Abstract
The human immunodeficiency virus (HIV) requires a programmed −1 ribosomal frameshift for Pol gene expression. The HIV frameshift site consists of a heptanucleotide slippery sequence (UUUUUUA) followed by a spacer region and a downstream RNA stem–loop structure. Here we investigate the role of the RNA structure in promoting the −1 frameshift. The stem–loop was systematically altered to decouple the contributions of local and overall thermodynamic stability towards frameshift efficiency. No correlation between overall stability and frameshift efficiency is observed. In contrast, there is a strong correlation between frameshift efficiency and the local thermodynamic stability of the first 3–4 bp in the stem–loop, which are predicted to reside at the opening of the mRNA entrance channel when the ribosome is paused at the slippery site. Insertion or deletions in the spacer region appear to correspondingly change the identity of the base pairs encountered 8 nt downstream of the slippery site. Finally, the role of the surrounding genomic secondary structure was investigated and found to have a modest impact on frameshift efficiency, consistent with the hypothesis that the genomic secondary structure attenuates frameshifting by affecting the overall rate of translation.
INTRODUCTION
Translation is a high-fidelity process in all organisms. Failure to maintain reading frame typically results in incorrect protein synthesis and/or early termination. However, a programmed change in reading frame can result in the translation of new proteins, thereby maximizing genomic coding capacity. Many retroviruses, including human immunodeficiency virus type 1 (HIV-1) (1), and some coronaviruses, such as severe acute respiratory syndrome (2) and infectious bronchitis virus (IBV) (3), use a programmed −1 ribosomal frameshift (−1 PRF) to control translation levels of their enzymatic proteins (4–7). In the retroviruses, the −1 PRF site lies between the gag and pol open reading frames (ORFs), with pol in the −1 reading frame relative to gag. The gag ORF encodes the viral structural proteins, whereas the pol ORF encodes the enzymatic proteins. During translation of HIV-1 mRNA, the majority of ribosomes terminate at a stop codon at the end of the gag ORF, producing the Gag polyprotein (2,8). However, the HIV −1 PRF induces ∼5% of ribosomes to shift into the −1 reading frame, thus producing the Gag–Pol polyprotein (1,9–11). The 5% frameshift efficiency determines the ratio of viral proteins produced and is important for viral replication and infectivity (10,12–15). A decrease in frameshift efficiency can inhibit viral replication (16,17).
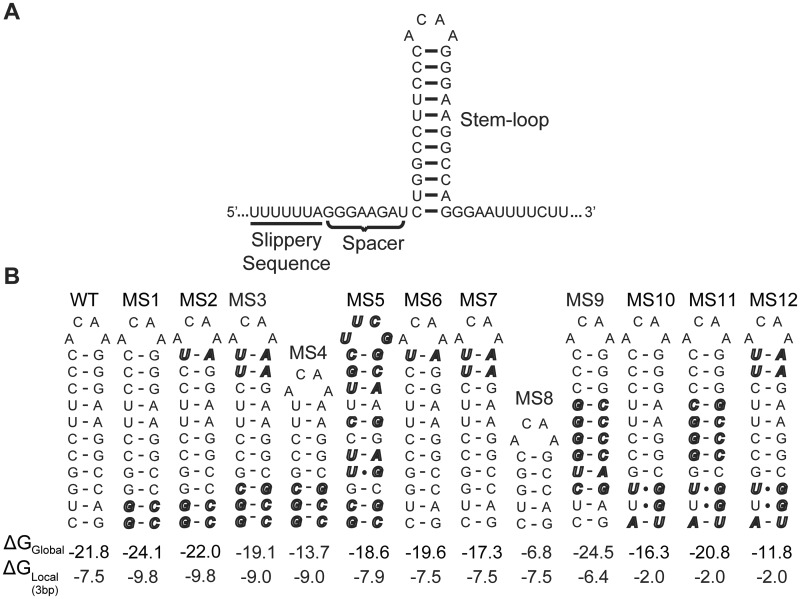
The HIV-1 frameshift site is composed of a heptanucleotide slippery sequence (UUUUUUA) followed by a downstream RNA stem–loop (Figure 1A). The slippery sequence follows a general XXXYYYZ consensus sequence, where X can be any nucleotide (nt) type, Y can be A or U and Z is not G in eukaryotes (15,18). This sequence allows near-cognate and cognate re-pairing of the A- and P-site tRNA anticodons, respectively, in the −1 reading frame. HIV-1’s slippery sequence is especially ‘slippery’, and in the absence of a downstream structure increases the basal level of ribosomal frameshifting from ∼0.0001% to 0.1% per codon (9,19,20). However, in order to further stimulate frameshifting to the levels required for viral replication, the slippery site must be followed by a stable RNA structure (9,21–30) (Figure 1A). Thus, frameshifting is achieved by the cis coupling of the slippery site and downstream structure (1,9–11,21).
Figure 1.
The HIV-1 frameshift site. (A) Two cis-acting RNA elements are separated by a single-stranded spacer. (B) Twelve MS RNA constructs were designed to discern the relative contributions of local and global RNA stability on HIV-1 frameshift efficiency. Sequence changes are indicated in bold and italicized. Predicted overall, ΔGGlobal and local, ΔGLocal, thermodynamic stabilities are in units of kcal/mol.
Multiple models have been proposed to explain the frameshift mechanism (1,6,31–38). Common among them are the following steps: (i) during translation, the ribosome pauses when the slippery sequence (UUU UUA in the 0 frame) is engaged in the ribosomal A- and P-sites (4,18,22,39). The pause is triggered by the downstream structure’s resistance to unwinding. (ii) While paused, ∼5% of ribosomes slip 1 nt in the 5′-direction and continue elongation in the −1 reading frame. The proposed models are differentiated by the exact step at which the frameshift occurs: during aminoacylated-tRNA accommodation (31), after accommodation, but before peptidyl transfer (1), after large subunit translocation (5) or after peptidyl transfer due to an incomplete translocation (37). Alternatively, the ‘many pathways model’ of −1 PRF suggests that frameshift efficiency is the sum of frameshift events occurring, each of which could occur at these different points in elongation (36).
An important role of the downstream structure is to induce ribosomal pausing on the slippery sequence, which is necessary but not sufficient to promote efficient levels of frameshifting (40–42). Interestingly, the pause length does not appear to correlate with frameshift efficiency (40). Interactions with the translational machinery have also been hypothesized to contribute to frameshifting (5,23,31,32,43). Previous studies have observed general trends between HIV-1 stem–loop thermodynamic stability and frameshift efficiency (30,44,45). However, a quantitative correlation between thermodynamic stability and frameshift efficiency has not been described, and the role of individual base pairs has not been systematically investigated.
For frameshift sites with a downstream pseudoknot structure, mechanical stability has been proposed to be a determinant of frameshift efficiency (46–48). It has been hypothesized that mechanical tension lowers the energy barrier for frameshifting (5), where the amount of tension sensed by the ribosome is proportional to the mechanical stability of the translocation barrier (46,47,49–51). However, a recent study found no correlation between pseudoknot mechanical stability and frameshift efficiency, but instead observed a correlation between frameshifting and the ability to form alternative structures (52).
Other factors can modulate the frameshift efficiency, such as translation initiation rates (37,53). Increased translation initiation rates lead to increased polysome density, which can cause ribosomes to stack at the frameshift site. This in turn affects the rate of mRNA refolding during translation and leads to a decrease in overall frameshift efficiency (37,53). Ribosome stacking can be promoted by RNA structure that precedes the frameshift site. Studies examining the secondary structure of the HIV-1 genomic RNA within capsids have revealed that the frameshift site is part of a conserved three-helix junction (3HJ) (54,55). It has been hypothesized that the role of this secondary structure is to decrease the rate of translation (54), which may affect frameshifting by facilitating pausing and inducing ribosome stacking.
Here, we investigate the role of the HIV-1 RNA structure in frameshifting, focusing on elucidating the relationships between frameshift efficiency and (i) the downstream RNA stem–loop thermodynamic stability, (ii) spacer length and (iii) surrounding genomic secondary structure. By systematically altering the base pair composition of the stem–loop, we dissect the contributions of global and local thermodynamic stability on frameshifting. These data reveal that the thermodynamic stability of the first 3–4 bp in the stem–loop is a primary determinant of frameshift efficiency. Our data further indicate that the base pairs important for frameshifting are located at a distance of 8 nt from the slippery site, which corresponds to the length of the spacer and is consistent with a structural model of the ribosome paused at the frameshift site. Finally, we find that the conserved genomic RNA secondary structure serves to attenuate the frameshift efficiency, likely by affecting the overall rate of translation. Importantly, our study describes the first quantitative and predictive model for frameshift inducing stem–loops, which can be generally applied to many −1 PRF viral systems.
MATERIALS AND METHODS
Plasmid construction
DNA templates used for the dual-luciferase frameshift assay were cloned into a p2luc vector between the rluc and fluc reporter genes. Briefly, complementary synthetic oligonucleotides [Integrated DNA Technologies (IDT), Inc.] with BamH I and Sac I compatible ends were cloned into the p2luc vector using the BamH I and Sac I sites between the rluc and fluc reporter genes. Oligonucleotides comprising the template sequences (Supplementary Table S1) and their complements were phosphorylated, annealed and ligated into the p2luc vector to produce the experimental constructs. This places the fluc gene in the −1 reading frame relative to rluc; analogous to the orientation of the gag and pol genes in the HIV-1 genome. For the spacer mutation constructs (MS13–17), a compensatory number of nts were added or removed downstream of the frameshift site to maintain the appropriate reading frame of the downstream reporter gene. The ‘wild-type’ (WT) sequence utilized here corresponds to the most frequently occurring sequence found in HIV-1 group M subtype B NL4–3 laboratory strain (56). Positive control sequences and their complements were also cloned into the p2luc vector and have two thymidine residues (Supplementary Table S1, bold) in the slippery sequence (Supplementary Table S1, underlined) replaced with cytidines, and an additional nt inserted immediately before the Sac I complementary sequence (GAGCT), which places the rluc and fluc genes in-frame. In all constructs, a Pml I restriction site was included at the end of the template to allow for run-off transcription after digestion with the Pml I enzyme (NEB). Resultant products were transformed into Escherichia coli competent cells (DH5α). Plasmid DNA was purified from cell cultures (Qiagen) and the sequences of all constructs were verified (University of Wisconsin-Madison Biotechnology Center).
RNA synthesis and purification
Microgram quantities of RNA for the frameshift assay were transcribed in vitro using linearized p2luc plasmid DNA, purified His6-tagged T7 RNA polymerase (10×), 11.25 mM NTPs and two units of RNasin Plus RNase Inhibitor (Promega), in 200 µl for 90 min at 37°C. Pyrophosphate was pelleted by centrifugation (10 min, 13 200 rpm, room temperature) and RNA was phenol/chloroform extracted. Unincorporated NTPs and salt were separated from the RNA using size-exclusion chromatography [two Econo-Pac P6 cartridges (Bio-Rad) in series]. Monomeric RNA folding was achieved by denaturation at 95°C for 5 min followed by incubation on ice for 30 min. RNAs were lyophilized to dryness and resuspended in water to a concentration of 1 µg/µl and stored in aliquots (∼25 µl) at −80°C. RNA integrity and purity were checked with 1% agarose gel electrophoresis. Finally, RNAs used for UV spectroscopy were purchased from IDT.
Frameshift assay
In vitro frameshift assays were completed with each RNA reporter (experimental and positive control) using a Rabbit Reticulocyte Lysate (RRL) System (Promega, nuclease treated, L416A). Differences from our previously described protocol (56) include the following: translation reactions contained 1.25 µg RNA, 10 units of RNasin Plus RNase Inhibitor (Promega, N2615), and 8.75 µl of RRL in 12.5 µl. Following a 90-min incubation at 37°C, reactions were quenched with the addition of 0.5 µl 0.156 M EDTA pH 8.0 (6 mM final), as described previously (28). For each reporter, a minimum of three independent frameshift assays were completed. Each independent assay included six replicate reactions.
Luminescence was measured using the dual-luciferase reporter assay (Promega) as previously described (57). Readings were taken with a Veritas microplate luminometer equipped with dual-injectors (Turner Bio-systems) for 10 s after 25 µl of the respective substrate was injected into the reaction mixture (2-s lag time prior to measurement). Ratios of firefly/Renilla luminescence were calculated for each of the experimental and control translation reactions. The frameshift efficiency was calculated by taking the ratio of the experimental/control luminescence (firefly/Renilla). Frameshift efficiencies were averaged and their standard deviations were propagated through to yield a standard error of the mean (SEM).
ΔGGlobal and ΔGLocal measurements
For the WT and a subset of the mutant stem–loop (MS) RNAs (Supplementary Table S2 and Figure 2), RNA overall thermodynamic stability, ΔGGlobal, was measured using UV absorbance at 260 nm as a function of temperature with a Cary Model 400 Bio UV-visible spectrophotometer equipped with a Peltier heating accessory and temperature probe. All samples contained 10 mM potassium phosphate buffer, pH 7.0, 2 µM RNA, in a volume of 1 ml. For RNAs that were too stable to measure ΔGGlobal under these conditions, urea was added to 4, 6 and 8 M, and the ΔGGlobal was deduced by extrapolating to 0 M urea as described below. Prior to data collection, samples were heated from 20°C to 95°C, at 10°C/min, held at 95°C for 5 min and cooled from 95°C back to 20°C at the same rate to ensure homogenous folding. Samples were heated at 1°C/min from 20°C to 95°C. Identical traces were obtained by cooling, indicating a lack of hysteresis. A260 data were collected in 0.5-min intervals and raw data were baseline corrected by subtraction of A320 values at each temperature. The average hyperchromicity [Equation (1)] and temperature were calculated from four curves and the SEM was determined for each average.
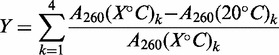 |
(1) |
Figure 2.
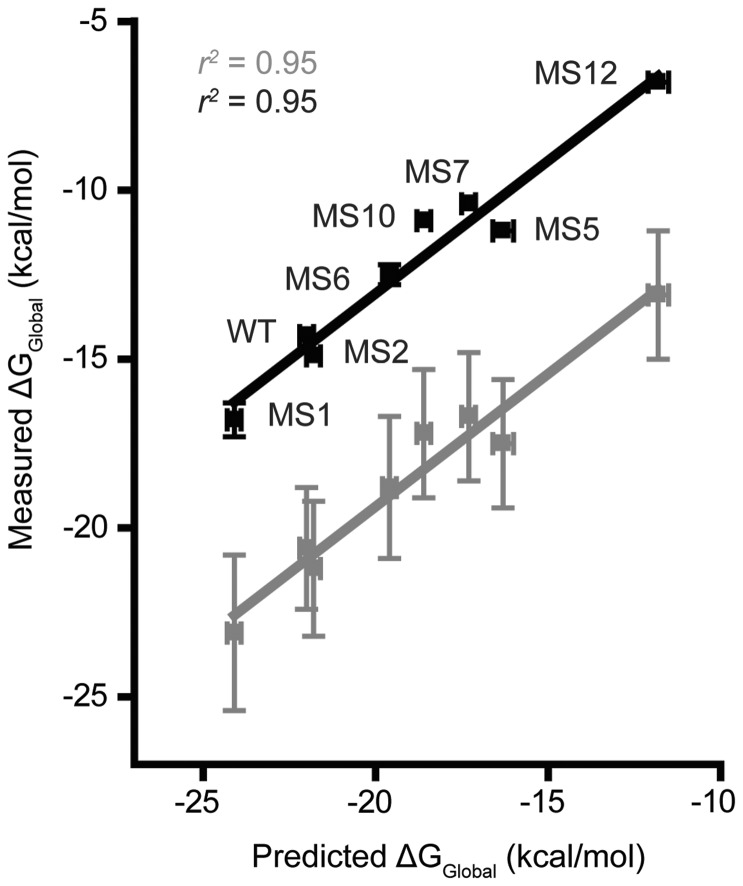
Measured ΔGGlobal values versus predicted values. Black symbols, data predicted with RNA structure (69) using revised UG parameters (61) (r2 = 0.95). Gray symbols, experimental data corrected to 1 M monovalent ionic strength (r2 = 0.95). A linear regression function was used to fit both data sets.
For RNA with a single melting transition, the average hyperchromicity can be fit by Equation (2) to measure ΔH and Tm.
| (2) |
Here, Af and Au are the temperature-dependent A260 of the folded and unfolded forms of the RNA, determined to be linear functions of the temperature. K is given by Equation (3), where T is the desired temperature (Kelvin, 310K for our calculations) for the ΔG calculation, and R is the gas constant in units of kcal/(mol × K).
| (3) |
With an average Tm and ΔH extrapolated using Equation (2), assuming a ΔCp of zero, the ΔGGlobal can be calculated using Equation (4). Error in ΔGGlobal is calculated using standard propagation of error (Supplementary ‘Materials and Methods’ section).
| (4) |
Average hyperchromicity data for each RNA were fit using Equation (2) and overall thermodynamic stabilities were calculated using Equation (4) (Prism 4.3, GraphPad). For RNAs with melting temperatures approaching or >95°C, a linear extrapolation of ΔGGlobal versus urea concentration was applied to determine the ΔGGlobal at 0 M urea (Supplementary Figure S1) (58). For all other RNAs, determination of ΔGGlobal at standard buffer conditions was sufficient to produce minimal error in ΔGGlobal. All the data used to calculate the reported ΔGGlobal values were established using a minimum of three independently prepared samples at each buffer condition. A small and non-cooperative transition was observed in the 40–50°C range for RNAs with less stable terminal base pairs (WT, MS2, MS6, MS7, MS10 and MS12). This transition was not present for RNAs with very stable terminal base pairs (e.g. MS1 and MS5) and can be attributed to helical fraying. For all RNAs, only the major cooperative unfolding transitions were used in data fitting. Starting values were determined by examining the first derivative plots for each set of averages. Local stabilities, ΔGLocal, for base pairs were calculated using nearest-neighbor parameters at 1 M NaCl, 37°C (59–61) (Supplementary Table S2).
Correlation between frameshift efficiency and RNA stability
Frameshift efficiency was plotted as a function of overall and local thermodynamic stability. Overall and local thermodynamic stabilities were predicted at 1 M NaCl. Data were fit to a one-phase exponential decay function [Equation (5)] (Prism 4.3, GraphPad). Here, the Amplitude, K and Plateau are variables and Xo is used to offset the exponential fit. Xo was set to the most negative X value in the data set. Errors were determined using a 95% confidence interval.
| (5) |
Modeling the HIV-1 frameshift site onto the eukaryotic ribosome
The 3.1 Å crystal structure of the Thermus thermophilus 30 S ribosomal subunit in complex with mRNA and tRNAs (62), PDB ID 3I8G, was aligned to the 3.0 Å crystal structure of the Saccharomyces cerevisiae 40S ribosomal subunit (63), PDB IDs 3U5B and 3U5C, over 20 conserved nts in the rRNA decoding center (63–66) (residues 780–800 in 3I8G and 991–1011 in 3U5B). The 5′-end of the HIV-1 frameshift site stem–loop, PDB ID 1Z2J (25) (residues 7–35), was connected to mRNA extending from the A-site, PDB ID 3I8G (62). The HIV-1 stem–loop was oriented to prevent steric clash using pymol (The PyMOL Molecular Graphics System, Version 1.5, Schrödinger, LLC).
RESULTS
Frameshift efficiency correlates with local stability of the HIV-1 frameshift site stem–loop
We utilized a well-established dual-luciferase frameshift assay (57,67,68) to quantitatively measure frameshift efficiency in RRL. The sequence of the frameshift site stem–loop was varied to dissect the relative contributions of local and overall RNA stability on HIV-1 frameshift efficiency. We hypothesized that once the ribosome is paused on the slippery sequence, the thermodynamic stability of the base pairs encountered at the base of the stem–loop should be a critical determinant for frameshifting. After the ribosome has completed one translocation step, it moves away from the slippery site and the reading frame is established. Therefore, we further hypothesized that downstream base pairs in the stem–loop should have a much lower impact on frameshifting. To test this hypothesis, 12 MS constructs (Figure 1B) were created using nearest-neighbor parameters (59–61,69) to systematically alter the stability of different regions of the stem–loop. We define local stability (ΔGLocal) as the thermodynamic stability of base pairs directly downstream of the spacer (Figure 1A), as determined by their nearest-neighbor interactions (59–61). Global stability (ΔGGlobal) is defined as the overall thermodynamic stability of the stem–loop.
The thermodynamic stabilities (ΔGGlobal) were experimentally determined for a subset or RNAs (WT, MS1-2, MS5-7, MS10 and MS12) using UV-monitored thermal denaturation (Figure 2, Supplementary Figure S1 and Supplementary Table S2). Owing to the extreme stabilities of the structures (25), thermal denaturation curves were measured at low ionic strength (10 mM potassium phosphate buffer) in the presence of varying concentrations of urea and extrapolated back to 0 M urea (58). Results followed the same trend as those predicted from nearest-neighbor parameters (59–61,69) (R2 = 0.95) (Figure 2). As expected, the measured stabilities were lower than the predicted values at 1 M NaCl (70). Upon correction of the experimental values to 1 M monovalent ionic strength (71), we observe an excellent agreement (R2 = 0.95) between experimental and predicted free energies (Figure 2). Indeed, free energy prediction is robust for small, stable RNAs with no competing suboptimal folds (20).
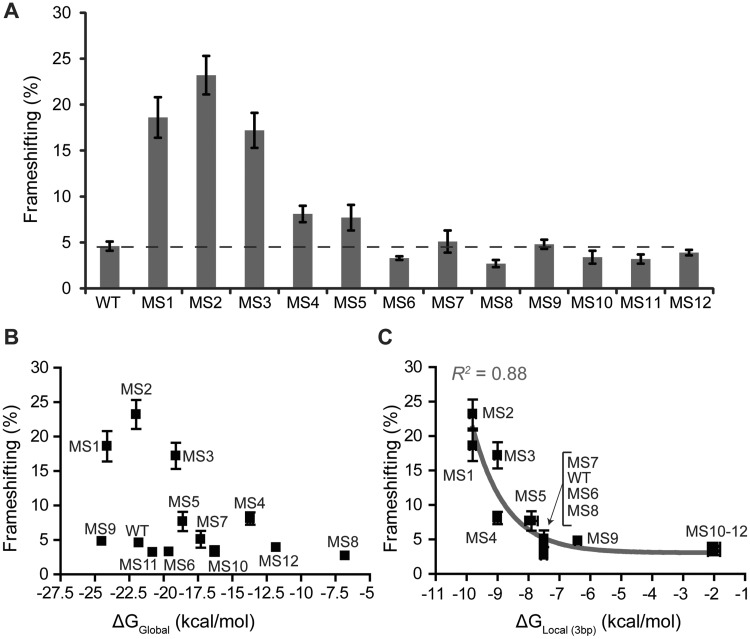
Frameshift efficiencies for the different RNA constructs were measured (Figure 3 and Table 1). Increases in the local stability of the first 3 bp resulted in significant increases in frameshift efficiency (Figure 3A, MS1–5). In contrast, sequence changes that significantly lowered the local stability of the first 3 bp resulted in decreased frameshift efficiencies (Figure 3A, MS10–12 and Table 1). No correlation between frameshift efficiency and overall thermodynamic stability is observed (Figure 3B). Instead, we observe a strong correlation (R2 = 0.88) between frameshift efficiency and local stability of the first 3 bp at the base of the stem–loop using a one-phase exponential decay function (Figure 3C and Supplementary Table S3). The frameshift efficiency for each variant frameshift site can be predicted using the parameters derived from the correlation, and each predicted frameshift efficiency falls within 1 SD of its measured value (data not shown). These results support the hypothesis that the stability of the base pairs at the base of the stem–loop is a primary determinant of frameshift efficiency.
Figure 3.
Frameshift efficiency is plotted as a function of RNA stability. Data represent an average of at least three replicates, with the SEM indicated. (A) In vitro frameshift assay results for WT and MS1-12 RNAs. Error bars representing the SEM are shown. The dashed line is indicative of the WT mean. (B) Overall thermodynamic stability (at 1 M NaCl) versus frameshift efficiency. (C) Local stability (at 1 M NaCl) of first 3 bp versus frameshift efficiency. A single exponential decay function was fit to the data [y = 18(±2)e−0.9(±0.3)(X + 9.8) + 3(±1), R2 = 0.88].
Table 1.
In vitro frameshift efficiencies
| RNA | Frameshift Efficiency (%) (±SEM) |
|---|---|
| WT | 4.6 (0.5) |
| MS1 | 18.6 (2.2) |
| MS2 | 23.2 (2.1) |
| MS3 | 17.2 (1.9) |
| MS4 | 8.1 (0.9) |
| MS5 | 7.7 (1.4) |
| MS6 | 3.3 (0.2) |
| MS7 | 5.1 (1.2) |
| MS8 | 2.7 (0.4) |
| MS9 | 4.8 (0.5) |
| MS10 | 3.4 (0.7) |
| MS11 | 3.2 (0.5) |
| MS12 | 3.9 (0.3) |
| MS13 | 5.4 (0.6) |
| MS14 | 7.8 (0.7) |
| MS15 | 13.8 (1.4) |
| MS16 | 5.6 (0.4) |
| MS17 | 5.2 (0.5) |
| 3HJ WT | 2.5 (0.4) |
| 3HJ Mut | 4.7 (0.5) |
| 3HJ MS1 | 12.2 (0.2) |
Extremely stable RNA structures can promote long-term ribosomal stalling or ‘roadblocking’, (35,72). In the dual-luciferase assay, roadblocking would result in decreased translation levels of the downstream firefly luciferase reporter gene product. However, the dual-luciferase assay controls for this, as frameshift efficiencies are normalized relative to in-frame control constructs (57,67,68). Nevertheless, we asked if differential degrees of roadblocking might occur for our various constructs. The luminescence data reveal a consistent ratio of firefly/Renilla activity (data not shown) for all constructs (Table 1). These values were calculated using the luminescence data from the positive control dual-luciferase constructs, where the Renilla and firefly genes are in frame and the slippery site is mutated such that the ribosome cannot frameshift. The consistency in the relative expression levels of the reporter genes indicates that roadblocking, if occurring, is uniform for all constructs.
Influence of spacer length in frameshifting
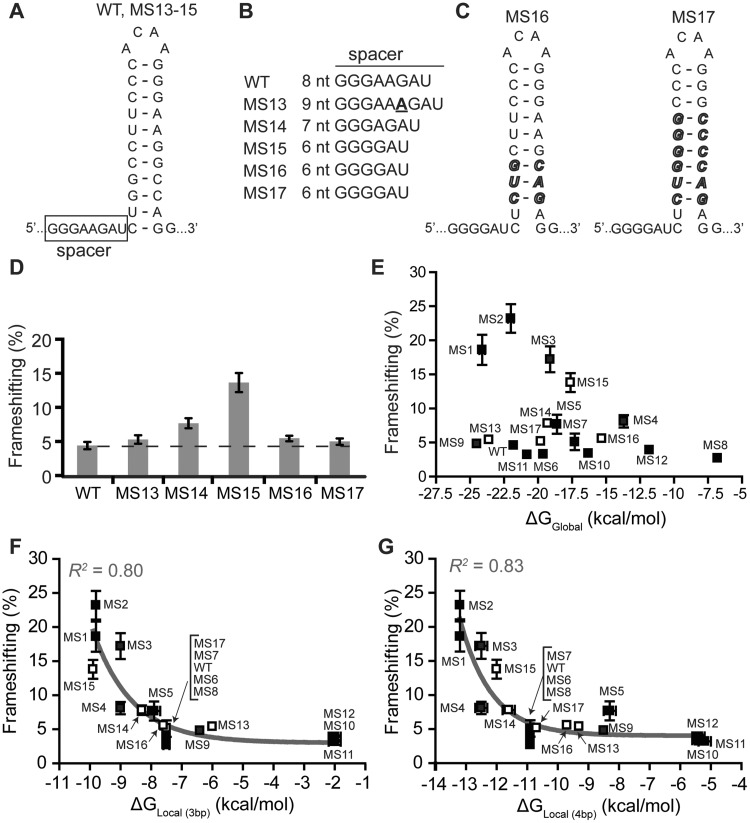
It has been hypothesized that during frameshifting, the mechanical force of translocation causes a build-up of tension that is transmitted through the spacer region (Figure 1A) and sensed at the anticodon–codon level (5,49). We therefore investigated the influence of nt deletions and insertion in the spacer region (Figure 4). The WT construct was compared to a version with an adenosine insertion that increases the spacer length by 1 nt (MS13) (Figure 4). Additionally, we created spacers with a single nt deletion (MS14) and 2-nt deletions (MS15–17) (Figure 4). The resulting frameshift efficiencies were measured (Figure 4D). Interestingly, MS15 shows a large increase in frameshift efficiency. This cannot be due to the 2-nt deletion, since MS16 and MS17 have the same spacers yet display WT-levels of frameshifting. We hypothesized that the 2-nt deletion in the spacer of MS15 increased frameshift efficiency by altering the base pairs in the stem–loop encountered by the ribosome during frameshifting. In other words, by deleting 2 nt in the spacer, a ribosome footprint may extend 2 nt further into the stem. In support of this hypothesis, a very stable set of base pairs are located 2 nt from the base of the stem (5′-GGC-3′/5′-GCC-3′). In MS16 and MS17, we replaced these base pairs with the less stable base pairs normally found at the base of the stem (5′-CUG-3′/5′-CAG-3′) (Figure 4C). Indeed, when these changes are made, the frameshifting efficiency is indistinguishable from WT, despite the apparent 2-nt spacer deletion (Figure 4D). Interestingly, the overall stability of MS17 is increased relative to MS16 (Figure 4C), yet the frameshift efficiency is unaltered (Figure 4D). These data indicate that changes in the spacer region correspondingly alter the base pairs encountered by the ribosome when it is engaged with the slippery site.
Figure 4.
The role of the spacer in frameshift stimulation. (A) Changes in spacer and stem–loop sequence are shown for MS13–17. (B) Spacer sequences for WT and MS13–17, with nt insertions shown in bold and underlined. (C) Changes in stem–loop sequence for MS16–17 are bold and italicized. (D) In vitro frameshift assay results for the WT and MS13–17 RNAs. The dashed line is indicative of the WT mean. Data represent an average of at least three replicates, with the SEM indicated. (E) Overall thermodynamic stability (at 1 M NaCl) versus frameshift efficiency. (F) Local stability (3 bp, at 1 M NaCl) versus frameshift efficiency. A single exponential decay function was fit to the data [y = 16(±2)e−0.8(±0.2)(X + 9.9) + 3(±1), R2 = 0.80]. (G) Local stability (4 bp, at 1 M NaCl) versus frameshift efficiency. A single exponential decay function was fit to the data [y = 9(±2)e−1.0(±0.3)(X + 12.5) + 4(±1), R2 = 0.83].
When plotting the data from all 18 RNA constructs studied (including the MS13–17 RNAs) as a function of overall RNA stability, no correlation is observed (Figure 4E). However, we observe a strong correlation between frameshifting and the thermodynamic stability of the first 3–4 bp 8 nt downstream of the slippery site (Figure 4F and G). Conversely, the correlations grow considerably weaker as more base pairs are considered in the analysis (Supplementary Figure S2). Likewise, no correlation is observed between local stability of base pairs at the top of the stem–loop and frameshift efficiency (Supplementary Figure S2L). The observed correlations are exponential functions with baselines of 2–4% frameshifting, which correspond to the lowest observed frameshift efficiencies in the presence of a stem–loop secondary structure downstream of the slippery site.
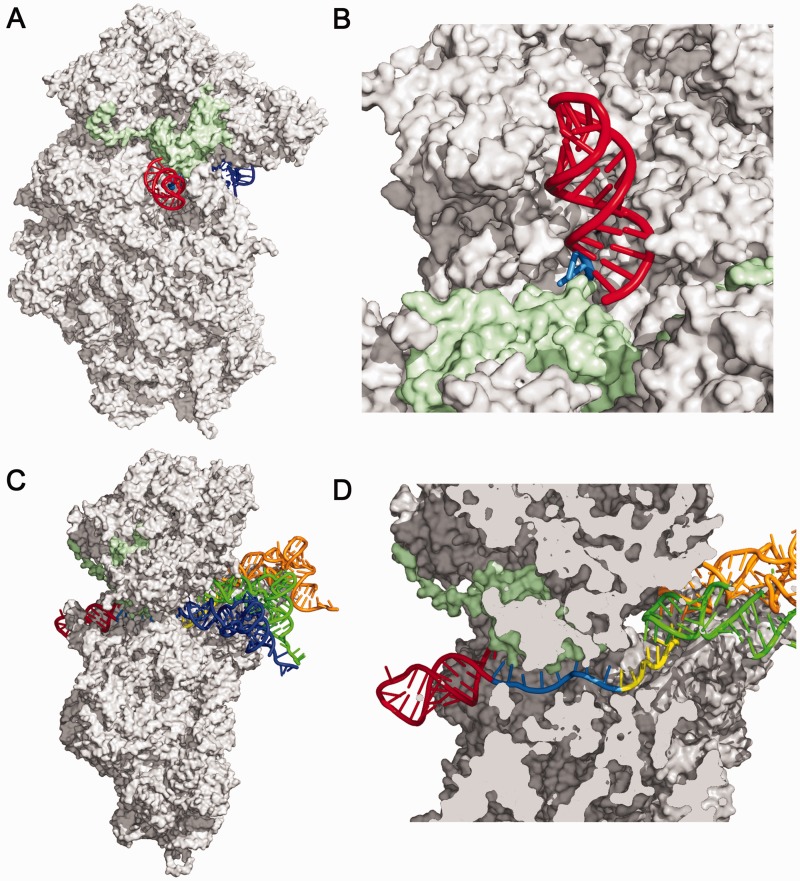
The stem–loop is flanked by a 5′-U and 3′-G (Figure 1A), that could potentially form a U–G wobble at the base of the stem. Inclusion of this wobble pair in the local stability term produces consistently weaker correlations between frameshift efficiency and local stability (Supplementary Figure S3). To further address whether or not this U–G wobble pair can form during frameshifting, we modeled the frameshift site stem–loop and spacer onto the eukaryotic ribosome (Figure 5). The spacer was connected to the terminal nt in the A-site, to recapitulate the position of the ribosome when it is engaged on the slippery sequence in the 0 reading frame. The model indicates that the minimal spacer distance between the slippery site and the stem–loop is 7 nt; however, formation of a U–G wobble at the base of the stem is blocked by steric clash with the ribosomal S3 protein (Figure 5B and D). Therefore, experimental data and structural modeling support an HIV-1 frameshift site spacer length of 8 nt.
Figure 5.
The HIV-1 frameshift site stem–loop modeled onto the eukaryotic 40S S. cerevisiae ribosomal subunit, PDB IDs 3U5B and 3U5C, using tRNA and mRNA coordinates from the 30S T. thermophilus structure, PDB ID 3I8G. (A) Ribosomal RNA and proteins are shown in gray, with ribosomal protein S3 surface in light green and the HIV-1 stem–loop in red. Part of the A site tRNA is visible in dark blue. (B) Close-up view of the HIV-1 stem–loop on the surface of the ribosome, tilted and rotated ∼180° relative to (A). The potential U–G wobble pair would occur between the last U in the spacer (light blue) and the G (red) at the base of the stem. (C) ‘Side’ view. tRNAs from the 30S T. thermophilus, PDB ID 3I8G, are blue, light green and orange for the A-, P- and E-sites, respectively. (D) Cut-away view showing the path of the mRNA. The mRNA corresponding to the slippery site is shown in yellow, and the 8-nt spacers are shown in light blue.
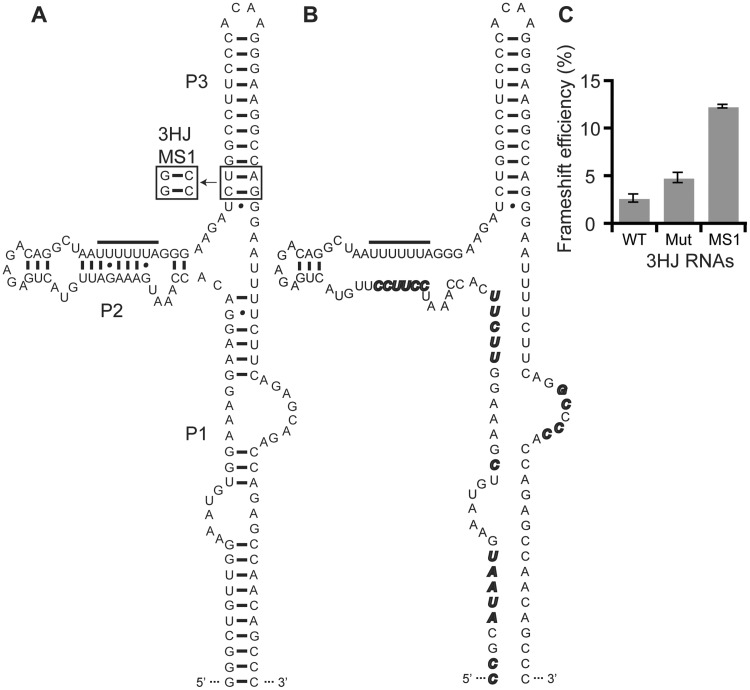
The HIV-1 3HJ modulates frameshift efficiency
Within viral capsids, the HIV-1 frameshift site RNA is part of a conserved 3HJ secondary structure (Figure 6A) (54,55). It has been hypothesized that the role of this secondary structure is to slow down the rate of translation (54), which in turn may modulate frameshift efficiency. We therefore compared the 3HJ secondary structure (3HJ WT) to a similar construct with mutations designed to disrupt secondary structure formation in the P1 and P2 helices (Figure 6B, 3HJ Mut). We observe a significant decrease in frameshift efficiency, from 4.6 ± 0.5% to 2.5 ± 0.4%, when the 3HJ secondary structure is present (Figure 6C and Table 1, compare 3HJ WT to 3HJ Mut). As expected, there is no significant difference between the observed frameshifting efficiencies of the 3HJ Mut and the WT construct used above (4.7 ± 0.5 and 4.6 ± 0.5, respectively). The observed frameshift efficiencies for our 3HJ WT and WT reporter constructs in RRL both fall within the range of previously measured frameshifting efficiencies for HIV-1 in vivo, which range from 2% to 5% (9,15,21,24).
Figure 6.
Effect of the 3HJ on frameshift efficiency. (A) The secondary structure of the 3HJ WT is shown. Sequence changes in 3HJ MS1 are boxed. (B) Sequence changes in the 3HJ mutant (3HJ Mut) are shown in bold and italicized. (C) In vitro frameshift assay results for the 3HJ RNAs. Data represent an average of at least three replicates, with the SEM indicated.
Next, we tested the local stability hypothesis in the context of the 3HJ secondary structure by increasing the local stability of 2 bp in the P3 helix (Figure 6A). The 2-bp change (3HJ MS1) results in a large, 5-fold increase in frameshift efficiency (Figure 6C and Table 1). This increase is similar to the 4-fold difference between the MS1 and WT RNAs (Figure 3A and Table 1). We conclude that the 3HJ secondary structure indirectly modulates frameshifting, likely by altering the kinetics of translation, as previously hypothesized (54). This effect must happen prior to frameshifting, as the ribosome disrupts the 3HJ secondary structure as it encounters the slippery site. Once the ribosome is engaged with the slippery site, local stability is the primary determinant of frameshifting efficiency, as illustrated by comparison of 3HJ WT to 3HJ MS1 (Figure 5C).
DISCUSSION
In this work, we report a strong correlation between the thermodynamic stability of the first 3–4 bp at the base of the stem–loop and frameshift efficiency in HIV-1. We therefore hypothesize that the frameshift mechanism involves a thermodynamic block to translocation, determined by the local stability of base pairs positioned directly at the mRNA entrance channel. This is in agreement with previous studies investigating antisense-induced frameshifting using either mixed locked nucleic acid/DNA (73) or morpholino/RNA (38) oligonucleotides. When these oligonucleotides were used to direct antisense-induced frameshifting, the local stability of the duplex was also critical to frameshift stimulation (38,73).
In light of the local stability hypothesis, we can re-examine data from prior studies that investigated trends between thermodynamic stability of the HIV-1 stem–loop and frameshift efficiency (30,44,45). Indeed, we find that these results are generally consistent with local stability being the primary determinant in frameshift efficiency. For example, Bidou et al. (30) investigated the frameshift efficiencies of five MSs in both yeast and mouse NIH3T3 cells and observed decreased frameshift efficiency when local stability was decreased. Telenti et al. (44) used a yeast frameshift reporter assay to test naturally occurring stem–loop variants and observed the greatest decrease in frameshift efficiency for a point mutation that disrupts the C–G base pair at the base of the stem, while little to no effect was observed for mutations in the upper regions of the stem–loop. Hill et al. (45) investigated mutant HIV-1 virus replicating in 293T cells. Of the four MSs investigated in this study, two eliminated base pairing at the base of the stem and these showed large reductions in frameshifting; the other two mutants had similar local stabilities to WT and similar levels of frameshifting.
Mutations in the frameshift site that arise in response to cytotoxic T-cell escape (74) and protease inhibitor resistance (75,76) are also consistent with our results. Prado et al. (74) investigated the frameshift efficiency of four HIV-1 strains with mutations in the frameshift site. In this study, the only mutation that produced a significant change (decrease) in frameshift efficiency was one with a decreased local stability due to disruption of the first base pair in the stem–loop. Nijhuis et al. (75) examined the effects of three-point mutations in the spacer region, all of which do not impact frameshift efficiency. Knops et al. (76) investigated mutations in the frameshift site that either mutate the ACAA tetraloop to CCAA or change a C–G base pair in the upper stem–loop to a U–G wobble (C2108U). Consistent with the local stability hypothesis, none of these mutations alters the frameshift efficiency.
If the local stability of 3–4 bp is sufficient to determine frameshift efficiency, why does the HIV-1 frameshift site stem–loop contain 11 Watson–Crick base pairs? We can see two possible explanations for this. First, the additional base pairs ensure that the stem–loop has a high probability of folding and cannot be out-competed by suboptimal folds, which can severely impact HIV replication (77). In the >9100-nt genome, there are a total of only 11 helices that are equal or larger in size (54). Second, the additional base pairs serve to cooperatively stabilize the base pairs at the base of the stem. These effects may explain why severely truncated constructs produce lower frameshift efficiencies compared to stem–loops with identical local stability (MS9 versus WT, MS16 versus MS17). Cooperative stabilization of local stability may also explain why severe truncations of a hairpin downstream of the Simian retrovirus type-1 (SRV-1) slippery site result in lower frameshift efficiency (78).
The observed frameshift efficiencies for the 3HJ WT and WT reporter constructs in RRL both fall within the range of previously measured frameshifting efficiencies for HIV-1 in vivo, which range from 2% to 5% (9,15,21,24). The wide range of observed frameshifting efficiencies in vivo is likely influenced by viral and cellular factors, for example, modulation of translation initiation by the HIV-1 TAR RNA structure (79) and polysome density (53). We find that the conserved 3HJ secondary structure in the HIV-1 genomic RNA (54) causes a significant decrease in frameshift efficiency (Figure 6). This observation is consistent with the previous hypothesis that the 3HJ secondary structure induces ribosomal pausing (54). Pausing at the upstream secondary structure may promote stacking of consecutive ribosomes (80), promoting a net decrease in frameshift efficiency because the mRNA would have less time to refold between ribosomes. Our data support this model and also indicate that local stability has a far greater impact on frameshift efficiency (Figure 6).
Prokaryotic ribosomes use two active mechanisms during translation to unwind RNA (49). In the first mechanism, the ribosomal helicase activity raises the free energy of an encountered base pair by +0.9 kcal/mol (49). This destabilizes the base pair, which can then be opened by the mechanical force generated by translocation. If the base pair is resistant to this force, tension may be created which is sensed at the codon–anticodon base pairs (31,32,43,49,51). Because G–C pairs require more force for unwinding (49), the tension sensed in the decoding center would be proportional to the local RNA stability (51). The mechanical tension may either cause the tRNAs to slip 1 nt in the 5′-direction (5,31,32,36,49) or, alternatively, cause the ribosome to translocate incompletely by 2 nt instead of 3 nt (37), which would also result in a −1 frameshift.
When the HIV-1 frameshift site is modeled onto the eukaryotic ribosome, base pairs critical for frameshifting are positioned at the entrance to the mRNA entry channel (Figure 5B and D), in agreement with chemical probing and toeprinting results with a prokaryotic ribosome stalled on the HIV-1 frameshift site (23). Interestingly, the decoding center and the mRNA channel are highly conserved between prokaryotes and eukaryotes (65) and a bacterial translation system produces similar levels and changes in frameshift efficiency in response to changes in the HIV-1 stem–loop sequence (11).
Our data support an 8-nt spacer length between the slippery site and the stem–loop, as the effect of deletions in the spacer correlates with corresponding changes in stem–loop local stability 8-nt downstream of the slippery site (Figure 4). Consistent with this idea, deletion of 1 nt in the spacer region of the Beet western yellow virus (BWYV) −1 PRF site promotes the melting of the first base pair in the downstream structure (81). If the mRNA channel length is maintained, it follows that spacer lengths in all −1 PRF sites should be ≥7 nt in length. Yet, some frameshift sites have been drawn with 5- to 6-nt spacers [reviewed in (4,5)], including that of BWYV (81). Our data suggest that these frameshift site structures may be partially unwound at the time of frameshifting, in order to accommodate the requisite spacer length and positioning of the slippery sequence in the ribosomal decoding center. Unfortunately, there are currently no high-resolution structural views of ribosomes engaged with frameshift site structures. In conjunction with functional studies such as the one presented here, high-resolution structural views will ultimately be required to define the frameshifting mechanism. Prior studies have observed relationships between spacer length and −1 PRF efficiency in various systems (22,39,82,83) and are consistent with a spacer length of 7–8 nt and local stability being the primary determinant in frameshift efficiency. Spacer lengths of 6–8 nt produced the highest level of −1 PRF for the antisense oligonucleotides, stem–loop and pseudoknot stimulatory structures (82). In agreement with our conclusions, these spacers position base pairs with strong local stabilities at the entrance to the mRNA channel.
HIV-1 group M subtype B is the dominant form of HIV-1 in North and South America, Europe, Japan, Thailand and Australia. The less common non-B subtypes (A, C, D, E, F, G, H, J and K) have decreased local stabilities; for example, a frequent C to U mutation in the first base pair of the stem–loop in these subtypes results in formation of a U–G wobble pair in place of a C–G (28,84). Interestingly, the exponential relationship we observe predicts that such a change would have little effect on frameshift efficiency. For instance, mutants MS10–12 all incorporate U–G wobble pairs at these positions, which significantly destabilize the local stability by +5.5 kcal/mol relative to WT (Figure 1). Nevertheless, these mutations result in near WT frameshift efficiencies (Figure 3A), owing to the exponential relationship between local stability and frameshift efficiency (Figures 3C, 4F and G). These results are consistent with the observed frameshift efficiencies of the less common subtypes (28). Finally, a randomized trial of HIV patients receiving protease inhibitor therapy examined mutations in the Gag–Pol frameshift site and found no relationship between overall stability of the stem–loop and virological response (85). This observation is consistent with our results that show no correlation between overall stability and frameshifting (Figures 3B and 4E).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary methods, Supplementary Tables 1–3 and Supplementary Figures 1–3.
FUNDING
The University of Wisconsin-Madison; NSF [BIR-9512577]; National Institutes of Health [S10 RR13790] to obtain UV data at the University of Wisconsin-Madison Biophysics Instrumentation Facility. National Institutes of Health [GM072447 to S.E.B.]. Funding for open access charge: NIH GM072447.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jordan Burke, Ashley Richie and Lauren Michael for helpful discussions. They also thank Raymond Gesteland (University of Utah) for the generous gift of the p2luc plasmid DNA and Prof. A.C. Palmenberg and her laboratory (University of Wisconsin-Madison) for equipment use.
REFERENCES
- 1.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 2.Brierley I, Dos Ramos FJ. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 2006;119:29–42. doi: 10.1016/j.virusres.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brierley I, Boursnell ME, Binns MM, Bilimoria B, Blok VC, Brown TD, Inglis SC. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brierley I. Ribosomal frameshifting on viral RNAs. J. Gen. Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 5.Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farabaugh PJ. Programmed translational frameshifting. Microbiol. Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brierley I, Gilbert RJ, Pennell S. RNA pseudoknots and the regulation of protein synthesis. Biochem. Soc. Trans. 2008;36:684–689. doi: 10.1042/BST0360684. [DOI] [PubMed] [Google Scholar]
- 8.Brierley I, Pennell S. Structure and function of the stimulatory RNAs involved in programmed eukaryotic-1 ribosomal frameshifting. Cold Spring Harb. Symp. Quant. Biol. 2001;66:233–248. doi: 10.1101/sqb.2001.66.233. [DOI] [PubMed] [Google Scholar]
- 9.Reil H, Kollmus H, Weidle UH, Hauser H. A heptanucleotide sequence mediates ribosomal frameshifting in mammalian cells. J. Virol. 1993;67:5579–5584. doi: 10.1128/jvi.67.9.5579-5584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dulude D, Berchiche YA, Gendron K, Brakier-Gingras L, Heveker N. Decreasing the frameshift efficiency translates into an equivalent reduction of the replication of the human immunodeficiency virus type 1. Virology. 2006;345:127–136. doi: 10.1016/j.virol.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 11.Leger M, Sidani S, Brakier-Gingras LEA. A reassessment of the response of the bacterial ribosome to the frameshift stimulatory signal of the human immunodeficiency virus type 1. RNA. 2004;10:1225–1235. doi: 10.1261/rna.7670704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung M, Patel P, Davis S, Green SR. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 particle assembly and replication. J. Virol. 1998;72:4819–4824. doi: 10.1128/jvi.72.6.4819-4824.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cherry E, Liang C, Rong L, Quan Y, Inouye P, Li X, Morin N, Kotler M, Wainberg MA. Characterization of human immunodeficiency virus type-1 (HIV-1) particles that express protease-reverse transcriptase fusion proteins. J. Mol. Biol. 1998;284:43–56. doi: 10.1006/jmbi.1998.1968. [DOI] [PubMed] [Google Scholar]
- 14.Shehu-Xhilaga M, Crowe SM, Mak J. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 2001;75:1834–1841. doi: 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biswas P, Jiang X, Pacchia AL, Dougherty JP, Peltz SW. The human Immunodeficiency virus type 1 ribosomal frameshifting site is an invariant sequence determinant and an important target for antiviral therapy. J. Virol. 2004;78:2082–2087. doi: 10.1128/JVI.78.4.2082-2087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karacostas V, Wolffe EJ, Nagashima K, Gonda MA, Moss B. Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology. 1993;193:661–671. doi: 10.1006/viro.1993.1174. [DOI] [PubMed] [Google Scholar]
- 17.Park J, Morrow CD. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J. Virol. 1991;65:5111–5117. doi: 10.1128/jvi.65.9.5111-5117.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brierley I, Jenner AJ, Inglis SC. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim Y-G, Maas S, Rich A. Comparative mutational analysis of cis-acting RNA signals for translational frameshifting in HIV-1 and HTLV-2. Nucleic Acids Res. 2001;29:1125–1131. doi: 10.1093/nar/29.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stahl G, McCarty GP, Farabaugh PJ. Ribosome structure: revisiting the connection between translational accuracy and unconventional decoding. Trends Biochem. Sci. 2002;27:178–183. doi: 10.1016/S0968-0004(02)02064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cassan M, Delaunay N, Vaquero C, Rousset JP. Translational frameshifting at the gag-pol junction of human immunodeficiency virus type 1 is not increased in infected T-lymphoid cells. J. Virol. 1994;68:1501–1508. doi: 10.1128/jvi.68.3.1501-1508.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kollmus H, Honigman A, Panet A, Hauser H. The sequences of and distance between two cis-acting signals determine the efficiency of ribosomal frameshifting in human immunodeficiency virus type 1 and human T-cell leukemia virus type II in vivo. J. Virol. 1994;68:6087–6091. doi: 10.1128/jvi.68.9.6087-6091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazauric M-H, Seol Y, Yoshizawa S, Visscher K, Fourmy D. Interaction of the HIV-1 frameshift signal with the ribosome. Nucleic Acids Res. 2009;37:7654–7664. doi: 10.1093/nar/gkp779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dulude D, Baril M, Brakier-Gingras L. Characterization of the frameshift stimulatory signal controlling a programmed -1 ribosomal frameshift in the human immunodeficiency virus type 1. Nucleic Acids Res. 2002;30:5094–5102. doi: 10.1093/nar/gkf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staple DW, Butcher SE. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J. Mol. Biol. 2005;349:1011–1023. doi: 10.1016/j.jmb.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 26.Staple DW, Butcher SE. Solution structure of the HIV-1 frameshift inducing stem-loop RNA. Nucleic Acids Res. 2003;31:4326–4331. doi: 10.1093/nar/gkg654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaudin C, Mazauric M-H, Traikia M, Guittet E, Yoshizawa S, Fourmy D. Structure of the RNA signal essential for translational frameshifting in HIV-1. J. Mol. Biol. 2005;349:1024–1035. doi: 10.1016/j.jmb.2005.04.045. [DOI] [PubMed] [Google Scholar]
- 28.Baril M, Dulude D, Gendron K, Lemay G, Brakier-Gingras L. Efficiency of a programmed -1 ribosomal frameshift in the different subtypes of the human immunodeficiency virus type 1 group M. RNA. 2003;9:1246–1253. doi: 10.1261/rna.5113603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkin NT, Chamorro M, Varmus HE. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J. Virol. 1992;66:5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bidou L, Stahl G, Grima B, Liu H, Cassan M, Rousset JP. In vivo HIV-1 frameshifting efficiency is directly related to the stability of the stem-loop stimulatory signal. RNA. 1997;3:1153–1158. [PMC free article] [PubMed] [Google Scholar]
- 31.Plant EP, Jacobs KLM, Harger JW, Meskauskas A, Jacobs JL, Baxter JL, Petrov AN, Dinman JD. The 9-A solution: how mRNA pseudoknots promote efficient programmed -1 ribosomal frameshifting. RNA. 2003;9:168–174. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Namy O, Moran SJ, Stuart DI, Gilbert RJC, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leger M, Dulude D, Steinberg SV, Brakier-Gingras L. The three transfer RNAs occupying the A, P and E sites on the ribosome are involved in viral programmed -1 ribosomal frameshift. Nucleic Acids Res. 2007;35:5581–5592. doi: 10.1093/nar/gkm578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moran SJ, Flanagan JFt, Namy O, Stuart DI, Brierley I, Gilbert RJ. The mechanics of translocation: a molecular “spring-and-ratchet” system. Structure. 2008;16:664–672. doi: 10.1016/j.str.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plant EP, Rakauskaite R, Taylor DR, Dinman JD. Achieving a golden mean: mechanisms by which coronaviruses ensure synthesis of the correct stoichiometric ratios of viral proteins. J. Virol. 2010;84:4330–4340. doi: 10.1128/JVI.02480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao P-Y, Choi YS, Dinman JD, Lee KH. The many paths to frameshifting: kinetic modelling and analysis of the effects of different elongation steps on programmed −1 ribosomal frameshifting. Nucleic Acids Res. 2011;39:300–312. doi: 10.1093/nar/gkq761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brakier-Gingras L, Charbonneau J, Butcher SE. Targeting frameshifting in the human immunodeficiency virus. Expert Opin. Ther. Targets. 2012;16:249–258. doi: 10.1517/14728222.2012.665879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dinman JD. Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip. Rev. RNA. 2012;3:661–673. doi: 10.1002/wrna.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: Requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kontos H, Napthine S, Brierley I. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell. Biol. 2001;21:8657–8670. doi: 10.1128/MCB.21.24.8657-8670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tu C, Tzeng TH, Bruenn JA. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc. Natl Acad. Sci. USA. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Somogyi P, Jenner AJ, Brierley I, Inglis SC. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plant EP, Dinman JD. Torsional restraint: a new twist on frameshifting pseudoknots. Nucleic Acids Res. 2005;33:1825–1833. doi: 10.1093/nar/gki329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Telenti A, Martinez R, Munoz M, Bleiber G, Greub G, Sanglard D, Peters S. Analysis of natural variants of the human immunodeficiency virus type 1 gag-pol frameshift stem-loop structure. J. Virol. 2002;76:7868–7873. doi: 10.1128/JVI.76.15.7868-7873.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hill MK, Shehu-Xhilaga M, Crowe SM, Mak J. Proline residues within spacer peptide p1 are important for human immunodeficiency virus type 1 infectivity, protein processing, and genomic RNA dimer stability. J. Virol. 2002;76:11245–11253. doi: 10.1128/JVI.76.22.11245-11253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hansen TM, Reihani SN, Oddershede LB, Sorensen MA. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 2007;104:5830–5835. doi: 10.1073/pnas.0608668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen G, Chang K-Y, Chou M-Y, Bustamante C, Tinoco I. Triplex structures in an RNA pseudoknot enhance mechanical stability and increase efficiency of -1 ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 2009;106:12706–12711. doi: 10.1073/pnas.0905046106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tinoco I, Chen G, Qu X. RNA reactions one molecule at a time. Cold Spring Harb. Perspect. Biol. 2010;2:a003624. doi: 10.1101/cshperspect.a003624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qu X, Wen J-D, Lancaster L, Noller HF, Bustamante C, Tinoco I. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475:118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Green L, Kim CH, Bustamante C, Tinoco I., Jr Characterization of the mechanical unfolding of RNA pseudoknots. J. Mol. Biol. 2008;375:511–528. doi: 10.1016/j.jmb.2007.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao S, Chen S-J. Predicting ribosomal frameshifting efficiency. Phys. Biol. 2008;5:16002. doi: 10.1088/1478-3975/5/1/016002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ritchie DB, Foster DA, Woodside MT. Programmed -1 frameshifting efficiency correlates with RNA pseudoknot conformational plasticity, not resistance to mechanical unfolding. Proc. Natl Acad. Sci. USA. 2012;109:16167–16172. doi: 10.1073/pnas.1204114109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gendron K, Charbonneau J, Dulude D, Heveker N, Ferbeyre G, Brakier-Gingras L. The presence of the TAR RNA structure alters the programmed -1 ribosomal frameshift efficiency of the human immunodeficiency virus type 1 (HIV-1) by modifying the rate of translation initiation. Nucleic Acids Res. 2008;36:30–40. doi: 10.1093/nar/gkm906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marcheschi RJ, Mouzakis KD, Butcher SE. Selection and characterization of small molecules that bind the HIV-1 frameshift site RNA. ACS Chem. Biol. 2009;4:844–854. doi: 10.1021/cb900167m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marcheschi RJ, Staple DW, Butcher SE. Programmed ribosomal frameshifting in SIV is induced by a highly structured RNA stem-loop. J. Mol. Biol. 2007;373:652–663. doi: 10.1016/j.jmb.2007.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greene RF, Pace CN. Urea and guanidine hydrochloride denaturation of ribonuclease, lysozyme, α-chymotrypsin, and β-lactoglobulin. J. Biol. Chem. 1974;249:5388–5393. [PubMed] [Google Scholar]
- 59.Xia T, SantaLucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 60.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 61.Chen JL, Dishler AL, Kennedy SD, Yildirim I, Liu B, Turner DH, Serra MJ. Testing the nearest neighbor model for canonical RNA base pairs: revision of GU parameters. Biochemistry. 2012;51:3508–3522. doi: 10.1021/bi3002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 63.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 64.Yusupova GZ, Yusupov MM, Cate JHD, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 65.Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M. One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 2012;19:560–567. doi: 10.1038/nsmb.2313. [DOI] [PubMed] [Google Scholar]
- 66.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 67.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 68.Harger JW, Dinman JD. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 2003;9:1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reuter J, Mathews D. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinf. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mathews DH, Turner DH, Zuker M. Current Protocols in Nucleic Acid Chemistry. 2007. RNA secondary structure prediction. John Wiley & Sons, Inc., Wiley Online Library. Vol. 28, pp. 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.SantaLucia J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tholstrup J, Oddershede LB, Sorensen MA. mRNA pseudoknot structures can act as ribosomal roadblocks. Nucleic Acids Res. 2012;40:303–313. doi: 10.1093/nar/gkr686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu C-H, Noteborn MHM, Olsthoorn RCL. Stimulation of ribosomal frameshifting by antisense LNA. Nucleic Acids Res. 2010;38:8277–8283. doi: 10.1093/nar/gkq650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prado JG, Honeyborne I, Brierley I, Puertas MC, Martinez-Picado J, Goulder PJR. Functional consequences of human immunodeficiency virus escape from an HLA-B*13-restricted CD8+ T-cell epitope in p1 Gag protein. J. Virol. 2009;83:1018–1025. doi: 10.1128/JVI.01882-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nijhuis M, van Maarseveen NM, Lastere S, Schipper P, Coakley E, Glass B, Rovenska M, de Jong D, Chappey C, Goedegebuure IW, et al. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med. 2007;4:e36. doi: 10.1371/journal.pmed.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Knops E, Brakier-Gingras L, Schulter E, Pfister H, Kaiser R, Verheyen J. Mutational patterns in the frameshift-regulating site of HIV-1 selected by protease inhibitors. Med. Microbiol. Immunol. 2012;201:213–218. doi: 10.1007/s00430-011-0224-z. [DOI] [PubMed] [Google Scholar]
- 77.Das AT, Vrolijk MM, Harwig A, Berkhout B. Opening of the TAR hairpin in the HIV-1 genome causes aberrant RNA dimerization and packaging. Retrovirology. 2012;9:59. doi: 10.1186/1742-4690-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu C-H, Noteborn MH, Pleij CWA, Olsthoorn RCL. Stem–loop structures can effectively substitute for an RNA pseudoknot in −1 ribosomal frameshifting. Nucleic Acids Res. 2011;39:8952–8959. doi: 10.1093/nar/gkr579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Charbonneau J, Gendron K, Ferbeyre G, Brakier-Gingras L. The 5′ UTR of HIV-1 full-length mRNA and the Tat viral protein modulate the programmed −1 ribosomal frameshift that generates HIV-1 enzymes. RNA. 2012;18:519–529. doi: 10.1261/rna.030346.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wolin SL, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mazauric MH, Leroy JL, Visscher K, Yoshizawa S, Fourmy D. Footprinting analysis of BWYV pseudoknot-ribosome complexes. RNA. 2009;15:1775–1786. doi: 10.1261/rna.1385409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin Z, Gilbert RJ, Brierley I. Spacer-length dependence of programmed -1 or -2 ribosomal frameshifting on a U6A heptamer supports a role for messenger RNA (mRNA) tension in frameshifting. Nucleic Acids Res. 2012;40:8674–8689. doi: 10.1093/nar/gks629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morikawa S, Bishop DH. Identification and analysis of the gag-pol ribosomal frameshift site of feline immunodeficiency virus. Virology. 1992;186:389–397. doi: 10.1016/0042-6822(92)90004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chang SY, Sutthent R, Auewarakul P, Apichartpiyakul C, Essex M, Lee TH. Differential stability of the mRNA secondary structures in the frameshift site of various HIV type 1 viruses. AIDS Res. Hum. Retroviruses. 1999;15:1591–1596. doi: 10.1089/088922299309892. [DOI] [PubMed] [Google Scholar]
- 85.Larrouy L, Chazallon C, Landman R, Capitant C, Peytavin G, Collin G, Charpentier C, Storto A, Pialoux G, Katlama C, et al. Gag mutations can impact virological response to dual-boosted protease inhibitor combinations in antiretroviral-naive HIV-infected patients. Antimicrob. Agents Chemother. 2010;54:2910–2919. doi: 10.1128/AAC.00194-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.