Abstract
DNA transposases facilitate genome rearrangements by moving DNA transposons around and between genomes by a cut-and-paste mechanism. DNA transposition proceeds in an ordered series of nucleoprotein complexes that coordinate pairing and cleavage of the transposon ends and integration of the cleaved ends at a new genomic site. Transposition is initiated by transposase recognition of the inverted repeat sequences marking each transposon end. Using a combination of solution scattering and biochemical techniques, we have determined the solution conformations and stoichiometries of DNA-free Mos1 transposase and of the transposase bound to a single transposon end. We show that Mos1 transposase is an elongated homodimer in the absence of DNA and that the N-terminal 55 residues, containing the first helix-turn-helix motif, are required for dimerization. This arrangement is remarkably different from the compact, crossed architecture of the dimer in the Mos1 paired-end complex (PEC). The transposase remains elongated when bound to a single-transposon end in a pre-cleavage complex, and the DNA is bound predominantly to one transposase monomer. We propose that a conformational change in the single-end complex, involving rotation of one half of the transposase along with binding of a second transposon end, could facilitate PEC assembly.
INTRODUCTION
Transposable elements are significant components of most eukaryotic genomes, shaping genome architecture and gene regulatory networks (1,2). By moving from one location to another, they can alter gene expression, create mutations or promote genome evolution by creating novel genes (3,4). DNA transposons of the mariner/Tc1 family are particularly widespread in nature (5) and are active in a broad range of species, including vertebrates. Because of this, they are being exploited successfully as tools for genetic engineering and gene delivery (6,7).
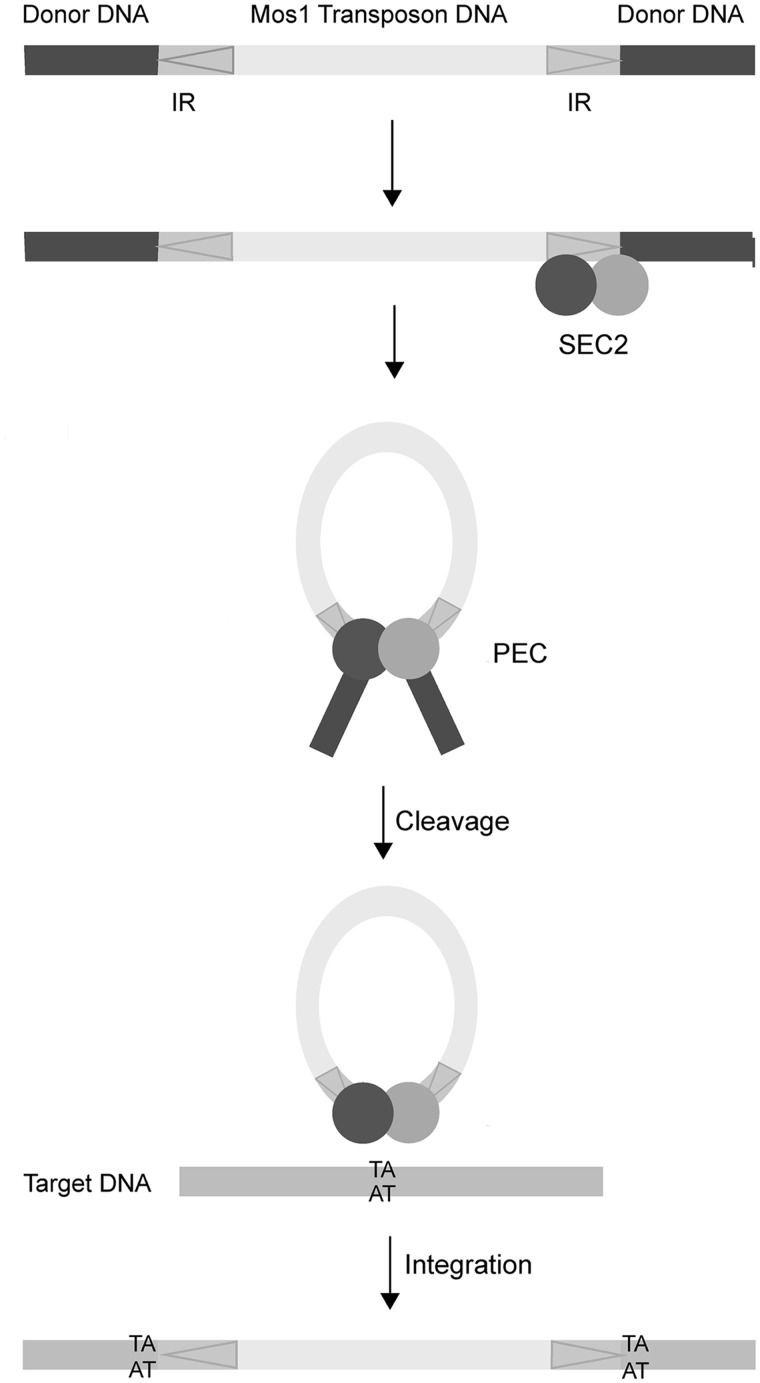
Mariner/Tc1 elements move via a DNA intermediate, in a cut-and-paste mechanism that brings the two ends of the transposon together (Figure 1). The ends are separated by 1–2 kb and marked by inverted repeat (IR) DNA sequences. Transposition is orchestrated by a transposase enzyme (encoded by the transposon sequence itself) within an ordered series of nucleoprotein complexes. Initially the specific sequence of the IR DNA is recognized by the transposase DNA-binding domain. The two transposon ends are then brought together, in a paired-end complex (PEC), and precisely cleaved at the IRs before being inserted at a new genomic site. DNA transposases act as oligomers to bring the two transposon ends together. This provides at least two catalytic sites for the excision reactions at each end as well as the insertion reactions to covalently join the cleaved transposon to a new target site. Various strategies for looping of DNA to assemble the paired-end transposition complex are possible (8). A pre-formed transposase oligomer can bind to one transposon end before recruiting the second naked end, as occurs during P-element transposition (9), or the transposase can oligomerize after initially binding separately to each IR or to just one IR.
Figure 1.
Schematic of Mos1 transposition. The 1.3 kB transposon has 28 bp inverted repeats (IR) indicated with arrows. Binding of a transposase dimer to one end forms a single-end complex (SEC2). The transposon is fully excised from donor DNA in the paired-end complex (PEC). Mariner/Tc1 transposons integrate at TA sequences, resulting in signature duplications either side of the inserted transposon.
The mechanism of transposition of the active mariner/Tc1 transposon Mos1 (from Drosophila mauritiana) has been studied from both biochemical and structural perspectives (10–14). The order and requirements for DNA excision have been established (10,11), and crystal structures have been determined of the transposase C-terminal catalytic domain (11) and the paired-end complex of transposase and cleaved transposon DNA (14). The PEC structure, representing the Mos1 transposition machinery after DNA cleavage and before transposon insertion into target DNA, contains a transposase dimer bound to two cleaved transposon ends in a crossed arrangement: each 28 bp IR sequence is recognized by the N-terminal DNA binding of one transposase monomer and by the C-terminal catalytic domain of the other monomer. The bipartite DNA-binding domain contains two helix-turn-helix (HTH) motifs. The N-terminal 55 residues (containing HTH1) also form part of the transposase dimerization interface within the PEC (14).
The molecular events occurring earlier in the Mos1 transposition pathway, leading to PEC formation, remain poorly understood. Mos1 transposase forms a dimer in solution without DNA (11), and it has been proposed that initially a transposase dimer binds rapidly to one transposon end, in a single-end complex (SEC2) (13,15); the N-terminal 35 residues were implicated in cis-Mos1 transposase dimerization, leading to binding of a single IR (12). The PEC could then form by recruitment of the other naked IR to the complex. Divalent Mg2+ or Mn2+ ions are required for PEC formation, as well as DNA cleavage of mariner transposons (10,13,14,16,17). The metal ions are coordinated by a triad of aspartic acid residues in the RNase-H-like catalytic core of the C-terminal catalytic domain (11). Metal ions may play an additional role in stabilizing protein–DNA interactions in the PEC and/or promoting the correct conformation of this complex (15). It has been suggested that nicking of one DNA strand can occur before pairing of the transposon ends (10,16) and that this cleavage event may promote a conformational change that facilitates PEC assembly. However, this proposal is controversial; the paradigm, established for bacterial DNA transposases, is for pairing of transposon ends before DNA catalysis. More recently Carpentier et al. (18) argued that PEC assembly is indeed required for first strand cleavage of Mos1 inverted repeats.
We sought to establish the architecture of the full-length Mos1 transposase prior to DNA binding, and the conformation of the pre-cleavage SEC2. Attempts to crystallize the full-length transposase without DNA have not been successful. However, solution scattering methods can provide structural parameters and low resolution conformations of proteins and complexes. Neutron scattering contrast variation in particular can distinguish the conformations and relative spatial arrangements of the constituents of a nucleoprotein complex. This technique exploits the significantly different scattering length densities of protein molecules compared with DNA, and of hydrogen compared with deuterium (19). The contrast between the solute and the solvent can be modulated by altering the solvent D2O:H2O ratio. The contrast is matched, at a particular D2O:H2O ratio, when the scatter from a solute molecule equals that of the solvent; the scattering from solute is therefore eliminated when the scattering from the solvent is subtracted. Thus, it is possible to mask the protein component of a complex, so that only the DNA component contributes to the overall scattering, and vice versa. Incorporation of deuterium-labelled proteins in a complex extends the range of contrasts available beyond those originally developed by Stuhrman (20) and reduces the incoherent background scatter from hydrogen in the sample. Dedicated facilities for the production of deuterated macromolecules have been developed within ILL’s Life Science Group (the D-LAB), and there are now numerous examples illustrating the power of deuteration approaches in small angle neutron scattering (SANS) (21–24), neutron crystallography (25) and dynamics (26).
Here we have combined complementary methods to obtain low resolution solution structures of two early molecular intermediates in Mos1 transpositon: Mos1 transposase and its pre-cleavage, single-end complex with transposon DNA. We show that the DNA-free transposase homodimer adopts an extended conformation in solution, in contrast to the more compact arrangement of the crossed dimer in the PEC crystal structure. The first 55 residues of the transposase DNA-binding domain are essential for dimerization. The elongated conformation is slightly extended in the SEC2 formed under non-catalytic conditions, and the DNA is bound predominantly to one half of the transposase dimer. We propose that rotation of the one transposase monomer about the DNA-binding domain, coupled to binding of a second transposon DNA end, could provide a mechanism to form the compact PEC architecture previously determined by X-ray crystallography.
MATERIALS AND METHODS
Expression and purification of hydrogenated and deuterated Mos1 transposases
Mos1 transposase (containing the solubilizing mutation T216A), and hereafter referred to as H-Mos1, was expressed and purified as previously described (27). Partially deuterated Mos1 transposase (D-Mos1) was expressed in Escherichia coli BL21(DE3) cells in minimal medium (28) containing 85% (v/v) recycled D2O. Media was inoculated with hydrogenated cell culture to 1% (v/v) and incubated at 37°C for 2–4 days. Protein expression was induced at 30°C with isopropylthiogalactoside (1 mM), and the cells were harvested after 5 h. D-Mos1 purification was performed as described previously (27) except that cation exchange was performed on a SP-Sepharose FF column. The yield of D-Mos1 was 0.75 mg per litre of culture. Proteins were stored at −20°C in 25 mM Tris pH 7.5, 250 mM NaCl, 1 mM DTT and 50% (v/v) glycerol.
Mass spectrometry of deuterated Mos1 transposase
The mass of D-Mos1 was measured as 42 166 Da by MALDI mass spectrometry. Thus, the level of deuterium incorporation was 52.2% in H2O and 77.7% in D2O (assuming exchange of all labile hydrogens and the loss of the first methionine in the sequence of the peptide).
Cloning of the N-terminal truncation mutant delta-55 Mos1
The sequence coding for Mos1 transposase lacking the N-terminal 55 amino acids was amplified from the full-length Mos1 gene using forward (5′-TACGTCATATGGGCGATTTTGATGTGGATG-3′) and reverse primers (5′-TACGGCTCGAGTTATTCAAAGTACTTGCC-3′), where the restriction sites are underlined and the stop codon is in italics. The 895 bp PCR product was digested with NdeI and XhoI enzymes (NEB), gel-purified and ligated (Rapid Ligation Kit, Roche) with similarly digested, gel-purified and de-phosphorylated pET30a or pET28a vector. The integrities of the resulting clones (pET30-D55 and pET28-D55) were confirmed by Sanger sequencing.
Expression and purification of deletion mutants
The delta-55 Mos1 mutant without a tag was expressed from the pET30a-D55 plasmid. The 290 residue protein (34.34 kDa) was purified by cation exchange and gel-filtration chromatography, as for full-length transposase. The N-terminal 130 amino acids of Mos1 were expressed as previously described (29), and purified by elution from an IMAC FF column (GE Healthcare) with Imidazole, followed by cation exchange chromatography. The N-terminal six His tag was removed by thrombin cleavage (5 units/mg of purified protein) in PBS at 17°C for 16 h prior to gel-filtration. The resulting protein contained 133 amino acids (with a molecular mass of 15.5 kDa).
Preparation of transposase samples for SANS experiments
H-Mos1 and D-Mos1 protein samples were exchanged multiple times (in a viva spin concentrator or by dialysis, respectively) into fully hydrogenated and fully deuterated buffers containing 25 mM Tris (pH or pD 7.5), 350 mM KCl and 1 mM DTT. Samples of D-Mos1 in buffer containing 30% (v/v) D2O or 65% (v/v) D2O were then prepared by mixing together appropriate volumes of D-Mos1 in fully hydrogenated and fully deuterated buffer. The concentration of each sample was calculated from triplicate measurements of absorbance at 280 nm on a Shimadzu UV-2401PC UV-Visible spectrophotometer.
DNA substrates
The DNA duplex for SEC2 was prepared by annealing two complementary 50-mers containing the 28 base IR sequence specifically recognized by transposase, surrounded by 8 bases of transposon sequence and 14 bases of flanking DNA. These had the sequences (5′ ttt aaa aa AAA CGA CAT TTC ATA CTT GTA CAC CTG A tag ttt cta tat tc) and (5′ gaa tat aga aac taT CAG GTG TAC AAG TAT GAA ATG TCG TTT ttt tta aa), where the IR sequence is in upper case. Oligonucleotides were synthesized and PAGE purified by Integrated DNA Technologies. Lyophilized samples were dissolved in either H2O or D2O and annealed by heating molar equivalents of complementary strands to 90°C for 10 min and cooling to 25°C in steps of 2°C per 30 s.
Preparation of transposase DNA complexes
Single-end complexes were prepared by adding either H-Mos1 or D-Mos1 to DNA in the molar ratio 2:1 in 20 μl aliquots. For contrast variation experiments, four complexes were prepared, with different ratios of H2O:D2O in the buffer. Two complexes (H-SEC2) contained H-Mos1 and 0% (v/v) or 100% (v/v) D2O and two contained D-Mos1 in buffers with 65% (v/v) or 100% (v/v) D2O (D-SEC2). Sample concentrations were calculated from the average of three absorbance measurements at 280 nm and 260 nm.
Gel-Filtration of transposase–DNA complexes
Samples were separated at room temperature on a Superdex 200 10/300GL column (GE Healthcare). Isocratic elution with buffer containing 20 mM Tris pH 7.5, 0.3 M KCl and 1 mM DTT was monitored by absorbance at 260 nm and 280 nm. Eluted fractions containing SEC2 were loaded onto a 15% SDS-PAGE along with 6 internal standards containing 150 ng, 300 ng or 450 ng of Mos1 transposase or DNA. The gel was silver-stained for protein and DNA, and scanned using a BioRad GelDoc EZImager (GE Healthcare). The intensity of bands was quantified with EZImager using a uniform box size and mean background subtraction.
Size-Exclusion Chromatography Multi-Angle Laser Light Scattering (SEC-MALLS)
The molar mass of transposases, duplex DNA and the SEC2 complex was determined by SEC-MALLS. Samples were separated at room temperature on a Superdex 200 10/300GL column linked to either an ÄKTA Ettan (GE Healthcare) or Shimadzu HPLC system. The column was pre-equilibrated with at least two column volumes of buffer (as above). Elution was performed isocratically at 0.5 ml/min and monitored by absorbance at 280 nm. The HPLC system was connected to a DAWN HELIOS II™ MALLS instrument (Wyatt Technology) and Optilab T-rEX refractometer (Wyatt Technology). On-line measurement of the intensity of the Rayleigh scattering as a function of the angle of the eluting peaks was used to determine the weight average molecular masses (Mw) of the eluted samples, using the ASTRA™ (Wyatt Technologies) software.
Small angle X-ray scattering (SAXS) data collection and processing
SAXS data were collected at the European Synchrotron Radiation Facility (ESRF) (beam line ID14-3, λ = 0.931 Å) on a Pilatus 1 M detector. The sample to detector distance was 2.43 m, and scattering data were collected within the momentum transfer (q) range 0.001–0.35 Å−1. To avoid radiation damage, the sample (30 μl) was pushed through the capillary cell during data collection. Data were collected in multiple 30 s frames, inspected and averaged in PRIMUS(30) normalized to the incident beam intensity and the scattering of the buffer subtracted. To check for concentration-dependent effects, scattering data were collected from H-Mos1 samples at 1.45 mg/ml, 2.9 mg/ml and 4.4 mg/ml. SAXS data were calibrated against bovine serum albumin (BSA) at 5.6 mg/ml. The molecular masses of the transposase and the complex were calculated from the extrapolated intensity at zero angle I(0), obtained with GNOM, by comparison with the I(0) of BSA (molecular mass 66 kDa).
SANS data collection and processing
All SANS data were collected at the high neutron flux reactor at Institut Laue Langevin on beam line D22 equipped with a 3He Reuter-Stokes® multi-detector. Samples (minimum volume 200 μl) were placed in 1.0 mm path length quartz cuvettes, sealed and transferred to a sample changer maintained at 23°C. Scattering data were collected at sample-detector distances of 2 m and 8 m for 900 s and 3600 s respectively. Transmission data were collected at a sample-detector distance of 8 m for 180 s. In each case the collimation distance was equal to the sample-detector distance.
SANS data were reduced in GRASansP v6.01 beta (www.ill.fr/lss/grasp). Data were normalized to the incident beam intensity and corrected for transmission, cuvette thickness and detector efficiency. Scattering from the sample holder and instrument background were then subtracted and the data averaged radially around the beam centre and normalized. Data collected at 2 m and 8 m sample-detector distances were then scaled and merged in MS-EXCEL. The range of momentum transfer sampled was 0.007 < q < to 0.35 Å−1. The Rg and I(0) values for each solute were extrapolated from a Guinier plot of the very low angle scattering data in PRIMUS and GNOM. Molecular masses of the hydrogenated solutes were estimated using water scattering as a reference (31,32).
Molecular modelling of Mos1 transposase
Low resolution protein structure models were reconstructed by ab initio simulated annealing, representing the protein as a chain-like ensemble of dummy residues in GASBOR (33). Ten models of Mos1 transposase were generated in separate runs, and aligned pair-wise with SUPCOMB (34) to compute the normalized spatial discrepancy (NSD) between models. The quaternary structure of Mos1 transposase was modelled with SASREF (35) on the ATSAS server (http://www.embl-hamburg.de/biosaxs/atsas-online). A dimeric arrangement of subunits was constructed by simulated annealing using the atomic structure of a transposase monomer from the PEC crystal structure (PDB ID: 3HOT) and minimized against the experiment SAXS data.
MONSA analysis
Low resolution models of the SEC2 complex were calculated using MONSA (36) at the ATSAS server. Five transposase–DNA complex data sets were included: one SAXS data set of H-SEC2, and four SANS data sets of H-SEC2 or D-SEC2 at different H2O:D2O ratios. Contrasts for each component of the complex were based on the isotopic composition of the sample for a given D2O buffer composition (Supplementary Table S1).
RESULTS
H-Mos1 transposase structural parameters
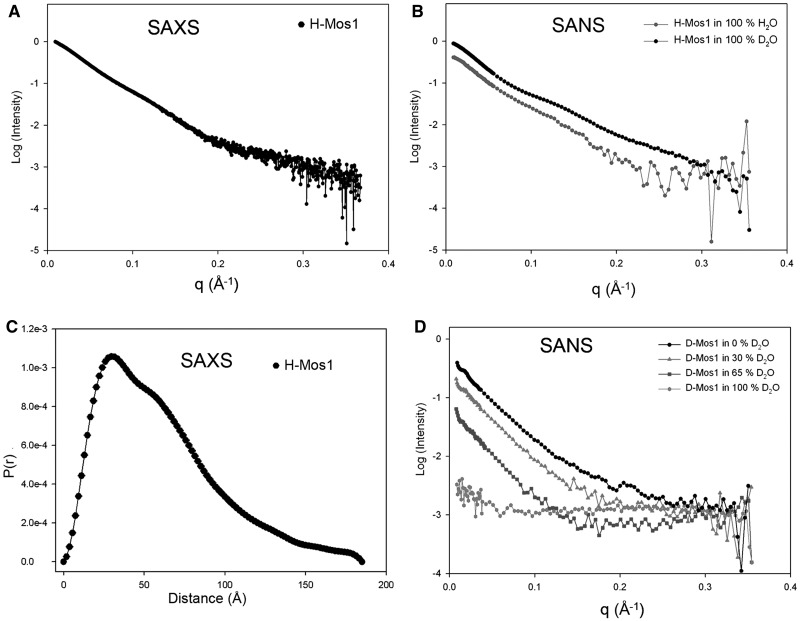
SAXS data were collected from a solution of hydrogenated transposase (H-Mos1) at 1.8 mg/ml (Figure 2A), and SANS data were collected from solutions of H-Mos1 in 100% H2O and 100% D2O (Figure 2B). By Guinier analysis, the average radius of gyration (Rg) of the transposase was 50.3 ± 2.3 Å (Table 1). The molecular weight of the H-Mos1 transposase was calculated to be 87.5 ± 7.5 kDa, from the scattering intensity extrapolated to zero angle (I(0)). This compares with a theoretical molecular mass of the transposase monomer of 40.7 kDa. Thus the SAXS and SANS data confirm that Mos1 transposase is a dimer in solution, consistent with previous conclusions from gel-filtration analysis (11).
Figure 2.
Small angle scattering of Mos1 transposase. (A) SAXS data of H-Mos1. (B) SANS of H-Mos1 in 100% H2O and 100% D2O. (C) Pair distribution function, P(r), computed from the SAXS data. (D) SANS of D-Mos1 in 0, 30, 65 and 100% D2O.
Table 1.
Structural parameters for Mos1 extracted from experimental SANS and SAXS data and the PEC crystal structure
| Sample | % D2O | Conc (mg/ml) | I(0) | MW (kDa) | Rg (Å) Guinier | Rg (Å) Gnom | Dmax (Å) | |
|---|---|---|---|---|---|---|---|---|
| SAXS | H-Mos1 | 0 | 1.8 | 13.5 | 79.9 | 49.2 ± 0.1 | 49.5 ± 0.2 | 185 |
| SANS | H-Mos1 | 0 | 7.5 | 0.433 | 96.2 | 51.9 ± 0.7 | 53.4 ± 0.5 | 180 |
| H-Mos1 | 100 | 8.8 | 0.913 | 89.5 | 51.2 ± 0.4 | 55.9 ± 0.2 | 180 | |
| SANS | D-Mos1 | 0 | 1.1 | 0.38 | 86.4a | 51.5 ± 0.7 | 55.1 ± 0.6 | 180 |
| D-Mos1 | 30 | 1.1 | 0.15 | 51.1 ± 1.5 | 53.7 ± 0.9 | 180 | ||
| D-Mos1 | 65 | 1.2 | 0.049 | 53.7 ± 2.1 | 53.0 ± 2.1 | 180 | ||
| D-Mos1 | 100 | 1.2 | 0.01 | |||||
| Crystal Structure | Mos1 | 81.3 | 38b | 110 |
The coordinates for the Mos1 dimer in the PEC were extracted from the PDB file 3HOT.
aRenormalized for mass of H-Mos1.
bCaculated using CRYSOL.
The SAXS and SANS data were transformed using GNOM into a distribution of paired distances, P(r), of all inter-atomic vectors in H-Mos1; the P(r) distribution calculated from the SAXS data is shown in Figure 2C. The maximum dimension (Dmax) of the transposase dimer was in the range 180–185 Å (Table 1).
SANS of D-Mos1 transposase
The use of deuterated transposase (D-Mos1) in neutron scattering extends the range of contrasts available for contrast variation experiments; proteins with ∼75% deuterium incorporation are contrast matched in 100% (v/v) D2O. We prepared partially deuterated D-Mos1 as described in the methods. To determine experimentally the contrast match point of the sample, and to confirm its integrity, we measured neutron scattering curves of D-Mos1 in buffer containing 0, 30, 65 or 100% (v/v) D2O (Figure 2D). The structural parameters Rg and Dmax were consistent with those measured for H-Mos1 (Table 1). The scattering amplitude at zero angle was calculated for each curve and plotted against the % D2O content of the buffer (Supplementary Figure S1). The linear best fit to these points intercepted the x-axis at 92% (v/v) D2O, establishing the contrast match point for D-Mos1.
Comparison of the DNA-free and PEC Mos1 dimers
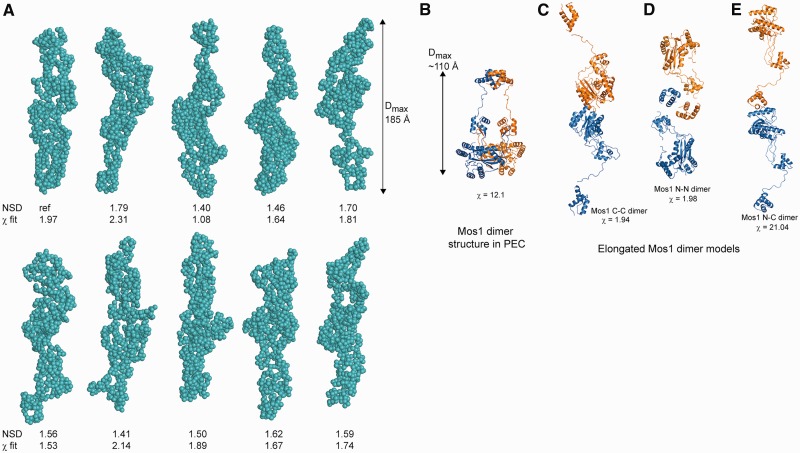
We compared the experimentally measured structural parameters Rg and Dmax for the H-Mos1 homodimer in solution with those values calculated using the crystallographic atomic coordinates of the transposase dimer within the Mos1 PEC. Coordinates for all but the N-terminal 3 residues of the sequence (which are likely to be disordered) are defined in the crystal structure. The theoretical Rg for the compact and crossed arrangement of the PEC dimer was 38 Å (compared with the measured value of 50.3 Å), and the Dmax was significantly shorter at ∼110 Å (Table 1). A theoretical scattering curve for the transposase was calculated with CRYSOL using the crystallographic atomic coordinates (Supplementary Figure S2A), but the agreement between the theoretical, PEC model-based scattering curve and the experimentally measured SAXS data was poor (χ = 12.1). This indicates that the conformation of the transposase dimer when free in solution is significantly different to that when bound to two transposon DNA molecules in the PEC.
Shape of the Mos1 Dimer
Next we performed ab initio reconstruction of the shape of the transposase dimer in solution from the SAXS data. The protein was considered as a chain-like ensemble of dummy residues in the program GASBOR (37), with no symmetry restraints imposed. A gallery of 10 solutions are shown in Figure 3A. Calculations were also performed with P2 symmetry imposed and these gave similar results (Supplementary Figure S3). The molecular envelopes each have a markedly elongated, narrow shape, which contrasts with the more compact, crossed architecture of the transposase dimer in the Mos1 PEC (Figure 3B).
Figure 3.
Solution conformations of Mos1 transposase. (A) Gallery of ten spherical bead models of the H-Mos1 dimer calculated from the SAXS data in GASBOR, with no symmetry imposed. The χ of the fit to the experimental SAXS data and the NSD between models is indicated below each model. (B) Compact structure of the Mos1 dimer in the PEC crystal structure (from PDB ID: 3HOT); one Mos1 monomer is blue and the other orange. (C) An elongated tail-to-tail Mos1 dimer with a catalytic domain dimerization interface (C-C model). (D) Alternative elongated head-to-head model with a DNA-binding domain interface (N-N model). (E) The elongated head-to-tail (N-C) model fitted poorly to the scattering data.
Relative orientation of domains
To establish how the monomers are arranged in the DNA-free transposase homodimer, we modelled the elongated quaternary structure by multi-subunit rigid body calculations. The atomic coordinates of a transposase monomer from the PEC were used as the starting point. An exhaustive grid search of homodimer configurations, and their fit to the experimental SAXS data, was performed. To allow for potential variations in the orientations of domains about flexible linkers, the monomer structure was split into three separate domains: HTH1 contained residues Val 5 to Gly 56, HTH2 comprised residues Asp 57 to Gly 117 and the catalytic domain spanned Arg 118 to Glu 345. Conditional distance restraints of 7 Å were defined between terminal residues of the three separated subunits adjacent in the protein sequence.
Two possible models for the homodimer emerged from this analysis, each fitting reasonably to the SAXS curve. In the first tail-to-tail model, shown in Figure 3C, the two catalytic domains are in contact at the centre of the elongated homodimer, with the DNA-binding domains distal from each other at the peripheries. This model fitted to the SAXS curve with χ = 1.94 (Supplementary Figure S2B). In the second head-to-head model, the DNA-binding domains are in close proximity at the centre of the homodimer (Figure 3D), with the HTH1 domains in contact. This model fitted to the SAXS data equally well, with χ = 1.98 (Supplementary Figure S2C). Head-to-tail models of the homodimer, in which the DNA-binding domain of one monomer contacts the catalytic domain of the second monomer (Figure 3E), had significantly lower quality fits to the experimental data, with χ = 21.0 (Supplementary Figure S2D).
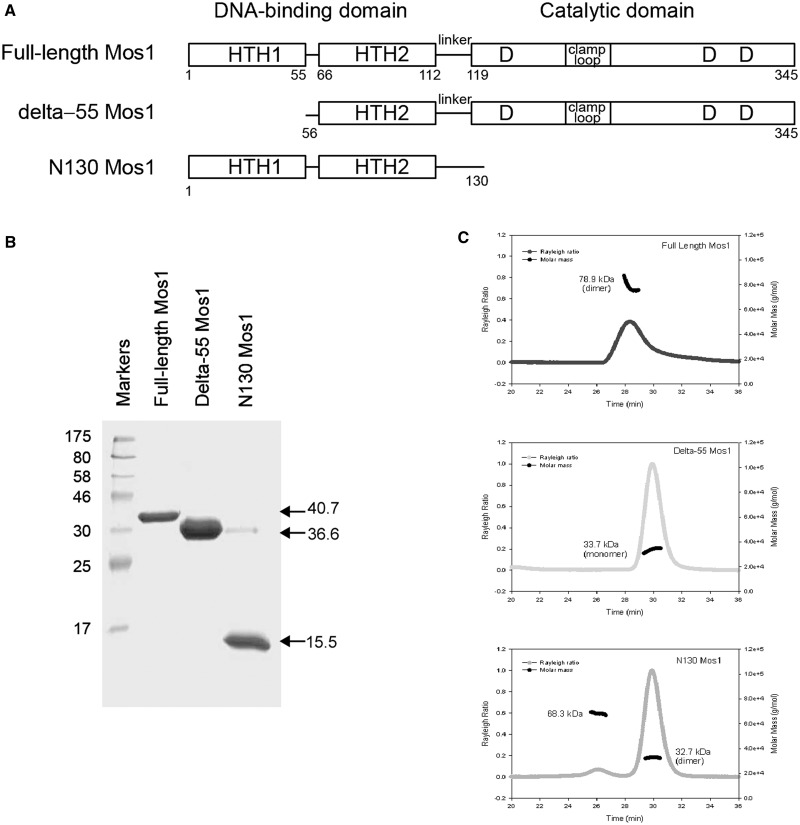
To distinguish between the tail-to-tail and head-to-head models, we created deletion mutants of Mos1 transposase (Figure 4A). We hypothesized that deletion of HTH1 would have little effect on transposase dimerization in the tail-to-tail model of the elongated dimer (Figure 3C) but would disrupt dimerization if the DNA-binding domains were in contact, as in the head-to-head model (Figure 3D). The converse would be true for mutants in which the catalytic domain had been deleted.
Figure 4.
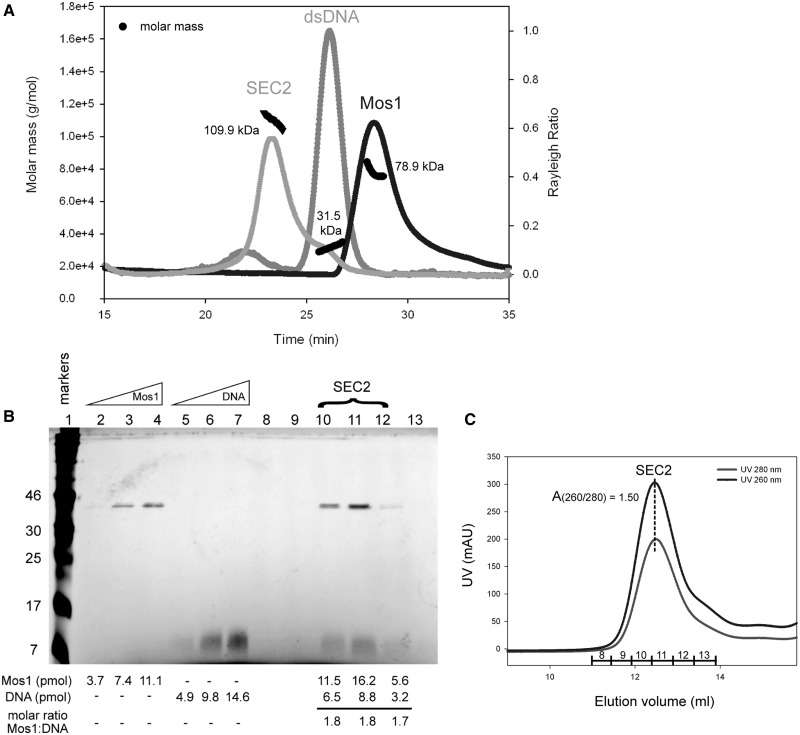
Analysis of Mos1 transposase deletion mutants. (A) Domain arrangements of full-length Mos1, the HTH1 domain deletion mutant delta-55 Mos1 and the C-terminal deletion mutant N130 Mos1. (B) SDS-PAGE of purified transposases. The molecular weights of the markers are indicated in kDa to the left of the gel and the mass by sequence of each sample is shown on the right of the gel. (C) SEC-MALLS analysis. The scattering intensity (Rayleigh Ratio) is plotted against elution time of full-length Mos1, delta-55 Mos1 and N130 Mos1. The molar mass distribution across each peak is shown in black.
A deletion mutant of Mos1 transposase lacking the N-terminal 55 residues (delta-55 Mos1) was expressed and purified (Figure 4B). Analysis by SEC-MALLS established that delta-55 Mos1 eluted from the gel-filtration column after 30 min with an average Mw of 33.7 kDa ± 2%, (Figure 4C). This mass corresponds to a monomeric species. By comparison, the full-length H-Mos1 homodimer eluted at 28.2 min and had an average Mw of 78.9 kDa ± 2%, consistent with a dimer. Thus deletion of the N-terminal 55 residues (containing HTH1) results in loss of transposase dimerization.
Next we performed the converse experiment. We expressed and purified the DNA-binding domain of Mos1, containing only the N-terminal 130 amino acids, as described previously (29) (Figure 4B). SEC-MALLS analysis showed that this deletion mutant eluted at 29.9 min with a molecular mass of 32.7 kDa (Figure 4C) and thus formed a dimer, like the full-length transposase. A minor proportion of the sample (<1.5%) eluted after only 26.1 min and had a molecular mass of 68.3 kDa. A silver-stained SDS-PAGE of the eluted fractions confirmed the minor component is a tetramer of transposase (Supplementary Figure S4). Taken together these results are consistent with the head-to-head model of the Mos1 homodimer in which the DNA-binding domains are at the centre of the elongated molecule, with the HTH1 domains in close contact as shown in Figure 3D.
Stoichiometry of the pre-cleavage single-end complex
Previously we proposed that Mos1 transposition is initiated by a transposase dimer binding to the inverted repeat sequence at one transposon end (14), forming a single-end complex (SEC2). This complex has been observed by EMSA under non-catalytic conditions: for example, when the Mg2+ ions required for PEC formation and DNA cleavage are excluded from the reaction, or if one of the catalytic Asp residues that coordinates Mg2+ is mutated to Ala [Supplementary Figure S6 in ref (14)]. We prepared a single-end complex (H-SEC2) by mixing H-Mos1 with a 50 mer DNA duplex containing the 28 bp transposase inverted repeat recognition sequence (as described in the methods). SEC-MALLS analysis was performed to confirm the homogeneity and composition of H-SEC2. Samples of the DNA duplex and H-Mos1 were also analysed separately as controls. The SEC2 was loaded onto the gel-filtration column at an initial concentration of 7 μM, and eluted in a single peak with an average molecular mass, measured by MALLS, of 109.9 kDa ± 0.5% (Figure 5A). This is close to the expected mass of 112 kDa for a complex containing a dimer of transposase (81 kDa) and a single ds DNA molecule (31 kDa). By comparison, the predicted mass of SEC1 (transposase monomer bound to one DNA duplex) is 71.4 kDa and the PEC (dimer bound to two DNA molecules) is 143 kDa.
Figure 5.
Mass and stoichiometry of SEC2. (A) Elution profile from SEC-MALLS. The scattering intensity (expressed as the Rayleigh ratio) is plotted for SEC2, Mos1 and ds DNA. The average measured Mw for each sample is indicated in kDa, and the molar mass distribution across each peak shown in black. The DNA duplex eluted earlier than the H-Mos1 dimer, despite its lower mass, due to its long and narrow shape. (B) Silver-stained SDS-PAGE confirms the stoichiometry of SEC2. Protein molecular weight markers (lane 1); internal standards containing 150, 300 and 450 ng of Mos1 transposase (lanes 2-4, respectively) or 50mer DNA (lanes 5–7, respectively); SEC2 gel-filtration fractions (lanes 8–13). The number of pmols of protein and DNA in each lane is shown below each lane. The molar ratio of Mos1 to DNA in SEC2 is also given below lanes 10–12. (C) Gel-filtration elution profile of SEC2 monitored by UV absorption at 260 nm and 280 nm. The UV absorbance at the peak maximum is 307 mAU at 260 nm and 205 mAU at 280 nm. The SEC2 fractions are labelled across the peak in correspondence with the SDS-PAGE lane numbers in (B).
To confirm the content and stoichiometry of the complex, we analysed fractions of SEC2, eluted by gel-filtration chromatography, by SDS-PAGE silver-stained for both protein and DNA (Figure 5B). We quantified the number of pmoles of transposase and DNA seen in each lane containing SEC2, by comparing the band intensities with those of three internal protein controls or three DNA controls, respectively. The molar ratio of transposase to DNA in the complex was estimated to be 1.8, compared with a ratio of 2 expected for SEC2.
Further confirmation of the stoichiometry of SEC2 was obtained from the ratio of UV absorbance at 260 nm and 280 nm during gel-filtration (Figure 5C). At the gel-filtration peak maximum this ratio was 1.50. We calculated the theoretical UV absorbance ratios for SEC1, SEC2 and PEC by summing the extinction coefficients of the Mos1 and DNA components according to the stoichiometry of each complex (Supplementary Table S2). The theoretical absorbance ratios are 1.49 for SEC2 and 1.63 for both SEC1 and the PEC. Thus the experimentally measured ratio of 1.50 is consistent only with SEC2.
Solution scattering of SEC2 and contrast variation
To establish the solution conformation of the pre-cleavage complex, we measured the small angle scattering from five SEC2 samples, prepared with a 50 mer DNA duplex (containing the 28 bp IR DNA sequence) and either H-Mos1 or D-Mos1 (Table 2). A range of contrasts between the components of the complex and the buffer were used in the neutron scattering experiments to enable the structural parameters, shape and relative spatial arrangement of the protein and DNA to be extracted. The use of deuterated transposase extended the range of contrasts available and provided enhanced scattering from the transposase, despite the lower concentration available.
Table 2.
Analysis of SANS and SAXS data of transposase–DNA single-end complexes
| Sample | % D2O | Conc (mg/ml) | I(0) | MW | Rg (Å) Guinier | Rg (Å) GNOM | Dmax (GNOM) (Å) | χ fit to data MONSA |
|---|---|---|---|---|---|---|---|---|
| SAXS | ||||||||
| H-SEC2 | 0 | 2.3 | 0.53 | 57.8 ± 3.5 | 62.5 ± 2.1 | 220 | 0.74 | |
| SANS | ||||||||
| H-SEC2 | 0 | 4.3 | 0.38 | 109.2 | 58.1 ± 0.7 | 60.1 ± 0.8 | 220 | 2.03 |
| H-SEC2 | 100 | 2.6 | 0.22 | 105.6 | 58.2 ± 0.5 | 60.6 ± 0.5 | 220 | 2.41 |
| D-SEC2 | 65 | 1.3 | 0.051 | 59.1 ± 3.5 | 60.37 ± 2.2 | 190 | 2.18 | |
| D-SEC2 | 100 | 1.4 | 0.009 | 42.4 ± 3.9 | 40.8 ± 0.4 | 125 | 0.85 |
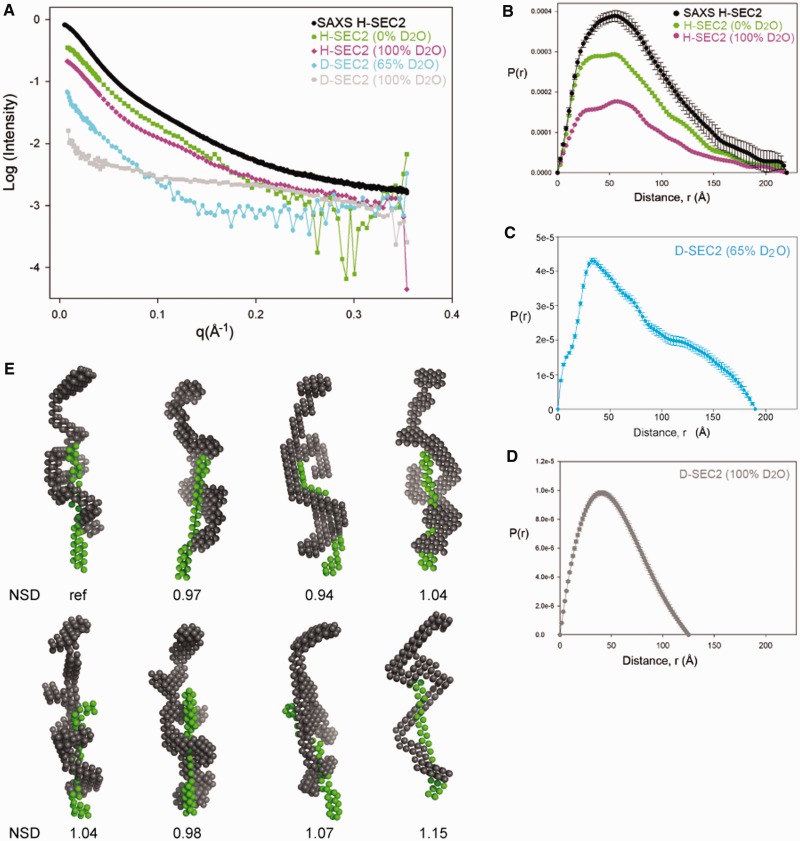
The scattering data collected are shown in Figure 6A. In the SANS experiments, the relative contribution of each component of the complex to the total scattering intensity is dependent on the H2O:D2O ratio of the solvent in each sample, as well as the deuteration level of the protein (Supplementary Table S2). In particular, at 100% (v/v) D2O solvent, the transposase component of D-SEC2 is close to the contrast match point and the measured scattering is dominated by the DNA component. Conversely, at 65% (v/v) D2O solvent content, the DNA is contrast matched and the measured scattering intensity is primarily from the protein in the complex. The scattering intensities measured from H-SEC2 in buffer containing 0% (v/v) or 100% (v/v) D2O reflect the parameters and conformation of the whole complex. We also collected SAXS data on the H-SEC2 complex and in this case the contrast of the DNA relative to the solvent was twice that of the protein.
Figure 6.
Small angle scattering and solution conformations of SEC2. (A) SAXS and SANS data of SEC2 complexes prepared with H-Mos1 and D-Mos1. (B) Pair distribution functions, P(r), of H-SEC2 complexes measured by SAXS (black) and SANS in 0% D2O (green) and SANS in 100% D2O (purple). The Dmax reflects the maximum dimension of the complex. (C) P(r) of D-SEC2 in 65% D2O reflects the shape and Dmax of the protein component of the complex only (DNA contrast matched). (D) P(r) of D-SEC2 at 100% D2O is dominated by the contribution from DNA. (E) Gallery of 8 bead models of SEC2 generated using MONSA, containing a DNA phase (green) and the protein phase (grey). Models were aligned in DAMAVER and the NSD for each model is indicated.
SEC2 model-independent structural parameters
We measured the molecular mass of H-SEC2 from the SANS experiment in 100% (v/v) H2O, and the value of 109.2 kDa was in agreement with the mass measured by SEC-MALLS. Consistent with this, the average Rg of H-SEC2 was 60.4 ± 0.8 Å (an increase from 50.3 Å for the protein in the absence of DNA) and the average Dmax was 220 Å (Figure 6B), suggesting that the complex is ∼35–40 Å longer than the DNA-free transposase dimer. From the scattering in 65% (v/v) D2O, where the DNA is masked, the Rg of the transposase within D-SEC2 was measured as 59.1 ± 3.5 Å. This is larger than the Rg of the DNA-free transposase dimer (53.7 ± 2.1 Å) and indicates that the protein conformation has opened up upon binding DNA and forming SEC2. This is also reflected by the larger Dmax of the transposase in D-SEC2 (Figure 6C): 190 Å compared with 180 Å for the DNA-free Mos1 dimer. From the scattering in 100% D2O, the Dmax of the DNA was 125 Å (Figure 6D). This compares with a Dmax of 140 Å and Rg of 40.2 Å for the DNA duplex alone (SAXS data not shown). Taken together these structural parameters indicate that the SEC2 has an elongated shape and that the transposase dimer has a slightly more elongated conformation within the complex than in its DNA-free state.
Next we used the Rg values for the complex and its components to establish the placement of the DNA relative to the transposase in H-SEC2. The distance between the centres of mass of the transposase and DNA can be related to the measured Rg values by the parallel axes theorem (38), given by the equation:
where L is the distance in Å between the centres of mass of the transposase and the DNA, and x is the fraction of total scattering contributed from Mos1 transposase (38). The Mos1 dimer contributes 80% of the volume of H-SEC2. Because the protein has a contrast of 2.5 in 100% H2O and the contrast of the DNA is 4.5, the fraction (x) of the scattering from Mos1 is 0.69 (Supplementary Table S1). Using the measured Rg values of the complex and its protein and DNA components (58.1, 59.1 and 42.4 Å, respectively), the distance L is ∼44 Å. Thus the centre of the DNA is displaced from the transposase centre of mass by a distance corresponding to approximately one fifth of the Dmax of the complex. This indicates that the DNA is bound predominately to one half of the transposase dimer.
Conformation of the pre-cleavage complex
Next we used multi-phase analysis of the scattering curves, collected at the different phase contrasts, to calculate a low-resolution envelope for the whole complex in which the transposase and DNA components are also distinguished. A gallery of the average bead models from eight separate simulations, performed using MONSA, is shown in Figure 6E. Models were aligned to each other using DAMAVER and the NSD is indicated below each model. The DNA phase has a long narrow shape consistent with the conformation of linear B-form DNA and is localized predominantly to one half of the complex. A B-form DNA duplex of 50 nucleotides can be superimposed on this phase (Figure 7). The transposase phase adopts an elongated conformation which is extended compared with the DNA-free Mos1 dimer, with a small dimerization interface. Two monomers of Mos1 can fit into the envelope (fitting performed by eye) so that the DNA is predominantly associated with one monomer, while the other monomer is presumed to be more flexible.
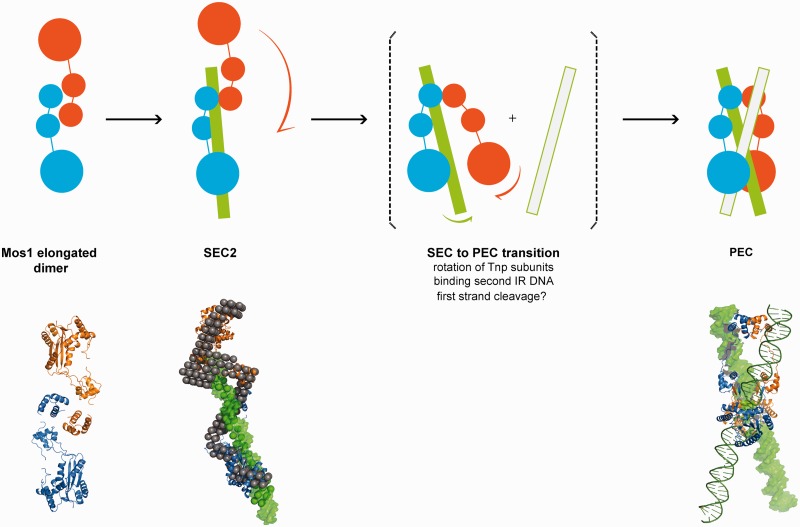
Figure 7.
Schematic mechanism of early steps in Mos1 transposition. Transposase (blue and orange monomers) exits as an elongated dimer in solution with the N-terminal domains at the central dimerization interface. When bound to one IR DNA in SEC2, the transposase dimer becomes more extended and the DNA (green) is predominantly associated with one monomer (blue); the other monomer (orange) may be more flexible. The phase model was interpreted by docking Mos1 monomers (ribbon) by eye to the protein phase and a 50 mer DNA duplex (green surface) to the DNA phase. The PEC could form by rotation of the flexible monomer about the HTH1 domain, accompanied by binding of the second transposon end. There may also be exchange of DNA between the two catalytic domains. Solution structures of the Mos1 dimer and SEC2, and the PEC crystal structure (3HOT) are shown below the schematic.
DISCUSSION
Cut-and-paste DNA transposition is orchestrated by transposase, which promotes the DNA cleavage and joining reactions. The initial steps of Mos1 transposition involve sequence-specific binding of transposase to the inverted repeat sequences at the transposon ends. Full excision of the transposon from the donor site occurs only after the two ends are brought together in a PEC containing a transposase dimer. To investigate how the PEC may form, we have established the stoichiometry and low resolution conformations of the early intermediates in the pathway: that is, the DNA-free full-length transposase and the pre-cleavage complex of transposase and a single transposon end (SEC2).
Using solution scattering techniques, we have confirmed that the transposase is a homodimer in solution without DNA and established that it adopts an extended conformation. This arrangement differs markedly from the more compact, crossed architecture of the dimer when bound to two pre-cleaved DNA molecules in the PEC crystal structure. The elongation is reflected by the larger measured radius of gyration (50 Å) and maximum dimension (180 Å) of the transposase dimer in solution compared with values calculated for the protein component of the PEC (Rg = 38 Å and Dmax = 110 Å). These results suggest that the transposase dimer changes conformation prior to PEC formation, either upon binding to one transposon DNA end or during the synapsis of the two ends.
To establish how the transposase domains are arranged within the elongated dimer, and which domains are essential for transposase dimerization in the absence of DNA, we prepared deletion mutants of Mos1 transposase. These lacked either the N-terminal 55 amino acids (containing HTH1) or the C-terminal catalytic domain. We found that the transposase was a monomer when the N-terminal 55 residues were deleted, whereas the mutant lacking the catalytic domain could still form dimers in solution. The elongated transposase dimer is therefore likely to be arranged with the DNA-binding domain at the central interface (as proposed in Figure 3D). An alternative model in which the catalytic domains interact and the DNA-binding domains extend to the dimer peripheries is not consistent with these results and is discounted.
Transposase dimerization via the N-terminal 55 residues may be a molecular feature that is maintained throughout the Mos1 transposition pathway. In the PEC structure (14), this domain establishes a hydrophobic dimerization interface as well as binding sequence specifically to the inner 8 bp of IR DNA, via the HTH1 motif. The N-terminal 30 residues are essential for sequence-specific IR DNA binding (12,29), and deletion of the first 34 or 50 residues resulted in the loss of transposition activity in a plasmid-based transposition assay in E. coli (18). Weak transposition activity was regained, however, when the first 34 residues were replaced by either the dimerization domain of Gal4 or a leucine zipper dimerization domain (18). Taken together these data suggest that sequence-specific interaction of the HTH1 motif with IR DNA is not vital for Mos1 transposition, whereas transposase dimerization via the N-terminal residues is required.
Oligomerization prior to sequence-specific DNA binding has been observed for other eukaryotic DNA transposases, and may be a general feature regulating early steps in eukaryotic transposition. The P element transposase from Drosophila melanogaster exists as a pre-formed tetramer that binds to one of the two P element ends (9). The core RAG1 recombinase is either a dimer or trimer and associates with two RAG2 subunits to assemble on one recombination signal sequence, as a hetero-oligomer, before synapsis (39,40). The N-terminally truncated hAT transposase Hermes, from Musca domestica, exists as a hexamer both in solution and in crystals (41). In the mariner/Tc1 family, the N-terminal 57 amino acids of Sleeping Beauty transposase have been implicated in tetramerization of the DNA-binding domain in complex with transposase-binding sites (42), and the N-terminal region of Himar1 is involved in a protein–protein interaction interface (43). By contrast, the bacterial Tn5 transposase is most likely a monomer when not bound to DNA (44). It is thought that a conformational change in the free transposase monomer could allow for DNA binding and transposase dimerization (45) in the context of the synaptic complex containing two transposon ends (46).
Mos1 transposition is initiated by sequence-specific binding of transposase to a transposon end. Here we used SEC-MALLS, small angle scattering, gel analysis and UV absorbance measurements to establish that the in vitro single-end complex contains a transposase dimer bound to one transposon DNA duplex. Using contrast variation neutron scattering experiments, we measured the structural parameters Rg and Dmax for the complex and its two constituent molecules. These revealed model-independent information about the shape, dimensions and relative spatial arrangement of the transposase and DNA in SEC2. We found that, like the transposase dimer, SEC2 has an elongated architecture, but the transposase in the complex has become slightly more extended upon binding DNA. The dimensions of the DNA phase within SEC2 are consistent with a linear B-form DNA duplex, and the 51 Å separation between the centres of mass of the DNA and transposase suggests that the DNA is bound predominantly to one half of the transposase dimer.
The low resolution structural models of the SEC2 in solution illustrate the elongated shape and the relative spatial arrangement of the transposase and DNA within the complex. As predicted from analysis of the structural parameters, the DNA phase is seen to associate primarily with one half of the transposase dimer; the other half of the protein phase has fewer contacts with the DNA and is presumed to have more conformational freedom. The similarities in the architectures of the transposase in SEC2 and in solution suggest that binding of the pre-formed transposase dimer to the first transposon end occurs without major conformational changes in either the protein or the IR DNA.
Comparison of the solution conformations of SEC2 and the PEC crystal structure suggests that a major conformational change in the transposase would occur during a transition from SEC2 to PEC. The data and models presented here provide a basis to propose a mechanism for this conformational change. We speculate that the transposase transitions from the elongated open dimer form when bound to one transposon end to the more closed compact arrangement seen in the PEC, by rotation of one monomer of transposase by ∼180° with respect to the other. The N-terminal 55 residues of the DNA-binding domain may be the pivot for this rotation. As depicted in Figure 7, rotation of the free half of the transposase dimer about the DNA-binding domain could promote rearrangement of the complex. This rotation may be in concert with or triggered by binding of a second transposon DNA end, which would promote DNA looping.
One controversial issue regarding the Mos1 transposition mechanism remains outstanding: does nicking of the first DNA strand occur before or after synapsis of the Mos1 transposon ends? Dawson and Finnegan (10) reported that first strand cleavage was not dependent on prior PEC formation. Similarly, catalysis of Himar1 without synapsis of the ends was reported (16). Nicking of one strand prior to PEC formation could trigger a conformational change in SEC2, which then facilitates pairing of the two ends. This could provide one molecular explanation for metal ion dependence of PEC formation.
The SEC2 to PEC model raises questions about how the crossed (or trans) arrangement of transposase and IR DNA observed in the PEC could arise. The modular nature of the transposase, with long flexible linkers between the DNA-binding and catalytic domains, could allow for sub-unit exchange from one IR DNA molecule to the other, during major conformational changes in the transposase. However, it remains to be established whether this exchange occurs before or after PEC formation or between cleavage events.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–2 and Supplementary Figures 1–4.
FUNDING
Wellcome Trust [085176/Z/08/Z to J.M.R.]; the EPSRC [EP/C015452/1 to V.T.F. providing postdoctoral support to M.G.C.]; Darwin Trust of Edinburgh (PhD scholarship to M.T.); JRA activities of EU award 226 507–NMI3. The EPPF is funded by the Wellcome Trust and BBSRC. Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Deuterated transposase was prepared in the Deuteration Laboratory (D-LAB) within ILL’s Life Sciences Group (Grenoble). Mass spectrometry analysis was performed in the IBS (Grenoble). SANS experiments were performed at the ILL beam line D22. The SAXS data were collected at the ESRF, beam line ID14-3, and we thank Cyril Dian for his assistance. The SEC-MALLS analysis was performed with Martin Wear (EPPF, Edinburgh) and by Andrew Leech (University of York). We sincerely thank Giuseppe Zaccai and David Finnegan for their insightful and helpful comments on the manuscript.
REFERENCES
- 1.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Donnell KA, Burns KH. Mobilizing diversity: transposable element insertions in genetic variation and disease. Mob. DNA. 2010;1:21. doi: 10.1186/1759-8753-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 4.Casola C, Hucks D, Feschotte C. Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol. Biol. Evol. 2008;25:29–41. doi: 10.1093/molbev/msm221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plasterk RH, Izsvak Z, Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999;15:326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- 6.Ammar I, Izsvak Z, Ivics Z. The Sleeping Beauty transposon toolbox. Methods Mol. Biol. 2012;859:229–240. doi: 10.1007/978-1-61779-603-6_13. [DOI] [PubMed] [Google Scholar]
- 7.Robert VJ. Engineering the Caenorhabditis elegans genome by Mos1-induced transgene-instructed gene conversion. Methods Mol. Biol. 2012;859:189–201. doi: 10.1007/978-1-61779-603-6_11. [DOI] [PubMed] [Google Scholar]
- 8.Halford SE, Welsh AJ, Szczelkun MD. Enzyme-mediated DNA looping. Annu. Rev. Biophys. Biomol. Struct. 2004;33:1–24. doi: 10.1146/annurev.biophys.33.110502.132711. [DOI] [PubMed] [Google Scholar]
- 9.Tang M, Cecconi C, Bustamante C, Rio DC. Analysis of P element transposase protein-DNA interactions during the early stages of transposition. J. Biol. Chem. 2007;282:29002–29012. doi: 10.1074/jbc.M704106200. [DOI] [PubMed] [Google Scholar]
- 10.Dawson A, Finnegan DJ. Excision of the Drosophila mariner transposon Mos1. Comparison with bacterial transposition and V(D)J recombination. Mol. Cell. 2003;11:225–235. doi: 10.1016/s1097-2765(02)00798-0. [DOI] [PubMed] [Google Scholar]
- 11.Richardson JM, Dawson A, O'Hagan N, Taylor P, Finnegan DJ, Walkinshaw MD. Mechanism of Mos1 transposition: insights from structural analysis. EMBO J. 2006;25:1324–1334. doi: 10.1038/sj.emboj.7601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auge-Gouillou C, Brillet B, Germon S, Hamelin MH, Bigot Y. Mariner Mos1 transposase dimerizes prior to ITR binding. J. Mol. Biol. 2005;351:117–130. doi: 10.1016/j.jmb.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 13.Auge-Gouillou C, Brillet B, Hamelin MH, Bigot Y. Assembly of the mariner Mos1 synaptic complex. Mol. Cell Biol. 2005;25:2861–2870. doi: 10.1128/MCB.25.7.2861-2870.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richardson JM, Colloms SD, Finnegan DJ, Walkinshaw MD. Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote. Cell. 2009;138:1096–1108. doi: 10.1016/j.cell.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claeys BC, Chalmers R. Transposition of the human Hsmar1 transposon: rate-limiting steps and the importance of the flanking TA dinucleotide in second strand cleavage. Nucleic Acids Res. 2010;38:190–202. doi: 10.1093/nar/gkp891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipkow K, Buisine N, Lampe DJ, Chalmers R. Early intermediates of mariner transposition: catalysis without synapsis of the transposon ends suggests a novel architecture of the synaptic complex. Mol. Cell Biol. 2004;24:8301–8311. doi: 10.1128/MCB.24.18.8301-8311.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu D, Bischerour J, Siddique A, Buisine N, Bigot Y, Chalmers R. The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol. Cell Biol. 2007;27:1125–1132. doi: 10.1128/MCB.01899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carpentier G, Jaillet J, Pflieger A, Adet J, Renault S, Auge-Gouillou C. Transposase-transposase interactions in MOS1 complexes: a biochemical approach. J. Mol. Biol. 2011;405:892–908. doi: 10.1016/j.jmb.2010.11.032. [DOI] [PubMed] [Google Scholar]
- 19.Timmins PA, Zaccai G. Low resolution structures of biological complexes studied by neutron scattering. Eur. Biophys. J. 1988;15:257–268. doi: 10.1007/BF00256476. [DOI] [PubMed] [Google Scholar]
- 20.Stuhrman HB. Neutron small-angle scattering of biological macromolecules in solution. J. Appl. Crystallogr. 1974;7:173–178. [Google Scholar]
- 21.Kennaway CK, Taylor JE, Song CF, Potrzebowski W, Nicholson W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, et al. Structure and operation of the DNA-translocating type I DNA restriction enzymes. Genes Dev. 2012;26:92–104. doi: 10.1101/gad.179085.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor JE, Swiderska A, Artero JB, Callow P, Kneale G. Structural and functional analysis of the symmetrical type I restriction endonuclease R.EcoR124I(NT) PLoS One. 2012;7:e35263. doi: 10.1371/journal.pone.0035263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vijayakrishnan S, Callow P, Nutley MA, McGow DP, Gilbert D, Kropholler P, Cooper A, Byron O, Lindsay JG. Variation in the organization and subunit composition of the mammalian pyruvate dehydrogenase complex E2/E3BP core assembly. Biochem. J. 2011;437:565–574. doi: 10.1042/BJ20101784. [DOI] [PubMed] [Google Scholar]
- 24.Laux V, Callow P, Svergun DI, Timmins PA, Forsyth VT, Haertlein M. Selective deuteration of tryptophan and methionine residues in maltose binding protein: a model system for neutron scattering. Eur. Biophys. J. 2008;37:815–822. doi: 10.1007/s00249-008-0280-5. [DOI] [PubMed] [Google Scholar]
- 25.Blakeley MP, Ruiz F, Cachau R, Hazemann I, Meilleur F, Mitschler A, Ginell S, Afonine P, Ventura ON, Cousido-Siah A, et al. Quantum model of catalysis based on a mobile proton revealed by subatomic x-ray and neutron diffraction studies of h-aldose reductase. Proc. Natl Acad. Sci. USA. 2008;105:1844–1848. doi: 10.1073/pnas.0711659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood K, Grudinin S, Kessler B, Weik M, Johnson M, Kneller GR, Oesterhelt D, Zaccai G. Dynamical heterogeneity of specific amino acids in bacteriorhodopsin. J. Mol. Biol. 2008;380:581–591. doi: 10.1016/j.jmb.2008.04.077. [DOI] [PubMed] [Google Scholar]
- 27.Richardson JM, Zhang L, Marcos S, Finnegan DJ, Harding MM, Taylor P, Walkinshaw MD. Expression, purification and preliminary crystallographic studies of a single-point mutant of Mos1 mariner transposase. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:962–964. doi: 10.1107/S0907444904003798. [DOI] [PubMed] [Google Scholar]
- 28.Meilleur F, Weiss KL, Myles DA. Deuterium labeling for neutron structure-function-dynamics analysis. Methods Mol. Biol. 2009;544:281–292. doi: 10.1007/978-1-59745-483-4_18. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L, Dawson A, Finnegan DJ. DNA-binding activity and subunit interaction of the mariner transposase. Nucleic Acids Res. 2001;29:3566–3575. doi: 10.1093/nar/29.17.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 31.Jacrot B. Study of biological structures by neutron-scattering from solution. Rep. Prog. Phys. 1976;39:911–953. [Google Scholar]
- 32.Jacrot B, Zaccai G. Determination of molecular-weight by neutron-scattering. Biopolymers. 1981;20:2413–2426. [Google Scholar]
- 33.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozin MB, Svergun DI. A software system for rigid-body modelling of solution scattering data. J. Appl. Crystallogr. 2000;33:775–777. [Google Scholar]
- 35.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svergun DI, Petoukhov MV, Koch MHJ. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serdyuk IN. A method of joint use of electromagnetic and neutron scattering: a study of internal ribosomal structure. Methods Enzymol. 1979;59:750–775. doi: 10.1016/0076-6879(79)59123-x. [DOI] [PubMed] [Google Scholar]
- 39.Bailin T, Mo XM, Sadofsky MJ. A RAG1 and RAG2 tetramer complex is active in cleavage in V(D)J recombination. Mol. Cell. Biol. 1999;19:4664–4671. doi: 10.1128/mcb.19.7.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swanson PC, Desiderio S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hickman AB, Perez ZN, Zhou L, Musingarimi P, Ghirlando R, Hinshaw JE, Craig NL, Dyda F. Molecular architecture of a eukaryotic DNA transposase. Nat. Struct. Mol. Biol. 2005;12:715–721. doi: 10.1038/nsmb970. [DOI] [PubMed] [Google Scholar]
- 42.Izsvak Z, Khare D, Behlke J, Heinemann U, Plasterk RH, Ivics Z. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in sleeping beauty transposition. J. Biol. Chem. 2002;277:34581–34588. doi: 10.1074/jbc.M204001200. [DOI] [PubMed] [Google Scholar]
- 43.Butler MG, Chakraborty SA, Lampe DJ. The N-terminus of Himar1 mariner transposase mediates multiple activities during transposition. Genetica. 2006;127:351–366. doi: 10.1007/s10709-006-6250-x. [DOI] [PubMed] [Google Scholar]
- 44.Braam LA, Goryshin IY, Reznikoff WS. A mechanism for Tn5 inhibition - Carboxyl-terminal dimerization. J. Biol. Chem. 1999;274:86–92. doi: 10.1074/jbc.274.1.86. [DOI] [PubMed] [Google Scholar]
- 45.Reznikoff WS. Transposon Tn5. Annu. Rev. Genet. 2008;42:269–286. doi: 10.1146/annurev.genet.42.110807.091656. [DOI] [PubMed] [Google Scholar]
- 46.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.