Abstract
Translesion synthesis (TLS) employs low fidelity polymerases to replicate past damaged DNA in a potentially error-prone process. Regulatory mechanisms that prevent TLS-associated mutagenesis are unknown; however, our recent studies suggest that the PCNA-binding protein Spartan plays a role in suppression of damage-induced mutagenesis. Here, we show that Spartan negatively regulates error-prone TLS that is dependent on POLD3, the accessory subunit of the replicative DNA polymerase Pol δ. We demonstrate that the putative zinc metalloprotease domain SprT in Spartan directly interacts with POLD3 and contributes to suppression of damage-induced mutagenesis. Depletion of Spartan induces complex formation of POLD3 with Rev1 and the error-prone TLS polymerase Pol ζ, and elevates mutagenesis that relies on POLD3, Rev1 and Pol ζ. These results suggest that Spartan negatively regulates POLD3 function in Rev1/Pol ζ-dependent TLS, revealing a previously unrecognized regulatory step in error-prone TLS.
INTRODUCTION
Genomic DNA can be damaged by environmental factors and endogenous metabolic processes. If not repaired in a timely manner, DNA lesions can stall replicative DNA polymerases, because the high fidelity polymerases are unable to accommodate damaged bases in their active sites. Such polymerase-blocking lesions can be bypassed by specialized DNA polymerases in a process called translesion synthesis (TLS) (1). When replicative polymerases encounter DNA lesions, the ubiquitin ligase Rad18 monoubiquitinates PCNA, a ring-shaped sliding clamp (2). Mono-ubiquitinated PCNA in turn recruits TLS polymerases via interactions with their ubiquitin-binding domains, thereby inducing a polymerase switch at the lesion. Many TLS polymerases are members of the Y family, which are characterized by their large active sites that accommodate distorted bases (3). Because these TLS polymerases exhibit low fidelity, TLS is a potentially error-prone process.
The process of TLS involves at least two steps: insertion of nucleotides opposite lesions and subsequent extension. Although some types of DNA lesions can be bypassed by a single TLS polymerase, many lesions require separate polymerases for the insertion and extension steps. In this two-polymerase mechanism, Y-family TLS polymerases insert nucleotides opposite lesions and then the B family polymerase Pol ζ extends from them (4,5). Pol ζ, composed of Rev7 and the catalytic subunit Rev3, has a unique ability to extend from mispaired primer termini (6). This activity makes Pol ζ a suitable extension polymerase in TLS because nucleotides inserted at lesions may not form base pairs.
Pol ζ recruitment to sites of TLS requires Rev1, which interacts with Pol ζ and the insertion polymerases from the Y family including Pol η, Pol κ and Pol ι (7–11). Accordingly, Rev1 is believed to play a pivotal role in the two-polymerase mechanism by coordinating the insertion and extension steps. Importantly, TLS involving Rev1 and Pol ζ can accommodate a wide variety of lesions, albeit at the expense of reduced fidelity. As a result, TLS involving Rev1 and Pol ζ is associated with damage-induced mutagenesis and therefore has been referred to as an error-prone TLS pathway (12). Although the error-prone TLS pathway is important for cell survival after DNA damage (13,14), its usage needs to be tightly regulated because it is potentially mutagenic.
Recently, we and others reported that the ubiquitin-binding protein Spartan (also known as DVC1 and C1orf124) is recruited to sites of DNA damage via ubiquitinated PCNA (15–18) and showed that Spartan is important to prevent mutations associated with TLS across UV-induced DNA damage (15,19,20). Herein, we report that Spartan negatively regulates error-prone TLS that involves the Pol δ subunit POLD3, Rev1 and Pol ζ. We show that the putative zinc metalloprotease domain SprT in Spartan is required for suppression of mutagenic TLS. Collectively, our results reveal an unexpected role of POLD3 in TLS and implicate Spartan in a previously unrecognized regulatory step in error-prone TLS.
MATERIALS AND METHODS
Plasmids
Full-length Spartan was cloned into the transient mammalian expression vectors pEFF-N (N-terminal Flag tag) and lentiviral expression vectors pLVX3-CMV-puro (N-terminal 3xFlag tag) and pLVX6-IRES-Neo (N-terminal EGFP tag). To express the Spartan SprT domain (amino acids 1-219), fragments of Spartan cDNA were amplified by polymerase chain reaction and subcloned into transient expression vectors pEFF-N/nuc/myc (N-terminal Flag and C-terminal 3xNLS/myc tags) and a lentivirus vector pLVX4-CMV-puro (N-terminal 3xFlag and C-terminal His8 tags). cDNAs encoding Pol δ subunits were subcloned from pVL1393 (a gift from Ellen Fanning) (21) to pEF4H (N-terminal 4HA tag). POLD3 was also cloned in pBABE-puro/3xFlag-His8. cDNA for Rev1 was obtained from Larry Karnitz (Mayo Clinic) and cloned into a lentivirus vector pLVX3-IRES-Neo (N-terminal 3xFlag tag). Full-length Rev3 cDNA was purchased from Open Biosystems and expressed from pLVX6-IRES-Neo (N-terminal EGFP tag). Baculovirus vector pFastBac SHT was used to express recombinant POLD3 with a C-terminal S/Strep-tagII/His6 tag. For production of recombinant GST-fusion proteins in Escherichia coli, cDNAs encoding the SprT domain were cloned in pGexHis. Site-directed mutagenesis was performed using pfu Turbo (Agilent Technologies) and all introduced mutations were confirmed by sequencing.
Cell culture, RNAi and SupF mutation assays
Human embryonic kidney cell lines 293 A (Invitrogen) and 293 T cells (ATCC) were cultured in DMEM containing 10% fetal bovine serum (FBS) and in RPMI 1640 containing 10% FBS, respectively. Cells were transfected with siRNA oligos using RNAiMAX (Invitrogen) and analysed 48–72 hr later. The target sequences of siRNAs are as follows: siControl, CGUACGCGGAAUACUUCGA; siSpartan.664, GACCCUGUGUGCUGGGAUA; siSpartan.915, ACGAUGAGGUGGAUGAGUA; siPOLD3.750, CAACAAGGCACCAGGGAAA; siPOLD3.1309, GAUAGUGAAGAGGAGCUUA; siRev1, CAGAAGAAGUAGAUGAUUU; and siRev3, GAAAUGGAAUGGAGUGAUA. Mutation frequencies were measured using the SupF shuttle vector system as described previously (15,22).
Western blotting and immunoprecipitation
Cells were lysed in NP-40 lysis buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 0.1% Nonidet P-40, 5 mM EDTA, 50 mM NaF, 1 mM Na3VO4, 10% Glycerol) supplemented with protease inhibitor mix (Sigma). For Western blotting, 30 µg of protein were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membrane, and probed with antibodies. Anti-POLD1, anti-Rev1, anti-EGFP and anti-HA antibodies were from Santa Cruz Biotechnology. Anti-POLD2 and anti-Rev7 antibodies were obtained from ProteinTech Group and anti-POLD3 antibody was from Bethyl Laboratory. Anti-β-actin and anti-Flag antibodies were purchased from Sigma-Aldrich. Anti-S-tag antibody was obtained from Scott Kaufmann (Mayo Clinic) and mouse monoclonal anti-Spartan antibody was reported previously (15).
Immunoprecipitation of Flag-tagged proteins was performed by incubating cell lysates containing 1–2 mg protein with anti-Flag M2 affinity agarose beads (Sigma) for 4 hr at 4°C. Beads were washed five times with NP-40 lysis buffer and precipitated proteins were eluted by boiling in 2 × SDS-PAGE sample buffer. For purification of proteins associated with 3xFlag-SprT, anti-Flag immunoprecipitates were eluted with 0.5 mg/ml 3xFlag peptide (Sigma), precipitated with trichloroacetic acid and analysed by mass spectrometry as described (23).
Recombinant proteins and GST pull-down assays
GST-tagged proteins were produced in E. coli BL21(DE3) and purified on glutathione agarose beads (Sigma-Aldrich). Full-length His6-tagged POLD3 proteins were produced in Sf9 cells infected with recombinant baculoviruses and purified on TALON metal affinity resins (Clontech). GST pull-down assays were performed as described (15).
Protein structure prediction of a truncated SprT domain
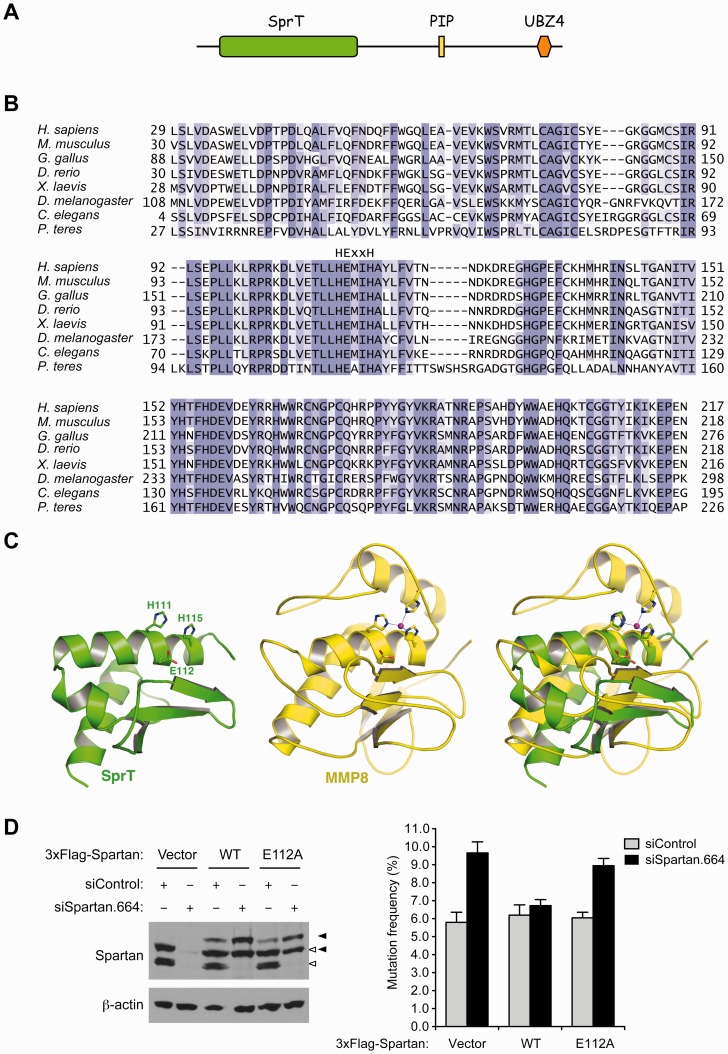
Because the sequence similarity between the human SprT domain and any of the protein structures reported to date is less than 40%, we submitted the SprT domain amino acid sequence to the Phyre server (http://www.sbg.bio.ic.ac.uk/∼phyre/) for protein-fold recognition and obtained 10 different models of the SprT domain fragments. Visual inspection of the 10 models generated by the server identified the model of residues 43–121 as a plausible structure of a truncated SprT domain. This model as shown in Figure 1C was generated on the basis of the crystal structure of the putative zinc-dependent peptidase Q74D82 (Protein Data Bank ID: 3C37) and resembles part of the active site of many zinc proteins including MMP1, MMP2, MMP3, MMP8, MMP12 and MMP13 that we computationally studied before. Subsequent 98 10-ns molecular dynamics simulations in explicit water using our published simulation protocol (24,25) showed that the model of residues 43–121 remained folded during the 10-ns simulation period and suggested that residues 43–121 in the human SprT domain adopt the conformation shown in Figure 1C.
Figure 1.
The SprT domain of Spartan is a putative zinc metalloprotease and Glu112 is required for Spartan to suppress UV-induced mutagenesis. (A) Schematic representation of the domain structure of human Spartan. PIP, PCNA-interacting peptide; UBZ4, ubiquitin-binding zinc-finger 4. (B) Alignment of the SprT domain sequences from Spartan homologs in selected species. The HExxH motif is indicated above the sequences. (C) Model of a truncated human SprT domain (residues 43–121; generated by the Phyre server) showing the three residues of the HExxH motif (left); crystal structure of MMP8 (Protein Data Bank ID: 2OY4) showing the three residues of the HExxH motif and the zinc divalent cation coordinating three histidines (middle); overlay of the truncated SprT and MMP8 revealing the structural similarity in the active site between the two proteins (right). (D) Glu112 is required for suppression of UV-induced mutagenesis by Spartan. UV-induced mutagenesis was measured using the SupF shuttle vector system in 293 T cells transfected with the indicated siRNA oligos. Mutation frequencies are presented as percentage of mutant SupF genes. Error bars represent SD (n = 3). Wild-type or the E112A mutant of Spartan was expressed with 3xFlag tag in an siRNA-resistant form. Spartan proteins were analysed by Western blotting. Positions of endogenous and exogenous Spartan proteins are indicated by white and black arrowheads, respectively.
RESULTS
Requirement of an intact SprT domain for suppression of UV-induced mutagenesis by Spartan
We and others have shown that UV-induced mutagenesis is increased in Spartan-depleted cells (15,19,20). To understand how Spartan suppresses UV-induced mutagenesis, we investigated the role of the uncharacterized SprT domain in the N-terminal region of Spartan (Figure 1A). This domain contains an HExxH motif (x indicates any amino acid) that is highly conserved among Spartan in different species (Figure 1B). The HExxH motif is commonly found in zinc-dependent metalloprotease active sites (26), where the two histidine residues coordinate the zinc ion, and the glutamic acid plays a direct role in peptide bond cleavage by facilitating activation of a zinc-coordinating water molecule (27). Our protein fold recognition (threading) study using the Phyre server (28) suggested that residues 43–121 in the SprT domain of human Spartan fold into three helices and a three-strand beta sheet (Figure 1C) that resemble part of the active site of zinc metalloproteases such as matrix metalloproteinases, botulinum neurotoxin endopeptidases, and anthrax lethal factor (29–31). These results suggest that the SprT domain of Spartan is a putative zinc metalloprotease.
To assess the importance of the putative metalloprotease domain in Spartan function, we created a presumably inactive mutant of the Spartan, in which Glu112 of the HExxH motif was mutated to alanine (E112A). We then expressed either wild-type Spartan or the E112A mutant in an RNAi-resistant form and tested their ability to suppress UV-induced mutagenesis when endogenous Spartan was depleted by RNAi. We confirmed that, like wild-type Spartan, the E112A mutant localized to nuclear foci in hydroxyurea-treated cells (Supplementary Figure S1). As shown in Figure 1D, wild-type Spartan reduced mutagenesis whereas Spartan E112A did not. These results indicate that an intact SprT domain is necessary for suppression of damage-induced mutagenesis by Spartan and imply that the putative zinc metalloprotease activity is required for the process.
Interaction of the SprT domain with the DNA polymerase δ subunit POLD3
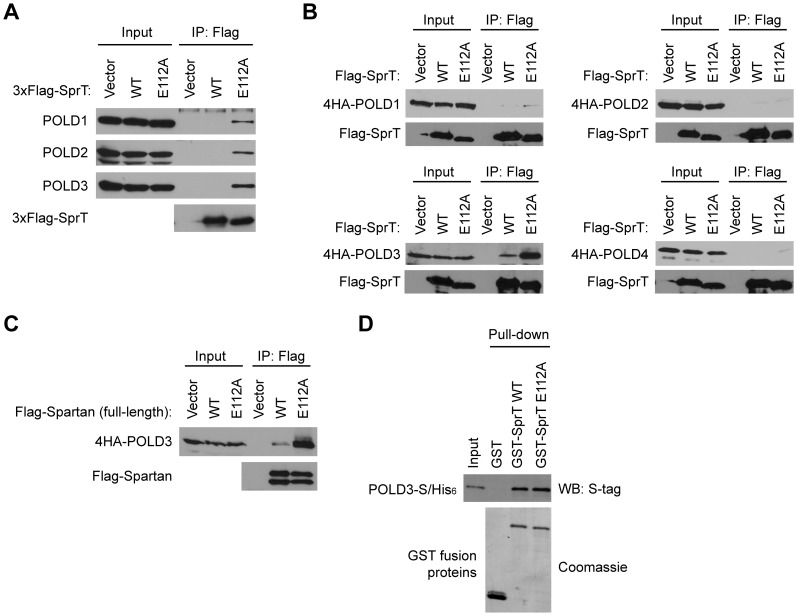
To explore how the SprT domain of Spartan might participate in the suppression of mutations, we first searched for its interacting proteins in vivo. After stably expressing either wild-type or E112A SprT domain tagged with 3xFlag and two nuclear localization signals, we purified associated proteins by anti-Flag immunoprecipitation (Supplementary Figure S2A) and analysed the precipitated proteins by mass spectrometry. Peptides corresponding to all four subunits of the replicative polymerase Pol δ, namely POLD1, POLD2, POLD3, and POLD4, were identified in the immunoprecipitate from E112A SprT, but interestingly not from wild-type SprT (Supplementary Table S1). Pull-down of the Pol δ complexes by SprT E112A in this screen was subsequently confirmed by immunoblotting using specific antibodies against POLD1, POLD2 and POLD3 (Figure 2A).
Figure 2.
The SprT domain of Spartan interacts with Pol δ subunit POLD3. (A) The E112A mutant of SprT captures Pol δ in vivo. Wild-type or the E112A mutant of SprT proteins were stably expressed in 293A and purified on anti-Flag beads. Copurified endogenous proteins were analysed by Western blotting for the indicated Pol δ subunits. (B) The SprT domain captures co-expressed POLD3 in vivo. Wild-type or the E112A mutant of SprT proteins were transiently expressed in 293T with each Pol δ subunit tagged with 4HA and immunoprecipitated with anti-Flag beads. Precipitated proteins were analysed by Western blotting for the indicated proteins. (C) Interactions between full-length Spartan and POLD3. Experiments were performed as in (B) except that full-length Spartan was expressed. (D) Direct interaction of the Spartan SprT domain with POLD3. In vitro GST-pull down assays were performed using the indicated recombinant proteins.
In the DNA polymerase δ complex, POLD1 is the catalytic subunit and others are accessory subunits. Because these subunits form a stable complex, we assessed which subunit is primarily responsible for association with SprT E112A. When immunoprecipitation was performed with individually expressed Pol δ subunits, POLD3 was preferentially pulled down by the SprT domain (Figure 2B), suggesting a more enduring interaction with this subunit. Importantly, comparison of wild-type and E112A SprT domains in these assays indicated greater recovery of POLD3 with the E112A mutant, although POLD3 was also recovered with the wild-type SprT domain (Figure 2B). Similarly, when full-length Spartan and POLD3 proteins were co-expressed, POLD3 was co-immunoprecipitated with Spartan, again with preferential pull-down with the E112A mutant (Figure 2C).
In contrast, when interactions between the SprT domain and POLD3 were examined in vitro by GST pull-down assays using purified recombinant proteins, the wild-type and E112A SprT domains bound POLD3 equally (Figure 2D and Supplementary Figure S2B), providing evidence that the interaction between the SprT domain and POLD3 is direct, and arguing against the possibility that the binding results from a conformational change caused by the E112A mutation. Moreover, the observation that wild-type SprT captures POLD3 less efficiently than the E112A mutant in immunoprecipitation from intact cells (Figure 2B and C) raises the possibility that the enzymatic activity of Spartan might limit the ability of the wild-type enzyme to capture POLD3 in vivo.
Requirement of POLD3, Rev1 and Pol ζ for mutagenesis induced by Spartan depletion
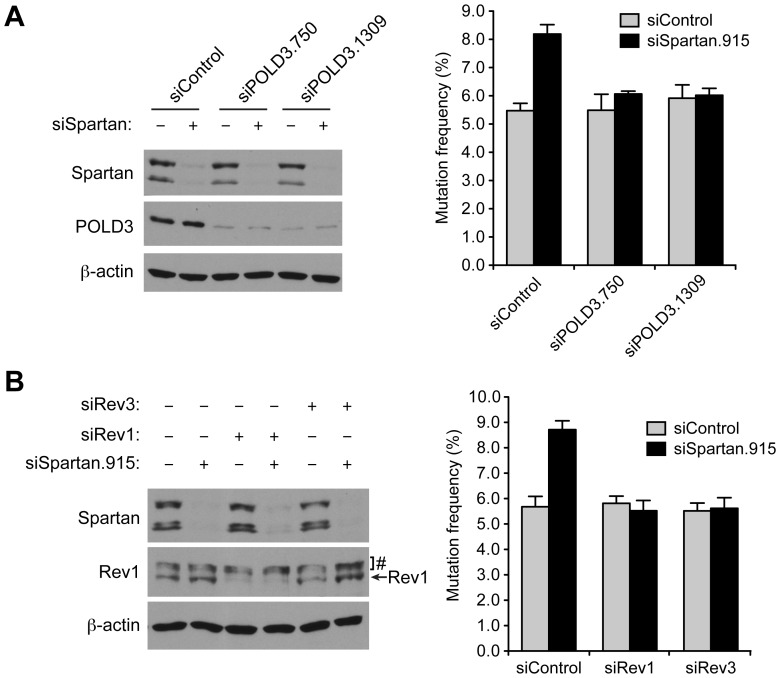
The S. cerevisiae POLD3 ortholog Pol32 has been implicated in mutagenic TLS (32). Given that Spartan interacts with POLD3 (Figure 2) and suppresses mutagenic TLS (15), we next investigated whether POLD3 is involved in the increased mutagenesis observed in Spartan-depleted cells. Specifically, we examined UV-induced mutagenesis after knockdown of POLD3 and Spartan either individually or in combination. POLD3 depletion had no effect on UV-induced mutagenesis by itself, but suppressed the increase in UV-induced mutations observed after Spartan knockdown (Figure 3A). Cell-cycle profiles after knockdown of Spartan or POLD3 were largely unaffected (Supplementary Figure S3A and S3B), suggesting that the effects on mutagenesis were not due to enrichment of cells in a particular phase of the cell cycle. In summary, these results indicate that POLD3 is required for the increase in UV-induced mutagenesis observed in Spartan-depleted cells and suggest a possible link between Spartan, POLD3, and mutagenesis.
Figure 3.
POLD3 is involved in error-prone TLS in Spartan-depleted cells. (A) POLD3 is required for mutagenesis in Spartan-depleted cells. Cells were treated as in Figure 1D. Error bars represent SD (n = 3). Knockdown of Spartan and POLD3 was confirmed by Western blotting. (B) Mutagenesis in Spartan-depleted cells is dependent on Rev1 and Pol ζ. Error bars represent SD (n = 3). Knockdown of Spartan and Rev1 was confirmed by Western blotting. #Cross-reactive bands. Action of Rev3 siRNA was confirmed in Figure S4.
To obtain further insight into the mechanism of POLD3-dependent mutagenesis in Spartan-depleted cells, we examined whether error-prone TLS is responsible for the increased mutations. Using siRNAs that are characterized in Figure 3B (left panel) and Supplementary Figure S4, we codepleted Rev1 or Rev3 (the catalytic subunit of Pol ζ) along with Spartan and again assessed UV-induced mutagenesis. Depletion of Rev1 or Rev3 did not markedly change cell-cycle distributions (Supplementary Figure S3C) nor did it affect UV-induced mutagenesis, indicating that the basal levels of UV-induced mutagenesis are not dependent on Rev1 and Rev3. In contrast, codepletion of Rev1 or Rev3 suppressed the increase in mutations induced by Spartan depletion (Figure 3B, right panel). These data suggest that the error-prone TLS pathway is responsible for the increased mutagenesis in Spartan-depleted cells. Because such mutagenesis is also dependent on POLD3 (Figure 3A), these results also revealed a role for POLD3 in mutagenic TLS in mammalian cells.
Increased interaction of POLD3 with Rev1 and Pol ζ in Spartan-depleted cells
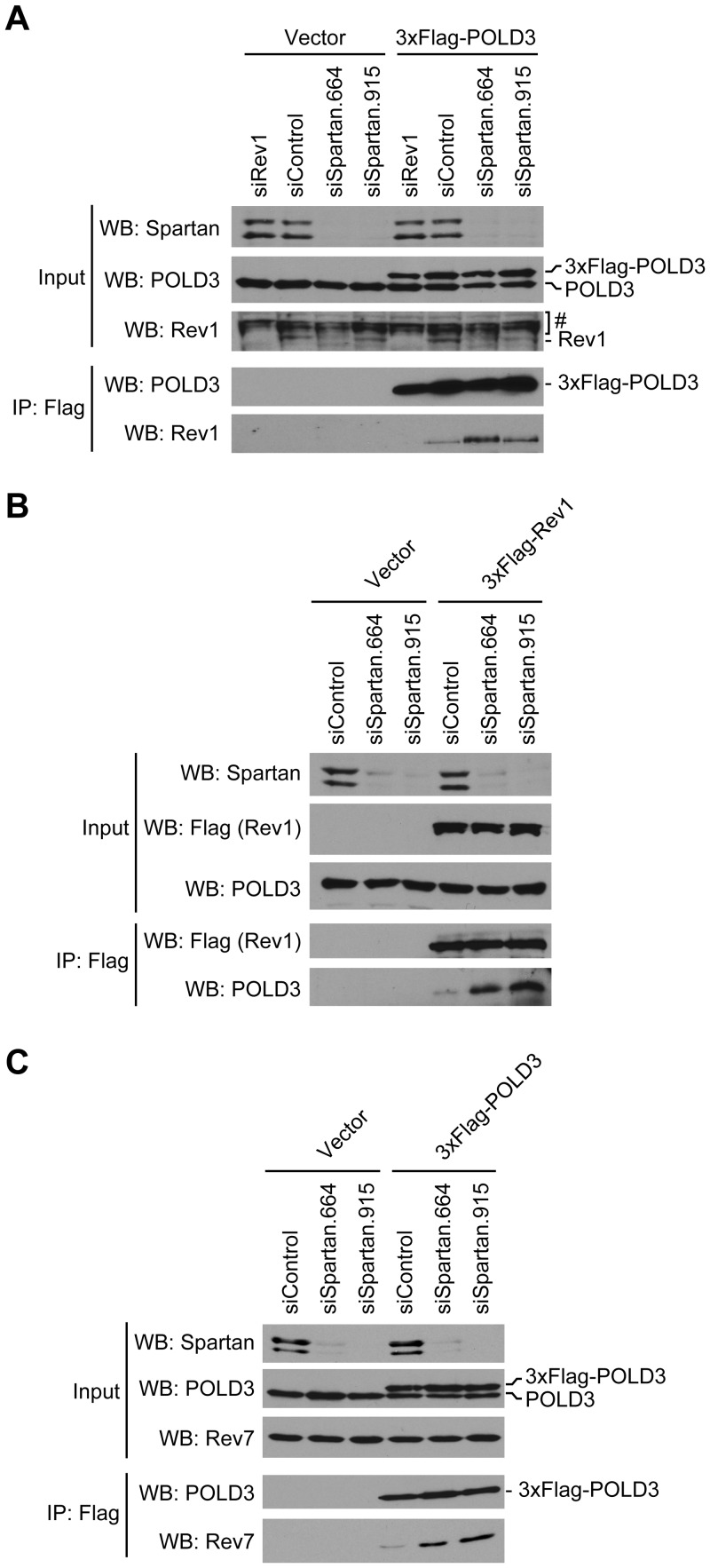
After finding that the increased mutagenesis observed in Spartan-depleted cells is dependent on both POLD3 and the error-prone TLS pathway, we explored a possible biochemical link between them. Interestingly, previous studies demonstrated a physical interaction between recombinant S. cerevisiae Pol32 and Rev1 proteins in vitro (33). To test the hypotheses that (i) mammalian POLD3 and Rev1 might similarly interact during error-prone TLS and (ii) Spartan might regulate this interaction, we established cells stably expressing 3xFlag-POLD3, at a level similar to that of endogenous POLD3 (Figure 4A, input). Immunoprecipitation with immobilized anti-Flag antibody revealed that Rev1 was coprecipitated with 3xFlag-POLD3, indicating that POLD3 and Rev1 form a complex in vivo. Importantly, this interaction was increased in Spartan-depleted cells (Figure 4A). Similar results were obtained in a reciprocal immunoprecipitation experiment performed in cells stably expressing 3xFlag-Rev1 (Figure 4B). Interaction between 3xFlag-Rev1 and POLD3 was again increased in Spartan-depleted cells, suggesting that Spartan is a negative regulator of the interaction.
Figure 4.
Spartan depletion induces complex formation of POLD3 with Rev1 and Pol ζ. (A) Spartan depletion induces interaction of POLD3 with Rev1. Cells stably expressing 3xFlag-POLD3 at near physiological levels were transfected with the indicated siRNAs. 3xFlag-POLD3 proteins were immunoprecipitated and Western blotting was performed for the indicated proteins. RNAi of Rev1 shows the specificity of anti-Rev1 antibodies. #Cross-reactive bands. (B) Spartan depletion induces interaction of Rev1 and POLD3. Experiments were performed similarly in cells stably expressing 3xFlag-Rev1 after transfection with the indicated siRNAs. (C) Spartan depletion induces interaction of POLD3 with Rev7. Experiments were performed as in (A) and Western blotting was performed for the indicated proteins.
Because Rev1 is known to regulate Pol ζ through direct interaction with Rev7, the non-catalytic subunit of Pol ζ (10), we also examined whether Rev7 was present in the immunoprecipitates of 3xFlag-POLD3. Rev7, like Rev1, was also recovered with 3xFlag-POLD3; and this interaction was again increased in Spartan-depleted cells (Figure 4C).
Taken together, these data not only provide a biochemical link between POLD3 and the error-prone TLS pathway but also suggest that Spartan negatively regulates the interaction of POLD3 with Rev1 and Pol ζ.
DISCUSSION
In this study, we show that Spartan negatively regulates POLD3-dependent error-prone TLS. Our data indicate that the SprT domain of Spartan interacts with POLD3 (Figure 2) and plays a role in suppression of damage-induced mutagenesis by Spartan (Figure 1D). Conversely, Spartan depletion promotes complex formation of POLD3 with Rev1 and Pol ζ (Figure 4), and concomitantly elevates UV-induced mutagenesis that is dependent on POLD3, Rev1 and Pol ζ (Figure 3). These data collectively imply that (i) a protein complex of POLD3, Rev1 and Pol ζ is involved in mutagenic TLS studied here, and (ii) Spartan modulates this error-prone TLS process by regulating the interaction of POLD3 with Rev1 and Pol ζ. In summary, our study not only identifies Spartan as a negative regulator of POLD3-dependent error-prone TLS but also reveals a previously uncharacterized regulatory step in this process.
To our knowledge the present results are the first to implicate mammalian POLD3 and Spartan in the regulation of error-prone TLS. Consistent with a role for POLD3 in this process, several groups demonstrated recently that POLD2 and POLD3 are also the components of Pol ζ complex (34,35). Our study showed that, in Spartan-depleted cells, POLD3 associates with the Pol ζ subunit Rev7 (Figure 4C), raising an interesting possibility that Spartan might negatively regulate polymerase switch from Pol δ to Pol ζ. Recent reports have implicated Spartan in multiple aspects of TLS including PCNA ubiquitination and regulation of the insertion TLS polymerase Pol η (16–20). Our study indicates that Spartan plays yet another role in TLS: regulation of the extension step. Collectively, Spartan might play a central role in the two-polymerase mechanism of TLS by coordinating multiple steps in this process.
Our results with the Spartan E112A mutant (Figure 1D) clearly show a role for the putative metalloprotease activity of the SprT domain in suppression of damage-induced mutagenesis. While this manuscript was in preparation, Ghosal et al. (17) reported interaction between Spartan and POLD3 in vivo. In our study, we demonstrated that the SprT domain of Spartan binds to POLD3 directly in vitro (Figure 2D), and that the mutant SprT domain harboring the E112A mutation (in the putative catalytic motif) captures POLD3 more efficiently than wild-type in vivo (Figure 2). These observations suggest that POLD3 might be a direct target of regulation by the Spartan SprT domain. In conclusion, this study clearly establishes Spartan as an important regulator of replication fidelity during TLS, and raises the intriguing possibility that its putative metalloprotease activity controls error-prone TLS by regulating POLD3.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–4.
FUNDING
American Cancer Society Research Scholar (to Y.J.M.); Minnesota Partnership for Biotechnology and Medical Genomics (to Y.J.M.); NIH [R01 GM089778 to J.A.W.]; the Jonsson Cancer Center at UCLA (to J.A.W.); NIH [5 U01AI082120-04 to Y.-P.P.]. Funding for open access charge: American Cancer Society.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank S. Kaufmann and L. Karnitz for critical reading of the manuscript.
REFERENCES
- 1.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 3.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 4.Livneh Z, Ziv O, Shachar S. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle. 2010;9:729–735. doi: 10.4161/cc.9.4.10727. [DOI] [PubMed] [Google Scholar]
- 5.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 6.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 7.Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohashi E, Murakumo Y, Kanjo N, Akagi J, Masutani C, Hanaoka F, Ohmori H. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 9.Tissier A, Kannouche P, Reck MP, Lehmann AR, Fuchs RP, Cordonnier A. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol eta and REVl protein. DNA Repair. 2004;3:1503–1514. doi: 10.1016/j.dnarep.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 10.Murakumo Y, Ogura Y, Ishii H, Numata S, Ichihara M, Croce CM, Fishel R, Takahashi M. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J. Biol. Chem. 2001;276:35644–35651. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 11.Andersen PL, Xu F, Ziola B, McGregor WG, Xiao W. Sequential assembly of translesion DNA polymerases at UV-induced DNA damage sites. Mol. Biol. Cell. 2011;22:2373–2383. doi: 10.1091/mbc.E10-12-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunz BA, Straffon AF, Vonarx EJ. DNA damage-induced mutation: tolerance via translesion synthesis. Mutat. Res. 2000;451:169–185. doi: 10.1016/s0027-5107(00)00048-8. [DOI] [PubMed] [Google Scholar]
- 13.Jansen JG, Tsaalbi-Shtylik A, Hendriks G, Verspuy J, Gali H, Haracska L, de Wind N. Mammalian polymerase zeta is essential for post-replication repair of UV-induced DNA lesions. DNA Repair. 2009;8:1444–1451. doi: 10.1016/j.dnarep.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Okada T, Sonoda E, Yoshimura M, Kawano Y, Saya H, Kohzaki M, Takeda S. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol. Cell. Biol. 2005;25:6103–6111. doi: 10.1128/MCB.25.14.6103-6111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Machida Y, Kim MS, Machida YJ. Spartan/C1orf124 is important to prevent UV-induced mutagenesis. Cell Cycle. 2012;11:3395–3402. doi: 10.4161/cc.21694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Centore RC, Yazinski SA, Tse A, Zou L. Spartan/C1orf124, a reader of PCNA ubiquitylation and a regulator of UV-induced DNA damage response. Mol. Cell. 2012;46:625–635. doi: 10.1016/j.molcel.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosal G, Leung JW, Nair BC, Fong KW, Chen J. PCNA-binding protein C1orf124 is a regulator of translesion synthesis. J. Biol. Chem. 2012;287:34225–34233. doi: 10.1074/jbc.M112.400135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juhasz S, Balogh D, Hajdu I, Burkovics P, Villamil MA, Zhuang Z, Haracska L. Characterization of human Spartan/C1orf124, an ubiquitin-PCNA interacting regulator of DNA damage tolerance. Nucleic Acids Res. 2012;40:10795–10808. doi: 10.1093/nar/gks850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mosbech A, Gibbs-Seymour I, Kagias K, Thorslund T, Beli P, Povlsen L, Nielsen SV, Smedegaard S, Sedgwick G, Lukas C, et al. DVC1 (C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat. Struct. Mol. Biol. 2012;19:1084–1092. doi: 10.1038/nsmb.2395. [DOI] [PubMed] [Google Scholar]
- 20.Davis EJ, Lachaud C, Appleton P, Macartney TJ, Nathke I, Rouse J. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat. Struct. Mol. Biol. 2012;11:1093–1100. doi: 10.1038/nsmb.2394. [DOI] [PubMed] [Google Scholar]
- 21.Podust VN, Chang LS, Ott R, Dianov GL, Fanning E. Reconstitution of human DNA polymerase delta using recombinant baculoviruses: the p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J. Biol. Chem. 2002;277:3894–3901. doi: 10.1074/jbc.M109684200. [DOI] [PubMed] [Google Scholar]
- 22.Parris CN, Levy DD, Jessee J, Seidman MM. Proximal and distal effects of sequence context on ultraviolet mutational hotspots in a shuttle vector replicated in xeroderma cells. J. Mol. Biol. 1994;236:491–502. doi: 10.1006/jmbi.1994.1160. [DOI] [PubMed] [Google Scholar]
- 23.Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J. Biol. Chem. 2009;284:34179–34188. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pang Y-P. Three-dimensional model of a substrate-bound SARS chymotrypsin-like cysteine proteinase predicted by multiple molecular dynamics simulations: catalytic efficiency regulated by substrate binding. Proteins. 2004;57:747–757. doi: 10.1002/prot.20249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pang Y-P, Dai H, Smith A, Meng XW, Schneider PA, Kaufmann SH. Bak conformational changes induced by ligand binding: insight into BH3 domain binding and Bak homo-oligomerization. Sci. Rep. 2012;2:257. doi: 10.1038/srep00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jongeneel CV, Bouvier J, Bairoch A. A unique signature identifies a family of zinc-dependent metallopeptidases. FEBS Lett. 1989;242:211–214. doi: 10.1016/0014-5793(89)80471-5. [DOI] [PubMed] [Google Scholar]
- 27.Rawlings ND, Barrett AJ. Evolutionary families of metallopeptidases. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 28.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 29.Agarwal R, Binz T, Swaminathan S. Structural analysis of botulinum neurotoxin serotype F light chain: implications on substrate binding and inhibitor design. Biochemistry. 2005;44:11758–11765. doi: 10.1021/bi0510072. [DOI] [PubMed] [Google Scholar]
- 30.Pannifer AD, Wong TY, Schwarzenbacher R, Renatus M, Petosa C, Bienkowska J, Lacy DB, Collier RJ, Park S, Leppla SH, et al. Crystal structure of the anthrax lethal factor. Nature. 2001;414:229–233. doi: 10.1038/n35101998. [DOI] [PubMed] [Google Scholar]
- 31.Bertini I, Calderone V, Fragai M, Luchinat C, Maletta M, Yeo KJ. Snapshots of the reaction mechanism of matrix metalloproteinases. Angew. Chem. Int. Ed. Engl. 2006;45:7952–7955. doi: 10.1002/anie.200603100. [DOI] [PubMed] [Google Scholar]
- 32.Huang ME, Rio AG, Galibert MD, Galibert F. Pol32, a subunit of Saccharomyces cerevisiae DNA polymerase delta, suppresses genomic deletions and is involved in the mutagenic bypass pathway. Genetics. 2002;160:1409–1422. doi: 10.1093/genetics/160.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Acharya N, Johnson RE, Pages V, Prakash L, Prakash S. Yeast Rev1 protein promotes complex formation of DNA polymerase zeta with Pol32 subunit of DNA polymerase delta. Proc. Natl Acad. Sci. USA. 2009;106:9631–9636. doi: 10.1073/pnas.0902175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baranovskiy AG, Lada AG, Siebler HM, Zhang Y, Pavlov YI, Tahirov TH. DNA polymerase delta and zeta switch by sharing accessory subunits of DNA polymerase delta. J. Biol. Chem. 2012;287:17281–17287. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proc. Natl Acad. Sci. USA. 2012;109:12455–12460. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.