Abstract
Detection of low-level DNA variations in the presence of wild-type DNA is important in several fields of medicine, including cancer, prenatal diagnosis and infectious diseases. PCR-based methods to enrich mutations during amplification have limited multiplexing capability, are mostly restricted to known mutations and are prone to polymerase or mis-priming errors. Here, we present Differential Strand Separation at Critical Temperature (DISSECT), a method that enriches unknown mutations of targeted DNA sequences purely based on thermal denaturation of DNA heteroduplexes without the need for enzymatic reactions. Target DNA is pre-amplified in a multiplex reaction and hybridized onto complementary probes immobilized on magnetic beads that correspond to wild-type DNA sequences. Presence of any mutation on the target DNA forms heteroduplexes that are subsequently denatured from the beads at a critical temperature and selectively separated from wild-type DNA. We demonstrate multiplexed enrichment by 100- to 400-fold for KRAS and TP53 mutations at multiple positions of the targeted sequence using two to four successive cycles of DISSECT. Cancer and plasma-circulating DNA samples containing traces of mutations undergo mutation enrichment allowing detection via Sanger sequencing or high-resolution melting. The simplicity, scalability and reliability of DISSECT make it a powerful method for mutation enrichment that integrates well with existing downstream detection methods.
INTRODUCTION
Cancer treatment is gradually moving toward personalized therapy, allowing patients to avoid unnecessary or ineffective treatments (1). Tailoring therapies to the genetic profile of tumors requires effective diagnostic tools that would accurately detect clinically relevant genetic alterations. The detection of mutated DNA is often masked by an abundance of wild-type DNA present in stromal tissue or in bodily fluids such as plasma, urine or sputum (2–4). These low abundance mutations are particularly important for early cancer detection, assessment of residual disease post treatment, disease staging and monitoring of therapy following remission/relapse (2). To date, several PCR-based technologies have been developed for enrichment of low-abundance mutations at known sequence positions including methods that modify the targeted sequence (e.g. restriction fragment length polymorphism or PCR-RFLP, restriction endonuclease-mediated selective PCR or REMS-PCR and allele-specific PCR), peptide/locked nucleic acid approaches and co-amplification at lower denaturation temperature-PCR or COLD-PCR (2,5–7). COLD-PCR enables enrichment of any mutation on the sequence, without prior knowledge of the mutation type or position within the PCR amplicon. Accordingly, COLD-PCR can be used in conjunction with downstream sequencing methods to reveal low-level unknown mutations (5,7,8). This is achieved by using preferential denaturation of mismatch-forming mutations at critical denaturation temperature. Upon two consecutive COLD-PCRs, mutations can be enriched by 100-fold or more (7–11). Further mutation enrichment can be a challenge since polymerase errors and mis-priming events increase upon additional PCR cycling.
As an alternative to PCR-based approaches, a bead-based method known as DNA enrichment by allele-specific hybridization (DEASH) has been described to enrich for low-abundance mutations (12). DEASH uses allele-specific biotinylated oligonucleotides that hybridize competitively to the location of the targeted base substitution followed by capture on streptavidin-coated paramagnetic beads. The simplicity of DEASH is powerful, as it circumvents the use of enzymatic steps and the potential generation of artifacts in the course of genotypic selection. Nevertheless, the approach is limited to the enrichment of known mutations since it requires two carefully designed probes that match both the mutant and the wild-type alleles.
Here, we describe a novel method based on Differential Strand Separation at Critical Temperature (DISSECT) to enrich unknown mutations within mixed DNA populations. Similar to COLD-PCR, DISSECT uses differential denaturation of DNA heteroduplexes and can therefore enrich mutations at any position on the sequence, enabling mutation scanning and discovery via downstream sequencing. At the same time DISSECT avoids polymerase extension or other enzymatic steps, as it is entirely based on repeated cycles of hybridization and preferential denaturation on solid support (streptavidin-coated magnetic beads). Since the target sequence remains unmodified during DISSECT, the resulting mutation-enriched DNA pool can be combined with any existing downstream detection method. Thus, DISSECT can generate DNA template enriched for mutations by 200-fold or more, resulting in radically enhanced ability to identify low-level nucleic acid alterations. Here, we validate DISSECT for a variety of mutated DNA targets of clinical interest (KRAS and TP53) and demonstrate its simplicity, scalability and application to low-level mutation enrichment in clinical cancer samples.
MATERIALS AND METHODS
Cell lines and tumor specimens
Human cancer cell lines SW480 and PFSK-1 were purchased from American Type Culture Collection (ATCC), and genomic DNA was extracted from cultured cells using a DNeasy™ Blood and Tissue kit (Qiagen, Valencia, CA, USA) as per the manufacturer’s protocol. Genomic DNA for A549, NCI-H69, SNU-182 and HCC1008 were purchased directly from ATCC. To demonstrate mutation enrichment, genomic DNA from human cancer cell lines SW480, NCI-H69, SNU-182, HCC1008, A549 and PFSK-1 were serially diluted into WT DNA as a test panel for various mutations (Supplementary Table S1). Human male genomic DNA (Promega Corporation, Madison, WI, USA) served as the wild-type control for all experiments. DNA mutation abundances examined were 10, 5, 3, 1, 0.1 and 0.05% mutant-to-WT ratios. For the capture of multiple mutations in a single-tube, genomic DNA of SW480, NCI-H69 and SNU-182 were combined to generate a 5 and 1% mixed mutant DNA dilution. All experiments were carried out in parallel with wild-type controls and mutant mixtures and were replicated at least three times for validation.
Clinical colorectal tumor and lung tumor samples containing low-level mutations previously identified using COLD-PCR-sequencing, dHPLC and REMS-PCR-sequencing (7,9,13,14) were also used to validate DISSECT. Additionally, plasma-circulating DNA donated by a radiation therapy patient following informed consent and institutional review board approval was used to demonstrate mutation enrichment using DISSECT. Plasma-circulating DNA was previously screened and validated to contain a TP53 mutation (15). DNA obtained from both plasma and clinical samples were pre-amplified using 20 cycles of multiplex PCR prior to DISSECT, as described below.
Multiplex pre-amplification
Multiplex PCR primers (Supplementary Table S2) were designed for 50 most commonly mutated exonic regions identified in lung and esophageal cancers according to the COSMIC database (16). These regions contain a mutant count of greater than 1 for any single point mutation. Multiplex pre-amplification from ∼20 ng genomic DNA or 10 ng plasma-circulating DNA was performed in a total volume of 25 µl using a mixture of 100 primers (50 paired sets) at a final concentration of 0.3 µM for each primer with 0.3 mM dNTPs, 3 mM MgCl2, 1× Kapa HiFi buffer and 0.5 U of Kapa HiFi HotStart DNA polymerase (Kapa Biosystems, Woburn, MA, USA) reported to have an error rate of 2.8 × 10−7 mis-incorporations/bp. Multiplex PCR cycling was performed according to the manufacturer’s recommendations (Kapa Biosystems) for a total of 15–25 PCR cycles using 63°C as optimal annealing temperature. Amplicon lengths ranged from 120 to 190 bp in size depending on the amplicon (Supplementary Table S2). Following multiplex cycling, 1 µl of exonuclease I (New England Biolabs, Ipswich, MA, USA) was added to each reaction and incubated at 37°C for 30 min and 80°C for 15 min to remove unincorporated primers.
DISSECT procedure and downstream analysis
Probe design and preparation. Streptavidin-coated Dynabeads (Life Technologies, Grand Island, NY, USA) were coupled to biotinylated probe sequences of 30–95 nt long matching the wild-type target sequences of TP53, KRAS and EGFR genes (Supplementary Table S3). Probes for TP53 exons 5–9 and EGFR exons 19–21 were synthesized by Integrated DNA Technologies Inc. (Coralville, IA, USA). The probe for KRAS exon 2 was synthesized to contain three locked nucleic acid bases, LNA (Exiqon, Woburn, MA, USA). The LNA design for KRAS exon 2 was based on previous reports for LNA-assisted detection of KRAS mutation hotspots (17,18). ‘Dual-biotin’ was used for the design of capture probes to strengthen the bond between the streptavidin-coated beads and the immobilized oligonucleotides during the thermal cycling procedure (19). Dual-biotinylated probes were conjugated to streptavidin beads as per the manufacturer’s protocol (Invitrogen Life Technologies, Grand Island, NY). Dynabead probes were finally resuspended in a stock buffer containing 6× SSPE (0.9 M NaCl, 60 mM Na2HPO4, 6 mM EDTA) to a final concentration of 10 μg/μl and kept at 4°C.
DISSECT protocol. 5–10 μl of Dynabead probes were incubated with a 1:50 dilution of pre-amplified DNA in a total volume of 50 μl of 1× Phusion Buffer (New England Biolabs). To hybridize DNA onto beads, DNA was denatured at 95°C for 2 min, then annealed at 60°C for 2 min, 58°C for 2 min, 56°C for 2 min, 54°C for 1 min and 25°C for 5 min using an Eppendorf Mastercycler EP machine (Eppendorf Inc., Haupage, NY, USA). Beads were then separated using a Dynamag-PCR magnet (Invitrogen Life Technologies, Grand Island, NY) and washed twice with 50 μl of 1× Phusion buffer to remove unbound DNA fragments. Washed beads were resuspended in 55 μl 1× Phusion buffer, and a 5-μl aliquot was collected for testing DNA bound to beads. Single-stranded mutant DNA was preferentially denatured by heating the bead solution at a critical denaturation temperature (e.g. 75°C) for 2 min followed by magnetization, thereby collecting enriched mutated DNA sequences in the supernatant. Critical denaturation temperature (Tc) of the shorter biotinylated probe sequences was predicted using IDT’s Oligo Analyzer that predicts the melting temperature (Tm) of the matched (wild-type) sequence at the buffer conditions used for DISSECT. A range of three to four potential Tc values were then experimentally tested to derive the optimal Tc for the probe tested, ranging from 3°C below the predicted Tm and increasing temperature in 0.5°C steps until ∼Tm-1. We found that adequate mutation enrichments are obtained for denaturation temperature within ∼2°C for most 30–45 nt probes, and optimal values for Tc can be arrived at using the approximation Tc = Tm(wild-type)-2 (data not shown). For longer 75–95 nt probes, the publicly available UMelt software developed by the Wittwer laboratory (http://dna.utah.edu/umelt/umelt.html) was initially used to predict Tm. The optimal Tc was experimentally derived using high-resolution melting (HRM) analysis on the LightScanner HR96 system (Idaho Technologies Inc.). Following elution from beads, a 5-μl aliquot of the supernatant enriched for mutant DNA was saved for analysis, while the remaining supernatant was used for additional rounds of DISSECT to increase the mutation enrichment as described in Figure 1. These additional rounds of DISSECT required a single 50 μl 1× Phusion buffer replacement following the step-down anneal procedure to eliminate unbound DNA material in the supernatant. The same Tc was used for each denaturation step following capture of target DNA from supernatant. For the multiplexed DISSECT (combined bead capture), the same DISSECT procedure was carried out, but with 2–4 µl of each kind of Dynabead probe in a total bead volume of 8–16 µl per sample. The capture probes used for multiplexed DISSECT were designed to have similar Tm so that a common critical denaturation temperature could be used for multiplexed mutation enrichment.
Figure 1.
DISSECT flowchart. Following multiplexed pre-amplification from genomic DNA (Step 1), enriched DNA target regions are captured on complementary probes immobilized to magnetic beads resembling the wild-type form of the target sequence (Step 2). Mutation-containing target DNA forms heteroduplexes that are subsequently denatured at a critical temperature leaving preferentially wild-type DNA bound to beads (Step 3). Eluted DNA is separated and re-bound to a fresh aliquot of beads. Steps 2 and 3 are repeated to increase the mutation enrichment. Aliquots of eluted DNA from each round are analyzed by downstream assays (Sanger sequencing and HRM were used in this work).
Downstream analysis. DNA eluted from beads following DISSECT was amplified by conventional PCR or COLD-PCR (5,14) followed by HRM analysis (9) and Sanger Sequencing (Eton Bioscience, Cambridge, MA, USA) (15). For post DISSECT amplification, nested primers were used for each target region amplified in the original pre-amplification reaction (Supplementary Table S4).
RESULTS
The principle of DISSECT
The DISSECT procedure presented in Figure 1 illustrates a bead-based method by which low-level mutations can be enriched using repeated cycles of annealing and denaturation at a critical temperature. Following multiplex pre-amplification (Step 1) from genomic DNA, amplicons are captured on magnetic beads using sequence-specific binding to bead-immobilized DNA probes (Step 2). Target DNA sequences are captured on beads by denaturation of the double-stranded DNA at 95°C, followed by step-down annealing procedure that allows single-stranded DNA to anneal to complementary oligonucleotides (probes) attached to streptavidin-coated magnetic beads. These probes contain the wild-type sequence of the targeted DNA regions (Supplementary Table S3), and therefore the presence of a mutation at any sequence position of the target DNA leads to a mismatch that lowers the melting temperature (Tm) of the duplex. In Step 3 of Figure 1, mutation-containing DNA is preferentially released from beads by denaturation at a critical denaturation temperature (Tc) corresponding to the probe-target duplex. Tc was defined as detailed in the ‘Materials and Methods’ section. The critical denaturation step preferentially elutes mutant DNA strands into the supernatant, while a fairly large fraction of the wild-type sequences remains substantially bound to the beads and are subsequently removed by magnetization. Steps 2 and 3 can be repeated multiple times to enable further enrichment of mutation-containing DNA regions. In the final step, the released mutation-enriched sequences are PCR-amplified and analyzed. HRM and Sanger sequencing were used for downstream analysis. It should also be noted that although the amplicons generated from the pre-amplification reaction range from 120 to 190 bp, only the region of the amplicons that corresponds to the immobilized probes actually becomes enriched for mutations during DISSECT.
KRAS and TP53 mutation enrichment
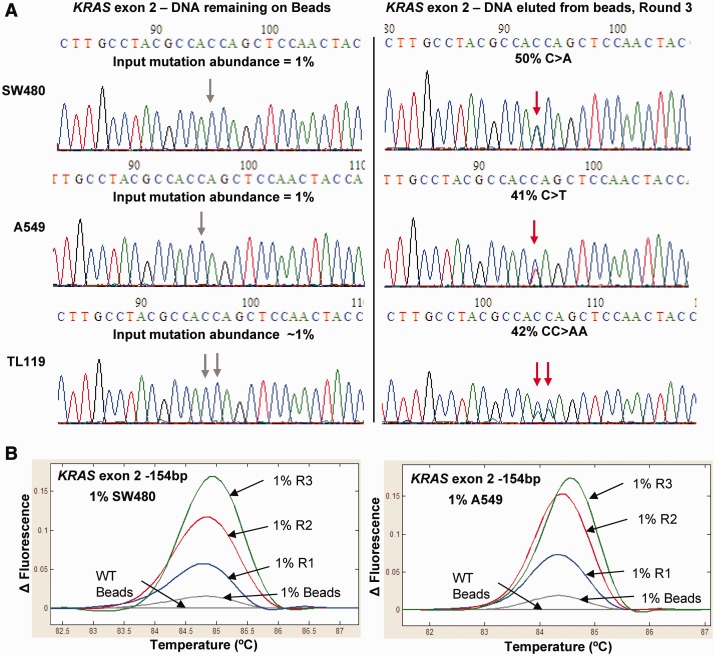
As a large portion of lung cancer mutations occur in KRAS (20) and TP53 genes (21,22), we implemented DISSECT to analyze and enrich point mutations within these two genes. Three successive rounds of DISSECT were performed from a 1% mutation abundance of both SW480 and A549 cell lines using a KRAS exon 2 probe at a critical denaturation temperature of 75°C. Figure 2A shows a 40- to 50-fold enrichment of two KRAS mutations (c.35C>A, p.G12V and c.C>T, p.G12S) after DNA elution following three rounds of DISSECT. Similarly, tumor sample TL119 containing a double KRAS mutation (c.34_35CC>AA, p.G12F) showed the same degree of enrichment (Figure 2A). PCR-amplified DISSECT products were also analyzed by HRM analysis showing incremental mutant enrichment for every additional round of DISSECT for both SW480 and A549 mutation-containing DNA (Figure 2B). Beads containing a non-LNA probe designed for TP53 exon 8 were equally effective in enriching for low-level mutations from an original 1 and 0.1% mutant abundance (Supplementary Figures S1–S4). DISSECT was able to successfully enrich low-level TP53 exon 8 Tm-reducing mutations (c.818C>T, Supplementary Figure S1), Tm-retaining mutations (c.841C>G, Supplementary Figure S2) and Tm-increasing mutations (c.823A>C, Supplementary Figure S3). All three mutations were enriched with a single bead-bound oligonucleotide probe, indicating the ability to enrich different types of single-base substitutions within the length of the DNA sequence covered by the probe. These experiments yielded up to 100- to 200-fold mutation enrichment after three rounds of DISSECT, equivalent to mutant enrichments of ∼5-fold/cycle, as inferred by comparing the apparent mutation abundance in the Sanger chromatographs to the initial abundance of 0.1%. The gradual increase in mutation enrichment of TP53 exon 8 after each round of DISSECT was demonstrated via HRM analysis following DISSECT, using 5, 1 or 0.1% original mutation abundances (Supplementary Figure S4).
Figure 2.
KRAS mutation enrichment. KRAS exon 2 enrichment following three rounds of DISSECT at a critical denaturation temperature of 75°C, using immobilized KRAS oligonucleotide (30 nt, wild-type sequence). (A) Sanger sequencing chromatographs following conventional PCR amplification of DISSECT products for three different KRAS mutations from an initial mutation abundance of 1% (c.35C>A, p.G12V and c.C>T, p.G12S from SW480 and A549, respectively, and c.34_35CC>AA, p.G12F from a lung tumor sample, TL119). The DNA remaining on beads (left) is compared with DNA eluted from beads following three rounds of DISSECT. (B) HRM analysis showing increasing mutation enrichment after every round of DISSECT (R1-R3) for SW480 and A549 samples with 1% input mutation abundance.
Next, to evaluate longer probes for use with DISSECT, a 90-nt long TP53 exon 8 probe was used. In Supplementary Figure S5A, serial dilutions of SW480 DNA into wild-type DNA (0.3, 1, 5 and 10% abundance) were used to enrich for a c.818G>A mutation using two rounds of DISSECT. Furthermore, a 1% mutation abundance using DNA with a C>G mutation (HCC1008 DNA) within the same exon 8 target region resulted in similar mutation enrichment (Supplementary Figure S5B). Finally, a single-tube, multiplexed mutation enrichment tested using 76, 95 and 90 nt probes for exons 5, 7 and 8, respectively, are depicted in Supplementary Figure S5C. The data demonstrated that two rounds of DISSECT in conjunction with the 76- to 95-nt probes result in ∼20- to 60-fold mutation enrichments, depending also on the mutation type and position. The longer probes provide a broader ‘footprint’ for the use of DISSECT in the enrichment of unknown mutations.
Limits of mutation detection via Sanger sequencing and HRM following DISSECT
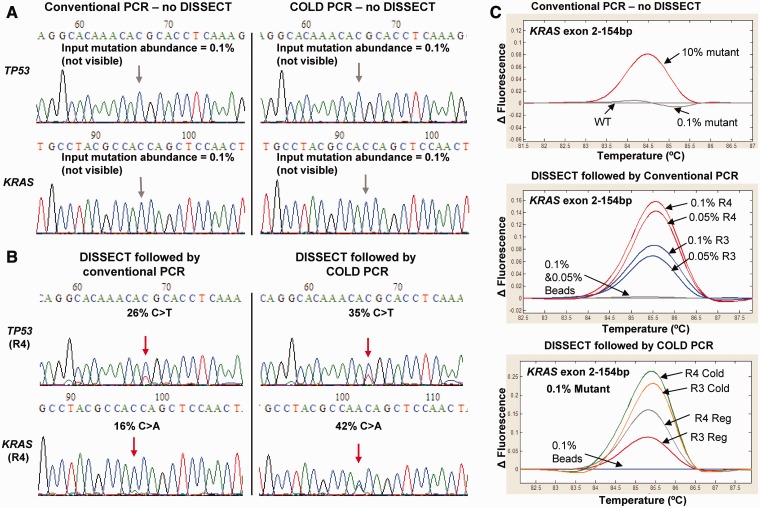
Using an initial mutation abundance of 0.1%, conventional PCR or single-round COLD-PCR-Sanger sequencing failed to detect the mutation (Figure 3A). In contrast, four rounds of DISSECT followed by conventional PCR resulted to an apparent mutation abundance of 26% for TP53 exon 8 and 16% for KRAS exon 2, corresponding to 260- and 160-fold mutation enrichment, respectively, or ∼4- to 5-fold mutation enrichment per DISSECT cycle (Figure 3B, left). The final mutation enrichment was higher when COLD-PCR was applied to the eluted DNA following four rounds of DISSECT, resulting in a mutation enrichment of ∼350- to 400-fold (Figure 3B, right). The increased overall enrichment reflects the additional enrichment provided by COLD-PCR itself. Three or four rounds of DISSECT followed by conventional PCR altered the HRM profiles for both 0.1 and 0.05% mutant KRAS compared with wild-type DNA (Figure 3C, middle), and these HRM differences were greater when COLD-PCR was adopted following DISSECT (Figure 3C, bottom). Similar results were shown for mutations in TP53 exon 8 and KRAS exon 2 after four rounds of DISSECT from an initial mutation of 0.05% (Supplementary Figure S6). In summary, three or four rounds of DISSECT enrich low-level mutations, originally present at 0.05–0.1% mutation abundance, to levels of about 15–25%, allowing their subsequent identification via COLD-PCR, HRM and Sanger sequencing.
Figure 3.
Mutation enrichment following four rounds of DISSECT. (A) Sanger sequencing following direct conventional PCR or COLD-PCR in the absence of DISSECT for TP53 exon 8 (C>T mutation) and KRAS exon 2 (C>A mutation) from an original 0.1% mutation abundance (from SW480 mutant DNA diluted into wild-type DNA). (B) Sanger sequencing following direct conventional PCR or COLD-PCR after four rounds (R4) of DISSECT. Resulting mutation abundance of 26% for TP53 and 16% for KRAS (corresponding to a final 160- to 260-fold mutation enrichment) are depicted. DISSECT in combination with COLD-PCR increased the enrichment to 35% for TP53 and 42% for KRAS (corresponding to a final ∼400-fold mutation enrichment). (C) HRM analysis shows differential melting curves for KRAS exon 2, before and after DISSECT, from original input mutation abundances of 0.05–0.1%. Data are representative of at least three independent experiments.
Multiplexing DISSECT
Bead-based processes are inherently scalable and amenable to multiplexing. For a proof of principle in multiplexing DISSECT, we prepared a 5 and 1% mixtures of genomic DNA from cell lines SW480, SNU-182 and NCI-H69, containing mutations in four different exon regions (Supplementary Table S1) diluted in a wild-type DNA background. A 50-plex pre-amplification was performed using this DNA mixture and processed for the simultaneous mutation enrichment of four mutated DNA targets using a combination of beads in a single tube. The oligonucleotide probes attached to the beads were designed to have similar melting temperatures, within a ±0.5°C temperature range. During the single-tube multiplexed DISSECT, a single denaturation temperature of 75°C was used for the enrichment of all four mutated DNA targets.
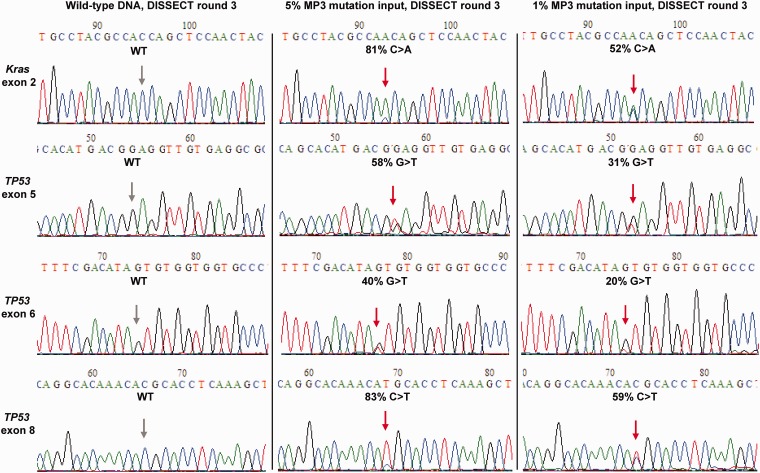
Figure 4 shows a combined mutation enrichment of ∼60-fold following three rounds of DISSECT for KRAS exon 2, TP53 exon 5, TP53 exon 6 and TP53 exon 8 amplicons, using initial mutation abundances of 1–5%. HRM analysis also depicts melting curves with increasing differences for all four mutated amplicons (KRAS exon 2 and TP53 exons 5, 6 and 8) following each round of DISSECT (Supplementary Figure S7). These results illustrate that DISSECT can be readily multiplexed provided the Tm of the bead-bound probes are matched.
Figure 4.
Multiplexed, single-tube DISSECT. Sanger sequencing analysis from a combined bead capture experiment showing simultaneous mutation enrichment for four independent regions, KRAS exon 2 and TP53 exons 5, 6 and 8 following three rounds of DISSECT, from an original mutation abundance of 1 and 5% (mixture of SW480, SNU-182 and NCI-H69 mutant DNA diluted into wild-type DNA, MP3). Data are representative of at least three independent experiments.
Analysis of clinical tumor samples and circulating DNA from plasma
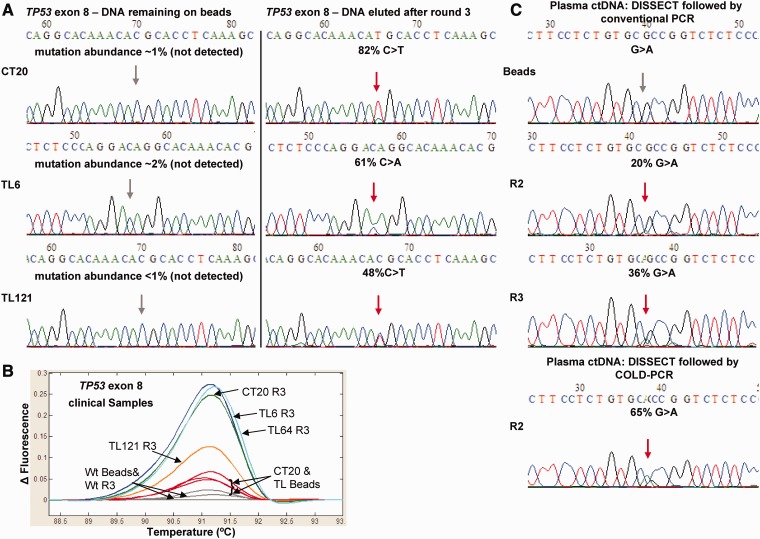
To validate the use of DISSECT with clinical specimens, we used previously tested DNA from lung and colorectal tumor samples containing mid- to low-abundance TP53 and KRAS mutations (7,14). Figure 5A shows strong mutation enrichment following three rounds of DISSECT for low-abundance TP53 exon 8 mutations in tumor samples CT20, TL6 and TL121. HRM analysis also demonstrates differences between the original DNA captured on TP53 exon 8 beads versus DNA eluted after three rounds of DISSECT followed by conventional PCR-HRM (Figure 5B). Additional tumor specimens were tested for enrichment of mutations from other hotspot regions including TP53 exons 7 and 9 and KRAS exon 2 (Supplementary Figure S8). A list of the clinical samples, mutation positions and enrichment following DISSECT is listed in Supplementary Table S5.
Figure 5.
Clinical specimen analysis using DISSECT. Conventional PCR-Sanger sequencing (A) and conventional PCR-HRM (B) showing antisense TP53 exon 8 mutations from lung (c.830C>A, TL6 and c.818C>T, TL121) and colorectal (c.818C>T, CT20) clinical tumor samples, after three rounds of DISSECT. Results from DNA remaining on beads versus DNA eluted from beads are depicted (C) Plasma-circulating DNA from an esophageal cancer patient with a low-level mutation (c.847G>A, R283C) was analyzed by DISSECT followed by conventional or COLD-PCR-Sanger sequencing.
As an additional test, we used DISSECT to enrich mutations from plasma-circulating DNA obtained from an esophageal cancer patient undergoing radiation therapy. The circulating DNA was purified from plasma and pre-amplified using 20 cycles of multiplex PCR. A biotinylated probe for DISSECT, p53Ex8-FC39 (Supplementary Table S3), was designed to enrich for a previously identified mutation at an abundance of ∼0.5% on TP53 exon 8 (c.847C>T, p.R823C) (15). Three rounds of DISSECT resulted in a final mutation abundance of 36% (G>A, antisense) (Figure 5C). Alternatively, a single round of COLD-PCR following two rounds of DISSECT was performed, which resulted in mutation abundance of 65% according to the chromatograph. In summary, DISSECT can readily be applied to enrich low-abundance mutations from clinical tumor specimens and circulating tumor DNA prior to downstream mutation detection.
DISCUSSION
DISSECT is a novel method for enriching DNA samples for low-level unknown mutations that is purely based on the thermal denaturation properties of DNA, without the need for enzymatic reactions during the enrichment process. The process is ‘minimally disruptive’ to the input DNA material since no enzymatic modification such as RFLP or sequence alterations by PCR primers and polymerase extension occur during the mutation enrichment, thus minimizing the probability for artifacts. By using selective denaturation of mismatch-forming target sequences bound to immobilized wild-type sequence at critical temperature, DISSECT enriches for most types of mutations, enabling mutation scanning over the entire length of the probe. This was not possible using the previously described DEASH competitive hybridization method (12) because (a) special oligonucleotides matching the specific targeted mutation were needed and (b) selective denaturation confers superior selectivity and faster kinetics over selective hybridization, as our experience with COLD-PCR has demonstrated (5).
To date, few methods have been able to provide multiplexing capability in enriching low-level mutations in diverse targets (2). Therefore, an advantage of DISSECT lies in the ability to enrich multiple mutant targets in a single tube without compromising accuracy or selectivity. This can inherently enable multiplexed downstream analysis platforms to be used with DISSECT, such as matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, pyrosequencing and next-generation sequencing, in addition to Sanger sequencing and HRM implemented here.
In the present work, a pre-amplification step using a 50-plex PCR from genomic DNA was used to generate target pre-amplification prior to applying DISSECT. While this step combines conveniently with DISSECT, the pre-amplification can potentially be omitted by performing direct target capture from genomic DNA, as performed with DEASH (12). This would eliminate potential artifacts introduced during the initial 15–25 cycles of pre-amplification. A limitation of DISSECT is that the original DNA content is gradually reduced following each round since part of the bead-bound DNA is removed after the critical denaturation step. Thus, following three to four consecutive rounds of DISSECT that produced up to ∼100- to 400-fold mutation enrichment, an amplification was applied prior to downstream analysis by Sanger sequencing (or, potentially, prior to additional rounds of DISSECT to increase mutation enrichment further).
Here, we have shown that DISSECT can simultaneously enrich diverse targets for multiple mutations in the same tube using a single denaturation temperature. The mutation enrichment varies somewhat since the different DNA duplexes may not have the same optimal critical denaturation temperature and also because mutation enrichment depends on the specific mismatches and the sequence context.
DISSECT is simple and amenable to scaling and automation, e.g. using liquid-handling robots, microfluidics or micro-arrays. Accordingly, we anticipate using DISSECT in routine genetic screening, especially in cancer-related applications where it is important to identify low-level mutations, such as EGFR resistance mutations (23) or predictive KRAS mutations (24,25). KRAS mutations at an initial abundance of 0.05–0.1% are capable of being directly detected via Sanger sequencing following DISSECT (Figure 3), which illustrates a detection limit improvement over previously reported methodologies used to improve KRAS Sanger sequencing limits (13,26). In the notable reports by Misale et al. (27) and Diaz et al. (28), BEAM-ing was used to identify KRAS mutations at the 0.1% level in plasma, as an indicator of developing resistance to cetuximab treatment in colorectal cancer patients. The BEAM-ing detection capability is similar to that provided by DISSECT when followed by PCR-Sanger sequencing (Figure 3). In addition, DISSECT coupled with sequencing provides information on adjacent positions on the same amplicon, thus enabling ‘unknown’ mutation detection and surveying of multiple ‘hotspots’ along the sequence. Accordingly, DISSECT may also be used for detecting biomarker mutations in tumor suppressor genes with multiple hotspot mutations present in bodily fluids such as plasma-circulating DNA, as was shown for the TP53 gene mutation in Figure 5. In addition to cancer, DISSECT applications in prenatal diagnosis or infectious diseases can also be envisioned.
In summary, DISSECT is a novel method for enriching DNA target sequences for low-level unknown mutations based on thermal denaturation properties of DNA heteroduplexes, without the need for excessive amplification or other enzymatic reactions. The process is readily amenable to scaling and automation and can be combined with existing downstream DNA sequencing platforms. The simplicity of DISSECT makes it a powerful method to screen for multiple mutation targets in a single tube without compromising accuracy or sensitivity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5 and Supplementary Figures 1–8.
FUNDING
NCI [R21 CA-111994 and R21 CA-155615, in part]. The contents of this article do not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. Funding for open access charge: Departmental funds.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Sadowska AM, Nowe V, Janssens A, Boeykens E, De Backer WA, Germonpre PR. Customizing systemic therapy in patients with advanced non-small cell lung cancer. Ther. Adv. Med. Oncol. 2011;3:207–218. doi: 10.1177/1758834011409000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milbury CA, Li J, Makrigiorgos GM. PCR-based methods for the enrichment of minority alleles and mutations. Clin. Chem. 2009;55:632–640. doi: 10.1373/clinchem.2008.113035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sidransky D, Von Eschenbach A, Tsai YC, Jones P, Summerhayes I, Marshall F, Paul M, Green P, Hamilton SR, Frost P, et al. Identification of p53 gene mutations in bladder cancers and urine samples. Science. 1991;252:706–709. doi: 10.1126/science.2024123. [DOI] [PubMed] [Google Scholar]
- 4.Tada M, Omata M, Kawai S, Saisho H, Ohto M, Saiki RK, Sninsky JJ. Detection of ras gene mutations in pancreatic juice and peripheral blood of patients with pancreatic adenocarcinoma. Cancer Res. 1993;53:2472–2474. [PubMed] [Google Scholar]
- 5.Li J, Wang L, Mamon H, Kulke MH, Berbeco R, Makrigiorgos GM. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat. Med. 2008;14:579–584. doi: 10.1038/nm1708. [DOI] [PubMed] [Google Scholar]
- 6.Milbury CA, Li J, Makrigiorgos GM. Ice-COLD-PCR enables rapid amplification and robust enrichment for low-abundance unknown DNA mutations. Nucleic Acids Res. 2010;39:e2. doi: 10.1093/nar/gkq899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milbury CA, Correll M, Quackenbush J, Rubio R, Makrigiorgos GM. COLD-PCR enrichment of rare cancer mutations prior to targeted amplicon resequencing. Clin. Chem. 2011;58:580–589. doi: 10.1373/clinchem.2011.176198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Milbury CA, Li C, Makrigiorgos GM. Two-round coamplification at lower denaturation temperature-PCR (COLD-PCR)-based sanger sequencing identifies a novel spectrum of low-level mutations in lung adenocarcinoma. Hum. Mutat. 2009;30:1583–1590. doi: 10.1002/humu.21112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milbury CA, Li J, Makrigiorgos GM. COLD-PCR-enhanced high-resolution melting enables rapid and selective identification of low-level unknown mutations. Clin. Chem. 2009;55:2130–2143. doi: 10.1373/clinchem.2009.131029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Wang L, Janne PA, Makrigiorgos GM. Coamplification at lower denaturation temperature-PCR increases mutation-detection selectivity of TaqMan-based real-time PCR. Clin. Chem. 2009;55:748–756. doi: 10.1373/clinchem.2008.113381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boisselier B, Marie Y, Labussiere M, Ciccarino P, Desestret V, Wang X, Capelle L, Delattre JY, Sanson M. COLD PCR HRM: a highly sensitive detection method for IDH1 mutations. Hum. Mutat. 2010;31:1360–1365. doi: 10.1002/humu.21365. [DOI] [PubMed] [Google Scholar]
- 12.Jeffreys AJ, May CA. DNA enrichment by allele-specific hybridization (DEASH): a novel method for haplotyping and for detecting low-frequency base substitutional variants and recombinant DNA molecules. Genome Res. 2003;13:2316–2324. doi: 10.1101/gr.1214603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song C, Milbury CA, Li J, Liu P, Zhao M, Makrigiorgos GM. Rapid and sensitive detection of KRAS mutation after fast-COLD-PCR enrichment and high-resolution melting analysis. Diagn. Mol. Pathol. 2011;20:81–89. doi: 10.1097/PDM.0b013e3181fde92f. [DOI] [PubMed] [Google Scholar]
- 14.Milbury CA, Chen CC, Mamon H, Liu P, Santagata S, Makrigiorgos GM. Multiplex amplification coupled with COLD-PCR and high resolution melting enables identification of low-abundance mutations in cancer samples with low DNA content. J. Mol. Diagn. 2011;13:220–232. doi: 10.1016/j.jmoldx.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castellanos-Rizaldos E, Liu P, Milbury CA, Guha M, Brisci A, Cremonesi L, Ferrari M, Mamon H, Makrigiorgos GM. Temperature-tolerant COLD-PCr reduces temperature stringency and enables robust mutation enrichment. Clin. Chem. 2012;58:1130–1138. doi: 10.1373/clinchem.2012.183095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr. Protoc. Hum. Genet. 2008 doi: 10.1002/0471142905.hg1011s57. Chapter 10, Unit 10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Efrati E, Elkin H, Peerless Y, Sabo E, Ben-Izhak O, Hershkovitz D. LNA-based PCR clamping enrichment assay for the identification of KRAS mutations. Cancer Biomark. 2010;8:89–94. doi: 10.3233/CBM-2011-0203. [DOI] [PubMed] [Google Scholar]
- 18.Huang Q, Wang GY, Huang JF, Zhang B, Fu WL. High sensitive mutation analysis on KRAS gene using LNA/DNA chimeras as PCR amplification blockers of wild-type alleles. Mol. Cell. Probes. 2010;24:376–380. doi: 10.1016/j.mcp.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat. Methods. 2006;3:551–559. doi: 10.1038/nmeth898. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki Y, Orita M, Shiraishi M, Hayashi K, Sekiya T. Detection of ras gene mutations in human lung cancers by single-strand conformation polymorphism analysis of polymerase chain reaction products. Oncogene. 1990;5:1037–1043. [PubMed] [Google Scholar]
- 21.Wistuba II, Gazdar AF, Minna JD. Molecular genetics of small cell lung carcinoma. Semin. Oncol. 2001;28:3–13. [PubMed] [Google Scholar]
- 22.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuang Y, Rogers A, Yeap BY, Wang L, Makrigiorgos M, Vetrand K, Thiede S, Distel RJ, Janne PA. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin. Cancer Res. 2009;15:2630–2636. doi: 10.1158/1078-0432.CCR-08-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molinari F, Felicioni L, Buscarino M, De Dosso S, Buttitta F, Malatesta S, Movilia A, Luoni M, Boldorini R, Alabiso O, et al. Increased detection sensitivity for KRAS mutations enhances the prediction of anti-EGFR monoclonal antibody resistance in metastatic colorectal cancer. Clin. Cancer Res. 2011;17:4901–4914. doi: 10.1158/1078-0432.CCR-10-3137. [DOI] [PubMed] [Google Scholar]
- 25.Molinari F, Frattini M. KRAS mutational test for metastatic colorectal cancer patients: not just a technical problem. Expert Rev. Mol. Diagn. 2012;12:123–126. doi: 10.1586/erm.11.94. [DOI] [PubMed] [Google Scholar]
- 26.Araki T, Shimizu K, Nakamura K, Nakamura T, Mitani Y, Obayashi K, Fujita Y, Kakegawa S, Miyamae Y, Kaira K, et al. Usefulness of peptide nucleic acid (PNA)-clamp smart amplification process version 2 (SmartAmp2) for clinical diagnosis of KRAS codon 12 mutations in lung adenocarcinoma: comparison of PNA-clamp SmartAmp2 and PCR-related methods. J. Mol. Diagn. 2010;12:118–124. doi: 10.2353/jmoldx.2010.090081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz LA, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.