Abstract
Recent developments in next-generation sequencing have enabled whole-genome profiling of nucleosome organizations. Although several algorithms for inferring nucleosome position from a single experimental condition have been available, it remains a challenge to accurately define dynamic nucleosomes associated with environmental changes. Here, we report a comprehensive bioinformatics pipeline, DANPOS, explicitly designed for dynamic nucleosome analysis at single-nucleotide resolution. Using both simulated and real nucleosome data, we demonstrated that bias correction in preliminary data processing and optimal statistical testing significantly enhances the functional interpretation of dynamic nucleosomes. The single-nucleotide resolution analysis of DANPOS allows us to detect all three categories of nucleosome dynamics, such as position shift, fuzziness change, and occupancy change, using a uniform statistical framework. Pathway analysis indicates that each category is involved in distinct biological functions. We also analyzed the influence of sequencing depth and suggest that even 200-fold coverage is probably not enough to identify all the dynamic nucleosomes. Finally, based on nucleosome data from the human hematopoietic stem cells (HSCs) and mouse embryonic stem cells (ESCs), we demonstrated that DANPOS is also robust in defining functional dynamic nucleosomes, not only in promoters, but also in distal regulatory regions in the mammalian genome.
Nucleosomes are fundamental building blocks of primary chromatin structure. Each nucleosome is composed of a 147-bp DNA wrapping around a histone octamer, which contains two copies of each of the core histones: H2A, H2B, H3, and H4 (Luger et al. 1997). The chemical composition and physical organization of nucleosomes in living cells have been implicated in the regulation of multiple chromatin-associated cellular functions, such as replication (Eaton et al. 2010), DNA repair (Ying et al. 2010), transcription (He et al. 2010; Jin et al. 2010), and RNA splicing (Tilgner et al. 2009), as well as disease processes, such as cancer and aging (Chi et al. 2010; Feser and Tyler 2011; Zhang and Pugh 2011). The chemical composition of nucleosomes includes histone variants, such as H3.3 and H2A.Z, and modifications of histone tails, such as methylation, acelytation, and ubiquitination (Wang et al. 2008; Gutiérrez et al. 2011). The physical organization of nucleosomes can be described as an array of nucleosome units across the genome, e.g., there are ∼60,000 nucleosome units in the yeast genome (Jiang and Pugh 2009). In different cells, the exact positions of the nucleosomes within each unit may deviate more or less while centering around a most preferred position. This deviation of nucleosome positions within each unit in a cell population is referred to as fuzziness. Thus, each nucleosome unit can be described by a most preferred position (hereafter referred to as nucleosome position) and its fuzziness, along with an occupancy value referring to the frequency with which the unit is occupied in a cell population (Kaplan et al. 2010; Pugh 2010). The nucleosome chemical composition is highly enriched in biological information, although systematic decoding of this complex information could be overwhelming (Henikoff and Shilatifard 2011). On the other hand, nucleosome physical organization provides the background for histone modifications and variant turnover; more importantly, nucleosome physical organization directly regulates the accessibility of DNA to binding proteins such as transcription factors (He et al. 2010).
Micrococcal nuclease (MNase) preferably digests chromatin at DNA sites that are not occupied by nucleosomes while leaving nucleosomal DNA largely intact. MNase digestion of chromatin followed by high throughput sequencing (MNase-seq) has facilitated the investigation of nucleosome physical organization across the whole genome (Zhang and Pugh 2011). Although some details are still under debate (Zhang et al. 2009), the physical organization of nucleosomes is likely determined in cis by the underlying DNA sequences (Chen et al. 2008; Kaplan et al. 2009) and in trans (Radman-Livaja and Rando 2009) by regulatory proteins in responding to dynamic environmental factors such as heat shock or hormonal treatment (Schones et al. 2008; Shivaswamy et al. 2008; He et al. 2010). The nucleosome dynamics regulated by these trans factors can happen at the position level, including position shifts and fuzziness changes, and/or at the occupancy level. Studying the dynamic nucleosomes carrying these changes will facilitate greater understanding of how chromatin and transcription factors cooperate to regulate cellular responses to environmental or physiological changes.
Algorithms devoted to nucleosome calling and scoring from a single experimental condition have been made publicly available (Zhang et al. 2008b; Flores and Orozco 2011). However, systematically analyzing dynamic nucleosomes remains a challenge. As dynamic nucleosomes are often associated with functional consequences, there is an urgent need to develop methods for systematic comparison between nucleosome maps. Furthermore, given that technical variations in nucleosome library preparation and sequencing are usually inevitable; many data filtering and processing steps are normally required to compensate for technical variations. However, the contribution of each step toward the functional interpretation of nucleosome dynamics has never been systematically studied; therefore, previous nucleosome analyses always omitted some important steps. Finally, despite the high resolution offered by MNase-seq data at the single-nucleotide resolution, most previous MNase-seq data analysis depended on a sliding window approach or peak calling algorithms for identifying the nucleosome unit before applying a statistical test to compare each unit between different conditions (He et al. 2010; Hu et al. 2011; Fu et al. 2012). However, the exact locations of nucleosomes are still poorly defined, especially in more fluid regions, where nucleosomes become fuzzy. Meanwhile, at the nucleosome unit resolution, subtle nucleosome changes such as position shifts and fuzziness changes are hardly detectable.
In this paper, we describe an integrated bioinformatics pipeline, DANPOS, explicitly designed for comparative analysis of nucleosome physical organization at single-nucleotide resolution. Based on information extracted from modeling the distribution of reads in each sample, the calculation of nucleosome occupancy is significantly enhanced by minimizing experimental variation. Optimized data normalization and statistical testing are used to infer reliable dynamic nucleosome changes. A side-by-side comparison with other leading algorithms shows that the DANPOS pipeline significantly improves the identification of dynamic nucleosomes. We explored the methods to classify different categories of nucleosome dynamics, such as position shift, fuzziness change, and occupancy change. Pathway analysis of genes associated with these changes indicates that each category can be involved in specific biological functions. Furthermore, the saturation analysis in DANPOS suggests that, even at the current sequencing depth of ∼200-fold, dynamic nucleosome analysis can still be improved significantly by further increasing the sequencing depth. Finally, we showed that DANPOS is robust in defining dynamic nucleosomes, not only in promoters, but also in distal regulatory regions in the mammalian genome.
Results
Flowchart of the DANPOS algorithm
Starting from sequencing reads that have been mapped to the reference genome, there are five steps in the DANPOS workflow (Fig. 1). The first step is preliminary data processing that calculates nucleosome occupancy from the mapped reads (Fig. 1A). DANPOS provides an option to remove clonal reads with identical sequences resulting from possible overamplification during sample preparation. We determine clonal reads based on their extremely high coverage relative to the mean coverage across the genome based on a Poisson P-value cutoff. Although nucleosome size is 147 bp in higher eukaryotes, the real size of DNA fragments after MNase digestion can vary from ∼120 bp to 170 bp (Kaplan et al. 2009; Shu et al. 2011). This variation can be compensated for by shifting each read toward the 3′ direction for half of the estimated fragment size. For single-end sequencing, the average fragment size can be estimated by the distribution of distances between reads on the positive and negative strands; whereas for paired-end sequencing, the fragment size is determined directly by the distance between the paired-end reads. The read length is then adjusted to half of the nucleosome size (i.e., 74 nt) to enhance the signal-to-noise ratio (Zhang et al. 2008b). Finally, nucleosome occupancy is calculated as the count of adjusted reads covering each base pair in the genome.
Figure 1.
Flowchart of the nucleosome dynamic analysis pipeline in DANPOS. (A) Schematic illustration of nucleosome occupancy calculation. (Purple line) DNA sequence. (Red arrows) Normal reads at both ends of a nucleosome; (gray arrows) clonal reads. (Red areas) Calculated nucleosome occupancy. (B) The alternative data normalization methods. (C) Calculation of differential signals at single nucleotide resolution. (Red and sky-blue areas) Nucleosome occupancies in samples A and B, respectively. (Blue area) Differential signal at single nucleotide resolution between samples A and B. Five statistical tests that can be used to calculate differential signal are listed on the right. (D) Differential nucleosome peaks. (Blue dashed curves) Differential signals at single nucleotide resolution. (Blue bars) Differential nucleosome peaks called based on the differential signals. (E) A cartoon to show the three categories of dynamic nucleosomes. (Red and sky-blue dashed curves) Nucleosome occupancy in samples A and B, respectively. (Red and sky-blue blocks) Baseline nucleosome peaks with differential signals and peaks displayed as in D.
The sequencing depths of different samples also vary in MNase-seq experiments. Therefore, to make the occupancy levels comparable, DANPOS provides several normalization options in its second step (Fig. 1B), including quantile normalization (default), global scaling, and bootstrap sampling. In the third step (Fig. 1C), DANPOS calculates the nucleosome differential signal at single-nucleotide resolution between control and treatment samples based on a Poisson test (default), which has been widely used in sequencing data analysis (Xi et al. 2011). Alternative statistical methods are also supported, such as Pearson's χ2 test (Robinson and Parkin 2008), negative binomial test (Robinson et al. 2009), fold-change, and numerical subtraction. In the fourth step, DANPOS performs peak calling on the single-nucleotide-resolution differential signal to identify differential peaks (Fig. 1D). Finally, in the fifth step, the differential peaks are classified into three categories including nucleosome position shifts, fuzziness changes, and occupancy changes (Fig. 1E).
DANPOS defines accurate nucleosome maps
Although DANPOS is explicitly designed for dynamic nucleosomes, several lines of evidence suggest that its peak-calling algorithm can also be used to accurately infer the nucleosome map from a single experimental condition. First, DANPOS analysis of pubic yeast nucleosome sequencing data (Kaplan et al. 2009) can reproduce well-known nucleosome positions that have been extensively mapped through conventional techniques (Supplemental Fig. S1A) and alternative high-throughput nucleosome-mapping methods such as tiling array hybridization (Supplemental Fig. S1B). Second, based on simulated nucleosome data (Supplemental Fig. S2; see Methods) that can serve as the gold standard to assess the performance of different algorithms, DANPOS performs significantly better than another nucleosome calling algorithm, nucleR (Flores and Orozco 2011), in defining nucleosome position, occupancy, and fuzziness (Supplemental Note S1; Supplemental Fig. S3).
Simulated nucleosome data reveals the superior performance of DANPOS
To achieve an optimal solution for analysis of nucleosome dynamics, we evaluated various alternative methods for each step based on simulated nucleosome data that can serve as the gold standard to assess the performance of different algorithms and chose the best method as the default in the DANPOS workflow. For each alternative method, we rank the defined dynamic nucleosomes by P-value from the lowest to highest and plot the true positive rate relative to the false positive rate as ROC curves (Fig. 2).
Figure 2.
ROC curves indicate that the default DANPOS algorithm improves the identification of dynamic nucleosomes. Each column assesses the rate of true and false positive dynamic nucleosomes identified by different preliminary data processing methods (left), data normalization methods (middle), or statistical tests (right). Each row presents the results from simulation of nucleosome occupancy changes (top), fuzziness change (middle), and position shift (bottom). The known nucleosome changes were simulated from a nucleosome reference map (see Methods).
We first determined how the detection of dynamic nucleosomes could be influenced by the preliminary data processing steps that use mapped reads to determine nucleosome occupancy (Fig. 1A). We compared the default DANPOS pipeline to three alternative pipelines, each of which lacks one of DANPOS' preliminary data processing steps: (1) a pipeline that does not remove clonal reads; (2) a pipeline that does not adjust average fragment sizes to be the same across samples; and (3) a pipeline that adjusts each read to a full nucleosome size rather than the default half nucleosome size. A pipeline specifically designed to correct fragment size based on paired-end reads was also compared with the default pipeline. The results revealed that the default DANPOS pipeline always performed the best compared to the three alternatives (Fig. 2, left column), suggesting that all three preliminary data processing steps contribute significantly to nucleosome dynamic analysis. The performance of DANPOS was even better when paired-end information was utilized.
We also compared three data normalization options, including quantile normalization, global scaling, and bootstrap sampling (Fig. 2, middle column). For the analysis of nucleosome occupancy change, quantile normalization performs significantly better than the other two methods, whereas the three methods show little difference in analyzing nucleosome fuzziness change or position shift.
Several popular statistical methods have been used for detecting differential signals in sequencing data; these include, but may not be limited to, Poisson test (Xi et al. 2011), Pearson's χ2 test (Robinson and Parkin 2008), and negative binomial test (Robinson et al. 2009). Their efficiencies in the analysis of MNase-seq data remain to be explored. In addition to these established statistical methods, bench researchers are also interested in simple fold change or direct numerical subtraction for differential signal detection. We compared the efficiencies of these methods in detecting dynamic nucleosomes (Fig. 2, right column,). The results indicate that the Poisson test always performs the best, especially for analyzing nucleosome occupancy change. Among the other methods, the negative binomial test is good for nucleosome position shift, whereas the χ2 test is acceptable for fuzziness change and position shift. In contrast, neither fold change nor direct subtraction appears to be a good measurement for dynamic nucleosomes. Altogether, our comparison of methods clearly indicate that the optimized methodology of DANPOS significantly improves the accuracy in defining dynamic nucleosomes. We have to acknowledge that the amount of improvement depends on the degree of noise simulated in the nucleosome data, e.g., the random sampling error and the variation in nucleosome fragment sizes.
Dynamic nucleosomes detected by DANPOS reflect environmental changes
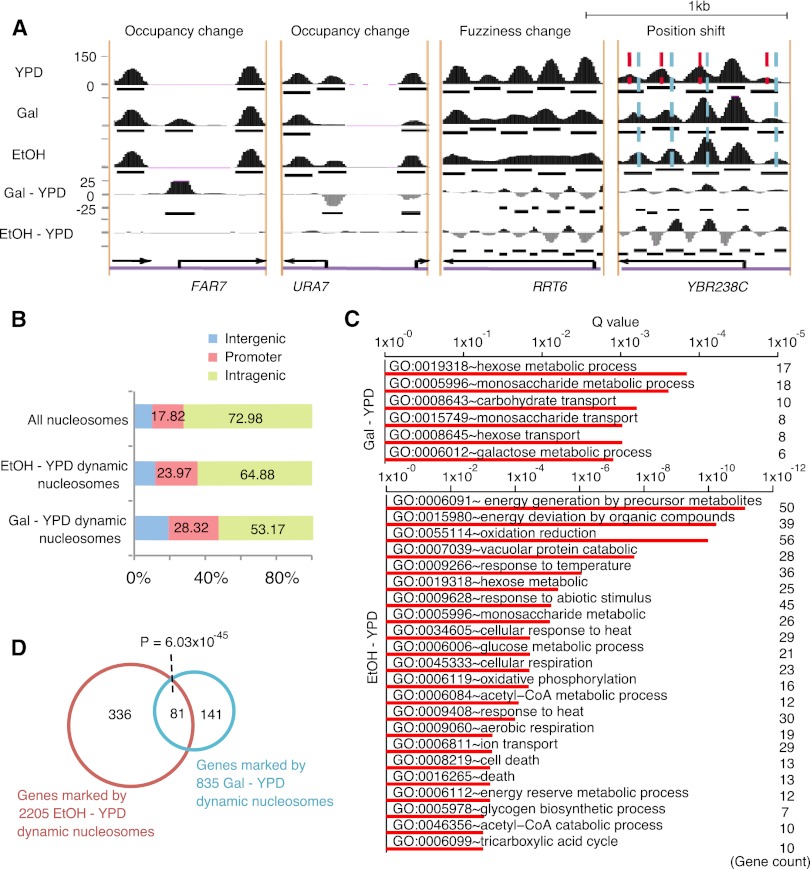
We further tested DANPOS using published MNase-seq data sets of yeast grown in rich media (YPD), YPD supplemented with galactose (Gal), or YPD supplemented with ethanol (EtOH), containing 24, 12, and 15 million reads, respectively (Kaplan et al. 2009). Using YPD as a baseline and based on a P-value cutoff of 1 × 10−7 (or a false discovery rate of ∼0.001), DANPOS identified 835 Gal-YPD and 2205 EtOH-YPD total dynamic nucleosomes carrying changes in either position, fuzziness, or occupancy. Visual inspection of individual dynamic nucleosomes indicates that they are more likely to be located close to the promoter regions (Fig. 3A). Statistical analysis confirms that the dynamic nucleosomes are enriched in the promoter regions encompassing −350 bp to +50 bp flanking the transcription start site (Fig. 3B). For example, although only 17.82% of total nucleosomes are in promoter regions, 28.32% of Gal-YPD dynamic nucleosomes are located in promoter regions, with an enrichment P-value < 2.2 × 10−16 based on Fisher's exact test. This observation suggests that dynamic nucleosomes are likely to play important roles in transcription regulation.
Figure 3.
Dynamic nucleosomes identified by DANPOS explain environmental changes. (A) Snapshots of individual dynamic nucleosome regions. Categories of dynamic nucleosomes were labeled on top of each region. Nucleosome occupancies in YPD, Gal, and EtOH conditions were plotted as black areas in the top three tracks, where black bars in each track represent baseline nucleosome peaks. Nucleosome differential signals between YPD and Gal (Gal-YPD) or EtOH (EtOH-YPD) are plotted as black (positive) or gray (negative) areas in the bottom two tracks, and black bars in these tracks represent dynamic nucleosome peaks. (Black arrows) Position and direction of genes. Red and sky-blue dashed lines in the position shift plot indicate the dyads of nucleosomes in the YPD and EtOH conditions, respectively. (B) The percentages of dynamic nucleosomes in intragenic, promoter, and intergenic regions. Promoter is defined as the −350 to +50-bp regions flanking each TSS. Intragenic region is defined as the region from 50 bp downstream from the TSS to the 3′ end of the gene, and all remaining regions are defined as intergenic. (C) Enrichment of biological processes in genes associated with dynamic nucleosomes on promoters. (D) Counts of genes associated with dynamic nucleosomes on promoters.
After mapping dynamic nucleosomes to promoters, we sought to evaluate the functional enrichment of associated genes using the DAVID pathway analysis (Fig. 3C; Huang da et al. 2009). As expected, Gal-YPD dynamic nucleosomes are associated with genes involved in the transport or metabolism of galactose (GO:0006012), hexose (GO:0008645, GO:0019318), and monosaccharides (GO:0015749, GO:0005996). In contrast, EtOH-YPD dynamic nucleosomes are associated with genes involved in biological processes related to ethanol metabolism, including energy derivation by oxidation of organic compounds (e.g., GO:0006091, GO:0015980, GO:0055114), acetyl-CoA metabolic processes (e.g., GO:0006084), and heat response (e.g., GO:00034605). Thus, the medium-specific dynamic nucleosomes reflect regulation of transcription programs needed for adaption to environmental change.
Yeast is adept at bioconversion of sugar to ethanol (Bai et al. 2008). This raises the question of whether culturing yeast in the presence of ethanol affects sugar-related processes. We observed that biological processes enriched in gene promoters associated with Gal-YPD dynamic nucleosomes are also associated with EtOH-YPD dynamic nucleosomes (e.g., GO:0019318, GO:0005996). In agreement with this observation, there are 81 genes in common between the 222 genes associated with Gal-YPD dynamic nucleosomes and the 417 genes associated with EtOH-YPD dynamic nucleosomes, with a significant Fisher's exact test P-value of 6.03 × 10−45 supporting the overlap (Fig. 3D).
DANPOS improves the functional interpretation of dynamic nucleosomes
The function enrichment of dynamic nucleosomes provides us an alternative strategy to further evaluate various methods at each step based on real nucleosome data. For each alternative method, we mapped the identified dynamic nucleosomes to gene promoters and ranked the associated genes by the P-value of their nucleosome change. The same number of top ranked genes from each method was then subjected to DAVID functional annotation. We anticipated that the most effective methods would provide us with the most significant functional enrichment related to the underlying environmental change.
The results confirm that the default DANPOS preliminary data processing pipeline performs much better than the other three pipelines that either did not remove clonal reads, did not adjust average fragment sizes, or adjusts read length to a full nucleosome size rather than the default half size. For each of the six biological processes enriched in Gal-YPD dynamic nucleosomes (Supplemental Fig. S4, top left), DANPOS achieves the most significant enrichment Q-values. For 10 of the 15 biological processes enriched in EtOH-YPD dynamic nucleosomes (Supplemental Fig. S4, bottom left), DANPOS also achieves the most significant Q-values, whereas the pipeline that sets the read length to the full nucleosome size performs best for four other biological processes.
In contrast to the large performance variations observed based on simulated data sets, modest differences were observed among the three normalization methods in terms of the functional enrichments of genes associated with dynamic nucleosomes (Supplemental Fig. S4, middle column). Consistent with the conclusion from the simulated nucleosome data, among the different statistical tests (Supplemental Fig. S4, right column), the Poisson test always performs the best, yielding the highest functional enrichment for genes associated with dynamic nucleosomes.
DANPOS distinguishes categories of dynamic nucleosomes
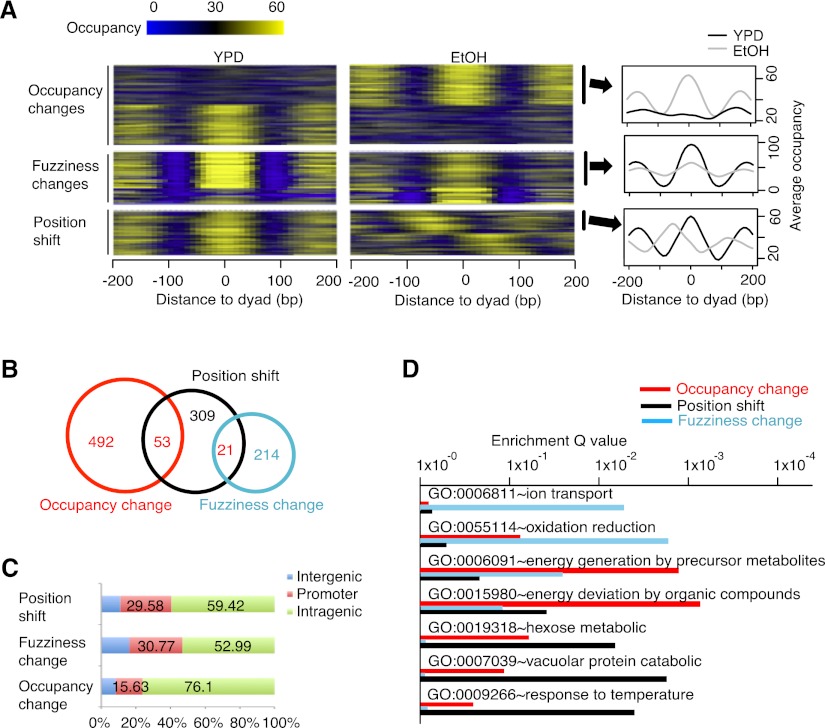
Nucleosome changes can generally be classified into three categories: position shifts, fuzziness changes, and occupancy changes. These categories can be observed either individually or together on each nucleosome (Fig. 1E). The differential nucleosome peaks called from single nucleotide resolution differential signals include all three categories (Fig. 3A). For each dynamic nucleosome, DANPOS estimates the degree to which each category applies, based on baseline nucleosome peaks from the control and treatment samples. We estimated the position shift by calculating the relative distance between the control nucleosome peak and the treatment nucleosome peak. Technically, fuzziness refers to the degree that read positions in each nucleosome peak deviate from the most preferred nucleosome position. We thus used the standard deviation of read positions in each peak as an estimate of nucleosome fuzziness (Jiang and Pugh 2009). The P-value of difference in fuzziness was then calculated using the F-test. Nucleosome occupancy change was determined by a Poisson test on the difference between the maximum values of the control and treatment nucleosome peaks. Some nucleosome occupancy changes are also coupled with fuzziness changes to some degree, i.e., reads move from the nucleosome dyad to the neighboring linker regions (Figs. 1E, 4A). To better explore the difference between pure fuzziness and pure occupancy changes, we decided to exclude those nucleosomes carrying both fuzziness and occupancy changes because we were not able to assign those nucleosomes to either category for the downstream functional analysis.
Figure 4.
DANPOS classifies dynamic nucleosomes as position shifts, fuzziness changes, and occupancy changes. (A) Heatmaps (left and middle) and average profile plot (right) to show nucleosome occupancy in regions containing EtOH-YPD dynamic nucleosomes. Each line in the heatmaps represents a 400-bp region flanking the dyad of a dynamic nucleosome; for position shifts, the heatmap for the EtOH condition is plotted flanking the dyads defined in the YPD condition. Nucleosomes showing EtOH-YPD occupancy increase (top right), fuzziness increase (middle right), or position shift toward the 5′ direction on positive strand (bottom right) were pooled to plot average occupancy at each base pair flanking dyads. (B) Venn diagram for the overlap between different categories of EtOH-YPD dynamic nucleosomes. (C) The percentage of EtOH-YPD dynamic nucleosomes in intergenic, promoter, and intragenic regions, as defined in Figure 2B. (D) Enrichment of biological processes in each category of EtOH-YPD dynamic nucleosomes.
The large number of EtOH-YPD dynamic nucleosomes provides an opportunity to determine the biological functions of each category as well as the relationship among them (Fig. 4). Of the 2205 EtOH-YPD dynamic nucleosomes, 383 show position shifts between 50 bp and 90 bp, 235 show fuzziness changes based on F-test P-value cutoff 1 × 10−7, 545 show strong occupancy changes by a Possion test P-value cutoff 1 × 10−7 with little fuzziness changes (F-test P-value > 1 × 10−5) (Fig. 4A,B), 126 carry both fuzziness and occupancy changes and thus were not included in the downstream analysis. The remaining dynamic nucleosomes cannot be categorized into any group. Nucleosome position shifts show little overlap with occupancy changes or fuzziness changes with Fisher's exact P-values greater than 0.99. The fuzziness changes and position shifts are highly enriched on promoters (Fig. 4C), with 29.58% and 30.77% located in the region from −350 bp to +50 bp flanking TSS, whereas occupancy changes are more likely to be enriched on gene bodies. Each nucleosome dynamic category shows unique specificity in its association with biological processes (Fig. 4D). The fuzziness changes have strong specificity to mitochondrial functions, e.g., the ion transport (GO:0006811) and oxidation (GO: 0055114) processes, whereas the occupancy changes are more specific to energy derivation in the metabolic processes (GO:0006091 and GO: 0015980). The major functions of nucleosome position shifts appear to be the metabolic processes of hexose (GO:0019318) and vacuolar proteins (GO:0007039). Interestingly, nucleosome position shifts seem to be more related to environmental factors such as temperature (GO:0009266). Collectively, these dynamic nucleosomes are all involved in functions related to energy deviation by oxidation of organic compounds, supporting the fact that they are all dynamic nucleosomes and are functionally related to the shift between EtOH and YPD media.
One caveat in the detection of the fuzziness change is that the single nucleotide resolution Poisson test initially used to define total differential peaks was not a direct test of fuzziness per se. Therefore, we asked whether there are “false negative” fuzziness changes that might have been missed by the initial identification of total differential peaks but could be rescued by the nucleosome resolution F-test. We found 75 genes associated with 117 such nucleosomes. However, there was no function term enriched with these genes (Supplemental Table S1, top). Adding these rescued nucleosomes back to our original analysis impeded the functional interpretation of fuzziness changes (Supplemental Table S1, middle). A careful examination of these 117 nucleosomes indicated that they only exhibit marginal fuzziness changes, which may not be biologically significant. Thus, we conclude that DANPOS can faithfully detect all major fuzziness changes. Taken together, these results indicate that DANPOS is able to distinguish different categories of dynamic nucleosomes, each of which may have unique biological functions.
The current sequencing depth does not reach saturation for nucleosome dynamic analysis
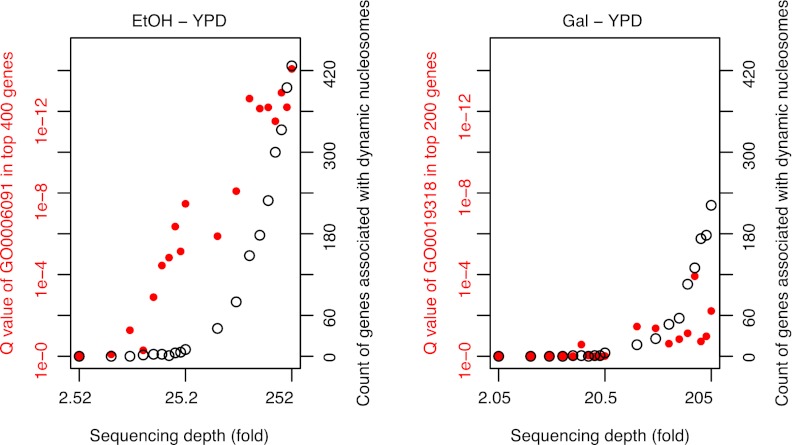
Although the number of reads required to detect static nucleosomes is generally known (Zhang and Pugh 2011), the required depth of sequencing for detecting dynamic nucleosomes between different conditions is largely unexplored. An important question is whether the sequencing depth has reached a saturation point beyond which no additional functional dynamic nucleosome can be detected. Considering that each nucleosome plus its linker sequence is ∼200 bp (Zhang and Pugh 2011), the 12, 15, and 24 million sequencing reads obtained for the Gal, EtOH, and YPD samples have covered the yeast genome (12 Mb) at 200-, 250-, and 400-fold, respectively. We randomly sampled 20 subsets with read counts ranging from 1% to 100% of the total reads. DANPOS then produced a saturation plot to report the proportion of dynamic nucleosomes that could still be detected when using a subset of the reads (Fig. 5). If the sequencing depth exceeded saturation, the number of dynamic nucleosomes will reach a plateau with an increasing number of reads. However, we found that neither the Gal-YPD nor the EtOH-YPD data set was saturated. The number of dynamic nucleosomes keeps increasing with an increasing number of reads.
Figure 5.
The current sequencing depth is not saturated for the detection of dynamic nucleosomes. Counts or functional enrichment Q-values of genes containing dynamic nucleosomes on promoters were plotted as a function of sequencing depth. The sequencing depth is estimated by the read counts multiplied by the nucleosome unit size (200 bp) and divided by the yeast genome size (12 Mb). The biological processes “energy generation by precursor metabolites” (GO:0006091) and “hexose metabolic process” (GO:0019318) were used to estimate the function enrichment of EtOH-YPD and Gal-YPD dynamic nucleosomes, respectively.
One caveat in the saturation analysis is that “fuzzy” nucleosomes or background noise might give us more “false positive” dynamic nucleosomes with an increasing number of reads. In order to distinguish genuine dynamic nucleosomes from background noise, we decided to use the degree of functional enrichment of the same number of top-ranked dynamic nucleosomes in the saturation analysis (Fig. 5). The rationale is that the background noise should be randomly distributed across the genome and thus has no functional enrichment. Our analysis demonstrated that for EtOH-YPD, increasing the sequencing depth significantly increases the degree of functional enrichment of dynamic nucleosomes, strongly indicating lack of saturation; whereas for Gal-YPD, the same analysis indicated a slightly saturated sequencing depth, probably because there are less dynamic nucleosomes involved in the Gal-YPD environmental change (Fig. 3D). Altogether, these results suggest that, even at the current sequencing depth of ∼200-fold, dynamic nucleosome analysis can still be improved significantly by further increasing the sequencing depth.
DANPOS analysis of dynamic nucleosomes in the mammalian genome
The proximal promoter is largely composed of well-positioned nucleosomes, e.g., more than 10 well-positioned nucleosomes are observed flanking yeast TSS (Fig. 6A), whereas chromatin in distal regulatory regions is in a more fluid organization where nucleosomes become fuzzier. Although it may not be as much of an issue for the yeast genome, the definition and functional interpretation of intergenic dynamic nucleosomes is a largely unexplored problem in the mammalian genome.
Figure 6.
DANPOS defines functional dynamic nucleosomes in both promoter proximal and distal regions in the human genome. (A) Average nucleosome occupancy plotted as a function of distance to the nearest yeast TSS. (B) Average nucleosome occupancy plotted as a function of distance to the nearest human TSS. (C) Boxplots showing relative distance of human dynamic nucleosome to the nearest TSS. The dynamic nucleosomes during differentiation of human hematopoietic stem cells (HSCs) to CD36+ cells were divided into three groups. (D) Pie chart showing the number of human dynamic nucleosomes in each group. (E) Enrichment of function terms for each group. GREAT (Mclean et al. 2010) was used to analyze the functional significance of dynamic nucleosomes in each group.
To demonstrate the broad utility of DANPOS, we analyzed dynamic nucleosomes during differentiation of human hematopoietic stem cells (HSCs) to CD36+ erythrocytes (Hu et al. 2011). There are 156,737,891 and 158,096,184 uniquely mapped MNase-seq reads for HSCs and erythrocytes cells, respectively. We observed four well-positioned nucleosomes around the TSS, whereas nucleosomes in more distal regions become fuzzier (Fig. 6B). Based on a Poisson test P-value cutoff of 1 × 10−50, we defined in total 1896 dynamic nucleosomes in the human genome. To test the functional relevance of dynamic nucleosomes in promoter proximal and distal regions, we further divided them into three groups located within 15 kb (group I), between 15 kb and 50 kb (group II), and beyond 50 kb (group III) from the nearest TSS (Fig. 6C), each containing 574, 566, and 756 dynamic nucleosomes, respectively (Fig. 6D). Functional annotation using GREAT (Mclean et al. 2010) indicates that dynamic nucleosomes in all three groups are significantly enriched in functions associated with blood cell differentiation and changes in phenotypes of the immune system (Fig. 6E).
We further validated the broad utility of DANPOS with nucleosome data from mouse embryonic stem cell (ESC) differentiation (Li et al. 2012). The data contain 144,096,443 and 177,233,717 MNase-seq reads in embryonic stem (ES) cells and endoderm/hepatic progenitor (EHP) cells that are differentiated from ES cells, respectively. Based on a Poisson test P-value cutoff of 1 × 10−50, we used DANPOS to define 5505 dynamic nucleosomes in total, of which 781 (14.19%), 977 (17.75%), and 3747 (68.07%) were mapped to the promoters, enhancers, and the other genomic regions in the mouse genome, respectively (Supplemental Fig. S7A). As expected, the GREAT functional annotation indicates that all three groups are significantly enriched in functions associated with stem cell differentiation or development of tissues derived from endoderm, e.g., embryonic pattern specification (GO:0009880) (Supplemental Fig. S7B). Taken together, these results suggest that DANPOS is robust in defining functional dynamic nucleosomes, not only in promoters, but also in distal regulatory regions in the mammalian genome.
Discussion
Here we report a comprehensive bioinformatics pipeline, DANPOS, for dynamic analysis of nucleosome position and occupancy. Using public nucleosome data from yeast, human, and mouse, we demonstrated that DANPOS is very effective for detecting functionally relevant dynamic nucleosomes, not only in promoters in yeast, but also in distal regulatory regions with more fuzzy nucleosomes, in complex genomes such as that of mammalian.
Our results are novel and significant in three major aspects: (1) DANPOS is a statistical pipeline explicitly designed for the analysis of dynamic nucleosomes. Most of the previous analysis programs have been devoted to nucleosome calling from only one experimental condition. However, much less attention has been paid to the dynamic changes of nucleosomes between different conditions, such as disease vs. control. (2) DANPOS is independent of any nucleosome peak-calling program and provides single-nucleotide resolution nucleosome dynamic analysis. This allows us to detect nucleosome fuzziness changes and location shifts in addition to occupancy changes using a uniform statistical framework. Previous MNase-seq analyses largely focused on occupancy change at nucleosome unit resolution. With resolution compromised at this level, subtle nucleosome changes, such as position shifts and fuzziness changes, are hardly detectable. (3) Provide a performance benchmark with other leading algorithms for nucleosome dynamics analysis. There are many steps in the nucleosome analysis workflow (Fig. 1). However, the contribution of each step toward the functional interpretation of nucleosome dynamics has never been systematically studied. Therefore, previous nucleosome analyses always omitted some important steps. We provided simulated nucleosome data, which can serve as the gold standard to assess the performance of different algorithms. We also used the significance of functional enrichment related to the underlying environmental change for the algorithm evaluation on real nucleosome data. The results indicate that DANPOS achieves the highest accuracy and the most significant functional enrichment, which can faithfully explain the environmental changes.
The comparison of different statistical testing methods reveals considerable variation in the ability to detect functional dynamic nucleosomes. The dynamic nucleosomes detected by Poisson test or Pearson's χ2 test show the most significant functional enrichments. Surprisingly, the negative binomial test, which has been shown to be superior to the Poisson test for modeling RNA sequencing data (Robinson et al. 2009), appears to be much less efficient for MNase-seq data. Unlike the Poisson distribution, which has equal mean and variance, the negative binomial distribution uses different mean and variance to address the so-called overdispersion problem. However, the number of replicates in our MNase-seq data sets might have been too small to estimate both parameters for the negative binomial. Thus, we would suggest that the comparison between the Poisson and negative binomial tests in modeling MNase-seq data may need to be revisited when more replicates are available.
The ability to evaluate and predict the sequencing depth requirement is an important aspect of dynamic nucleosome analysis. Our results indicate that none of the three data sets used in this work (200- to 400-fold) is saturated. Deeper sequencing depth not only increases the number of dynamic nucleosomes and associated genes but also improves functional enrichment of the same number of most dynamic nucleosomes. We propose that functional enrichment of dynamic nucleosomes can be used as a more biologically meaningful metric to assess the degree of sequencing saturation in detecting nucleosome dynamics.
In summary, because nucleosome dynamics is increasingly recognized as a key regulator of genomic function, deciphering dynamic nucleosomes between different conditions will continue to be a major research interest. Although exclusively tested on nucleosome data from yeast and humans in this study, DANPOS is well suited to studying nucleosome dynamics in other organisms. We believe that DANPOS' superior performance will greatly facilitate the understanding of the roles of chromatin, and particularly nucleosome organization, in various cellular functions and disease processes.
Methods
Public nucleosome data
The uniquely mapped nucleosome sequencing reads of yeast grown in YPD, Gal, and EtOH media were downloaded from GEO using accession numbers GSM351492, GSM351493, and GSM351494, respectively. Human and mouse nucleosome data were downloaded from GEO with accession numbers GSM651558 and GSM651559. Gene-related analyses were based on the yeast Ensembl Genes and human RefSeq Genes downloaded from the UCSC Genome Browser (http://genome.ucsc.edu).
MNase-seq data simulation
We generated a yeast nucleosome reference map by pooling three public data sets (GSM351492, GSM351493, and GSM351494) together. For each of the 54,668 nucleosomes in the reference map, occupancy and fuzziness were determined by total number of reads within the nucleosome unit (147 bp) and standard deviation of read positions relative to the nucleosome center, respectively. To simulate dynamic nucleosome changes in the treatment sample, we assigned occupancy change up to 100%, fuzziness change from 1/8 to eightfold, and position shift up to 70 bp away from the nucleosome center to 50% of the total nucleosomes.
For each nucleosome unit in the reference or treatment maps, multiple nucleosome fragments were randomly sampled with fragment center following a normal distribution (mean=nucleosome center and SD=nucleosome fuzziness). Paired-end MNase-seq reads were then simulated from each nucleosome fragment, allowing variable fragment size (normal distribution mean = 147 bp and SD = 20 bp). Single-end reads were also assigned to a small portion (0.05%) of nucleosomes to accommodate the fact that sometimes only one end of the paired-end reads can be mapped. Clonal reads that were exceptionally preferred in PCR amplification were also added to 1200 genomic sites, allowing up to 1000 clonal reads at each site. In order to simulate a treatment nucleosome map that represents a library derived from more extensively digested chromatin. The nucleosome fragment size in the treatment map was further shortened by a size following a normal distribution (mean = 40 bp and SD = 20 bp). The mean value of 40 bp represents the global difference between samples, whereas the standard deviation of 20 bp indicates the difference between nucleosomes within the same sample. Finally, to simulate variation in sequencing depth, we randomly sampled 50% and 25% of the total 47,548,776 reads for the reference and treatment nucleosome maps, respectively.
Removing clonal reads
“Clonal reads” refers to a group of reads derived from the same fragment that has been preferentially over-amplified in the preparation of a high-throughput sequencing library. These reads will be mapped to exactly the same position on the reference genome and can cause the read counts at such positions to become extremely large. When we check the distribution of read count per nucleotide in the whole genome, the read counts at such positions would appear to be outliers with significant Poisson test P-values. With the current sequencing throughput, Poisson distribution can be used to model the read count per nucleotide (Zhang et al. 2008a; Xi et al. 2011). DANPOS removes the per-nucleotide read counts that are extremely large compared to the whole genome average by a Poisson P-value cutoff of 1 × 10−10.
Calculating nucleosome occupancy from sequencing reads
The nucleosome DNA fragment size is 147 bp in higher eukaryotes; therefore, the nucleosome data analysis pipelines always simply adjust each read to 147 bp. However, due to technical variations in sample preparation, the real DNA fragment sizes generated in each experiment could range from ∼120 bp to 170 bp (Kaplan et al. 2009; Shu et al. 2011). If the average DNA fragment sizes in sample A and sample B are different, simply adjusting each read to 147 bp in the two samples can introduce extensive false positives in differential analysis (Supplemental Fig. S5). This variation can be compensated for by shifting each read toward the 3′ direction for half of the estimated fragment size. For single-end sequencing, the average fragment size can be estimated by the distribution of distances between reads on the positive and negative strands, whereas for paired-end sequencing the fragment size is determined directly by the distance between the paired-end reads. DANPOS assesses the average DNA fragment size in single-end MNase-seq sample by calculating the Pearson correlation coefficient between the read densities on the positive and negative strands (Kharchenko et al. 2008). The coefficient would reach its maximum value when the distance equals the average fragment size in a sample. Although the nucleosome DNA fragment size is 147 bp, DANPOS adjusts read lengths to 74 bp by default, as it was reported that an adjustment to 74 bp rather than 147 bp could improve the signal-to-noise ratio (Supplemental Fig. S6; Zhang et al. 2008b). Finally, nucleosome occupancy at each base pair is calculated as local read coverage.
Calculating differential signals
DANPOS calculates nucleosome differential signals at single-nucleotide resolution based on Poisson distribution by default. When nucleosome occupancy is higher in sample A than in sample B, we calculate the P-value of nucleosome occupancy in sample A based on a Poisson distribution with λ defined by the nucleosome occupancy in sample B. DANPOS also supports Pearson χ2 test based on a 2 × 2 contingency table (see Table 1).
Table 1.
The 2 × 2 contingency table for the χ2 test in DANPOS

To compare with the negative binomial model as implemented in the EdgeR package (http://www.bioconductor.org/packages/2.3/bioc/html/edgeR.html), we generated a read count table in which columns represent samples and rows represent every nucleotide in the genome. EdgeR takes the table as input and calculates a differential P-value for each nucleotide. To allow for peak calling and visualization of nucleosome changes at single-nucleotide resolution in the genome browser, DANPOS transforms a differential P-value to a –log10(P-value) (e.g., a P-value of 1 × 10−10 will be transformed to 10). Fold change and numerical subtraction are also provided as alternative differential test options in DANPOS.
Peak calling
Nucleosome peaks can be defined by peak width (w bp) and peak height (q reads). Because nucleosomes are arranged as an array of beads along a DNA strand, another important feature of nucleosome peaks is the distance between neighboring peaks (d bp). Peaks can be first called using a w-bp sliding window to identify a bell-shaped curve, with the occupancy maximum higher than q and located in the middle of the sliding window. Neighboring bell-shaped curves that are closer than d could be merged together. Each peak could then be determined by the bell-shaped curve and its edges, which are determined by searching for the lowest flanking occupancy troughs. Differential peaks were called with the same algorithm, except that the peak height cutoff q was replaced by a differential P-value cutoff.
Peak calling by nucleR
The following R scripts were used to call nucleosomes by nucleR version 1.4.0:
library(nucleR);
d=readAligned(‘control.simu.reads.frag80.bam',type=”BAM”);
t=processReads(d, type=”single”,fragmentLen=80, trim=80);
# average fragment size has been adjusted to 80 before;
c = coverage.rpm(t);
f = filterFFT(c, pcKeepComp=0.02);
peaks = peakDetection(f, score=TRUE, width=148); and
export.bed(peaks, name=”nuc”, splitByChrom=FALSE).
Data access
The source code is provided in the Supplemental Material, and new versions will be frequently released at http://code.google.com/p/danpos/.
Acknowledgments
This work was partially supported by Cancer Prevention and Research Institute of Texas grant RP110471-C3, DOD grant PC094421, and 973 project 2010CB944900 of China (to W.L.); and NIH grant HG004840 (to X.P.). We thank Bo Qin for help with the Negative Binomial Test. We would also like to thank all the reviewers and users of DANPOS for their helpful feedback.
Author contributions: K.C. and W.L. conceived the project, designed the algorithms, analyzed the data, and wrote the manuscript. K.C. implemented the algorithms and wrote the software package. All authors participated in the discussions and edited the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.142067.112.
References
- Bai FW, Anderson WA, Moo-Young M 2008. Ethanol fermentation technologies from sugar and starch feedstocks. Biotechnol Adv 26: 89–105 [DOI] [PubMed] [Google Scholar]
- Chen K, Meng Q, Ma L, Liu Q, Tang P, Chiu C, Hu S, Yu J 2008. A novel DNA sequence periodicity decodes nucleosome positioning. Nucleic Acids Res 36: 6228–6236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG 2010. Covalent histone modifications–miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10: 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM 2010. Conserved nucleosome positioning defines replication origins. Genes Dev 24: 748–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feser J, Tyler J 2011. Chromatin structure as a mediator of aging. FEBS Lett 585: 2041–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores O, Orozco M 2011. nucleR: A package for non-parametric nucleosome positioning. Bioinformatics 27: 2149–2150 [DOI] [PubMed] [Google Scholar]
- Fu K, Tang Q, Feng J, Liu XS, Zhang Y 2012. DiNuP: A systematic approach to identify regions of differential nucleosome positioning. Bioinformatics 28: 1965–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez L, Oktaba K, Scheuermann JC, Gambetta MC, Ly-Hartig N, Müller J 2011. The role of the histone H2A ubiquitinase Sce in Polycomb repression. Development 139: 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. 2010. Nucleosome dynamics define transcriptional enhancers. Nat Genet 42: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A 2011. Histone modification: Cause or cog? Trends Genet 27: 389–396 [DOI] [PubMed] [Google Scholar]
- Hu G, Schones DE, Cui K, Ybarra R, Northrup D, Tang Q, Gattinoni L, Restifo NP, Huang S, Zhao K 2011. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res 21: 1650–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA 2009. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF 2009. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol 10: R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Bai L, Johnson DS, Fulbright RM, Kireeva ML, Kashlev M, Wang MD 2010. Synergistic action of RNA polymerases in overcoming the nucleosomal barrier. Nat Struct Mol Biol 17: 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Hughes TR, Lieb JD, Widom J, Segal E 2010. Contribution of histone sequence preferences to nucleosome organization: Proposed definitions and methodology. Genome Biol 11: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ 2008. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26: 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, Kaestner YH 2012. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Mclean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh BF 2010. A preoccupied position on nucleosomes. Nat Struct Mol Biol 17: 923. [DOI] [PubMed] [Google Scholar]
- Radman-Livaja M, Rando OJ 2009. Nucleosome positioning: How is it established, and why does it matter? Dev Biol 339: 258–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SJ, Parkin IA 2008. Differential SAGE analysis in Arabidopsis uncovers increased transcriptome complexity in response to low temperature. BMC Genomics 9: 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK 2009. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR 2008. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol 6: e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu XS, Geng H, Li L, Ying J, Ma C, Wang Y, Poon FF, Wang X, Ying Y, Yeo W, et al. 2011. The epigenetic modifier PRDM5 functions as a tumor suppressor through modulating WNT/β-catenin signaling and is frequently silenced in multiple tumors. PLoS ONE 6: e27346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R 2009. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol 16: 996–1001 [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Yao J, Chen R, Li W, He X 2011. Nucleosome fragility reveals novel functional states of chromatin and poises genes for activation. Genome Res 21: 718–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Epps J, Williams R, Huttley G 2010. Evidence that localized variation in primate sequence divergence arises from an influence of nucleosome placement on DNA repair. Mol Biol Evol 27: 637–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Pugh BF 2011. High-resolution genome-wide mapping of the primary structure of chromatin. Cell 144: 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008a. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shin H, Song JS, Lei Y, Liu XS 2008b. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics 9: 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K 2009. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol 16: 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]