Abstract
In germ cells, early embryos, and stem cells of animals, PIWI-interacting RNAs (piRNAs) have an important role in silencing retrotransposons, which are vicious genomic parasites, through transcriptional and post-transcriptional mechanisms. To examine whether the piRNA pathway can be used to silence genes of interest in germ cells, we have generated knock-in mice in which a foreign DNA fragment was inserted into a region generating pachytene piRNAs. The knock-in sequence was transcribed, and the resulting RNA was processed to yield piRNAs in postnatal testes. When reporter genes possessing a sequence complementary to portions of the knock-in sequence were introduced, they were greatly repressed after the time of pachytene piRNA generation. This repression mainly occurred at the post-transcriptional level, as degradation of the reporter RNAs was accelerated. Our results show that the piRNA pathway can be used as a tool for sequence-specific gene silencing in germ cells and support the idea that the piRNA generating regions serve as traps for retrotransposons, enabling the host cell to generate piRNAs against active retrotransposons.
Approximately 40% of the mouse genome consists of retrotransposons, a class of transposable elements replicating via RNA intermediates (Waterston et al. 2002). Although transposable elements may contribute to genome evolution (Kazazian 2004), they can disrupt genes at their insertion sites (Han et al. 2004). To secure the genome of future generations from de novo insertion mutations, transposable elements should be tightly controlled in germ cells. The PIWI family proteins and their associated small RNAs, called PIWI-interacting RNAs (piRNAs), constitute an evolutionarily conserved defense system to control transposable elements in germ cells (Klattenhoff and Theurkauf 2008; Siomi et al. 2011; Pillai and Chuma 2012). In Drosophila, zebrafish, and Xenopus, a majority of piRNAs has sequences corresponding to transposable elements including retrotransposons (Vagin et al. 2006), and mutations in genes encoding the piRNA pathway components result in their derepression. The PIWI-piRNA complexes recognize their target RNAs based on sequence complementarity and cause RNA degradation (post-transcriptional gene silencing or PTGS) (Lim et al. 2009; De Fazio et al. 2011; Reuter et al. 2011). In fetal mouse testes, the complexes also repress retrotransposons transcriptionally by introducing DNA methylation (transcriptional gene silencing or TGS) (Kuramochi-Miyagawa et al. 2008).
Two piRNA biogenesis pathways exist, namely the primary and secondary pathways (Aravin et al. 2007a; Kim et al. 2009). In the primary pathway, piRNAs are derived from genomic regions termed piRNA clusters, of which size ranges from 1 to 100 kb in mice. These regions are transcribed into long single-stranded precursor RNAs, and they are processed to yield piRNAs by a yet-unidentified enzyme(s) (Kim et al. 2009). These primary piRNAs are predominantly antisense with respect to the open reading frames of retrotransposon-derived RNAs (Brennecke et al. 2007). Then, long sense RNAs derived from retrotransposons are cleaved presumably with the slicer activity of the PIWI proteins using the primary piRNAs as guides, and this results in the generation of secondary piRNAs (the secondary pathway) (Brennecke et al. 2007; Gunawardane et al. 2007). This eventually constitutes the “ping-pong cycle” that specifically amplifies the piRNAs derived from active retrotransposons.
In fetal mouse testes, prospermatogonia express piRNAs of 25–28 nucleotides (nt) in length, which we call prenatal piRNA (also called prepachytene piRNA). They are derived from retrotransposon-rich regions and loaded onto either of the two PIWI proteins, PIWIL2 (also known as MILI) or PIWIL4 (MIWI2) (Kuramochi-Miyagawa et al. 2001; Kuramochi-Miyagawa et al. 2004; Carmell et al. 2007; Aravin et al. 2008). Targeted disruptions of the two genes resulted in decreased DNA methylation in retrotransposon sequences and increased levels of their RNAs (Aravin et al. 2007b, 2008; Kuramochi-Miyagawa et al. 2008). In postnatal testes, meiotic spermatocytes and round spermatids express piRNAs of 25–32 nt in length, which are called pachytene piRNAs (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006a; Lau et al. 2006; Watanabe et al. 2006). They interact with PIWIL2 or PIWIL1 (MIWI) (Kuramochi-Miyagawa et al. 2001; Deng and Lin 2002) and have diverse sequences that are mapped in clusters in unannotated regions. PIWIL1 is localized to perinuclear structures called chromatoid bodies (Grivna et al. 2006b; Kotaja et al. 2006; Aravin et al. 2009; Chuma et al. 2009), which are the presumed sites of PTGS, analogous to the processing bodies in somatic cells (Liu et al. 2005). However, since only 17%–20% of the total pachytene piRNAs corresponds to retrotransposons (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006a; Lau et al. 2006; Watanabe et al. 2006), their role in retrotransposon silencing is less clear. A partial deletion of a pachytene piRNA cluster, however, was reported to result in increased expression of LINE-1 ORF1 proteins (Xu et al. 2008). A more recent study showed that disruption of PIWIL1 catalytic activity causes accumulation of LINE1 retrotransposon transcripts in postnatal germ cells (Reuter et al. 2011).
RNA interference (RNAi) is a mechanism by which double-stranded RNAs (dsRNAs) silence genes in a sequence-specific manner. It uses the microRNA/small interfering RNA (miRNA/siRNA) pathway of the host cells. To silence or knockdown genes of interest, long dsRNAs, siRNAs, or hairpin RNA expression cassettes, having sequences homologous or complementary to the target sequences, are introduced into cells (Fire et al. 1998). They enter the RNAi pathway, and the resulting small RNAs cause target-specific PTGS. However, targeted gene silencing by the piRNA pathway has not been reported. In this study, we have generated mice carrying a foreign DNA insertion in a pachytene piRNA cluster and show that piRNAs generated from this fragment silence reporter genes in a sequence-specific manner at the post-transcriptional level. Our results show that the piRNA pathway can be used for targeted PTGS and support the idea that piRNA cluster regions serve as a trap for transposable elements (Girard and Hannon 2008), providing an opportunity for host cells to generate complementary piRNAs.
Results
Generation of EGFP-neo knock-in mice
A major pachytene piRNA-generating region lies on mouse chromosome 17 (chr. 17), which contains a bidirectional promoter and expresses two divergent piRNA precursors (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006a; Lau et al. 2006; Watanabe et al. 2006). Thus, piRNAs mapped in this region form two clusters on different strands (the Watson cluster and the Crick cluster). To examine whether a foreign DNA can serve as a source for piRNAs, we introduced a DNA fragment containing both EGFP and neo genes into the Watson cluster. We expected that the knock-in sequence would be used to generate piRNAs that are distinguishable from the endogenous piRNAs. Here, EGFP was inserted in an orientation identical to the Watson precursor RNA, and neo was inserted in an opposite orientation (Supplemental Fig. S1A). Thus, EGFP and neo would serve as sources for sense and antisense piRNAs, respectively. Correct targeting was confirmed in embryonic stem (ES) cells by Southern blotting (Supplemental Fig. S1B). Chimeric mice were generated with the targeted ES cells and then germline transmission was confirmed. Male mice heterozygous for EGFP-neo (EGFP-neo/+) were fertile with no discernible defect in spermatogenesis (Supplemental Fig. S1C) and thus were used for further studies.
Interestingly, we never observed mice or embryos of EGFP-neo/EGFP-neo genotype in heterozygous crosses (Supplemental Table S1). It is possible that the homozygotes are early embryonic lethal, perhaps before implantation. However, the ratio of mice with heterozygotes versus wild-type was 4 to 1 in heterozygous crosses (2 to 1 is expected) (Supplemental Table S1). Therefore, we now consider other possibilities, such as the duplication of the chr17 piRNA generating region in J1 ES cells: In this case, the homozyotes may be viable but misidentified as heterozygotes because of the existence of one extra copy of the wild-type region. This is consistent with the report that the rate of piRNA cluster expansion is higher than that of any known gene family (Assis and Kondrashov 2009). A more detailed study is now ongoing to resolve this issue.
Biogenesis of primary and secondary piRNAs from EGFP-neo
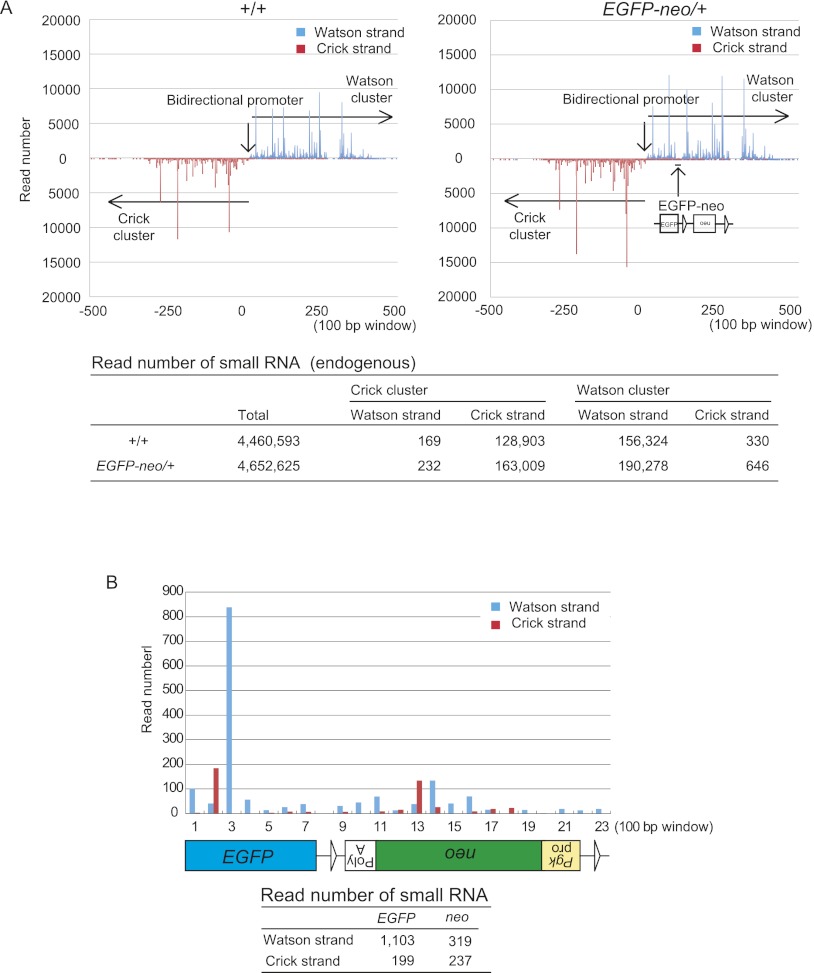
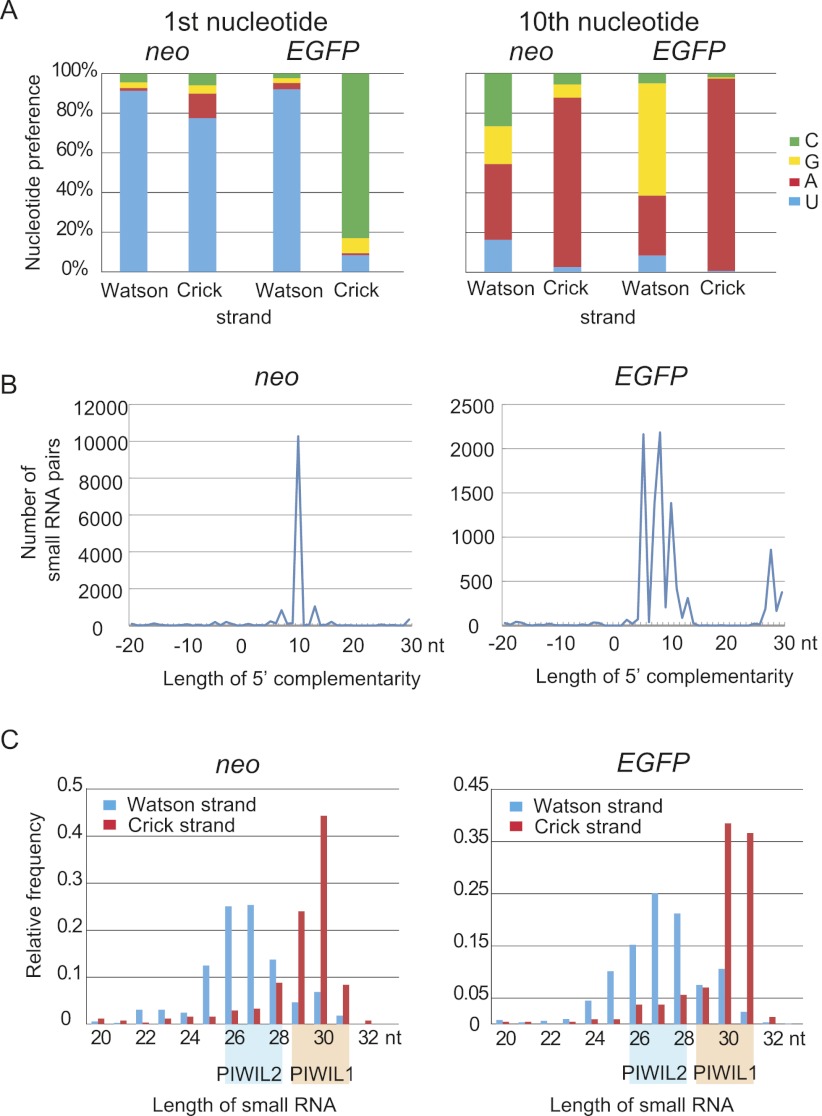
To examine whether piRNAs are generated from EGFP-neo inserted into the Watson cluster, small RNA libraries were prepared from EGFP-neo/+ and wild-type testes at 5 wk after birth and subjected to high-throughput sequencing. The distribution and frequency of the endogenous piRNAs from both upstream and downstream regions of the knock-in site were not detectably altered in EGFP-neo/+ testes (Fig. 1A; Supplemental Fig. S2A), suggesting that piRNA precursor transcription is not affected by the insertion. We detected small RNAs from EGFP-neo with the features of piRNA (see below) (Fig. 1B; Supplemental Tables S2, S3). Contrary to our initial expectation, however, the piRNAs from EGFP-neo were mapped on both strands (Fig. 1B), suggesting that the knock-in sequence is transcribed in both orientations. The piRNAs mapped on the Watson strand tended to have uridine at the first position, which is a feature of the primary piRNA, and those mapped on the Crick strand often had adenine at the 10th position, a feature of the secondary piRNA (Fig. 2A; Supplemental Table S3). These results suggest that the Watson strand piRNAs are the primary piRNAs, whereas the Crick strand piRNAs are the secondary piRNAs. Furthermore, the Watson and Crick strand piRNAs frequently showed 10-nt complementarity in their 5′ portions (Fig. 2B), suggesting that the ping-pong cycle is active. The length distribution of the piRNAs was bimodal (Fig. 2C) and, based on the previous reports, we speculated that the peak at 27 nt corresponds to PIWIL2-bound piRNA and the peak at 30 nt to PIWIL1-bound piRNA (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006a; Lau et al. 2006; Watanabe et al. 2006). The Watson strand piRNAs (EGFP sense and neo antisense) were more likely PIWIL2-bound, whereas the Crick strand piRNAs (EGFP antisense and neo sense) were likely PIWIL1-bound. We speculate that this preference of incorporation was made dependent on the expression timing of PIWIL2 and PIWIL1. PIWIL2 appears first and acquires primary piRNAs from the chr17 piRNA precursor, and PIWIL1 comes later and acquires secondary piRNAs from the transcript driven by the Pgk1 promoter.
Figure 1.
Identification of small RNAs generated from the EGFP-neo knock-in sequence. A small RNA library prepared from 5-wk-old EGFP-neo/+ testes was subjected to high-throughput sequencing. (A) Distribution of endogenous small RNA sequences across the Watson and Crick clusters (mm9 chr.17: 27,409,752-27,511,260) in wild-type control (+/+) and EGFP-neo/+ testes. The data is shown for 100-bp windows across the region (top). The precise read numbers are also shown (bottom). The overall patterns in the wild-type and EGFP-neo+/– testes were very similar. (B) Distribution of small RNA sequences mapped within EGFP-neo. The small RNA read number for each 100-bp window is shown across the EGFP-neo fragment (top). The total read number for each strand of each gene is also shown (bottom).
Figure 2.
Small RNAs from EGFP-neo have features of piRNA. (A) Nucleotide preference at the first and 10th positions. Note that Watson strand small RNAs are enriched for U at the first position (a feature of the primary piRNA), and Crick strand small RNAs for A at the 10th position (a feature of the secondary piRNA). (B) Frequent 10-nt complementarity between the 5′ regions of Watson and Crick strand piRNAs. Small RNA pairs with 5′ complementarity for indicated lengths are counted. If eight reads of Watson strand small RNA and five reads of Crick strand small RNA had the same complementary sequences, it was counted as 8 × 5 = 40. The prevailing 10-nt complementarity suggests an active ping-pong cycle. (C) Size distributions of small RNAs from different strands. The Watson strand piRNAs are 25–28 nt in length and presumably bound by PIWIL2, and the Crick strand piRNAs are 29–32 nt in length and presumably bound by PIWIL1.
In both EGFP-neo/+ and wild-type testes, we obtained evidence for an active ping-pong cycle for the endogenous piRNAs throughout the Watson and Crick clusters (Supplemental Fig. S3). Since the ratio of the piRNAs from the Crick strand to those from the Watson strand was higher for EGFP-neo (see above) than for the endogenous Watson cluster (Supplemental Fig. S2B), the Pgk1 promoter (Fig. 1A,B) may have driven transcription of neo and EGFP for generation of the secondary piRNAs.
Gene repression by the knock-in sequences
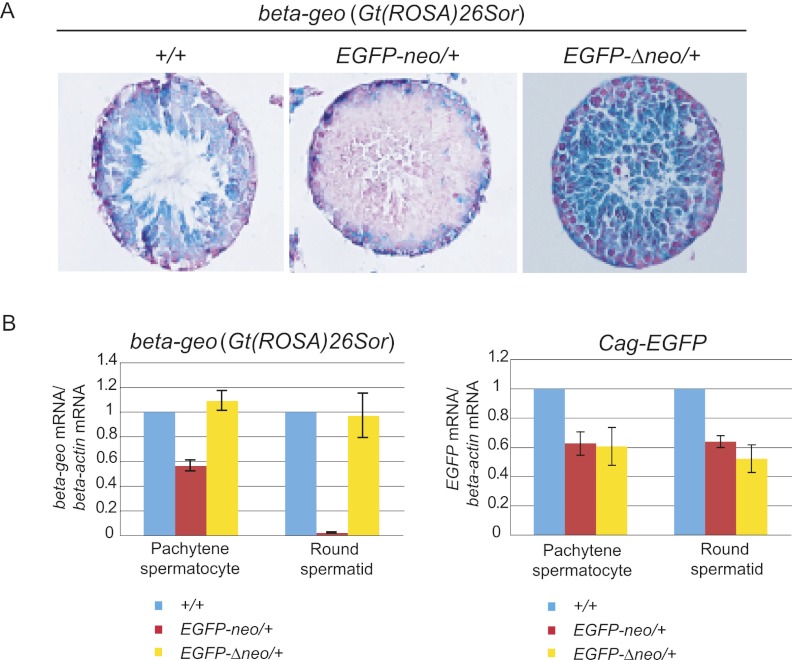
To examine whether the piRNAs generation from the knock-in sequence can cause gene repression, we introduced reporter genes with a sequence homology with EGFP-neo. For this, EGFP-neo/+ mice were crossed with Gt(ROSA)26Sor (carrying beta-geo, a fusion gene comprising both Glb1 and neo) (Zambrowicz et al. 1997) or Cag-EGFP transgenic mice (Kawamoto et al. 2000). We examined the expression of the reporter genes in EGFP-neo/+ testes at 5 wk after birth by X-gal staining (beta-geo) or immunostaining (EGFP). Whereas beta-geo expression was observed in a range of germ cells from spermatogonia to elongated spermatid in control (+/+) testes, it was not detected in round spermatids or more differentiated germ cells in EGFP-neo/+ testes (Fig. 3A). This suggests that gene repression occurred after the stage of pachytene piRNA expression. Down-regulation of EGFP was not evident in EGFP-neo/+ testes by immunostaining under the condition that we used (data not shown) (but see below).
Figure 3.
Repression of reporter genes by the EGFP-neo knock-in sequence. (A) Activity of beta-geo [Gt(ROSA)26Sor] monitored by X-gal staining (blue). Testes were obtained at 5 wk after birth from Gt(ROSA)26Sor mice with either EGFP-neo or EGFP-Δneo, or without these sequences. Tissue sections were counterstained with nuclear fast red (red). Note that beta-geo is repressed only in the presence of neo at the knock-in locus and that the repression is observed in spermatocytes and spermatids, but not in spermatogonia or Sertoli cells (located in the periphery of seminiferous tubules). (B) qRT-PCR of beta-geo and EGFP mRNAs. Germ cells were isolated from Gt(ROSA)26Sor and Cag-EGFP mice with either EGFP-neo or EGFP-Δneo, or without these sequences. The level of beta-actin mRNA was used as a reference. Control (+/+) samples were obtained from the same litters. Error bars indicate the standard deviations (SDs) of five biological replicates.
For more accurate evaluation, we collected pachytene spermatocytes and round spermatids from EGFP-neo/+ and control testes by fluorescence-activated cell sorting (FACS) (Supplemental Fig. S4) and performed Western blot analyses using antibodies against GLB1 and EGFP proteins. Consistent with the results from the histological studies, the level of beta-geo was greatly reduced in round spermatids from EGFP-neo/+ testes while that of EGFP showed a very small reduction (Supplemental Fig. S5). Pachytene spermatocytes showed almost no change in levels of the reporter proteins (Supplemental Fig. S5). Quantitative RT-PCR (qRT-PCR) assays showed that the level of beta-geo mRNA is dramatically reduced (about 50-fold) in round spermatids and, to a lesser extent, in pachytene spermatocytes from EGFP-neo/+ testes (Fig. 3B). This milder reduction in the level of beta-geo mRNA could be due to the presence of not enough functional PIWIL2 and/or PIWIL1 proteins in early pachytene spermatocytes, as they are not produced in the previous stages. This is supported by the fact that overexpression of PIWIL2 in pachytene spermatocytes enhanced repression (see below). The assays also showed that EGFP mRNA is reduced in EGFP-neo/+ pachytene spermatocytes and round spermatids (Fig. 3B). This is most likely due to the unexpected generation of the piRNAs complementary to EGFP mRNA, as described above (Fig. 1A; Supplemental Tables S2, S3).
To confirm that the observed repression is, indeed, due to the knock-in sequence, we deleted the Pgk-neo sequence flanked by two loxP sites by crossing with Cag-Cre mice (Supplemental Fig. S1A). Both X-gal staining and qRT-PCR showed that beta-geo expression has been recovered in EGFP-Δneo pachytene spermatocytes and round spermatids (Fig. 3A). As expected, expression of EGFP was not changed by the deletion, confirming that the repression is due to the presence of a homologous or complementary sequence in the knock-in locus.
Involvement of the piRNA pathway in repression
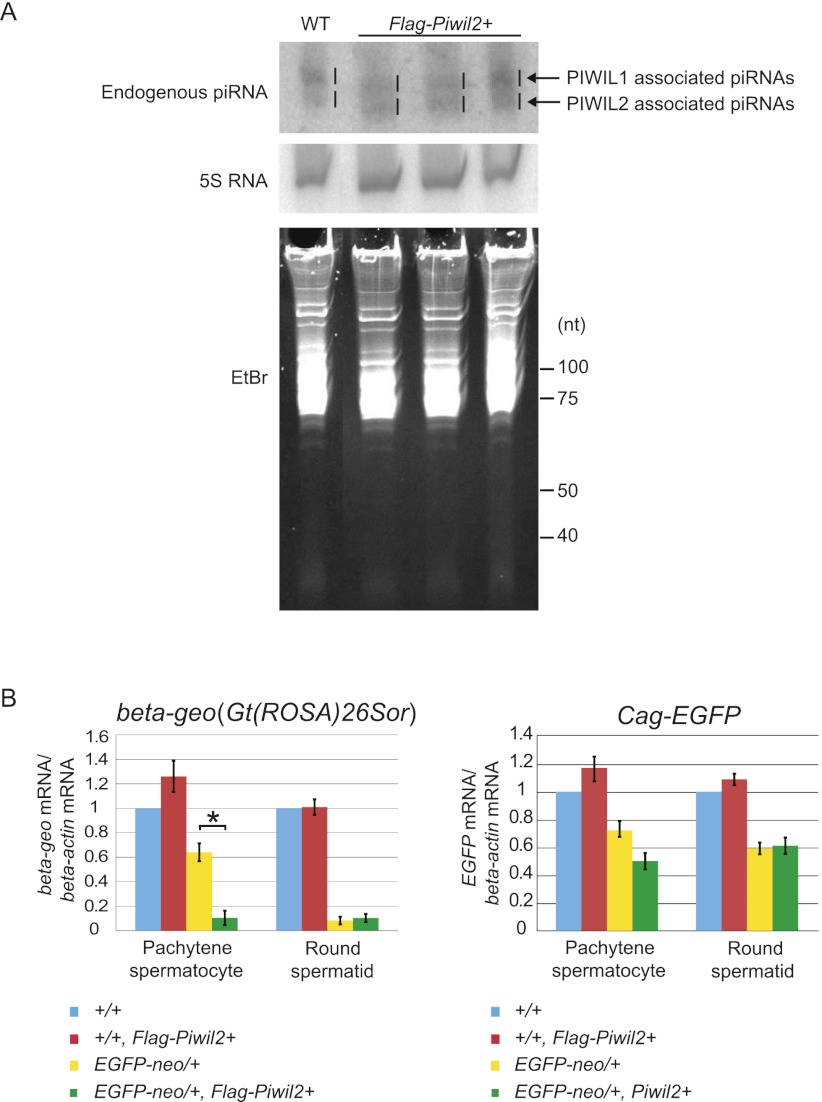
We next attempted to examine whether the repression of the reporter genes involves the piRNA pathway. Since mice with a mutation in any piRNA pathway component gene show spermatogenesis failure, they are not appropriate for such a study. We, therefore, overexpressed a tagged PIWIL2 protein in EGFP-neo/+ mice and examined whether it has an effect on the repression activity. We generated Flag-Piwil2 transgenic mice, in which the transgene carried by a bacterial artificial chromosome (BAC) was driven by the canonical Piwil2 promoter (Supplemental Fig. S6A). In these mice, the level of total PIWIL2 proteins, including both the endogenous and tagged one, was elevated (Supplemental Fig. S6B), and expression of FLAG-PIWIL2 was highest in pachytene spermatocytes just as that of endogenous PIWIL2 (Supplemental Fig. S6C). Furthermore, the transgene successfully rescued the meiotic arrest phenotype of the Piwil2 knockout mice (Supplemental Fig. S6D). A Northern blot analysis showed that an endogenous piRNA from the Watson cluster was increased in the Flag-Piwil2 testes (Fig. 4A). When the Flag-Piwil2 transgene was introduced into EGFP-neo/+ mice with either Gt(ROSA)26Sor (beta-geo) or Cag-EGFP reporter genes by crossing, the reduction in beta-geo mRNA expression caused by EGFP-neo was significantly enhanced in pachytene spermatocytes (Fig. 4B). The enhancement of the repression was not observed in round spermatids, perhaps due to a lower expression of Flag-Piwil2 (Supplemental Fig. S6C) or already saturated repression activity in this cell type. Similar but not significant changes were detected for EGFP mRNA, consistent with the fact that the number of piRNAs antisense to EGFP was small. These results suggest that beta-geo gene repression involves PIWIL2 and thus, the piRNA pathway.
Figure 4.
Enhancement of the reporter gene repression by a Flag-Piwil2 transgene. (A) Demonstration of increased PIWIL2-associated piRNAs in Flag-Piwil2 BAC transgenic testes at 5 wk. PIWIL2- and PIWIL1-bound piRNAs are indicated. Northern blotting was performed with a probe detecting a pachytene piRNA from the Watson cluster. 5S RNA and ethidium bromide staining are the loading controls. (B) qRT-PCR of beta-geo [Gt(ROSA)26Sor] and EGFP mRNAs. Germ cells were isolated from Gt(ROSA)26Sor and Cag-EGFP mice with or without EGFP-neo and also with or without Flag-Piwil2. A significant difference in beta-geo expression is observed between EGFP-neo/+ germ cells with or without Flag-Piwil2. The level of beta-actin mRNA was used as a reference. (*) P < 0.05, Student's t-test. Error bars indicate the standard deviations (SDs) of biological replicates.
Repression at the post-transcriptional level
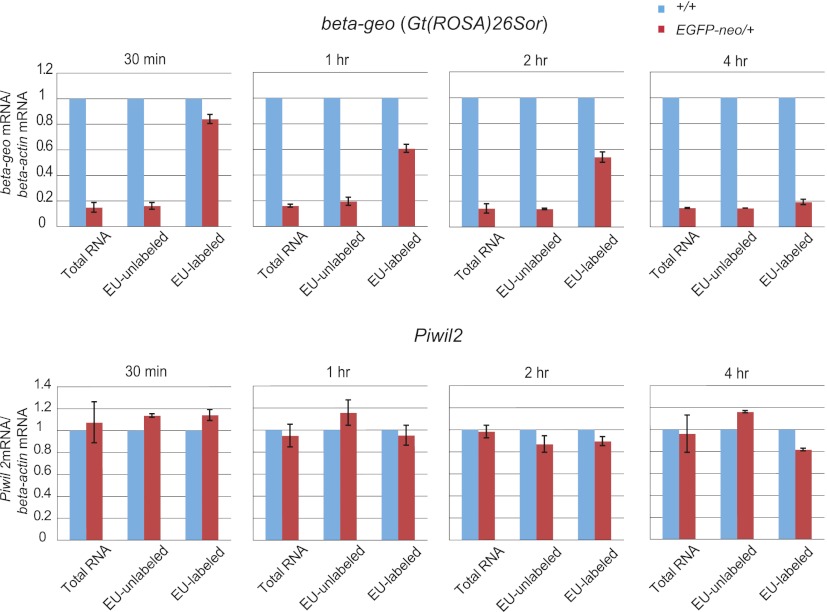
It is generally believed that the PIWI-piRNA complexes regulate gene expression by cleaving target RNAs (PTGS) (Brennecke et al. 2007; Gunawardane et al. 2007; De Fazio et al. 2011; Reuter et al. 2011). In round spermatids, PIWIL2 and PIWIL1 proteins are localized to chromatoid bodies implicated in RNA processing and PTGS (Chuma et al. 2009). To determine whether the observed reporter gene repression is PTGS, we compared the rate of degradation of beta-geo RNA between EGFP-neo+/– and control testicular germ cells. After pulse-labeling of newly synthesized RNAs with 5-ethyluridine (EU), the level of EU-labeled beta-geo mRNA was measured at different time points. At 30 min after withdrawal of EU, the level of EU-labeled beta-geo mRNA was not much different between EGFP-neo/+ and control germ cells (Fig. 5). Then, the level of EU-labeled beta-geo mRNA gradually decreased in EGFP-neo/+ germ cells and, at four hours after EU withdrawal, reached to only 20% of that of control cells (Fig. 5). By contrast, the level of EU-labeled endogenous Piwil2 mRNA (a control) showed no difference between EGFP-neo/+ and control germ cells. These results indicate that the degradation of beta-geo mRNA is accelerated in EGFP-neo/+ testes and suggest that the reporter gene repression occurs, at least in part, through PTGS. Accumulating evidence indicates that the piRNA pathway of the fetal testis is involved in de novo DNA methylation (Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008). Therefore, DNA methylation at the beta-geo transgene was analyzed by bisulfite sequencing. No detectable methylation change was observed at this locus in EGFP-neo/+ testes (Supplemental Fig. S7).
Figure 5.
Reduced stability of beta-geo mRNA in EGFP-neo/+ testes. Newly synthesized RNA was pulse-labeled with EU in testicular germ cells collected at 5 wk after birth, and RNA was isolated for qRT-PCR at the indicated time point after the withdrawal of EU. The level of beta-actin mRNA was used as a reference. Error bars indicate SDs of four biological replicates.
Discussion
In this study, we have introduced a foreign DNA fragment into a pachytene piRNA generating region in mice and showed successful piRNA generation from the knock-in sequence in the testis. This is most likely due to the transcription of the knock-in fragment as a part of the piRNA precursor transcript and provides evidence for the first time that a foreign DNA can be appropriately processed to yield piRNAs in vivo. Furthermore, these piRNAs successfully repressed reporter genes with a sequence homologous to the knock-in sequence in pachytene spermatocytes and spermatids. The repression was orientation-dependent, and the reporter gene complementary to the piRNAs was more effectively repressed, suggesting that it was, indeed, dependent on piRNA generation. Considering the studies on other transgenes in the mouse testis (Wang et al. 2006; Suzuki et al. 2007), we believe that transgene insertions outside of piRNA generating regions would not result in a silencing effect. The repression was, at least in part, due to accelerated degradation of the target RNA (PTGS) and clearly dependent on the piRNA pathway, as overexpression of PIWIL2 enhanced the repression. However, we do not exclude the involvement of TGS at present.
We propose that the piRNA system can be used as a mediator of germ cell-specific gene knockdown. Targeted knock-in of a DNA fragment homologous to the target gene (or RNA) into a piRNA cluster region in an opposite orientation should cause specific knockdown in vivo without a side effect in somatic tissues. When this manuscript was in preparation, Muerdter et al. (2011) reported successful production of artificial piRNAs from BAC transgene in mouse testis, although they did not analyze their effect on target mRNA. Mouse BAC clones containing a piRNA precursor region with its promoter elements will be particularly useful, because recombination in host bacteria combined with pronuclear injection of the BAC transgene can lead to generation of the desired piRNAs in germ cells and quickly provide knockdown mice. Alternatively, if artificially synthesized piRNA precursors (Kawaoka et al. 2011) or piRNA mimics of 23–30 nt in length are effectively introduced by transfection or microinjection into cells with an active piRNA pathway, they could also cause specific silencing. The piRNA-mediated gene knockdown might also be useful in early embryos or oocytes of some animals such as Xenopus since RNAi and miRNA biogenesis are restricted in these species due to the limited amount of the Argonaute proteins (Lund et al. 2011).
Although we used a pachytene piRNA cluster as a source in this study, there are regions generating piRNAs in prospermatogonia in the fetal testis. These prenatal piRNAs are involved not only in PTGS but also in TGS through de novo DNA methylation, because retrotransposons and at least one imprint control region are hypomethylated in Piwil2, Piwil4, and Pld6 (also known as mitoPLD) knockout prospermatogonia (Aravin et al. 2007b; Carmell et al. 2007; Kuramochi-Miyagawa et al. 2008; Watanabe et al. 2011). Thus, in the future, it might become possible to introduce epigenetic marks such as DNA methylation to specific genomic regions in germ cells, which could then be inherited by the progeny. So far, there is no evidence that pachytene piRNAs are involved in TGS, and we did not observe a difference in DNA methylation between EGFP-neo/+ and control germ cells, at beta-geo.
Active retrotransposons increase their copy number via RNA intermediates, and the piRNA pathway provides a defense mechanism against these genomic parasites. Although pachytene piRNAs are relatively depleted of retrotransposons (Watanabe et al. 2006; Reuter et al. 2011), a partial deletion of a pachytene piRNA cluster was reported to derepress LINE-1 ORF1 proteins (Xu et al. 2008). Furthermore, disruption of PIWIL1 catalytic activity causes increased accumulation of LINE1 retrotransposon transcripts (Reuter et al. 2011). It has been proposed that piRNA clusters may serve as a trap for new retrotransposon copies (Girard and Hannon 2008; Khurana et al. 2011), thus providing an opportunity for the host cells to generate piRNAs with sequences identical to, or most similar to, the active copies. Our knock-in insertion of EGFP-neo into a piRNA cluster mimicked a retrotransposon insertion, and the resulting piRNA generation and silencing of the reporter genes support this hypothesis. Furthermore, we observed that beta-geo, but not EGFP, was silenced, most likely because only neo, which has a sequence identical to beta-geo, lay in an orientation opposite to the piRNA precursor transcript and thus is expected to generate antisense piRNAs. Consistent with this, a majority of transposable elements in the piRNA precursors are found to be orientated in an antisense orientation (Aravin et al. 2007a; Brennecke et al. 2007). It is unknown, however, how the promoters of the precursor RNAs are regulated during the course of germ cell development to ensure piRNA generation at the time of retrotransposon expression.
Methods
Generation of EGFP-neo knock-in mice
The knock-in scheme is outlined in Supplemental Figure S1A. Two regions (mm9 chr.17: 27,471,793-27,474,015 and 27,463,197-27,471,823) within the Watson cluster were amplified by polymerase chain reaction (PCR) using KOD Plus High-fidelity Polymerase (Toyobo). The targeting vector was constructed by integrating the two PCR products and an EGFP fragment into a lox-neo diphtheria toxin A vector. The vector was linearized using NotI and introduced into J1 ES cells by electroporation. Targeted ES cells were used to generate chimeric mice, and one of the mice showed germline transmission of the targeted allele. Heterozygous EGFP-neo/+ mice were crossed with CAG-Cre transgenic mice to remove the neo cassette. Genotyping was done by PCR of DNA from tail snips or embryos. Primers used for vector construction and genotyping are listed in Supplemental Table S4.
Generation of Flag-Piwil2 BAC transgenic mice
A BAC clone containing the mouse Piwil2 gene (RP23-366F18) was obtained from Invitrogen. The transgene was constructed as outlined in Supplemental Figure S5A by recombination-based methodology as previously described (Oginuma et al. 2008). Transgeneic mice were generated by pronuclear injection. Primers used for transgene construction and genotyping are listed in Supplemental Table S4.
Small RNA sequencing
Total RNA was extracted from testes using Isogen (Wako) and used to construct small RNA libraries using the Digital Gene Expression for Small RNA Sample Prep Kit (Illumina). The libraries were subjected to high-throughput sequencing using Genome Analyzer II (Illumina). After removing the linker sequences in silico, sequence reads of 20–32 nt in length were extracted. The small RNA sequences were mapped to the mouse genome using SeqMap (Jiang and Wong 2008), and only perfectly matched sequences were used for analysis.
FACS isolation of germ cells
Testes were dissociated in 1 mg/mL collagenase type IV and then in 0.25% trypsin with 1 mM EDTA. The activity of trypsin was terminated by addition of a 10%–20% volume of fetal bovine serum. Large pieces of undigested tubules were removed, and cell suspensions were filtered through a nylon mesh (40 μm, BD Falcon) to remove cell clumps. Single-cell suspensions were incubated in a fluorescent dye solution (Vybrant Dyecycle Ruby Stain, Invitrogen) for 15 min at 37°C. After washing in PBS, pachytene spermatocytes and round spermatids were purified based on DNA content and cell size using a desktop cell sorter JSAN (Bay Bioscience).
Western blotting
Total protein was prepared from FACS-sorted germ cells, denatured, electrophoresed, and transferred to a nitrocellulose membrane (Amersham). After washing in Tris-buffered saline with 0.1% Tween 20 (TBST), membranes were blocked with skimmed milk and then incubated at room temperature for 4 h with anti-GLB1 (Millipore) or anti-tubulin (Abcam) antibodies. After washing with TBST, membranes were incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit (for GLB1) or anti-mouse (for tubulin) IgG (Millipore). Bands were visualized using the ECL Advance Western Blotting Detection Kit (Amersham).
X-gal staining and immunostaining
For X-gal staining, testes were fixated in 0.1 M phosphate buffer with 0.2% glutaraldehyde, 1% formaldehyde, 5 mM EGTA, 0.2% Nonidet P-40, and 2 mM MgCl2 for 1 h at room temperature. After thorough washing in phosphate-buffered saline (PBS), tissues were stained overnight at 37°C in 0.1 M phosphate buffer with 0.5 mg/mL X-gal, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 2 mM MgCl2, 0.01% deoxycholic acid, and 0.02% Nonidet P-40. After staining, tissues were fixed overnight in 4% paraformaldehyde, embedded in paraffin, and sliced. For immunostaining, testes were fixed in 4% paraformaldehyde for 5 h. After washing in PBS, tissues were dehydrated, embedded in paraffin, and sliced. After blocking, sections were incubated overnight with anti-GFP (Nacalai), anti-PIWIL2 (Abcam), or anti-FLAG M2 (Sigma) antibodies. After washing in PBS, sections were incubated with Alexa 488-conjugated goat anti-rat IgG (Invitrogen) (for GFP) or HRP-conjugated anti-rabbit (for PIWIL2) or anti-mouse IgG (for FLAG). Detection of the HRP activity was done using a VectaStain Kit (Vector Laboratories). FACS-isolated germ cells were fixed in 4% paraformaldehyde at room temperature for 10 min and attached on a side glass using CytoSpin (Thermo Fisher Scientific). After washing in PBS, the cells were permeabilized in 0.5% Triton X-100 in PBS at room temperature for 15–30 min. After blocking, cell specimens were incubated overnight with anti-MVH (Abcam), anti-SYCP3 (Abcam), or anti-γH2AX (Millipore) antibodies. After washing in PBS with 0.05% Tween 20, sections were incubated with Alexa 555-conjugated goat anti-rabbit IgG (Invitrogen) (for MVH, SYCP3, PIWIL2, or PIWIL1) or Alexa 488-conjugated goat anti-mouse IgG (Invitrogen) (for γH2AX).
qRT-PCR and piRNA Northern blotting
Total RNA of 1 μg was treated with DNase I (Invitrogen) and reverse-transcribed by PrimeScript (Takara) using random hexamers as primers. PCR was performed using SYBR Premix EX Taq and the Thermal Cycler Dice Real Time PCR Detection System (Takara). The primers are described in Supplemental Table S4. Northern blotting was performed as described (Watanabe et al. 2007) using total RNA of 20 μg from testes. Hybridization was performed at 42°C in 0.2 M NaHPO4 (pH 7.2), 1 mM EDTA, 1% BSA, and 7% SDS.
Pulse-chase analysis
Testes were dissociated in 1 mg/mL collagenase type IV and then in 0.25% trypsin with 1 mM EDTA. Single cells were suspended in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. 5-ethynyluridine was added to the culture medium in a final concentration of 0.2 mM, and cells were incubated at 37°C for 1 h. After withdrawal of EU, the cells were cultured for a certain period and harvested. Newly synthesized RNA labeled with EU was isolated from total RNA using the Nascent RNA Capture Kit (Invitrogen) according to the manufacturer's directions. The RNA levels were analyzed by qRT-PCR.
Bisulfite sequencing
Bisulfite treatment of DNA was performed as described previously (Hirasawa et al. 2008). In brief, DNA was denatured with NaOH and then treated with 9 M sodium bisulfite from a BisulFAST kit (TOYOBO). DNA was collected using a microcolumn, desulphonated, and eluted using an EZ DNA methylation kit (Zymo Research).
Data access
Small RNA sequence data have been deposited in the DDBJ Sequence Read Archive (DRA) (http://trace.ddbj.nig.ac.jp/) under accession no. DRA000480.
Acknowledgments
We thank Hiroyasu Furuumi and Takehisa Sakaguchi in the Sasaki laboratory and Yumiko Saga (National Institute of Genetics) for scientific and technical advice. We also thank Jun-ichi Miyazaki (Osaka University) for Cag-EGFP mice. This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and a Research Grant from the Naito Foundation to H. Sasaki.
Author contributions: Y.Y. and T.W. carried out the experiment and analyzed the data. Y.H., S.M., M.O., H. Suzuki, T.S., and T.N. were involved in the generation of mice. A.T. and A.F. sequenced small RNA libraries. Y.Y., T.W., and H. Sasaki designed the study. H. Sasaki supervised the study. Y.Y., T.W., and H. Sasaki wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.137224.112.
References
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J 2007a. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ 2007b. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A 2009. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 5: e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R, Kondrashov AS 2009. Rapid repetitive element-mediated expansion of piRNA clusters in mammalian evolution. Proc Natl Acad Sci 106: 7079–7082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Tanaka T, Nakatsuji N 2009. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: Germinal granules in mammals. Mol Cell Endocrinol 306: 17–23 [DOI] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O'Carroll D 2011. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480: 259–263 [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H 2002. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830 [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Girard A, Hannon GJ 2008. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol 18: 136–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202 [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H 2006a. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 20: 1709–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H 2006b. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci 103: 13415–13420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Han JS, Szak ST, Boeke JD 2004. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 429: 268–274 [DOI] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H 2008. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev 22: 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wong WH 2008. SeqMap: Mapping massive amount of oligonucleotides to the genome. Bioinformatics 24: 2395–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto S, Niwa H, Tashiro F, Sano S, Kondoh G, Takeda J, Tabayashi K, Miyazaki J 2000. A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett 470: 263–268 [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Izumi N, Katsuma S, Tomari Y 2011. 3′ end formation of PIWI-interacting RNAs in vitro. Mol Cell 43: 1015–1022 [DOI] [PubMed] [Google Scholar]
- Kazazian HH Jr 2004. Mobile elements: Drivers of genome evolution. Science 303: 1626–1632 [DOI] [PubMed] [Google Scholar]
- Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC, Nowosielska A, Li C, Zamore PD, Weng Z, Theurkauf WE 2011. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell 147: 1551–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W 2008. Biogenesis and germline functions of piRNAs. Development 135: 3–9 [DOI] [PubMed] [Google Scholar]
- Kotaja N, Lin H, Parvinen M, Sassone-Corsi P 2006. Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. J Cell Sci 119: 2819–2825 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y, Fujita Y, Sato M, Matsuda Y, Nakano T 2001. Two mouse piwi-related genes: miwi and mili. Mech Dev 108: 121–133 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. 2004. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE 2006. Characterization of the piRNA complex from rat testes. Science 313: 363–367 [DOI] [PubMed] [Google Scholar]
- Lim AK, Tao L, Kai T 2009. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J Cell Biol 186: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 7: 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Sheets MD, Imboden SB, Dahlberg JE 2011. Limiting Ago protein restricts RNAi and microRNA biogenesis during early development in Xenopus laevis. Genes Dev 25: 1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Olovnikov I, Molaro A, Rozhkov NV, Czech B, Gordon A, Hannon GJ, Aravin AA 2011. Production of artificial piRNAs in flies and mice. RNA 18: 42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oginuma M, Hirata T, Saga Y 2008. Identification of presomitic mesoderm (PSM)-specific Mesp1 enhancer and generation of a PSM-specific Mesp1/Mesp2-null mouse using BAC-based rescue technology. Mech Dev 125: 432–440 [DOI] [PubMed] [Google Scholar]
- Pillai RS, Chuma S 2012. piRNAs and their involvement in male germline development in mice. Dev Growth Differ 54: 78–92 [DOI] [PubMed] [Google Scholar]
- Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS 2011. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480: 264–267 [DOI] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA 2011. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258 [DOI] [PubMed] [Google Scholar]
- Suzuki A, Tsuda M, Saga Y 2007. Functional redundancy among Nanos proteins and a distinct role of Nanos2 during male germ cell development. Development 134: 77–83 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Wang YL, Liu W, Sun YJ, Kwon J, Setsuie R, Osaka H, Noda M, Aoki S, Yoshikawa Y, Wada K 2006. Overexpression of ubiquitin carboxyl-terminal hydrolase L1 arrests spermatogenesis in transgenic mice. Mol Reprod Dev 73: 40–49 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H 2006. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev 20: 1732–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Sasaki H, Minami N, Imai H 2007. Analysis of small RNA profiles during development. Methods Enzymol 427: 155–169 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, et al. 2011. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332: 848–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Xu M, You Y, Hunsicker P, Hori T, Small C, Griswold MD, Hecht NB 2008. Mice deficient for a small cluster of Piwi-interacting RNAs implicate Piwi-interacting RNAs in transposon control. Biol Reprod 79: 51–57 [DOI] [PubMed] [Google Scholar]
- Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P 1997. Disruption of overlapping transcripts in the ROSA β geo 26 gene trap strain leads to widespread expression of β-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci 94: 3789–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]