Abstract
The vast majority of prenatal genetic testing requires invasive sampling. Since this poses a risk to the fetus, one must make a decision that weighs the desire for genetic information against the risk of an adverse outcome due to hazards of the testing process. These issues are not required to be coupled, and it would be desirable to discover genetic information about the fetus without incurring a health risk. Here we demonstrate that it is possible to noninvasively sequence the entire prenatal genome. Our results show that molecular counting of parental haplotypes in maternal plasma by shotgun sequencing of maternal plasma DNA allows the inherited fetal genome to be deciphered noninvasively. We also applied the counting principle directly to each allele in the fetal exome by performing exome capture on maternal plasma DNA prior to shotgun sequencing. This approach enables noninvasive exome screening of clinically relevant and deleterious alleles that were paternally inherited or had arisen as de novo germline mutations, and complements the haplotype counting approach to provide a comprehensive view of the fetal genome. Noninvasive determination of the fetal genome may ultimately facilitate the diagnosis of all inherited and de novo genetic disease.
Our work is based on the phenomenon of circulating cell free DNA, whose existence and role in pregnancy was first investigated in 19481. A portion of the cell-free DNA in a pregnant woman’s blood is derived from the fetus2, and this fact has enabled the development of a number of noninvasive prenatal diagnostic techniques3. A prominent example is the non-invasive detection of Down syndrome and other aneuploidies, which was first demonstrated by our group4, validated by clinical trials5–10, and is now available in the clinic. We describe here how the chromosome counting principle we invented for aneuploidy detection can be applied to non-invasive fetal genome analysis by directly counting haplotypes and even individual alleles. Others have studied the relationship between maternal and fetal cell-free DNA11, but their approach required invasively sampled fetal material, did not determine the fetal genome, and also needed knowledge of paternal genetic data.
Measuring the fetal genome by counting parental haplotypes
Maternal plasma DNA is a mixture of maternal and fetal DNA; the fraction of fetal DNA ranges from a few percent or lower early in pregnancy to as high as ~50%2,7, and generally increases with gestational age. Since the fetal genome is a combination of the four parental chromosomes, or haplotypes, as a result of random assortment and recombination during meiosis, three haplotypes exist in maternal plasma per genomic region: the maternal haplotype that is transmitted to the fetus, the maternal haplotype that is not transmitted, and the paternal haplotype that is transmitted. If the relative copy number of the untransmitted maternal haplotype is 1 - ε, where ε is the fetal DNA fraction, then the relative copy number of the transmitted maternal haplotype is 1, and the relative copy numbers of the transmitted and untransmitted paternal haplotypes are ε and 0, respectively (Figure 1). Therefore, within each pair of parental haplotypes, the transmitted haplotype is over-represented relative to the untransmitted one. By measuring the relative amount of parental haplotypes through counting the number of alleles specific to each parental haplotype (referred to as ‘markers’), one can deduce the inheritance of each parental haplotype and hence build the full inherited fetal genome.
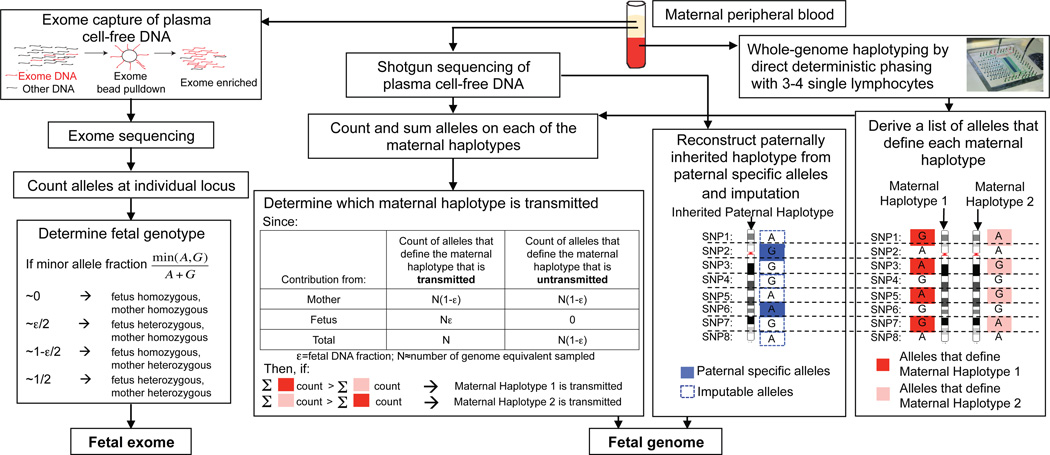
Figure 1.
Molecular counting strategies for measuring the fetal genome noninvasively from maternal blood only. Genome-wide, chromosome length haplotypes of the mother are obtained using direct deterministic phasing. The inheritance of maternal haplotypes is revealed by sequencing maternal plasma DNA and summing the count of the alleles specific to each haplotype at heterozygous loci and determining the relative representation of the two alleles. The inherited paternal haplotypes are defined by the paternal specific alleles (i.e. those that are different from the maternal ones at positions where the mother is homozygous). The allelic identity at loci linked to the paternal specific alleles on the paternal haplotype can be imputed. Alternatively, molecular counting can be applied directly to count alleles at individual locus to determine fetal genotypes via targeted deep sequencing, such as exome enriched sequencing of maternal plasma DNA. For illustrative purpose, each locus is biallelic and carries the ‘A’ or ‘G’ alleles.
Strictly speaking, the markers that define each maternal haplotype are the alleles that are present in one maternal haplotype but not in the other maternal haplotype and the two paternal haplotypes. However, since it is rare that two unrelated persons share the same long-range haplotype, that is, a haplotype much longer than the usual length of haplotype blocks observed in the population (~100kb), the presence of alleles contributed by the transmitted paternal haplotype at these loci would not interfere with the measurement of representation of maternal haplotypes as long as the haplotype being considered is sufficiently long (>1 Mb). Thus all the maternal heterozygous loci can be used to define the two maternal haplotypes (Figure 1). This enables the measurement of relative representation of the two maternal haplotypes without the knowledge of paternal haplotypes. The relative representation of the two maternal haplotypes is the difference in the counts of markers specific to each haplotype. Even if the over-representation of the transmitted maternal haplotype is small, the over-represented haplotype can be identified provided that the counting depth exceeds the counting noise, which is governed by Poisson statistics. Table S1 and Figure S1 provide estimations of counting requirement as a function of confidence of measurement and fetal DNA percentage in the clinically observed range. Because the number of markers that define each parental haplotype increases with haplotype length, the longer the phased haplotypes, the lower the average number of sampling per individual marker is required for confident determination of the over-represented parental haplotypes.
If paternal haplotypes are known, it is straightforward to determine the inherited paternal haplotypes by comparing the sum of count of alleles specific to each paternal haplotype (Figure S2), thereby revealing the entire inherited fetal genome. Figure S3 and the accompanied supplemental text show how this could be achieved using sequencing data of a synthetic mixture of DNA from a mother and daughter within a fully phased family trio12. However, it is not always possible to obtain paternal information; the incidence of non-paternity is estimated to be between 3% and 10%13,14, making this a particularly delicate issue. In the absence of paternal information, the paternally inherited haplotypes can be reconstructed via linkage to observed non-maternal (i.e. paternal specific) alleles (Figure 1).
We verified this approach on samples collected from two pregnancies. Pregnant woman P1 carried a female fetus with normal karyotype, while pregnant woman P2 is an individual with a ~2.85 Mb heterozygous deletion on chromosome 22 that is associated with DiGeorge syndrome. To obtain phased maternal chromosomes, we performed ‘direct deterministic phasing’ (DDP)15 on 3 or 4 maternal metaphase cells obtained by culturing maternal whole blood (Table S2, Figure S4). DDP involves microfluidic separation and amplification of individual metaphase chromosomes from single cells followed by genome-wide genotyping analysis of amplified materials, and enables each chromosome in the genome to be phased along its full length. Genomic DNA of cord blood collected at delivery was also genotyped to serve as the true reference for fetal genotypes. The true inheritance of maternal haplotypes was determined by aligning the homozygous SNPs of the fetus by cord blood genotyping against the two maternal haplotypes defined by the phased maternal heterozygous SNPs (Figure 2). The analysis here concerns the ~1 million positions across the genome present on Omni1-Quad genotyping array. Phase information of the remaining genomic positions, particularly those that carry rare variants of clinical importance, can be obtained by broader array coverage or direct sequencing of amplified chromosome materials, as demonstrated previously15.
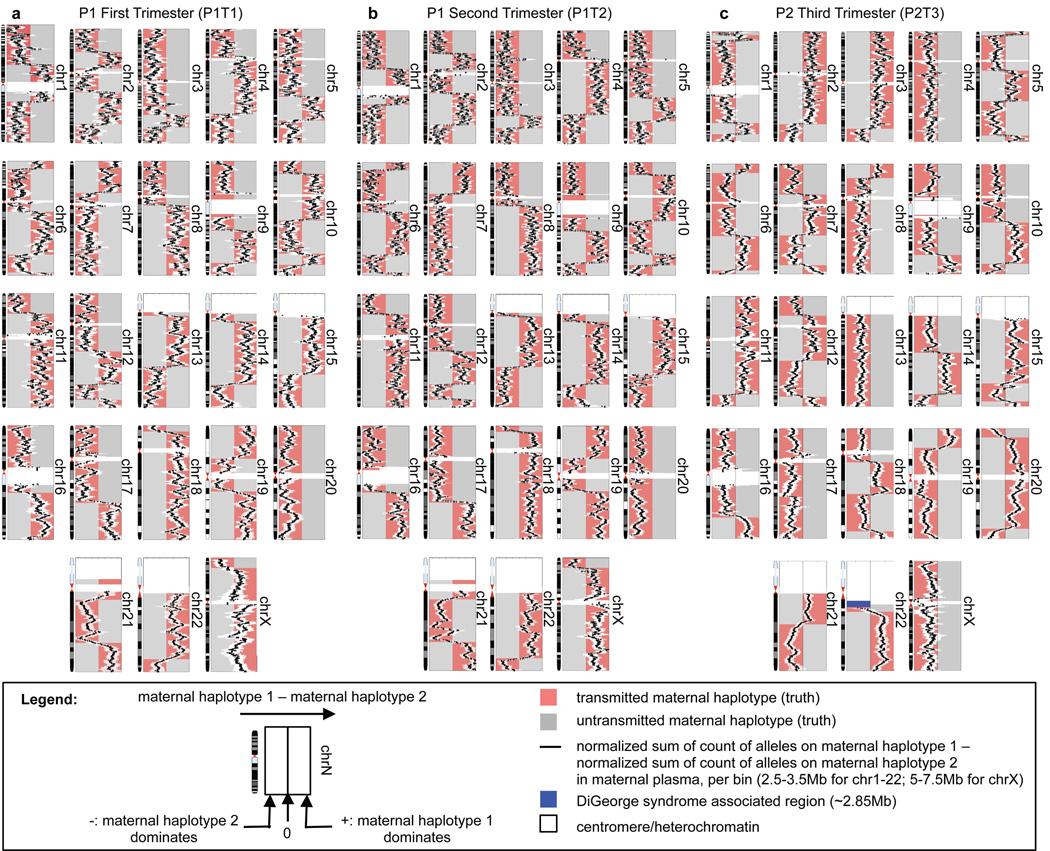
Figure 2.
Noninvasively determining genome-wide fetal inheritance of maternal haplotypes via haplotype counting of maternal plasma DNA with at least 99.8% accuracy over 99% of the genome in three maternal plasma samples (A-C). Each point on a black line represents the relative amount of the two maternal haplotypes evaluated using the markers lying within a bin centered at the point, and is accompanied by a white bar that corresponds to the 95% confidence interval for each measurement. The maternal haplotypes are colored pink or grey according to the true transmission states, as determined by fetal cord blood genotypes. Over-representation of ‘maternal haplotype 2’ in P2T3 maternal plasma immediately adjacent to the DiGeorge syndrome associated deletion (blue) indicates fetal inheritance of the deletion, which agrees with fetal cord blood genotype.
Maternal cell-free DNA samples were shotgun sequenced on the Illumina platform to a final depth of ~52.7x (151Gb), ~20.8x (59.7Gb), and ~1.3x (30.8Gb) haploid genome coverage for P1T1 (P1, 1st trimester), P1T2 (P2, 2nd trimester), and P2T3 (P3, 3rd trimester) respectively (Table S2). To determine fetal inheritance of maternal haplotypes, we divided each chromosome into bins of 2.5–3.5Mb for autosomal chromosomes and 5Mb–7.5Mb for chromosome X (Table S2), with sliding steps of 100kb, and compared the counts of alleles specific to each of the two haplotypes. Bin sizes were chosen according to the estimated sampling requirement (Table S1) based on the sequencing depth, density of markers, and fetal DNA fraction, which was estimated, by comparing relative representation of maternal haplotypes, to be ~5%, ~18%, and ~43% for P1T1, P1T2, and P2T3, respectively. The lower SNP array density on chromosome X required larger bin sizes for that chromosome. The over-represented maternal haplotype over the entire genome was apparent and corresponded to the maternal haplotype transmitted to the fetus (Figure 2). Taking into account the uncertainty surrounding regions of cross-overs (median ~350–450kb per cross-over, Figure S5), maternal inheritance of at least 99% of the SNPs could be deduced with at least 99.8% accuracy for all samples. Less sequencing depth also allowed the inherited maternal haplotypes to be deduced (Figure S6) with lower resolution of cross-overs (Figure S5).
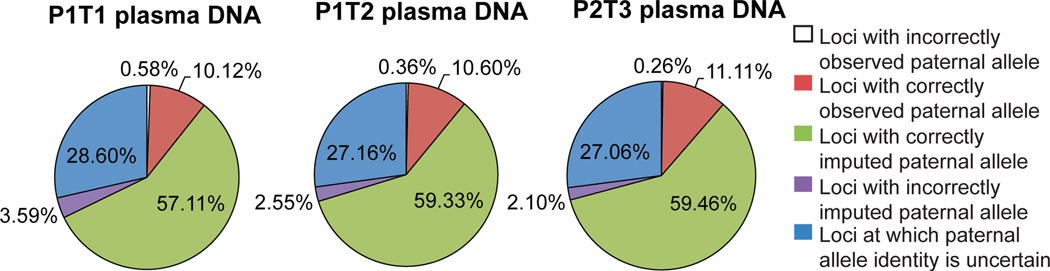
The paternally inherited haplotypes were reconstructed by detection of paternal specific alleles, followed by imputation at linked positions. We used the haplotypes of normal population documented by the 1000 Genome Project16 as reference haplotypes for imputation. Imputation accuracy is dependent on the density of markers, and the number of identified non-maternal alleles is dependent on sequencing depth and fetal DNA fraction. At the final sequencing depth, we detected ~66–70% of the paternal specific alleles at least once (Table S2, Figure S7). Approximately 3.4%–5.6% of the non-maternal alleles were sequencing noise. Using the non-maternal markers, we deduced ~70% of the paternally inherited haplotypes with ~94–97% accuracy via imputation (Figure 3). The loci that could not be confidently imputed reside in regions where paternal specific alleles were not detected, in regions that lack paternal specific alleles, or where the paternal alleles are associated with more than one haplotype observed in the population. In principle these regions could be completely determined by deeper sequencing and application of the counting principle directly to the local regions or the individual alleles at every genomic position, as shown below.
Figure 3.
Reconstruction of paternally inherited chromosomes noninvasively based on imputation using observed non-maternal alleles. The paternally inherited haplotypes were reconstructed by detection of paternal specific alleles, followed by imputation at linked positions. At the final sequencing depth, ~66–70% of all the paternal specific alleles were detected at least once. Using those markers, ~70% of the paternally inherited haplotypes were imputed with ~94–97% accuracy. The loci that could not be confidently imputed could in principle be completely determined by deeper sequencing and application of the counting principle directly to the individual alleles at every genomic position.
Measuring fetal exome by counting alleles at individual locus
We sought to determine clinically relevant portions of the fetal genome in maternal plasma DNA by applying the counting principle to each allele at all positions in the exome. Because the exome is two orders of magnitude smaller than the genome, less sequencing throughput is required to provide deep sequencing at individual loci and thus allows sensitive and specific detection of clinically relevant and deleterious polymorphisms that either were paternally inherited alleles or de novo mutations. We performed exome capture and sequencing on maternal plasma DNA samples of P1 in all three trimesters (Figure 1, Figure S9). We obtained a median coverage of 194x, 221x, and 631x per position in the exome for the first, second, and third trimester respectively (Figure 4D). After stringent data filtering to eliminate miscalled paternal specific alleles due to limited sampling and mis-mapping to the reference genome (Figure S10), 75%, 78%, and 90% of all exomic positions in the first, second, and third trimester samples, respectively, had >100x coverage and were retained for analysis (Table S2).
Figure 4.
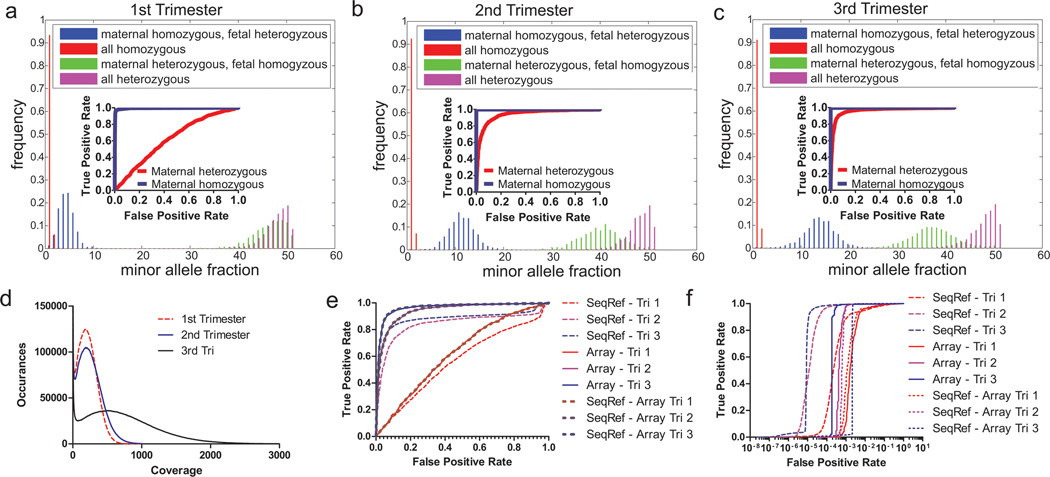
Exome sequencing of P1 maternal plasma DNA in all three trimesters to determine maternal and fetal genotypes.. A-C. Histograms of minor allele fraction in maternal plasma from all three trimesters of P1 at positions that are confidently called in both plasma sequencing data and pure fetal/maternal DNA genotyping data. Insets: ROC curves of positions detecting fetal genotypes differing from maternal genotype when the maternal position is either homozygous or heterozygous. The higher the fetal fraction (~6, 20, 26% for Trimester 1–3), the more the distributions are separated, and the easier it is to distinguish between the two distributions of fetal genotype. D. Histogram of per-position coverage, with bin size of 5. Exome positions >100X are 75%, 78%, and 90% respectively for Trimester 1–3 and >200X are 48%, 56%, and 84%. E-F. ROCs curves at genomic positions where mother is heterozygous (E) or homozygous (F), using either sequencing or SNP array of pure DNA as references for maternal and fetal genotypes. ‘SeqRef’ uses a sequenced reference, ‘Array’ uses a SNP array, and ‘SeqRef-Array’ uses a sequenced reference only at positions on a SNP array.
We calculated minor allele fraction, defined as the second largest nucleotide fraction divided by the sum of the two largest nucleotide fractions, at positions that are confidently called in genotyping data within the exome (Figure 4A-C) or exome sequencing data (Figure S11–13) of fetal cord blood DNA and pure maternal DNA. In all three trimesters, fetal genotypes could be assigned robustly at loci where the mother is homozygous based on the separation in minor allele fraction at a depth of 200x. Paternal specific alleles were detected with sensitivity of 96–99.8% at the specificity threshold of 99% (Figure 4E-F, Table 1). Since the minor allele fraction at loci with paternal specific alleles is theoretically half of the fetal DNA fraction, we estimated fetal DNA percentage to be 6.6%, 20.1%, 26.3% for the three trimesters, respectively (Table S2). For the second and third trimester samples with higher fetal DNA fraction, fetal genotypes could be extracted for most loci at which the mother is heterozygous, as the separation in minor allele fraction for fetal homozygous and fetal heterozygous SNPs was apparent (Figure 4A-C, E-F). For these loci, the ability to differentiate fetal heterozygosity from homozygosity depended on sequencing depth and fetal DNA fraction (Figure S1).
Table 1.
Exome diagnostic cutoffs and the resulting sensitivity and specificity
| Specificity Cutoffs | ||||||
|---|---|---|---|---|---|---|
| Maternal Homozygous | Maternal Heterozygous | |||||
| Trimester | 95% | 99% | 85% | 90% | 95% | |
| Sensitivity | 1 | 98% | 96% | 25% | 16% | 8% |
| 2 | 99.8% | 99.8% | 89% | 85% | 71% | |
| 3 | 99.7% | 99.6% | 96% | 93% | 87% | |
Discussion
The molecular counting methods described here offer a gateway to comprehensive non-invasive prenatal diagnosis of genetic disease. There are substantial ethical issues associated with noninvasive prenatal genome determination, which we have not attempted to address. We will note however that there are numerous clinical scenarios where this approach would be useful. In the first or second trimester, it is possible to test for conditions that are not survivable or lead to medical complications. As technologies for pharmaceutical and surgical intervention improve, it may be possible to develop prenatal treatment or even cures for these congenital conditions.
This is illustrated by our data on P2, who is an individual with DiGeorge syndrome. Haplotyping of the maternal genome identified a ~2.85 Mb deletion on 22q11.1 that is associated with the syndrome on one copy of the maternal chromosome 22 (denoted as ‘maternal haplotype 2’ in Figure 2C). Haplotype counting in maternal plasma indicated an over-representation of ‘maternal haplotype 2’ of the region immediately adjacent to that deletion, indicating fetal inheritance of the DiGeorge syndrome associated deletion (Figure 2C, deletion indicated in blue). This result was confirmed by quantitative PCR of cord blood DNA (Figure S8). In this clinical scenario, confirmation of the deletion would argue for a fetal echocardiogram and neonatal assessment of calcium levels.
Knowledge of the fetal genotypes obtained in the third trimester enables diagnosis of conditions that would benefit from treatment immediately after delivery; these include metabolic and immunological disorders such as phenylketonurea, galactosemia, maple syrup urine disease, and severe combined immunodeficiency. Currently, newborns with these conditions suffer as symptoms manifest themselves in the time it takes to determine the proper diagnosis and treatment, which is often as simple as diet change. In summary, we anticipate that there is no technical barrier and many practical applications to having the entire fetal genome determined noninvasively in clinical settings.
Additional Methods
Prediction of counting depth requirement for determination of over-representation of transmitted maternal haplotypes
Given two distributions of Poisson random variables, one with mean of N, and the other with mean of N(1-ε), where N is the cumulative sum of the count of all usable markers on the transmitted maternal haplotype, the sampling requirement of N to differentiate the two distributions can be estimated from the following expression, using the normal approximation of the Poisson distribution for large values of N:
where zα is the z-score associated with the confidence level of α. Thus,
Table S1 present the estimated requirement of N for different values of fetal DNA fraction (ε) and level of confidence (α).
Patient samples
Two subjects – referred to as P1 and P2, were recruited to the study under approval of the Internal Review Board of Stanford University. For P1, peripheral blood was obtained during the first, second, third trimesters, and postpartum (Table S2). For P2, peripheral blood was obtained during the third trimester and postpartum (Table S2). Cord blood was obtained at delivery for both patients.
Whole-genome haplotyping of patient subjects
Postpartum maternal whole blood was collected into sodium heparin coated Vacutainer. Postpartum blood was used in this study because blood samples collected during pregnancy were not cyropreserved based on blood culture requirement. One milliliter of whole blood was cultured with PB Max Karyotyping medium for 4 days. Direct deterministic phasing (DDP)15 was performed on 3 to 4 single cells. Each haplotype was genotyped with Illumina’s Omni1-Quad genotyping array. About 92% to 96% of the ~1 million SNPs present on the Omni1Quad BeadChip array (Illumina) (~25% are heterozygous within each individual) were phased (Figure S3), yielding 250–350 heterozygous markers per 3.5Mb window. In addition to genotyping array analysis, PCR was performed on amplified materials from separated chromosome 22 of P2 to determine which maternal haplotype carried the DiGeorge syndrome associated deletion. Two regions, dgs37 and dgs40, within the deletion were tested. Primer sequences are listed in Table S3. Other rare SNPs not present on the genotyping array or not linked to loci on the array could also be phased by PCR or sequencing.
Whole-genome genotyping of the study subjects and their infants
Genomic DNA was extracted from 200µl of postpartum maternal blood and 200µl cord blood using QIAamp Blood Mini Kit (Qiagen), and subjected to genome-wide genotyping on Illumina’s Omni1-Quad genotyping array.
Quantitative PCR confirmation of fetal inheritance of DiGeorge associated deletion
The inheritance of the maternal haplotype carrying the deletion on chromosome 22q11.1 by the fetus of P2 was independently confirmed by quantitative real-time PCR performed on cord blood genomic DNA. The quantity of an amplicon within the deletion region (Table S3) was compared to that of an amplicon on chromosome 1 (E1F2C1). A ratio of ~0.5 indicated that the maternal deletion was inherited.
Extraction of cell-free DNA from maternal plasma
Maternal blood were collected into EDTA coated Vacutainers. Blood was centrifuged at 1600g for 10min at 4C, and the plasma was centrifuged again at 16000g for 10min at 4C to remove residual cells. Cell-free DNA was extracted from plasma using QIAamp Blood Mini Kit (Qiagen) or QIAamp Circulating Nucleic Acid Kit (Qiagen).
Whole genome shotgun sequencing of cell-free DNA extracted from maternal plasma
DNA was extracted from 1 to 2 ml of plasma, and subsequently converted into Illumina sequencing libraries4 and quantified by digital PCR18. Sequencing was performed on the GAII and the HiSeq instruments (Table S2). Sequences were aligned to the human genome (hg19) using CASVA version 1.7.0. Only alleles called with quality scores > 30 were used. In addition, only alleles that match previously reported variants in dbSNP were used for analyses.
Identifying the inherited parental haplotypes
Each chromosome was divided into equally sized bins with sliding window of 100kb. The bin size was chosen such that the total number of count of markers within the bin was at least that required to overcome counting noise specifically when determining relative representation of the two maternal haplotypes.
The relative representation of the haplotype pairs of each parent was calculated using the expression (Np1/np1 - Np2/np2), where Np1 is the number of occurrences of markers defining ‘maternal or paternal haplotype 1’ within the bin counted by sequencing, np1 is the total number of usable markers that define ‘maternal or paternal haplotype 1’ within the bin, Np2 is the number of occurrences of markers defining ‘maternal or paternal haplotype 2’ within the bin counted by sequencing, np2 is the total number of usable markers that define ‘maternal or paternal haplotype 2’ within the bin. If the expression was positive over a continuous region of 5Mb, parental haplotype 1 was considered as inherited. If the expression was negative over a continuous region of 5Mb, parental haplotype 2 was considered as inherited. The 95% confidence interval of relative maternal haplotype representation calculated within each bin was estimated by simulating the distribution of reads assuming the count of each maternal haplotype was the mean of a Poisson random variable.
Determining locations of recombination
The true recombination events on the maternally inherited sets of chromosomes were determined by comparing the genotype of the fetus and to the allele on each of the two maternal haplotypes at locations where the fetus is homozygous and the mother is heterozygous. In maternal plasma, a cross-over event between the two maternal haplotypes giving rise to the maternally inherited chromosome in the fetus was called if in plasma DNA if two criteria were met: 1. A continuous increase or decrease in the relative representation of haplotype 1 over haplotype 2 (i.e. the expression Np1/np1 - Np2/np2), accompanied by a sign change, as one scanned in the direction from the p arm to the q arm of a chromosome. 2. The sign of the expression remained the same for the sliding bins 5Mb downstream, based on the fact of cross-overs are rarely close to each other (positive interference).
Imputation of untyped loci on experimentally measured haplotypes
Imputation was performed using Impute v117, using the –haploid option. Imputation was performed using August 2010 data from the 1000 Genome Project of the CEU population. For maternal genomes, imputation was based on the ~1 million markers phased by DDP. For paternal haplotypes, imputation was based on non-maternal alleles observed in shotgun sequencing data at locations where mother is observed and predicted based on imputation (>99% confidence) to be homozygous. Only loci with confidence of imputation >99% were considered; the allele identity for the rest were deemed uncertain. The results were compared to the true paternal haplotypes derived based on the comparison of the phased maternal genome and the cord blood genotyping array data. Imputation was performed in 5Mb segments along each chromosome.
Estimating fetal DNA fraction from maternal plasma sequencing by comparing maternal haplotype representation
Fetal DNA fraction was estimated from the over-representation of one of the maternal haplotypes. Precisely, fetal DNA fraction (ε) was estimated as 2x/(2-x), where x is the median absolute value of the expression (Np1/np1 - Np2/np2) for all bins evaluated on either the maternal haplotypes, divided by the average marker density of the two maternal haplotypes.
Exome enrichment from maternal genomic DNA, fetal genomic DNA, and cell-free DNA extracted from maternal plasma
Exome capture was performed with the SeqCap EZ v2.0 Kit (Roche Nimblegen) according to manufacturer’s protocol with modifications. There are several commercially available exome kits available with varying degrees of coverage for exons, untranslated region, and microRNA regions19. We chose the Nimblegen platform due to its ability to capture efficiently on targeted regions and our desire for cost-efficient deep sequencing, but other platforms may perform similarly when sequenced at enough depth.
For exome enriched directly from genomic DNA extracted from maternal blood cells and cord blood, DNA was first sheared using Covaris S220 using the recommended settings for 200 bp fragments. End repair and dA tailing reactions were cleaned up by QIAquick PCR Purification Kit (Qiagen) whereas ligation and PCR were cleaned by Agencourt Ampure XP beads (Beckman Coulter) at a 1.8X ratio of bead reagent to input volume to discard shorter adaptors, primers, and ligation/PCR byproducts.
Cell-free DNA extracted from approximately 3 mL, 4 mL, 4 mL, and 2.5 mL of plasma were extracted from P1T1, P1T2, P1T3, and P2 respectively, was used for exome capture. For exome capture from cell-free DNA, sequencing libraries were first prepared following the NEBNext Master Mix 1 Kit (NEB). Extracted DNA was end repaired and dA tailed using the NEBNext kit and subsequently cleaned up with QIAquick Nucleotide Removal Kit (Qiagen) in both steps. Ligation to typical Illumina paired end adaptors was performed at a 1:10 concentration ratio of the initial sample DNA to the adaptors. The first PCR prior to hybridization was carried for 18 cycles as detailed in the SeqCap protocol. Both ligation and PCR were cleaned up with Agencourt Ampure XP beads as described in the Nimblegen protocol. Prepared non-exome sequencing libraries were incubated with SeqCap kit reagents and the exome-rich sequencing library was amplified for 18 cycles in the second PCR. Libraries were quantified with digital PCR18.
Analysis of exome sequencing data
Figure S10 outlined the informatics pipeline for analyzing exome data. Paired end sequencing for 100 bases on each end was performed on the HiSeq 2000 (Illumina) using v3 chemistry. Illumina’s native software provided image analysis and base calling to provide FASTQ files. Those files were aligned via BWA’s ‘sampe ‘function.
Exome sequencing yielded 332, 344, and 930 million aligned reads for first, second, and third trimesters respectively (Table S2). Because exome preparation involved more procedural steps and cycles of PCR than whole genome shotgun sequencing preparation, we imposed a set of filters on the exome data. To remove or at least minimize bias, we opted to remove PCR duplicates based on aligned location with the Picard MarkDuplicates program (the Broad Institute)20. In this deduplication procedure, reads with ends aligned to the exact same locations are considered PCR duplicates and amplified from same original single molecule. Deduplication helps substantially reduce bias when using paired end and sequencing depths exceeding the sample library size. For single end reads 100 bases long, there is only a maximum unique identification of 200 (for both directions). However for paired end reads both ends of a DNA fragment are aligned and if fragments lengths are varied equally by 50 bases then the maximum identification library size can be 10000, which is at least an order of magnitude above the highest coverage seen in this study. In theory it is possible to remove nearly all PCR bias if sequencing is deep enough to discover under-amplified DNA and if the theoretical identification library size is well above the actual molecular library size.
After deduplication, reads were piped through GATK (the Broad Institute) local realigner. Samtools mpileup was used to stack per position counts of different nucleotides within the exome tiles provided by the manufacturer of the SeqCap exome kit. The nucleotide count of each position was analyzed against pure fetal and maternal DNA genotyping and sequencing data using custom python and MATLAB code. The minor allele fraction at each position was calculated to be the second largest nucleotide fraction divided by the sum of the two largest nucleotide fractions.
Given that fetal heterozygous genotypes at positions where maternal is homozygous can have a minor allele fraction as low as 1% on the lower end of the distribution, it is important to have more than 100X coverage to avoid classification errors occurring by chance. Beyond 100X coverage, there are also marginal improvements in sensitivity and specificity (Figure S14A). In addition, we filtered out misaligned regions by detecting regions with several excessively high minor allele fractions in close proximity. We filtered out 3–4 positions 40 bases apart with minor allele fractions greater than 1–5% and were able to achieve marked reduction in specificity (Figure S14B). While filtering removes up to 4% of all positions (Figure S14C), it can reduce false positives by an order of magnitude at approximately the same level of sensitivity (Figure S14B).
Fetal DNA fraction was estimated from exome data based on minor allele fraction. The theoretical minor allele fractions are 0 for group 1 SNPs at which both mother and fetus are homozygous, ε/2 for group 2 SNPs of which fetus is heterozygous and mother is homozygous, 1-ε/2 for group 3 SNPs at which fetus is homozygous and mother is heterozygous, and ½ for group 3 SNPs at which both mother and fetus are heterozygous, where ε is the fetal DNA fraction. We used the median of the distribution of minor allele fraction for group 2 SNPs to provide an estimate of fetal DNA fraction.
Supplementary Material
Acknowledgements
The authors would like to thank Elizabeth Kogut and staff of the Division of Perinatal Genetics and the General Clinical Research Center of Stanford University for coordination of patient recruitment; Ron Wong for initial sample processing of clinical samples; Norma Neff, Gary Mantalas, Ben Passarelli, and Winston Koh for their help in sequencing library preparation and data analysis.
Footnotes
Supplementary Information
Supplementary information is provided in a separate document.
Author Contributions
H.C.F., W.G., S.R.Q. conceived the study. H.C.F., W.G., J.W. performed experiments. H.C.F., W.G., and J.W. analyzed the data. Y.J.B. and Y.Y. E coordinated patient recruitment. H.C.F., W.G., J.W., and S.R.Q. wrote the manuscript. All authors discussed the results and commented on the manuscript.
The authors declare competing financial interests. S.R.Q. is a founder and shareholder of Fluidigm Corporation and Helicos BioSciences. S.R.Q. and H.C.F. are shareholders of Verinata Health.
References
- 1.Mandel P, Metais P. Les acides nucleiques du plasma sanguin chez l’homme. C R Acad Sci Paris. 1948;142:241–243. [PubMed] [Google Scholar]
- 2.Lo YM, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodurtha J, Strauss JF., 3rd Genomics and perinatal care. N Engl J Med. 2012;366:64–73. doi: 10.1056/NEJMra1105043. doi: 10.1056/NEJMra1105043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sehnert AJ, et al. Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell-free fetal DNA from maternal blood. Clinical chemistry. 2011;57:1042–1049. doi: 10.1373/clinchem.2011.165910. doi: 10.1373/clinchem.2011.165910. [DOI] [PubMed] [Google Scholar]
- 6.Bianchi DW, et al. Genome-Wide Fetal Aneuploidy Detection by Maternal Plasma DNA Sequencing. Obstetrics and gynecology. 2012 doi: 10.1097/AOG.0b013e31824fb482. doi: 10.1097/AOG.0b013e31824fb482. [DOI] [PubMed] [Google Scholar]
- 7.Palomaki GE, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genetics in medicine : official journal of the American College of Medical Genetics. 2012;14:296–305. doi: 10.1038/gim.2011.73. doi: 10.1038/gim.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palomaki GE, et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med. 2011;13:913–920. doi: 10.1097/GIM.0b013e3182368a0e. doi: 10.1097/GIM.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- 9.Ehrich M, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am J Obstet Gynecol. 2011;204:205, e201–e211. doi: 10.1016/j.ajog.2010.12.060. doi: S0002-9378(11)00018-4 [pii] 10.1016/j.ajog.2010.12.060. [DOI] [PubMed] [Google Scholar]
- 10.Chiu RW, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. Bmj. 2011;342:c7401. doi: 10.1136/bmj.c7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo YM, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2:61ra91. doi: 10.1126/scitranslmed.3001720. doi: 2/61/61ra91 [pii] 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 12.Fan HC, Quake SR. In principle method for noninvasive determination of the fetal genome. Available from Nature Proceedings. 2010 doi: < http://dx.doi.org/10.1038/npre.2010.5373.1>. [Google Scholar]
- 13.Macintyre S, Sooman A. Non-paternity and prenatal genetic screening. Lancet. 1991;338:869–871. doi: 10.1016/0140-6736(91)91513-t. doi: 01406736(91)91513-T [pii] [DOI] [PubMed] [Google Scholar]
- 14.Bellis MA, Hughes K, Hughes S, Ashton JR. Measuring paternal discrepancy and its public health consequences. J Epidemiol Community Health. 2005;59:749–754. doi: 10.1136/jech.2005.036517. doi: 59/9/749 [pii] 10.1136/jech.2005.036517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan HC, Wang J, Potanina A, Quake SR. Whole-genome molecular haplotyping of single cells. Nat Biotechnol. 2011;29:51–57. doi: 10.1038/nbt.1739. doi: nbt.1739 [pii] 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Consortium TGP. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. doi: nature09534 [pii] 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchini J, et al. A comparison of phasing algorithms for trios and unrelated individuals. Am J Hum Genet. 2006;78:437–450. doi: 10.1086/500808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White RA, 3rd, Blainey PC, Fan HC, Quake SR. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genomics. 2009;10:116. doi: 10.1186/1471-2164-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark MJ, et al. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol. 2011;29:908–914. doi: 10.1038/nbt.1975. doi: nbt.1975 [pii] 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. doi: 1105422108 [pii]10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.