Abstract
Highly malignant glioblastoma (GBM) is characterized by high genetic heterogeneity and infiltrative brain invasion patterns, and aberrant miRNA expression has been associated with hallmark malignant properties of GBM. The lack of effective GBM treatment options prompted us to investigate whether miRNAs would constitute promising therapeutic targets toward the generation of a gene therapy approach with clinical significance for this disease. Here, we show that microRNA-21 (miR-21) is upregulated and microRNA-128 (miR-128) is downregulated in mouse and human GBM samples, a finding that is corroborated by analysis of a large set of human GBM data from The Cancer Genome Atlas. Moreover, we demonstrate that oligonucleotide-mediated miR-21 silencing in U87 human GBM cells resulted in increased levels of the tumor suppressors PTEN and PDCD4, caspase 3/7 activation and decreased tumor cell proliferation. Cell exposure to pifithrin, an inhibitor of p53 transcriptional activity, reduced the caspase activity associated with decreased miR-21 expression. Finally, we demonstrate for the first time that miR-21 silencing enhances the antitumoral effect of the tyrosine kinase inhibitor sunitinib, whereas no therapeutic benefit is observed when coupling miR-21 silencing with the first-line drug temozolomide. Overall, our results provide evidence that miR-21 is uniformly overexpressed in GBM and constitutes a highly promising target for multimodal therapeutic approaches toward GBM.
INTRODUCTION
Glioblastoma (GBM) is the most common and aggressive type of glioma, a class of tumors arising from glial cells. Despite the increasing knowledge about this malignancy at genetic and molecular levels, and the considerable advances in cancer therapy, patient outcome has slowly improved over the past decade. Standard treatment for GBM includes surgical resection of the tumor, when possible, followed by single-agent adjuvant therapy with temozolomide and radiotherapy (1). However, these procedures lack effective long-term impact on disease control and patient survival, and clinical recurrence is nearly universal (1–3). Hence, there is an urgent need to explore new treatment options that can prove to be effective for brain tumors, as well as to better understand the molecular and cellular alterations that occur in GBM.
The discovery of miRNAs, a class of small non-coding RNAs that regulate gene expression through imperfect pairing with the target mRNAs (4,5), has revealed an additional level of fine tuning of the genome that integrates with transcriptional and other regulatory mechanisms to expand the complexity of eukaryotic gene expression. MiRNAs regulate posttranscriptionally the expression of over 30% of protein-coding genes (6), and in silico data indicate that each miRNA can control hundreds of gene targets, including oncogenes and tumor suppressors, underscoring the influence of miRNAs in key cellular processes that define the cell phenotype (6,7). Accumulated evidence has shown that miRNAs are differentially expressed in normal tissues and cancers, and aberrant miRNA expression is associated with tumor development and progression (8,9), including GBM pathogenesis (10,11).
In the present work, we analyzed the expression of miR-128, miR-21 and miR-221 in human GBM samples and in mouse GBM models and in several GBM cell lines. Our results demonstrate that miR-21 is upregulated and miR-128 is downregulated in GBM tissue samples and cell lines screened, a finding that is corroborated by analysis of a large set of human GBM data from The Cancer Genome Atlas (TCGA) Research Network. Furthermore, we identified a group of miRNAs, including the cluster miR-221/222 and oncogenic miR-106a/miR-20a, whose alterations may be correlated with different molecular subtypes of GBM described in the literature (12).
The classic genetic alterations that occur in GBM are found in pathways governing cellular proliferation and survival, including epidermal growth factor receptor (EGFR) and PTEN-regulated pathways, as well as invasion and angiogenesis (13). However, the therapeutic intervention with inhibitory agents targeting EGFR and other transduction pathways has yet to demonstrate a clear survival benefit for patients (14,15). Due to their small size and pivotal roles in the cell, certain microRNAs may be of direct therapeutic utility, as single agents or in combinations with other regimens (16). Studies performed by Silber and colleagues revealed that overexpression of miR-124 and miR-137, which are found to be downregulated in human GBM samples, induce GBM multiforme cell cycle arrest and differentiation of brain tumor stem cells (17). Similarly, overexpression of miR-128 has been shown to reduce tumor cell proliferation, both in glioma cell lines and a glioma-bearing animal model (18).
Here, we tested a therapeutic strategy for GBM that combines gene therapy through silencing of miR-21, found to be overexpressed in this type of brain tumor, with sunitinib, an inhibitor of platelet-derived growth factor (PDGF) and vascular endothelial growth factor (VEGF) receptors, (19) that is being currently evaluated in clinical trials for GBM.
Our results demonstrate that lipoplex-mediated miR-21 silencing in U87 human and F98 rat glioma cells significantly enhances cell sensitivity to the cytotoxic effect of sunitinib, which may represent an attractive and effective therapeutic approach toward GBM.
RESULTS
MiR-21 is overexpressed and miR-128 is downregulated in human and mouse GBM samples and GBM cell lines
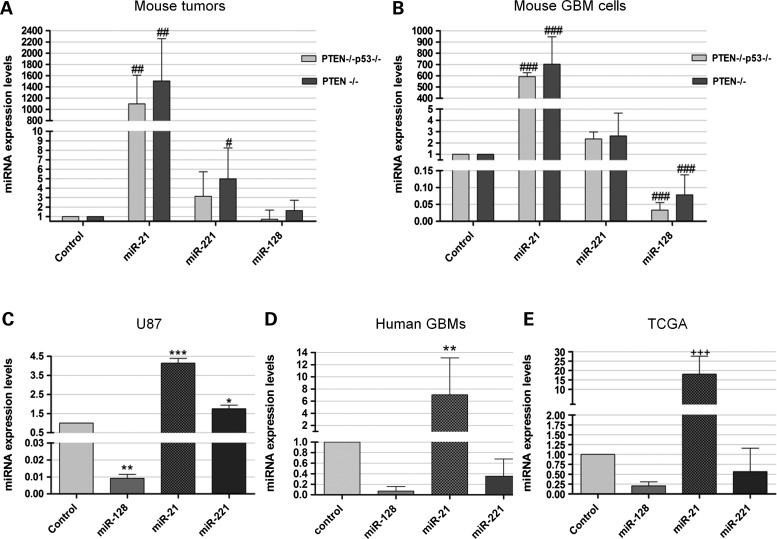
We have recently developed retrovirally induced mouse GBM models, characterized by the overexpression of the oncogenic ligand PDGF-B and conditional deletion of tumor suppressor genes (PTEN−/− and PTEN−/−p53−/−) that display molecular and histopathologic features that closely resemble human GBM (20). Real-time PCR (qPCR) quantification of miR-128, miR-21 and miR-221 expression in RNA extracts from six brain tumors (of each genotype) and three control samples, obtained from double-floxed mouse brains following animal injection with a control vector (no PDGF), revealed that miR-21 was highly overexpressed in all tumor samples when compared with control mouse brain, whereas miR-221 was moderately overexpressed in all samples from PTEN-floxed (3.15 ± 2.59) and double-floxed (4.99 ± 3.27) tumor samples (both values representing the relative miRNA expression value to control). MiR-128 was slightly downregulated in 83% of the double-floxed mouse tumors (0.70 ± 0.97), whereas slightly increased levels of miR-128 were observed in 67% of PTEN-floxed mouse tumors (1.63 ± 1.09, P > 0.05), when compared with those observed in samples from control mouse brain. MiRNA expression levels, evaluated in mouse GBM cell lines derived from the retrovirally induced double-floxed and PTEN-floxed mouse models (Fig. 1B), were consistent with the values observed in the tumor models, despite a significant decrease in miR-128 levels being observed in both double-floxed and PTEN-floxed mouse cells (0.033 ± 0.022 and 0.078 ± 0.0589, respectively). Increased expression of miR-21 and decreased miR-128 expression were also observed in the F98 rat glioma cell line, when compared with that observed in primary rat astrocytes or P19 embryonic carcinoma cells (Supplementary Material, Fig. S1A and B), previously demonstrated to express low levels of miR-21 (21).
Figure 1.
MiRNA expression levels in mouse and human GBM tissue samples and cell lines. MiR-21, miR-128 and miR-221 expression levels in (A) double-floxed (PTEN−/−p53−/−) and PTEN-floxed (PTEN−/−) mouse tumor samples (n = 6) and (B) double-floxed (PTEN−/−p53−/−) and PTEN-floxed (PTEN−/−) mouse GBM cells, when compared with control sample (n = 7), obtained from double-floxed mouse brains following animal injection with a control vector (no PDGF). MiR-21, miR-128 and miR-221 expression levels in human (C) U87 cells and (D) tumor samples from the Tissue Bank (n = 22), when compared with control epileptic brain tissue (n = 4). (E) MiR-21, miR-128 and miR-221 expression levels obtained from the analysis of the TCGA data on 188 human GBMs. *P < 0.05, **P < 0.01, ***P < 0.001 when compared with control epileptic tissue. #P < 0.05, ##P < 0.01, ###P < 0.001 when compared with control mouse brain. +++P < 0.001 when compared with normal human brain.
To evaluate whether the results observed for the mouse GBM models would replicate in the human disease, we measured the expression of miR-128, miR-21 and miR-221 in RNA extracts from 22 human GBM samples and from the widely used U87 human GBM cell line. Two different brain tissues (epileptic and tumor-adjacent brain tissue) and total RNA extracted from human brain (see Materials and Methods) were used as references. Similarly to the results obtained in the mouse models and in accordance with published data (10,18,21), miR-21 overexpression and miR-128 downregulation were observed in U87 cells (Fig. 1C). More importantly, miR-21 was significantly overexpressed in 80% of the human tumors when compared with control epileptic tissue (7.05 ± 6.07; P < 0.01), whereas miR-128 was downregulated in all tumor samples (0.07 ± 0.08) (Fig. 1D). However, as opposed to our results in the mouse models and U87 cells, as well as to those previously reported (10,22), miR-221 was found to be downregulated in 91% of the tumors (Fig. 1D). For comparison, the miRNA expression levels in U87 cells and human tumor samples relative to tumor-adjacent brain tissue and total RNA from human brain are presented in (Supplementary Material, Fig. S1C and D).
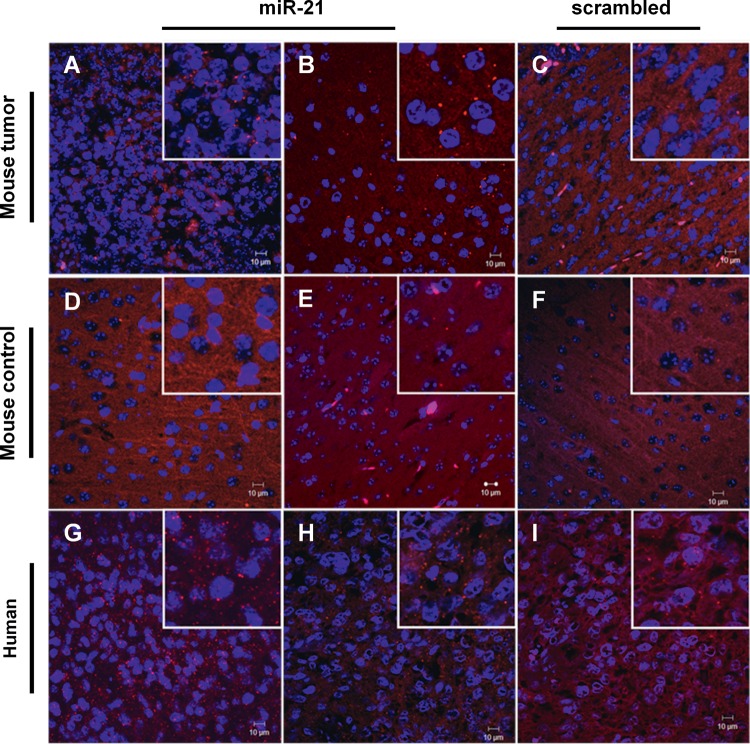
Aiming at increasing the biologic significance of our results, we analyzed the miRNA expression in a large number of human GBM samples from the TCGA, a publicly available repository that has accumulated comparative genomic hybridization, gene expression and miRNA expression analyses for ∼200 human GBM samples (23). The analysis of a specific subset of GBMs from this set of samples (Supplementary Material, Table S1) revealed that miR-21 was significantly overexpressed and miR-128 was downregulated in 98% of the tumors (185/188) when compared with control normal samples (Fig. 1E). Remarkably, miR-221 was found to be downregulated in 48% (90/188) of the tumors (0.57 ± 0.59), thus corroborating our experimental data from the studies on the human tumor samples. MiR-21 overexpression in the double-floxed mouse model was also evident from fluorescence in situ hybridization (FISH) experiments performed in formalin-fixed paraffin embedded (FFPE) tissue sections. Figure 2 displays typical images obtained from these experiments showing miR-21 staining in two different GBM samples (Fig. 2A and B), whereas residual staining was detected in control brains (Fig. 2D and E) or using a control scrambled probe (Fig. 1C and F). Similarly, analysis of tissue distribution of miR-21, assessed by FISH in FFPE human tumor sections, showed an increase in miR-21 staining when compared with that using a control scrambled probe (Fig. 2G–I).
Figure 2.
FISH staining in mouse and human GBM tissue samples. FISH staining in (A–C) double-floxed mouse GBM sections and (D–F) control mouse brain tissue (n = 2) stained with (A, B, D and E) miR-21-specific or (C and F) scrambled LNA probe. Nuclear staining was obtained using DNA-binding Hoechst 33342. MiR-21 staining (red dots) is observed in the GBM sections, surrounding the cell nucleus (blue), whereas residual staining is observed in control brain sections. Human GBM sections (n = 2) were stained with (G and H) miR-21-specific or (I) non-coding (scrambled) LNA probe. MiR-21 staining (red dots) is predominantly cytoplasmic, surrounding the cell nucleus (blue). Control experiments targeting the endogenous U6snRNA (positive control) and without LNA probe (negative control) were performed in parallel (not shown). Images were obtained by confocal microscopy with a 40× EC Plan-Neofluar. Scale corresponds to 10 µm.
MiR-106a, miR-130b, miR-20a, miR-221, miR-222, miR-155 and let-7i dysregulation correlates with different subtypes of GBM
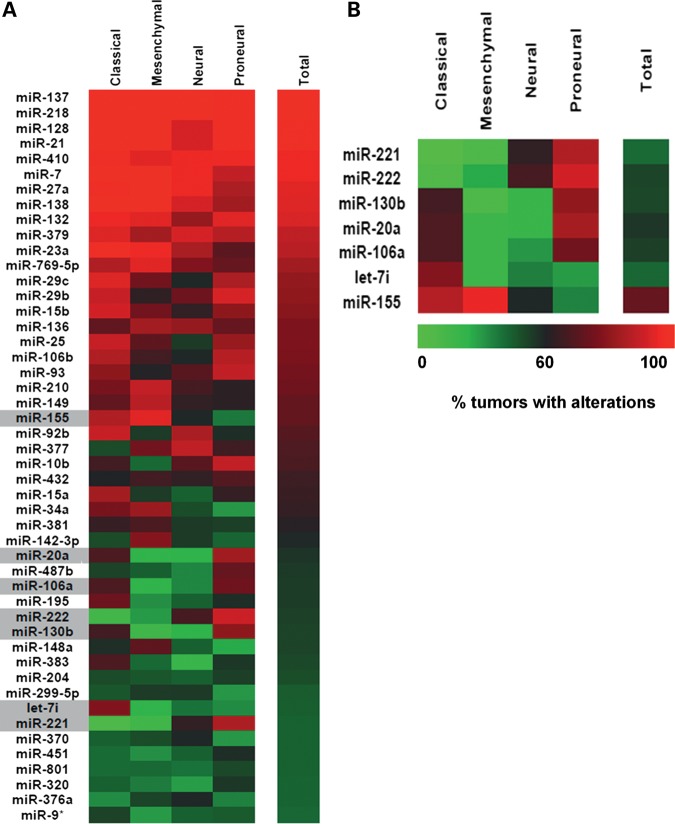
Recent analysis of the set of gliomas available from TCGA has shown that they can be divided into four different subtypes (classical, mesenchymal, neural and proneural) that reflect the common signaling abnormalities found in these tumors (12). As miR-21 and miR-128 dysregulation constitutes a common molecular alteration among the four GBM subtypes (Fig. 3 and Supplementary Material, Table S1), we sought to identify miRNAs whose dysregulation might correlate with the different subtypes of GBM. Considering those miRNAs whose expression is altered in at least 45% of the tumors (Fig. 3A), excluding uniformly altered miRNAs (>90% tumors with alterations in miRNA levels), and employing a significance threshold of 0.05 and a 5% false discovery rate (FDR) to correct for multiple comparisons (Supplementary Material, Table S2), we identified a small group of miRNAs whose alterations are associated with specific GBM subtypes. Our findings, presented in Figure 3B, suggest that concurrent alterations in miR-106a, miR-130b, miR-20a, miR-221 and miR-222 are predominant in the proneural subtype of GBM, whereas alterations occurring only in miR-155, miR-221 and miR-222 are predominant in the neural group. Furthermore, alterations in miR-106a, miR-130b, miR-20a and let-7i concomitant with the absence of alterations in miR-221/222 are predominant in the classical subtype of GBM . This characterization is potentially very useful as a molecular classification tool.
Figure 3.
Analysis of TCGA data on 188 human GBMs. (A) Heat map representation of the miRNAs dysregulated in at least 45% (85/188) of the tumors (n = 188), displayed as percentage of tumors with alterations in each of the four GBM subtypes (classical, mesenchymal, neural and proneural). Highlighted in gray are the miRNAs represented in (B). (B) MiRNAs whose dysegulation is associated with specific GBM subtypes, by applying a significance threshold of 0.05 and a FDR of 5% (0.05). Statistical analysis was performed as described in the Materials and Methods section.
Delivery liposomal system-based lipoplexes efficiently deliver anti-miR-21 oligonucleotides to glioma cells
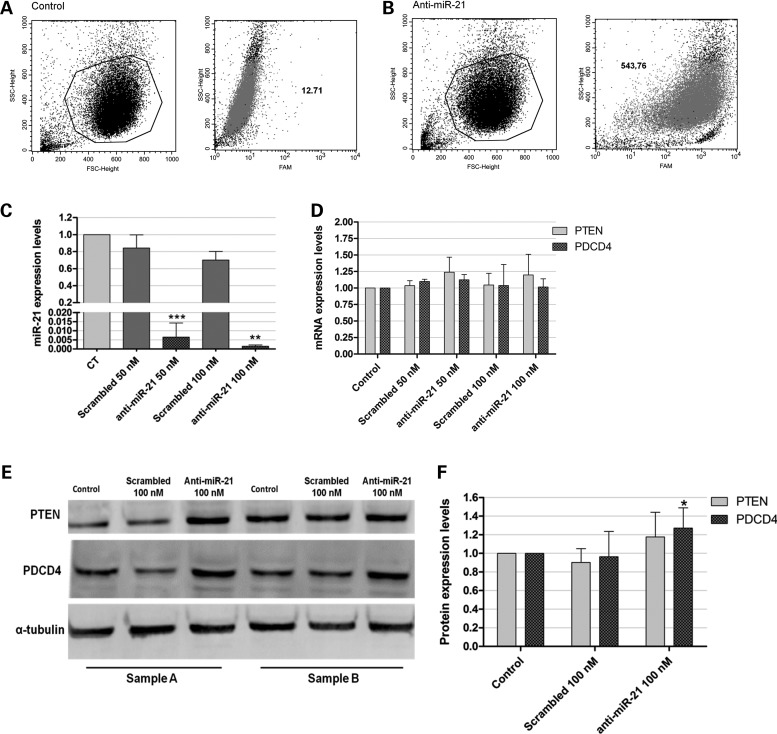
Accumulated evidence from in vitro and in vivo studies strongly suggests a key role for miR-21 in tumorigenesis (24,25). In this context, our findings of invariant overexpression of miR-21 in GBM cells prompted us to examine whether silencing of miR-21 in GBM cells, through delivery of anti-miR-21 oligonucleotides, would result in an antitumoral effect. In this regard, we have recently shown that delivery liposomal system (DLS) liposomes enhance intracellular delivery of single-stranded antisense oligonucleotides while improving release from endocytic vesicles (26). Therefore, our initial studies addressed the capacity of DLS liposomes to deliver anti-miR-21 oligonucleotides to GBM cells. Using flow cytometry, extensive lipoplex-cell association was observed 4 h after cell transfection with lipoplexes (∼87% transfected cells), as illustrated by the huge increase (∼42-fold) in the fluorescence intensity of the transfected cells (Fig. 4B), when compared with that of untreated cells (control) (Fig. 4A). Internalization of the lipoplexes carrying 6-carboxyfluorescein (FAM)-labeled anti-miR-21 oligonucleotides was also monitored in U87 cells, by confocal microscopy. The results presented in Supplementary Material, Figure S2 reveal that 4 h after lipoplex-mediated delivery of FAM-labeled oligonucleotides, green fluorescent particles were found in a large extent in the cytoplasm of transfected U87 cells (Supplementary Material, Fig. S2C), whereas no fluorescence was observed in untreated control cells (Supplementary Material, Fig. S2A). A similar observation was made when F98 glioma cells were incubated with lipoplexes carrying FAM-labeled anti-miR-21 oligonucleotides (Supplementary Material, Fig. S2B and D). In contrast, the delivery of a similar amount of naked FAM-labeled oligonucleotides did not allow the detection of green fluorescence within the cells (data not shown).
Figure 4.
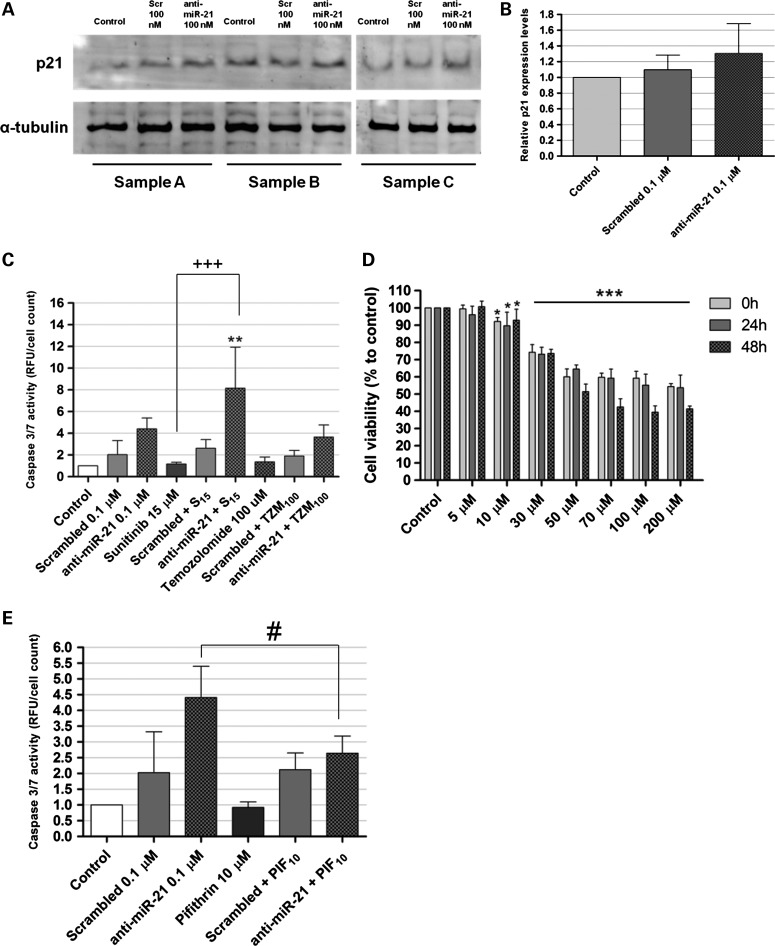
Lipoplex-mediated delivery of anti-miR-21 oligonucleotides in U87 human GBM cells and miR-21, PTEN and PDCD4 expression after oligonucleotide-mediated miR-21 silencing. For the assessment of lipoplex-cell association, U87 cells were incubated with lipoplexes for 4 h, rinsed with PBS and prepared for flow cytometry analysis (as described in Supplementary Materials and Methods). Viable cells were gated based on morphologic features, including cell volume (given by the forward scatter, FSC) and cell complexity (given by the side scatter, SSC) (left plots in [A and B]). Fluorescent intensity plots of U87 cells (A) untreated or (B) transfected with lipoplexes at a final concentration of 100 nm anti-miR-21 oligonucleotides are shown in the right plots in (A) and (B). Mean fluorescence values (geometric mean) are indicated for each plot. (C) miR-21 and (D) PTEN and PDCD4 mRNA expression levels in U87 cells 48 h after transfection with anti-miR-21 or scrambled oligonucleotides (n = 3), at a final oligonucleotide concentration of 50 or 100 nm. MiR-21 expression levels, normalized to the reference U6snRNA, as well as PTEN and PDCD4 expression levels, normalized to the reference HPRT1, are presented as relative expression values to control untreated cells. (E) Representative gel showing PTEN and PDCD4 protein levels in U87 cells 48 h after transfection with anti-miR-21 or scrambled oligonucleotides (n = 3) at a final oligonucleotide concentration of 100 nm. (F) Quantification of PTEN and PDCD4 bands observed in (E), corrected for individual α-tubulin signal intensity. Results are presented as PTEN and PDCD4 expression levels relative to control. *P < 0.05, **P < 0.01, ***P < 0.001 when compared with cells transfected with a similar amount of scrambled oligonucleotides.
MiR-21 silencing increases PTEN and PDCD4 expression in U87 human GBM cells
Following the demonstration of the feasibility of DLS-based lipoplexes to mediate efficient delivery of oligonucleotides into GBM cells, we evaluated the effect of intracellularly delivered anti-miR-21 oligonucleotides on the levels of miR-21 and mRNA of their target genes. The transfection of U87 cells with anti-miR-21 oligonucleotides (50 nm) resulted in a significant decrease in miR-21 levels (0.0065 ± 0.007), which was further enhanced when cells were transfected with 100 nm anti-miR-21 oligonucleotides (0.0015 ± 0.0007), as compared to those obtained upon transfection with a non-coding (scrambled) oligonucleotide sequence (Fig. 4C). Parallel experiments demonstrated that miR-21 silencing did not significantly affect the levels of other miRNAs (Supplementary Material, Fig. 3).
We further evaluated the effect of miR-21 silencing on the expression of the tumor suppressors PDCD4 and PTEN that have been previously identified as miR-21 targets (25,27,28). As shown in Figure 4D, a moderate, even though not significant, increase in PTEN mRNA levels was observed in cells transfected with 50 or 100 nm anti-miR-21 oligonucleotides, when compared with those observed in cells transfected with a scrambled sequence (∼20 and 15% increase, respectively), whereas no significant changes were observed in PDCD4 mRNA levels. Nevertheless, a considerable increase in PTEN (∼34%, P > 0.05) and PDCD4 (∼33%, P < 0.05) protein expression was observed in U87 cells transfected with anti-miR-21 oligonucleotides, when compared with that observed in cells transfected with a scrambled sequence, as shown in Figure 4E and F.
MiR-21 silencing increases apoptotic activity in U87 GBM cells
MiR-21 has been previously shown to target several components of important apoptotic pathways, including the p53-dependent apoptotic pathway (29). For this reason, we evaluated whether a decrease in the levels of this miRNA would have any effect on the activity of the tumor suppressor p53. Western blot quantification of p53 using two different antibodies failed to detect the protein in U87 cells (data not shown). The fact that U87 cells express wild-type p53, an isoform with extremely short half-life (30), may explain the absence of detection of this protein. Therefore, we assessed p53 activity indirectly, by measuring the levels of p21/WARF1, a cyclin-dependent kinase inhibitor that is a direct transactivation target of p53 (31). In this regard, a moderate increase (∼20%) in the levels of p21 was observed in cells transfected with anti-miR-21 oligonucleotides, when compared with that observed in cells transfected with scrambled oligonucleotides (Fig. 5A and B; P > 0.05). Moreover, we investigated whether miR-21 silencing would increase the apoptotic activity in U87 GBM cells, by determining the activity of the effector caspases 3 and 7, crucial components of the apoptotic cell death. As shown in Figure 5C, transfection of U87 cells with 100 nm anti-miR-21 oligonucleotides resulted in a 2-fold increase in caspase 3/7 activity (P > 0.05), when compared with that observed for cells transfected with a scrambled sequence. More importantly, silencing of miR-21 followed by cell exposure to 15 µm of the tyrosine kinase inhibitor sunitinib resulted in a dramatic increase in caspase 3/7 activity (8.15 ± 3.773), when compared with that observed for cells exposed to sunitinib, either per se (1.171 ± 0.1539, P < 0.001) or in combination with transfection mediated by scrambled oligonucleotides (2.62 ± 0.800, P < 0.01). In contrast, miR-21 silencing followed by treatment of U87 cells with 100 µm of the alkylating drug temozolomide, currently used in the clinic as frontline therapy, did not result in significant enhancement of cell apoptotic activity.
Figure 5.
Western blot detection of p21 and caspase activation in U87 human GBM cells. (C) Caspase 3/7 activity in U87 cells transfected with anti-miR-21 or scrambled oligonucleotides (0.1 μm), either per se or in combination. (A) Representative gel showing p21 levels in U87 cells 48 h after transfection with anti-miR-21 or scrambled (Scr) oligonucleotides (n = 3) at a final oligonucleotide concentration of 100 nm. (B) Quantification of p21 bands observed in (A), corrected for individual α-tubulin signal intensity bination with exposure to 15 μm sunitinib or 100 μm temozolomide (n = 3). Twenty-four hours after transfection, cells were exposed to 15 μm sunitinib or 100 μm temozolomide for 24 h, rinsed with PBS, after which cells were either prepared for caspase detection (cells incubated with sunitinib) or further cultured for 24 h and then prepared for caspase detection (cells incubated with temozolomide). Results, presented as relative fluorescence units with respect to control untreated cells, were normalized for the number of cells in each condition. Scrambled/anti-miR-21 + S15/TZM100 cells were transfected with scrambled or anti-miR-21 oligonucleotides and further incubated with 15 μm sunitinib or 100 μm temozolomide. **P < 0.01 when compared with cells transfected with scrambled oligonucleotides and further incubated with 15 μm sunitinib. +++ P < 0.001 when compared with cells exposed to 15 μm sunitinib. #P < 0.01 when compared with cells transfected with scrambled oligonucleotides (one-way analysis of variance with Benferroni's posthoc test). (D) Cell viability was evaluated by the Alamar Blue assay, immediately (0), 24 h (24 h) or 48 h (48 h) after incubation with pifithrin-α. *P < 0.05, ***P < 0.001 when compared with untreated cells. (E) Caspase 3/7 activity in U87 cells transfected with anti-miR-21 or scrambled oligonucleotides (0.1 μm), either per se or in combination with exposure to 10 μm pifithrin-α (n = 3). Four hours after transfection, cells were incubated with 10 μm pifithrin-α for 24 h, and further incubated for 24 h in fresh DMEM medium. Caspase activity was assessed as described in the Materials and Methods section. Results, presented as relative fluorescence units with respect to control untreated cells, were normalized for the number of cells in each condition. Scrambled/anti-miR-21 + PIF100 cells were transfected with scrambled or anti-miR-21 oligonucleotides and further incubated with 10 μm pifithrin-α. #P < 0.01 when compared with cells transfected with anti-miR-21 oligonucleotides.
Pifithrin-α-mediated p53 inhibition reduced the caspase activation associated with decreased miR-21 expression levels
To evaluate whether the increase in apoptosis observed for U87 cells following miR-21 silencing could be mediated by p53, a transient inhibition of p53 function was induced in both untreated and anti-miR-21 transfected U87 cells (wild-type p53), using pifithrin-α, a DNA-binding inhibitor of p53 transcriptional activity (32,33). In accordance with previously reported studies (34,35), reduced signs of toxicity were detected when U87 cells were incubated with 10 μm of pifithrin-α for up to 48 h while a marked decrease in viability could be observed immediately after cell treatment with 30 μm or higher concentrations of the drug (Fig. 5D). As demonstrated in Figure 5E, a small, but significant, decrease in the levels of caspase 3/7 activity was observed in cells transfected with anti-miR-21 oligonucleotides and further incubated with 10 μm of pifithrin-α (2.645 ± 0.54), when compared with those of cells transfected with anti-miR-21 oligonucleotides without drug treatment (4.41 ± 0.99, P < 0.05).
Lipoplex-mediated miR-21 silencing enhances the cytotoxic effect of sunitinib in U87 and F98 glioma cells
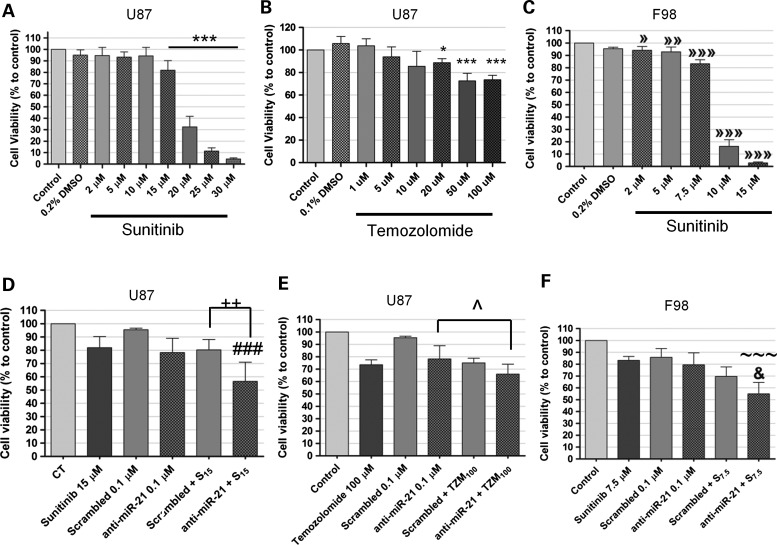
To evaluate whether the caspase activation associated with decreased miR-21 expression in U87 cells would correlate with changes in cell proliferation, cell viability was measured after transfection with anti-miR-21 oligonucleotides, either per se or in combination with sunitinib or temozolomide. Initial experiments were performed by exposing U87 cells to different concentrations of sunitinib or temozolomide for 24 h (Fig. 6A and B), to determine the optimal concentration of drug to be used in the assay. A moderate decrease in cell viability was observed when U87 cells were transfected with anti-miR-21 oligonucleotides (78.2 ± 10.8), as compared to that observed for cells transfected with a scrambled sequence (95.4 ± 1.2, P > 0.05) (Fig. 6D). Remarkably, a significant decrease in the percentage of viable cells was observed when cells were transfected with anti-miR-21 oligonucleotides and further exposed to sunitinib (56.52 ± 14.48), as compared to that observed upon exposure to sunitinib, either per se (78.20 ± 10.78, P < 0.001) or in combination with the transfection of scrambled oligonucleotides (80.26 ± 7.78, P < 0.01). Conversely, lipoplex-mediated miR-21 silencing in U87 cells did not improve significantly the reduction in cell viability associated with exposure to temozolomide (Fig. 6E). A significant decrease in the percentage of viable cells was also observed when F98 rat glioma cells were transfected with anti-miR-21 oligonucleotides and further exposed to sunitinib (54.95 ± 9.611), as compared to that observed upon exposure to sunitinib, either per se (83.16 ± 3.317, P < 0.001) or in combination with the transfection of scrambled oligonucleotides (69.6 ± 8.109, P < 0.05) (Fig. 6F). Similarly to what was observed with the U87 cells, lipoplex-mediated miR-21 silencing of F98 cells did not improve significantly the reduction in cell viability associated with exposure to temozolomide (data not shown).
Figure 6.
Cell viability after incubation with sunitinib or temozolomide, either per se or in combination with the transfection of anti-miR-21 or scrambled oligonucleotides. Twenty-four hours before any experiment, U87 and F98 cells were plated onto 24-well plates at a density of 3.5 × 104 and 3 × 104 cells/well (respectively). Cells were transfected for 4 h with anti-miR-21 or scrambled oligonucleotides, rinsed with PBS, cultured for 24 h in fresh DMEM and then exposed to sunitinib or temozolomide for 24 h. Cell viability was evaluated by the Alamar Blue assay (as described in the Materials and Methods section) immediately (sunitinib) or 24 h (temozolomide) after incubation with the drug. Cell viability in (A and B) U87 and (C) F98 glioma cells after incubation with different concentrations of (A and C) sunitinib or (B) temozolomide for 24 h. Cell viability in (D and E) U87 and (F) F98 cells after incubation with sunitinib (D and F) or temozolomide (E), either per se or in combination with the transfection of anti-miR-21 or scrambled oligonucleotides. Scrambled/anti-miR-21 + S7.5/S15/TZM100 cells were transfected with scrambled or anti-miR-21 oligonucleotides and further incubated with 7.5 or 15 μm sunitinib or 100 μm temozolomide. *P < 0.05, ***P < 0.001 when compared with control untreated U87 cells. »P < 0.05, »»P < 0.01, »»»P < 0.001 when compared with control untreated F98 cells. ++P < 0.01 when compared with U87 cells transfected with scrambled oligonucleotides and further incubated with 15 μm sunitinib. ###P < 0.001 when compared with U87 cells exposed to 15 μm sunitinib. ^P < 0.05 when compared with U87 cells transfected with anti-miR-21 oligonucleotides. &P < 0.05 when compared with F98 cells transfected with scrambled oligonucleotides and further incubated with 7.5 μm sunitinib. ∼∼∼P < 0.001 when compared with F98 cells transfected with anti-miR-21 oligonucleotides or incubated with 7.5 μm sunitinib.
Sunitinib exposure decreases NF-kB activation in U87 GBM cells
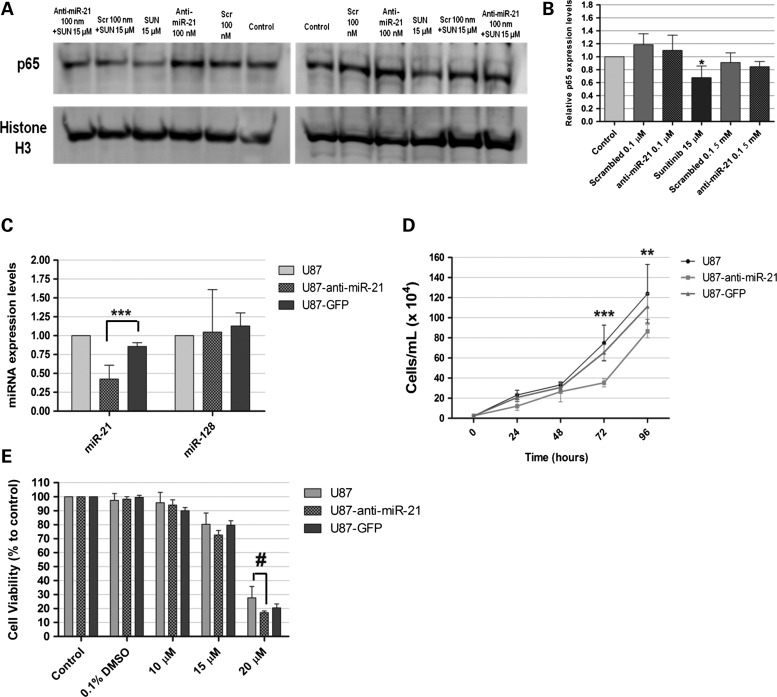
A study by Perkins and colleagues has shown that the p53-inhibitor pifithrin-α may indirectly enhance NF-kB signaling in tumor cells (36). Moreover, several reports have demonstrated the existence of reciprocal regulation between p53 and NF-kB (30,37). The observation of increased p53 activity and sunitinib cytotoxicity in cells with decreased miR-21 levels prompted us to test whether miR-21 silencing and/or sunitinib exposure would have any effect on NF-kB, a transcription factor whose activity has been linked to inflammation and cancer (38). In response to cellular stimuli, the p65/RelA subunit of NF-kB is translocated to the nucleus, where it binds specific DNA motifs and initiates transcription. The nuclear expression of p65/RelA was assessed in U87 cells transfected with scrambled or anti-miR-21 oligonucleotides, and compared to that of untransfected cells. As shown in Figure 7A and B, no significant decrease in p65 expression was observed in U87 cells transfected with anti-miR-21 oligonucleotides, when compared with that observed for cells transfected with scrambled oligonucleotides. Importantly, p65 expression was reduced by ∼32% in U87 cells exposed to sunitinib, when compared with normal untreated cells (P < 0.05). Decreased p65 expression was also observed in U87 cells transfected with anti-miR-21 oligonucleotides and further exposed to sunitinib, when compared with that observed for untreated cells or cells transfected with scrambled oligonucleotides and further exposed to sunitinib (∼10 and 5%, respectively; P > 0.05).
Figure 7.
Western blot detection of NF-kB (p65 subunit) in U87 GBM cells and evaluation of miR-21 expression, proliferation and viability in U87, U87-anti-miR-21 and U87-GFP GBM cells after incubation with sunitinib. Twenty-four hours after cell transfection with scrambled (Scr) or anti-miR-21 oligonucleotides, at a final concentration of 100 nm, cells were exposed to 15 μm sunitinib (SUN 15 μm) for 24 h, rinsed with PBS, after which nuclear extracts were prepared. (A) Representative gel showing p65 levels in U87 cells 48 h after transfection with anti-miR-21 or scrambled (Scr) oligonucleotides (n = 3) at a final oligonucleotide concentration of 100 nm, either per se or in combination with exposure to 15 μm sunitinib. (B) Quantification of p65 bands observed in (A), corrected for individual histone H3 signal intensity. *P < 0.05 when compared with control untreated U87 cells. Modified U87 cell lines, expressing either an anti-miR-21 short hairpin (U87-anti-miR-21) or a GFP-coding short hairpin (U87-GFP), were developed as described in the Materials and Methods section. (A) miR-21 and miR-128 expression in U87-anti-miR-21 and U87-GFP cells, normalized to the reference U6snRNA. Results are presented as relative expression values to control U87 cells. Two-way analysis of variance combined with the Bonferroni posthoc test was used for statistical analysis. ***P < 0.001 when compared with U87-GFP cells. (B) U87, U87-anti-miR-21 and U87-GFP cells were plated onto 48-well plates at a density of 2.5 × 104 cells/well. At defined time points, cells were washed with PBS, trypsinized and resuspended in culture medium. The number of viable cells was determined by Trypan blue exclusion. Two-way analysis of variance combined with the Bonferroni posthoc test was used for statistical analysis. **P < 0.01, ***P < 0.001 when compared with U87-GFP cells. (C) Twenty-four hours before incubation with sunitinib, cells were plated onto 24-well plates at a concentration of 5 × 104 cells/well. Cells were exposed to different concentrations of sunitinib for 24 h and further grown for 24 h in fresh culture medium, after which the cell viability was evaluated by the Alamar Blue assay (as described in the Materials and Methods section). Two-way analysis of variance combined with Bonferroni's posthoc test was used for statistical analysis. #P < 0.001 when compared with U87 cells.
Lentivirally mediated miR-21 silencing does not significantly affect the sensibility of U87 cells toward sunitinib
Aiming at evaluating whether a permanent decrease in EmiR-21 expression in U87 GBM cells could sensitize these cells toward sunitinib, we developed a lentivirally modified U87 cell line expressing a short hairpin against the mature form of miR-21 and green fluorescent protein (GFP) (U87-anti-miR-21). Lentivirally modified U87 cells expressing only GFP were used as a control (U87-GFP). Viral transduction of U87 cells was very efficient, as concluded by the observation of ∼85% (U87-anti-miR-21) and 90% (U87-GFP) of cells expressing GFP (data not shown). As shown in Figure 7C, miR-21 expression levels were significantly reduced in U87-anti-miR-21 cells (0.426 ± 0.184, P < 0.001), as compared with those observed in U87-GFP cells expressing GFP (0.858 ± 0.05) or parental (non-transduced) U87 cells. Parallel experiments demonstrated that miR-21 silencing did not significantly affect the levels of miR-128 (Fig. 7C), thus indicating that the observed reduction in miR-21 levels is sequence specific. Furthermore, U87-anti-miR-21 cells displayed decreased proliferation rate, when compared with that of U87-GFP and parental U87 cells (Fig. 7D). A small, although not significant, decrease in cell viability was observed in U87-anti-miR-21 cells following incubation with 15 μm sunitinib for 24 h (72.68 ± 7.85), as compared with that observed in U87-GFP cells expressing GFP (79.64 ± 7.03) (Fig. 7E). A smaller decrease was observed when cells were incubated with 20 μm sunitinib (∼3%), whereas no differences were observed when cells were incubated with 10 μm sunitinib.
DISCUSSION
Over the last decade, accumulated evidence has shown that miRNAs play an important role in relevant molecular and cellular mechanisms governing GBM tumorigenesis, including cell proliferation, invasion and stem cell renewal (16,18,39). Due to their small size and influence in a broad range of biologic processes, miRNAs are very attractive therapeutic targets for GBM. In the present study, we demonstrate that a combination of a liposome-based gene therapy approach targeting miR-21, uniformly overexpressed in the different subtypes of GBM, with sunitinib, an inhibitor of tyrosine kinase receptors that include PDGFR and VEGF, enhances the cytotoxic effect of this drug in U87 human and F98 glioma cells.
Alterations in miRNA signaling have been associated with several aspects of carcinogenesis, including tumor invasion and metastasis and patient outcome (40,41). Here, by demonstrating that miR-21 is overexpressed and miR-128 is downregulated in mouse GBM models and in a large number of human GBM samples, our results not only support those from previously reported studies (10,21,22), but also underline the universal character of these alterations in the global GBM profile. Recently, an interesting study from the TCGA Research Network divided GBMs into four clinically relevant subtypes characterized by abnormalities in PDGF receptor alpha, isocitrate dehydrogenase 1, EGFR and neurofibromin 1 (12), and among the two main subtypes identified, the ‘classical’ GBMs predominantly harbor EGFR amplification and rearrangement, whereas the ‘proneural’ GBMs are predominantly driven by PDGF signaling (12). By identifying a small group of miRNAs whose alterations are associated with the different GBM subclasses, we extended the molecular characterization of this group of tumors to non-coding RNAs. The results here presented suggest that alterations in the expression of the miR-221/222 cluster occur predominantly in the proneural and neural subtypes (Fig. 3B and Supplementary Material, Table S1), whereas alterations in let-7i are predominant in the classical group, and alterations in the oncogenic miR-20a and miR-106a are common to both subtypes (Fig. 3B). However, as none of the miRNAs analyzed are strictly altered in a unique GBM subtype (under the defined exclusion criteria), our data do not implicate such miRNAs as additional direct biomarkers, but rather complementary molecular markers that may help in the correct identification of the different GBM subclasses.
The biologic effects of miR-21 are most likely associated to the simultaneous repression of multiple tumor suppressor genes, including tropomyosin 1, PDCD4 and PTEN (28,42,43), as well as of several invasion/metastasis suppressors (25). Here, and in accordance with previous studies (42,44), we provide evidence that oligonucleotide-mediated miR-21 silencing increases the expression levels of the tumor suppressors PTEN and PDCD4. PTEN is a frequently disrupted tumor suppressor in glioma and capable of restricting growth and survival signals by limiting the activity of the phosphoinositide-3-kinase (PI3K) pathway, a signaling cascade that controls cell proliferation, growth, differentiation and survival (45,46). PDCD4 is also frequently lost in glioma (47) and has been shown to regulate important cellular processes affecting cell phenotype, including protein translation (48), promoter activation (49) and cell cycle regulation (50). The observation of increased levels of the PDCD4 protein (and not mRNA) following miR-21 silencing suggests that miR-21 regulates PDCD4 expression in U87 GBM cells by inhibiting its translation, rather than promoting PDCD4 mRNA degradation. A similar mechanism has previously been reported by Chen and colleagues (51).
MiR-21 has also been shown to target multiple components of important apoptosis-related pathways, including p53, TGF-β and mitochondrial apoptosis (29). Knockdown of miR-21 in cultured GBM cells and glioma-bearing mice has been previously shown to trigger caspase activation and lead to increased apoptotic cell death and decreased tumor cell viability (21,52). In this regard, our results also reveal an increase in caspase 3/7 activation following miR-21 silencing in U87 cells, thus confirming an antiapoptotic role of miR-21 in human GBM cells. The observation of increased p21 expression in U87 cells with decreased miR-21 levels, as well as reduced caspase activation following miR-21 silencing in cells with repressed p53 transcriptional activity, strongly suggests that miR-21 silencing increases p53 activity in U87 GBM cells. The increased expression of the tumor suppressors and the activation of caspases observed following miR-21 silencing may not only explain the reduction observed in the proliferation of U87 cells, but also render the cells susceptible to drugs targeting other signaling pathways governing GBM tumorigenesis. In this regard, several in vitro studies have already shown that miR-21 modulation potentiates the cytotoxic effect of antineoplastic drugs. The codelivery of anti-miR-21 oligonucleotides significantly improved the cytotoxicity of fluorouracil (5-FU) in U251 human GBM cells while increasing apoptosis and decreasing tumor cell migration (53). MiR-21 inhibition was also shown to enhance the chemosensitivity of human GBM cells to taxol, independent of PTEN status (54). Here, we are the first to demonstrate that lipoplex-mediated miR-21 silencing enhances significantly the cytotoxic effect of sunitinib in both U87 human and F98 rat glioma cells. Sunitinib is currently approved by the FDA for the treatment of gastro-intestinal stromal tumors (GIST) and renal cell carcinoma, and several phase II clinical trials are underway for GBM (55). Moreover, we demonstrate that sunitinib decreases the nuclear expression of p65, the gene regulatory subunit of the NF-kB transcription factor, which suggests that sunitinib decreases the activation of the oncogenic NF-kB pathway in U87 GBM cells. The oncogenic NF-kB and the tumor suppressor p53 have reciprocal activities and functions. P53 can regulate the levels of NF-kB to promote apoptosis and cell death, whereas NF-kB-mediated negative regulation of p53 can contribute to tumorigenesis (37). In this regard, the simultaneous induction of p53-related activity and repression of NF-kB, achieved by the silencing of miR-21 concomitant with sunitinib exposure, may explain the synergistic cytotoxic effect observed with the simultaneous application of both strategies. In this context, the results obtained in this study reveal a miRNA-based therapeutic approach with potential to complement a primary drug-based therapy that may prove to be of great importance for clinical application, particularly considering the drug resistance and toxic side effects usually associated with the use of chemotherapy. Experiments in an animal model of GBM are currently in progress in our laboratory to address the in vivo efficacy of the generated combined therapeutic approach and will constitute a separate study.
The results obtained in this study also indicate that a permanent decrease in miR-21 levels, achieved by lentiviral transduction of U87 cells, does not significantly improve the sensitivity of U87 cells toward sunitinib, as opposed to what was achieved with the transient oligonucleotide-based approach. In this regard, the moderate decrease in the levels of miR-21 obtained in cells transduced with lentivirus (∼2-fold), when compared with the pronounced decrease in the levels of miR-21 in oligonucleotide-transfected cells (∼150-fold), may help in explaining the observed differences. Experiments involving an optimized lentiviral vector, able to produce a more efficient anti-miR-21 short hairpin RNA (shRNA), may clarify whether the sensitivity of U87 cells toward sunitinib relates with the expression levels of miR-21.
In summary, the observation of an invariable overexpression of miR-21 in GBM associated with the highly promising results achieved in lipoplex-mediated miR-21-antagonism studies suggests that miR-21 is an ideal candidate for a multimodal therapeutic approach, combining miRNA-based gene therapy with antiangiogenic activity toward GBM.
MATERIALS AND METHODS
Materials
Sunitinib malate (Sutent®) was kindly offered by Pfizer (Basel, Switzerland) and temozolomide (Temodar®, Merck) was acquired from Sigma (Munich, Germany). Stock solutions were prepared in DMSO (Sigma, Germany) and stored at −20°C or 4°C (respectively). The miRZip anti-miR-21 construct was acquired from System Biosciences (Mountain View, CA, USA). The locked nucleic acid (LNA)-modified anti-miR-21 oligonucleotides and a non-coding (scrambled) sequence, as well as digoxigenin (DIG)-labeled LNA detection probes for miR-21, scrambled and U6snRNA, were acquired from Exiqon (Vedbaek, Denmark). All sequences are displayed in Supplementary Material, Table S3. All other reagents were obtained from Sigma unless stated otherwise.
Human and mouse tissue samples
Human GBM samples and non-neoplastic brain tissue were obtained from the Bartoli Brain Tumor Bank at the Columbia University Medical Center (CUMC) and were used in accordance with the policies of the CUMC review board. Briefly, tissue samples were flash frozen in liquid nitrogen immediately after surgical resection, and further stored at −80°C. Neoplastic tissue samples were obtained from viable areas of tumor while trying to avoid necrotic areas. Non-neoplastic brain tissue samples were derived from the temporal lobes of patients surgically treated for temporal lobe epilepsy. Tissue adjacent to the tumors was also obtained from postmortem specimens. Commercially available total RNA from adult human brain was acquired from Clontech (Mountain View, CA, USA) (Catalog No: 636530, Lot number: 9022522A).
Mouse GBM samples were obtained from established GBM models developed by injection of the retroviral vector PDGF-IRES-CRE in the subcortical white matter of double-floxed (PTEN−/−p53−/−) or PTEN-floxed (PTEN−/−) mice, as described previously (20). Control samples were obtained from brains of double-floxed mice injected with Cre-only retrovirus (no PDGF).
Cell lines and culturing conditions
Mouse GBM cell lines, derived from brain tumors of double-floxed (PTEN−/−p53−/−) or PTEN-floxed mice, were maintained in culture as described previously (20). The F98 rat glioma cell line was a kind offer from Dr Hélène Elleaume (European Synchrotron Radiation Facility, Grenoble, France), and the U87 human GBM cell line was obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were maintained in DMEM (Invitrogen, Carlsbad, CA, USA), supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco, Paisley, Scotland), 100 U/ml penicillin (Sigma), 100 µg/ml streptomycin (Sigma) and cultured at 37°C under a humidified atmosphere containing 5% CO2. Undifferentiated P19 embryonal carcinoma cell line was a kind gift from Dr Richard Cerione (Cornell University, NY, USA) and was maintained in α-MEM (Gibco), and supplements and growth conditions were similar to those used for glioma cells. Primary rat cortical astrocyte cultures were prepared from the cerebral cortices of newborn pups according to established protocols (56).
For all the experiments, cell plating densities are indicated in Supplementary Material.
Lentiviral production and cellular transdution
Lentiviruses encoding the anti-miR-21 shRNA and GFP or a control shGFP were produced in 293T cells with a four-plasmid system, as previously described (57,58). The lentiviral particles were produced and resuspended in phosphate-buffered saline (PBS) containing 1% bovine serum albumin. The viral particle content of batches was determined by assaying HIV-1 p24 antigen (RETROtek, Gentaur, Paris, France). The stocks were stored at −80°C until use.
For the lentiviral transduction of U87 cells, cells were plated onto six-well plates at a final concentration of 1.6 × 105 cells/well. Twenty-four hours after plating, 10 ng of virus coding for either anti-miR-21 shRNA or control shGFP were added per 1 × 105 cells, and 8 μg of polybrene (hexadimethrine bromide) were also added to each well, to increase the efficiency of infection. Cell culture medium was replaced 6 h after infection, and cells were further grown for 48 h, after which were plated onto 10 cm dishes. Infected cells were selected by growing cells in a culture medium containing 1 ug/ml of puromycin.
Lipoplex preparation and cell transfection
For transfection of anti-miR-21 oligonucleotides, lipoplexes were prepared with DLS liposomes, as described previously (26,59). DLS liposomes were prepared by mixing 1 mg of dioctadecylamidoglycylspermidine (DOGS) (Promega, Madison, WI) and 1 mg of dioleoyl phosphatidylethanolamine (DOPE) (Sigma, Munich) in 40 μl of 90% ethanol, followed by the addition of 360 μl of sterile H2O, and the mixture was further incubated for 30 min to allow liposome formation. The final lipid concentration was 5 mg/ml (2.5 mg of DOGS and 2.5 mg of DOPE). Lipoplexes were prepared by gently mixing 12 μg of oligonucleotides with 125 μg of lipid in a final volume of 125 μl, followed by incubation for 30 min at room temperature. DLS lipoplexes were prepared fresh for every experiment. Lipoplexes were added to cells, maintained in OptiMEM (Gibco), at a final concentration of 50 or 100 nm oligonucleotides/well. After a 4 h incubation period, cells were washed with PBS and further cultured in fresh DMEM medium for 48 h.
RNA extraction in tissue samples and cDNA synthesis
Total RNA from human and mouse tissue samples was extracted using the miRNAeasy Mini kit (Qiagen, Valencia, CA, USA) following the manufacturer's instructions. After RNA quantification, cDNA conversion was performed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) and miRNA-specific primers. For each sample, cDNA was produced from 10 ng of total RNA in an TC-PLUS SAT/02 thermocycler (VWR, Radnor, PA, USA), by applying the following protocol: 30 min at 16°C, 30 min at 42°C and 5 min at 85°C. The cDNA was further diluted 1:20 with RNase-free water prior to quantification by qPCR. RNA extraction for cultured cells and cDNA synthesis are described in the Supplementary Material.
qPCR quantification of miRNA expression in tissue samples
MiRNA quantification in human and mouse tissue samples was performed in an ABI Prism 7300 qPCR System (Applied Biosystems) using 96-well microtiter plates and the TaqMan® Universal PCR Master Mix (Applied Biosystems). The primers for the target miRNAs (miR-128, miR-21 and miR-221) and the reference RNA (U6snRNA) were also acquired from Applied Biosystems (60). A master mix was prepared for each primer set, containing a fixed volume of TaqMan master mix and the appropriate amount of each primer to yield a final concentration of 150 nm. For each reaction, performed in duplicate, 17.67 μl of master mix were added to 1.33 μl of template cDNA. The reaction conditions consisted of enzyme activation at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s (denaturation) and 60 s at 60°C (annealing and elongation). Threshold values for threshold cycle determination (Ct) were generated automatically by the SDS Optical System software. Relative miRNA levels were determined following the Pfaffl method for relative miRNA quantification in the presence of target and reference genes with different amplification efficiencies (61). The amplification efficiency for each target or reference gene was determined according to the formula: E = 10(−1/S), where S is the slope of the standard curve obtained for each gene.
MiRNA and mRNA quantification in cultured cells is described in the Supplementary Material.
Western blot analysis
Total protein extracts were prepared from cultured U87 cells, homogenized at 4°C in radioimmunoprecipitation assay lysis buffer (50 mm Tris pH 8.0, 150 mm NaCl, 50 mm EDTA, 0.5% sodium deoxycholate, 1% Triton X-100) containing a protease inhibitor cocktail (Sigma), 2 mm dithiothreitol and 0.1 mm phenylmethylsulfonyl fluoride. The concentration of protein lysates was determined using the Bio-Rad Dc protein assay (Bio-Rad), and 25 μg of total protein were resuspended in loading buffer (20% glycerol, 10% SDS, 0.1% bromophenol blue), incubated for 5 min at 95°C and loaded onto a 10% polyacrylamide gel for electrophoretic separation. After electrophoresis, the proteins were blotted onto a polyvinylidene fluoride (PVDF) membrane, blocked in 5% non-fat milk for 1 h, incubated overnight at 4°C with an anti-PTEN (#9552, Cell Signaling; 1:1000), anti-PDCD4 (clone D29C6, Cell signaling; 1:1000), anti-p53 (clone Pab240, Millipore; 1:500 and clone 1C12, Cell signaling; 1:1000) and anti-p21 (Ab7960, Abcam; 1:200) antibody and with the appropriate alkaline phosphatase labeled-secondary antibody (1:20 000) (Amersham, Uppsala, Sweden) for 2 h at room temperature. Equal protein loading was verified by reprobing the membrane with an anti-α-tubulin antibody (1:10 000) (Sigma) and with the same secondary antibody. After antibody incubation, the blots were washed several times with TBS-T (Bio-Rad), incubated with the enzyme substrate ECF (Amersham Biosciences, UK) for 5 min at room temperature and then subjected to fluorescence detection at 570 nm using a VersaDoc Imaging System Model 3000 (Bio-Rad). The analysis of band intensity was performed using the Quantity One software (Bio-Rad).
NF-kB activation analysis
To detect NF-kB activation, the nuclear translocation of the regulatory subunit p65/RelA was evaluated by western blotting. The protocol for extraction of nuclear and cytoplasmic fractions, described in Supplementary Material, was adapted from the protocol described by Ferreira and colleagues (62). Ten micrograms of nuclear protein were separated on a 10% polyacrylamide gel. After electrophoresis, the proteins were blotted onto a PVDF membrane, blocked in 5% non-fat milk for 1 h, incubated overnight at 4°C with an anti-N (#9552, Cell Signaling; 1:1000), anti-PDCD4 (clone D29C6, Cell signaling; 1:1000), anti-p53 (clone Pab240, Millipore; 1:500 and clone 1C12, Cell signaling; 1:1000) and anti-p21 (Ab7960, Abcam; 1:200) antibody and with the appropriate alkaline phosphatase labeled-secondary antibody (1:20 000) (Amersham, Uppsala, Sweden) for 2 h at room temperature.
FISH in tissue sections
FISH was performed in human and mouse FFPE tissue sections as described by Pena and colleagues (63), with a few modifications. The detailed protocol is supplied in the Supplementary Material.
Cell viability
Cell viability was evaluated by a modified Alamar Blue assay (64). Briefly, 10% (v/v) resazurin dye in complete DMEM medium was added to each well, and cells were incubated at 37°C until the development of a pink coloration. Two hundred microliters of supernatant were collected from each well, transferred to clear 96-well plates, and the absorbance at 570 (reduced form) and 600 nm (oxidized form) was measured in a microplate reader (SpectraMax Plus 384, Molecular Devices). Cell viability was calculated as percentage of control cells using the equation: (A570−A600) of treated cells × 100/(A570–A600) of control cells.
Apoptosis assay
Caspase-3/7 activity was assessed using the SensoLyte homogenous AMC caspase-3/7 assay (AnaSpec, San Jose, CA, USA). Briefly, 48 h after lipoplex-mediated oligonucleotide transfection, 24 h after temozolomide incubation or immediately after exposure to sunitinib, cells were collected and lyzed, according to the manufacter's instructions. Cell supernatant and caspase substrate (Ac-DEVD-AMC) were mixed, according to the manufacturer's recommendation, and further incubated in a black 96-well plate for 40 min at room temperature (with shaking). The production of the AMC fluorophore, released as a result of caspase action on the substrate, was measured for a period of 8 h, using a microplate reader (SpectraMax Plus 384, Molecular Devices) at excitation/emission of 354/442 nm. Results, presented as relative fluorescence units to control untreated cells, were normalized for the number of cells in each condition.
Confocal microscopy and flow cytometry studies
To assess the extent of cellular internalization of the DLS-based lipoplexes, confocal microscopy and flow cytometry studies in proliferating cells were performed. The detailed protocols are described in the Supplementary Material.
Analysis of the TCGA GBM data
The TCGA miRNA expression data were obtained from the public access Data Portal (http://tcga-data.nci.nih.gov/tcga/) through the Data Browser tool. Expression values were obtained in a logarithmic scale (log 2 tumor/normal ratio) and further converted into tumor/normal ratio (a threshold value of one was used to identify dysregulated miRNAs in tumor samples). Tumor IDs and derived data values are given in Supplementary Material, Table S1. Multiple comparison analysis was performed using the commercially available Microsoft Excel and P-values were adjusted by controlling the FDR (65). Changes were considered significant (P < 0.05), if the FDR was smaller than 0.05 (Supplementary Material, Table S2).
Statistical analysis
All data are presented as means ± standard deviation of at least three independent experiments, each performed in triplicate, unless stated otherwise. One-way analysis of variance combined with the Tukey posthoc test was used for multiple comparisons in cell culture experiments (unless stated otherwise) and considered significant when P < 0.05. Statistical differences are presented at probability levels of P < 0.05, P < 0.01 and P < 0.001. Calculations were performed with standard statistical software (Prism 5, GraphPad, San Diego, CA, USA).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Portuguese Foundation for Science and Technology. P.M.C. is a recipient of a fellowship from the Portuguese Foundation for Science and Technology (SFRH/BD/45902/2008). Work performed at Columbia University was supported by a grant from the National Institute of Health/National Institute of Neurological Disorders and Stroke (1R01NS066955-01 to P.C.).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Tao Su for the assistance in the RNA extraction from human GBM samples, Dr Adam Sonabend for the helpful advices on the statistical analysis of the TCGA data and Dr Orlando Gil for technical support (all from Columbia University, NY, USA), Dr Luisa Cortes and Dr Isabel Nunes (Center for Neuroscience and Cell Biology) for the assistance with the confocal microscopy imaging and flow cytometry, respectively and Dr Cristina Rego, Dr João Malva and Dr Cláudia Cavadas (Center for Neuroscience and Cell Biology) for supplying some of the antibodies used for western blot detection.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J., Belanger K., Brandes A.A., Marosi C., Bogdahn U., et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Khasraw M., Lassman A.B. Advances in the treatment of malignant gliomas. Curr. Oncol. Rep. 2010;12:26–33. doi: 10.1007/s11912-009-0077-4. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R., Hegi M.E., Mason W.P., van den Bent M.J., Taphoorn M.J., Janzer R.C., Ludwin S.K., Allgeier A., Fisher B., Belanger K., et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 4.Bartel D.P., Chen C.Z. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 5.Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Esquela-Kerscher A., Slack F.J. Oncomirs–microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 8.Calin G.A., Ferracin M., Cimmino A., Di Leva G., Shimizu M., Wojcik S.E., Iorio M.V., Visone R., Sever N.I., Fabbri M., et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 9.Volinia S., Calin G.A., Liu C.G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciafre S.A., Galardi S., Mangiola A., Ferracin M., Liu C.G., Sabatino G., Negrini M., Maira G., Croce C.M., Farace M.G. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 11.Hank N.C., Shapiro J.R., Scheck A.C. Over-representation of specific regions of chromosome 22 in cells from human glioma correlate with resistance to 1,3-bis(2-chloroethyl)-1-nitrosourea. BMC Cancer. 2006;6:2. doi: 10.1186/1471-2407-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohgaki H., Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am. J. Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lena A., Rechichi M., Salvetti A., Bartoli B., Vecchio D., Scarcelli V., Amoroso R., Benvenuti L., Gagliardi R., Gremigni V., et al. Drugs targeting the mitochondrial pore act as cytotoxic and cytostatic agents in temozolomide-resistant glioma cells. J. Transl. Med. 2009;7:13. doi: 10.1186/1479-5876-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prados M.D., Lamborn K.R., Chang S., Burton E., Butowski N., Malec M., Kapadia A., Rabbitt J., Page M.S., Fedoroff A., et al. Phase 1 study of erlotinib HCl alone and combined with temozolomide in patients with stable or recurrent malignant glioma. Neuro. Oncol. 2006;8:67–78. doi: 10.1215/S1522851705000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawler S., Chiocca E.A. Emerging functions of microRNAs in glioblastoma. J. Neurooncol. 2009;92:297–306. doi: 10.1007/s11060-009-9843-2. [DOI] [PubMed] [Google Scholar]
- 17.Silber J., Lim D.A., Petritsch C., Persson A.I., Maunakea A.K., Yu M., Vandenberg S.R., Ginzinger D.G., James C.D., Costello J.F., et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godlewski J., Nowicki M.O., Bronisz A., Williams S., Otsuki A., Nuovo G., Raychaudhury A., Newton H.B., Chiocca E.A., Lawler S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 19.Morris P.G., Abrey L.E. Novel targeted agents for platelet-derived growth factor receptor and c-KIT in malignant gliomas. Target Oncol. 2010;5:193–200. doi: 10.1007/s11523-010-0160-7. [DOI] [PubMed] [Google Scholar]
- 20.Lei L., Sonabend A.M., Guarnieri P., Soderquist C., Ludwig T., Rosenfeld S., Bruce J.N., Canoll P. Glioblastoma models reveal the connection between adult glial progenitors and the proneural phenotype. PLoS One. 2011;6:e20041. doi: 10.1371/journal.pone.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan J.A., Krichevsky A.M., Kosik K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 22.Conti A., Aguennouz M., La Torre D., Tomasello C., Cardali S., Angileri F.F., Maio F., Cama A., Germano A., Vita G., et al. miR-21 and 221 upregulation and miR-181b downregulation in human grade II-IV astrocytic tumors. J. Neurooncol. 2009;93:325–332. doi: 10.1007/s11060-009-9797-4. [DOI] [PubMed] [Google Scholar]
- 23.Network C.G.A.R. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selcuklu S.D., Donoghue M.T., Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem. Soc. Trans. 2009;37:918–925. doi: 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- 25.Zhu S., Wu H., Wu F., Nie D., Sheng S., Mo Y.Y. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 26.Trabulo S., Resina S., Simoes S., Lebleu B., Pedroso de Lima M.C. A non-covalent strategy combining cationic lipids and CPPs to enhance the delivery of splice correcting oligonucleotides. J. Control Release. 2010;145:149–158. doi: 10.1016/j.jconrel.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 27.Lu Z., Liu M., Stribinskis V., Klinge C.M., Ramos K.S., Colburn N.H., Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- 28.Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S.T., Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papagiannakopoulos T., Shapiro A., Kosik K.S. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 30.Ak P., Levine A.J. p53 and NF-kappaB: different strategies for responding to stress lead to a functional antagonism. FASEB J. 2010;24:3643–3652. doi: 10.1096/fj.10-160549. [DOI] [PubMed] [Google Scholar]
- 31.Stiewe T. The p53 family in differentiation and tumorigenesis. Nat. Rev. Cancer. 2007;7:165–168. doi: 10.1038/nrc2072. [DOI] [PubMed] [Google Scholar]
- 32.Kim S., Han J., Lee S.K., Hur S.M., Koo M., Cho D.H., Bae S.Y., Choi M.Y., Shin I., Yang J.H., et al. Pifithrin-alpha, an inhibitor of p53 transactivation, up-regulates COX-2 expression through an MAPK-dependent pathway. Pharmacology. 2010;86:313–319. doi: 10.1159/000321327. [DOI] [PubMed] [Google Scholar]
- 33.Murphy P.J., Galigniana M.D., Morishima Y., Harrell J.M., Kwok R.P., Ljungman M., Pratt W.B. Pifithrin-alpha inhibits p53 signaling after interaction of the tumor suppressor protein with hsp90 and its nuclear translocation. J. Biol. Chem. 2004;279:30195–30201. doi: 10.1074/jbc.M403539200. [DOI] [PubMed] [Google Scholar]
- 34.Fan S., Qi M., Yu Y., Li L., Yao G., Tashiro S., Onodera S., Ikejima T. P53 activation plays a crucial role in silibinin induced ROS generation via PUMA and JNK. Free Radic. Res. 2012;46:310–319. doi: 10.3109/10715762.2012.655244. [DOI] [PubMed] [Google Scholar]
- 35.Strosznajder R.P., Jesko H., Banasik M., Tanaka S. Effects of p53 inhibitor on survival and death of cells subjected to oxidative stress. J. Physiol. Pharmacol. 2005;56(Suppl. 4):215–221. [PubMed] [Google Scholar]
- 36.Rocha S., Campbell K.J., Roche K.C., Perkins N.D. The p53-inhibitor pifithrin-alpha inhibits firefly luciferase activity in vivo and in vitro. BMC Mol. Biol. 2003;4:9. doi: 10.1186/1471-2199-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tergaonkar V., Perkins N.D. p53 and NF-kappaB crosstalk: IKKalpha tips the balance. Mol. Cell. 2007;26:158–159. doi: 10.1016/j.molcel.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Ben-Neriah Y., Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat. Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 39.Gabriely G., Wurdinger T., Kesari S., Esau C.C., Burchard J., Linsley P.S., Krichevsky A.M. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol. Cell Biol. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heinzelmann J., Henning B., Sanjmyatav J., Posorski N., Steiner T., Wunderlich H., Gajda M.R., Junker K. Specific miRNA signatures are associated with metastasis and poor prognosis in clear cell renal cell carcinoma. World J. Urol. 2011;29:367–373. doi: 10.1007/s00345-010-0633-4. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H., Li Y., Lai M. The microRNA network and tumor metastasis. Oncogene. 2010;29:937–948. doi: 10.1038/onc.2009.406. [DOI] [PubMed] [Google Scholar]
- 42.Asangani I.A., Rasheed S.A., Nikolova D.A., Leupold J.H., Colburn N.H., Post S., Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 43.Zhu S., Si M.L., Wu H., Mo Y.Y. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J. Biol. Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 44.Gaur A.B., Holbeck S.L., Colburn N.H., Israel M.A. Downregulation of Pdcd4 by mir-21 facilitates glioblastoma proliferation in vivo. Neuro. Oncol. 2011;13:580–590. doi: 10.1093/neuonc/nor033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lino M.M., Merlo A. PI3Kinase signaling in glioblastoma. J. Neurooncol. 2010;103:417–427. doi: 10.1007/s11060-010-0442-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou X., Ren Y., Moore L., Mei M., You Y., Xu P., Wang B., Wang G., Jia Z., Pu P., et al. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab. Invest. 2010;90:144–155. doi: 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]
- 47.Gao F., Zhang P., Zhou C., Li J., Wang Q., Zhu F., Ma C., Sun W., Zhang L. Frequent loss of PDCD4 expression in human glioma: possible role in the tumorigenesis of glioma. Oncol. Rep. 2007;17:123–128. [PubMed] [Google Scholar]
- 48.Yang H.S., Jansen A.P., Komar A.A., Zheng X., Merrick W.C., Costes S., Lockett S.J., Sonenberg N., Colburn N.H. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol. Cell. Biol. 2003;23:26–37. doi: 10.1128/MCB.23.1.26-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang H.S., Jansen A.P., Nair R., Shibahara K., Verma A.K., Cmarik J.L., Colburn N.H. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene. 2001;20:669–676. doi: 10.1038/sj.onc.1204137. [DOI] [PubMed] [Google Scholar]
- 50.Goke R., Barth P., Schmidt A., Samans B., Lankat-Buttgereit B. Programmed cell death protein 4 suppresses CDK1/cdc2 via induction of p21(Waf1/Cip1) Am. J. Physiol. Cell Physiol. 2004;287:C1541–C1546. doi: 10.1152/ajpcell.00025.2004. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y., Liu W., Chao T., Zhang Y., Yan X., Gong Y., Qiang B., Yuan J., Sun M., Peng X. MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett. 2008;272:197–205. doi: 10.1016/j.canlet.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 52.Corsten M.F., Miranda R., Kasmieh R., Krichevsky A.M., Weissleder R., Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 53.Ren Y., Kang C.S., Yuan X.B., Zhou X., Xu P., Han L., Wang G.X., Jia Z., Zhong Y., Yu S., et al. Co-delivery of as-miR-21 and 5-FU by poly(amidoamine) dendrimer attenuates human glioma cell growth in vitro. J. Biomater. Sci. Polym. Ed. 2010;21:303–314. doi: 10.1163/156856209X415828. [DOI] [PubMed] [Google Scholar]
- 54.Ren Y., Zhou X., Mei M., Yuan X.B., Han L., Wang G.X., Jia Z.F., Xu P., Pu P.Y., Kang C.S. MicroRNA-21 inhibitor sensitizes human glioblastoma cells U251 (PTEN-mutant) and LN229 (PTEN-wild type) to taxol. BMC Cancer. 2010;10:27. doi: 10.1186/1471-2407-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neyns B., Sadones J., Chaskis C., Dujardin M., Everaert H., Lv S., Duerinck J., Tynninen O., Nupponen N., Michotte A., et al. Phase II study of sunitinib malate in patients with recurrent high-grade glioma. J. Neurooncol. 2011;103:491–501. doi: 10.1007/s11060-010-0402-7. [DOI] [PubMed] [Google Scholar]
- 56.Banker G., Goslin K. Culturing Nerve Cells. 2nd edn. Cambridge, MA: The MIT Press; 1998. pp. 499–542. [Google Scholar]
- 57.de Almeida L.P., Ross C.A., Zala D., Aebischer P., Deglon N. Lentiviral-mediated delivery of mutant huntingtin in the striatum of rats induces a selective neuropathology modulated by polyglutamine repeat size, huntingtin expression levels, and protein length. J. Neurosci. 2002;22:3473–3483. doi: 10.1523/JNEUROSCI.22-09-03473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alves S., Nascimento-Ferreira I., Dufour N., Hassig R., Auregan G., Nobrega C., Brouillet E., Hantraye P., Pedroso de Lima M.C., Deglon N., et al. Silencing ataxin-3 mitigates degeneration in a rat model of Machado-Joseph disease: no role for wild-type ataxin-3? Hum. Mol. Genet. 2010;19:2380–2394. doi: 10.1093/hmg/ddq111. [DOI] [PubMed] [Google Scholar]
- 59.Cardoso A.L., Guedes J.R., Pereira de Almeida L., Pedroso de Lima M.C. miR-155 modulates microglia-mediated immune response by down-regulating SOCS-1 and promoting cytokine and nitric oxide production. Immunology. 2012;135:73–88. doi: 10.1111/j.1365-2567.2011.03514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee H., Choi H.J., Kang C.S., Lee H.J., Lee W.S., Park C.S. Expression of miRNAs and PTEN in endometrial specimens ranging from histologically normal to hyperplasia and endometrial adenocarcinoma. Mod. Pathol. 2012;25:1508–1515. doi: 10.1038/modpathol.2012.111. [DOI] [PubMed] [Google Scholar]
- 61.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferreira R., Xapelli S., Santos T., Silva A.P., Cristovao A., Cortes L., Malva J.O. Neuropeptide Y modulation of interleukin-1{beta} (IL-1{beta})-induced nitric oxide production in microglia. J. Biol. Chem. 2010;285:41921–41934. doi: 10.1074/jbc.M110.164020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pena J.T., Sohn-Lee C., Rouhanifard S.H., Ludwig J., Hafner M., Mihailovic A., Lim C., Holoch D., Berninger P., Zavolan M., et al. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat. Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O'Brien J., Wilson I., Orton T., Pognan F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000;267:5421–5426. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- 65.Benjamini Y., Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.