Abstract
Neuronal activity elicits changes in synaptic composition that play an important role in experience-dependent plasticity (Choquet and Triller, 2003; Lisman and Raghavachari, 2006; Bourne and Harris, 2008; Holtmaat and Svoboda, 2009). We used a modified version of stable isotope labeling by amino acids in cell culture to identify activity-dependent modifications in the composition of postsynaptic densities (PSDs) isolated from rat primary neuronal cultures. We found that synaptic activity altered ∼2% of the PSD proteome, which included an increase in diverse RNA binding proteins (RNABPs). Indeed, 12 of the 37 identified proteins whose levels changed with synaptic activity were RNABPs and included the heterogeneous nuclear ribonucleoproteins (hnRNPs) G, A2/B1, M, and D. Knockdown of hnRNPs M and G using shRNAs resulted in altered numbers of dendritic spines, suggesting a crucial role for these proteins in spine density. Synaptic activity also resulted in a concomitant increase in dendritic and synaptic poly(A) mRNA. However, this increase was not affected by knockdown of hnRNPs M or G. Our results suggest that hnRNP proteins regulate dendritic spine density and may play a role in synaptodendritic mRNA metabolism.
Introduction
The postsynaptic density (PSD) is a protein complex specialized in receiving and transducing information at neuronal synapses (Banker et al., 1974; Blomberg et al., 1977; Kennedy, 1993; Ziff, 1997). Proteomic analyses of the PSD have shown that it contains diverse downstream signaling components including regulators of cell polarity, protein translation and degradation, phosphorylation, G-protein signaling, and nuclear function (Walsh and Kuruc, 1992; Walikonis et al., 2000; Vinade et al., 2003; Jordan et al., 2004, 2007; Li et al., 2004; Peng et al., 2004; Yoshimura et al., 2004; Collins et al., 2006; Dosemeci et al., 2006; Jordan and Ziff, 2008; Jordan and Kreutz, 2009). Among PSD components identified by mass spectrometry are members of the RNA binding proteins (RNABP) family (Jordan et al., 2004, 2006). RNABPs regulate all aspects of RNA metabolism, including mRNA transport, translation, and decay. The presence of RNABPs at synapses is consistent with current hypotheses regarding the role of selective mRNA transport and translation in the synapse-specific regulation of dendritic spine morphology and function (Martin and Zukin, 2006; Richter and Klann, 2009).
Changes in PSD protein composition are intimately linked with short- and long-term regulation of synaptic function. For example, stable increases in AMPA receptors associated with the PSD result in long-term potentiation (LTP) (Malenka, 2003; Malenka and Bear, 2004), which is a physiologically relevant increase in synaptic efficacy and a model for learning and memory (Milner et al., 1998; Miller and Mayford, 1999). However, little is known about the dynamics of PSD composition given the technical challenges associated with proteomic quantitation methods as applied to neurons. To overcome these obstacles, we used a variation of stable isotope labeling by amino acids in cell culture (SILAC) (Ong et al., 2002) that corrects for incomplete stable isotopic labeling of nondividing primary neurons (Zhang et al., 2011) to quantitatively investigate PSD protein dynamics in an unbiased manner. We compared PSDs isolated from cultured rat cortical primary neurons stimulated using a modified chemical LTP inducing protocol (cLTP) to PSDs isolated from neurons inhibited with tetrodotoxin (TTX) and found that synaptic activity resulted in increased abundance of diverse RNABPs, including heterogeneous nuclear ribonucleoproteins (hnRNPs) A, A2/B1, A3, M, D, G, and L. In fact, 12 of the 37 proteins whose expression were altered in the PSD were RNABPs, suggesting that neuronal activity regulates synaptic RNA metabolism. We found that knockdown of hnRNPM and hnRNPG, but not hnRNPA2/B1 or hnRNPD, resulted in marked changes in dendritic spine density. These results suggest a crucial role for hnRNP proteins in synaptic function and provide support for the notion that local regulation of RNA metabolism underlies the synapse-specific and activity-dependent regulation of dendritic spine morphology.
Materials and Methods
Lentiviral vectors.
We used pTRIP vectors to generate lentiviral shRNA vectors for knockdown following methods previously described (Janas et al., 2006). pTRIP contains an H1 promoter derived from pSUPER vectors to drive shRNA expression, and a GFP construct driven by an elongation factor 1α promoter to identify transduced neurons. At least three shRNA sequences for each rat-derived sequence of hnRNPD, hnRNPG, hnRNPA2/B1, and hnRNPM were selected using the SFOLD online software prediction algorithms (Wadsworth Center, Albany, NY) and tested. We ended up using shRNAs targeting the rat coding sequences of hnRNPD [base pair 385: 5′GATCCTATCACAGGGCGAT (sh1); base pair 509: 5′CCAAAGCCATGAAAACAAA (sh2)], hnRNPG [base pair 200: 5′-GAGATATGAATGGAAAGTC (sh1); base pair 700: 5′CCACCAAGAGATTATACTT (sh2)], hnRNPM (base pair 1992: 5′ATGGAAGATGCTAAAGGACAA (sh1); base pair 87: 5′AGAACGACCUACUCAGAAU (sh2)],and hnRNPA2/B1 [base pair 84: 5AAGCTTTGAAACCACAGAAGA (sh1); base pair 864: 5′CAACTATGGAGGAGGAAAT (sh2)]. Viruses were generated by triple transfection of pTRIP-shRNA, pCMV-dR8.2, and pMD2.G (which provide structural viral proteins) into 293FT cells (Invitrogen). The media from these cells were harvested and concentrated. We determined viral titers by transducing primary neuronal cortical cultures plated on 24-well plates with increasing amounts of lentivirus-containing 293FT-derived media.
Cell cultures, neuronal stimulation.
Rats were killed using CO2 in compliance with the Albert Einstein College of Medicine Institutional Animal Care and Use Committee. Primary neuronal cultures from E19 embryos of either sex were prepared as described previously (Osten et al., 2000). Cells at DIV 14–21 were treated with either bicuculline (20 μm) or TTX (500 nm). For cLTP-Gly-treated cells, neurons were washed once in artificial CSF (aCSF) without Mg2+ (in mm: 140 NaCl, 5 KCl, 26 NaHCO3, 1.25 NaH2PO4, 1.8 CaCl2, 33 glucose, 25 HEPES, 0.001 strychnine) and then incubated in aCSF + 200 μm glycine for 5 min followed by 15 min incubation in aCSF without glycine (with 2.5 mm Mg2+). cLTP-cAMP-treated cells were washed once with aCSF and then incubated with aCSF + Rolipram (100 nm), forskolin (50 μm), and picrotoxin (100 μm) for 30 min. BDNF-treated cells were treated with 50 ng/ml BDNF for 30 min and cLTP+BDNF was cLTP-cAMP treated in the presence of BDNF (50 ng/ml) for 30 min.
Neuronal imaging, ELISAs, Western blots, and fluorescent in situ hybridizations.
All immunocytochemistry (ICC) was performed using standard conditions. Briefly, cells were fixed using 4% paraformaldehyde (PFA) for 5 min and permeabilized using 0.2% Triton X-100 for 5 min. Antibodies used were as follows: hnRNPD (ab61193; 1:1000; Abcam), hnRNPG (NBP1-67923; 1:500; Novus), hnRNPM (NBP1-70822; 1:500; Novus), hnRNPA2/B1 (sc-32316; 1:200; Santa Cruz Biotechnology), PSD-95 (K28/43; 1:2000; University of California Davis/NIH NeuroMab Facility), MAP2 (CPCA-MAP2; 1:5000; EnCor Bio), fragile X mental retardation protein (FMRP; 7G1-1-s; 0.5 μgs/ml), and SV2 (0.5 μgs/ml; Developmental Studies Hybridoma Bank). Western blots were performed using standard conditions; primary antibodies (above) were incubated overnight at 4°C with rocking and used at the same concentration as used for ICC. For ELISA assays, 7 × 104 rat primary cortical neurons were plated in 24-well plates. At DIV 14–17, the cells were treated as described, fixed with PFA, and immunostained with GluR1 Ab targeting the extracellular domain (PC246; Calbiochem). Neurons were then washed, incubated in HRP-labeled secondary antibody, and incubated with o-phenylenediamine for the colorimetric assay. After the substrate was collected and measured, the cells were lysed in SDS-PAGE buffer, boiled, and subjected to SDS-PAGE for protein normalization. Fluorescent in situ hybridizations (FISH) with ICC was performed by fixing and permeabilizing primary hippocampal neurons as described above, washing once with wash buffer (10% formamide, 2× SSC), and incubating neurons with 0.5 ngs/μl Oligo dT(50) probe coupled to Cy3 (Genelink) in hybridization buffer (10% dextran sulfate, 1 μg/μl E. coli tRNA, 2 mm vanadyl ribonucleoside complex, 0.02% acetylated BSA, and 10% formamide) for 2 h at 37°C. Cells were then washed twice (30 min per wash) with wash buffer, then washed once with DEPC-treated PBS, blocked with 1% acetylated BSA/PBS for 1 h, and incubated with primary and secondary antibodies as per standard conditions for ICC. Quantitative analysis of ICC and FISH experiments were performed blind for the identity of the viruses used for transduction.
SILAC and isolation of PSDs from neuronal cultures.
For the SILAC assays, cortical neurons were grown in 15 cm plates at 9 × 106 cells/plate using the following modified Neurobasal medium: Neurobasal medium lacking Arg and Lys (Specialty Media; Invitrogen) was supplemented with either L-Arg:HCl (U-13C6) and L-Lysine:2HCl (4,4,5,5-D4) (medium) or L-Arg:HCl (U-13C6; U-15N4) and L-Lys:2HCl (U-13C6; U-15N2) (heavy). All isotope-labeled amino acids were from Cambridge Isotope Laboratories. Lys and Arg were supplemented at 146 and 84 mg/l, respectively (as in regular unlabeled Neurobasal medium). One half of the media was changed every 3–4 d. To obtain a relatively pure neuronal population, we added 0.2 μm cytosine arabinoside (AraC) at DIV 5 to inhibit glial growth. PSDs were enriched according to the methods of Cohen and Carlin (Cohen et al., 1977; Carlin et al., 1980) using the long method (Jordan et al., 2004). Briefly, at DIV 15, heavy- and medium-labeled neuronal cultures were scraped off and mixed in ice-cold Solution A (0.32 m sucrose, 1 mm NaHCO3, 1 mm MgCl2, 0.5 mm CaCl2) containing protease and phosphatase inhibitors. Cells were Dounce homogenized in 4 ml of Solution A and diluted to 10% weight/volume. This initial homogenate represents the equivalent of the total lysate (WB) fraction shown in Figure 5, A and B (right). Following centrifugation at 1400 g for 10 min, the supernatant fluid was saved and the pellet rehomogenized to 10% weight/volume in Solution A and centrifuged at 710 g for 10 min. Supernatants were pooled and subjected to a second centrifugation at 710 g for 10 min. The resulting supernatant was then centrifuged at 30,000 g for 15 min to collect the P2 pellet. The P2 pellet was resuspended in Solution B (0.32 m sucrose, 1 mm NaHCO3) and layered on top of a 0.85 m sucrose, 1 m sucrose, and 1.2 m sucrose step sucrose gradient and centrifuged at 82,500 g for 2 h. The synaptosomes (Syn fraction; Fig. 5A) were collected at the 1/1.2 m sucrose layer interface and pelleted down at 45,000 g for 30 min after sixfold dilution with solution B. The pellet was resuspended in pure ice-cold water (with protease and phosphatase inhibitors), incubated at 4°C for 30 min to induce hypo-osmotic shock, and repelleted. The lysed synaptosomes were then resuspended in Solution B and placed on top of a second 0.85, 1.0, 1.2 m sucrose step gradient. The enriched synaptosomes at the 1–1.2 m interface were diluted with Solution B and an equal volume of 1% Triton X-100 and 0.32 m sucrose; 12 mm Tris, pH 8.1 was added. Synaptosomes were lysed at 4°C for 15 min, centrifuged at 32,800 g for 25 min, and the pellet resuspended in Solution B. The suspension was layered on a final sucrose step gradient of 1 m sucrose, 1.5 m sucrose, and 2 m sucrose and centrifuged at 200,000 g for 2 h. The PSD fraction was collected at the 1.5 m and 2 m sucrose interface and diluted with Solution B. An equal volume of 1% Triton X-100, 150 mm KCl was added to obtain the two-Triton PSD fraction. These enriched PSDs represent the PSD fraction in Figure 5, A and B, and were collected by centrifugation at 200,000 g for 20 min and lysed in a 2% SDS-Tris buffer with brief sonication.
Figure 5.
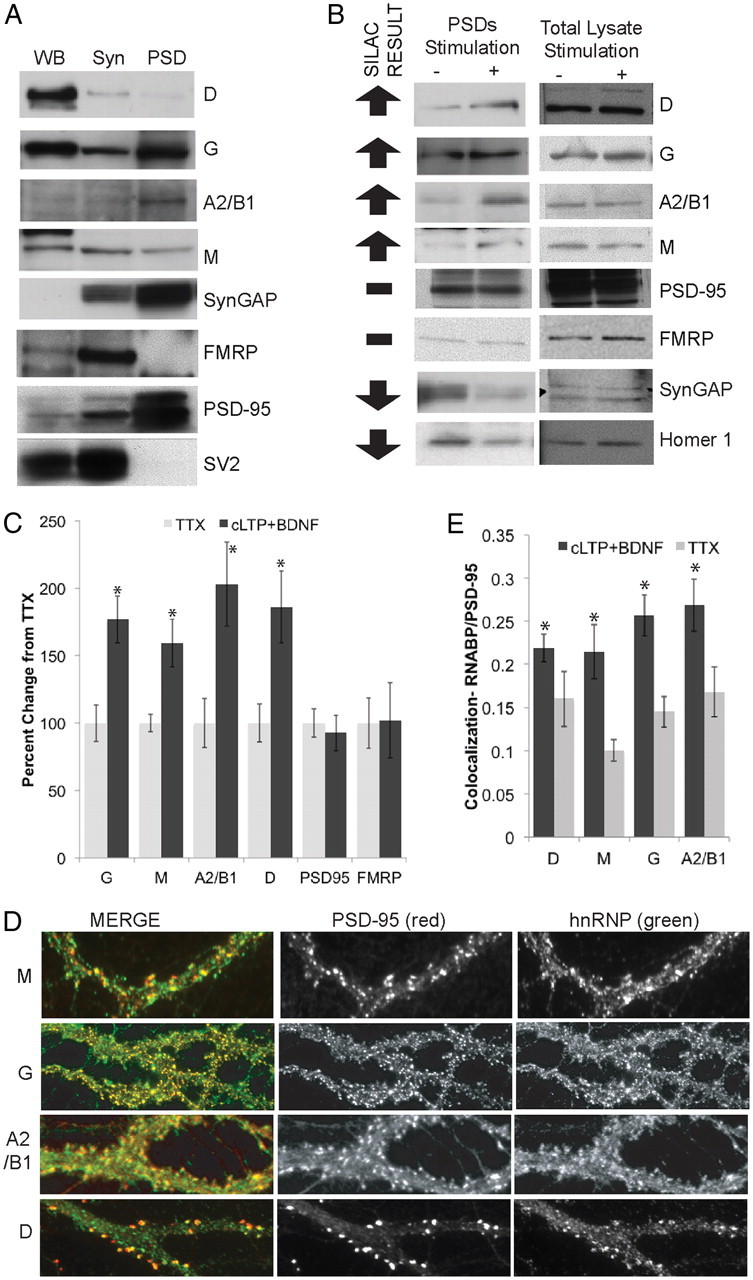
Distribution of RNABPs and other proteins in neurons and verification of SILAC results. A, Total lysate [10 μg; Western blot (WB)], synaptosomes (Syn), or PSDs purified from rat brains were separated by SDS-PAGE and immunoblotted using antibodies against the indicated proteins. The presynaptic and postsynaptic markers SV2 and PSD95, respectively, were used to evaluate the quality of the PSD fractions. B, Rat cortical neurons (DIV 18) were pretreated with cycloheximide (1 h) and then treated with cLTP+BDNF (+) or TTX (−) and PSDs were isolated. Cycloheximide was present throughout the treatment. Western blots confirm the SILAC results obtained (arrows). Right, Western blots of total lysates confirm that activity did not induce any overall changes in the total levels of these proteins. C, Quantitation of Western blots in B by gel densitometry (ImageJ) and normalizing using Western blots against tubulin (data not shown). N = 3 gels, *p < 0.05; t test comparing cLTP+BDNF to TTX for each protein. D, Primary hippocampal neurons (DIV 21) were immunostained with hnRNPM, hnRNPG, hnRNPA2/B1, or hnRNPD antibodies (green) and costained for the synaptic marker PSD95 (red) showing the synaptic distribution of these proteins. E, cLTP+BDNF resulted in increased colocalization between the RNABPs and PSD-95, as determined using ICC and image quantitation (Mander's coefficient, ImageJ). N = 3 biological replicates, 250–300 puncta taken from 10 μm patches that were 20 μm from the cell body; 8–11 neurons per replicate. *p < 0.05, t test between cLTP+BDNF and TTX for each protein.
SDS-PAGE and LC-MS/MS.
Enriched PSD proteins were separated by 10% SDS-PAGE and Coomassie stained. Gel lanes were cut into ∼20 bands. Gel bands were cut into small pieces and destained in 25 mm NH4HCO3/50% acetonitrile, dehydrated with acetonitrile, and dried (Shevchenko et al., 1996). The gel pieces were then rehydrated with 12.5 ng/μl trypsin solution in 25 mm NH4HCO3 and incubated overnight at 37°C. Peptides were extracted twice with 5% formic acid/50% acetonitrile followed by a final extraction with acetonitrile. Samples were concentrated by vacuum centrifugation to dryness and redissolved with 2% acetonitrile in 0.1% formic acid before further analysis. For all LC-MS/MS analysis, an LTQ-Orbitrap hybrid mass spectrometer (Thermo Fisher Scientific) equipped with a nanoelectrospray ionization source (Jamie Hill Instrument Services) was used. A nanoLC system (Eksigent Technologies) equipped with a self-packed 750 μm × 12 cm reverse phase column (Reprosil C18, 3 μm; Dr. Maisch HPLC) was coupled to the mass spectrometer. Peptides were eluted by a gradient of 3–40% acetonitrile in 0.1% formic acid over 110 min at a flow rate of 300 nl/min. Mass spectra were acquired in data-dependent mode with one 60,000 resolution MS survey scan by the Orbitrap and up to eight MS/MS scans in the LTQ for the most intense peaks selected from each survey scan. Automatic gain control target value was set to 1,000,000 for Orbitrap survey scans and 5000 for LTQ MS/MS scans. Survey scans were acquired in profile mode and MS/MS scans were acquired in centroid mode.
Peptide identification and quantitation.
The raw mass spectra files were processed using MaxQuant software (version 1.0.13.8) (Cox and Mann, 2008). Protein and peptide identification was performed using the Mascot search engine (version 2.2.1; Matrix Science) by searching the concatenated forward and reverse International Protein Index protein database (version 3.52) containing 55,303 mouse and 39,906 rat protein sequences and 175 commonly observed contaminants (in total, 190,768 sequences including reverse and contaminant sequences). The minimum required peptide length was 6 a.a., and trypsin cleavage specificity was applied with up to two missed cleavages allowed. Variable modifications included oxidation (M) and N-acetylation of protein N termini. Initially, the maximum allowed mass deviations for precursor and fragment ions were set to 20 ppm and 0.5 Da, respectively. The results of the database search were further processed and statistically evaluated by MaxQuant. For protein and peptide identification, the initial maximum protein and peptide false discovery rates were set to 1%. To further improve confidence in protein identification and quantitation, the following filters were applied to the MaxQuant output: (1) at least two peptides from each protein, (2) >30% SILAC label incorporation, (3) protein was quantified by at least three SILAC ratio measurements from peptides in both forward and reverse experiments, (4) variability of peptide ratio measurements of the protein was <100% in both forward and reverse experiments, and (5) total peptide signal intensity of the protein was >1,000,000. After filtering, there were 1637 protein identifications matched to the forward protein database and three matched to the reverse database. Thus, the false discovery rate was estimated to be 0.2%. Proteins were considered changing if their significance B values were <0.05 in both forward and reverse experiments. Significance B values are p values for detection of significant outlier protein ratios calculated on the protein subsets obtained by intensity binning (Cox and Mann, 2008).
Results
Cell-surface and synaptic GluR1-containing AMPA receptor levels are often used as general markers of synaptic activity and plasticity (Lu et al., 2001; Malenka, 2003). We first tested how different neuronal activation protocols altered the abundance of cell-surface and synaptic GluR1 receptors in cortical neuronal cultures. While the protocols tested [bicuculline (Bic) (Ehlers, 2003), ChemLTP using glycine (cLTP-Gly) (Lu et al., 2001), ChemLTP using forskolin and rolipram (cLTP-cAMP) (Boehm et al., 2006; Oh et al., 2006), and BDNF (Kang and Schuman, 1996; Ji et al., 2010)] (Fig. 1A) have been previously shown to elicit LTP and morphological changes in hippocampal primary cultures and hippocampal slices, to our knowledge they have not been used on primary cortical cultures. We used cortical neurons because we required large quantities of neurons to obtain sufficient material for PSD purification. We therefore performed ELISA assays on cortical neurons plated at high density and found that cLTP-cAMP in the presence of BDNF (cLTP+BDNF) resulted in the largest increase in cell surface GluR1 (Fig. 1A). To determine whether these changes were synaptic, we quantified the colocalization of GluR1 with the synaptic marker PSD-95 (by fluorescence imaging) and found that cLTP+BDNF drives GluR1 into synapses, which is indicative of synaptic potentiation (Fig. 1B). We obtained similar results in primary hippocampal neurons (data not shown).
Figure 1.
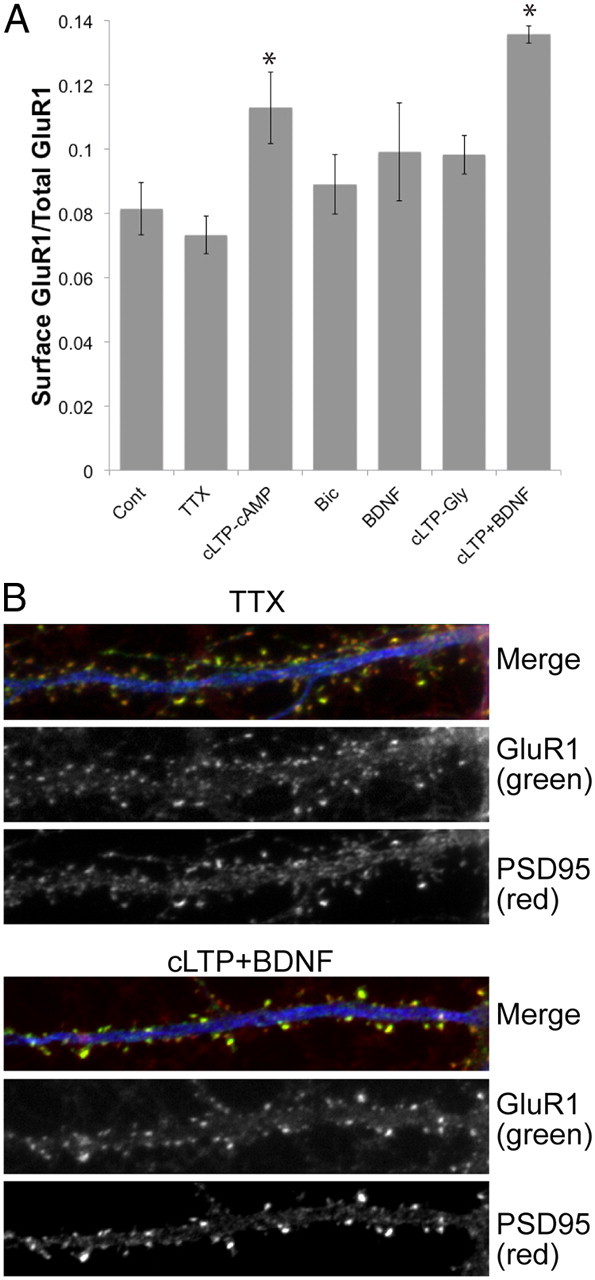
cLTP+BDNF stimulation results in an increase in synaptic GluR1 receptors. A, Quantitation of ELISA results for surface GluR1 levels in response to different neuronal stimulation paradigms and normalized using total GluR1 levels. No changes in total GluR1 levels were observed (data not shown). N = 4 biological replicates in triplicate technical measurements. *p < 0.005, t tests comparing each treatment to untreated cells [Control (Cont)]. B, cLTP+BDNF stimulation results in an increase of GluR1 receptors (green) at synapses labeled by PSD-95 (red) in rat primary cortical neurons (DIV 21). Dendrites are labeled using MAP2 antibodies (blue). Quantification of these results using ImageJ (Mander's coefficients) shows GluR1/PSD95 colocalization was 56.6 ± 5.2% in response to TTX-mediated inhibition and increased to 75.5 ± 7.4% in response to cLTP+BDNF. N = 3 biological replicates, 250–300 puncta from 12 to 15 neurons were measured in 10 μm patches 20 μm away from the cell body; p < 0.05, t test comparing cLTP+BDNF with TTX values.
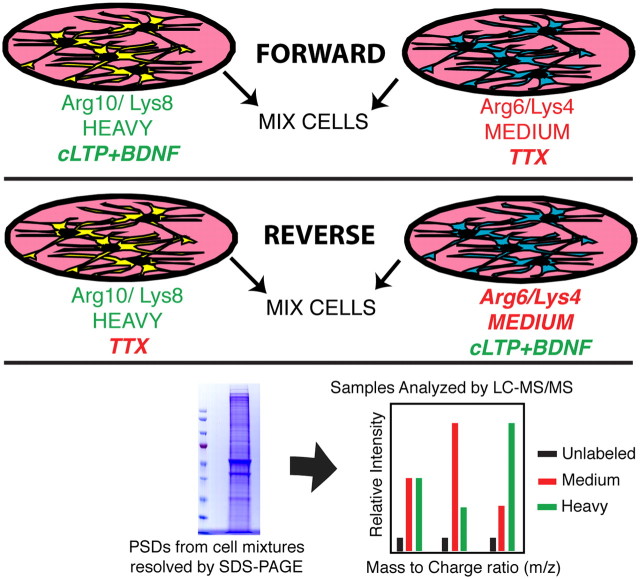
We therefore chose to compare PSDs isolated from neurons treated with cLTP+BDNF to neurons treated with TTX, speculating that these conditions would maximize the changes in PSD composition. To perform the SILAC experiment, two populations of primary cortical neurons were grown in either Neurobasal A medium lacking Arg and Lys and supplemented with Arg6 (containing six C13 atoms) and Lys4 (containing four deuterium atoms) (medium) or Neurobasal A medium lacking Arg and Lys and supplemented with Arg10 (containing six C13 and four N15 atoms) and Lys8 (containing six C13 and two N15 atoms) (heavy). These two sets of labeling amino acids are incorporated into the neuronal proteome at the same rate, which allows for a comparison between the two labeled groups and thus eliminates the need to correct for unlabeled peptides (Zhang et al., 2011). At day 15 in culture, we treated all neurons with cycloheximide (100 μm) for 1 h and then stimulated heavy neurons using cLTP+BDNF and inhibited medium neurons with 1 μm TTX. After 30 min, the media were replaced and neurons were allowed to recover for 15 min (cycloheximide was present at every step). This represented the forward experiment. We also performed a separate reverse experiment in which medium neurons were treated with cLTP+BDNF and heavy neurons were inhibited with TTX. The reverse experiment is a biological replicate that should reveal changes opposite to those found in the forward experiment (Fig. 2). After treatment, PSDs were isolated from the neuronal cultures and analyzed by mass spectrometry (see Materials and Methods, above).
Figure 2.
SILAC methodology. Neurons were grown in Neurobasal A containing either Arg6/Lys4 amino acids (medium) or Arg10/Lys8 amino acids (heavy) to label the neuronal proteome. Forward and reverse experiments are replicates intended to reveal reciprocal changes to ensure that labeling by heavy or medium amino acids did not affect the experimental outcome. After 15 d in culture, cells were treated with cLTP+BDNF or TTX. The PSDs were isolated, solubilized, and the proteins fractionated by SDS-PAGE. After tryptic digestion, peptides were extracted from gel slices and analyzed by LC-MS/MS. The activity-dependent changes in PSD protein composition were determined by comparing the intensity of adjacent heavy- and medium-labeled peptide peaks. Peptides were then identified by MS/MS and subsequent bioinformatic analysis.
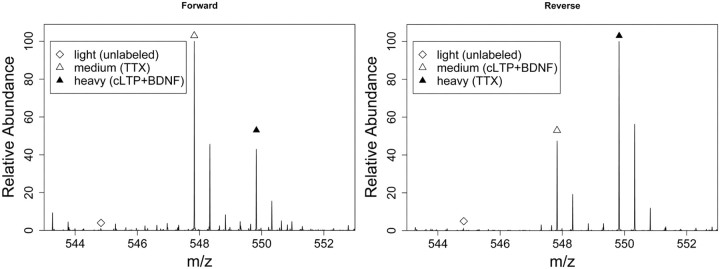
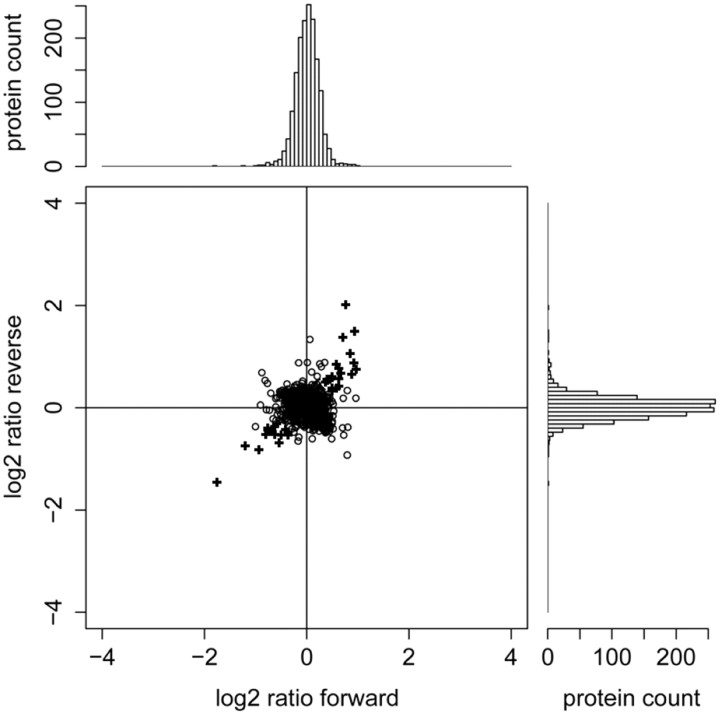
Figure 3 shows an example of how SILAC ratios were measured by MS in the forward and reverse experiments. Consistent with previous studies (Spellman et al., 2008; Zhang et al., 2011), the average label incorporation level was >90% after 15 d in SILAC culture, allowing for quantitation of almost all identified proteins. From the two biological SILAC replicates, 1637 proteins were identified in purified PSD fractions that were quantified in both forward and reverse experiments. The estimated false discovery rate for protein identification was 0.2%. This list of proteins included 69% of the 466 proteins listed in a consensus PSD protein database, which was based on the results of different proteomic studies of the PSD (Collins et al., 2006; Jordan et al., 2006). Gene ontology analysis using the DAVID functional annotation tool 6.7 (Huang et al., 2009) revealed that our SILAC PSD list contained 55 proteins with synapse annotation for GO cellular component, while the consensus PSD list analyzed previously contained 44 (Collins et al., 2006). This suggests that our PSD analysis was more comprehensive than previous proteomic studies. SILAC quantitation showed that most of the protein ratios (i.e., the ratio of the abundance of PSD proteins in stimulated and inhibited neurons) are clustered tightly around 1 (Fig. 4), indicating that our SILAC screen exhibited a high accuracy of quantitation, as incorrectly identified peptide signal pairs would have ratios deviating significantly from 1.
Figure 3.
Representative MS spectra showing SILAC quantitation of protein abundance changes induced by cLTP+BDNF treatment. The data show the MS analysis of a tryptic peptide (LLSDISTALR) from the protein SynGAP, which had decreased abundance in PSDs after cLTP+BDNF treatment. The peptide ratio is calculated by comparing the intensities of the medium signal to the heavy signal. The light signal, which represents the unlabeled fraction of the protein, was not used for protein quantitation. In the forward and reverse experiments, the ratio change is in opposite directions because the SILAC labeling was flipped in these two experiments.
Figure 4.
Identification of proteins whose abundance were significantly changed upon LTP treatment. The histogram on top shows protein ratios in the forward experiment. The histogram on the right shows protein ratios in the reverse experiment. Protein ratios from the two replicate experiments are shown in the central scatter plot. All ratios are presented as cLTP+BDNF/TTX. Proteins with consistent significant ratio changes are marked with a plus sign (+) (p value <0.05; Significance B; MaxQuant).
Stringent criteria were used to identify proteins whose abundance was significantly changed upon treatment (see Materials and Methods, above). After applying this filter to the 1637 identified proteins, we found that 37 proteins (∼2% of the PSD proteome) displayed significant changes in response to cLTP+BDNF (Table 1, Fig. 4). Among the identified changes, we found a large decrease in PSD-associated SynGAP and Homer1, as well as changes in cytoskeletal regulators such as actin-binding LIM protein 1 and ArgBP2 (Table 1). Surprisingly, 12 of the 37 (>30%) identified proteins with altered expression were RNABPs, with the largest component being members of the hnRNPs. Less than 8% of all proteins identified at the PSD were RNABPs, suggesting a substantial enrichment of these proteins among the activity-dependent changes identified in the PSD. We found that hnRNPs A1, A2/B1, A3, G, L, M, and D showed increased abundance at synaptic junctions in the forward experiments and decreased abundance in the reverse experiment, as expected. Some of these proteins, such as hnRNPA2/B1, have been previously identified in dendrites (Hoek et al., 1998; Munro et al., 1999; Kanai et al., 2004). However, since most of these RNABPs have been previously implicated in nuclear functions (Venables et al., 2000; Dobi et al., 2006; Jain and Gavis, 2008; Lee et al., 2008), we selected four of these to verify their synaptic distribution. Western blots showed that, unlike classical synaptic markers such as SynGAP and Homer1, these hnRNPs were present but not enriched in PSD fractions (Fig. 5A). Western blots for postsynaptic (PSD95) and presynaptic (SV2) markers confirmed that we had isolated an enriched PSD fraction (Fig. 5A). To confirm our SILAC results, we performed Western blots for several proteins in PSD fractions isolated from cultured cortical neurons stimulated with cLTP+BDNF or inhibited with TTX (Fig. 5B). These immunoblots confirmed the SILAC predicted results (Fig. 5B, arrows), showing that hnRNPs M, D, G, and A2/B1 accumulated in PSDs after cLTP+BDNF treatment, while SynGAP and homer1 were decreased at PSDs in response to this stimulation (Fig. 5B,C). No changes in abundance were detected for PSD95 or FMRP (FMRP was barely detectable in the PSD fraction; Fig. 5B,C). Using immunocytochemical analysis, we found that hnRNPs G, M, D, and A2/B1 were present in the nucleus (data not shown), as well as in neuronal dendrites and dendritic spines where they colocalized with PSD95 (Fig. 5D). Moreover, using image quantitation, we found that cLTP+ BDNF resulted in an increase in the colocalization of hnRNPs M, A2/B1, G, and D with PSD-95, which further confirmed our biochemical analysis and SILAC results (Fig. 5E).
Table 1.
Proteins of the PSD that displayed significant changes in abundance between the cLTP+BDNF treated and the TTX treated neurons; proteins have been grouped into categories based on their reported functions
| Protein ID | Protein name | Gene name | Forward cLTP/TTX | Reverse TTX/cLTP |
|---|---|---|---|---|
| RNA binding | ||||
| IPI00553777 | Heterogeneous nuclear ribonucleoprotein A1 | Hnrnpa1 | 1.94 | 0.59 |
| IPI00867934 | Heterogeneous nuclear ribonucleoprotein A2/B1 | Hnrnpa2b1 | 1.84 | 0.55 |
| IPI00382376 | Heterogeneous nuclear ribonucleoprotein A3 | Hnrnpa3 | 1.59 | 0.63 |
| IPI00124979 | Heterogeneous nuclear ribonucleoprotein G | Hnrnpg | 1.93 | 0.36 |
| IPI00765134 | Heterogeneous nuclear ribonucleoprotein L | Hnrnpl | 1.47 | 0.67 |
| IPI00876603 | Heterogeneous nuclear ribonucleoprotein M | Hnrnpm | 1.33 | 0.70 |
| IPI00755892 | Heterogeneous nuclear ribonucleoprotein D | Hnrpdl | 1.30 | 0.71 |
| IPI00828741 | HnRNP-associated with lethal yellow | C130057N11Rik | 1.56 | 0.59 |
| IPI00653307 | DEAD box polypeptide 17 | Ddx17 | 1.76 | 0.48 |
| IPI00366249 | ATP-dependent RNA helicase A/ DEAD box 9 | Dhx9 | 1.52 | 0.74 |
| IPI00400578 | Heterochromatin protein 1-binding protein 3 | Hp1bp3 | 1.36 | 0.68 |
| IPI00117063 | RNA-binding protein FUS | Fus | 1.50 | 0.56 |
| Cytoskeletal | ||||
| IPI00199101 | Serine/threonine-protein kinase MRCK alpha | Cdc42bpa | 0.64 | 1.39 |
| IPI00368911 | CTTNBP2 N-terminal-like protein | Cttnbp2 nl | 1.35 | 0.67 |
| IPI00844646 | Echinoderm microtubule-associated protein-like 1 | Eml1 | 0.77 | 1.42 |
| IPI00474002 | Stathmin-2 | Stmn2 | 0.66 | 1.45 |
| IPI00309223 | Stathmin-3 | Stmn3 | 0.68 | 1.60 |
| IPI00125328 | Dematin | Epb49 | 0.43 | 1.68 |
| IPI00364284 | LIM and calponin homology domains-containing 1 | Limch1 | 0.71 | 1.45 |
| GAPs | ||||
| IPI00558902 | ADP-ribosylation factor GTPase-activating protein 2 | Arfgap2 | 0.76 | 1.36 |
| IPI00339023 | Ras GTPase-activating protein SynGAP | Syngap1 | 0.29 | 2.73 |
| IPI00469012 | TBC1 domain family member 10B | Tbc1d10b | 1.47 | 0.65 |
| IPI00758248 | IQ motif and SEC7 domain-containing protein 2 | Iqsec2 | 1.43 | 0.78 |
| Synaptic | ||||
| IPI00556925 | Protein bassoon | Bsn | 1.55 | 0.67 |
| IPI00420725 | Calcium/calmodulin-dep. protein kinase II, alpha | Camk2a | 0.58 | 1.42 |
| IPI00649778 | Calcium/calmodulin-dep. protein kinase II, beta | Camk2b | 0.67 | 1.27 |
| IPI00196730 | Calcium/calmodulin-dep. protein kinase II, gamma | Camk2g | 0.67 | 1.25 |
| IPI00204418 | ERC protein 2 | Erc2 | 1.32 | 0.72 |
| IPI00209336 | Sorbin and SH3 domain-containing protein 2 | Sorbs2 | 0.52 | 1.76 |
| IPI00358533 | Actin-binding LIM protein 1 | Ablim1 | 1.45 | 0.77 |
| IPI00210570 | Homer protein homolog 1 | Homer1 | 0.58 | 1.42 |
| Kinases | ||||
| IPI00196987 | Beta-adrenergic receptor kinase 1 | Adrbk1 | 1.83 | 0.64 |
| Other | ||||
| IPI00198101 | Uncharacterized protein | C530008M17Rik | 0.74 | 1.39 |
| IPI00190176 | Golgin subfamily A member 3 | Golga3 | 0.70 | 1.44 |
| IPI00407835 | Ubiquitin-associated protein 2-like | Ubap2l | 0.60 | 1.35 |
| IPI00230394 | Lamin-B1 | Lmnb1 | 1.70 | 0.25 |
| IPI00652934 | Core histone macro-H2A0.2 | H2afy2 | 1.62 | 0.39 |
Translation of dendritically localized mRNAs (Miyashiro et al., 1994; Eberwine et al., 2002; Zhong et al., 2006) plays an important role in synaptogenesis (Lyles et al., 2006) and dendritic spine morphology (Engert and Bonhoeffer, 1999; Maletic-Savatic et al., 1999; Okamoto et al., 2002; Fukazawa et al., 2003; Matsuzaki et al., 2004; Tanaka et al., 2008; Holtmaat and Svoboda, 2009). We therefore speculated that these RNABPs might regulate the metabolism of RNAs that control synapse morphology. To explore this possibility, we generated lentiviral shRNA particles using pTRIP vectors to perform knockdown experiments (Janas et al., 2006). We generated at least two different shRNA viruses for each RNABP (shRNA#1 and shRNA#2; see Materials and Methods, above). We found that transduction of rat primary hippocampal neurons at DIV 7–10 using these viruses (incubation, 5–7 d) resulted in ≥70% specific downregulation of these proteins compared with a scrambled shRNA sequence or untransduced cells (Fig. 6A). There was no effect of these shRNAs on cell viability. RNABP knockdown using shRNAs led to a loss of their immunoreactivity in immunocytochemistry experiments on primary hippocampal neurons, confirming the specificity of the antibodies used (Fig. 6C). To determine the effects of loss of these hnRNPs on dendritic spine density, we transduced rat primary hippocampal neurons at DIV 14 with lentiviral shRNAs at a low multiplicity of infection (<1) to be able to identify transduced and untransduced cells in the same coverslip (Fig. 6B). Five days later (DIV 19), we fixed and immunostained neurons for GFP and PSD-95 (to identify dendritic spines). We found that there was a significant decrease in the number of dendritic spines in neurons lacking hnRNPG but a significant increase in dendritic spines in neurons lacking hnRNPM (Fig. 6C,D). No changes were observed in neurons lacking hnRNPs D or A2/B1. These results suggest that hnRNPG and hnRNPM play a crucial role in maintaining normal spine density.
Figure 6.
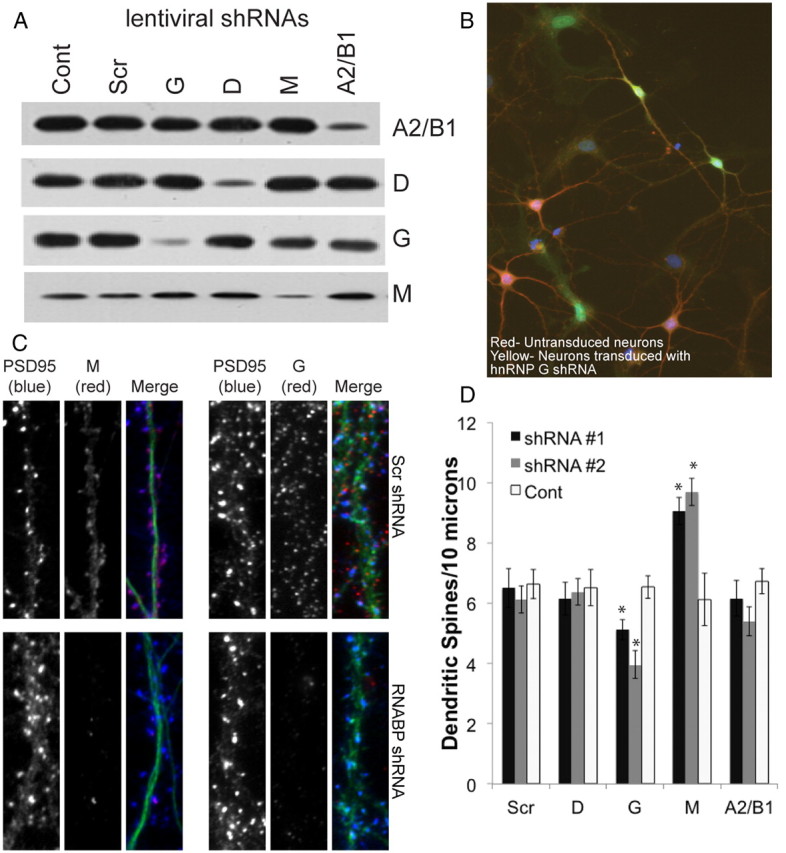
Knockdown of RNABPs alters dendritic spine density. A, Western blots of cortical neurons transduced at DIV 7–10 with RNABP specific shRNA viruses (G, M, D, or A2/B1), viruses with control scrambled shRNAs (Scr) or untransduced neurons [control (Cont)]. Shown are Western blots for shRNA#1 for each protein. Downregulation was 70.4 ± 6.4% (hnRNPA2/B1), 88.4 ± 2.4% (hnRNPD), 90.5 ± 5.4% (hnRNPG), and 72.4 ± 4.9% (hnRNPM) quantified using gel densitometry (ImageJ) and normalized by dividing the intensities of the RNABP with tubulin immunoreactivity (data not shown), which was used as a loading control (n = 3 gels). The second set of shRNAs for each protein (shRNA#2) exhibited similar amounts of downregulation (data not shown). B, Primary hippocampal cultures transduced with lentiviral shRNAs at low multiplicity of infections show that transduced neurons (yellow) can be compared with untransduced control neurons in the same coverslip (red). C, Knockdown of hnRNPG or hnRNPM results in decreased RNABP immunoreactivity (demonstrating antibody specificity) and results in altered spine density at DIV 19–21. D, Quantitation of dendritic spine numbers in primary hippocampal neurons by counting the number of spines (identified manually by GFP and PSD95 immunoreactivity) in a 10 μm dendritic patch, 20 μm away from the cell body. We found that loss of hnRNPG and hnRNPM using two different shRNAs for each results in differential changes in dendritic spine numbers. Controls represent untransduced neurons in the same coverslip (as shown in B). N = 4 biological replicates, 350–400 puncta from 9 to 14 neurons for each condition. *p < 0.05, t test comparing transduced and untransduced cells in the same coverslip for each protein studied.
To determine whether the synaptic upregulation of RNABPs resulted in a concomitant upregulation of synaptic RNA, we treated DIV 17–21 hippocampal neurons with cLTP+BDNF or TTX and imaged poly(A) mRNA using FISH using an oligo(dT) probe (Fig. 7A). We found that cLTP+BDNF resulted in a substantial increase in poly(A) mRNA overall and in dendrites (Fig. 7A). To determine whether this mRNA was synaptic, we quantified colocalization of oligo(dT)-Cy3 reactive puncta with PSD-95 immunoreactivity (Fig. 7B). We found that cLTP+ BDNF resulted in an increase in the extent of their colocalization (Fig. 7C, Cont), suggesting that poly(A) mRNAs are delivered into synapses in response to the same treatment that resulted in synaptic upregulation of hnRNPs. To determine whether hnRNPG and hnRNPM played a role in this process, we knocked down their expression using lentiviral shRNAs. However, loss of these proteins had no effect on the cLTP-dependent upregulation of mRNA into synapses (Fig. 7C, Scr, G, M). Moreover, loss of these proteins had no effect on the activity-dependent upregulation of poly(A) mRNA at dendrites (data not shown). These results suggest that hnRNPG and hnRNPM do not play a role in activity-dependent mRNA transport.
Figure 7.
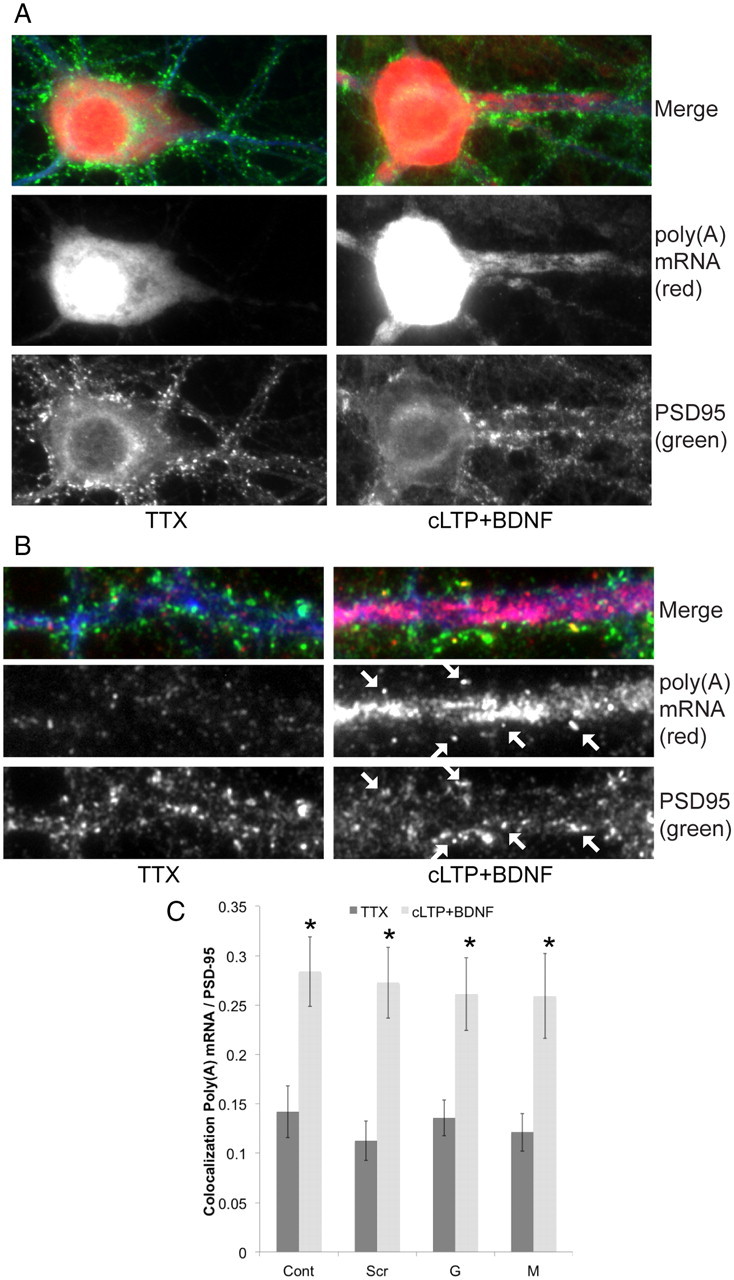
cLTP+BDNF stimulation results in increased dendritic and synaptic poly(A) mRNA in rat primary hippocampal neurons (DIV 17–21). A, cLTP+BDNF results in increased poly(A) mRNA labeled by FISH using oligo(dT)(50)-Cy3 probe staining in cell body and dendrites. Dendrites are marked using MAP2 immunostaining (blue). B, cLTP+BDNF results in increased poly(A) mRNA in dendrites and dendritic spines identified using PSD95 immunostaining (arrows). C, Quantitation of imaging results (ImageJ Mander's coefficients) shows cLTP+BNDF results in an increase in mRNA/PSD95 colocalization [control (cont)]. There was no statistical difference in the cLTP-induced increase in synaptic mRNA between RNABP knockdowns (G, M) or control neurons [scrambled shRNA (Scr), untransduced neurons (Cont)]. N = 250–370 puncta from at least eight neurons in three biological replicates. *p < 0.005 for the statistically significant increase in synaptic poly(A) mRNA at in response to cLTP+BDNF compared with TTX for each condition.
Discussion
Here we identified changes in PSD composition induced by cLTP+BDNF treatment using a modified version of SILAC. SILAC is an efficient method for precisely measuring the relative abundance of proteins in cell samples. One advantage of SILAC is that it allows for combining differentially labeled protein samples early in the sample preparation, thus minimizing variations in quantitation caused by parallel sample handling. This is important for this study since the cellular fractionation required for PSD purification from neuronal cell cultures is complex and could introduce substantial variability if the samples were processed separately. Standard SILAC requires complete metabolic labeling of proteins and is therefore difficult to apply to postmitotic cells such as primary neurons. To overcome this limitation, we have recently shown that by using two different sets of heavy amino acids for labeling, straightforward SILAC quantitation can be performed using partially labeled cells because the two cell populations are always equally labeled (Zhang et al., 2011). This enabled us to take full advantage of the high precision of SILAC to measure changes in the abundance of PSD components after cLTP induction.
In this study, we found that several RNABPs, including hnRNPs A1, A2/B1, A3, G, L, M, DEAD-box 17 and DEAD-box 9, demonstrated increased abundance at synapses in response to synaptic activity. RNABPs regulate all aspects of RNA metabolism by binding to and incorporating them into specialized ribonucleoparticles (Sossin and DesGroseillers, 2006). To date, a few RNABPs, including Staufen, CPEB, FMRP, ZBP1 (Martin and Zukin, 2006), and hnRNP A2/B1 (Hoek et al., 1998; Munro et al., 1999), have been implicated in neuronal mRNA transport and translation. While we identified FMRP, Staufen, and Pur α and β in this study, the levels of these proteins were either not altered in response to synaptic activity, or their level of change fell below our statistical cutoff (Fig. 5B,C). This could suggest that they either do not participate in activity-dependent processes, or that their function does not rely on changes in their abundance at synapses. Since we have included cycloheximide throughout the stimulation paradigms, the synaptic upregulation of the identified proteins could reflect an increase in the trafficking of these proteins into synapses. However, this change could also result from other cellular processes such as protein turnover via ubiquitination. Overall, our results suggest that members of the hnRNP class of RNABPs may play an important role in dendritic RNA metabolism and are worthy of further scientific research.
Interestingly, 10 of the 12 RNABPs identified have been previously shown to be components of RNA granules/transport particles in neurons (Kanai et al., 2004; Elvira et al., 2006). This suggests that synaptic activity results in the translocation of these ribonucleoparticles into synapses. Indeed, we find an increase in mRNA at synaptic sites in response to the same synaptic stimulation paradigm used in our proteomic screen (Fig. 7). Our results are consistent with previous findings showing that synaptic activity results in the trafficking of diverse mRNAs into dendrites and dendritic spines, including transcripts of BDNF, TrkB (Tongiorgi et al., 1997), CaMKIIα (Thomas et al., 1994; Rook et al., 2000; Håvik et al., 2003), ZBP1, Beta actin (Tiruchinapalli et al., 2003), and Arc (Link et al., 1995; Lyford et al., 1995; Schuman et al., 2006). However, hnRNPG or hnRNPM knockdown did not affect the cLTP-induced increase in synaptic mRNA. This suggests that either these proteins do not participate in mRNA transport, or that there are redundant pathways. Alternatively, given the number of RNABPs identified in this study, it is possible that the individual contribution of each RNABP to the total transport of mRNA into dendrites is not detectable.
The control of RNA distribution, stability, and translation is a fundamental mechanism for the local regulation of synaptic morphology and function. Translation of dendritically localized mRNAs (Miyashiro et al., 1994; Eberwine et al., 2002; Zhong et al., 2006) plays an important role in synaptogenesis (Lyles et al., 2006), synaptic plasticity (Miniaci et al., 2008; Wang et al., 2009), and regeneration after nerve injury (Weragoda et al., 2004). Indeed, activity-dependent changes in dendritic spines and neuronal morphology require protein synthesis (Engert and Bonhoeffer, 1999; Maletic-Savatic et al., 1999; Okamoto et al., 2002; Fukazawa et al., 2003; Matsuzaki et al., 2004; Tanaka et al., 2008; Holtmaat and Svoboda, 2009). We found that hnRNPG or hnRNPM knockdown altered spine density, suggesting that they regulate RNAs that play a role in cytoskeletal remodeling. Unfortunately, little is known about the RNA targets of hnRNPs G and M, and even less about their function outside the nucleus. One caveat to this study is that while we speculate that the change in spine density in response to hnRNPs G or M knockdown is due to the cytoplasmic and synaptic function of these proteins, we cannot rule out that the observed change is due to perturbations in their nuclear functions. Indeed, the X-linked RNABP hnRNPG has been shown to participate in alternative splicing of the Tra2B transcription factor (Venables et al., 2000) and may be required for neural development (Dichmann et al., 2008). HnRNPM has also been implicated in differential splicing by regulating exon-splice site selection, although a Drosophila homolog (Rumpelstiltskin) has been shown to regulate the localization of nanos mRNA and play a role in anterior/posterior patterning (Jain and Gavis, 2008). However, hnRNP A2/B1 (A2) has been implicated in RNA transport as a component of RNA transport particles in brain (Kanai et al., 2004) and plays a role in the trafficking of myelin basic protein mRNA (Hoek et al., 1998; Munro et al., 1999). We did not see any changes in spine density in response to knockdown of hnRNPs A2/B1 or D. Our SILAC results and verification of their synaptic distribution by immunocytochemistry suggest that these hnRNPs play a role in synaptic RNA metabolism. One interesting possibility is that these hnRNPs regulate RNA splicing outside the nucleus, as suggested by the growing evidence of intron-containing RNAs in neuronal dendrites (Bell et al., 2008, 2010; Buckley et al., 2011).
In this study, we initially expected to see changes in proteins traditionally associated with synaptic transmission, including glutamate receptors and their scaffolding molecules. For example, we found an activity-dependent upregulation of GluR1 at the PSD (25%) and a decrease of NMDA receptors NR1 (∼19%) and NR2B (∼13%) in the forward experiment that were reciprocated in our reverse experiment. However, these results did not pass our statistical filter. Therefore, we cannot rule out the possibility that our statistical criteria removed proteins displaying biologically meaningful changes after cLTP stimulation. This points to the difficulties in characterizing the dynamics of PSD composition using quantitative proteomics and identifies our limits of detection.
There is considerable interest in the mechanisms that underlie local regulation of protein abundance at synapses given their importance in synaptic function and dysfunction. Neuronal stimulation has been shown to target mRNAs selectively into activated spines (Steward et al., 1998; Steward and Worley, 2001; Moga et al., 2004), with translation occurring specifically at activated synapses (Wang et al., 2009). Improper targeting of mRNAs into dendrites results in unstable synaptic potentiation and deficits in spatial memory tasks in mice (Miller et al., 2002; Tzingounis and Nicoll, 2006). In humans, dysregulation of RNA distribution is the primary cause of several neurodegenerative disorders including myotonic dystrophy type 2; spinal cerebellar ataxias types 8, 10 and 12; and fragile X tremor ataxia syndrome (Ranum and Cooper, 2006). Some of these disorders result from expanded trinucleotide repeats at noncoding regions of RNA transcripts that cause their toxic accumulation at subnuclear structures (Ranum and Cooper, 2006). Other disorders, such as fragile X syndrome, result from null mutations of RNABPs involved in RNA transport/translation (Ranum and Cooper, 2006). Our results showing that RNABPs are upregulated at synapses in response to cLTP induction are consistent with these previous studies highlighting the importance of local RNA metabolism during plasticity and suggest that hnRNPs provide a layer of local regulation during plasticity-related events.
Notes
Supplemental material for this article is available at http://saturn.med.nyu.edu/files/mylab/neubert/Supplementary_table_1.xlsx. Complete list of proteins identified in isolated PSD fractions from the SILAC experiments. This material has not been peer reviewed.
Footnotes
This work was supported by grants from the NIH (K01-MH073759 and R01-AG039521 to B.A.J; P30 NS050276 and S10 RR017990 to T.A.N.). We thank Matthew Klein and Jaafar Tindi for their critical review of this manuscript.
The authors declare no financial conflicts of interest.
References
- Banker G, Churchill L, Cotman CW. Proteins of the postsynaptic density. J Cell Biol. 1974;63:456–465. doi: 10.1083/jcb.63.2.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell TJ, Miyashiro KY, Sul JY, McCullough R, Buckley PT, Jochems J, Meaney DF, Haydon P, Cantor C, Parsons TD, Eberwine J. Cytoplasmic BK(Ca) channel intron-containing mRNAs contribute to the intrinsic excitability of hippocampal neurons. Proc Natl Acad Sci U S A. 2008;105:1901–1906. doi: 10.1073/pnas.0711796105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell TJ, Miyashiro KY, Sul JY, Buckley PT, Lee MT, McCullough R, Jochems J, Kim J, Cantor CR, Parsons TD, Eberwine J. Intron retention facilitates splice variant diversity in calcium-activated big potassium channel populations. Proc Natl Acad Sci U S A. 2010;107:21152–21157. doi: 10.1073/pnas.1015264107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg F, Cohen RS, Siekevitz P. The structure of postsynaptic densities isolated from dog cerebral cortex. II. Characterization and arrangement of some of the major proteins within the structure. J Cell Biol. 1977;74:204–225. doi: 10.1083/jcb.74.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm J, Kang MG, Johnson RC, Esteban J, Huganir RL, Malinow R. Synaptic incorporation of AMPA receptors during LTP is controlled by a PKC phosphorylation site on GluR1. Neuron. 2006;51:213–225. doi: 10.1016/j.neuron.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Bourne JN, Harris KM. Balancing structure and function at hippocampal dendritic spines. Annu Rev Neurosci. 2008;31:47–67. doi: 10.1146/annurev.neuro.31.060407.125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley PT, Lee MT, Sul JY, Miyashiro KY, Bell TJ, Fisher SA, Kim J, Eberwine J. Cytoplasmic intron sequence-retaining transcripts can be dendritically targeted via ID element retrotransposons. Neuron. 2011;69:877–884. doi: 10.1016/j.neuron.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin RK, Grab DJ, Cohen RS, Siekevitz P. Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J Cell Biol. 1980;86:831–845. doi: 10.1083/jcb.86.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet D, Triller A. The role of receptor diffusion in the organization of the postsynaptic membrane. Nat Rev Neurosci. 2003;4:251–265. doi: 10.1038/nrn1077. [DOI] [PubMed] [Google Scholar]
- Cohen RS, Blomberg F, Berzins K, Siekevitz P. The structure of postsynaptic densities isolated from dog cerebral cortex. I. Overall morphology and protein composition. J Cell Biol. 1977;74:181–203. doi: 10.1083/jcb.74.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MO, Husi H, Yu L, Brandon JM, Anderson CN, Blackstock WP, Choudhary JS, Grant SG. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J Neurochem. 2006;97(Suppl 1):16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Dichmann DS, Fletcher RB, Harland RM. Expression cloning in Xenopus identifies RNA-binding proteins as regulators of embryogenesis and Rbmx as necessary for neural and muscle development. Dev Dyn. 2008;237:1755–1766. doi: 10.1002/dvdy.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobi A, Szemes M, Lee C, Palkovits M, Lim F, Gyorgy A, Mahan MA, Agoston DV. AUF1 is expressed in the developing brain, binds to AT-rich double-stranded DNA, and regulates enkephalin gene expression. J Biol Chem. 2006;281:28889–28900. doi: 10.1074/jbc.M511858200. [DOI] [PubMed] [Google Scholar]
- Dosemeci A, Tao-Cheng JH, Vinade L, Jaffe H. Preparation of postsynaptic density fraction from hippocampal slices and proteomic analysis. Biochem Biophys Res Commun. 2006;339:687–694. doi: 10.1016/j.bbrc.2005.11.069. [DOI] [PubMed] [Google Scholar]
- Eberwine J, Belt B, Kacharmina JE, Miyashiro K. Analysis of subcellularly localized mRNAs using in situ hybridization, mRNA amplification, and expression profiling. Neurochem Res. 2002;27:1065–1077. doi: 10.1023/a:1020956805307. [DOI] [PubMed] [Google Scholar]
- Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- Elvira G, Wasiak S, Blandford V, Tong XK, Serrano A, Fan X, del Rayo Sánchez-Carbente M, Servant F, Bell AW, Boismenu D, Lacaille JC, McPherson PS, DesGroseillers L, Sossin WS. Characterization of an RNA granule from developing brain. Mol Cell Proteomics. 2006;5:635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- Engert F, Bonhoeffer T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature. 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- Fukazawa Y, Saitoh Y, Ozawa F, Ohta Y, Mizuno K, Inokuchi K. Hippocampal LTP is accompanied by enhanced F-actin content within the dendritic spine that is essential for late LTP maintenance in vivo. Neuron. 2003;38:447–460. doi: 10.1016/s0896-6273(03)00206-x. [DOI] [PubMed] [Google Scholar]
- Håvik B, Røkke H, Bårdsen K, Davanger S, Bramham CR. Bursts of high-frequency stimulation trigger rapid delivery of pre-existing alpha-CaMKII mRNA to synapses: a mechanism in dendritic protein synthesis during long-term potentiation in adult awake rats. Eur J Neurosci. 2003;17:2679–2689. doi: 10.1046/j.1460-9568.2003.02712.x. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Kidd GJ, Carson JH, Smith R. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry. 1998;37:7021–7029. doi: 10.1021/bi9800247. [DOI] [PubMed] [Google Scholar]
- Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci. 2009;10:647–658. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jain RA, Gavis ER. The drosophila hnRNP M homolog Rumpelstiltskin regulates nanos mRNA localization. Development. 2008;135:973–982. doi: 10.1242/dev.015438. [DOI] [PubMed] [Google Scholar]
- Janas J, Skowronski J, Van Aelst L. Lentiviral delivery of RNAi in hippocampal neurons. Methods Enzymol. 2006;406:593–605. doi: 10.1016/S0076-6879(06)06046-0. [DOI] [PubMed] [Google Scholar]
- Ji Y, Lu Y, Yang F, Shen W, Tang TT, Feng L, Duan S, Lu B. Acute and gradual increases in BDNF concentration elicit distinct signaling and functions in neurons. Nat Neurosci. 2010;13:302–309. doi: 10.1038/nn.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan BA, Kreutz MR. Nucleocytoplasmic protein shuttling: the direct route in synapse-to-nucleus signaling. Trends Neurosci. 2009;32:392–401. doi: 10.1016/j.tins.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Jordan BA, Ziff EB. To the nucleus with proteomics. In: Dudek SM, editor. Transcriptional regulation by neuronal activity. New York: Springer; 2008. pp. 27–51. [Google Scholar]
- Jordan BA, Fernholz BD, Boussac M, Xu C, Grigorean G, Ziff EB, Neubert TA. Identification and verification of novel rodent postsynaptic density proteins. Mol Cell Proteomics. 2004;3:857–871. doi: 10.1074/mcp.M400045-MCP200. [DOI] [PubMed] [Google Scholar]
- Jordan BA, Fernholz BD, Neubert TA, Ziff EB. The dynamic synapse: molecular methods in ionotropic receptor biology. Boca Raton: CRC/Taylor and Francis; 2006. [PubMed] [Google Scholar]
- Jordan BA, Fernholz BD, Khatri L, Ziff EB. Activity-dependent AIDA-1 nuclear signaling regulates nucleolar numbers and protein synthesis in neurons. Nat Neurosci. 2007;10:427–435. doi: 10.1038/nn1867. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- Kennedy MB. The postsynaptic density. Curr Opin Neurobiol. 1993;3:732–737. doi: 10.1016/0959-4388(93)90145-o. [DOI] [PubMed] [Google Scholar]
- Lee C, Gyorgy A, Maric D, Sadri N, Schneider RJ, Barker JL, Lawson M, Agoston DV. Members of the NuRD chromatin remodeling complex interact with AUF1 in developing cortical neurons. Cereb Cortex. 2008;18:2909–2919. doi: 10.1093/cercor/bhn051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li KW, Hornshaw MP, Van Der Schors RC, Watson R, Tate S, Casetta B, Jimenez CR, Gouwenberg Y, Gundelfinger ED, Smalla KH, Smit AB. Proteomics analysis of rat brain postsynaptic density: implications of the diverse protein functional groups for the integration of synaptic physiology. J Biol Chem. 2004;279:987–1002. doi: 10.1074/jbc.M303116200. [DOI] [PubMed] [Google Scholar]
- Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci U S A. 1995;92:5734–5738. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J, Raghavachari S. A unified model of the presynaptic and postsynaptic changes during LTP at CA1 synapses. Sci STKE. 2006;2006:re11. doi: 10.1126/stke.3562006re11. [DOI] [PubMed] [Google Scholar]
- Lu W, Man H, Ju W, Trimble WS, MacDonald JF, Wang YT. Activation of synaptic NMDA receptors induces membrane insertion of new AMPA receptors and LTP in cultured hippocampal neurons. Neuron. 2001;29:243–254. doi: 10.1016/s0896-6273(01)00194-5. [DOI] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Lyles V, Zhao Y, Martin KC. Synapse formation and mRNA localization in cultured Aplysia neurons. Neuron. 2006;49:349–356. doi: 10.1016/j.neuron.2005.12.029. [DOI] [PubMed] [Google Scholar]
- Malenka RC. Synaptic plasticity and AMPA receptor trafficking. Ann N Y Acad Sci. 2003;1003:1–11. doi: 10.1196/annals.1300.001. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Maletic-Savatic M, Malinow R, Svoboda K. Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science. 1999;283:1923–1927. doi: 10.1126/science.283.5409.1923. [DOI] [PubMed] [Google Scholar]
- Martin KC, Zukin RS. RNA trafficking and local protein synthesis in dendrites: an overview. J Neurosci. 2006;26:7131–7134. doi: 10.1523/JNEUROSCI.1801-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Mayford M. Cellular and molecular mechanisms of memory: the LTP connection. Curr Opin Genet Dev. 1999;9:333–337. doi: 10.1016/s0959-437x(99)80050-1. [DOI] [PubMed] [Google Scholar]
- Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36:507–519. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- Milner B, Squire LR, Kandel ER. Cognitive neuroscience and the study of memory. Neuron. 1998;20:445–468. doi: 10.1016/s0896-6273(00)80987-3. [DOI] [PubMed] [Google Scholar]
- Miniaci MC, Kim JH, Puthanveettil SV, Si K, Zhu H, Kandel ER, Bailey CH. Sustained CPEB-dependent local protein synthesis is required to stabilize synaptic growth for persistence of long-term facilitation in Aplysia. Neuron. 2008;59:1024–1036. doi: 10.1016/j.neuron.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro K, Dichter M, Eberwine J. On the nature and differential distribution of mRNAs in hippocampal neurites: implications for neuronal functioning. Proc Natl Acad Sci U S A. 1994;91:10800–10804. doi: 10.1073/pnas.91.23.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moga DE, Calhoun ME, Chowdhury A, Worley P, Morrison JH, Shapiro ML. Activity-regulated cytoskeletal-associated protein is localized to recently activated excitatory synapses. Neuroscience. 2004;125:7–11. doi: 10.1016/j.neuroscience.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Munro TP, Magee RJ, Kidd GJ, Carson JH, Barbarese E, Smith LM, Smith R. Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J Biol Chem. 1999;274:34389–34395. doi: 10.1074/jbc.274.48.34389. [DOI] [PubMed] [Google Scholar]
- Oh MC, Derkach VA, Guire ES, Soderling TR. Extrasynaptic membrane trafficking regulated by GluR1 serine 845 phosphorylation primes AMPA receptors for long-term potentiation. J Biol Chem. 2006;281:752–758. doi: 10.1074/jbc.M509677200. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Li Z, Ju C, Scholzke MN, Mathews E, Cui J, Salvesen GS, Bossy-Wetzel E, Lipton SA. Dominant-interfering forms of MEF2 generated by caspase cleavage contribute to NMDA-induced neuronal apoptosis. Proc Natl Acad Sci U S A. 2002;99:3974–3979. doi: 10.1073/pnas.022036399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Osten P, Khatri L, Perez JL, Köhr G, Giese G, Daly C, Schulz TW, Wensky A, Lee LM, Ziff EB. Mutagenesis reveals a role for ABP/GRIP binding to GluR2 in synaptic surface accumulation of the AMPA receptor. Neuron. 2000;27:313–325. doi: 10.1016/s0896-6273(00)00039-8. [DOI] [PubMed] [Google Scholar]
- Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M. Semi-quantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J Biol Chem. 2004;279:21003–21011. doi: 10.1074/jbc.M400103200. [DOI] [PubMed] [Google Scholar]
- Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- Richter JD, Klann E. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 2009;23:1–11. doi: 10.1101/gad.1735809. [DOI] [PubMed] [Google Scholar]
- Rook MS, Lu M, Kosik KS. CaMKIIalpha 3′ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuman EM, Dynes JL, Steward O. Synaptic regulation of translation of dendritic mRNAs. J Neurosci. 2006;26:7143–7146. doi: 10.1523/JNEUROSCI.1796-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Sossin WS, DesGroseillers L. Intracellular trafficking of RNA in neurons. Traffic. 2006;7:1581–1589. doi: 10.1111/j.1600-0854.2006.00500.x. [DOI] [PubMed] [Google Scholar]
- Spellman DS, Deinhardt K, Darie CC, Chao MV, Neubert TA. Stable isotopic labeling by amino acids in cultured primary neurons: application to brain-derived neurotrophic factor-dependent phosphotyrosine-associated signaling. Mol Cell Proteomics. 2008;7:1067–1076. doi: 10.1074/mcp.M700387-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Worley PF. Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron. 2001;30:227–240. doi: 10.1016/s0896-6273(01)00275-6. [DOI] [PubMed] [Google Scholar]
- Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- Tanaka J, Horiike Y, Matsuzaki M, Miyazaki T, Ellis-Davies GC, Kasai H. Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science. 2008;319:1683–1687. doi: 10.1126/science.1152864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KL, Laroche S, Errington ML, Bliss TV, Hunt SP. Spatial and temporal changes in signal transduction pathways during LTP. Neuron. 1994;13:737–745. doi: 10.1016/0896-6273(94)90040-x. [DOI] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongiorgi E, Righi M, Cattaneo A. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J Neurosci. 1997;17:9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzingounis AV, Nicoll RA. Arc/Arg3.1: linking gene expression to synaptic plasticity and memory. Neuron. 2006;52:403–407. doi: 10.1016/j.neuron.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Venables JP, Elliott DJ, Makarova OV, Makarov EM, Cooke HJ, Eperon IC. RBMY, a probable human spermatogenesis factor, and other hnRNP G proteins interact with Tra2beta and affect splicing. Hum Mol Genet. 2000;9:685–694. doi: 10.1093/hmg/9.5.685. [DOI] [PubMed] [Google Scholar]
- Vinade L, Chang M, Schlief ML, Petersen JD, Reese TS, Tao-Cheng JH, Dosemeci A. Affinity purification of PSD-95-containing postsynaptic complexes. J Neurochem. 2003;87:1255–1261. doi: 10.1046/j.1471-4159.2003.02091.x. [DOI] [PubMed] [Google Scholar]
- Walikonis RS, Jensen ON, Mann M, Provance DW, Jr, Mercer JA, Kennedy MB. Identification of proteins in the postsynaptic density fraction by mass spectrometry. J Neurosci. 2000;20:4069–4080. doi: 10.1523/JNEUROSCI.20-11-04069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MJ, Kuruc N. The postsynaptic density: constituent and associated proteins characterized by electrophoresis, immunoblotting, and peptide sequencing. J Neurochem. 1992;59:667–678. doi: 10.1111/j.1471-4159.1992.tb09421.x. [DOI] [PubMed] [Google Scholar]
- Wang DO, Kim SM, Zhao Y, Hwang H, Miura SK, Sossin WS, Martin KC. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science. 2009;324:1536–1540. doi: 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weragoda RM, Ferrer E, Walters ET. Memory-like alterations in Aplysia axons after nerve injury or localized depolarization. J Neurosci. 2004;24:10393–10401. doi: 10.1523/JNEUROSCI.2329-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y, Yamauchi Y, Shinkawa T, Taoka M, Donai H, Takahashi N, Isobe T, Yamauchi T. Molecular constituents of the postsynaptic density fraction revealed by proteomic analysis using multidimensional liquid chromatography-tandem mass spectrometry. J Neurochem. 2004;88:759–768. doi: 10.1046/j.1471-4159.2003.02136.x. [DOI] [PubMed] [Google Scholar]
- Zhang G, Deinhardt K, Chao MV, Neubert TA. Study of neurotrophin-3 signaling in primary cultured neurons using multiplex stable isotope labeling with amino acids in cell culture. J Proteome Res. 2011;10:2546–2554. doi: 10.1021/pr200016n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Zhang T, Bloch LM. Dendritic mRNAs encode diversified functionalities in hippocampal pyramidal neurons. BMC Neurosci. 2006;7:17. doi: 10.1186/1471-2202-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziff EB. Enlightening the postsynaptic density. Neuron. 1997;19:1163–1174. doi: 10.1016/s0896-6273(00)80409-2. [DOI] [PubMed] [Google Scholar]