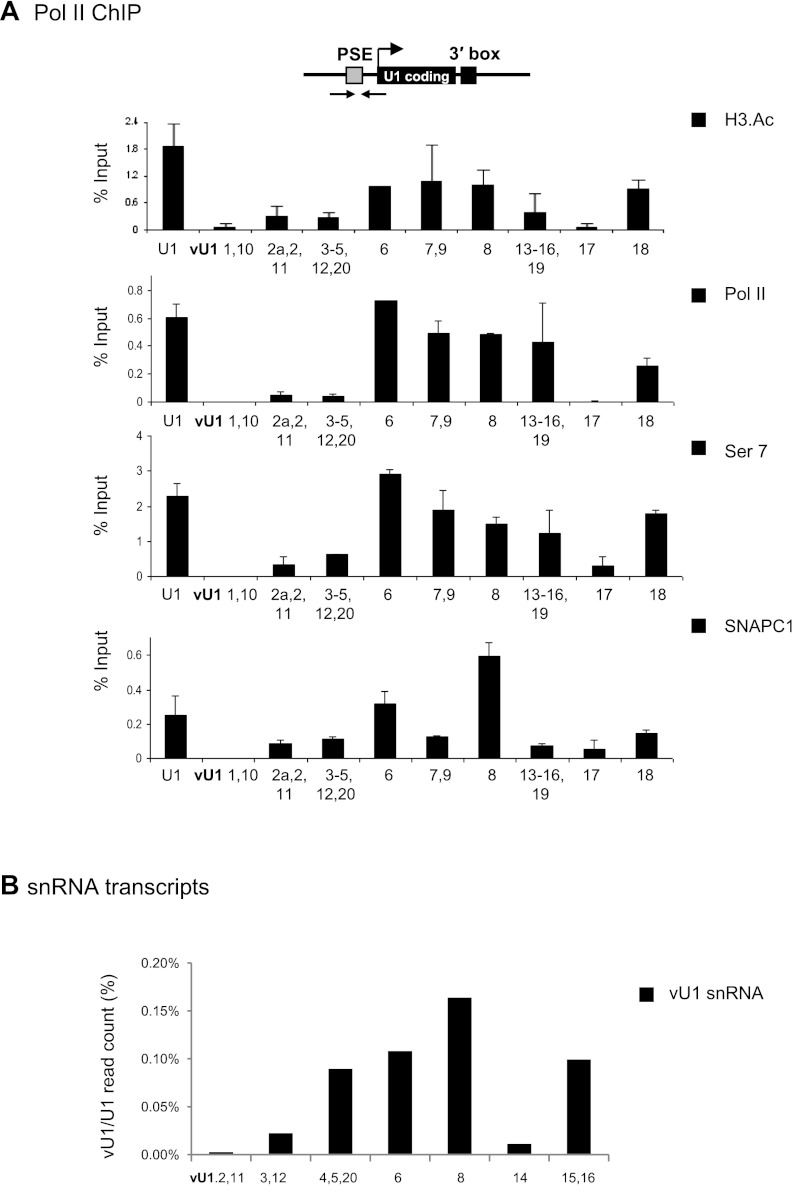
To investigate whether these vU1 snRNA genes are active in vivo, we performed chromatin immunoprecipitation (ChIP) experiments in HeLa cells with antibodies to factors involved in expression of the pol II-transcribed UsnRNA genes. These include the multisubunit transcription factor PTF/PBP/SNAPc (PSE-binding transcription factor/PSE-binding protein/snRNA-activating protein complex), which recognizes the PSE element located ∼50 bp upstream of the transcription start site, and pol II phosphorylated on serine 7 (Ser7) of the carboxy-terminal domain (CTD) heptapeptide (Egloff et al. 2008). In addition, we used antibodies to pol II and acetylated H3 core histone proteins, which are primarily associated with promoter regions of actively transcribing genes (Shahbazian and Grunstein 2007). As shown in Figure 2A, almost half of the vU1 snRNA promoters contain acetylated H3 and the same vU1 snRNA genes (vU1.6, vU1.7 + 9, vU1.8, vU1.13-16 + 19, and vU1.18) are occupied by pol II at levels similar to those observed at U1 snRNA gene promoters. The snRNA gene-specific transcription factor SNAPC and phosphorylation of Ser7 of the CTD of the large subunit of pol II, which is found on active snRNA genes (Egloff et al. 2007), are also detected on the same vU1 snRNA genes (Fig. 2A).
Figure 2.
Several vU1 snRNA are transcribed in HeLa cells. (A) ChIP analysis of U1 snRNA and vU1 snRNA genes in HeLa cells with antibodies indicated at right. Error bars in this and subsequent figures represent standard error of at least three repeats. Location of the primers used is indicated on the diagram. The proximal sequence element (PSE), coding and 3′ box are specified by gray and black boxes, respectively, and start of transcription by an arrow. (B) Quantification of vU1 snRNA steady-state levels in HeLa cells, normalized to U1 snRNA. Only reads with two or more nucleotide differences from U1 snRNA were counted toward the vU1 snRNA levels. Experimental data from Fejes-Toth et al. (2009).
With the exception of the vU1.6, vU1.1 + 10, and vU1.8 snRNA genes, the levels of SNAPC1 (also known as PTFgamma) are similar for all vU1 snRNA genes tested. Recent data indicate that SNAPC1 may be required to maintain a persistently open chromatin state at snRNA promoters to facilitate rapid transcription reinitiation, as failure to remove this factor results in chromosome fragility at metaphase (Pavelitz et al. 2008). The finding that SNAPC1 is associated with vU1 snRNA gene promoters with low pol II occupancy (vU1.2a-2 + 11, vU1.3-5,12 + 20, and vU1.17 snRNA genes, respectively) suggests that some genes are regulated at a step downstream from SNAPC recruitment to the PSE.
vU1 snRNA genes that appear transcriptionally active in HeLa cells also generate transcripts that continue beyond the 3′ box, as detected by qRT-PCR analysis, with transcripts from vU1.7 + 9, vU1.13-16 + 19, and vU1.18 snRNA genes detected at levels comparable to the level of nascent transcripts from U1 snRNA genes (Supplemental Fig. S4). Primer specificity, for ChIP and qRT-PCR analysis, was confirmed using cloned U1 or vU1 snRNA genes as templates (Supplemental Fig. S5). To determine the steady-state levels of vU1 snRNAs in HeLa cells, we analyzed the small RNA sequencing data from Fejes-Toth et al. (2009). Our analysis confirms expression of vU1 snRNAs in HeLa cells and estimates their level to be <0.2% the level of U1 snRNA (Fig. 2B). Interestingly, vU1.8 snRNA, which contains numerous base changes and a deletion of the U1.A binding site (Fig. 1B; Pomeranz Krummel et al. 2009) is the most abundant vU1 snRNA. Taken together, our results indicate that up to 50% of the previously considered U1 snRNA class I pseudogenes are transcriptionally active in HeLa cells and could therefore contribute to regulation of mRNA production.
Some vU1 snRNA genes are differentially expressed
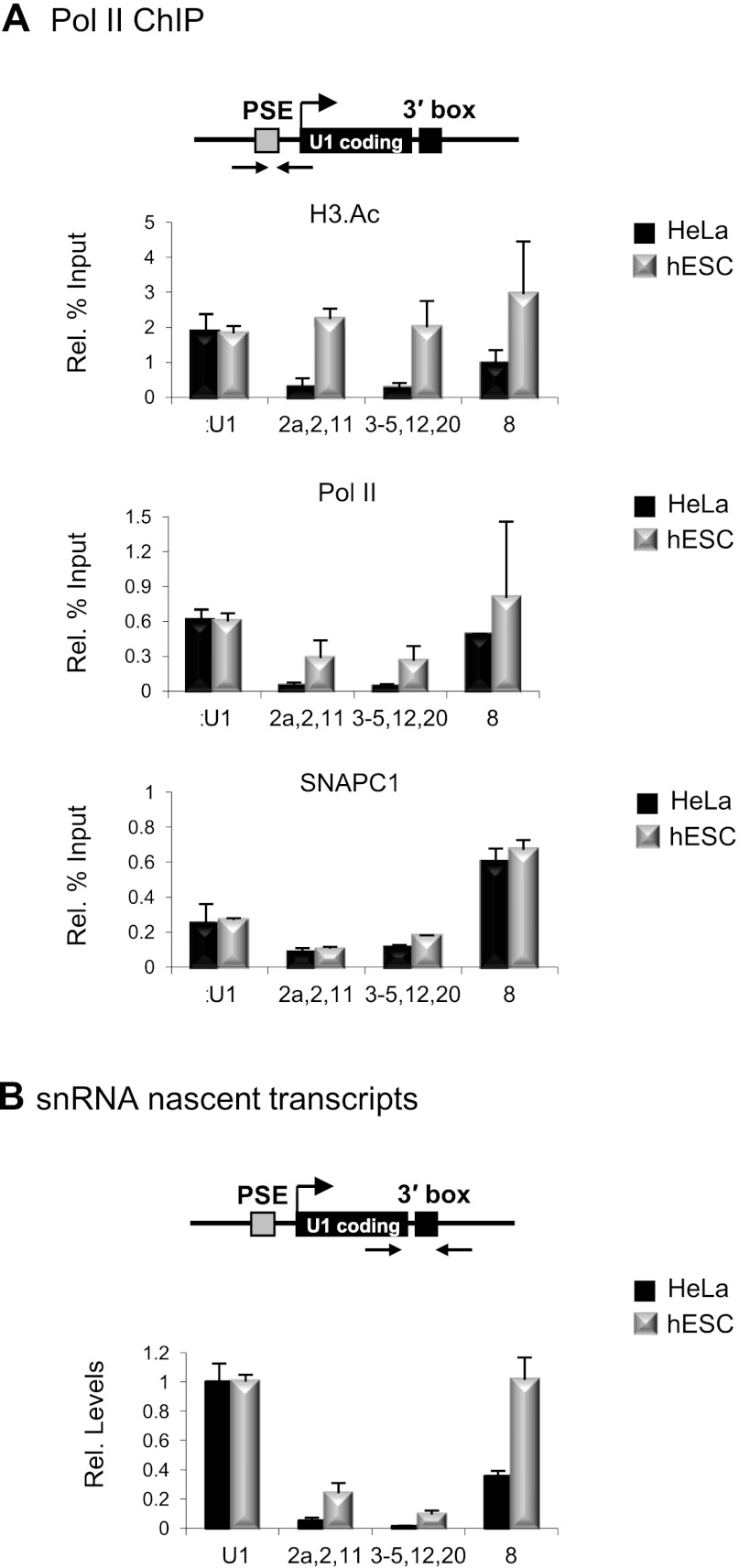
Numerous reports have documented the expression of variant U1 snRNAs during development of mouse, sea urchin, frog, and fly embryos (Forbes et al. 1984; Santiago and Marzluff 1989; Lo and Mount 1990; Cheng et al. 1997), which differ from their corresponding U1 snRNAs by only a few bases. U1 snRNA is generally constitutively expressed at all stages of development, whereas the U1 snRNA variants appear to be restricted to early embryo development (Lund et al. 1985). To determine whether expression of any human vU1 snRNA gene is restricted to early developmental stages, pol II, SNAPC1 and H3.Ac association, and vU1 snRNA levels were compared in HeLa and human embryonic stem cells (hESCs). Pol II association and the levels of nascent transcripts are comparable in both cell types for the vU1 snRNA genes active in HeLa cells (vU1.6, vU1.7 + 9, vU1.13–16 + 19, and vU1.18 snRNA genes) (Supplemental Figs. S6, S7). Thus, several vU1 snRNAs may be constitutively expressed. Interestingly, some vU1 snRNA genes (vU1.2a,2,+11, vU.3–5,12 + 20, and vU1.8) have marks of active transcription and generate up to 30-fold more nascent transcript in hESCs than in HeLa, with a corresponding increase in pol II occupancy at their promoters (Fig. 3A,B). These may therefore represent a distinct class of vU1 snRNA genes that are differentially regulated. Furthermore, as noted above, the levels of SNAPC1 do not correlate with the transcriptional status of vU1 snRNA genes in the different cell types (Fig. 3A; Supplemental Fig. S6), reinforcing the notion that control of expression is regulated at a step downstream from SNAPC1 association.
Figure 3.
vU1 snRNA genes are up-regulated in hESCs. (A) ChIP analysis of the U1 snRNA and some vU1 snRNA genes in hESCs and HeLa cells with antibodies indicated at the right. (B) qRT-PCR analysis of nascent transcripts from the corresponding U1 snRNA and vU1 snRNA genes in HeLa and hESCs. (Rel. levels) The levels of vU1 snRNA nascent transcripts are expressed relative to U1 snRNA, normalized to 1.0 in each cell type.
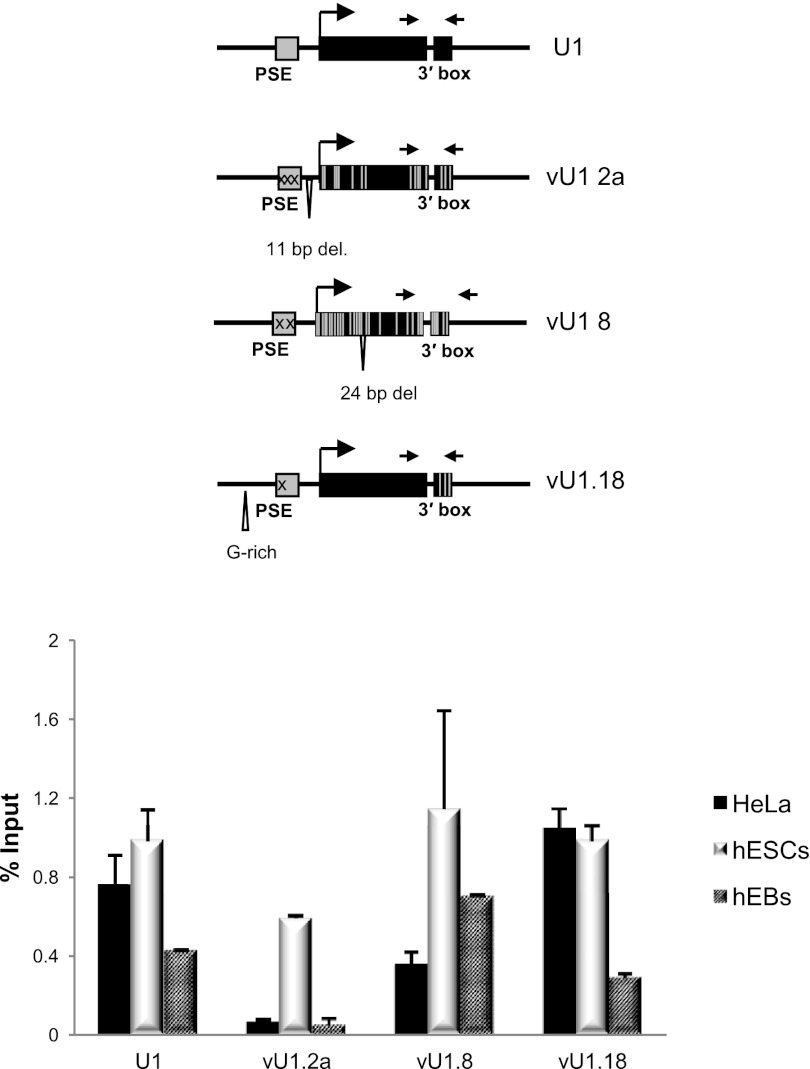
More extensive analysis of the transcription units of vU1.2a snRNA, vU1.8 snRNA, and vU1.18 snRNA genes confirms that the vU1.2a snRNA gene is inactive in HeLa cells but transcribed in hESCs (Fig. 4). When hESCs are differentiated into Embryoid Bodies (EB) there is a drastic reduction in pol II occupancy within the snRNA encoding region of the vU1.2a snRNA gene relative to the other U1/vU1 snRNA genes analyzed. In contrast to the pol II ChIP data, however, qRT-PCR analysis of steady-state levels of the vU1 snRNAs in the 4-d-old hEBs indicate a marginal reduction in snRNA levels in these cells as compared with levels in hESCs (Supplemental Fig. S8). This is in agreement with previous studies in mice, which show that a reduction in the embryo-specific U1b snRNA level is only detectable in 11 to 19-d-old embryos (Cheng et al. 1997). This might reflect that vU1 snRNAs are as stable as the U1 snRNA, which has an estimated half-life of 4–5 d (Sauterer et al. 1988). This data indicates that regulation of vU1 snRNA gene activity occurs early in the differentiation process, possibly allowing time for steady-state levels of vU1 snRNAs to reduce before the hEBs differentiate into different cell types. Importantly, down-regulation of the expression of some vU1 snRNA genes during differentiation suggests a stage-specific function of these vU1 snRNAs during development.
Figure 4.
Some vU1 snRNA genes are differentially regulated. Pol II ChIP analysis of the U1, vU1.2a, vU1.8, and vU1.18 snRNA genes in HeLa and hESCs, before and after differentiation to embryoid bodies (EB). The position of the primers is indicated in the schematics.
vU1 snRNAs are in ribonucleoprotein complexes
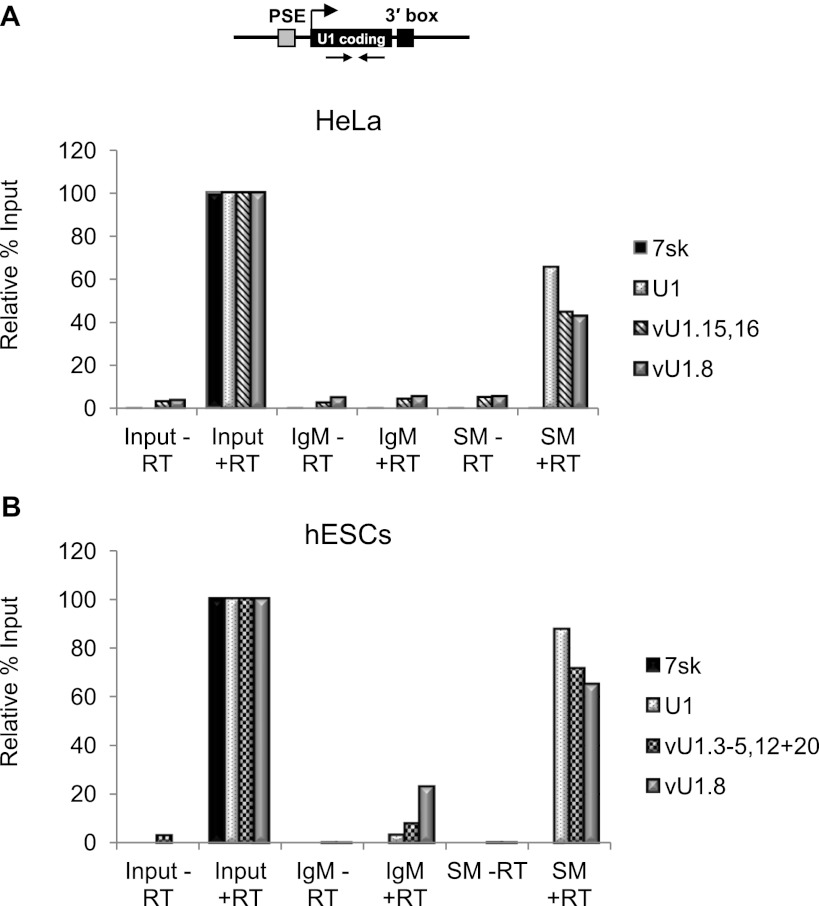
U1 snRNA is synthesized as a precursor in the nucleus and exported to the cytoplasm for 3′ end processing and maturation (Will and Luhrmann 2001). While in the cytoplasm, seven core proteins, known as the Smith (Sm) proteins, assemble onto the 5′-AUUUGUG-3′ motif located toward the 3′ end of the U1 snRNA (see Fig. 1B). The association of U1 snRNP-specific proteins (U1-70K, U1-A, and U1-C) and nuclear import completes the biogenesis and enables the U1 snRNP to participate in pre-mRNA processing. To determine whether vU1 snRNAs are also associated with RNPs, RNA was isolated from RNP complexes purified from both HeLa and hESC whole-cell extracts by immunoprecipitation with anti-Sm antibodies. Anti-Sm was the antibody of choice, as this is the only conserved sequence motif across the representative groups of vU1 snRNA genes (Fig. 1B). Immunoprecipitation with human anti-IgM was used as a negative control. The presence of vU1 snRNAs was determined by qPCR amplification using gene-specific primers that target the snRNA encoding region. vU1.8 and vU1.16 snRNAs, and vU1.3–5,12 + 20 snRNAs were chosen for further analysis as they are the only vU1 snRNAs expressed in HeLa and hESCs, respectively, that are sufficiently divergent from U1 snRNA to ensure specific amplification of the mature snRNA. The results of qPCR analysis indicate that these vU1 snRNA transcripts are present in the purified Sm-containing RNP complexes (Fig. 5A,B). Using a genomic DNA standard to normalize primer efficiencies between the different primer sets, the vU1 snRNAs were estimated to be present at 0.2% and 0.5% the level of U1 snRNA in Hela and hESCs, respectively. As expected, the 7SK snRNA is not specifically immunoprecipitated.
Figure 5.
vU1 snRNAs are packaged into RNP complexes. qRT-PCR analysis of U1 snRNA and vU1 snRNA levels in HeLa (A) and hESC (B) extracts, before and after immunoprecipitation with anti-Sm (Y-12) antibody. The non-Sm-containing 7SK RNA level was used as a negative control. Location of the primers is indicated in A. The level of the U1 snRNA and vU1 snRNAs, enriched in the Sm-immunoprecipitate, is expressed relative to input at 100%. RT (reverse transcription) reactions without “–” or with “+” the addition of Superscript III. Note: Although vU1.15/vU1.16 snRNA was enriched in RNP complexes, molecular cloning, followed by sequencing, indicated that this variant was not properly processed at the 3′ end. The levels of the vU1 snRNAs, enriched in the Sm immunoprecipitates, relative to the U1 snRNA were estimated using a genomic DNA standard to normalize primer efficiency.
To determine whether the vU1 snRNAs are fully processed at the 3′ end, a poly(A) tail was added to the Sm-purified snRNAs and individual vU1 snRNAs were amplified using gene-specific forward primers together with an oligo dT reverse primer. Sequencing of the cloned transcripts confirmed that at least vU1.7 + 9 and vU1.8 snRNAs, and vU1.3 and vU1.20 snRNAs in HeLa and hESCs, respectively, are fully 3′ end processed (Supplemental Fig. S9). Notably, transcripts from vU1.7 + 9 snRNA genes, which differ from the U1 snRNA by one base at position 150, comprise 10%–15% of the clones when primers specific to U1 snRNA are used for amplification. In addition, vU1.18 snRNA encodes an snRNA identical to U1 snRNA and could therefore contribute to the pool of U1 snRNA. A significant proportion of the total U1 snRNA in mammalian cells may therefore derive from vU1 snRNA genes.
To further characterize the vU1 snRNP complexes in HeLa cells, immunoprecipitation was performed with antibodies against the U1 snRNA-specific RNP proteins, U1.70K, U1-A, U1-C, and an antibody that recognizes the UsnRNA-specific trimethyl cap structure. U1 snRNA was specifically immunoprecipitated with all antisera tested (Supplemental Fig. S10). Importantly, the 7SK was not enriched in any of the immunoprecipitation reactions, as expected. qRT-PCR analysis of RNA purified from the immunoprecipitates, using primers specific for vU1.8 snRNA and vU1.3–5,12 + 20 snRNAs, indicates that both of these snRNAs contain a trimethyl cap structure and are associated with the U1-C protein (Supplemental Fig. S10). Anti-U1-70K and anti-U1-A precipitates very little vU1.8 snRNA (1.4% and 4.7%, respectively) and vU1.3–5,12 + 20 snRNA (5.7% and 6.3%, respectively), although this is significantly more than for the 7SK RNA negative control (0.43% and 0.37%, respectively). This may indicate that these proteins are not associated with these vU1 snRNAs or that the epitopes are not readily accessible. The absence of U1-70K and U1-A would suggest that vU1 snRNAs may exist in novel RNP complexes with unique roles in vivo. Nonetheless, the presence of the U1-C protein in all vU1 snRNAs tested indicates a potential role for vU1 snRNAs in pre-mRNA processing (Du and Rosbash 2002).
vU1 snRNAs regulate gene expression in vivo
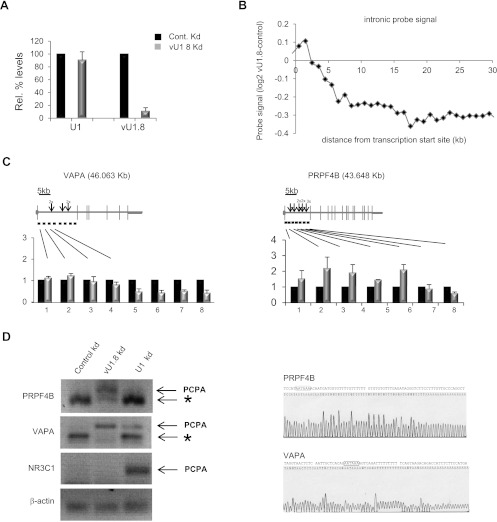
In addition to vU1 snRNAs, variant U5 snRNAs have also previously been identified in fly and purified from spliceosomal complexes extracted from HeLa cells (Sontheimer and Steitz 1992; Chen et al. 2005). However, analysis of the potential function of vUsnRNAs has been hampered, in part, by the fact that they all deviate from the major species by only a few base changes, making specific knockdown and/or target-gene identification difficult. In contrast, 52% of the vU1 snRNAs described here have base changes in the region complementary to the 5′ss, and 54% of vU1 snRNAs diverge significantly from U1 snRNA. To explore the contribution that vU1 snRNAs make to the regulation of gene expression in vivo, we measured global transcriptome changes following knockdown of the vU1.8 snRNA. This vU1 snRNA was chosen as it is the most abundant vU1 snRNA expressed from a single-copy gene in HeLa cells, is properly 3′ end processed, packaged into an RNP complex, and importantly for this analysis, the only vU1 snRNA expressed in HeLa cells that contains a number of consecutive base changes, which enables functional knockdown with specific antisense oligonucleotides (Figs. 1B, 2B, 5; Supplemental Figs. S9, S10). As illustrated in Figure 6A, the 2′O Methyl (2′OMe) RNA/DNA antisense oligonucleotide targeting positions 11–33 of vU1.8 snRNA specifically reduces the level of this snRNA in vivo by 89% following transfection into HeLa cells. Importantly, the control oligonucleotide has no effect on vU1.8 snRNA levels.
Figure 6.
Altering vU1.8 snRNA levels cause global transcriptome changes in HeLa cells. (A) qRT-PCR analysis of vU1.8 and U1 snRNA levels in HeLa cells transfected with control or vU1.8 snRNA antisense oligonucleotides. (B) Average profile of intronic probe signal difference between vU1.8 snRNA knockdown and control cells across expressed nonoverlapping genes relative to the annotated transcription start sites. Each point represents an average of more than 100 probes in the corresponding region. (C) Validation of array results by qRT PCR, confirming the reduction in VAPA and PRPF4B pre-mRNA levels toward the 3′ end. Gene structures are illustrated above the graphs; horizontal and vertical lines indicate introns and exons, respectively. Location of primers is denoted as horizontal lines below the schematics. (Arrow) Cryptic poly(A) sites within the first intron of VAPA and PRPF4B genes. Sizes of genes are indicated in kilobases. (D) 3′ RACE, using nested PCR, on total RNA from HeLa cells transfected with control, vU1.8, or U1 snRNA antisense oligonucleotides was performed on the endogenous PRPF4B, VAPA, and NR3C transcripts to detect polyadenylated mRNAs. PCPA denotes a premature cleaved and polyadenylated mRNA product. (*) Mispriming of the oligo dT primer at an A-rich region. Primers targeting the endogenous beta-actin mRNA was used as a normalization control. Sequencing results of the PCPA products for the PRPF4B and VAPA gene are shown on the right. The putative poly(A) sites are indicated with black bordered boxes.
Having established the specificity of the chimeric RNA/DNA oligo, we were now in a position to address whether degradation of the vU1.8 snRNA in vivo would lead to global transcriptome changes. We analyzed total RNA prepared from HeLa cells transfected with either control or vU1.8 snRNA antisense oligonucleotides using the Affymetrix Human Exon ST 1.0 array. This array contains, on average, 40 probes per gene, targeting exonic, intronic, and untranslated (UTR) regions of all well-annotated protein-coding genes. Variations in expression levels, alternative 3′ and 5′ UTR usage, and changes in pre-mRNA splicing events can all be measured, allowing the full effect of vU1.8 snRNA knockdown on gene expression to be analyzed. Four independent biological repeats were analyzed. We initially chose to look for novel changes in alternative splicing events (ASE) using the Splicing Index (SI) approach (Yamamoto et al. 2009), since this is thought to be the primary function of U1 snRNA in vivo. A total of 444 hits were identified, representing 214 genes that exceed the following thresholds: fold change ≥ 2, P-value < 0.01 (data not shown). However, the vast majority of these hits (>92%) indicated a change in probe intensity over intronic and/or intergenic regions, with each gene represented by more than one hit. Following manual examination of all differentially expressed probesets using the UCSC Genome Browser, it became apparent that additional probesets flanking these hits were also deregulated, but outside the strict selection criteria. Furthermore, there appeared to be a clear bias in signal intensities at the start of the transcript compared with the end. Consequently, we felt the results of this analysis do not support a major role for vU1.8 snRNA in pre-mRNA splicing, but instead indicate a potential regulatory role in gene expression at the nascent transcript level. To address this bioinformatically, we profiled the average intronic probe signal relative to the distance from the transcription start site (TSS) genome-wide and revealed that 140 of the expressed genes (2%) (Supplemental Table S2) exhibited more than a twofold drop in nascent signal after ∼5 kb from TSS in the vU1.8 snRNA knockdown cells. This result is exemplified in Figure 6B, and Supplemental Figure S11 shows the validation of some of the array predictions by reverse transcription, followed by qPCR. These data suggest that the major role of vU1.8 snRNA in vivo is to regulate expression of a subset of target genes at the level of transcription elongation and not pre-mRNA splicing, as previously predicted. Interestingly, from the 8834 genes detected as expressed, 116 genes (∼1.3%) showed significant (more than twofold, individual P-value < 0.01) change in expression across the entire gene, as measured by signals from exonic probesets following knockdown of the vU1.8 snRNA (Supplemental Table S3). Although we chose to exclude these genes from further analysis, the GeneOntology terms enriched for this group of genes include female pregnancy, regulation of proliferation, and/or response to hypoxia (Supplemental Table S4), potentially consistent with a role for vU1 snRNA genes in cell differentiation during development.
A loss in nascent transcript toward the 3′end of the gene is consistent with the recent reports showing a similar loss of the 3′ end of pre-mRNAs following functional knockdown of the U1 snRNA (Kaida et al. 2010; Berg et al. 2012). In the absence of U1 snRNA recruitment, the cleavage/polyadenylation machinery is able to cleave the pre-mRNA ∼20 bp downstream from a cryptic polyadenylation site and subsequently stabilize the shortened RNA by polyadenylation. To assess whether similar events are occurring following degradation of vU1.8 snRNA, we first characterized cDNA fragments for a number of transcripts that show this pattern of expression to more precisely define the region within which RNA processing is affected. As illustrated in Figure 6C, using the VAPA ([vesicle-associated membrane protein]-associated protein A) and PRPF4B (pre-mRNA processing factor 4 homolog B) transcripts as examples, the loss in nascent transcript typically occurs within the first intron, ∼5–10 kb from the start of transcription. To determine whether these shortened transcripts are polyadenylated, we located the potential polyadenylation sites within the first intron of both of these genes using Polyadj (http://rulai.cshl.org/tools/polyadq/polyadq_form.html) (Fig. 6C). 3′ RACE products were generated from control and vU1.8 snRNA knockdown cells using gene-specific forward primers within the first intron together with an oligo dT reverse primer. The endogenous NR3C1 (nuclear receptor subfamily 3, group C, member 1) pre-mRNA was used as a control since it has been shown to be a target of U1 snRNA (Kaida et al. 2010). Only one predicted poly(A) site within the first intron of both the PRPF4B and the VAPA pre-mRNAs generated specific products, following cleavage of the vU1.8 snRNA (Fig. 6D). Sequence analysis confirmed that these 3′ RACE products are polyadenylated (Fig. 6D). Functional knockdown of U1 snRNA, using a specific 2′OMe RNA/DNA antisense oligonucleotide targeting bases 1–25 (Supplemental Fig. S12) results in a major premature cleavage and polyadenylation defect for the NR3C1 transcript only (Fig. 6D). These results suggest that vU1.8 snRNA protects a subset of pre-RNAs from premature cleavage at cryptic poly(A) sites within introns, in a manner analogous to that reported for U1 snRNA.
Discussion
The human genome contains upward of 20,000 pseudogenes (Torrents et al. 2003; Harrow et al. 2012; Kalyana-Sundaram et al. 2012; Kapranov and St Laurent 2012), which are generally classified as nonfunctional sequences within our genome. However, wherever studied, it seems that the inability to produce a protein does not necessarily imply nonfunctionality, as recent studies have demonstrated that pseudogenes often exert their activity through RNA-based mechanisms, generating antisense transcripts that either interfere with translation of real genes or sense transcripts that act as miRNA decoys or compete for mRNA stabilizing factors (Korneev et al. 1999; Chiefari et al. 2010; Poliseno et al. 2010; Muro et al. 2011). For the past 30 yr, the class I U1 snRNA pseudogenes were also considered to be nonfunctional since they would encode imperfect or incomplete copies of the U1 snRNA (Denison et al. 1981; Lindgren et al. 1985). However, the results presented here challenge this notion and suggest that these “imperfect” copies have a biological function. We have focused our attention on the entire U1 snRNA class I pseudogene locus and shown that 50% of the vU1 snRNA genes annotated within this 6.8-Mb region express an snRNA in HeLa cells and that all 21 vU1 snRNA genes are active, at varying levels, in hESCs. In addition, three other vU1 snRNAs (U1A5, U1A6, and U1A7) were also previously isolated from HeLa cells (Kyriakopoulou et al. 2006). A “BLAT” search of their sequences indicates that they map to different locations on the human genome. Interestingly, U1A5 snRNA and U1A7 snRNA have features characteristic of class II and class III, including a lack in homology with the U1 snRNA gene in the 3′ and 5′ flanking regions. This is a strong indication that many U1 snRNA pseudogenes may have been misclassified, and our results therefore highlight a novel aspect of snRNA gene expression, which has previously been overlooked.
The incorporation of fully processed vU1 snRNAs in snRNP complexes suggests that vU1 snRNAs are likely to participate in pre-mRNA processing events. In agreement with this, high-throughput analysis of total RNA extracted from control and vU1.8 snRNA knockdown cells revealed a primary role for vU1.8 snRNA in 3′ end processing of pre-mRNAs rather than pre-mRNA splicing, as initially predicted. Knocking down U1 snRNA, at varying levels, has a profound effect on both pre-mRNA splicing and mRNA length (Supplemental Fig. S12) (Kaida et al. 2010; Berg et al. 2012). In contrast, knocking down vU1.8 snRNA levels in HeLa cells affects only 3′ end processing of a small subset of genes (Fig. 6D), which is consistent with the fact that the level of vU1.8 snRNA in HeLa cells is 0.2% of the U1 snRNA level. Although much less abundant than the U1 snRNA, vU1.8 snRNA is present at two to 500 copies per HeLa cell, which is comparable to many highly abundant mRNAs (Wickrema et al. 1991). Given that there are significant differences between vU1.8 snRNA and U1 snRNA at their 5′ends, and likely also in their RNP composition (Supplemental Figs. S3, S10), we propose that vU1.8 snRNA is selectively recruited to a subset of pre-mRNAs. Previous data have shown that inhibition of polyadenylation by U1 snRNP requires interaction of the U1-70K protein with the Poly(A) Polymerase (PAP) (Gunderson et al. 1998). Mutational and biochemical analysis indentified a number of sequence motifs within the SR region of the U1-70K that are necessary for this U1-70K–PAP interaction. However, such motifs are also present in a number of different SR-containing and non-SR-containing proteins. Since data presented in this study suggest that little U1-70K protein is associated with the vU1.8 snRNA, we suggest that a different SR-factor associates with this snRNA and inhibits the PAP activity in a manner analogous to the U1-70K. Moreover, since U1 snRNA can marginally affect the cleavage and polyadenylation of the same cryptic Poly(A) in the VAPA gene as the vU1.8 snRNA (Fig. 6D), we propose that in some instances the vU1.8 snRNP can act cooperatively with the U1 snRNP.
The presence of the U1-C protein in the vU1.8 snRNP complex suggests that vU1.8 snRNA could also function in pre-mRNA splicing (Du and Rosbash 2002; Du et al. 2004; Rosel et al. 2011). Of the 444 ASE (214 genes) that we initially identified using the splicing index algorithm, only 8% (∼17 genes) indicated a potential ASE. However, further analysis of some of these ASEs did not support a direct role for vU1.8 snRNA in splicing regulation (data not shown). Moreover, GO:TERM analysis of transcripts subject to premature 3′ end processing following knockdown of vU1.8 indicates an enrichment in factors involved in RNA binding and pre-mRNA processing. It is therefore plausible that the identified splicing defects are an indirect consequence of changes in expression of one or a combination of these factors. The absence of evidence supporting a role for vU1.8 snRNA in splicing regulation is possibly not surprising considering that this variant is the most divergent in sequence conservation from the U1 snRNA (see Fig. 1B). In addition to numerous base changes, vU1.8 snRNA also contains a 24-bp deletion encompassing part of the U1-A binding site. Although U1 snRNP is active in splicing assays in the absence of U1-A (Heinrichs et al. 1990; Pomeranz Krummel et al. 2009), lack of this factor on the vU1.8 snRNA may favor an alternative function in vivo. Moreover, U1A7 snRNA was unable to rescue mutations at complementary noncanonical 5′ss in vivo, further supporting the notion that vU1 snRNAs are involved in processes other than splicing (Roca and Krainer 2009). However, we cannot rule out the possibility that the vU1.8 snRNA or other variants characterized in this and other studies do participate in pre-mRNA splicing events.
Profiling the pattern of expression of vU1 snRNA genes in different cell types indicates that the majority of these genes are active in hESCs and ∼50% of them are specifically expressed in this cell type. Pervasive transcription is considered a common feature of ESCs, and this could explain why most of the vU1 snRNA genes are expressed in the HUES 2 cell line tested in this study. However, the finding that some vU1 snRNA genes are specifically down-regulated upon differentiation to EBs suggests that the control of vU1 snRNA gene expression is a crucial step during the early stages of embryogenesis (Fig. 4). Since pre-mRNA processing is known to play an important role during lineage differentiation of stem cells, promoting the production of different isoforms of mRNAs in different cell types, it is plausible that developmental control of vU1 snRNA expression is a critical mechanism controlling gene expression patterns throughout development (Wu et al. 2010). For example, vU1.2a expression is down-regulated as hESCs differentiate into hEBs. Further examination of late-stage embryos and/or embryo-derived differentiated cells, coupled with high-throughput transcriptome analysis of control and vU1-specific knockdowns, should allow for a better understanding of the functional importance of this group of vU1 snRNAs in gene expression. Multiple vU1 snRNAs expressed at low levels in different cells at different stages in development could have a significant impact on proteome diversity. A 5-nt deletion in one of the multicopy mouse U2 snRNA genes causes ataxia and neurodegeneration, and four point mutations in the human U4ATAC is associated with brain and bone malformations and unexplained postnatal death (microcephalic osteodysplastic primordial dwarfism type 1), emphasizing the potential for variant snRNAs to exert tissue-specific effects. (Edery et al. 2011; He et al. 2011; Jia et al. 2012). Moreover, deletion of the chromosome region (1q12-21), which harbors the vU1 snRNA genes, causes severe neurological dysfunction (Mefford et al. 2008), reinforcing the notion that these snRNAs may be important for normal development.
Methods
ChIP analysis
Chromatin immunoprecipitation (ChIP) analysis was carried out as previously described (O'Reilly et al. 2003). On average, 0.5 × 106 HeLa and 1 × 105 human embryonic stem cells (hESCs) were used per immunoprecipitation. Anti-pol II (N-20), anti H3 (ab1791), anti-H3.Ac (06.598), and anti-PTFγ antibodies were obtained from Santa Cruz Biotechnology, Inc., Abcam, Upstate laboratories, and Robert G Roeder and Jong-Bok Yoon (Yoon et al. 1995), respectively. Refer to Supplemental Table S5 for primer sequences. Typical quantitative PCR reactions included a 95°C step for 15 min, 40 cycles at 95°C/15 sec, 57°C/15 sec, and 72°C/25 sec, followed by a 10-min melt. % Input was calculated by (IP average—no antibody average) × 100/Input.
Plasmid construction
U1 snRNA and vU1 snRNA genomic constructs
Genomic U1 and vU1 snRNA clones were generated to control for U1 snRNA and vU1 snRNA primer specificity in ChIP and qRT-PCR. Human whole-blood genomic DNA (Promega) was amplified with snRNA gene-specific primers (Supplemental Table S6) to generate 1.3-kb fragments containing ∼100 bp of the 5′ flanking sequence, the RNA-encoding region and ∼1 kb of the 3′ flanking sequence. The fragments, which contain EcoRI and XhoI restriction sites at the 5′ and 3′ends, respectively, were cloned into pcDNA3 (Invitrogen) and sequenced. Genomic constructs were generated for the U1 snRNA and vU1 snRNAs, including vU1.2a snRNA, vU1.7 snRNA, vU1.8 snRNA, vU1.13 snRNA, and vU1.18 snRNA.
Cell culture and transfection
HeLa cells were grown in Dulbecco's Modified Eagle Media (Invitrogen) supplemented with 10% fetal calf serum, 50 μg of Penicillin per mL and 4 μM L-glutamine. Human ESC, HUES-2 (passages 16–38), was obtained from the HUES Facility, University of Harvard (Cowan et al. 2004). Work with this line was reviewed and approved by the UK Stem Cell Bank Steering Committee. hESCs were cultured in mTeSR1 medium (Stem Cell Technologies) on hESC-qualified Matrigel (Becton Dickinson). For differentiation to Embryoid Bodies (EBs), hESCs were dissociated with TrypLE Express (Invitrogen) and aggregated in mTeSR1 medium with 10 μM ROCK inhibitor Y-27632 (Calbiochem) by spinning in an AggreWell (Stem Cell Technologies) according to the manufacturers' manual to form EBs of 4000 cells/EB. The EBs were analyzed at day 4 post-aggregation.
Transient transfections in HeLa cells were performed with lipofectAMINE 2000 reagent (Invitrogen) according to the manufacturers' instructions.
Knockdown experiments
2′-O-Methyl (2′OMe) RNA/DNA oligonucleotides were purchased from Integrated DNA Technologies (IDT). Five nucleotides at the 5′ and 3′ ends were substituted with 2′-O-Methyl ribonucleotides. All bases were phosphorothioate converted. The control oligonucleotide is sense to the U1 snRNA at position 11 to 32, the vU1.8 snRNA-specific and U1 snRNA-specific oligonucleotide are antisense to vU1.8 snRNA at position 11 to 33, and U1 snRNA at position 1 to 25, respectively, where 1 is the first base of the U1/vU1 snRNA sequence (Supplemental Table S8). Knockdown experiments were performed in HeLa cells with lipofectAMINE 2000 reagent (Invitrogen) according to manufacturers' instructions. Typically, 1 × 107 cells were transfected with 600 pmoles of the 2′-OMe oligonucleotide and cells harvested 18 h later.
RT-PCR and amplification of cDNA
Total RNA was isolated from HeLa and hESCs using TRIzol (Invitrogen) according to the manufacturers' instructions. cDNA was generated at 50°C from 0.5 μg of total RNA using Superscript III (Invitrogen), random hexamers (Roche), or oligo dT in a total volume of 20 μL. The reverse transcriptase enzyme was omitted in cDNA preparations to control for genomic DNA contamination. Real-time quantitative PCR was performed using a QuantiTect SYBER Green master mix (Qiagen) on the Corbett Rotor-gene 3000 system. Typical reactions included 0.4 μL of cDNA and 1 μM concentration for each primer (Supplemental Table S7) in a total volume of 10 μL amplified for 40 cycles at 15 sec at 94°C, 15 sec at 57°C, and 45 sec at 72°C.
3′ RACE and amplification of PCPA products
Total RNA was isolated from HeLa cells using TRIzol (Invitrogen), according to manufacturers' instructions. cDNA was generated at 50°C with 1.0 μg of total RNA from control and knockdown cells using Superscript III (Invitrogen) and an oligo dT_1 primer in a total volume of 20 μL. Premature cleavage and polyadenylation products (PCPA) products were amplified by nested PCR with gene-specific forward primers and the oligo dT_2 reverse primer for 25 cycles at 15 sec at 94°C, 15 sec at 57°C, and 45 sec at 72°C. This reaction was diluted 10-fold and 1 μL was reamplified with another gene-specific forward primer and the oligo dT_2 reverse primer for a further 25 cycles at 15 sec at 94°C, 15 sec at 57°C, and 45 sec at 72°C. See Supplemental Table S7 for primer sequences.
Purification of ribonucleoprotein (RNP) complexes and subsequent cloning of vU1 snRNAs
Anti-IgM (Sigma; A-4540) or anti-Sm (abcam; ab3138) antibodies were incubated overnight at 4°C with Protein G-agarose. Total cell extracts from 1 × 107 HeLa cells or 1 × 107 hESCs were prepared as follows. Confluent monolayers of cells, in 9-cm dishes, were washed twice in ice-cold PBS and twice in ice-cold TBS (150 μM NaCl, 40 μM Tris-HCl at pH7.4). The cell pellet was resuspended in 1 mL ice-cold NET-2 buffer (150 μM NaCl, 0.05% NP-40, 50 μM Tris-HCl at pH7.4), sonicated on ice—three times for 30 sec with 1-min intervals, followed by centrifugation at 15,000 rpm at 4°C for 15 min. U1-RNPs were isolated from the cleared lysate by pre-incubation with the antibody/agarose suspension overnight at 4°C. Beads were washed, treated with DNase 1 and Proteinase K and extracted with phenol. RNA was recovered by precipitation.
To facilitate cloning, a poly(A) tailing kit (Ambion) was used to add a poly(A) tail to the 3′ end of the purified RNA, according to manufacturers' instructions. cDNA was generated by reverse transcription using Superscript III (Invitrogen) and oligo dT(12-20)(Roche) primers, according to manufacturers' instructions. U1 snRNA sequences were PCR amplified using gene-specific U1 snRNA or vU1 snRNA coding primers (forward) together with an oligo dT reverse primer (Supplemental Table S6). The resulting amplicons were cloned into the pCR 2.1 TOPO cloning vector (Invitrogen) and sequenced.
U1-70K, U1-A, and U1-C immunoprecipitation were carried out as follows: 300 μL of monoclonal hybridoma supernatant (kindly provided by Cindy Will and Reinhard Luhrmann), 1 μg of U1-A (abcam), or 1 μg U1-C (abcam) were incubated with 10 μL of Protein G Dynabeads in a total volume of 300 μL of ice-cold NET-2 buffer for 1 h at 4°C rotating. Following two washes in ice-cold NET-2 buffer, 25 μL of HeLa nuclear extract (CilBiotech) were added to the Dynabead-antibody complexes in a total volume of 500 μL of ice-cold NET-2 buffer, and reincubated for a further 3 h at 4°C rotating. Beads were washed six times in ice-cold NET-2 buffer, RNA extracted with TRIzol, treated with DNase 1, and re-extracted with phenol. RNA was recovered by precipitation. U1 and vU1 snRNA sequences were PCR amplified using gene-specific primers.
The immunoprecipitation of trimethyl capped U1 and vU1 snRNAs were carried out as outlined above for the U1-70K immunoprecipitation with the exception that total RNA from HeLa extract (10 μg) was used. The anti-m3 antibody (R1131) was kindly received from Cindy Will and Reinhard Luhrmann.
Affymetrix human exon ST 1.0 array analysis
Array processing was carried out by Source Bioscience Plc. Data from four biological repeats (each for vU1.8 snRNA knock-down and control) were processed by Affymetrix Power tools (http://www.affymetrix.com) using DABG and RMA algorithms for background correction and quantile normalization. Signal values were expressed in log2-scale. Individual probe sets were called “present” if detected above background (P-value < 0.01) in one group in three or more repeats and “differential” when mean signal differed by more than 1 (approximately twofold change in linear scale) with a t-test P value of <0.01 between the groups. The transcript was called “expressed” if more than two-thirds of probes overlapping its annotated exons were “present” and “differentially expressed” when their mean signal changed by more than 1 (approximately twofold) with a t-test P-value of <0.01 between the groups.
Splicing index analysis was performed as in Exon Array Analyzer (Gellert et al. 2007), with minor modifications. To generate the average profile of nascent transcription across genes (Fig. 6B), signals from “present” probes in intronic regions were averaged according to the distance from the annotated transcription start site (TSS) and plotted in 1-kb bins. Only a subset of transcripts, which are expressed, nonoverlapping, and longer than 10 kb (3889 genes) was used for this analysis. See Supplemental Table S9 for primer sequences used in validation experiments.
Gene annotations
All analysis was performed using RefSeq annotation of NCBI36 (hg18) human genome assembly, downloaded from UCSC Genome Browser (http://genome.ucsc.edu/). For genes with multiple transcript annotations, only the first transcript is presented in analysis outputs. GeneOntology term enrichment analysis (Supplemental Table S4) was performed using the DAVID functional annotation tool (Huang et al. 2009).
vU1 snRNA quantification by RNA-seq
To find out whether stable vU1 snRNAs can be detected (Fig. 2B) in RNA-seq experiments, we analyzed HeLa cell small RNA sequencing data from Fejes-Toth et al. (2009), downloaded from the GEO archive (accession no. GSE14262). Reads from six sequencing experiments were mapped to the human genome using Bowtie aligner, allowing mapping to multiple locations, but no sequence mismatch. Reads matching with 0 or 1 (to count for the potential sequencing errors) mismatch to U1 snRNA genes were subsequently removed from vU1 snRNA counts, thus potentially underestimating vU1 snRNA abundance. Considering mapability as well as sequencing biases, the resulting values need to be understood as very rough estimates. New algorithms dealing with sequencing error-rates and mapability of individual variants are currently being developed, which will allow more precise vU1 snRNA profiling using RNA-seq in different cell-lines.
Data access
The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE37623. GenBank (http://www.ncbi.nlm.nih.gov/genbank/) accession numbers assigned to the vU1 snRNA genes are: vU1.2 JQ988795, vU1.2a JQ988796, vU1.7 JQ988801, vU1.8 JQ988802, vU1.9 JQ988803, vU1.11 JQ988805, vU1.13 JQ988807, vU1.18 JQ988812.
Acknowledgments
D.O'R., M.D., S.M., and W.S.J. were supported by grants from the Wellcome Trust. Marek Drożdż was supported by a FEBS summer fellowship, and S.A.C. by a Wellcome Trust Career Re-entry Fellowship and the Oxford Martin School. We are grateful to Fernando Martinez for help with bioinformatic analysis.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.142968.112.
Freely available online through the Genome Research Open Access option.
References
- Balakirev ES, Ayala FJ 2003. Pseudogenes: Are they “junk” or functional DNA? Annu Rev Genet 37: 123–151 [DOI] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, et al. 2012. U1 snRNP determines mRNA length and regulates isoform expression. Cell 150: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein LB, Manser T, Weiner AM 1985. Human U1 small nuclear RNA genes: Extensive conservation of flanking sequences suggests cycles of gene amplification and transposition. Mol Cell Biol 5: 2159–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Lullo DJ, Ma E, Celniker SE, Rio DC, Doudna JA 2005. Identification and analysis of U5 snRNA variants in Drosophila. RNA 11: 1473–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Lund E, Kahan BW, Dahlberg JE 1997. Control of mouse U1 snRNA gene expression during in vitro differentiation of mouse embryonic stem cells. Nucleic Acids Res 25: 2197–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiefari E, Iiritano S, Paonessa F, Le Pera I, Arcidiacono B, Filocamo M, Foti D, Liebhaber SA, Brunetti A 2010. Pseudogene-mediated posttranscriptional silencing of HMGA1 can result in insulin resistance and type 2 diabetes. Nat Commun 1: 1–7 [DOI] [PubMed] [Google Scholar]
- Cowan CA, Klimanskaya I, McMahon J, Atienza J, Witmyer J, Zucker JP, Wang S, Morton CC, McMahon AP, Powers D, et al. 2004. Derivation of embryonic stem-cell lines from human blastocysts. N Engl J Med 350: 1353–1356 [DOI] [PubMed] [Google Scholar]
- Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J 2008. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell 29: 271–278 [DOI] [PubMed] [Google Scholar]
- Denison RA, Weiner AM 1982. Human U1 RNA pseudogenes may be generated by both DNA- and RNA-mediated mechanisms. Mol Cell Biol 2: 815–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison RA, Van Arsdell SW, Bernstein LB, Weiner AM 1981. Abundant pseudogenes for small nuclear RNAs are dispersed in the human genome. Proc Natl Acad Sci 78: 810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Rosbash M 2002. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 419: 86–90 [DOI] [PubMed] [Google Scholar]
- Du H, Tardiff DF, Moore MJ, Rosbash M 2004. Effects of the U1C L13 mutation and temperature regulation of yeast commitment complex formation. Proc Natl Acad Sci 101: 14841–14846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery P, Marcaillou C, Sahbatou M, Labalme A, Chastang J, Touraine R, Tubacher E, Senni F, Bober MB, Nampoothiri S, et al. 2011. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science 332: 240–243 [DOI] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S 2007. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318: 1777–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Murphy S 2008. Expression of human snRNA genes from beginning to end. Biochem Soc Trans 36: 590–594 [DOI] [PubMed] [Google Scholar]
- Fejes-Toth K, Satirova V, Sachidanandam R, Assof G, Hannon GJ, Kapranov P, Foissac S, Willingham AT, Duttaupta R, Dumais E, et al. 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457: 1028–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes DJ, Kirschner MW, Caput D, Dahlberg JE, Lund E 1984. Differential expression of multiple U1 small nuclear RNAs in oocytes and embryos of Xenopus laevis. Cell 38: 681–689 [DOI] [PubMed] [Google Scholar]
- Fortes P, Cuevas Y, Guan F, Liu P, Pentlicky S, Jung SP, Martinez-Chantar ML, Prieto J, Rowe D, Gunderson SI 2003. Inhibiting expression of specific genes in mammalian cells with 5′ end-mutated U1 small nuclear RNAs targeted to terminal exons of pre-mRNA. Proc Natl Acad Sci 100: 8264–8269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert P, Uchida S, Braun T 2007. Exon Array Analyzer: a web interface for Affymetrix exon array analysis. Bioinformatics 25: 3323–3324 [DOI] [PubMed] [Google Scholar]
- Gunderson SI, Polycarpou-Schwarz M, Mattaj IW 1998. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell 1: 255–264 [DOI] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res 22: 1760–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Liyanarachchi S, Akagi K, Nagy R, Li J, Dietrich RC, Li W, Sebastian N, Wen B, Xin B, et al. 2011. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science 332: 238–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs V, Bach M, Winkelmann G, Luhrmann R 1990. U1-specific protein C needed for efficient complex formation of U1 snRNP with a 5′ splice site. Science 247: 69–72 [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Jia Y, Mu JC, Ackerman SL 2012. Mutation of a U2 snRNA gene causes global disruption of alternative splicing and neurodegeneration. Cell 148: 296–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson DR, Wu YM, Cao X, Asangani IA, Kothari V, Prensner JR, Lonigro RJ, et al. 2012. Expressed pseudogenes in the transcriptional landscape of human cancers. Cell 149: 1622–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, St Laurent G 2012. Dark Matter RNA: Existence, function, and controversy. Front Genet 3: 60 doi: 10.3389/fgene.2012.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneev SA, Park JH, O'Shea M 1999. Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J Neurosci 19: 7711–7720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwek KY, Murphy S, Furger A, Thomas B, O'Gorman W, Kimura H, Proudfoot NJ, Akoulitchev A 2002. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat Struct Biol 9: 800–805 [DOI] [PubMed] [Google Scholar]
- Kyriakopoulou C, Larsson P, Liu L, Schuster J, Soderbom F, Kirsebom LA, Virtanen A 2006. U1-like snRNAs lacking complementarity to canonical 5′ splice sites. RNA 12: 1603–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren V, Bernstein LB, Weiner AM, Francke U 1985. Human U1 small nuclear RNA pseudogenes do not map to the site of the U1 genes in 1p36 but are clustered in 1q12-q22. Mol Cell Biol 5: 2172–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo PC, Mount SM 1990. Drosophila melanogaster genes for U1 snRNA variants and their expression during development. Nucleic Acids Res 18: 6971–6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E 1988. Heterogeneity of human U1 snRNAs. Nucleic Acids Res 16: 5813–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE 1984. True genes for human U1 small nuclear RNA. Copy number, polymorphism, and methylation. J Biol Chem 259: 2013–2021 [PubMed] [Google Scholar]
- Lund E, Kahan B, Dahlberg JE 1985. Differential control of U1 small nuclear RNA expression during mouse development. Science 229: 1271–1274 [DOI] [PubMed] [Google Scholar]
- Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, et al. 2008. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med 359: 1685–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro EM, Mah N, Andrade-Navarro MA 2011. Functional evidence of post-transcriptional regulation by pseudogenes. Biochimie 93: 1916–1921 [DOI] [PubMed] [Google Scholar]
- Naylor SL, Zabel BU, Manser T, Gesteland R, Sakaguchi AY 1984. Localization of human U1 small nuclear RNA genes to band p36.3 of chromosome 1 by in situ hybridization. Somat Cell Mol Genet 10: 307–313 [DOI] [PubMed] [Google Scholar]
- O'Gorman W, Thomas B, Kwek KY, Furger A, Akoulitchev A 2005. Analysis of U1 small nuclear RNA interaction with cyclin H. J Biol Chem 280: 36920–36925 [DOI] [PubMed] [Google Scholar]
- O'Reilly D, Quinn CM, El-Shanawany T, Gordon S, Greaves DR 2003. Multiple Ets factors and interferon regulatory factor-4 modulate CD68 expression in a cell type-specific manner. J Biol Chem 278: 21909–21919 [DOI] [PubMed] [Google Scholar]
- Patton JG, Wieben ED 1987. U1 precursors: Variant 3′ flanking sequences are transcribed in human cells. J Cell Biol 104: 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelitz T, Bailey AD, Elco CP, Weiner AM 2008. Human U2 snRNA genes exhibit a persistently open transcriptional state and promoter disassembly at metaphase. Mol Cell Biol 28: 3573–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465: 1033–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K 2009. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458: 475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca X, Krainer AR 2009. Recognition of atypical 5′ splice sites by shifted base-pairing to U1 snRNA. Nat Struct Mol Biol 16: 176–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosel TD, Hung LH, Medenbach J, Donde K, Starke S, Benes V, Ratsch G, Bindereif A 2011. RNA-Seq analysis in mutant zebrafish reveals role of U1C protein in alternative splicing regulation. EMBO J 30: 1965–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago C, Marzluff WF 1989. Expression of the U1 RNA gene repeat during early sea urchin development: Evidence for a switch in U1 RNA genes during development. Proc Natl Acad Sci 86: 2572–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauterer RA, Feeney RJ, Zieve GW 1988. Cytoplasmic assembly of snRNP particles from stored proteins and newly transcribed snRNA's in L929 mouse fibroblasts. Exp Cell Res 176: 344–359 [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M 2007. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem 76: 75–100 [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ, Steitz JA 1992. Three novel functional variants of human U5 small nuclear RNA. Mol Cell Biol 12: 734–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiluttini B, Gu B, Belagal P, Smirnova AS, Nguyen VT, Hebert C, Schmidt U, Bertrand E, Darzacq X, Bensaude O 2010. Splicing-independent recruitment of U1 snRNP to a transcription unit in living cells. J Cell Sci 123: 2085–2093 [DOI] [PubMed] [Google Scholar]
- Torrents D, Suyama M, Zdobnov E, Bork P 2003. A genome-wide survey of human pseudogenes. Genome Res 13: 2559–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorlova S, Rocco G, Lefave CV, Jodelka FM, Hess K, Hastings ML, Henke E, Cartegni L 2011. Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol Cell 43: 927–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wickrema A, Bondurant MC, Krantz SB 1991. Abundance and stability of erythropoietin receptor mRNA in mouse erythroid progenitor cells. Blood 78: 2269–2275 [PubMed] [Google Scholar]
- Will CL, Luhrmann R 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 13: 290–301 [DOI] [PubMed] [Google Scholar]
- Wu JQ, Habegger L, Noisa P, Szekely A, Qiu C, Hutchison S, Raha D, Egholm M, Lin H, Weissman S, et al. 2010. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. Proc Natl Acad Sci 107: 5254–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto ML, Clark TA, Gee SL, Kang JA, Schweitzer AC, Wickrema A, Conboy JG 2009. Alternative pre-mRNA splicing switches modulate gene expression in late erythropoiesis. Blood 113: 3363–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JB, Murphy S, Bai L, Wang Z, Roeder RG 1995. Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol Cell Biol 15: 2019–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Frankish A, Baertsch R, Kapranov P, Reymond A, Choo SW, Lu Y, Denoeud F, Antonarakis SE, Snyder M, et al. 2007. Pseudogenes in the ENCODE regions: Consensus annotation, analysis of transcription, and evolution. Genome Res 17: 839–851 [DOI] [PMC free article] [PubMed] [Google Scholar]