Abstract
Escherichia coli can rapidly switch to the metabolism of l-arabinose and d-xylose in the absence of its preferred carbon source, glucose, in a process called carbon catabolite repression. Transcription of the genes required for l-arabinose and d-xylose consumption is regulated by the sugar-responsive transcription factors, AraC and XylR. E. coli represents a promising candidate for biofuel production through the metabolism of hemicellulose, which is composed of d-xylose and l-arabinose. Understanding the l-arabinose/d-xylose regulatory network is key for such biocatalyst development. Unlike AraC, which is a well-studied protein, little is known about XylR. To gain insight into XylR function, we performed biochemical and structural studies. XylR contains a C-terminal AraC-like domain. However, its N-terminal d-xylose-binding domain contains a periplasmic-binding protein (PBP) fold with structural homology to LacI/GalR transcription regulators. Like LacI/GalR proteins, the XylR PBP domain mediates dimerization. However, unlike LacI/GalR proteins, which dimerize in a parallel, side-to-side manner, XylR PBP dimers are antiparallel. Strikingly, d-xylose binding to this domain results in a helix to strand transition at the dimer interface that reorients both DNA-binding domains, allowing them to bind and loop distant operator sites. Thus, the combined data reveal the ligand-induced activation mechanism of a new family of DNA-binding proteins.
INTRODUCTION
Bacteria rapidly switch to the use of the most accessible energy source by inhibiting the synthesis of proteins involved in the catabolism of unavailable carbon metabolites. This preferential pattern of sugar metabolism has been termed carbon catabolite repression (CCR) or diauxie (1). In the absence of adequate stores of the preferred carbon source, glucose, Escherichia coli can rapidly change to the metabolism of l-arabinose and d-xylose. These sugars are transported into E. coli by the transporters AraFGH and XylFGH, respectively, and metabolized by similar gene clusters encoding isomerases and kinases (araBA and xylAB) (1–5). Transcription of the genes necessary for the consumption of each sugar is regulated by a sugar-responsive transcription factor: AraC regulates arabinose-responsive operons and XylR activates d-xylose-responsive genes (3,4). Interestingly, recent data have shown that AraC binds to both the l-arabinose and d-xylose responsive promoters and acts as an activator in the former and repressor in the latter (4,5). As a result, l-arabinose is metabolized before d-xylose.
E. coli has potential as a biocatalyst for production of biofuels because it can metabolize all sugars in plant materials. The two most abundant sugars in lignocellulosic sources are glucose and d-xylose. E. coli mutants with a constitutively active cAMP receptor protein (CRP) are able to simultaneously consume glucose and d-xylose (6). With the broader goal of generating an E. coli biocatalyst that can co-metabolize all biomass sugars, it would be necessary to also eliminate the diauxie between d-xylose and l-arabinose, as these two sugars comprise 95% of the total sugar hemicellulose (6,7). Indeed, the fact that E. coli consumes d-xylose only after it consumes l-arabinose, prevents it from affecting a complete bioconversion of sugar mixtures into fuels. Notably, recent studies showed that this hierarchy could be disrupted, allowing for the equal consumption of the both l-arabinose and d-xylose, if the intracellular levels of XylR were increased (5). The resultant engineered bacteria were able to produce 36% more ethanol compared with wild-type E. coli. Thus, understanding how XylR functions at the atomic level would not only provide insight into CCR but also ways to alter the efficacy of XylR as a transcription activator, which may lead to the development of an improved E. coli biocatalyst.
Unlike AraC, which is a well-studied protein, little is known about the XylR protein. Data show that XylR activates transcription by binding to a 37 bp consensus DNA operator sites found in the promoters of the genes it regulates (2,3). AraC inhibits metabolism of d-xylose by binding the same promoter sites. XylR is a 392 amino acid protein, and residues 304–392 show weak similarity to the DNA-binding domain of AraC proteins (8–15). However, its N-terminal domain is nearly twice the size of most AraC proteins and shows no homology to any characterized protein. The AraC family of transcriptional regulators is defined by a 100-residue region of sequence similarity that forms an independently folding DNA-binding domain composed of two helix-turn-helix (HTH) motifs (8). Members fall into three functional groups depending on the types of genes that they regulate. Those that regulate carbon metabolism, such as E. coli AraC, are active as dimers and respond to small molecule effectors that bind to the protein N-terminal domain. Those members that are involved in stress responses, such as SoxS, Rob and MarA typically function as monomers. The third group is involved in regulating virulence gene expression and includes the Vibrio cholera ToxT protein. Structures of the DNA-binding domains of AraC, Rob and MarA have been determined, including structures of the Rob and MarA domains in complex with cognate DNA (9–12). In the MarA-DNA structure, the recognition helices of each motif inserts into adjacent major grooves on the same face of the DNA, making base-specific contacts (9). The tandem arrangement of two helix-turn-helix DNA-binding domains allows for the recognition of ∼20 bp.
Dimerization by AraC proteins further enhances DNA-binding specificity, as it permits the insertion of four helix-turn-helix motifs onto the DNA. The N-terminal domain of AraC binds l-arabinose and also functions in dimerization. This domain is flexibly attached to its C-terminal DNA-binding domain. Structures of the N-terminal domains have been obtained for AraC, E. coli Rob and V. cholerae ToxT (11–14). The AraC N-terminal domain contains a flexible N-terminal arm connected to an eight-stranded antiparallel β-barrel. l-arabinose binds in a pocket within the β-barrel. The N-terminal domains of ToxT and Rob are structurally similar to the AraC l-arabinose-binding domain. Indeed, all contain an eight-stranded antiparallel β-sheet. In ToxT, this domain binds fatty acids, whereas its function in Rob is as yet unknown. Surprisingly, despite the wealth of data on AraC proteins, the molecular details by which the signal of ligand binding to the N-terminal domains of these proteins is communicated to their DNA-binding regions to effect transcription regulation are still unclear. To gain insight into the function of the atypical AraC protein, XylR, we performed biochemical and structural studies. These combined studies reveal a new structural family of DNA-binding proteins and also how ligand binding is communicated from a separate N-terminal ligand-binding domain to an AraC-like DNA-binding domain to activate it for DNA binding. These data also provide structural insight that may aid in the development of more efficient biocatalysts.
MATERIALS AND METHODS
Purification of E. coli XylR
The xylR gene was amplified from DH5α genomic DNA by polymerase chain reaction (PCR) and cloned into the pET28a expression vector such that a C-terminal his-tag was added for purification purposes. BL21(DE3) competent cells were transformed with the xylRpET28a vector, and the resultant protein was expressed by inducing with 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) for 16 hrs at 15°C. Cells were lysed in a buffer consisting of 20 mM Tris pH 8.0, 300 mM NaCl and 10 mM imidazole by a microfluidizer, and the lysate was loaded onto a Ni-NTA column. After extensive washing, XylR protein was eluted using 20 mM Tris pH 8.0, 300 mM NaCl, 300 mM imidazole and then dialysed into 20 mM Tris pH 8.0, 300 mM NaCl.
Crystallization and data collection of XlyR crystals
Apo XylR crystals were grown by mixing the protein at a concentration of 3 mg/ml with 25% PEG 3350, 0.1 M Tris pH 8.0 at a protein to drop ratio of 1:1. The crystals grew to maximum size in 2 weeks. The crystals were cryo-protected by dipping them into a solution consisting of the crystallization solution supplemented with 25% glycerol for 2–5 sec followed by direct placement in the liquid nitrogen stream. The crystals are trigonal, space group P3221 with a = b = 124.5 Å and c = 189.8 Å and diffracted to 3.4 Å at synchrotron sources. Data were successfully collected on only one crystal; the crystals were fragile and typically diffracted to only 5.0 Å. The crystal contains three subunits in the crystallographic asymmetric unit (ASU); two subunits form a dimer and the third subunit forms a dimer with itself via crystallographic symmetry. Crystals of the XylR–d-xylose complex were obtained via hanging drop vapor diffusion using 100 mM Tris pH 8.0, 1.3 M Li2SO4, 50 mM NaSCN, 4 mM 1,4-Dithio-dl-threitol (DTT) and 4 mM d-Xylose as a crystallization condition. The crystals took several days to grow and reached their maximum size in a week. The crystals were cryo-preserved by a quick dip (<1 sec) in a solution containing the crystallization reagent supplemented with 25% sucrose and maintaining the d-xylose concentration at 4 mM. These crystals were tetragonal, space group P42212 with a = b = 70.0 Å and c = 215.4 Å, contain 1 subunit in the ASU. X-ray intensity data were collected at 100 K at beamline 8.3.1 at Advanced Light Source (ALS) in Berkeley. Data were integrated with MOSFLM, merged and scaled with SCALA in CCP4 (16).
Structure determination and refinement of apo XylR and the XylR–d-xylose complex
The structure of XylR–d-xylose complex was solved to 2.90 Å by multi-wavelength anomalous diffraction (MAD) using crystals grown with selenomethionine-substituted protein. Selenomethionine-substituted XylR was produced using the methionine inhibitory pathway method. The selenomethionine-substituted protein was purified as for wild type, with the exception that 5 mM β-mercapto-ethanol (β-ME) was included in all buffers. MAD data were collected on these crystals, and the selenium sites located with SOLVE, resulting in a figure of merit of 0.67 (17). The phases were improved by density modification using the program RESOLVE. The structure was then manually built using Coot (18) and refined in REFMAC5 (19). The final XylR–d-xylose structure includes residues 1–46, 55–389, 1 d-xylose molecule and 53 solvent molecules and has Rwork/Rfree values of 21.9%/27.9% to 2.90 Å resolution. The apo XylR structure was solved by molecular replacement (using MolRep) with the XylR structure (minus the d-xylose). This structure contains residues 1–46, 55–389 of each of the three subunits and was minimally refined in CNS to Rwork/Rfree values of 28.9%/31.6% to 3.40 Å resolution (20). Selected data collection and refinement statistics are given in Table 1.
Table 1.
Selected crystallographic data for XylR structures
| Selenomethionine MAD data for XylR–d-xylose | |||
| Energy (keV) | 12678.8/peak | 12676.8/inflection | 12979.6/remote |
| Resolution (Å) | 107.83-2.90 | 107.83-2.90 | 107.83-2.90 |
| Overall Rsym(%)a | 7.5 (36.6)b | 7.5 (36.2) | 7.9 (38.5) |
| Overall I/σ(I) | 30.2 (8.0) | 29.9 (8.0) | 29.2 (7.6) |
| #Total reflections | 196 548 | 196 226 | 196 403 |
| #Unique reflections | 12 701 | 12 729 | 12 704 |
| Multiplicity | 8.6 | 8.5 | 8.6 |
| Overall Figure of Meritc | 0.67 | ||
| Refinement statistics | |||
| Structure/pdb ID code | XylR–d-xylose/4FE7 | apo XylR/4FE4 | |
| Resolution (Å) | 66.67-2.90 | 107.83-3.40 | |
| Overall Rsym(%)a | 7.9 (38.5) | 11.7 (49.6) | |
| Overall I/σ(I) | 29.2 (7.6) | 7.0 (1.7) | |
| #Total reflections | 196 403 | 22 188 | |
| #Unique reflections | 12 022 | 2286 | |
| % complete | 100 (100) | 97.0 (98.5) | |
| Rwork/Rfree(%)d | 21.9/27.9 | 28.9/31.6 | |
| Rmsd | |||
| Bond angles (°) | 1.275 | 1.60 | |
| Bond lengths (Å) | 0.010 | 0.011 | |
| Ramachandran analysis | |||
| Most favoured (%) | 89.4 | 78.9 | |
| Additional allowed (%) | 10 | 20.2 | |
| Generously allowed (%) | 0.6 | 0.9 | |
| Disallowed (%) | 0.0 | 0.0 | |
aRsym = ΣΣ|Ihkl−Ihkl(j)|/ΣIhkl, where Ihkl(j) is observed intensity and Ihkl is the final average value of intensity.
bValues in parentheses are for the highest resolution shell.
cFigure of Merit = <|ΣP(α)eiα/ΣP(α)|>, where α is the phase and P(α) is the phase probability distribution.
dRwork = Σ||Fobs| − |Fcalc||/Σ|Fobs| and Rfree = Σ||Fobs| − |Fcalc||/Σ|Fobs|; where all reflections belong to a test set of 5% randomly selected data.
Atomic force microscopy (AMF) sample preparation and imaging
The 371 bp DNA construct (xyl Promoter) used encompasses the xyl promoter and spans the region between the start codons of xylA and xylF (21,22). This region contains two outwardly directed promoters, IA and IF. The xyl Promoter DNA construct was amplified from E. coli MG1655 genomic DNA with the following primers: 5′-ATATTGAACTCCATAATCAGGTAATGC-3′ (forward) and 5′-CATGGTGTAGGGCCTTCTGT-3′ (reverse). The 900 bp DNA construct (IFIF900) encodes two IF promoter sites separated by 500 bp with 200 bp flanking each termini. The latter construct was used to clearly visualize DNA looping. AFM samples were prepared with 20 uM of XylR-DNA, with a ratio of 2:1 (protein:DNA), in binding buffer (75 mM NaCl, 20 mM Tris-HCl pH 7.5 and 2 mM Xylose). For sample deposition, specially modified 1-(3-aminopropyl)silatrane (APS) mica surface was used. The APS mica was obtained by incubation of freshly cleaved mica in 167 nM 1-(3-aminopropyl)silatrane. The details of APS mica surface modification are described in (23,24). The sample droplet (5–10 µl) was deposited on APS mica and after 2-min incubation, sample excess was washed with deionized water (AquaMaxTm Ultra, LabWater.com) and dried with an Argon gas flow. AFM images in air were acquired using MultiMode AFM NanoScope IV system (Veeco/Bruker Iinstruments, Santa Barbara, CA, USA) operating in tapping mode. Regular tapping Mode Silicon Probes (Olympus from Asylum Research, Santa Barbara, CA, USA) with a spring constant of 42 N/m and a resonant frequency between 300–320 kHz were used.
Fluorescence Polarization assays
Fluorescence Polarization assays of XylR-DNA binding were performed as described (25). All oligonucleotides used in the assays were 5′-fluorescein labelled. In each assay, 1 nM oligonucleotide was added to the binding buffer (75 mM NaCl, 20 mM Tris pH 7.5, 2 μg/ml poly[dI-dC], ± 2 mM d-xylose), and increasing concentrations of protein were titrated into the binding mixture. The excitation and emission wavelengths were 490 nm and 530 nm, respectively. The data were fit using Kaleidagraph as previously described (25).
Isothermal titration calorimetry (ITC) experiments
All ITC experiments were performed using a VP-ITC system (MicroCal Inc., Northampton, MA, USA). The ITC experiments were performed with either d-xylose or l-arabinose (in the syringe) at the concentration of 1 mM and 100 uM of XylR (in the sample cell). Ligand was titrated into the sample cell containing XylR at 25°C, and the resulting isotherm was fitted with Origin. The XylR sample was placed into the ITC buffer (75 mM NaCl, 20 mM Tris-HCl pH 7.5) via dialysis, and the two ligands sample were dissolved in the dialysis buffer.
RESULTS AND DISCUSSION
Overall structure of E. coli apo XylR and the XylR–d-xylose complex
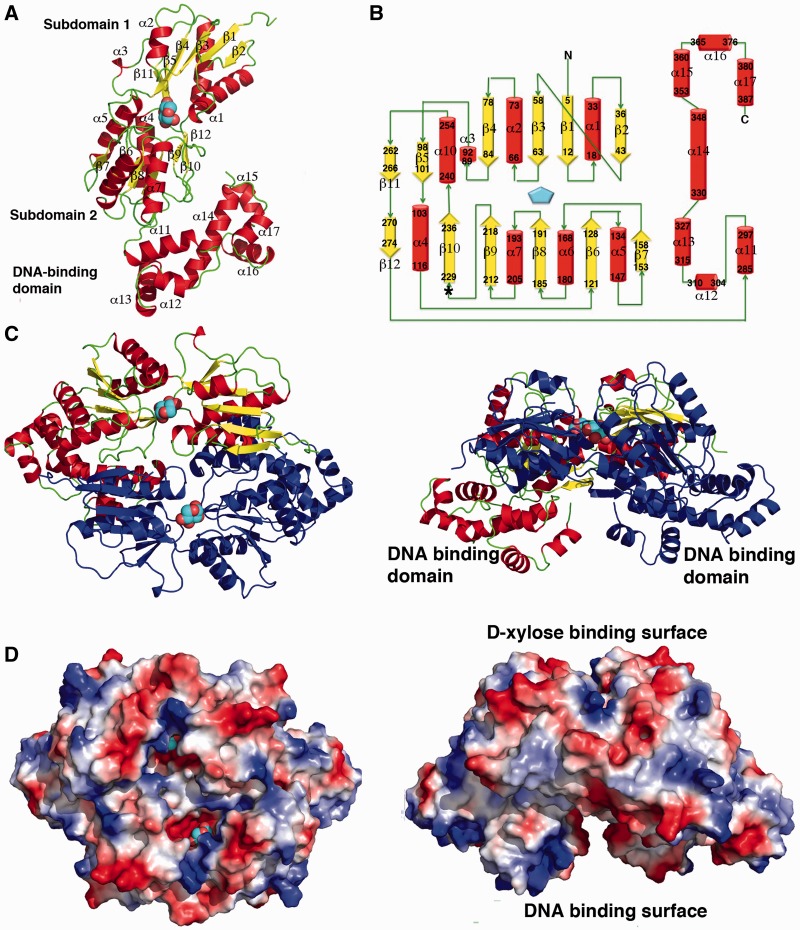
To gain insight into the molecular functions of XylR, we determined structures of apo XylR and a XylR–d-xylose complex. The XylR–d-xylose complex structure was solved by MAD to 2.90 Å and refined to Rwork/Rfree of 21.9%/27.9%. The apo XylR structure was solved by molecular replacement using the XylR structure from the XylR–d-xylose complex as a search model (‘Materials and Methods’ section). The final apo XylR model was refined to Rwork/Rfree of 28.9%/31.6% to 3.40 Å resolution (Table 1; ‘Materials and Methods’ section). The structures show that XylR is composed of two domains, an N-terminal domain (residues 1–274) and a C-terminal domain (residues 285–389). The N-terminal domain has a α−β fold, whereas the C-terminal domain is all helical and composed of 7 helices. The two domains are connected by a long, but structured, linker formed by residues 275–284 (Figure 1A and B). As predicted, the C-terminal domain shows structural similarity to AraC proteins. This domain of XylR shows the strongest structural homology with the corresponding domain of the MarA protein, with the Cα superimposition resulting in a root mean squared deviations (rmsds) of 2.1 Å. However, the N-terminal domain of XylR is distinct from the β-barrel like ligand-binding domain in other AraC proteins (11–13). Structural homology searches revealed that this domain shows the strongest similarity to the periplasmic-binding protein (PBP) domains of the LacI/GalR family of transcription regulator (27–33). In particular, the XylR N-terminal domain is the most similar to the PBP domain of the Purine Repressor, PurR (27,28). One subunit of XylR can be superimposed onto a PurR subunit with an rmsd of 2.5 Å for 212 corresponding Cα atoms (Supplementary Figure S1).
Figure 1.
Structure of E. coli XylR defines a new DNA-binding family. (A) Overall structure of a XylR subunit. α-helices and β-strands are coloured red and yellow, respectively, and labelled, and loops are coloured green. The bound d-xylose is shown as cpk, with carbons and oxygens coloured cyan and red, respectively. The DNA-binding domain and PBP subdomains 1 and 2 are labelled. (B) Topology diagram of the XylR subunit with α-helices and β-strands coloured as in Figure 1A. The residues contained within each secondary structural element are also indicated. The asterisk indicates the region, which encompasses residues 221–229, which is a helix in the apo form and a strand in the d-xylose bound form (the latter of which is shown here). (C) Two views of the XylR dimer (rotated by 90°). One subunit is coloured as in Figure 1A and other is coloured dark blue. (D) Electrostatic surface representation of the XylR dimer (shown in the same orientations as Figure 1C). Electropositive and electronegative regions are coloured blue and red, respectively. This Figure, Figures 2A–B, 3A–B and 4F were made with PyMOL (26).
The E. coli XylR structure combines two domains previously not found in the same protein, an N-terminal d-xylose-binding domain that is homologous to those of the LacI/GalR family and a C-terminal DNA-binding domain with an AraC-like DNA-binding fold. Thus, the XylR structure defines a new family of DNA-binding proteins. BLAST searches, which revealed >100 proteins with strong sequence homology to E. coli XylR, suggest that this ‘XylR family’, which consists of an N-terminal effector binding domain with a PBP fold connected to an AraC-like DNA-binding domain, is wide spread in Gram-negative bacteria. The Caulobacter crescentus XylR protein, however, represents an exception. Recent sequence homology analyses predict that this protein is a bone fide member of the LacI/GalR proteins, with an N-terminal HTH domain followed by a hinge helix and C-terminal PBP-like domain (34). These findings suggest a possible role in domain swapping during the evolution of XylR proteins. Like LacI/GalR and periplasmic-binding proteins, the N-terminal region of XylR is composed of two α−β subdomains (herein called subdomains 1 and 2), which are connected by short crossover regions that, in the PBPs and LacI proteins, permit rotation between subdomains (Figure 1A). This subdomain movement allows the protein to trap a ligand once it has entered the binding cavity. The resulting subdomain movement can be transmitted to attached regions to elicit other effects, such as conformational changes or folding of attached domains.
XylR contains a periplasmic binding fold used in antiparallel dimerization
Examination of the packing of the XylR crystal structures suggests that, like the LacI/GalR proteins, XylR dimerizes via its PBP-like domain (Figure 1C and D). The XylR dimer interface buries an extensive 2960 Å2 of protein surface from solvent. Size exclusion chromatography experiments revealed molecular weights consistent with a XylR dimer (Supplementary Figure S2). Like LacI/GalR proteins, the dimerization interface of XylR is formed primarily by interactions between the PBP domains. However, in sharp contrast to the LacI/GalR proteins, which dimerize via parallel interactions between PBP regions, the XylR PBP domains interact in an antiparallel fashion. Also distinct from LacI/GalR oligomers, the PBP folds of XylR also make extensive interactions with the C-terminal DNA-binding domain of the other subunit in the XylR dimer (Figure 1C and D). These contacts are made between the DNA-binding domain and subdomain 1 from the other subunit.
The only other PBP-containing proteins that use a form of antiparallel dimerization are the LysR family of transcription regulators (35). LysR proteins contain an N-terminal winged helix DNA-binding domain, which is unrelated to the DNA-binding domains of LacI/GalR proteins or XylR. Although both PBP domains of LysR proteins and XylR both dimerize in an antiparallel mode, the subunit structure/PBP folds of these proteins are significantly different; the PBPs of XylR and LysR proteins superimpose with rmsds of 4.6–5.5 Å (35). Also, XylR dimerization involves interactions between its DNA-binding domain and subdomain 1 of the other PBP subunit in its dimer, which is not observed in LysR proteins. The XylR antiparallel PBP dimerization mode results in an arrangement in which the DNA-binding domains are found on opposite ends of the dimer. The long linker allows for the formation of an oligomer with two faces, one containing the DNA-binding domains and the other, the PBP antiparallel dimer (Figure 1C and D). This arrangement leaves both the DNA-binding and ligand-binding domains unobstructed such that each domain can bind its ligand without impediment from the other domain.
d-xylose binding to XylR
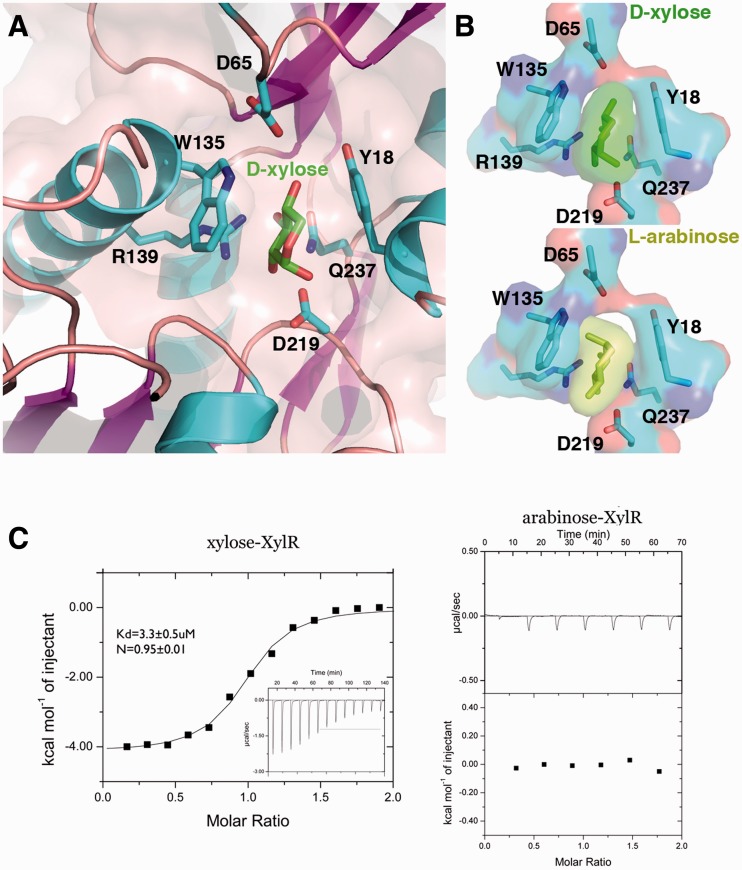
To ascertain how d-xylose affects XylR function, we co-crystallized XylR in the presence of d-xylose. Clear density was observed for a d-xylose molecule in the pocket between the subdomains of the XylR PBP domain (Supplementary Figure S3). Stacking interactions to the d-xylose molecule are provided by Tyr18 from subdomain 1 and Trp135 from subdomain 2 (Figure 2A and B). Every oxygen moiety of the d-xylose sugar is contacted by one, or several XylR residues, with the exception of the xylose O5 atom. Asp65 from subdomain 1 contacts the d-xylose O4 hydroxyl and the remaining hydrogen bonds are provided by subdomain 2 residues. Gln237 contacts the O3 hydroxyl, the Oδ atoms of Asp219 hydrogen bonds with the O1 and O2 hydroxyls, while both Nε atoms of Arg139 contact the O2 and O3 hydroxyls. Previous studies on periplasmic-binding proteins and LacI/GalR members have shown that ligand binding captures or stabilizes the closed conformation of the PBP clamshell fold. Indeed, d-xylose binding requires the specific arrangement of residues only found in the closed state. It is also interesting to note that XylR residue Gln237 is located on one of the three cross-overs that connect the subdomains. Thus, this Gln237-d-xylose contact likely also stabilizes the closed state.
Figure 2.
d-xylose binding by XylR. (A) Close up of the d-xylose–XylR interaction. XylR residues that make key interactions with d-xylose are shown as sticks and labelled. (B) Comparison of XylR–d-xylose complex with a model of a XylR–l-arabinose complex. As indicated by the transparent surface representations, l-arabinose binding in this mode would result in significant clash with Trp135. (C) ITC studies on d-xylose (left) and l-arabinose (right) binding to XylR. The binding isotherm of XylR for d-xylose resulted in a Kd of 3.3 ± 0.5 uM with a stoichiometry of 1 XylR: 1 d-xylose. By contrast, l-arabinose showed no binding by XylR.
In vivo studies have shown that XylR does not respond to l-arabinose despite its structural similarity to d-xylose. Consistent with this, modelling of l-arabinose in the XylR binding pocket revealed significant clash between the l-arabinose O4 hydroxyl and the XylR Trp135 side chain (O4 to Trp135 Cε2 distance of 2.1 Å) (Figure 2B). However, these modelling exercises were carried out assuming that l-arabinose would bind in the same orientation in the pocket as d-xylose (see Figure 2B), and other binding modes can not be ruled out. Thus, to determine the binding affinities of XylR for d-xylose and l-arabinose, we performed ITC studies. Clear binding of XylR to d-xylose was observed, resulting in a Kd of 3.3 ± 0.5 µM (Figure 2C; Supplementary Figure S4). Consistent with the XylR–d-xylose structure, these experiments also revealed a binding stoichiometry of 1 subunit of XylR to 1 molecule of d-xylose. Also consistent with the structure, ITC experiments revealed no binding of l-arabinose by XylR (Figure 2C).
Only when l-arabinose is exhausted will d-xylose bound XylR activate the expression of the d-xylose metabolic genes in the presence of hemicellulose food sources. Recent studies have shown, however, that this diauxie can be altered leading to the design of a more efficient E. coli biocatalyst by controlled overexpression of XylR. This overexpression allows XylR–d-xylose to compete with AraC–l-arabinose for binding to xyl promoters (5). Additional strategies in the design of E. coli biocatalyst would be to engineer a XylR protein that is responsive to both d-xylose and l-arabinose or identify a small molecule that can readily cross the membrane (as high affinity d-xylose transporters are only highly expressed upon xyl transcription activation) and bind with nanomolar affinity to XylR. The use of such a high affinity small molecule ligand would also alleviate the need for a bioengineering step. The details of the d-xylose-binding pocket provided by the XylR–d-xylose structure should significantly facilitate such design efforts.
Comparison of apo XylR and XylR–d-xylose structures; d-xylose binding leads to a helix to strand transition and reorganization of the XylR dimer
The XylR DNA-binding domain′–subdomain 1 interface (where prime indicates other subunit in the dimer) is formed by contacts between β1, β2, α1 and α10 of subdomain 1 with helices α11′ and α13′ of the DNA-binding domain. Key hydrophobic and stacking interactions in this interface are formed between Phe2 with His297′ and Tyr29 and Ala32 with Met359′. In addition, there are numerous salt bridge and hydrogen bonding interactions. Specifically, Glu28 interacts with Thr349′, Glu36 with Lys300′, K248 and Tyr244 with Glu355′ (Supplementary Figure S5). As noted, however, the most extensive XylR interface is composed of antiparallel contacts between PBP subdomains. In particular, subdomain 1 residues from α1 and β2 interact with subdomain 2 residues from α6 (residues 162–170), α7 (residues 193–205) and β10.
To obtain insight into how d-xylose activates DNA binding by XylR, we compared structures of the apo and d-xylose bound states. The structures revealed the same overall dimer organization whereby the d-xylose-binding PBP faces are located on one side of the dimer and the DNA-binding domains on the other. However, d-xylose binding leads to significant structural changes as underscored by resulting rmsds of 1.5 Å for superimposition of individual subunits and 2.2 Å for overlays of both subunits in the dimer. Moreover, rmsds of 1.2 Å are obtained from superimpositions of individual PBP domains, whereas superimpositions of entire subunits, including the DNA-binding domain, results in rmsds of 1.7 Å. These findings indicate that d-xylose binding causes structural changes in the orientation of the subunits within the dimer as well as the relative orientation of the ligand binding to the DNA-binding domain within each subunit.
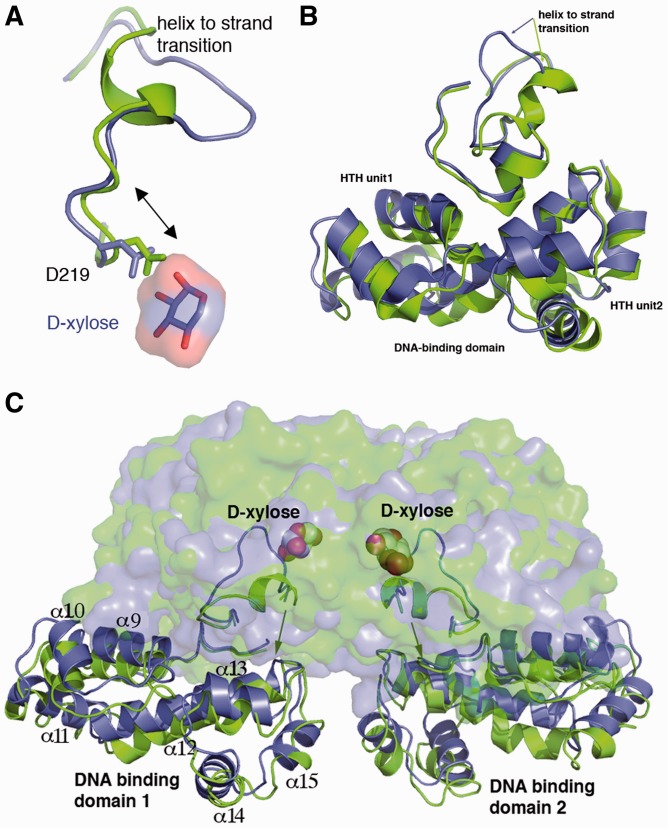
Examination of the residues near the ligand-binding pocket revealed the striking finding that d-xylose binding is accompanied by a transition in residues 221–229 from an α-helix in the apo form to a strand (β10) in the d-xylose bound state (Figure 3A–C; Supplementary Figure S6). This helix to strand conversion appears to be triggered by d-xylose interaction with residues 219–221, as modelling shows that the side chain of Asp219 is <1.4 Å from the d-xylose moiety in the apo state (Figure 3A). Hence, this residue must move to permit d-xylose binding. The presence of the rigid helix my impede this movement. As a result, in the d-xylose-bound structure residues 225–226 buckle out (Figure 3A and B), thus creating a binding pocket that permits d-xylose insertion (Supplementary Figures S7–S9). What is particularly striking about this structural transformation of residues 221–229 is that these residues lie in the dimerization interface. Notably, Tyr226 moves from its helical position where it hydrogen bonds with Arg240′ in the apo structure to its strand position in the d-xylose bound structure, where it interacts with Asp38′ (Supplementary Figure S6). In addition, there is a large shift in the position of α1 of the neighbouring subunit (Figure 3A and B). This helix lies at the nexus between the PBP and the DNA-binding domain and in fact inserts between the two HTH repeat elements within each DNA-binding domain subunit (Figure 3A; Supplementary Figure S6). As a result even minor shifts in the position of α1 are capable of producing significant structural changes within the tandem helix-turn-helix containing DNA-binding domain. Moreover, because these changes occur in both subunits of the dimer, the net movement of the DNA-binding domains is further amplified. This explains the large differences noted from superimposition of the apo and d-xylose bound structures. A notable result of these large conformational changes is an increase in the overall buried surface area of the dimer (the d-xylose bound dimer buries 2960 Å2 compared with 2446 Å2 for the apo bound form). Previous work indicated that d-xylose binding activates XylR for DNA binding (2–5). Hence, d-xylose-induced conformational changes presumably align the HTH elements properly for DNA binding; however, a complete understanding of the d-xylose-induced DNA-binding mode will require a XylR–d-xylose-DNA structure.
Figure 3.
d-xylose binding triggers helix to strand transition. (A) Superimposition of the apo (green) and d-xylose bound (blue) XylR structure showing a close up of the region undergoing a helix to coil transition. The overlay indicates that d-xylose triggers this response by forcing Asp219 and the accompanying N-terminal region of the helix to move, which requires the helix to unfold. (B) d-xylose binding leads to a helix to strand transition. For clarity the strand is shown as a thin ribbon in this Figure. (C) Overall result of the helix to strand transition upon d-xylose binding is a reorientation of the DNA-binding domains. d-xylose is shown as cpk, the d-xylose binding domain as transparent surfaces and the DNA-binding domains as ribbons.
d-xylose-induced conformational changes and DNA binding stoichiometry
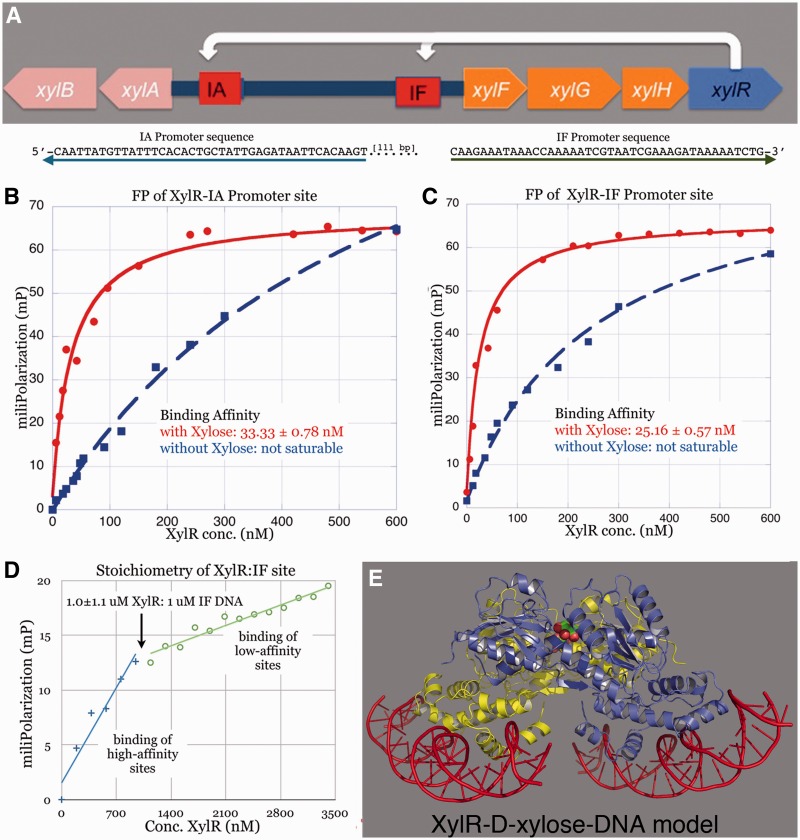
XylR regulates two co-transcribed operons by binding the ∼37 bp sites, IA and IF. These sites are each composed of two direct repeats (with consensus–-gaAa-a–a-AAT–-gaAa-a–a-AAT) (2,3). These binding sites each control a separate operon, one controls the xylAB cluster and the other, the xylFGH genes (2,3). Interestingly, these two gene clusters, which are separated by ∼360 bp, are transcribed in opposite directions (Figure 4A). In each promoter, the XylR-binding sites are located next to the −35 motif that specifies the σ70 subunit of RNA polymerase. How XylR regulates these two operons is unclear. Indeed, the initial studies did not ascertain the binding affinity of XylR for these sites nor did they deduce how many XylR molecules interact at each motif. Understanding how XylR regulates the two operons requires knowledge of the binding stoichiometry of XylR for each site, as the structures show that each XylR dimer contains four HTH motifs. Hence, four XylR molecules could potentially bind this region whereby each HTH interacts with one direct repeat. Thus, we performed FP experiments to determine the DNA-binding affinities and stoichiometry of XylR for its DNA site. These studies showed that d-xylose was required to achieve high affinity binding to the IA and IF operator sites (Figure 4B–C). In the presence of 2 mM d-xylose, XylR binds its operator sites with Kds of 25 nM and 33 nM. By contrast, in the absence of d-xylose, saturable binding was not observed. Strikingly, the data show that a XylR dimer binds two DNA duplexes or promoter sites (Figure 4D). Thus, to generate a XylR-DNA model, we docked two DNA duplexes onto each XylR subunit using the MarA-DNA structure as a guide (Figure 4E).
Figure 4.
XylR DNA binding and activation by d-xylose. (A) Top shows the organization of the two operons regulated by XylR. Below, sequences of the xyl promoters (IA and IF), which are transcribed in opposite directions. The arrows represent the 5′ to 3′ directions of the sequence motifs. (B) The binding affinity of XylR for the IA promoter. The resulting isotherm revealed a binding affinity of ∼33 nM, in the presence of d-xylose. In the absence of d-xylose, no significant binding is observed. (C) The XylR-IF promoter binding isotherm reveals an affinity of ∼25 nM in the presence of d-xylose, whereas in the absence of d-Xylose no binding is observed (monomer concentration). (D) Stoichiometric FP experiment carried out in the same manner as that shown in (C) with the exception that the IF DNA concentration was increased to 1 µM. This concentration is 40-fold higher than the Kd, thus ensuring stoichiometric binding. The transition from high- to low-affinity binding resulted in an inflection point of ∼1 uM XylR in the presence of 1 uM IF DNA. This indicated a binding stoichiometry of one XylR dimer to two DNA duplexes. (E) Model of XylR-operator DNA based on the stoichiometry study in (D) showing two duplexes binding a dimer.
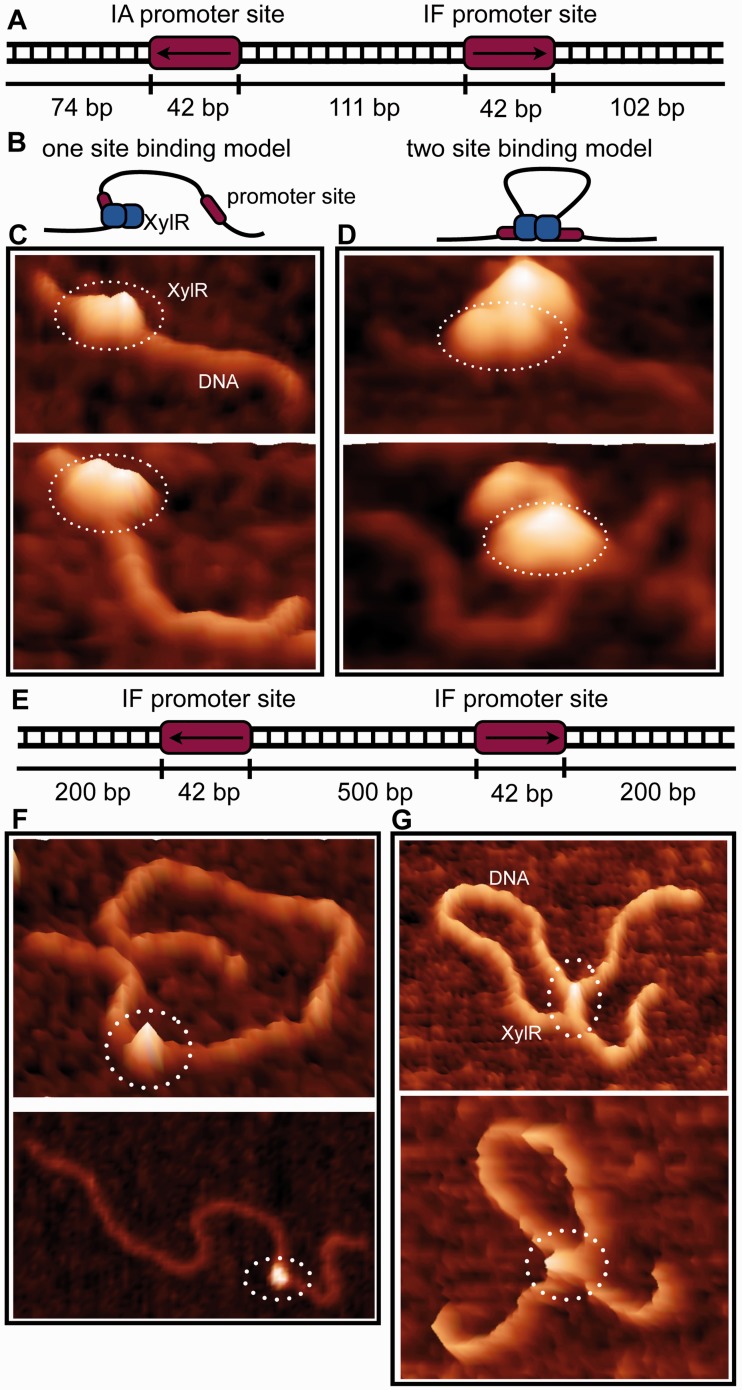
The fact that the two operons regulated by XylR are transcribed in opposite orientation with the transcription start sites of the first transcribed genes and the finding that one XlyR dimer binds two separate DNA sites suggested the intriguing possibility that one XylR dimer may interact with both IA and IF via a looping mechanism. To test this we performed AFM experiments. We first looked at XylR binding to the natural promoter region encompassing both XylR operator sites. Consistent with stoichiometry studies, these analyses (Figures 5A–D) strongly suggested that one XylR dimer binds between the promoter operator sites and mediates looping. However, the intervening DNA between operator sites was too short to readily visualize via AFM (Figure 5D). Thus, to clearly deduce whether one XylR dimer can bind between distant DNA sites, we constructed IF900, which encodes two IF promoter sites separated by 500 bp. When mixed with this construct, AFM studies revealed clear evidence for DNA looping by XylR (Figures 5E–G).
Figure 5.
The binding mode of XylR to the xyl promoter region observed by atomic force microscopy. (A) Schematic of the xyl promoter region, which contain two XylR binding sites (IA and IF). The arrow indicates the 5′ to 3′ direction of the XylR-binding site. (B) A cartoon representation of the two observed modes of XylR-DNA binding. The AFM data show that a XylR dimer binds first to one DNA site and, subsequently, the second site on the same DNA strand, looping the DNA. (C) AFM images of a XylR dimer bound to a single promoter site of the xyl promoter region. (D) A XylR dimer binding to both promoter sites creating a DNA loop. However, the intervening DNA between operator sites was too short to readily visualize via AFM using this DNA site. See Figure 5G. (E) Unnatural DNA substrate used to visualize DNA loop more clearly. (F) AFM images of a XylR dimer bound to a single promoter site of the longer DNA substrate shown in E. (G) AFM images of a XylR dimer binding to the two promoter sites, leading to clear DNA looping.
The -35 and -10 regions of both xyl promoters possess poor matches to the optimal σ70 consensus motifs, suggesting that XylR–d-xylose likely activates transcription by recruitment of RNA polymerase. DNA looping by XylR could be an integral mechanism by which XylR performs its transcription activation function, as it would closely juxtapose the two promoter sites perhaps allowing RNA Polymerase recruitment to both sites. This activation looping contrasts with the repressive outcome of DNA looping by AraC, which in its apo state, loops DNA and inhibits PBAD transcription (36,37). Transcription activation by AraC occurs when it binds l-arabinose and, in collaboration with CRP, stimulates loop opening (37). By contrast, apo XylR does not bind DNA. However, a repressive mode of XylR is not required as AraC–l-arabinose binds the xyl promoters mediating repression (5). Thus, the combined data indicate that under l-arabinose and d-xylose replete conditions, AraC–l-arabinose binds tightly to the xyl promoters and only when l-arabinose is depleted does d-xylose bound XylR bind and loop DNA to activate transcription.
In conclusion, our studies on XylR define a new family of DNA-binding proteins, which harbours a DNA-binding domain with an AraC-like fold and a ligand-binding domain with a LacI/GalR-like structure. The ligand-binding domain dimerizes in a distinct antiparallel mode. d-xylose binding causes a helix to strand transition in the dimer interface of the d-xylose-binding domain that results in dimer rearrangement, which is transmitted to the DNA-binding domains. XylR binds to two promoters that are transcribed in opposite directions. Strikingly, FP and AFM studies indicate that XylR binds to two DNA sites per dimer and loops DNA. Finally, the XylR–d-xylose structure reveals key determinants that explain its exquisite selectivity towards d-xylose. This knowledge could be used in design efforts towards the development of more efficient E. coli biocatalysts.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–9.
ACCESSION NUMBERS
Coordinates and structure factor amplitudes for the apo XylR and XylR-d-xylose structures have been deposited with the Protein Data Bank under the accession codes 4FE4 and 4FE7, respectively.
FUNDING
M.D. Anderson Trust Fellowship; National Institutes of Health [GM068453 to M.A.S.]; NIH (SIG program), UNMC Program of Excellence (POE) and Nebraska Research Initiative (NRI) (to The University of Nebraska Nanoimaging Core Facility). Funding for open access charge: Startup funds, Duke University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Advanced Light Source (ALS) and their support staff. The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, and Material Science Division of the US Department of Energy at the Lawrence Berkeley National Laboratory. We also acknowledge the University of Nebraska Nanoimaging Core Facility for AFM data collection.
REFERENCES
- 1.Bruckner R, Titgemeyer F. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 2002;209:141–148. doi: 10.1111/j.1574-6968.2002.tb11123.x. [DOI] [PubMed] [Google Scholar]
- 2.Song S, Park C. Utilization of D-ribose through D-xylose transporter. J. Bacteriol. 1997;179:7025–7032. doi: 10.1128/jb.179.22.7025-7032.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song S, Park C. Organization, regulation of the D-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. FEMS Microbiol. Lett. 1998;163:255–261. [Google Scholar]
- 4.Desai TA, Rao CV. Regulation of arabinose and xylose metabolism in Escherichia coli. Appl. Environ. Micobiol. 2010;76:1524–1532. doi: 10.1128/AEM.01970-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groff D, Benke PI, Batth TS, Bokinsky G, Petzold CJ, Adams PD, Keasling JD. Supplementation of intracellular XylR leads to co-utilization of hemicellulose sugars. Appl. Environ. Micobiol. 2012;78:2221–2229. doi: 10.1128/AEM.06761-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JH, Block DE, Mills DA. Simultaneous consumption of pentose and hexose sugars: an optimal microbial phenotype for efficient fermentation of lignocellulosic biomass. Appl. Microbiol. Biotechnol. 2010;88:1077–1085. doi: 10.1007/s00253-010-2839-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inada T, Kimata K, Aiba H. Mechanism responsible for glucose–lactose diauxie in Escherichia coli: challenge to the cAMP model. Genes Cells. 1996;1:293–301. doi: 10.1046/j.1365-2443.1996.24025.x. [DOI] [PubMed] [Google Scholar]
- 8.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhee S, Martin R, Josner J, Davies D. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl Acad. Sci. USA. 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodgers M, Schleif R. Solution structure of the DNA binding domain of AraC protein. Proteins. 2009;77:202–220. doi: 10.1002/prot.22431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon H, Bennik M, Demple B, Ellenberger T. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Mol. Biol. 2000;7:424–430. doi: 10.1038/75213. [DOI] [PubMed] [Google Scholar]
- 12.Lowden M, Skorupski K, Pellegrine M, Chiorazzo M, Taylor R, Kull F. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc. Natl Acad. Sci. USA. 2010;107:2860–2865. doi: 10.1073/pnas.0915021107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soisson S, MacDougall-Shackleton B, Schleif R, Wolberger C. Structural basis for ligand-regulated oligomerization of AraC. Science. 1997;276:421–425. doi: 10.1126/science.276.5311.421. [DOI] [PubMed] [Google Scholar]
- 14.Schleif R. AraC protein: Regulation of the L-arabinose operon in Escherichia coli and the light switch mechanism of AraC action. FEMS Microbiol. Rev. 2010;34:779–796. doi: 10.1111/j.1574-6976.2010.00226.x. [DOI] [PubMed] [Google Scholar]
- 15.Schleif R. AraC protein: a love-hate relationship. Bioessays. 2003;25:274–282. doi: 10.1002/bies.10237. [DOI] [PubMed] [Google Scholar]
- 16.Leslie AG. Integration of macromolecular diffraction data. Acta. Crystallogr. D. Biol. Crystallogr. 1999;D55:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- 17.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta. Crystallogr. D. Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features, development of coot. Acta. Crystallogr. D. Biol. Crystallogr. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vagin AA, Steiner RS, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organisation of prior chemical knowledge and guidelines for its use. Acta. Crystallogr. D. Biol. Crystallogr. 2004;D60:2284–2295. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 20.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Crosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta. Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 21.Shlyakhtenko LS, Gall AA, Filonov A, Cerovac Z, Lushnikov A, Lyubchenko YL. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003;97: 279–287. doi: 10.1016/S0304-3991(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 22.Lyubchenko YL, Shlyakhtenko LS, Aki T, Adhya S. Atomic force microscopic demonstration of DNA looping by GalR and HU. Nucleic Acids Res. 1997;25:873–876. doi: 10.1093/nar/25.4.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen CL, Bromley KM, Moradian-Oldak J, DeYoreo JJ. In situ AFM study of amelogenin assembly and disassembly dynamics on charged surfaces provides insights on matrix protein self-assembly. J. Am. Chem. Soc. 2011;133:17406–17413. doi: 10.1021/ja206849c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shlyakhtenko LS, Gall AA, Lyubchenko YL. Mica functionalization for imaging of DNA and protein-DNA complexes with atomic force microscopy. Methods Mol. Biol. 2013;931:295–312. doi: 10.1007/978-1-62703-056-4_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundblad JR, Laurance M, Goodman RH. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol. Endocrinol. 1996;10:607–612. doi: 10.1210/mend.10.6.8776720. [DOI] [PubMed] [Google Scholar]
- 26.Delano WL. The PyMOL Molecular Graphics System. (DeLano Scientific San Carlos California 2002) [Google Scholar]
- 27.Schumacher MA, Choi KY, Zalkin H, Brennan RG. Crystal structure of LacI member PurR bound to DNA: minor groove binding by alpha helices. Science. 1994;266:763–770. doi: 10.1126/science.7973627. [DOI] [PubMed] [Google Scholar]
- 28.Schumacher MA, Choi KY, Lu F, Zalkin H, Brennan RG. Mechanism of corepressor-mediated specific DNA binding by the purine repressor. Cell. 1995;83:147–155. doi: 10.1016/0092-8674(95)90243-0. [DOI] [PubMed] [Google Scholar]
- 29.Schumacher MA, Allen GS, Diel M, Seidel G, Hillen W, Brennan RG. Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P. Cell. 2004;118:731–741. doi: 10.1016/j.cell.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 30.Bell CE, Lewis M. A closer view of the conformation of the Lac repressor bound to operator. Nat. Struct. Biol. 2000;7:209–214. doi: 10.1038/73317. [DOI] [PubMed] [Google Scholar]
- 31.Lewis M, Chang G, Horton NC, Kercher MA, Pace HC, Schumacher MA, Brennan RG, Lu P. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science. 1996;271:1247–1254. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]
- 32.Felder CB, Graul RC, Lee AY, Merkle HP, Sadee W. The venus flytrap of periplasmic binding proteins: an ancient protein module present in multiple drug receptors. AAPS PharmSci. 1999;1:E2. doi: 10.1208/ps010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams MD, Oxender DL. Bacterial periplasmic binding protein tertiary structures. J. Biol. Chem. 1989;264:15739–15742. [PubMed] [Google Scholar]
- 34.Stephens C, Christen B, Watanabe K, Fuchs T, Jenal U. Regulation of D-xylose metabolism in Caulobacter crescentus by a LacI-type repressor. J. Bacteriol. 2007;189:8828–8834. doi: 10.1128/JB.01342-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maddocks SE, Oyston PC. Structure and function of the LysR-type transcription regulator (LTTR) family proteins. Microbiology. 2008;154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 36.Lobell RB, Schleif RF. DNA looping and unlooping by AraC protein. Science. 1990;250:528–532. doi: 10.1126/science.2237403. [DOI] [PubMed] [Google Scholar]
- 37.Schleif RF. Regulation of the L-arabinose operon of Escherichia coli. Trends Genet. 2000;16:559–565. doi: 10.1016/s0168-9525(00)02153-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.