Abstract
We have developed an approach termed PUB-NChIP (proximity utilizing biotinylation with native ChIP) to purify and study the protein composition of chromatin in proximity to a nuclear protein of interest. It is based on coexpression of (1) a protein of interest, fused with the bacterial biotin ligase BirA, together with (2) a histone fused to a biotin acceptor peptide (BAP), which is specifically biotinylated by BirA-fusion in the proximity of the protein of interest. Using the RAD18 protein as a model, we demonstrate that the RAD18-proximal chromatin is enriched in some H4 acetylated species. Moreover, the RAD18-proximal chromatin containing a replacement histone H2AZ has a different pattern of H4 acetylation. Finally, biotin pulse-chase experiments show that the H4 acetylation pattern starts to resemble the acetylation pattern of total H4 after the proximity of chromatin to RAD18 has been lost.
Chromatin immunoprecipitation (ChIP) (Orlando 2000; Weinmann and Farnham 2002; Ren and Dynlacht 2004) has been widely used for over a decade to study the in vivo association of a particular DNA sequence with regulatory proteins. When combined with high-throughput approaches in the form of ChIP-on-chip or ChIP-seq (Buck and Lieb 2004; Negre et al. 2006; Robertson et al. 2007; Park 2009), it complements gene expression profiling as a way to reconstruct and analyze gene regulatory networks.
Although both DNA and protein components are purified during the ChIP procedure, only the DNA is typically analyzed, and the protein part is discarded. This is because the cross-linking procedure used during ChIP, which is necessary to fix the protein of interest on DNA, irreversibly damages the protein component of chromatin (targeting mostly lysines), rendering it difficult to analyze by mass spectrometry or Western blot. On the other hand, given that it is increasingly recognized that epigenetic regulation is of utmost importance in both normal and disease states, the analysis of histone modifications and replacement histone variants could provide valuable information about the state of chromatin in the vicinity of a particular protein of interest.
With the aim of addressing this limitation of ChIP, we have developed a new technique called PUB-NChIP (proximity utilizing biotinylation and native chromatin immunoprecipitation). It is based on coexpression of (1) the protein of interest (transcription factor, or any other nuclear protein), fused to the bacterial biotin ligase BirA, together with (2) a histone fused to a biotin acceptor peptide (BAP), which is specifically biotinylated by BirA. With the incorporation of BAP-histones into chromatin, the chromatin located in the proximity of the BirA-fusion protein of interest is preferentially biotinylated. The subsequent steps of the procedure take advantage of the strong interaction between core histones and DNA, which allows one to avoid cross-linking for the purification of the biotinylated chromatin (so-called NChIP) (O'Neill and Turner 2003; Ooi et al. 2010). In the absence of cross-linking-induced modifications, analysis of the protein component of the chromatin (e.g., histone post-translational modifications [PTMs]) becomes feasible.
Another advantage of our technique is the possibility of using, instead of the canonical histones, their alternative variants expressed as BAP-fusions. Importantly, the presence of the histone variants was found to correlate with particular functional states of chromatin; for example, silenced chromatin is enriched in histone macroH2A and depleted of H2A.Bbd (Costanzi and Pehrson 1998; Chadwick and Willard 2001a,b). These features make the variant histones a convenient tool for purification of alternative chromatin states. Using the BAP-fusions of these histones instead of the canonical histones allows one to analyze the histone PTMs associated with the protein of interest, but now in the context of a particular functional state of chromatin—e.g., using BAP-macroH2A, which marks repressed chromatin, or BAP-H2A.Bbd, which marks mostly active chromatin, etc.
Finally, we also demonstrate that, given the introduction of a covalent mark into the protein of interest, one can follow the state of histone PTMs at defined times after it came into close proximity with the BirA-fusion. Thus, in addition to helping in direct analysis of histone PTMs in the proximity to the nuclear protein of interest, our approach adds a temporal dimension to such analyses.
Results
Analysis of post-translational modifications of a specific protein fraction using proximity utilizing biotinylation (PUB)
PUB-MS (proximity utilizing biotinylation mass spectrometry) has been developed in our laboratory, originally as a new method to study protein-protein proximity (PPP-networks) in vivo using mass spectrometry (the vectors are described elsewhere [see Fig. 1 in Kulyyassov et al. 2011]). A similar method, although not designed for mass spectrometry analysis, was also developed in the laboratory of Dr. A.Y. Ting (Fernandez-Suarez et al. 2008). The general principle of the approach is based on coexpression of two proteins of interest fused to a biotin acceptor domain in one case and a biotin ligase enzyme (BirA) in the other (Kulyyassov et al. 2011). The BAP domain was modified to weaken its affinity to BirA and thus to decrease the background biotinylation levels. Proximity between the two proteins (e.g., due to their interaction) leads to a more efficient biotinylation of the BAP domain, which can be either detected by Western blotting or quantified using proteomics approaches, such as a combination of multiple reaction monitoring (MRM) analysis and stable isotope labeling in vivo (SILAC) (Ong et al. 2002).
The same protein can exist in several complexes in the cell, and its properties can vary depending on the proximity partners (PP). Given that PUB allows a PP-dependent labeling of a fraction of the BAP-fusion of the protein of interest, one can expand the scope of the PUB approach beyond the mere study of protein-protein proximity. Namely, one can purify a particular PP-dependent subfraction of a BAP-fused protein biochemically, and then study its properties, e.g., its post-translational modifications.
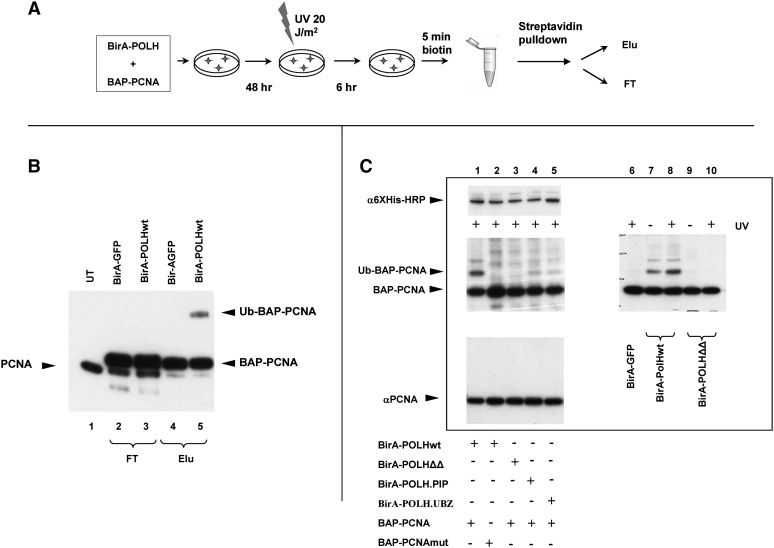
To test this possibility experimentally, we have analyzed the ubiquitination status of the subset of replication processivity factor PCNA (Moldovan et al. 2007) that is located in proximity to the translesion polymerase POLH after UV irradiation of cells (Haracska et al. 2001a,b). Ubiquitination is a convenient model to study PTMs because it is easy to detect as a shift in protein mobility on SDS-PAGE. UVC irradiation induces ubiquitination of PCNA, which then recruits POLH to the sites of DNA damage (Kannouche et al. 2004; Prakash et al. 2005). The POPH has PCNA-binding and ubiquitin-binding domains (PIP and UBZ, respectively), both believed to be involved in the interaction.
To analyze the ubiquitination status of PCNA located in proximity to POLH in vivo, the cells were transfected with plasmids expressing BAP-PCNA and BirA-POLH. Two days later, UV damage was induced, and 6 h after that, the cells were pulse-labeled with biotin. These cells were harvested, and the biotinylated BAP-PCNA was separated from the nonbiotinylated BAP-PCNA using streptavidin-sepharose pulldown (Fig. 1A). The ubiquitination status of the two fractions was analyzed by Western blotting with α-PCNA antibodies. As one can see from Figure 1B (lane 5), a significant part of the biotinylated BAP-PCNA is present in a ubiquitinated form, whereas this form is absent from the nonbiotinylated BAP-PCNA fraction remaining in the flow-through (lane 3). A control experiment, performed in parallel with a BirA-GFP fusion, showed no such enrichment in the biotinylated BAP-PCNA fraction (lanes 2 and 4).
Figure 1.
Analysis of post-translational modifications of a specific protein fraction using proximity utilizing biotinylation (PUB). (A) Experimental scheme. 293T cells are transfected with the plasmids expressing BAP-PCNA and BirA-POLH. Two days later, the cells are treated with UVC (20 J/m2), and 6 h afterward, the cells are pulse-labeled (5 min) with biotin. The cells are then harvested, and the biotinylated BAP-PCNA is separated from the nonbiotinylated BAP-PCNA using streptavidin-sepharose pulldown. (B) Ubiquitination status of POLH-proximal PCNA. Western blot with α-PCNA antibodies. (Lane 1) Untransfected cells; (lanes 2,4) BAP-PCNA cotransfected with BirA-GFP fusion; (lanes 3,5) BAP-PCNA cotransfected with Bir-POLH fusion; (lanes 2,3) flow-through fraction; (lanes 4,5) eluate. Note that the BAP-PCNA from the flow-through fraction was further purified via Ni-NTA chromatography in order to decrease the signal from endogenous PCNA. The endogenous PCNA, the BAP-PCNA fusion, and the ubiquitinated BAP-PCNA are indicated by arrows. (C) Ub-BAP-PCNA is preferentially biotinylated due to its specific interaction with POLH. Western blot analysis of nuclear extracts from transfected 293T cells. (Middle left and right panels) Strepatividin-HRP, detecting two forms of biotinylated BAP-PCNA in the presence of different Bir-POLH fusions, treated or untreated with UV; (top left) expression of different forms of BirA-POLH, detected by α-6×His Western blot; (bottom left) PCNA in the transfected 293 cells, showing that BAP-PCNA constitutes an undetectable fraction of the total PCNA in transfected cells. BirA-POLHwt, -.PIP, -.UBZ, -ΔΔ correspond, respectively, to wild-type POLH, mutant with deletion in the PCNA binding domain, point mutant D652A in the UBZ domain, and double mutant.
To confirm that the ubiquitinated fraction of BAP-PCNA is preferentially biotinylated due to its specific interaction with POLH, we used mutations in POLH lacking the PIP domain and/or mutated in the UBZ domain in our next experiments. This time, the ubiquitination status of the biotinylated BAP-PCNA was analyzed directly in nuclear lysates by Western blotting with streptavidin-HRP. As one can see in Figure 1C (middle), the higher-molecular-weight form of biotinylated PCNA (lanes 1,7,8) was decreased when we used the BirA-POLHΔPIP and BirA-POLHD652A mutated in the UBZ domain (lanes 4,5, respectively), and it practically disappeared when the double mutant BirA-POLHΔΔ was used (lanes 3,9,10). Consistent with the notion that the higher-molecular-weight fraction of BAP-PCNA corresponds to ubiquitinated BAP-PCNA, a BAP-PCNAK164R carrying a mutation in the ubiquitination site showed no such fraction when cotransfected with BirA-POLH in a parallel experiment.
We conclude that purification of the biotinylated fraction of a BAP-fusion of a protein of interest allowed us to study PTMs specific for the particular fraction of this protein that is located in proximity to another protein of interest, fused to BirA. Whereas in the case of ubiquitination, the PTM analysis could be accomplished by Western blot analysis of cellular extracts with streptavidin-HRP, in most other cases, it will require biochemical purification of the biotinylated fraction of the protein.
PUB-NChIP reveals a specific pattern of H4 acetylation in the RAD18-proximal chromatin
Next, we applied the same principle for the analysis of chromatin in close proximity to a protein of interest fused to BirA. For this purpose, we fused the BAP domain to the core histone proteins H3.1 and H2A. After their incorporation into chromatin, the biotinylation of the BAP-histone fusions by a protein of interest fused to BirA is expected to lead to specific labeling of chromatin located in the proximity of the BirA-fusion protein. Purification and analysis of such biotinylated chromatin is the principle behind the PUB-NChIP approach.
To verify that mainly the BAP-histones that are located in proximity to a BirA-fusion are biotinylated under such conditions, we used repair-related RAD18 protein as a convenient model. This protein forms characteristic foci in the nuclei after UV treatment (these foci are believed to correspond to specialized compartments in the nuclei, containing repair and replication proteins). HEK 293T cells were cotransfected with the BirA-RAD18 fusion together with BAP-H2A (see Supplemental Fig. S1 for expression levels of these proteins and the incorporation efficiency of BAP-H2A). Two days after transfection, the cells were irradiated with UVC and pulse-labeled with biotin 6 h later, i.e., at the time when RAD18 forms distinct foci in the nuclei. The cells were fixed immediately after the biotin pulse. Staining of the pulse-labeled cells with streptavidin-Cy3 revealed foci of biotinylation signal, which colocalized with RAD18 (Fig. 2A, top), detected by specific antibodies. Importantly, staining with α-H2A antibodies, which detect H2A regardless of its biotinylation status, and staining with α-His antibodies to detect BAP-H2A gave a very different pattern, consistent with the known homogeneous distribution of H2A (Fig. 2A, bottom; Supplemental Fig. S2A, respectively). Cotransfection of BAP-H2A with a control BirA-GFP construct also produced a homogeneous biotinylation pattern (Supplemental Fig. S2B). Western blot analysis (Fig. 2B, bottom left, lane 3) shows that only the BAP-H2A histone is biotinylated in cells under these conditions, hence the Cy3 signal that we observe in microscopy should reflect the localization of biotinylated BAP-H2A. On the other hand, labeling with biotin for different lengths of time shows that during the 5 min of exposure to biotin, only a small portion of the BAP-H2A is biotinylated (Fig. 2B, right, lane 6). Overall, these data clearly demonstrate that mainly the subfraction of BAP-H2A containing chromatin located in proximity to RAD18 was biotinylated in our experiments. Similar results were obtained when we used a BAP-H3.1 fusion instead of BAP-H2A (Fig. 2B; Supplemental Fig. S2C).
Figure 2.
PUB-NChIP reveals a specific pattern of H4 acetylation in the RAD18-proximal chromatin. (A) Chromatin is biotinylated at the sites of RAD18 foci. Confocal microscopy analysis of colocalization of RAD18 and biotinylated chromatin (top) or H2A histone and biotinylated chromatin (bottom). (B) Biotinylation signal is due to BAP-H2A in proximity to BirA-RAD18. (Left) Western blot analysis, showing that a specific biotinylation signal appears only after BirA-RAD18 and BAP-histone are cotransfected. (Lane 1) BAP-H2A alone; (lane 2) BAP-H3.1 alone; (lane 3) BirA-RAD18 + BAP-H2A; (lane 4) BirA-RAD18 + BAP-H3.1. The two forms of H2A (nonubiquitinated and ubiquitinated), both biotinylated in this experiment, are indicated by arrows. (Top) α−6×His antibodies; (bottom) streptavidin-HRP. (Right) Western blot analysis of the biotinylation status of BAP-H2A and BAP-H3.1 after different times of biotin-labeling. (Top) α-His antibodies; (bottom) streptavidin-HRP. (C) Purification of biotinylated chromatin. (Top) SDS-PAGE gel stained with Coomassie blue, with the identities of the histones indicated. The H4 band in the elution also contains streptavidin and thus is thicker than the other histone bands. (Middle) Western blot analysis of the same samples with streptavidin-HRP, showing that all biotinylated BAP-H2A was affinity-purified with immobilized streptavidin. (Bottom) Agarose gel electrophoresis and ethidium bromide staining for the same samples, showing that mainly the mononucleosome fractions were generated after micrococcal digestion. (Lane 1) Molecular weight marker; (lane 2) nuclei lysate after micrococcal digestion; (lane 3) supernatant after micrococcal digestion; (lane 4) nuclear pellet after salt extraction; (lane 5) chromatin extracted by 0.4M NaCl; (lane 6) chromatin (from fraction 5) unbound to streptavidin-sepharose; (lane 7) eluate (see Methods for details). Note that to make the final preparation visible on Coomassie SDS-PAGE (top panel), we have loaded larger amounts of the eluate fraction, compared to all other fractions. However, for the middle and bottom panels, all fractions are loaded in equal amounts, corresponding to the same number of cells. (D) Relative acetylation status of H4 N-terminal tails (aa 4–17). The presence of a particular peptide was monitored by MRM. For the mono-, di-, and triacetylated peptides, it was difficult to distinguish between modifications at different lysine positions. Hence, we present results without reference to the position. Due to the variations in ratio between the heavy and light isotope-labeled chromatin in each experiment, the H/L ratio for every peptide has been normalized by dividing it by the H/Lum ratio for the unmodified peptide (UM). The graph presents the average normalized H/M ratios and standard deviations for all modified peptides from two independent experiments. The standard deviation for the unmodified peptide represents the standard deviation for the H/L value for this peptide (as H/Lum) in two different experiments. For each pair of columns, 1 corresponds to heavy isotope-labeled cells transfected with BirA-GFP + BAP-H2A, and 2 corresponds to the heavy cells transfected with BirA-RAD18 + BAP-H2A. The light isotope-labeled cells were always transfected with BirA-GFP + BAP-H2A.
The biotinylated chromatin was purified on streptavidin sepharose without prior cross-linking (see Methods, NChIP). Judging by the DNA content of the input, flow-through, and eluate samples (Fig. 2C, bottom, lanes 5,6,7, respectively), only a small fraction of total chromatin was purified in this experiment. However, Western blot analysis of the same fractions with streptavidin-HRP (Fig. 2C, middle) shows that all of the biotinylated BAP-H2A was, in fact, purified. As one can see from Coomassie staining (Fig. 2C, top, lane 7), the composition of the purified histone octamers is consistent with the presence of a single BAP-tagged H2A/H2B protein. Whereas in both input and flow-through samples (lanes 5,6), all core histones are present in stoichiometric amounts, there is approximately twofold less H2A in the eluate. Confirming that not every BAP-H2A molecule is biotinylated in our experiment, the flow-through also contains a BAP-H2A band stained by Coomassie (Fig. 2B, top, lane 6). Note that after elution from streptavidin beads by boiling with Laemmli buffer, significant amounts of streptavidin appear in the sample, overlapping with the H4 histone band (Fig. 2C, top, lane 7).
Next, we wished to verify that PUB-NChIP-purified chromatin is enriched in the expected post-translational modifications of histones. As a well-characterized histone PTM, we have chosen γ-H2AFX (formerly known as γ-H2AX), which forms foci in the nucleus after ionizing irradiation. It was previously shown that RAD18 colocalizes with γ-H2AFX under these conditions (Huang et al. 2009; Watanabe et al. 2009), and we, too, observed that the chromatin that is proximity-biotinylated by BirA-RAD18 often colocalizes with γ-H2AFX after ionizing irradiation in 293T cells (Supplemental Fig. S3A, top). The control protein, chromobox homolog 5 (CBX5; also known as heterochromatin protein 1 α, or HP1 α), did not show any histone biotinylation that was colocalized with the γ-H2AFX foci (Supplemental Fig. S3A, bottom). Western blot analyses of histones prepared by PUB-NChIP have shown that, consistent with the observed colocalization between RAD18-induced biotinylation signal and γ-H2AFX, the RAD18 proximal chromatin is, indeed, enriched in γ-H2AFX (Supplemental Fig. S3B, bottom). To verify that the enrichment in γ-H2AFX is not affected by overexpression of RAD18, we also performed a conventional chromatin immunoprecipitation using α-RAD18 antibodies. Despite a weaker signal, we still could see reproducible enrichment of γ-H2AFX in RAD18-associated chromatin (Supplemental Fig. S3C, bottom), as compared to control HP1 α-associated chromatin.
Next, we returned to our principal experimental system, i.e., 293T cells transfected with “BirA-RAD18 + BAP-H2A” and irradiated with UVC. We set out to analyze the acetylation status of the N terminus of histone H4 in the RAD18-proximal chromatin by mass spectrometry and to compare it with control BirA-GFP biotinylated chromatin. We used SILAC (stable isotope labeling with amino acids in culture) (Ong et al. 2002) in order to quantitatively compare the presence of specific modifications in the chromatin in proximity to RAD18, as compared to that seen with the GFP-BirA fusion. The cells were grown in medium containing either a light version of lysine and arginine amino acids or their heavy counterparts (i.e., labeled with C13 and N15). The cells were cotransfected with BAP-H2A and either BirA-RAD18 (heavy labeled cells) or BirA-GFP (light labeled cells). Two days after transfection, the cells were UV-irradiated, and 6 h later, they were pulse-labeled with biotin for 15 min. Then, the cells were harvested and mixed, and biotinylated chromatin was purified as described above. The bands corresponding to histone H4 were excised and treated with trypsin; then, MRM analysis was performed to quantitatively detect the presence of different acetylated forms of H4 peptide (4-17), comparing the results obtained with experimental (RAD18) and control (GFP) samples (see Supplemental Fig. S4A–E for the MS/MS spectra of the light and heavy version of every peptide).
Figure 2D shows that the H4 histone in close proximity to BirA-RAD18 has a different pattern of acetylation of its N-terminal tail (aa 4–17) compared to the H4 histone contained in BirA-GFP biotinylated chromatin. Notably, the di- and tetra-acetylated forms are overrepresented (column 2 in 2Ac and 4Ac, respectively) compared to H4 from the control chromatin biotinylated by the GFP-BirA fusion (column 1 in 2Ac and 4Ac, respectively). Intriguingly, we have also observed underrepresentation of the triacetylated form of H4 in the RAD18 proximal chromatin (column 2 in 3Ac).
The RAD18-specific pattern of H4 acetylation changes after the proximity to RAD18 has been diminished
The distinctive feature of the PUB-NChIP method is that chromatin in proximity to the BirA-fusion is left with a permanent molecular mark (i.e., biotin), which can persist after the proximity between BAP- and BirA-fusions has been lost (Kulyyassov et al. 2011). We decided to take advantage of this property to ask what happens to the histone PTMs in the RAD18-labeled chromatin at a specified time after the biotinylation.
To confirm our previous findings (Kulyyassov et al. 2011), we used immunofluorescence microscopy to monitor the distribution of biotinylated chromatin 6 h after its labeling by BirA-RAD18. Up until the biotin labeling, the cells were treated as in the previous section. Then, they were either fixed immediately after the biotin pulse (pulse sample) or else intensively washed to eliminate biotin and then left in fresh medium for 6 h before fixation (chase sample). After biotin removal, the biotinylation levels of histones do not increase with time (Fig. 3B, bottom). The pulse-labeled cells showed strong colocalization of labeled BAP-H2A with RAD18 foci (Fig. 3A, top). However, after the chase (Fig. 3A, middle and bottom), the colocalization of the remaining biotinylated foci with the RAD18 foci was considerably less pronounced (Fig. 3C, scatter plot).
Figure 3.
The RAD18-specific pattern changes after the proximity with RAD18 has been diminished. (A) Decrease in colocalization between RAD18 and biotinylated chromatin after a 6-h chase. (Top) Confocal microscopy showing strong colocalization between RAD18 and biotinylated chromatin in the nucleus of 293T cells immediately after pulse-labeling. (Middle) Confocal microscopy showing different localization of RAD18 and biotinylated chromatin 6 h after pulse-labeling. (Bottom) Zoomed area from the chase sample showing an example of biotinylated foci that do not colocalize with the RAD18 foci, and vice versa (indicated by arrows). (B) No increase in biotinylation signal after chase. Western analysis showing that the level of biotinylation of BAP-H3 and BAP-H2A is not increased 6 h after the cells have been washed free of biotin. (Top) α-6×His antibody. (Bottom) Strepavidin-HRP. (P) Pulse sample, (Ch) chase sample. (C) Scatter plot with the coefficient of correlation (top left corner) between the intensities of the red and green signals for every pixel (above a background threshold), showing stronger colocalization of the biotin and RAD18 signals in the pulse sample. (D) Scheme of the pulse-chase experiment. Cells grown in light SILAC medium are transfected by BirA-GFP and BAP-H2A (reference sample). Cells grown in heavy SILAC medium are transfected with BirA-RAD18 (or BirA-GFP) and BAP-H2A. The heavy labeled cells are either harvested immediately after biotin labeling or washed to remove biotin and left for 6 h before harvest. The heavy and light labeled cells are then mixed, and chromatin is prepared as in Figure 2C. (E) Acetylation status of H4 N-terminal tails. The analysis was performed and is presented as described in the legend of Figure 2D. The heavy isotope-labeled cells were: (lane 1) BirA-GFP + BAP-H2A pulse; (lane 2) BirA-GFP + BAP-H2A chase; (lane 3) BirA-RAD18 + BAP-H2A pulse; (lane 4) BirA-RAD18 + BAP-H2A chase. The light isotope-labeled cells were always transfected with BirA-GFP + BAP-H2A and pulse-labeled with biotin, as in sample 1.
At the next step, we purified the PUB-labeled chromatin after a 6-h chase and analyzed the pattern of H4 acetylation. SILAC labeling and MRM were again used to quantitatively compare the modifications between different samples (Fig. 3D). All experimental samples were grown in heavy medium and, as a common reference sample (grown in the light medium), pulse-labeled “BirA-GFP + BAP-H2A” transfected cells were used. Judging by the more modest H/L ratios in the case of the chased sample (Fig. 3E), the characteristic underrepresentation of the tri-acetylated form, which was clearly seen in the RAD18-proximal chromatin at the time of the biotinylation pulse (i.e., 6 h after UV irradiation) (column 4, 3Ac), became less pronounced and began to resemble the modification pattern seen in the control chromatin (i.e., biotinylated by GFP-BirA) (column 3, 3Ac).
The pattern of H4 acetylation near RAD18 is different in the case of H2AZ-containing chromatin
Another advantage of the PUB-NChIP method resides in the fact that, instead of the canonical core histones (e.g., H3.1, H2A), one can use their alternative forms, which have been shown to preferentially associate with different functional states of chromatin.
Immunofluorescence microscopy together with Western blot analysis shows that, similar to the canonical histone H2A, cotransfection of BirA-RAD18 with BAP-H2AZ, BAP-macroH2A2 and BAP-H2AFB3 (also known as H2A-Bbd type 3) leads to the appearance of biotinylated foci in the nuclei that are colocalized with RAD18 and are due to the BAP-histone fusions (Fig. 4A,B). As expected, BAP-macroH2A2 foci coincide mostly with DNA-dense areas of the nucleus, whereas BAP-H2AFB3 foci associate with less densely stained areas. Given the more complicated distribution of H2AZ in the nucleus (Viens et al. 2006), no correlation with DNA density was detected for this protein. Overall, the results are consistent with the notion that using alternative histones in PUB-NChIP should lead to labeling of functional chromatin states associated with these histone variants.
Figure 4.
The pattern of H4 acetylation near RAD18 is different in the case of H2AZ-containing chromatin. (A) Alternative histones are biotinylated at the locations of RAD18. Confocal microscopy of 293T cells cotransfected by BirA-RAD18 and BAP-H2AFB3 (top), BAP-macroH2A2 (middle), and BAP-H2AZ (bottom). Shown are staining with Topro-3 (left), α−RAD18 (middle), and streptavidin (right). (B) Biotinylation signal is due to BAP-histone fusions. (Top and middle) Western blot analysis showing that a specific biotinylation signal appears only after BirA-RAD18 and BAP-macroH2A or BAP-H2AFB3 fusions are cotransfected. (Lanes 1,4) BAP-histone + BirA-RAD18; (lanes 2,5) BAP-histone; (lanes 3,6) control untransfected samples. (Top) α−6×His antibodies, (middle) streptavidin-HRP. (Bottom) Western blot analysis for the BAP-H2AZ fusion. The two forms of H2AZ (nonubiquitinated and ubiquitinated), both biotinylated in this experiment, are indicated by arrows. (C) Acetylation status of H4 N-terminal tails. The analysis was performed and is presented as described in the legend to Figure 2D. The heavy isotope-labeled cells were: (lane 1) BirA-GFP + BAP-H2A; (lane 2) BirA-RAD18 + BAP-H2A; (lane 3) BirA-GFP + BAP-H2AZ; (lane 4) BirA-RAD18 + BAP-H2AZ. The light isotope-labeled cells were always transfected with BirA-GFP + BAP-H2A.
To test the possibility of simultaneously studying histone PTMs both in the vicinity of a protein of interest and in the context of a particular alternative histone, we performed SILAC experiments with the alternative histone H2AZ, which is implicated in many cellular functions, including the cellular response to DNA damage (Kusch et al. 2004; Ahmed et al. 2007; Kalocsay et al. 2009; Morillo-Huesca et al. 2010; Bao 2011; Kotova et al. 2011).
As before, all experimental samples were labeled in heavy medium and, as a reference (light) sample, pulse-labeled “BirA-GFP + BAP-H2A” transfected cells were used. The relative abundance of heavy versus light peptides was analyzed by MRM. As Figure 4C shows, when BAP-H2A is replaced by BAP-H2AZ, levels of the diacetylated forms of H4 clearly increase in the RAD18-proximal chromatin (cf. columns 3 and 4, 2Ac). The same modification also appears to increase in the control GFP-BirA labeled chromatin containing BAP-H2AZ (cf. columns 1 and 3, 2Ac). The most parsimonious interpretation of this observation is that the two factors (RAD18-proximity vs. GFP, and presence of H2AZ vs. H2A) affect the diacetylated form of H4 independently, resulting in an additive effect in the case of ‘RAD18 + H2AZ' combination.
Strikingly, however, we have observed a nonadditive response to the two factors in the case of the tri- and tetra-acetylated forms of H4. Unlike in the case of ‘RAD18 + H2A,' there was no decrease in the triacetylated form in the case of the ‘RAD18 + H2AZ' combination (Fig. 4C, cf. columns 3 and 4, 3Ac). On the other hand, the tetra-acetylated form was underrepresented in the case of ‘RAD18 + H2AZ,' compared to ‘GFP + H2AZ' (columns 3 and 4, 4Ac).
These results show that the patterns of post-translational histone modifications in proximity to the RAD18 protein depend on the histone composition of chromatin.
Discussion
We describe here a new methodology to study chromatin in close proximity to a protein of interest in vivo. The well-established approach for such analysis is chromatin immunoprecipitation (Orlando 2000; Weinmann and Farnham 2002; Ren and Dynlacht 2004). However, it suffers from the following limitation. Due to the cross-linking of DNA to histones, only the DNA portion of chromatin purified by ChIP can be used, and the protein part is discarded, being irreversibly damaged by the cross-linking procedure. On the other hand, the analysis of histone modifications and replacement histone variants could provide valuable information about the state of chromatin around the particular protein of interest. Until now, the correlations between a protein and histone PTMs could be inferred only indirectly, after comparison of the results of parallel ChIP experiments that use antibodies against the protein and antibodies against particular PTMs. Here, a more direct method to access the chromatin state is proposed.
The principle of PUB-NChIP is somewhat similar to the DamID approach, an alternative to the ChIP way to map the DNA binding sites of nuclear proteins (van Steensel and Henikoff 2000; Greil et al. 2006). This methodology is based on expression of a fusion of bacterial Dam-methylase to a protein of interest that leads to labeling the protein's genomic binding site by adenine methylation. The label on DNA can be detected by various methods. Our approach is conceptually similar, but instead of Dam-methylase, we fuse the biotin-ligase BirA to the protein of interest, and, instead of DNA, we label the histone part of the chromatin. The scopes of these approaches are also different, as PUB-NChIP is mostly aimed at analyzing the histone PTMs. We cannot exclude that PUB-NChIP could be used for DNA analysis as well, although the resolution of PUB-NChIP could be lower than that of regular ChIP. On the other hand, one can point to several potential advantages of PUB-NChIP for DNA analysis. There are a wide variety of commercially available resources for detection and/or purification of biotinylated macromolecules, and NChIP with biotinylated chromatin has been shown to work (Ooi et al. 2010). In addition, by substituting the canonical core histone with its replacement variants, the DNA analysis can be made dependent on the context of particular functional states of chromatin.
As far as the histone PTM analysis is concerned, we present here a proof of principle that it can be done with the PUB-NChIP purified chromatin, focusing mostly on the acetylation status of H4 N termini. Our approach to analyzing histone PTMs is based on SILAC, which allows us to quantitatively analyze the histone modifications in the RAD18-proximal chromatin, their dynamics after labeling, and their dependence on histone variants. This particular approach is well suited for the type of mass spectrometer (ion-trap) we used. However, because it requires trypsin digestion, the information about correlations between different histone modifications on individual histone molecules is not accessible. On the other hand, the mass spectrometry instruments with higher resolution (such as FTICR and Orbitrap) can be used in the so-called top-down approach that targets histones without tryptic digestion (Pesavento et al. 2004; Zhang et al. 2004). Using these instruments for downstream analysis of the PUB-NChIP prepared chromatin, one will be able to detect correlations between PTMs on individual histone molecules. It should be noted that such correlation analysis is impossible with the traditional ChIP technique.
In addition to histones, some other proteins associate with DNA sufficiently strongly and thus are expected to be purified in PUB-NChIP (Hager et al. 2009; Lambert et al. 2009; Shen et al. 2010). However, efficient pull-down of nonhistone proteins requires changing conditions for micrococcal digestion. Instead of aiming to generate mononucleosomes (which requires rather extensive digestion), one has to generate larger chromatin fragments, which could accommodate nonhistone proteins, in addition to the biotinylated nucleosome. With these modifications of protocol, one can also use PUB-NChIP for analysis of the nonhistone fraction of chromatin, which is complicated in the case of regular ChIP due to the cross-linking.
With respect to the biological significance of our findings, their simplest interpretation would be that the chromatin in the vicinity of RAD18 is locally destabilized to facilitate the access of the repair machinery to DNA. This is consistent with the notion that acetylation of the N-terminal tails of H4 loosens up histone-DNA interactions and hence creates a relaxed nucleosome (Van Holde 1989). However, the fact that triacetylated H4 tails are underrepresented in RAD18 proximal chromatin suggests that this physicochemical model of the H4 acetylation might be an oversimplification. It is more consistent with the concept of histone code, i.e., protein-protein recognition of a specific PTM pattern mediating their downstream regulatory effects (Jenuwein and Allis 2001). Our observation of a less pronounced PTM pattern 6 h after labeling with biotin (pulse-chase experiment) is consistent with the notion that, after the repair is completed, the chromatin returns to its original state.
Another novelty of PUB-NChIP is in allowing one to vary two independent variables (protein of interest fused to BirA and histone-BirA fusion), thus providing a more detailed analysis of chromatin organization. In this respect, the acetylation pattern of the H2AZ-containing chromatin in proximity to RAD18 requires interpretation. The alternative histone H2AZ has been implicated in cellular response to DNA damage and repair (Kusch et al. 2004; Ahmed et al. 2007; Kalocsay et al. 2009; Morillo-Huesca et al. 2010; Bao 2011; Kotova et al. 2011). In mammals, H2AZ is deposited into chromatin by p400/Tip60 complexes (Svotelis et al. 2009), whereas in Drosophila, Tip60 acetylates nucleosomal phospho-H2AV and exchanges it with an unmodified H2AV replacement histone at DNA lesions (Kusch et al. 2004). Notably, the Drosophila H2AV is the only alternative H2A variant and is homologous to the H2AZ family of alternative histones (van Daal et al. 1988; Madigan et al. 2002), thus H2AV appears to play the role of both mammalian H2AX and H2AZ. This raises an intriguing possibility that the chromatin containing H2AZ histone in proximity to RAD18 corresponds to a specific stage of DNA-lesion processing, which could be reflected in the different histone PTM signature observed in our experiments. Regardless of the specific mechanistic interpretation of these observations, our data strongly suggest that one can analyze how changes in specific histone PTMs in the proximity of a nuclear protein of interest depend on the particular chromatin type, here represented by a specific histone variant. Alternatively to a variant histone, one can introduce a mutation in a canonical histone and use its BAP-fusion in PUB-NChIP in order to see how it affects the chromatin state in proximity to a particular protein of interest fused to BirA.
In our work, we used a transient transfection protocol. This might explain why the expression levels of BirA-RAD18 were higher than that of its endogenous counterpart. Importantly, using untransfected cells and conventional immunoprecipitation with α-RAD18, we have confirmed that RAD18-associated chromatin is enriched in γ-H2AFX, a well-defined histone PTM (single phosphorylation of H2AX of serine 139). Note that to perform similar validation would be significantly more challenging in the case of histone acetylation, as (1) lysines have more chance to be affected by formaldehyde cross-linking and (2) there are four different acetylated lysine residues at the N terminus of H4 (each detectable by specific antibodies) and (3) an overwhelming number of their combinations. In this regard, we also wish to emphasize that (just like with the downstream analysis of histone modifications, which can be performed in many different ways), the general principle of PUB-NChIP is compatible with stable cell line and transgenic organism approaches, which would allow better control of protein expression at more physiological levels.
Methods
Recombinant DNA
The vectors for expression of BAP- and BirA-fusions, as well as BirA-GFP, BirA-RAD18, BAP-H2A, BAP-macroH2A2, and BAP-H2AFB3 expression vectors have been described elsewhere (Kulyyassov et al. 2011). The H2AZ and H3.1 ORFs were subcloned from pOZ.FHH vectors (Saade et al. 2009), whereas the POLH ORFs (wild type and mutant) were amplified by PCR and inserted into the BirA-fusion vector using XhoI and NotI restriction sites.
Cell culture
HEK 293T cells were grown in Dulbecco's modified Eagle's medium with high glucose (PAA) and 10% fetal bovine serum (FCS, PAA). For transient transfection and Western analysis, a standard calcium phosphate precipitation method was used, and the cells were analyzed 1 or 2 d after transfection, as indicated. For the biotin labeling in vivo, cells were grown for several days before transfection in the DMEM supplemented with dialyzed FCS, and for the specified time of labeling, biotin (Sigma) was added to a final concentration of 5 μg/mL, while the pH was stabilized by addition of 50 mM HEPES (pH 7.35) to the medium. For SILAC experiments, the cells were grown in DMEM with 12C6 L-lysine and 12C6 14N4 L-arginine or 13C6 L-lysine 13C6 15N4 L-arginine (Thermo Scientific; cat # 89983) for at least five divisions before transfection and kept in the same medium until harvest. For the Western analysis, 3 × 105 cells (corresponding to one well in a 6-well plate) were used for transfection. For the mass spectrometry analysis, 3 × 106 cells were transfected per data point. DNA damage was induced with UVC (20 J/m2) or ionizing radiation (10 Gy). The time of incubation after various treatments is indicated in the results section.
Biochemistry and Western analysis
Except where indicated otherwise, cell nuclei were used for analysis. They were prepared by cell disruption in CSK buffer (100 mM NaCl, 300 mM sucrose, 10 mM Tris [pH 7.5], 3 mM MgCl2, 1 mM EGTA, 1.2 mM PMSF, 0.5% Triton X-100) and centrifugation for 5 min at 4000 rpm. For the analysis of chromatin-associated histones (chromatin fraction), the nuclei were first incubated in CSK buffer containing 450 mM NaCl by 30 min rotation at 4°C, then spun at 4000 rpm, and the supernatant containing soluble histones was discarded. For Western analysis, 1× NuPAGE LDS Sample buffer (Invitrogen; NP0007) with DTT (10 mM) was added, the nuclei were sonicated, boiled for 5 min at 96°C, and loaded on 4%–12% gradient Novex Tris-Glycine precast gels (Invitrogen; NP0315). After separation, the proteins were transferred to nitrocellulose membranes and probed with HRP-conjugated streptavidin (Sigma; # S5512) or HRP-conjugated α-PentaHis antibody (Qiagen; # 34460) according to the manufacturer's protocol, except that for the detection of biotinylated proteins by the HRP-conjugated streptavidin, 500 mM NaCl was added to the washing buffer (PBS + 0.1% Tween). For protein visualization, the gels were stained with PageBlue (Fermentas; # R0579). For the densitometric analysis of Western blots, the program ImageJ 1.42q (freely available online) was used. To compare the biotinylation levels between different samples, the value of the streptavidin-HRP signal was normalized to the value of the α-PentaHis signal, which reflects the amount of the transfected protein regardless of its biotinylation status. Every Western analysis was performed three times; a representative figure is shown in each case.
For Ni2+NTA purification of 6×His tagged proteins, the nuclei were solubilized in buffer A (10% glycerol, 250 mM NaCl, 6 M guanidine-HCl, 20 mM TrisHCl [pH 8.0], 0.1% Tween) by rotation for 30 min at 4°C. One tenth of the volume of Ni2+NTA agarose, prewashed in the same buffer, was added to the lysate, followed by rotation at 4°C for 3 h. The beads with bound proteins were washed twice with buffer A, then twice with buffer B (10% glycerol, 250 mM NaCl, 20 mM TrisHCl [pH 8.0], 0.1% Tween, 0.2 mM PMSF, and cOmplete protease inhibitor cocktail [Roche]). The bound proteins were eluted by incubating the beads in four volumes of buffer C (10% glycerol, 250 mM NaCl, 20 mM Tris HCl [pH 8.0], 0.1% Tween, 0.2 mM PMSF, 300 mM imidazole, 50 mM EDTA) and concentrating the sample by ultrafiltration with Microcon YM-10 (Millipore).
NChIP
Cells were washed with PBS and lysed in 1 mL of CSK buffer (with protease inhibitor cocktail [Roche;1697498], 10 mM Na-Butyrate, 2 mM PMSF, 5 mM nicotinamide [Sigma; N5535], 5 mM Na-orthovanadate [Sigma; S6508]) for 5 min at RT. Nuclei were centrifuged at 4000 rpm for 10 min and kept at 4°C or else at –20°C if not used immediately. For micrococcal digestion, the nuclei were resuspended in 500 μL of TM2 buffer (10 mM Tris-HCl, 2 mM MgCl2, 0.1% Triton, inhibitors), and 2.5 μL of 0.5 M CaCl2 and 3 μL of micrococcal nuclease (MNase) (1 U/μL; Sigma; N3755) were added before incubation at 37°C for 10 min. The reaction was stopped by adding 15 μL of 0.1 M EGTA. Nuclei were collected by spinning at 400g (2000 rpm in Eppendorf tabletop centrifuge) for 10 min at 4°C (see Fig. 2C, lanes 2,3 for analysis of pellet and supernatant after this separation). After MNase digestion, the nuclei pellet was resuspended in 500 μL of prechilled 0.4 M salt extraction buffer (385 mM NaCl, 10 mM Tris-HCl [pH 7.4], 2 mM MgCl2, 2 mM EGTA, 0.1% Triton X-100 with inhibitors). The tubes were rotated at 4°C for 30 min. Supernatant containing digested chromatin (Fig. 2C, lane 5) was separated from the remaining material (lane 4) by spinning at 400g (2000 rpm in Eppendorf tabletop centrifuge) for 10 min at 4°C. An equal volume of TM2 buffer was added to the extracted chromatin to arrive at a 0.2 M final salt concentration. The sample was centrifuged at maximum speed (13,000g, i.e., 16,000 rpm in an Eppendorf centrifuge) for 5 min at 4°C. Two hundred μL of a suspension of sepharose-streptavidin beads (GE Healthcare; 17-5113-01) were washed three times with 0.2 M salt extraction buffer containing Triton and finally resuspended in 100 μL of the same buffer. The bead suspension was combined with chromatin and rotated for 3 h at 4°C. Afterward, the beads were washed twice in 500 μL of 0.4 M salt extraction buffer containing Triton for 5 min at 4°C. The biotinylated chromatin was eluted from the beads by adding 100 μL of 1× LDS sample buffer (with reducing agent + 2% SDS), heating at 99°C for 10 min, and separating the beads by a quick spin in the tabletop centrifuge for 10 sec. Elution was repeated a second time, and the two eluates were combined (Fig. 2A, lane 7).
Mass spectrometry
The protein bands were excised from the gel and were chemically derivatized using propionic anhydride (Garcia et al. 2007). Briefly, 100 μL of freshly prepared 75% propionic anhydride in methanol was added to each tube and immediately supplemented by 40 μL of 50 mM ammonium bicarbonate. After 30 min at 37°C, the solution was discarded, and propionic anhydride derivatization was repeated again. The peptides were digested by trypsin as in Shevchenko et al. (1996). The gel slices were dehydrated with 300 μl of 50% acetonitrile followed by 300 μl of 100% acetonitrile, then rehydrated with 300 μl of 50 mM ammonium bicarbonate. A final dehydration was performed with two washes of 300 μl of 50% acetonitrile, followed by two washes of 300 μl of 100% acetonitrile. Each wash was carried out for 10 min at 25°C with shaking at 1400 rpm. The gel slices were dried in a SpeedVac at 35°C for 10 min. For trypsin digestion, the gel slices were preincubated with 7 mL of 15 ng/mL trypsin (Promega; # V5280) at room temperature for 10 min. Afterward, 25 μl of 50 mM ammonium bicarbonate was added, and the gel slices were incubated at 37°C for 16 h. The peptide-containing supernatants were dried at 56°C by SpeedVac for 30 min, then resuspended in 20 μl of solution containing 0.05% formic acid and 3% acetonitrile for mass spectrometry experiments. The resulting peptides were analyzed with a nano-HPLC (Agilent Technologies 1200) directly coupled to an ion-trap mass spectrometer (Bruker 6300 series) equipped with a nano-electrospray source. The separation gradient was 7 min from 5% to 90% acetonitrile. The fragmentation voltage was 1.3V. For quantification of the presence of a particular peptide in the sample, the ion trap was set in MRM mode, i.e., it was set to isolate, fragment, and MS/MS scan several parental ions, corresponding to the heavy and light un-, mono-, di-, tri- and tetra-acetylated H4 peptide (aa 4–17), having predetermined M/Z ratios (747.4/760.4; 740.4/757.4; 733.3/750.3; 726.3/743.3; 719.3/736.3, respectively). The relative quantity of each peptide in the different fractions was estimated by comparison between the sum of peak areas in the extracted ion chromatograms (EIC) for several most intensive daughter ions of this peptide obtained from MRM analysis of these fractions. The identities of the daughter ions used for comparison between different parental ions are now indicated by asterisks on Supplemental Figure S4A–E.
Immunofluorescent microscopy
HEK 293T cells were washed two times with cold PBS and fixed by immersion in –20°C methanol for 5 min. These fixed cells were washed with cold PBS and then permeabilized with 0.2% Triton X-100 in PBS for 10 min at 4°C and submitted to additional fixation in 4% formaldehyde for 20 min at RT. Cells were then washed twice with PBS at room temperature (RT). Blocking was done with 3% BSA for 30 min under mild agitation. Cells were then incubated with mouse monoclonal anti-RAD18 antibody (Abcam; ab57447) followed by three washes in PBS (1×) at RT. Cells were then incubated with goat anti-mouse secondary antibody conjugated with Alexa-488 (Invitrogen; A11017) for 1 h (1/1000). Streptavidin-Cy3 conjugate (Sigma; S-6402) and ToPro (Invitrogen) were also added (1/500 and 1/1000, respectively) to the same incubation mix. After three washes in PBS (1×) at RT, cover slips were mounted on glass slides using VectaShield mounting medium (Vector Laboratories; H-1000). Cells were visualized with a Zeiss LSM 510 Meta confocal scanning microscope, using a Plan-Apochromat 63× 1.4 oil immersion objective. Imaging was performed with sequential multitrack scanning using the 488- and 543-nm laser wavelengths separately. The colocalization analysis was performed with LSM Examiner software. Pearson's correlation coefficient (Fig. 3A) was calculated with setting the threshold signal common for all images compared (15 cells for both experiment and control samples). For histones H2A and H3 staining, cells were incubated with α-H2A antibody (Abcam; ab18255) or γ-H2AFX (Cell Signaling; 2577S) or α-His antibody (GE Healthcare; 27-4710-01) and α-H3 antibody (Abcam; ab1791), respectively.
Acknowledgments
We thank Dr. L.L. Pritchard for critical reading of the manuscript. This work was supported by grants from La Ligue Contre le Cancer (9ADO1217/1B1-BIOCE), the Institut National du Cancer (247343/1B1-BIOCE), and the Centre National de la Recherche Scientifique (CNRS-INCA-MSHE Franco-Pologne #3037987) to V.O., and by NCB Kazakhstan (0103_00404) to A.K. A.K. thanks Mr. Guillaume Giraudet for his help in organizing a visit from NCB to IGR.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.134874.111.
References
- Ahmed S, Dul B, Qiu X, Walworth NC 2007. Msc1 acts through histone H2A.Z to promote chromosome stability in Schizosaccharomyces pombe. Genetics 177: 1487–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y 2011. Chromatin response to DNA double-strand break damage. Epigenomics 3: 307–321 [DOI] [PubMed] [Google Scholar]
- Buck MJ, Lieb JD 2004. ChIP-chip: Considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics 83: 349–360 [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF 2001a. Histone H2A variants and the inactive X chromosome: Identification of a second macroH2A variant. Hum Mol Genet 10: 1101–1113 [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF 2001b. A novel chromatin protein, distantly related to histone H2A, is largely excluded from the inactive X chromosome. J Cell Biol 152: 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR 1998. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393: 599–601 [DOI] [PubMed] [Google Scholar]
- Fernandez-Suarez M, Chen TS, Ting AY 2008. Protein-protein interaction detection in vitro and in cells by proximity biotinylation. J Am Chem Soc 130: 9251–9253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, Hunt DF 2007. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc 2: 933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greil F, Moorman C, van Steensel B 2006. DamID: Mapping of in vivo protein-genome interactions using tethered DNA adenine methyltransferase. Methods Enzymol 410: 342–359 [DOI] [PubMed] [Google Scholar]
- Hager GL, McNally JG, Misteli T 2009. Transcription dynamics. Mol Cell 35: 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S 2001a. Physical and functional interactions of human DNA polymerase η with PCNA. Mol Cell Biol 21: 7199–7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Unk I, Phillips BB, Hurwitz J, Prakash L, Prakash S 2001b. Targeting of human DNA polymerase ι to the replication machinery via interaction with PCNA. Proc Natl Acad Sci 98: 14256–14261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J 2009. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol 11: 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD 2001. Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S 2009. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell 33: 335–343 [DOI] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR 2004. Interaction of human DNA polymerase η with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol Cell 14: 491–500 [DOI] [PubMed] [Google Scholar]
- Kotova E, Lodhi N, Jarnik M, Pinnola AD, Ji Y, Tulin AV 2011. Drosophila histone H2A variant (H2Av) controls poly(ADP-ribose) polymerase 1 (PARP1) activation in chromatin. Proc Natl Acad Sci 108: 6205–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulyyassov A, Shoaib M, Pichugin A, Kannouche P, Ramanculov E, Lipinski M, Ogryzko V 2011. PUB-MS: A mass-spectrometry-based method to monitor protein-protein proximity in vivo. J Proteome Res 10: 4416–4427 [DOI] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR III, Abmayr SM, Washburn MP, Workman JL 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306: 2084–2087 [DOI] [PubMed] [Google Scholar]
- Lambert JP, Mitchell L, Rudner A, Baetz K, Figeys D 2009. A novel proteomics approach for the discovery of chromatin-associated protein networks. Mol Cell Proteomics 8: 870–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan JP, Chotkowski HL, Glaser RL 2002. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res 30: 3698–3705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S 2007. PCNA, the maestro of the replication fork. Cell 129: 665–679 [DOI] [PubMed] [Google Scholar]
- Morillo-Huesca M, Clemente-Ruiz M, Andujar E, Prado F 2010. The SWR1 histone replacement complex causes genetic instability and genome-wide transcription misregulation in the absence of H2A.Z. PLoS ONE 5: e12143 doi: 10.1371/journal.pone.0012143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Lavrov S, Hennetin J, Bellis M, Cavalli G 2006. Mapping the distribution of chromatin proteins by ChIP on chip. Methods Enzymol 410: 316–341 [DOI] [PubMed] [Google Scholar]
- O'Neill LP, Turner BM 2003. Immunoprecipitation of native chromatin: NChIP. Methods 31: 76–82 [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1: 376–386 [DOI] [PubMed] [Google Scholar]
- Ooi SL, Henikoff JG, Henikoff S 2010. A native chromatin purification system for epigenomic profiling in Caenorhabditis elegans. Nucleic Acids Res 38: e26 doi: 10.1093/nar/gkp1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V 2000. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem Sci 25: 99–104 [DOI] [PubMed] [Google Scholar]
- Park PJ 2009. ChIP-seq: Advantages and challenges of a maturing technology. Nat Rev Genet 10: 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento JJ, Kim YB, Taylor GK, Kelleher NL 2004. Shotgun annotation of histone modifications: A new approach for streamlined characterization of proteins by top down mass spectrometry. J Am Chem Soc 126: 3386–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L 2005. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem 74: 317–353 [DOI] [PubMed] [Google Scholar]
- Ren B, Dynlacht BD 2004. Use of chromatin immunoprecipitation assays in genome-wide location analysis of mammalian transcription factors. Methods Enzymol 376: 304–315 [DOI] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. 2007. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 4: 651–657 [DOI] [PubMed] [Google Scholar]
- Saade E, Mechold U, Kulyyassov A, Vertut D, Lipinski M, Ogryzko V 2009. Analysis of interaction partners of H4 histone by a new proteomics approach. Proteomics 9: 4934–4943 [DOI] [PubMed] [Google Scholar]
- Shen Z, Sathyan KM, Geng Y, Zheng R, Chakraborty A, Freeman B, Wang F, Prasanth KV, Prasanth SG 2010. A WD-repeat protein stabilizes ORC binding to chromatin. Mol Cell 40: 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Svotelis A, Gevry N, Gaudreau L 2009. Regulation of gene expression and cellular proliferation by histone H2A.Z. Biochem Cell Biol 87: 179–188 [DOI] [PubMed] [Google Scholar]
- van Daal A, White EM, Gorovsky MA, Elgin SC 1988. Drosophila has a single copy of the gene encoding a highly conserved histone H2A variant of the H2A.F/Z type. Nucleic Acids Res 16: 7487–7497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Holde KE. 1989 Chromatin. Springer-Verlag, New York. [Google Scholar]
- van Steensel B, Henikoff S 2000. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol 18: 424–428 [DOI] [PubMed] [Google Scholar]
- Viens A, Mechold U, Brouillard F, Gilbert C, Leclerc P, Ogryzko V 2006. Analysis of human histone H2AZ deposition in vivo argues against its direct role in epigenetic templating mechanisms. Mol Cell Biol 26: 5325–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Iwabuchi K, Sun J, Tsuji Y, Tani T, Tokunaga K, Date T, Hashimoto M, Yamaizumi M, Tateishi S 2009. RAD18 promotes DNA double-strand break repair during G1 phase through chromatin retention of 53BP1. Nucleic Acids Res 37: 2176–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann AS, Farnham PJ 2002. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods 26: 37–47 [DOI] [PubMed] [Google Scholar]
- Zhang L, Freitas MA, Wickham J, Parthun MR, Klisovic MI, Marcucci G, Byrd JC 2004. Differential expression of histone post-translational modifications in acute myeloid and chronic lymphocytic leukemia determined by high-pressure liquid chromatography and mass spectrometry. J Am Soc Mass Spectrom 15: 77–86 [DOI] [PubMed] [Google Scholar]