Abstract
The concept of one-protein–multiple-function, i.e. moonlighting proteins, is an ever-expanding paradigm. We obtained compelling evidence that an array of ‘cytoplasmic’ metabolic enzymes can enter the nuclei to carry out moonlighting transcription functions; this phenomenon is conserved from Drosophila to humans. Of particular interest are the classical glycolytic enzymes GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and LDH (lactate dehydrogenase), which utilize NAD(H) as coenzymes and not only moonlight (in their nuclear forms) to regulate the transcription of S-phase-specific histone genes, but also act as metabolic/redox sensors that link histone gene switching to DNA replication and S-phase progression.
Keywords: chromosome, enzyme, GAPDH, histone 2B, S-phase
Abbreviations: ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia mutated- and Rad3-related; awd, abnormal wing disc; CBP, CREB (cAMP-response-element-binding protein)-binding protein; CDK, cyclin-dependent kinase; dm, Drosophila melanogaster; DSB, double-strand break; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; H2B, histone 2B; HAT, histone acetyl transferase; HDAC, histone deacetylase; HU, hydroxyurea; LDH, lactate dehydrogenase; MMC, mammalian metabolic cycle; nm23, non-metastasis 23; NPAT, nuclear protein, ataxia-telangiectasia locus; OCA-S, Oct-1 co-activator in the S-phase; PIKK, phosphoinositide 3-kinase-related kinase; RNAi, RNA interference; SLBP, stem–loop-binding protein; Tip60, Tat (transactivator of transcription)-interactive protein 60 kDa; YMC, yeast metabolic cycle
INTRODUCTION
There are two classes of histones, namely canonical histones and histone variants (known as replacement histones); canonical histones are encoded by genes that are expressed in a DNA-replication-dependent and S-phase-specific manner, whereas the replacement histones are encoded by genes that are constitutively expressed (reviewed in [1]). Canonical histone genes encode core histones H2A, H2B (histone 2B), H3, H4 and linker histone H1, and are clustered and multi-copied; in mammalian cells, there are two such clusters: one cluster (human chromosome 6, mouse chromosome 13) comprises ~80% of the genes and the other cluster (human chromosome 1, mouse chromosome 3) contains the remaining genes (reviewed in [2]). Despite the fact that their expression is regulated by diverse promoter regulatory elements and transcription factors, vertebrate canonical histone genes, those encoding the core histones in particular, are found to be expressed in a highly coordinated fashion (reviewed in [3]); these genes are colocalized in nuclear domains known as Cajal bodies (reviewed in [4]).
Division of a mother cell to two daughter cells occurs after a series of orchestrated events that constitute a cell cycle. The S-phase is marked by DNA replication that allows two daughter cells, upon mitosis, to each obtain a full complement of the mother's DNA. Newly synthesized (acidic) DNA must immediately interact with (basic) histone proteins to assemble the chromatin, thus the maintenance of genomic stability dictates that S-phase DNA replication ought to be accompanied by availability of histones such that the histone expression predominantly occurs in an S-phase- and DNA-replication-dependent manner. This links histone expression to S-phase progression. Because the regulation of histone expression is at least in part at the transcriptional level (reviewed in [2]), histone genes have become models for studying key mechanisms of S-phase- and gene-specific transcriptional regulation.
How is histone biosynthesis initiated? To understand this we must first know how a cell enters S-phase. Principal mechanisms have been revealed in great detail in the last two decades using yeast as a model system (reviewed in [5]); however, a wealth of information has also been accumulated regarding mammalian cell-cycle progression, in which the cyclin E/CDK (cyclin-dependent kinase) 2 signalling controls S-phase entry (reviewed in [4,6]). Cyclin E/CDK2 activation involves a cascade that includes phosphorylation of Rb by cyclin D/CDK4 in response to cell growth signal(s), followed by activation of Rb/E2F-regulated cyclin E promoter. Rb, along with the CDK inhibitor p27, is also among the substrates of cyclin E/CDK2, which inactivates them by phosphorylation – a committing step for S-phase entry. Importantly, this step is in concert with the initiation of DNA biosynthesis, presumably as a result of the activated cyclin E/CDK2 complex that in turn phosphorylates and inactivates certain inhibitor(s) of DNA biosynthesis [7].
Cyclin E/CDK2 also functions upstream, and is a regulator, of histone biosynthesis (reviewed in [4]); however, none of the earlier known cyclin E/CDK2 substrates serves downstream to promote histone biosynthesis. For example, although E2F-binding elements are present in some histone gene promoters and the tumour suppressor Rb as a transcription repressor is involved in E2F-mediated transcriptional regulation, vertebrate histone genes are largely regulated by transcriptional activation rather than de-repression mechanisms (reviewed in [3]). Moreover, histone genes are still expressed with a proper temporal pattern in cells without Rb. Therefore other substrates of cyclin E/CDK2 were being sought as candidates that promote histone expression and, as a result, NPAT (nuclear protein, ataxia-telangiectasia locus) was identified [4,8–10]; if cells were made NPAT-deficient by RNAi (RNA interference) [11] or conditional somatic knockout [12], the expression of all histone subtype genes was down-regulated with S-phase concomitantly stalked. These studies thus clearly establish NPAT as a key global regulator for histone expression and S-phase progression.
NPAT overexpression activates promoters of all tested core histone genes [10]; however, how NPAT regulates histone gene transcription and whether the regulation is via a direct mechanism remain unanswered. In the present paper, we first review certain breakthroughs made over the last decade by us and others in the field, and then offer perspectives based on the most recent discoveries.
OCA-S: LINKING NPAT TO H2B EXPRESSION
In 2003, we published a paper [13] that describes the isolation/characterization of OCA-S, a transcription cofactor complex that functions downstream of NPAT to directly stimulate the transcription of a human H2B gene in the S-phase. This discovery led us to believe that we had discovered an essential link between a global cell-cycle regulator and subtype-specific histone transcription machinery. The significance along with historic reasons of this discovery is described below.
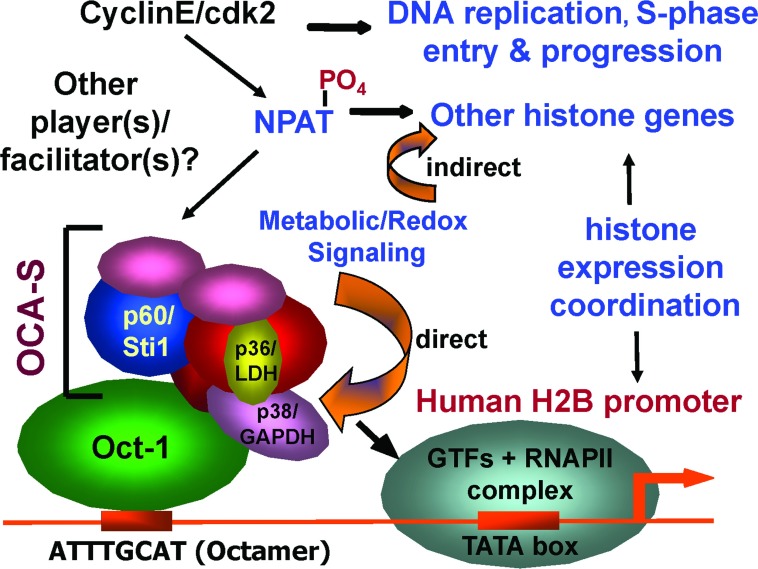
Activation of the H2B promoter requires an octamer element (5′-ATTTGCAT-3′) and its bound activator Oct-1 [14,15], but Oct-1 is not directly responsible for S-phase-specific H2B transcription [16]. This prompted us to search for an Oct-1 cofactor that abets its role, leading to the isolation of OCA-S (Oct-1 coactivator in the S-phase) [13]. OCA-S stimulates the H2B promoter activation in a reconstituted transcription system and copurifies with seven polypeptides. That these subunits interact to form a complex and that the putative complex interacts with Oct-1 in vitro [13] strongly suggest their relevance to the OCA-S activity and a complex formation in living cells. OCA-S also interacts with NPAT [13], a global regulator for histone expression. Hence, despite the suggestion by Zhao et al. [10] that NPAT, along with cyclin E/CDK2, is directly involved in the regulation of histone expression, our data suggest that NPAT acts upstream of histone transcription machineries, probably by facilitating promoter recruitment and/or activation of transcriptional (co)factors, e.g. OCA-S in the case of the H2B promoter (Figure 1).
Figure 1. OCA-S links Oct-1 and the H2B promoter to NPAT, and confers redox sensitivity to the target gene.
This Figure conveys three pieces of information: (i) activation of the cyclin E/CDK2 complex dictates S-phase entry, DNA replication and S-phase progression; (ii) NPAT, in conjunction with other (potential) partner(s)/facilitator(s), is downstream of the cyclin E/CDK2 signalling and upstream of transcription machineries of histone genes; (iii) the metabolic/redox signalling directly controls the transcription of the H2B gene via the redox sensors GAPDH and LDH in the OCA-S complex and indirectly regulates the expression of other histone genes, which ensures the coordinated and stoichiometric expression of all histone subtypes. RNAPII, RNA polymerase II.
How do NPAT and OCA-S meet? The answer may provide a clue regarding signalling pathway(s) between the global histone expression regulators and subtype-specific transcriptional machineries. Coimmunoprecipitation assays had clearly demonstrated an Oct-1–OCA-S–NPAT interaction [13], although whether the interaction is direct or via mediating protein(s) is yet to be known. At a minimum, however, available data suggest that NPAT is part of a larger and highly dynamic complex with OCA-S and Oct-1 in cells and that NPAT is capable of activating the OCA-S complex specifically in the S-phase [13]. Once activated, however, OCA-S might function independently of NPAT in gene coactivation, in line with the fact that purified OCA-S did not contain NPAT [13]. We thus speculate that in the S-phase the OCA-S activity or the assembly of the OCA-S complex is promoted by NPAT – possibly in conjunction with other mediating proteins – and that we can explore the mechanism(s) by which the OCA-S activity is activated by NPAT on receiving the cyclin E/CDK2 signalling (Figure 1); likewise, transcription activation of other histone subtype genes may also involve protein complexes, which function similarly with the large and highly dynamic NPAT–OCA-S–Oct-1 complex in receiving and forwarding cyclin E/CDK2 signalling to respective target promoters.
OCA-S: LINKING H2B EXPRESSION TO CELLULAR METABOLIC STATE/REDOX STATUS
Surprisingly, a key component of the OCA-S complex represents a nuclear form of GAPDH (we called it p38/glyceraldehyde-3-phosphate dehydrogenase [13]). The key roles for p38/GAPDH were established by multiple compelling lines of evidence, including: (i) a direct interaction between p38/GAPDH and Oct-1 that provides the anchor point for an OCA-S recruitment to the H2B promoter; (ii) the indispensability of p38/GAPDH for H2B transcription both in vitro and in vivo; and (iii) the occupancy of the H2B promoter in concert with transcriptional activation by p38/GAPDH (OCA-S) in an S-phase-dependent fashion [13]. Thus, the S-phase-specific H2B promoter activation is mediated by OCA-S, with p38/GAPDH being a central component.
The significance of the above finding is consistent with the ‘moonlighting proteins’ concept first proposed by Jeffery [17], in which she listed ~30 examples of multitasking proteins that along with a number of later identified proteins, ours included, support the ‘one-protein–multiple-function paradigm’. Probably of more significance, we found that the binding to Oct-1 and OCA-S function can be modulated by NAD(H) (stimulated by NAD+ but inhibited by NADH in initial testing [13]). These revelations not only confirm a novel moonlighting function for a glycolytic enzyme, but also suggest that coenzymes may play a significant role in modulating histone transcription. Taken together, these studies link the components of a gene-specific histone transcription machinery (Oct-1, OCA-S) to the global histone expression regulators (cyclin E/CDK2, NPAT), and possibly also to cellular metabolic state (redox status), and set a stage for exploring underlying mechanisms. In addition, given that silencing the p38/GAPDH expression by RNAi led to a blockage of cell cycle progression into S-phase and eventual inhibition of the histone H4 expression [13], our studies provide an important model to study the basis for coordinated histone expression and its coupling with DNA replication.
A GLOBAL LINKAGE BETWEEN CELLULAR REDOX AND OVERALL HISTONE EXPRESSION
In more detailed studies towards understanding redox-sensitive H2B transcription, we used a wider range of NAD(H) titrations and found that the H2B transcription in vitro responded in a biphasic fashion to NAD+ dosages or NAD+/NADH ratios [18]. Given that intracellular NAD+/NADH ratios determine the NAD(H) redox status, we figured that the H2B expression was confined to a proper NAD(H) redox status both in vivo and in vitro; indeed, perturbing the NAD(H) redox status in living cells also significantly reduced H2B expression [18,19].
Another glycolytic enzyme, LDH (we call it p36/lactate dehydrogenase [18]), is also an essential OCA-S component, and when in OCA-S can exercise the enzyme activity to reverse in vitro inhibition of H2B transcription by NADH via converting it into NAD+ in the presence of substrate pyruvate [18]. In the cytoplasm, relative enzyme activities of p38/GAPDH and p36/LDH contribute to sustaining a proper NAD+/NADH ratio, i.e. the redox status; a tantalizing possibility is that p38/GAPDH and p36/LDH are not only able to exercise moonlighting transcriptional functions in the nuclei, but also can sense the (local) redox status to modulate histone expression. We proposed that a proper redox status might allow proper configurations of the two proteins, which optimizes an OCA-S–Oct-1 interaction, a proper assembly of the OCA-S complex and capacities of the proteins as transcriptional regulators [18] (also Figure 2). This redox-mediated optimization process is most probably cell cycle regulated, given that the expression of canonical histone genes occurs only in the S-phase (also see below).
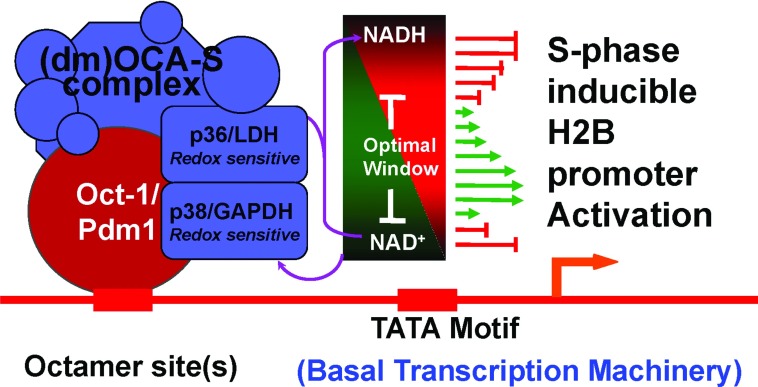
Figure 2. Conserved H2B gene expression regulation, redox-mediated modulation included, from Drosophila to humans.
Transcription from the human H2B gene promoter is stimulated by Oct-1 in conjunction with coactivator OCA-S; transcription from the fly H2B promoter is stimulated by Pdm-1 (a fly counterpart of human Oct-1) along with dmOCA-S (a dm version of the human OCA-S complex). In the nuclei of both cases, the transcriptional activation from the target genes is mediated by the essential coactivator components p38/GAPDH and p36/LDH, which also sense a local redox status to modulate histone expression. A proper local redox status in the nuclei might optimize the moonlighting functions, e.g. allowing proper configurations of the proteins by virtue of their capacities to associate with the coenzymes NAD(H). Note that while the expression of the human core histone genes is regulated by distinct transcriptional (co)activators, the expression of the Drosophila core histone genes is co-ordinately regulated in a concerted fashion by the Pdm-1/dmOCA-S regulatory module. See the text for further details.
In vitro, a redox-sensitivity is probably restricted to the H2B promoter, because the transcription from the H4 promoter was not subject to redox modulation [19]; however, perturbing the NAD(H) redox in cells significantly reduced not only H2B expression but also the expression of other core histone genes in a concerted fashion [19]. On redox perturbation, p38/GAPDH (OCA-S) was disengaged from the H2B promoter [18,19], thus eliminating an OCA-S function. Eliminating the OCA-S function by another method, i.e. silencing the p38/GAPDH expression, however, exhibited an H2B expression defect that was manifested prior to that of an H4 gene [13]. Taken together, these observations suggest the existence of a redox signalling system that has broader effects on S-phase progression as seen by the concertedly repressed expression of all core histone genes on redox perturbation.
OCA-S occupies the H2B promoter but not other histone promoters, and it is unlikely that non-H2B promoters employ other redox-sensitive transcriptional components [13,18,19]. Thus, the concerted histone expression defects upon redox perturbations [19] were most probably due to redox-sensitive machineries not directly regulating transcription of other core histone genes. Candidate machineries include those involved in regulating histone mRNA stability/maturation, which may feed back to regulate overall histone expression levels, or machineries involved in other S-phase event(s) such as DNA replication, which indirectly feed back to histone expression due to tight coupling of the DNA replication with histone expression (Figure 1).
AN MMC (MAMMALIAN METABOLIC CYCLE) REMINISCENT OF A YMC (YEAST METABOLIC CYCLE)
We proposed a redox signalling system that gates H2B expression, coordinates the expression of other histone genes and affects other redox-sensitive machineries to dictate S-phase progression; indeed, perturbing the redox eventually led to an activation of intra-S-phase checkpoint(s) to hamper S-phase progression [19].
Typified by the cell-cycle control through the cyclin/CDK signalling systems, a lot of signalling molecules exhibit oscillatory patterns along with pathways that they regulate, in either activities or levels. To explore whether the NAD+/NADH ratios might fluctuate in a cell cycle, with an optimal ratio(s) to gate or fine-tune histone expression during the S-phase progression, we used centrifugal elutriation to synchronize HeLa cells at G1-, S- and G2-phases, measured the intracellular NAD+/NADH ratios and found that G1- and G2-phase cells are more oxidative than the S-phase cells [19]. Recultured G1-phase cells exhibited lowered NAD+/NADH ratios when entering the G1/S border/S-phase, and synchronized S-phase cells as well as randomly growing cells were sensitive to reductive or oxidative redox perturbations with much reduced histone expression [19].
The fluctuating NAD+/NADH ratios in a mammalian cell cycle should reflect overall oscillatory intracellular metabolism, and was called MMC [19] that shares certain similarity with an earlier defined YMC [20]. In YMC, many biological/metabolic processes are compartmentalized in time, and YMC is characterized by oxidative, reductive/building and reductive/charging phases, which are in synchrony with the yeast cell cycle in that the S-phase overlaps with the most reductive stage of YMC (the early stage of the reductive/building phase) [20]. Given that the YMC is a redox cycle [20], the S-phase of a yeast cell cycle might be more reductive as compared with G1- or G2-phase, which is reminiscent of the MMC [19].
The logic behind the temporal segregation of metabolic processes in eukaryotic cells is clear [20]. A synchrony of S-phase with the most reductive stage of YMC may avoid oxidative damage to replicating DNA, thus providing genome protection. Indeed, in some yeast mutant stains, S-phase was shifted either left- or right-ward of the most reductive stage of YMC, and these strains accumulate a high level of spontaneous mutations [21]. Given a similarity between YMC and MMC, we envision that such a genome protection strategy exists in mammalian cells [19]. Thus the relatively reductive redox status in the S-phase of a mammalian cell cycle may constitute part of a redox signalling, which gates H2B transcription, coordinates concerted S-phase expression of diverse histone subtype genes via a yet-to-be explored mechanism, and ensures genome integrity [19].
CONSERVED HISTONE EXPRESSION PATHWAY(S) IN DROSOPHILA WITH A UNIQUE FEATURE
Of seven components of the OCA-S complex [13], the most prominent are p38/GAPDH and p36/LDH, and p18/nm23H1 and p20/nm23H2, which stand for human versions 1 and 2 of nm23 (non-metastasis 23) genes that are underexpressed in certain metastatic human cancer cells (reviewed by Hartsough and Steeg [22]); and proteins encoded by the nm23 genes possess (d)NDP kinase activities [23]. We found that the dm (Drosophila melanogaster) versions of the above proteins, namely dmGAPDH, dmLDH and awd (abnormal wing disc, a fly version of the nm23 proteins), were essential in conjunction with Pdm1 (a Drosophila version of Oct-1) for histone expression [24]. Thus, during the metazoan evolution, the signalling pathway from cyclin E/CDK2 to histone expression appears to be fundamentally conserved; however, there is a unique feature with fly histone expression in that all of the fly core histone genes have in their promoters multiple degenerate octamer elements, which provide the anchor points for Pdm-1 and the cognate coactivator, i.e. the dm version of OCA-S, dmOCA-S [24]. This is different from that of vertebrate histone genes, which contain SSRE (subtype-specific regulatory elements, reviewed in [3]) that are different and targeted by diverse transcription factors and/or cofactors. Hence, while in insects the coordinated expression of histone genes is realized in a more direct fashion by a common module [24], the expression of vertebrate histone genes is coordinated via a mechanism yet to be explored [13,18,19].
Given that both OCA-S and dmOCA-S contain classical enzymes and in the case of OCA-S the transcriptional activity on the H2B gene is modulated by coenzymes and/or other metabolites [13,18,19], the redox-modulated histone expression, in conjunction with metabolic cycles, may well be a conserved process during metazoan evolution and may provide the basis linking metabolism to cell-cycle gene switching (Figure 2).
FUNCTIONS OF OTHER OCA-S SUBUNITS
While we have not yet systematically analysed the functional relevance of each individual OCA-S subunit, preliminary studies may have already provided certain clues. For instance, the mammalian nm23 proteins have a counterpart (awd) in Drosophila known to be essential for histone expression [24], and are enzymes catalysing (d)NDP to (d)NTP conversions as (d)NDP kinases (reviewed in [25]). The nm23H1/nm23H2 proteins may function to regulate vertebrate histone expression. Given that S-phase may utilize a different NTP and dNTP homoeostasis, because the proper DNA replication requires a larger dNTPs pool for building blocks, the likelihood is that the (d)NDP kinase activities of these proteins are involved in regulating a proper NTP and dNTP homoeostasis, which might feed back to DNA replication and histone expression.
On the other hand, some other OCA-S component(s) or other independent components may be involved in linking NPAT-mediated cyclin E/CDK2 signalling to histone transcription machineries, functioning as the facilitator of this signalling pathway (Figure 1).
GENERAL TRANSCRIPTION COACTIVATOR(S) AND COREPRESSOR(S) AT THE CHROMATIN LEVEL
In eukaryotes, for transcription of a given gene to occur, the relevant transcription machinery must be recruited to the gene template DNA; however, the compact structure of chromatin restrains the accessibility of DNA to the transcription machinery. Chromatin can be modulated by two mechanisms to increase the accessibility of DNA templates to transcription machineries: alteration of nucleosome architecture by ATP-dependent chromatin remodelling complexes or covalent modifications of associated histones by diverse enzymes. The chromatin structure of a human H4 gene is subject to alteration in a cell cycle: a site sensitive to both DNase I and nuclease S1 was identified in the promoter region of this gene [26,27]. The length of this site is broader in S-phase and the level of nuclease S1 sensitivity is greatest in early S-phase [26]; in the promoter as well as coding region of this gene, a reversible disruption of nucleosomal organization was observed in early S-phase directly correlating with its active transcription [27].
We and others found that CBP [CREB (cAMP-response-element-binding protein)-binding protein], p300 (a transcriptional activator protein required to drive p53 expression) and Tip60 [Tat (transactivator of transcription)-interactive protein 60 kDa], the transcription coactivators possessing HAT (histone acetyl transferase) activity, are recruited to core histone gene promoters in an NPAT-dependent manner [28,29]. The association of CBP and p300 with histone promoters fluctuates through the cell cycle in a pattern similar to the cell cycle-regulated histone acetylation on histone promoters, which is directly correlated with RNA polymerase II recruitment and histone gene expression [28]. In line with these findings, down-regulation of CBP, p300 or Tip60 by RNAi diminishes histone H3/H4 acetylation on histone gene promoters and decreases histone gene transcription [28,29]. Histone acetylation contributes to relaxed chromatin structure in two ways: first, it reduces the positive charges of histone proteins, thus weakening the interactions between the basic histones and acidic DNA; secondly, the acetylated lysine residues in their N-terminal tails provide anchors for bromo-domain-containing chromatin remodelling complexes [30].
Histone acetylation on histone gene promoters is characterized to be dynamic and reversible. We demonstrated that SIRT1, a member of the NAD+-dependent HDAC (histone deacetylase) complexes, is associated with histone promoters [28]. Inhibition of SIRT1 HDAC activity with inhibitors or down-regulation of its expression with RNAi led to up-regulation of histone genes; conversely, cells treated with SIRT1 activator(s) exhibited repressed histone transcription, indicating that SIRT1 is a corepressor of histone genes. Recruitment of multiple transcriptional coactivators and corepressor(s) to histone gene promoters in a temporal fashion may contribute to rapid and efficient fine tuning of histone gene transcription in response to cell cycle signals and environmental cues. In particular, according to the concept of MMC [19], the relative NAD+ level may increase near the end of S-phase or the S/G2 border during which histone expression needs to slow down; the NAD+-dependent SIRT1 activity may correspondingly increase in order to participate in corepressing histone expression. This would link the cellular redox/metabolic state to histone expression at yet another level.
S-PHASE CHECK POINTS AND HISTONE EXPRESSION
During eukaryote evolution, molecular mechanisms were established that ensure accurate DNA replication and chromatin assembly. In the S-phase, DNA replication and histone biosynthesis are finely balanced to ensure genome integrity. Several checkpoints may be activated in the S-phase in response to certain genotoxic stresses, and can inhibit DNA replication and delay cell-cycle progression. In particular, the PIKK (phosphoinositide 3-kinase-related kinase) family members play a key role in S-phase checkpoints, of which ATR [ATM (ataxia telangiectasia mutated)- and Rad3-related] is activated by aberrant DNA structures induced by UV light or replication stress (see [31,32]). Downstream of ATR are Chk1 and Chk2, in which Chk1 functions in the activation of the homologous recombination repair machinery [33] and, in metazoans, in the activation of the origin firing and replisome integrity checkpoint [34,35]. DNA-PK, another PIKK family member, is activated by DSBs (double-strand breaks) caused by the collapse of stalled replication forks, and targets components of the DNA replication and repair machineries (reviewed in [36]; see also [37]). For stoichiometry (DNA compared with histones) considerations, S-phase checkpoint signals must be transmitted to histone biosynthesis machineries and vice versa (see below). For instance, the activation of G1 checkpoint in response to ionizing radiation in human cells results in inhibited histone gene expression; intra-S-phase checkpoints activated by replication stress also lead to inhibition of histone gene transcription as well as destabilization of histone mRNAs [38,39].
The chromatin structure not only enables the cell to fit its lengthy genome inside the nucleus, but also protects the genome integrity and provides highly regulated decoding of genetic information at the epigenetic level [40]; this necessitates coordination between histone synthesis and DNA replication. On the one hand, when histones are in excess, they may potentially lead to abnormal chromatin assembly by non-specifically interacting with DNA, or interfere with many other cellular processes. This scenario may be anticipated when DNA replication is inhibited by S-phase checkpoints. On the other hand, if the DNA replication is allowed to continue under a circumstance of repressed histone expression, the sub-stoichiometry (histones compared with DNA) might lead to accumulation of naked DNA. How S-phase cells avoid excessive production of free histones or avoid the accumulation of naked DNA is not completely understood, but must involve cross-talks between histone expression and DNA replication machineries under the control of S-phase checkpoints. Regulated S-phase histone expression is achieved at three levels: transcription, mRNA stability and proteolysis (e.g. [1–3]); all need to be tightly coupled with the S-phase checkpoints to ensure a proper DNA compared with histone pool stoichiometry. Additionally, for each unit of newly synthesized nucleosomal DNA (~180 bp), there must be stoichiometric availability of two molecules each of the core histones H2A, H2B, H3 and H4 (reviewed in [40]). This stoichiometry needs to be maintained via the coordinated expression of different histone subtypes; it is known that non-stoichiometric expression of core histones leads to genomic instability owing to defective chromosome segregation [41].
The above-mentioned redox signalling might provide not only a mechanism to fine tune the H2B gene transcription via OCA-S [13,18], but also exert certain wider effects on global core histone expression in living cells [19], thus coordinating stoichiometric histone expression during S-phase progression in concert with DNA replication. Known cell-cycle machineries such as cyclins/CDKs appear to only govern the entry and exit of S-phase and, normally, it is unlikely that cells suffer frequent replication arrest; thus tight coordination between DNA replication and histone biosynthesis during S-phase progression must be regulated by other mechanisms. Indeed, the coupling of histone expression to DNA replication after S-phase entry appeared to be independent of cyclin E/CDK2 [42].
Using HU (hydroxyurea), which inhibits the formation of dNDPs, thus reducing the dNTP pool [43], or ENA, a compound proposed to collapse the replication fork [44], we could achieve expected S-phase arrest; but an immediate response to these replication stresses was co-ordinately repressed expression of all core histone genes (S. Goh and Y. Luo, unpublished work). The physiological implication of this observation is discussed below.
HOW IS HISTONE EXPRESSION COUPLED TO DNA REPLICATION?
The above description brings up another, perhaps even more important, question that has puzzled us from the very beginning: how is histone expression coupled to DNA replication? This coupling is so tight such that there is virtually no free, extra-nucleosomal histone pool in the cells, which are capable of sensing the level of biosynthesis of each histone subtype to match the level of DNA biosynthesis. The cellular machineries that couple these processes must be precisely controlled, otherwise free histones might as well accumulate or DNA would become (partially) naked thus improperly packaged and less protected. It is hence extremely important to understand this coupling mechanism.
It is known that the initiation of both DNA replication and histone biosynthesis in concert with S-phase entry stringently requires cyclin E/CDK2 (reviewed in [4]); however, the coupling of histone expression to DNA synthesis following S-phase entry seems to be regulated by factors other than cyclin E/CDK2 [42]. In our previous paper [13], we speculated that some components of the multi-subunit OCA-S complex, in conjunction with NPAT, might play this role in addition to mediating the coordination of the expression of histone subtypes. Given the discussion in the previous section (see above) and earlier speculation [13], we take one step further to propose two additional exciting models. One is that NPAT could couple the histone transcription to DNA replication by interacting with both histone transcription and DNA replication machineries (Figure 3), and the other, as illustrated in Figure 4, is that the dynamic Oct-1/OCA-S complex could function as a regulatory machinery for histone transcription and DNA biosynthesis, which is initiated from certain mammalian replication origins with octamer element(s) in proximity. In this scenario, it is completely possible that some DNA replication origins are adjacent to the chromosomal loci where the histone genes are clustered (chromosomes 1 and 6 for humans). Although H2B genes are only localized to chromosomes 1 and 6, the OCA-S complex may send a signal from the H2B gene loci to the replication elongation complex through Oct-1 and this signal is in turn transmitted by the replication elongation complex to the DNA replication folks in other chromosomes and in other regions of replicating chromosomes 1 and 6. It will therefore be interesting to examine the interaction between the OCA-S complex and Oct-1 containing replication elongation complexes. Of high note is a suggestion that Oct-1 is an activator for the initiation of mammalian DNA replication [45,46]. If, in conjunction with Oct-1, OCA-S is also involved in DNA replication, then the mechanisms by which its transcription activity is regulated, notably the S-phase-specific occupancy of OCA-S with target DNA and its modulation by NAD(H) coenzymes [13,18,19], may well be applied to the regulation of DNA replication (see below). In this way, these two critical S-phase events can be intimately coupled by redox signalling.
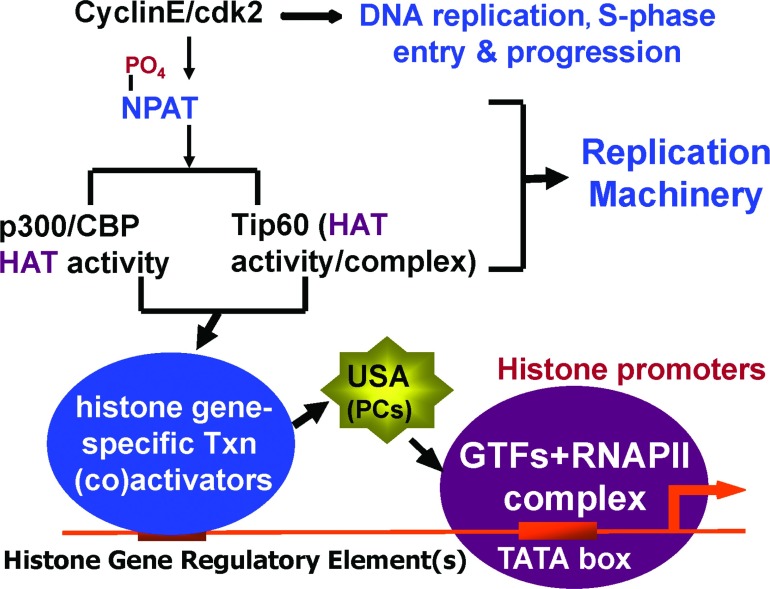
Figure 3. NPAT, in conjunction with general transcriptional coactivators, may be involved in regulating both histone expression and DNA replication during S-phase progression.
Given that both the general coactivator (HAT) activities p300/CBP and Tip60-containing complex are downstream of NPAT to stimulate the histone expression, and that both HAT activities have been implicated in DNA replication/repair, we propose that NPAT couples histone transcription to DNA replication by interacting with both histone transcription and DNA replication machineries in conjunction with these HAT activities. See the text for further details. RNAPII, RNA polymerase II; Txn, transcription.
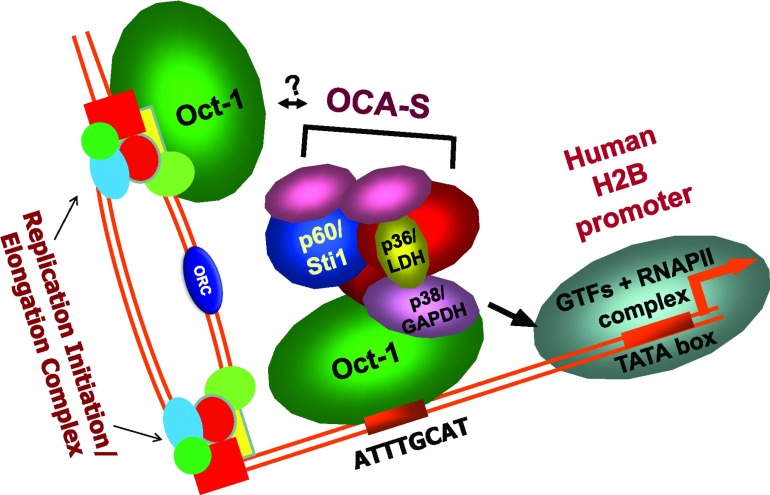
Figure 4. The dynamic Oct-1/OCA-S module may be involved in regulating both histone expression and DNA replication during S-phase progression.
In addition to the octamer sites on H2B gene promoters, which facilitate transcription, octamer elements in the proximity of certain mammalian replication origins may also be occupied by Oct-1; the interaction of Oct-1 with OCA-S complex could function as a regulatory machinery for DNA biosynthesis as well, which may in turn send a signal from corresponding transcription machineries or replication elongation complexes to other DNA replication forks in other regions of the same replicating chromosome(s) or other replicating chromosomes. See the text for details.
Given that NPAT also interacts with transcription factors and/or cofactors of other histone genes [8,47], and that one-protein–multiple-function is an expanding paradigm (McKnight, 2003) [48], the above two models may be applied to other (core) histone genes to explain a tight coupling of DNA replication to overall histone expression. It is imagined that, when replication stresses such as HU and ENA are imposed on cells, the slowed-down DNA replication is sensed immediately by histone expression machineries to maintain a stoichiometric expression of core histone genes in concert with a reduced DNA replication rate. The phenotype of the repressed histone expression may be manifested with a fast kinetics, before the replication stresses could bring about measurable defect in S-phase progression such as that scored by bromodeoxyuridine incorporation. Coupling DNA replication with coordinated and stoichimetric histone expression might constitute a fine-tune mechanism for ensuring genome integrity; this notion is in accordance with the observation made in yeast a quarter of a century ago [41].
COUPLING HISTONE EXPRESSION WITH DNA DAMAGE REPAIR
DNA replication is usually suspended in response to DNA damage, allowing cells to repair damaged DNA; concomitantly, histone gene transcription is correspondingly repressed [38,49]. The exact mechanism by which histone transcription is repressed in response to DNA damage is unknown. One hypothesis is that the coupled suppression of DNA replication and histone gene transcription is a result of inhibition of cyclin E/CDK2 by up-regulated p21 that is stimulated through the ATM/ATR–p53–p21 pathway [38,49]; however, in cells exposed to some genotoxic chemicals for prolonged periods, the activity of cyclin E/CDK2 is no longer inhibited by p21 [42], possibly due to concurrent activation of both cyclin E/CDK2 and p21 in the G1/S-phase-arrested cells (H. He and Y. Luo, unpublished work). Hence, the mechanism coupling the damage-induced suppression of DNA replication and repressed histone gene transcription is not always cyclin E/CDK2 dependent.
We and others showed that the HATs CBP, p300 and Tip60 function in histone gene transcription [28,29]. Interestingly, these transcription factors are found to participate in DNA damage repair as well. CBP/p300 catalyses the acetylation of histone H3 Lys56 that is critical for chromatin assembly following DNA damage repair [50,51]. In addition, CBP/p300 physically interacts with DNA damage repair proteins, such as ATR and DDB (damaged-DNA-binding protein), which suggests direct recruitment of CBP/p300 to DNA damage sites [52,53]. The importance of Tip60 in DNA DSB repair has been extensively revealed. At DSB sites, Tip60 is recruited to acetylate both histones and DNA repair proteins. For instance, it acetylates Lys3016 of ATM and activates ATM kinase activity [54]. Further, histone H4 acetylation by Tip60 is important for the alteration of nucleosome stability during DNA damage repair [55]; Tip60 also acetylates γ-H2A.X, a crucial modification for the turnover of γ-H2A.X after repair [56].
The histone deacetylase SIRT1, a corepressor for histone gene transcription (see above), is involved in DNA damage repair as well in view of the observation that SIRT1 mutant cells are more sensitive to ionizing radiation and exhibit much fewer γ-H2A.X foci upon irradiation; the ability of cells to repair broken DNA is weakened by SIRT1 mutation [57]. In response to DNA damage, the genomic association pattern of SIRT1 is changed, with more SIRT1 concentrated at damaged sites [58]. Furthermore, SIRT1 associates with and deacetylates certain DNA damage repair proteins such as Ku70 [59] and NBS1 [60], which partially explains a regulatory role of SIRT1 in DNA damage repair.
Taken together, the involvement of CBP/p300, Tip60 and SIRT1 in both histone gene transcription and DNA damage repair suggests a coordinator role of the HATs and HDAC in the two coupled events.
POST-TRANSCRIPTIONAL REGULATION OF HISTONE EXPRESSION
Owing to the simple fact that the elevated steady-state levels of histone mRNAs in S-phase are also attributable to increased pre-mRNA processing and the mRNA stability (half-lives), it is of equal importance to emphasize the post-transcriptional control of histone biosynthesis. The 3′ ends of histone mRNAs contain a stem–loop structure that is conserved among histone subtypes, and SLBP (stem–loop-binding protein) can both facilitate the histone pre-mRNA processing and increase the histone mRNA stability, thus is responsible for much of the post-transcriptional control of the histone mRNA levels. This layer of control is in concert with elevated expression of SLBP in the S-phase to meet its stoichiometric requirement for the production of histone mRNAs (see [2]). Thus there ought to be a mechanism that couples histone transcription to post-transcriptional control. Significantly, given that the control at both transcriptional and post-transcriptional levels would influence overall steady-state histone mRNA levels, and that cells ought to be able to sense the overall histone mRNA levels to match that of DNA biosynthesis and vice versa, the entire histone mRNA metabolic pathway (mRNA transcription, processing and stability) must be efficiently coupled to DNA replication in the S-phase; NPAT could be one of the critical components involved in this complicated regulation. In this regard, the dynamic Oct-1–OCA-S–NPAT connections [13], and the interactions between NPAT and factors and/or cofactors involved in transcriptional regulation of other histone genes [47], are of particular interest, for they may also prompt a search for dynamic interactions between NPAT and proteins involved in post-transcriptional regulation of histone mRNAs. We emphasize that all DNA-replication-dependent histone genes, cognate transcription factors and/or cofactors, cyclin E/CDK2, NPAT and proteins involved in histone mRNA maturation, are colocalized in the Cajal bodies (see [10]).
PERSPECTIVES
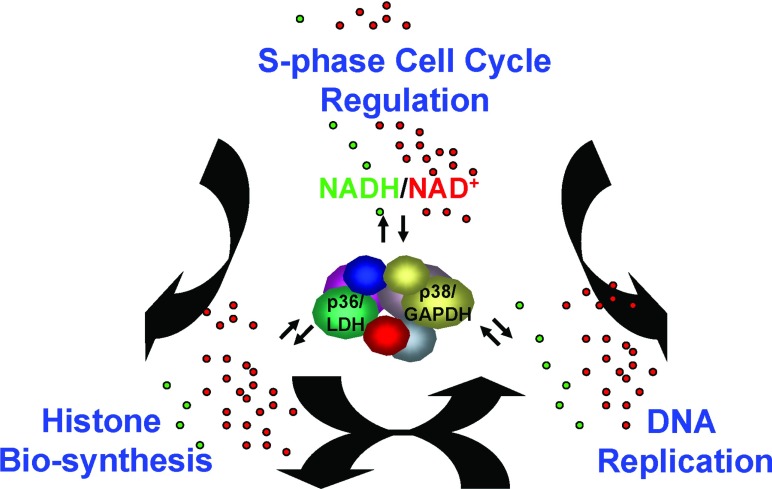
How are signals transmitted between the cell-cycle regulators (e.g. cyclin E/CDK2, NPAT), transcription factors and cofactors of the H2B gene (Oct-1, OCA-S) and those of other histone subtype genes, histone mRNA processing proteins (e.g. SLBP) and DNA replication/repair machineries? Our studies introduced the NAD(H) coenzymes into the picture and demonstrated that they can significantly modulate H2B transcription and the interaction between Oct-1 and OCA-S, and possibly other redox-sensitive machineries. Our bold speculation is that there is a connection between a local metabolic state of a cell and its capacity to progress through the S-phase. Hence, for instance, a local change in the NAD+/NADH concentration and/or the ratio of the redox status, perhaps just restricted to certain nuclear loci where histone transcription (and/or DNA replication) initiates, may simultaneously modulate histone transcription, DNA replication and other S-phase events. In living cells, it will be technically challenging to pursue this issue; however, our observed in vitro modulation of H2B promoter activity by NAD+/NADH may pave the way to explore whether histone expression and other S-phase events such as histone mRNA processing and DNA replication also undergo similar modulation(s) in vivo. We emphasize that the OCA-S complex contains an array of classical enzymes [13], that transcriptional activity of this complex is subject to modulation by coenzymes [13,18,19], and that the complex is thus probably multifunctional. In retrospect, these observations make total sense, especially should there indeed be a link between the (local) metabolic state of a cell and its capacity to progress through cell cycle (S-phase). This might render the aforementioned signalling pathways to be sensitive to or fine-tuned by NAD(H), or to be modulated by other types of coenzymes or relevant metabolites, to account for at least in part the highly coordinated histone expression and its tight coupling to DNA replication (Figure 5).
Figure 5. A broader role of the NAD(H) redox signalling in S-phase cell-cycle regulation.
Downstream of critical cell-cycle regulators (e.g. cyclin E/CDK2, NPAT) are transcription (co)factors for diverse histone subtype genes (e.g. Oct-1 and OCA-S), histone mRNA processing proteins (e.g. SLBP) and DNA replication machineries. The activities of these machineries are either directly redox-modulated through sharing common redox-sensitive components or indirectly redox-modulated through inter-machinery communications via redox-sensitive components. See the text for further details.
ACKNOWLEDGEMENTS
We express sincere apologies to our colleagues whose work was indirectly cited for space considerations.
FUNDING
Our own work was supported by the Agency for Science, Technology and Research, Singapore (to Y.L.), Zhejiang University College of Medicine [starting fund through the National 985 platform (to Y.L.)], and the Fundamental Research Funds for the Central Universities (to L.-L.Z.).
References
- 1.Heintz N. The regulation of histone gene expression during the cell cycle. Biochim. Biophys. Acta. 1991;1088:327–339. doi: 10.1016/0167-4781(91)90122-3. [DOI] [PubMed] [Google Scholar]
- 2.Marzluff W. F., Duronio R. J. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 2002;14:692–699. doi: 10.1016/s0955-0674(02)00387-3. [DOI] [PubMed] [Google Scholar]
- 3.Osley M. A. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 4.Ewen M. E. Where the cell cycle and histones meet. Genes Dev. 2000;14:2265–2270. doi: 10.1101/gad.842100. [DOI] [PubMed] [Google Scholar]
- 5.Hoyt M. A. Eliminating all obstacles: regulated proteolysis in the eukaryotic cell cycle. Cell. 1997;91:149–151. doi: 10.1016/s0092-8674(00)80396-7. [DOI] [PubMed] [Google Scholar]
- 6.Sherr C. J., Roberts J. M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 7.Zheng L., Lee W. H. The retinoblastoma gene: a prototypic and multifunctional tumor suppressor. Exp. Cell Res. 2001;264:2–18. doi: 10.1006/excr.2000.5129. [DOI] [PubMed] [Google Scholar]
- 8.Zhao J., Dynlacht B., Imai T., Hori T., Harlow E. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes Dev. 1998;12:456–461. doi: 10.1101/gad.12.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma T., Van Tine B. A., Wei Y., Garrett M. D., Nelson D., Adams P. D., Wang J., Qin J., Chow L. T., Harper J. W. Cell cycle-regulated phosphorylation of p220 (NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J., Kennedy B. K., Lawrence B. D., Barbie D. A., Matera A. G., Fletcher J. A., Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
- 11.Gao G., Bracken A. P., Burkard K., Pasini D., Classon M., Attwooll C., Sagara M., Imai T., Helin K., Zhao J. NPAT expression is regulated by E2F and is essential for cell cycle progression. Mol. Cell. Biol. 2003;23:2821–2833. doi: 10.1128/MCB.23.8.2821-2833.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye X., Wei Y., Nalepa G., Harper J. W. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol. Cell. Biol. 2003;23:8586–8600. doi: 10.1128/MCB.23.23.8586-8600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng L., Roeder R. G., Luo Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;114:255–266. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]
- 14.Fletcher C., Heintz N., Roeder R. G. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell. 1987;51:773–781. doi: 10.1016/0092-8674(87)90100-0. [DOI] [PubMed] [Google Scholar]
- 15.LaBella F., Sive H. L., Roeder R. G., Heintz N. Cell-cycle regulation of a human histone H2b gene is mediated by the H2b subtype-specific consensus element. Genes Dev. 1988;2:32–39. doi: 10.1101/gad.2.1.32. [DOI] [PubMed] [Google Scholar]
- 16.Segil N., Roberts S. B., Heintz N. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science. 1991;254:1814–1816. doi: 10.1126/science.1684878. [DOI] [PubMed] [Google Scholar]
- 17.Jeffery C. J. Moonlighting proteins. Trends Biochem. Sci. 1999;24:8–11. doi: 10.1016/s0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- 18.Dai R. P., Yu F. X., Goh S. R., Chng H. W., Tan Y. L., Fu J. L., Zheng L., Luo Y. Histone 2B (H2B) expression is confined to a proper NAD+/NADH redox status. J. Biol. Chem. 2008;283:26894–26901. doi: 10.1074/jbc.M804307200. [DOI] [PubMed] [Google Scholar]
- 19.Yu F. X., Dai R. P., Goh S. R., Zheng L., Luo Y. Logic of a mammalian metabolic cycle: an oscillated NAD+/NADH redox signaling regulates coordinated histone expression and S-phase progression. Cell Cycle. 2009;8:773–779. doi: 10.4161/cc.8.5.7880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu B. P., Kudlicki A., Rowicka M., McKnight S. L. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 21.Chen Z., Odstrcil E. A., Tu B. P., McKnight S. L. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science. 2007;316:1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- 22.Hartsough M. T., Steeg P. S. Nm23/nucleoside diphosphate kinase in human cancers. J. Bioenerg. Biomembr. 2000;32:301–308. doi: 10.1023/a:1005597231776. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y., Gallois-Montbrun S., Schneider B., Veron M., Morera S., Deville-Bonne D., Janin J. Nucleotide binding to nucleoside diphosphate kinases: X-ray structure of human NDPK-A in complex with ADP and comparison to protein kinases. J. Mol. Biol. 2003;332:915–926. doi: 10.1016/j.jmb.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Lee M. C., Toh L. L., Yaw L. P., Luo Y. Drosophila octamer elements and Pdm-1 dictate the coordinated transcription of core histone genes. J. Biol. Chem. 2010;285:9041–9053. doi: 10.1074/jbc.M109.075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nallamothu G., Dammai V., Hsu T. Developmental function of Nm23/awd: a mediator of endocytosis. Mol. Cell. Biochem. 2009;329:35–44. doi: 10.1007/s11010-009-0112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chrysogelos S., Riley D. E., Stein G., Stein J. A human histone H4 gene exhibits cell cycle-dependent changes in chromatin structure that correlate with its expression. Proc. Natl. Acad. Sci. U.S.A. 1985;82:7535–7539. doi: 10.1073/pnas.82.22.7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno M. L., Chrysogelos S. A., Stein G. S., Stein J. L. Reversible changes in the nucleosomal organization of a human H4 histone gene during the cell cycle. Biochemistry. 1986;25:5364–5370. doi: 10.1021/bi00367a003. [DOI] [PubMed] [Google Scholar]
- 28.He H., Yu F. X., Sun C., Luo Y. CBP/p300 and SIRT1 are involved in transcriptional regulation of S-phase specific histone genes. PLoS ONE. 2011;6:e22088. doi: 10.1371/journal.pone.0022088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeRan M., Pulvino M., Greene E., Su C., Zhao J. Transcriptional activation of histone genes requires NPAT-dependent recruitment of TRRAP-Tip60 complex to histone promoters during the G1/S phase transition. Mol. Cell. Biol. 2008;28:435–447. doi: 10.1128/MCB.00607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 31.Shiloh Y. ATM and ATR: networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 2001;11:71–77. doi: 10.1016/s0959-437x(00)00159-3. [DOI] [PubMed] [Google Scholar]
- 32.Abraham R. T. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 33.Sorensen C. S., Hansen L. T., Dziegielewski J., Syljuasen R. G., Lundin C., Bartek J., Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 34.Feijoo C., Hall-Jackson C., Wu R., Jenkins D., Leitch J., Gilbert D. M., Smythe C. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J. Cell Biol. 2001;154:913–923. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zachos G., Rainey M. D., Gillespie D. A. Chk1-dependent S-M checkpoint delay in vertebrate cells is linked to maintenance of viable replication structures. Mol. Cell. Biol. 2005;25:563–574. doi: 10.1128/MCB.25.2.563-574.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith G. C., Jackson S. P. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 37.Kurimasa A., Kumano S., Boubnov N. V., Story M. D., Tung C. S., Peterson S. R., Chen D. J. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell. Biol. 1999;19:3877–3884. doi: 10.1128/mcb.19.5.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su C., Gao G., Schneider S., Helt C., Weiss C., O’Reilly M. A., Bohmann D., Zhao J. DNA damage induces down-regulation of histone gene expression through the G1 checkpoint pathway. EMBO J. 2004;23:1133–1143. doi: 10.1038/sj.emboj.7600120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaygun H., Marzluff W. F. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- 40.Wu J., Grunstein M. 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- 41.Meeks-Wagner D., Hartwell L. H. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986;44:43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- 42.Nelson D. M., Ye X., Hall C., Santos H., Ma T., Kao G. D., Yen T. J., Harper J. W., Adams P. D. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol. Cell. Biol. 2002;22:7459–7472. doi: 10.1128/MCB.22.21.7459-7472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koc A., Wheeler L. J., Mathews C. K., Merrill G. F. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J. Biol. Chem. 2004;279:223–230. doi: 10.1074/jbc.M303952200. [DOI] [PubMed] [Google Scholar]
- 44.Krishnan V., Dirick L., Lim H. H., Lim T. S., Si-Hoe S. L., Cheng C. S., Yap K. L., Ting A., Schwob E., Surana U. A novel cell cycle inhibitor stalls replication forks and activates S phase checkpoint. Cell Cycle. 2007;6:1621–1630. doi: 10.4161/cc.6.13.4373. [DOI] [PubMed] [Google Scholar]
- 45.O’Neill E. A., Fletcher C., Burrow C. R., Heintz N., Roeder R. G., Kelly T. J. Transcription factor OTF-1 is functionally identical to the DNA replication factor NF-III. Science. 1988;241:1210–1213. doi: 10.1126/science.3413485. [DOI] [PubMed] [Google Scholar]
- 46.Matheos D. D., Ruiz M. T., Price G. B., Zannis-Hadjopoulos M. Oct-1 enhances the in vitro replication of a mammalian autonomously replicating DNA sequence. J. Cell. Biochem. 1998;68:309–327. [PubMed] [Google Scholar]
- 47.Miele A., Braastad C. D., Holmes W. F., Mitra P., Medina R., Xie R., Zaidi S. K., Ye X., Wei Y., Harper J. W., et al. HiNF-P directly links the cyclin E/CDK2/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol. Cell. Biol. 2005;25:6140–6153. doi: 10.1128/MCB.25.14.6140-6153.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKnight S. Gene switching by metabolic enzymes–how did you get on the invitation list? Cell. 2003;114:150–152. doi: 10.1016/s0092-8674(03)00563-4. [DOI] [PubMed] [Google Scholar]
- 49.Zhao J. Coordination of DNA synthesis and histone gene expression during normal cell cycle progression and after DNA damage. Cell Cycle. 2004;3:695–697. [PubMed] [Google Scholar]
- 50.Vempati R. K., Jayani R. S., Notani D., Sengupta A., Galande S., Haldar D. p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. J. Biol. Chem. 2010;285:28553–28564. doi: 10.1074/jbc.M110.149393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das C., Lucia M. S., Hansen K. C., Tyler J. K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stauffer D., Chang B., Huang J., Dunn A., Thayer M. p300/CREB-binding protein interacts with ATR and is required for the DNA replication checkpoint. J. Biol. Chem. 2007;282:9678–9687. doi: 10.1074/jbc.M609261200. [DOI] [PubMed] [Google Scholar]
- 53.Datta A., Bagchi S., Nag A., Shiyanov P., Adami G. R., Yoon T., Raychaudhuri P. The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone acetyltransferase. Mutat. Res. 2001;486:89–97. doi: 10.1016/s0921-8777(01)00082-9. [DOI] [PubMed] [Google Scholar]
- 54.Sun Y., Xu Y., Roy K., Price B. D. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol. Cell. Biol. 2007;27:8502–8509. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Y., Sun Y., Jiang X., Ayrapetov M. K., Moskwa P., Yang S., Weinstock D. M., Price B. D. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J. Cell Biol. 2010;191:31–43. doi: 10.1083/jcb.201001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kusch T., Florens L., Macdonald W. H., Swanson S. K., Glaser R. L., Yates, 3rd J. R., Abmayr S. M., Washburn M. P., Workman J. L. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 57.Wang R. H., Sengupta K., Li C., Kim H. S., Cao L., Xiao C., Kim S., Xu X., Zheng Y., Chilton B., et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oberdoerffer P., Michan S., McVay M., Mostoslavsky R., Vann J., Park S. K., Hartlerode A., Stegmuller J., Hafner A., Loerch P., et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jeong J., Juhn K., Lee H., Kim S. H., Min B. H., Lee K. M., Cho M. H., Park G. H., Lee K. H. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp. Mol. Med. 2007;39:8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 60.Yuan Z., Zhang X., Sengupta N., Lane W. S., Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol. Cell. 2007;27:149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]