Abstract
The incredible power of DNA technology as an identification tool had brought a tremendous change in crimnal justice . DNA data base is an information resource for the forensic DNA typing community with details on commonly used short tandem repeat (STR) DNA markers. This article discusses the essential steps in compilation of COmbined DNA Index System (CODIS) on validated polymerase chain amplified STRs and their use in crime detection.
Keywords: DNA fingerprinting, Forensic DNA profiling, Forensic DNA database, micro-satellites, mini-satellites, RFLP, STR, VNTR
Introduction
The advent of DNA fingerprinting identification has revolutionized the science of crime detection (1–3). This technique when performed according to strict guidelines is highly reliable in convicting criminals and, equally importantly, helps in exonerating innocent individuals (4). This short review will discuss the history and development of forensic DNA profiling and the role of DNA database in forensic investigations.
Deoxyribonucleic Acid (DNA)
DNA is an acronym, which stands for deoxyribonucleic acid. Every cell in an individual’s body, with the exception of red blood cells and eggs or sperm, contains the full genetic program for that individual in its DNA. The program is coded by four chemical compounds called, bases, or subunits - Guanine, Cytosine, Adenine and Thymine (usually abbreviated as G, C, A and T), that are arranged into extremely long sequences. Groups of three bases (known as codons) code for the 20 amino acids, the basic building blocks of life. The amino acids in turn are linked together to form proteins. There are also stop codons signaling termination of the amino acid sequence. Though the code is well understood, biologists are still a long way from understanding how the code is expressed; for example, although each cell in an individual contains identical genetic information, the way the information is expressed in a liver cell is very different from the way it is expressed in a brain cell.
The human genome which consists of about 3 billion base pairs harbours genetically relevant information which is essential for the characterization of each individual. It is believed that genetically relevant information represents less than 10 % of the human genome. This minor part of the gene-coding DNA has been subjected to evolutionary pressure and selection mechanisms ensuring the development of higher organized organisms. The other 90% of the genome is junk DNA, a term which is more of a misnomer since their functions are still unknown rather than useless. A part of this non-coding DNA is comprised of repetitive sequences. Highly polymorphic spots in these non-coding regions are referred to as mini- or micro-satellites characterized by repeated blocks of DNA. The single-locus satellites are localized at a specific site of a given human chromosome, while multi-locus satellite elements or short tandem repeats (STRs) are spread throughout the entire genome.
There exists a significant level of diversity within the genome. During evolution, the process of selection involves non-directed mutations, which may be maintained when generation of a neutral or improved ability is successful while negative mutations normally get lost. The non-coding regions of the human genome are not regulated by these rules of selection and maintenance as long as they are not affecting the survival capacities of the individual. This is the reason for the accumulation of mutations leading to the generation of genetic diversity within the non-coding genomic DNA. Exceptions are polymorphisms in gene-coding regions, which reveal a high genetic stability combined with a very low mutation frequency.
Restriction Fragment Length Polymorphisms (RFLP) Method of DNA Profiling
Restriction Fragment Length Polymorphisms (RFLP) is a technique wherein genomic DNA is treated with one or more restriction enzymes which cut the DNA whenever certain specific sequence of bases occurs (each restriction enzyme will cut in a unique restriction site); thus generating a number of fragments of the DNA of varying lengths. In some individuals, random changes in the DNA will cause one or more sites to be lost or may otherwise cause variation between individuals in these fragment lengths. If the DNA is placed on a gel, and an electric field applied, the differing sized fragments will move at varying distances across the gel. The DNA can then be rendered visible by a variety of methods, yielding a pattern of bands, sometimes described as similar to a supermarket bar code (5). It is relatively easy to determine that two samples are different, if one has a band that the other lacks, but it is far more difficult to determine, on the basis of identical banding patterns, that two samples must have come from the same individual.
Variable Number of Tandem Repeat Sequences (VNTR) Typing
Stretches of the human genome consist of short sequences of DNA which are repeated in tandem. The number of blocks of these short sequence repeats in a given locus is highly variable between unrelated individuals. These repeated sequences are known as variable number of tandem repeat sequences (VNTR). VNTRs are broadly characterized into mini- and micro-satellites based on the size of the repeated blocks. In micro-satellites, the sequence repeat unit consists of between 2 to 9 base pairs, while mini-satellites consist of between 9 to 100 base pairs. Micro-satellites or STRs are generally more practical to be used for individualization (see below). The RFLP method of DNA fingerprinting as described above has therefore been replaced by the much simpler STR typing which is coupled with the extremely sensitive technique of polymerase chain reaction (PCR) (6–8).
Polymerase Chain Reaction (PCR)
Extraction of DNA from cells is a relatively straightforward process. However DNA is frequently rapidly degraded once it is no longer within a living organism. A spectacular advance has been the discovery of the PCR, which permits potentially unlimited amplification of minute traces of DNA, such as may be found in small samples of dry bone or skin or that is contained in traces of body fluids. An inevitable consequence of this massive amplification potential is its sensitivity to contamination, particularly if the same forensic laboratory and technicians are handling samples from both the suspect and the crime scene. Some idea of the potential extent of this problem can be gained from the fact that technicians frequently amplify their own DNA. Thus strict guidelines must be adhered to when using this method. PCR is currently used for STR typing.
Short Tandem Repeat (STR) Typing
STRs are highly polymorphic, and alleles of the STR loci are differentiated by the number of copies of the repeat sequence within each of the STR locus. The more STR loci being used for typing, the greater the discrimination value since the likelihood that a single individual has an identical STR profile, that possesses the exact same number of repeat units for all the STR being analyzed, with another individual taken at random in the population becomes extremely rare.
The STRs chosen and validated for typing for personal identification contain tetranucleotide repeats comprising of alleles of discrete size. Commercially robust and validated STR multiplex kits are available. The kits also include allelic ladder for each STR locus, which incorporates all the alleles of the STR locus so far known. This helps in the precise assignment of each allele and also in assigning the allele number.
The microsatellite alleles for a particular locus are codominant. In a given individual there are 2 alleles which are inherited in a Mendelian fashion. This means that an individual receives one allele from the mother and the other allele from the father. The two alleles are either heterozygous - the alleles are different or, homozygous - both the alleles are of the same type. In the case of a heterozygous situation, the individual shows two bands indicating the two different alleles, and, in a homozygous situation the individual shows only one band since both the alleles are of the same type and are superimposed.
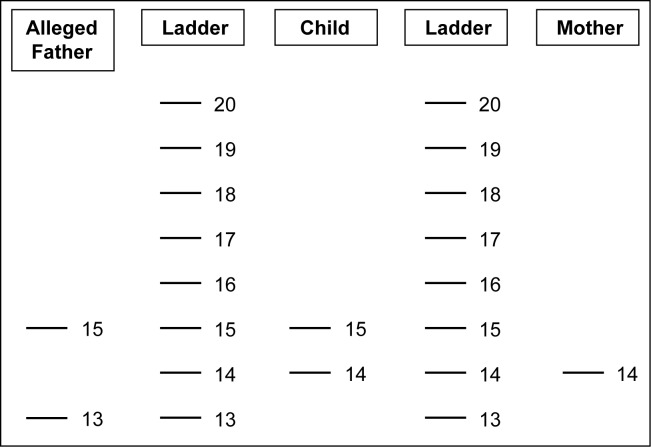
The following example of STR typing is to explain the above principle. Say in a given case of paternity dispute the alleged father, the mother and the child are tested for the STR locus vWA. The vWA locus – von Willebrand factor gene contains 8 alleles in the population and the alleles are numbered 13 to 20. Though 8 alleles are present in the population for this STR locus, only two alleles can be found in an individual. A hypothetical STR vWA locus typing result is as follows: Alleged Father – [13,15]; Child – [14,15]; Mother – [14,14]
In this case-example the child has received one allele [15] from the heterozygous alleged father [13, 15] and the other one allele [14] from the homozygous mother [14, 14] (Figure 1). It is evident that the bands indicating the alleles inherited by the child appear in the exact positions corresponding to the allelic ladder; and, there is no ambiguity in the allele number indicated by the bands of the ladders. Thus based on this one STR typing, the alleged father cannot be ruled out as the biological father. However, as mentioned above, the more the number of STRs being utilized for typing, the more discriminatory this method will be for personal identification. At present, 15 STRs are being used for typing, providing a level of discrimination as high as 1 in 30 to several hundred billion! This means that in the absence of identical twins, the probability of finding a matching DNA profile to an individual in a random population is, for example, 1 in 30 billion!
Figure 1 :
Schematic representation of a hypothetical case of paternity dispute showing the STR vWA locus typing result of the alleged father, the child and the mother with allelic ladders run adjacent to the test samples. Note that the allelic number assignment commences from the bottom and ascends by one unit increment to the top. Reading of the profile is easy and unambiguous - Alleged Father – [13,15]; Child – [14,15]; Mother – [14,14]. The alleged father cannot be ruled out as the biological father.
Forensic Science and DNA evidence
DNA fingerprinting was first used in forensic science in 1986 when police in the UK requested Dr. Alec J. Jeffreys, of University of Leicester, to verify a suspect’s confession that he was responsible for two rape-murders. Tests proved that the suspect had not committed the crimes.
The first person to be convicted on the basis of DNA evidence in the UK was Robert Melias in 1987 (9,10). In the same year in the US, Tommy Lee Andrews was convicted in a rape case based on DNA evidence (9), in which his DNA profile was matched with that of semen traces recovered from the victim. Two other important early cases gave much impetus to the use of DNA evidence: They were, the case of Glen Dale Woodal versus the State of West Virginia in 1992 and the multiple murder trial of Timothy Wilson Spencer versus the state of Virginia in 1994. The DNA evidence in the Woodal case exonerated him while that of the Spencer case resulted in his conviction and sentencing to the death penalty.
Admissibility of DNA evidence was seriously challenged for the first time in a case in the New York Supreme Court in 1989. Jose Castro was accused of murdering one Vimla Pence and her two year old daughter. Although a blood stain on Castro’s watch was matched to the victim, this evidence per se was not instrumental to his conviction. He was convicted after admitting to the crime. In this case, the DNA tests conducted by Life Code Corporation did not include a specific test for human blood and also did not include blind testing protocols in the attempt to link the stain to the victims. Furthermore, the laboratory in the above case had used contaminated probes and did not provide the worksheets and other manuscripts relating to the testing. Hence the court issued many directive guidelines regarding the test procedures and maintenance of laboratory results and reports as well as explanations for probability calculations and recording of observed defects or laboratory errors. The need to identify and document chain of custody and allowing access to data, methodology and actual results for an independent expert to review were also instructed.
In another case in 1989, the Supreme Court of Minnesota had also refused to admit the DNA evidence analyzed by a private forensic laboratory. The court noted that the laboratory did not comply with appropriate standards and controls. In particular the court castigated the laboratory for failure to reveal its underlying population data and testing methods. Such secrecy precluded replication of the test.
Thus, courts have denounced improper application of DNA scientific techniques to particular cases, especially when used to declare matches based on frequency estimates. However, DNA testing when properly applied is generally accepted as admissible and currently in many countries, DNA evidence is routinely used as evidence. As stated in the US National Research Council’s (NRC) 1996 report (10) on DNA evidence, “The state of the profiling technology and the methods for estimating frequencies and related statistics have progressed to the point where the admissibility of properly collected and analyzed DNA data should not be in doubt”(11,12,13).
Population data
Currently, time and expense limit an examination of an individual’s entire genome, which would show unique identity. Due to the fact that DNA typing is only an examination of a DNA sample’s sequence and/or length at discrete locations, a match in DNA typing is always a statistical exercise. In order to determine the probability that a particular genotype might occur at random in a population, population data must be compiled to make an estimate of the frequency of each possible allele and genotype. Usually a sample size of greater than 100 is sufficient to make reliable projections about a genotype’s frequency in a larger population (14)
Population databases are compiled based on ethnic or racial groups. Population subdivisions are not taken into account in the distribution of alleles. This can be illustrated by the following example. Let us assume the DNA profile is based on six separate loci or genes, and that the suspect possesses alleles or versions of these that are present respectively in 8 percent, 1 percent, 5 percent, 10 percent, 10 percent and 2 percent of the total population. Then the chance that a random member of the population would have all 6 of these particular alleles is 0.08 × 0.01 × 0.05 × 0.1 × 0.1× 0.02 = 0.000000008, or 8 in 1 billion.
The above calculation is valid when there are no associations among the alleles and they are distributed randomly throughout the population. In fact, there are many population subgroups in an ethnic group. A few geneticists proposed that the frequencies of genetic markers could differ widely from the frequencies estimated in larger groups. Hence any estimate calculated may vary considerably. Another group of geneticists advocated that although population sub-groups exist, the method currently in use then, were so conservative that they can compensate for small sub-group variations. Hence in 1992, the NRC proposed a compromise by recommending the so-called “ceiling principle” for making adjustments for the population subdivisions. In addition, the NRC report (1992) endorsed the use of DNA in courts, insisted for standardization proficiency tests and accreditation. Though there were many recommendations in the 1992 NRC report, the Forensic Science Laboratories (FSLs) did not implement the programme. The major reason for this reluctance was the advent of a new DNA typing method (namely, STR typing), which obviated the need for most of the recommendations proposed in the first NRC report. Hence a second NRC committee was convened in 1996.
This second NRC report (1996) endorsed the methods of DNA typing and statistical interpretation, then in use in the US. The report categorically stated that the technology for DNA and the methods used for estimation of gene frequencies and related statistics should not be doubted if properly collected. The 1996 NRC report addressed the issue of uniqueness of DNA typing and it stated that uniqueness (excluding identical twins) cannot be determined unless all members of the population are typed. The report further advocated that however, if a large number of loci are typed, the DNA profile obtained from the evidence can be so rare that it is highly likely that a suspect with a matching profile is the source of that evidence.
The report further stated that to ensure a high degree of confidence regarding the source of DNA, a threshold probability value (p) should be established. On this proposed suggestion an approach was developed at the FBI to determine a threshold value for examinations of DNA profile. The approach in brief is as follows:
An individual (excluding identical twins) can be identified as the source of the evidence DNA profile with a reasonable degree of certainty if the DNA profile satisfies the condition:
where a = 0.01 representing a confidence level of 99% and N = size of the entire population
These 1996 NRC recommendations are currently in use for DNA database compilation.
Federal Bureau of Investigation’s (FBI) Combined DNA Index System Program (CODIS)
The case behind the CODIS (20)
In 1989 one Mrs. Debbie Smith was abducted from her home and was raped in the woods behind her house. Police arrested a suspect and conventional serological tests excluded the suspect. However physical evidences from the victim were preserved.
In 1994 many sexual assaults and rapes were reported in the vicinity where Debbie Smith lived. Police arrested a suspect and used DNA technology to investigate the crime. The DNA profile of the physical evidence recovered from the victims as well as those from the Debbie Smith’s case were compared to the suspect’s profile. The suspect was however excluded.
However the Police began to routinely preserve and document DNA profiles of unsolved cases and compiled DNA databases of criminals involved in violent crimes. Police had periodically searched the DNA profiles in the unsolved cases with the convicted offenders profiles. Debbie Smith’s rapist was eventually identified from a match against this databank. The criminal, Norman Jimmerman, was already in prison for abduction and robbery and he is currently serving a 161-year sentence.
The CODIS Concept
The Combined DNA Index System (CODIS) blends computer and DNA technologies into an effective tool for comparing DNA profiles. The current version of CODIS uses two indices to generate investigative leads in crimes where biological evidence is recovered from the crime scene. The Convicted Offender index contains DNA profiles of individuals convicted of violent crimes, including sex offences. The Forensic Index contains DNA profiles developed from crime scene evidence. CODIS utilizes computer software to automatically search these indices for matching DNA profiles.
Profiles stored in CODIS contain a specimen identifier, the sponsoring laboratory’s identifier, the initials (or name) of DNA personnel associated with the analysis, and the actual DNA characteristics. CODIS does not store criminal history information, case-related information, social security numbers or dates-of-birth. Matches made among profiles in the Forensic Index can link crime scenes together; possibly identifying serial offenders. Based on a match, police can coordinate separate investigations, and share leads developed independently. Matches made between the Forensic and Convicted Offender indices ultimately provide investigators with the identity of the suspect(s). CODIS also supports a Population File. The Population File is a database of anonymous DNA profiles used to determine the statistical significance of a match.
CODIS is designed so that forensic laboratories have control over their own data. The system has three tiers (or levels): local, state, and national. The Forensic and Convicted Offender indices, and the population file may exist at each tier. Typically, the Local DNA Index System, or LDIS, is installed at crime laboratories operated by police departments or state police agencies. At the local level, DNA examiners use CODIS software on the bench when sizing autoradiograms. After sizing, examiners transfer unknown subject profiles into the local Forensic Index, where they are searched against other unknown subject profiles. The custodian of the local database can share this data with other CODIS laboratories within the state by forwarding it to the state level.
Each state participating in the CODIS program has a single State DNA Index System (SDIS). The SDIS is typically operated by the agency responsible for implementing the state’s convicted offender statute. At the state level, interlaboratory searching occurs. That is, the DNA profiles submitted by different laboratories within the state are compared against each other. Forensic profiles developed at local laboratories are also searched against the Convicted Offender index. The state custodian can share this data with the rest of the CODIS community by forwarding it to the national level. The National DNA Index System, or NDIS, is operated by the FBI. NDIS provides a mechanism for forensic crime laboratories located throughout the US to share and exchange DNA profiles. The DNA Identification Act of 1994 formalized the FBI’s authority to establish a national DNA index for law enforcement purposes. Today, CODIS is installed in 42 laboratories in twenty-two states in the US and the District of Columbia.
The FBI measures the success of the CODIS program by counting the crimes it helps to solve. The “cold hit” (20), is defined as a match which provides the police with an investigative lead that would not otherwise have been developed. The following two cases illustrate typical CODIS hits.
Case 1:
St. Paul, Minnesota, November 1994: A man wearing a nylon stocking over his face and armed with a knife jumped out from behind bushes and forced a woman who was walking by to perform oral sex. Semen recovered from the victim’s skirt and saliva was analyzed using DNA technology. The resulting profile was searched against Minnesota’s CODIS database. The search identified Terry Lee Anderson, who confessed and he is now in prison.
Case 2:
Tallahassee Florida, February 1995: The Florida Department of Law Enforcement linked semen found on a Jane Doe - rape-homicide victim to a convicted offender’s DNA profile. The suspect’s DNA was collected, analyzed, and stored in a CODIS database. A match was later identified to be of a convicted rapist was timely; it prevented the offender’s release on parole scheduled eight days later.
The organization and structure of CODIS can be a model for establishing such a system in Malaysia. The Malaysian parliament has recently passed an Act to collect and type DNA from convicted offenders hence paving the way for such a system to be implemented.
Discussion
The first DNA typing technology introduced in the mid 1980s was RFLP. The RFLP method of DNA typing involved core units of sequences consisting of 30 to 100 nucleotides which are present in many repeats (VNTR). The RLFP method of DNA typing requires intact genomic DNA in large quantities (20 to 30 mg). However, the biological specimens received in a forensic science laboratory are usually environmentally assaulted and occasionally only small amounts of DNA can be obtained. Hence in many situations, the RFLP method could not be applied.
The DNA typing method presently in use is STR typing. In this method many loci composed of core units of nucleotides repeated up to a length of 80 to 400 base pairs can be co-amplified and the results can be obtained in the same day by automated DNA fragment analyses. This technology is more superior than the RFLP method because it requires minute amounts of DNA (0.5 to 1 ng) and degraded samples can also be tested.
DNA analysis has been instrumental in securing convictions in hundreds of violent crimes, from homicides to assaults. It has also helped to eliminate suspects and has led to the exoneration and release of previously convicted individuals. DNA can focus investigations, and will likely shorten trials and lead to guilty pleas. It could also deter some offenders from committing serious offences. The increased use of forensic DNA evidence will lead to long-term savings for the criminal justice system.
Through storing DNA data in computer data banks, DNA analysis can be used to solve crimes without suspects. Forensic scientists can compare DNA profiles of biological evidence samples with a data bank to assist the police in detecting suspects. A data bank would also enable unsolved earlier offences where DNA evidence had been found but not linked with the offender, to be cleared up if DNA samples taken from a suspect in connection with a later offence matched the evidence found at the scene of the earlier crime. A national DNA data bank would also help police identify serial offenders both within and across the country.
Forensic DNA analysis is conducted throughout the world. Hence it is imperative on the part of the developing nations including Malaysia to develop and compile a national DNA database consisting of “crime scene DNA profile index”, “convicted offender DNA profile index”, and an index containing DNA profiles of unidentified bodies and body parts. This effort in turn will warrant appropriate amendments in criminal laws to help law enforcement agencies identify persons alleged to have committed serious and violent offences and empowering collection of samples for DNA profiling database. To date, there is already published data for 9 STRs for three ethnic population groups of Malaysia (Malay, Chinese and Indians) (21, 22) and efforts are currently underway to type subpopulations of Malays and to start the newly validated, 15 STR profiling kit in various populations in Malaysia. Extensive database and DNA profiling of criminals and indexing them will help to speed up crime detection.
References
- 1.Jeffreys AJ, Wilson V, Thein SL. Hypervariable ‘minisatellite’ regions in human nature. Nature. 1985;314:67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- 2.Jeffreys AJ, Wilson V, Thein SL. Individual-specific “fingerprints” of human DNA. Nature. 1985;316:76–79. doi: 10.1038/316076a0. [DOI] [PubMed] [Google Scholar]
- 3.Peter G, Jeffreys AJ, Werrett DJ. Forensic application of DNA fingerprints. Nature. 1985;318:577–579. doi: 10.1038/318577a0. [DOI] [PubMed] [Google Scholar]
- 4.National Research Council, National Academy of Sciences. DNA Technology in Forensic Science. Washington, DC: National Academy Press; 1992. p. 156. (Cited as NRC report.) [Google Scholar]
- 5.Lewontin RC. Comment: the use of DNA profiles in forensic contexts. Statistical Science. 1991;9:17–19. [Google Scholar]
- 6.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. 1986. Biotechnology. 1992;24:17–27. [PubMed] [Google Scholar]
- 7.Edwards A, Civitello A, Hammond HA, Caskey CT. DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am J Hum Genet. 1991;49:746–756. [PMC free article] [PubMed] [Google Scholar]
- 8.Polymeropoulos MH, Rath DS, Xieo H, Merril CR. Tetranucleotide repeat polymorphism at the human beta – actin related pseudogene H- beta – AC – Psi – 2 (ACTBP2) Nucleic Acids Res. 1992;20:432. doi: 10.1093/nar/20.6.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.State v. Andrews, 533 So.2d 841 (Dist. Ct. App. 1989)
- 10.The evaluation of Forensic DNA Evidence (NRCII) (1)1996 - http://www.dnalwyr.com/nrc [PubMed]
- 11.Frye v. United States, 293 F. 1013 (D.C. Cir. 1923)
- 12.Daubert v. Merrell Dow Pharmaceuticals, Inc.,113 S.Ct. 2786 (1993)
- 13.Kaye DH. The forensic debut of the National Research Council’s DNA report: Population structure, ceiling frequencies and the need for numbers. Jurimetrics Journal. 1994;34:369–382. [Google Scholar]
- 14.Chakraborty R. Sample size requirements for addressing the population genetic issues of forensic use of DNA typing. Hum Biol. 1992;642:141–159. [PubMed] [Google Scholar]
- 15.Lewontin RC. The use of DNA profiles in forensic contexts – comment. Statistical Science. 1994;9:259. [Google Scholar]
- 16.Lempert R. Comment: theory and practice in DNA fingerprinting. Statistical Science. 1994;9:255. [Google Scholar]
- 17.Macilwain C. Ceiling principle ‘not needed’ in DNA cases. Nature. 1996;103:381. doi: 10.1038/381103a0. [DOI] [PubMed] [Google Scholar]
- 18.Marshall E. Criminology – Academy’s about-face on forensic DNA. Science. 1996;272:803–804. doi: 10.1126/science.272.5263.803b. [DOI] [PubMed] [Google Scholar]
- 19.Balding DJ, Donelly P. How convincing is DNA evidence? Nature. 1994;368:285. doi: 10.1038/368285a0. [DOI] [PubMed] [Google Scholar]
- 20.CODIS –combined DNA index system-http://www.fbi.gov/hq/lab/codis/index1.html
- 21.Lim KB, Jeevan NH, Jaya P, Othman MI, Lee YH. STR data for the AmpFlSTR Profiler loci from the three main ethnic population groups (Malay, Chinese and Indian) in Malaysia. Forensic Sci Int. 2001;119:109–112. doi: 10.1016/s0379-0738(00)00386-8. [DOI] [PubMed] [Google Scholar]
- 22.Panneerchelvam S, Ravichandran M, Norazmi MN, Zainuddin ZF. Allele frequency distribution for 10 STR loci in the Malay population of Malaysia. J Forensic Sci. 2003;48:451–452. [PubMed] [Google Scholar]