Abstract
N6-threonylcarbamoyladenosine (t6A) is a modified nucleotide found in all transfer RNAs (tRNAs) decoding codons starting with adenosine. Its role is to facilitate codon–anticodon pairing and to prevent frameshifting during protein synthesis. Genetic studies demonstrated that two universal proteins, Kae1/YgjD and Sua5/YrdC, are necessary for t6A synthesis in Saccharomyces cerevisiae and Escherichia coli. In Archaea and Eukarya, Kae1 is part of a conserved protein complex named kinase, endopeptidase and other proteins of small size (KEOPS), together with three proteins that have no bacterial homologues. Here, we reconstituted for the first time an in vitro system for t6A modification in Archaea and Eukarya, using purified KEOPS and Sua5. We demonstrated binding of tRNAs to archaeal KEOPS and detected two distinct adenosine triphosphate (ATP)-dependent steps occurring in the course of the synthesis. Our data, together with recent reconstitution of an in vitro bacterial system, indicated that t6A cannot be catalysed by Sua5/YrdC and Kae1/YgjD alone but requires accessory proteins that are not universal. Remarkably, we observed interdomain complementation when bacterial, archaeal and eukaryotic proteins were combined in vitro, suggesting a conserved catalytic mechanism for the biosynthesis of t6A in nature. These findings shed light on the reaction mechanism of t6A synthesis and evolution of molecular systems that promote translation fidelity in present-day cells.
INTRODUCTION
All naturally occurring cellular RNA species carry numerous modified nucleosides. Of 100 different modified nucleosides reported to date, >80 are present in mature transfer RNAs (tRNAs). Each tRNA molecule has some subset of these modifications, altering nucleoside identity at an average of 11 positions. Modified nucleosides favour correct folding and stability of tRNA, and some of them are important for the efficiency and fidelity of translation (1–5). Notably, some tRNA positions are almost always modified. This is the case for the nucleotide at the position 37, which is modified in >70% of all tRNAs (6). In a vast majority of cases, the modifications introduced at position 37 are the universal N1-methyl-guanosine (m1G) and N6-threonylcarbamoyladenosine (t6A), or their derivatives. t6A is found exclusively at the position 37, 3′ adjacent to the third base of the anticodon, in all tRNAs reading ANN codons (where N corresponds to four canonical nucleotides A, C, G and T), tRNAIle, tRNAThr, tRNAAsn, tRNALys, tRNASer and tRNAArg (Figure 1). The t6A modification facilitates correct anticodon–codon pairing by enhanced base stacking and by preventing intraloop base pairing of U33 • A37, resulting in enhanced translation fidelity (7,8).
Figure 1.
t6A modification. On the left is a schematic representation of an anticodon loop of tRNA carrying NNU anticodon and an mRNA molecule carrying ANN codon. Adenosine at position 37 is indicated in white circle. On the right is the chemical structure of t6A-modified nucleoside. Chemical groups originating from threonine or carbonate are indicated in grey and bold, respectively.
t6A modification has been discovered, and its chemical structure was elucidated >40 years ago (9). Initial studies of t6A biosynthesis performed in vivo or with Escherichia coli cellular extracts demonstrated that the reaction required adenosine triphosphate (ATP), carbonate, magnesium and threonine (10,11). However, the proteins involved in the catalysis of this multistep reaction were not identified until recently. The breakthrough came when de Crecy-Lagard et al. made the link between the universality of t6A and a previous comparative genomic analysis that identified YrdC and YgjD and their archaeal and eukaryotic homologues (Sua5 and Kae1, respectively) as universal proteins of unknown function (12). In particular, they noticed that proteins HypF and NodU/CmcH, which catalyse chemical reactions similar to the ones expected in t6A biosynthesis, are formed by the fusion of domains homologous to YrdC and YgjD. Using genetic experiments, they demonstrated that YrdC/Sua5 and YgjD/Kae1 are responsible for the formation of t6A in vivo in E. coli and Saccharomyces cerevisiae (13,14).
Earlier this year, Deutsch et al. (15) successfully reconstituted the t6A synthesis reaction in vitro by combining YrdC, YgjD and two additional E. coli proteins, YeaZ and YjeE, in the presence of ATP, Mg2+, threonine and carbonate. Subsequently, the reconstitution of a Bacillus subtilis system using purified YwlC (YrdC), YdiB (YjeE), YdiC (YeaZ) and YdiE (YgjD) was reported (16). Notably, YeaZ and YjeE have no detectable orthologues in Archaea and Eukarya. YeaZ is a paralogue of YgjD (thus homologous but distantly related to Kae1), whereas YjeE is specific to Bacteria. This suggested either that Sua5 and Kae1 are sufficient to perform the reaction in these two domains of life or that the YeaZ and YjeE functions are performed by functional analogues evolutionarily unrelated to their bacterial counterparts. Obvious candidate proteins to fulfil this role are the partner proteins of Kae1 (the YgjD homologue) in the KEOPS (kinase, endopeptidase, and other proteins of small size) or EKC (endopeptidase-like and kinase associated with transcribed chromatin) complex. In addition to Kae1, the KEOPS/EKC complex (thereafter called KEOPS) contains three proteins conserved between Archaea and Eukarya, the protein kinase Bud32, Cgi121 and Pcc1 (17,18). Genetic and transcriptomic studies in S. cerevisiae demonstrated that, in addition to Kae1, Bud32 and Pcc1 are required for the formation of t6A modification (19,20). Recent genetic investigation of the KEOPS complex in halophilic archaeon Haloferax volcanii showed that Kae1, Bud32 and Cgi121 are essential, whereas Pcc1 knockout strain presented a decreased growth rate and reduced levels of t6A modification (21). Furthermore, using a whole cell yeast extract for in vitro t6A synthesis, Srinivasan et al. (20) showed that, as in the case of the bacterial system, the reaction requires Sua5, Kae1, threonine, bicarbonate and ATP.
We have previously characterized the archaeal homologues of Kae1 and Bud32, with the aim to get more insight into functions that were probably already present in the last universal common ancestor (LUCA; Kae1/YgjD) or in the last common ancestor of Archaea and Eukarya (Bud32). We have shown that Kae1 is not an O-sialoglycoprotein endopeptidase, as initially reported (22), but rather a metalloprotein that binds ATP and DNA in vitro, suggesting a role in some unknown universal function related to nucleic acid. In the course of our work, we have purified the archaeal KEOPS complex from the hyperthermophilic archaeon Pyrococcus abyssi (PaKEOPS, this work) and the S. cerevisiae KEOPS complex (ScKEOPS) (23) for structural and functional studies. Here, we report for the first time the in vitro formation of t6A37-modified tRNA catalysed by the archaeal and yeast complexes. We present in more detail the enzymatic properties of the more active PaKEOPS complex. The efficiency of the reconstituted in vitro system indicates that KEOPS and Sua5 are both necessary and sufficient for the reaction. Notably, PaKEOPS, but not Sua5, strongly binds tRNA in vitro, suggesting a role for KEOPS as a tRNA-binding platform in the t6A modification reaction. Finally, we identified two distinct ATP-dependent catalytic steps associated with Sua5 and KEOPS. Our data, together with results obtained with the bacterial system (15), show that the universal proteins Sua5/YrdC and Kae1/YgjD cannot synthesize t6A by themselves but require additional non-universal accessory proteins. Using interdomain complementation experiments, we demonstrated that the KEOPS complex and bacterial proteins YeaZ, YgjD and YjeE are functional analogues, suggesting mechanistic similarity of t6A synthesis in the three domains of life. We discuss evolutionary implications raised by these observations and alternative models they suggested on the nature of LUCA.
MATERIALS AND METHODS
Cloning procedures
A polycistronic sequence containing the genes encoding the PaKEOPS subunits PAB1159 (Kae1), PAB1047 (Bud32), PAB1522 (Cgi121) and PAB3073 (Pcc1) was synthesized by GenScript and cloned into pET28(a) plasmid (Novagen). PAB3073 (Pcc1) was fused to a hexa-histidine tag at its 3′-end. The gene PAB1159 (Kae1) was cloned into the pET28(a) as previously described (24).
The gene PAB1302 encoding PaSua5 was amplified by polymerase chain reaction (PCR) using total genomic DNA from the P. abyssi GE5 strain as template and cloned into pET26b (Novagen). The ScSua5 gene was amplified by PCR using genomic DNA from S. cerevisiae S288C strain (ATCC). A hexa-histidine tag was added at the 3′-end of the gene, which was subsequently cloned into pET21d vector (Novagen).
E. coli genes ygjD and yeaZ were cloned in pET9a with a histidine tag at their 3′-end; yrdC was cloned in pET26b with a histidine tag at its 3′-end. yjeE gene was synthesized (GenScript) and cloned in pET28a with a Strep-tag II on its 3′-end.
Media were supplemented with 50 µg/ml kanamycin, 34 µg/ml chloramphenicol or 100 µg/ml ampicillin when needed.
Recombinant protein expression and purification
Recombinant proteins were expressed in Rosetta2 (DE3) pLysS E. coli strains (Novagen). Protein overexpression was induced in Overnight Express Instant TB medium (Novagen) supplemented with 10% glycerol.
Cells were collected by centrifugation, resuspended in lysis buffer (LBf) and sonicated on ice. After centrifugation and in the case of archaeal protein purification, the supernatant was heated at 65°C for 15 min to precipitate bacterial proteins. His-tagged proteins from the soluble fraction were purified by gravity-flow chromatography on an Ni-NTA resin column (Qiagen) according to the manufacturer’s recommendations. Proteins carrying a strep-tag II were purified on a Strep-tactin Sepharose resin (IBA) following manufacturer advice. Fractions of interest were pooled, concentrated and injected on a Superdex™200 column (GE Healthcare) for KEOPS or Superdex™75 column for Sua5 and bacterial proteins. Fractions containing pure proteins were concentrated and stored at −20°C in LBf without β-mercaptoethanol and 10% glycerol.
tRNA expression plasmid construction
P. abyssi (Pa) and E. coli (Ec) tRNA gene sequences PaVal (GAC), PaLys (UUU), PaIle (GAU), PaAsn (GUU), PaMet (CAU), EcVal (GAC), EcLys (UUU), EcIle (GAU) and EcAsn (GUU) were retrieved from the Genomic tRNA Database (http://gtrnadb.ucsc.edu/) (25). Two complementary oligonucleotides corresponding to the forward and reverse sequence of tRNA genes were synthesized (IDT), annealed and cloned in a pBlue-Script vector yielding pB-tRNA, to allow their expression under the control of T5 promoter/lac operator sequence. All sequences are available on request.
tRNA production and purification
XL1 Blue E. coli cells (Agilent Technologies) were transformed with pB-tRNA, and tRNAs were overexpressed in Overnight Express Instant TB medium (Novagen). After centrifugation, total RNAs were extracted with Trizol (Sigma) using the manufacturer’s procedure.
Run-off transcription of tRNA genes was performed according to Mougin et al. (26): Pa_tRNALys and Ec_tRNAAsn genes were cloned in pB-tRNA and amplified by PCR using primers containing the sequence of the T7 promoter upstream of the gene to be transcribed. The transcription reaction was carried out in transcription buffer supplemented with 200 nmol of each rNTP, 40 units of RNasine (Thermo Scientific), 300 ng of PCR product and 0.2 µM of his-T7 RNA polymerase expressed and purified according to Arnaud et al. (27). Reaction occurred at 37°C for 4 h. Template DNA was then digested with DNase I (Thermo Scientific) for 30 min at 37°C.
For purification, RNAs were loaded on an 8 M urea, 12% polyacrylamide gel, and the dominant band corresponding to overexpressed tRNA was cut out. tRNA was eluted from gel in elution buffer, precipitated and resuspended in water. To favour correct folding of tRNAs, the solution was heated at 70°C, cooled slowly to room temperature and stored at −80°C.
In vitro assay for the synthesis of t6A-modified tRNA
The reaction was performed in a final volume of 50 µl containing archaeal (1 µM or 5 µM for complementation experiments), yeast (5 µM) or bacterial (5 µM) proteins, tRNA (2 µM) and 18.2 µM 14C-l-threonine (0.05 µCi, 55 Ci/mol, Hartmann Analytic) in reaction buffer. After 30-min incubation at 50°C (or other incubation times when indicated), macromolecules were precipitated by the addition of 1 ml of trichloroacetic acid 15% (TCA), and incubated at 4°C for 1 h. Precipitated material was applied on pre-wet glass microfibre filters GF/F (Whatmann) using vacuum apparatus (Millipore). Filters were washed with 2 ml of 5% TCA and 3 ml of 95% EtOH and dried. Radioactivity was recorded as average counts per minute (CPM) for 2 min, with a Packard liquid scintillation analyser.
The amount of incorporated threonine was calculated from a standard curve obtained with different concentrations of 14C-l-threonine spotted on filters (1 pmol = 102.8 CPM).
Yield was calculated as [CPM (assay) − CPM (background)/102.8]/(pmol tRNA) × 100.
2D thin layer chromatography for detection of t6A-modified nucleosides
tRNAs obtained after in vitro t6A reaction were extracted with phenol/chloroform and ethanol precipitated. The tRNA pellet was dissolved in 10 µl of 100 mM ammonium acetate supplemented with 1 µg of P1 nuclease from Penicillium citrinum (Sigma) and incubated overnight at 37°C. Digested tRNAs (2 µl) were mixed with 20 µg of cold 5′P-mononucleosides pA, pU, pG and pC and spotted on a CEL-300 cellulose plate (Merck). 5′P-mononucleosides were separated using chromatographic solvents A, B or C as previously described (28). First dimension chromatography was performed in solvent A, and the second in solvent B or C. The positions of the four major mononucleosides, pA, pG, pU and pC, were revealed by UV shadowing. The position of t6A nucleoside carrying radiolabelled threonine was revealed by phosphorimaging and compared with that of reference maps obtained under identical experimental conditions.
ATP hydrolysis assay
ATP hydrolysis reactions were carried out in a final volume of 25 µl containing archaeal or yeast proteins (2 µM, if not otherwise specified) in reaction buffer, supplemented with 100 µM of cold ATP and 1 µCi of α-32P-ATP (PerkinElmer). When indicated, tRNA, single- or double-strand random sequence RNA (1 µM) and other amino acids (50 µM) were added to the reaction.
Reaction was performed at 50°C during 30 min, stopped on ice, and 1 µl of the mix was spotted on a PEI cellulose plate (Merck), pre-run in distilled water. Radioactive nucleotides were separated by thin layer chromatography (TLC) using 0.5 M KH2PO3-, pH 3.5, as the solvent. Plates were dried and revealed by phosphorimaging.
Electrophoretic mobility shift assay
The binding reactions were carried out in a final volume of 20 µl containing 0.05 pmol (10 nM) of RNA probes and purified PaKEOPS or PaSua5 (0.5 nM to 50 µM) in binding buffer. Incubations were performed for 15 min at 4°C, the products were loaded onto a 10% non-denaturing acrylamide gel and electrophoresis was performed in 1 × TGE buffer at 4°C for 4 h at 7.5 V/cm. The RNA–protein complexes were visualized by autoradiography and phosphorimaging.
RESULTS
P. abyssi KEOPS binds tightly to tRNAs
The whole PaKEOPS complex (Kae1, Bud32, Cgi121 and Pcc1) was purified from an E. coli overproducing strain by exploiting the polyhistidine tag of the Pcc1 subunit. The complex eluted as a single peak with an apparent molecular mass of ∼200 kDa after gel filtration (Supplementary Figure S1A). The theoretical mass of PaKEOPS was 90.5 kDa; therefore, the apparent molecular mass of this peak could correspond to a dimer, in agreement with the 3D structural model proposed previously (29). Interestingly, PaKEOPS co-purified with nucleic acids that were resistant to DNaseI digestion but sensitive to RNaseA (not shown) and displayed a similar electrophoretic migration pattern as tRNAs isolated from E. coli expression cells (Supplementary Figure S1B). These observations suggested that purified recombinant PaKEOPS was bound to E. coli tRNAs. In contrast, recombinant PaSua5 did not co-purify with nucleic acids.
Electrophoretic mobility shift assays of PaSua5 and PaKEOPS in the presence of substrate, non-substrate tRNA and random sequence 65mer RNA showed binding of all RNAs only by PaKEOPS, whereas PaSua5 did not bind tRNA even at a high protein to tRNA molar ratio (Supplementary Figure S2). This suggested that the discrimination between substrate and non-substrate tRNAs occurs after the binding of tRNA to the complex and that specific tRNA-like structural and sequence features are not required for the binding of RNA by KEOPS. Taken together, these observations suggested that PaKEOPS might be mainly responsible for the binding of tRNA substrates during t6A synthesis.
PaSua5 and PaKEOPS together catalyse the biosynthesis of t6A
To determine whether PaSua5 and PaKEOPS were able to catalyse the biosynthesis of t6A, we needed an appropriate tRNA substrate lacking this modification. We initially anticipated that additional modifications might also be important for the formation of t6A because a previous in vivo study using Xenopus oocytes showed that t6A and several nucleotide modifications (D47, m5C48, m1A58) were formed almost simultaneously (30). To obtain tRNA substrates that most closely satisfied these criteria, we designed a recombinant strategy for tRNA production in vivo using E. coli cells that overexpressed various P. abyssi (Pa_tRNA) or E. coli (Ec_tRNA) tRNA genes under the control of an inducible promoter. Under these conditions, it is known that the resulting overexpressed tRNA is often hypomodified, thus generating a collection of so-called modivariants (RNA molecules that differ only by their modified nucleoside content) (31,32). Using this system, we typically obtained a major band on denaturing acrylamide gel corresponding to overexpressed, possibly undermodified tRNAs, species (Supplementary Figure S3A).
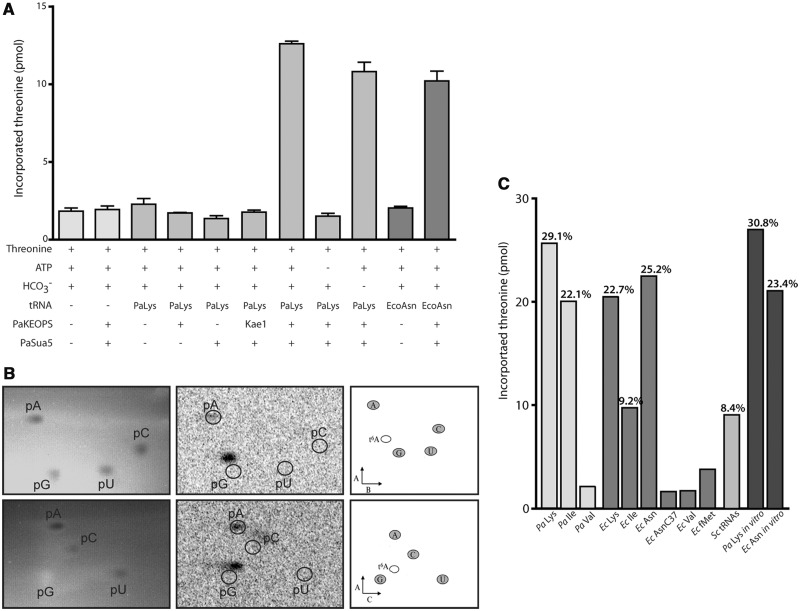
We incubated PaSua5 and PaKEOPS in presence of ATP, Pa_tRNALys, carbonate and 14C-threonine, and measured the incorporation of 14C-threonine into TCA-precipitated material. We observed a significant increase of radioactivity when both PaSua5 and PaKEOPS were present in the reaction mixture (Figure 2A). In contrast, no incorporation was observed with PaSua5 or PaKEOPS alone, indicating that both are necessary for the reaction to take place. Notably, PaSua5 and PaKae1 together do not catalyse the incorporation of threonine, indicating that PaKae1 cannot substitute for PaKEOPS. Threonine incorporation also required the addition of ATP and tRNA but, surprisingly, not carbonate. We hypothesized that carbonate was still present in the reaction mixtures despite boiling and extensive degassing of reaction solutions. For comparison, we performed the complete t6A assay using E. coli proteins (as explained later). As shown in Supplementary Figure S7C, added carbonate had a stimulating effect on the E. coli system, indicating that at least some of atmospheric carbonate was removed from reaction solutions. Thus, it appears that archaeal and also eukaryal systems (as explained later) function at lower carbonate concentrations as compared with bacterial systems.
Figure 2.
In vitro reconstitution of t6A-modified tRNAs. (A) Incorporation of radiolabelled threonine in tRNAs: reaction mixtures composed of the reactants indicated under the graph were precipitated with TCA, and the amount of incorporated 14C-threonine was measured. The tRNA species from P. abyssi (Pa) and E. coli (Ec) (purified from in vivo overexpression) are indicated. In one case, Kae1 was used instead of the whole KEOPS complex.  was added (+) or not (−) to the reaction. Error bars indicate standard deviation from the mean obtained for three independent experiments. (B) Detection of t6A-modified nucleotide using 2D TLC. Upper line shows chromatographic mobility in solvent system A + B, lower line in solvent system A + C. Left panels: abundant, non-modified nucleotides were detected by ultraviolet (UV) shadowing. Centre panel: 14C-threonine incorporated in modified nucleotides was detected by phosphorimaging, and the positions of unmodified nucleotides were superimposed by aligning with the UV image (white circles). Right panel: reference maps [from (28)] showing the theoretical position of t6A nucleoside in both solvent systems. (C) Incorporation of radiolabelled threonine using different tRNA species as substrates. Legend is the same as in (A); tRNAs were overexpressed in vivo, or produced in vitro when stated. fMet, bacterial initiator N-formyl-methionine; AsnC37 is Asn tRNA where A37 was mutated into a cytosine; Sc tRNAs are tRNAs purified from a Δsua5 S. cerevisiae strain. The percentage indicated on top of each bar is the yield of t6A-modified tRNA obtained for each reaction.
was added (+) or not (−) to the reaction. Error bars indicate standard deviation from the mean obtained for three independent experiments. (B) Detection of t6A-modified nucleotide using 2D TLC. Upper line shows chromatographic mobility in solvent system A + B, lower line in solvent system A + C. Left panels: abundant, non-modified nucleotides were detected by ultraviolet (UV) shadowing. Centre panel: 14C-threonine incorporated in modified nucleotides was detected by phosphorimaging, and the positions of unmodified nucleotides were superimposed by aligning with the UV image (white circles). Right panel: reference maps [from (28)] showing the theoretical position of t6A nucleoside in both solvent systems. (C) Incorporation of radiolabelled threonine using different tRNA species as substrates. Legend is the same as in (A); tRNAs were overexpressed in vivo, or produced in vitro when stated. fMet, bacterial initiator N-formyl-methionine; AsnC37 is Asn tRNA where A37 was mutated into a cytosine; Sc tRNAs are tRNAs purified from a Δsua5 S. cerevisiae strain. The percentage indicated on top of each bar is the yield of t6A-modified tRNA obtained for each reaction.
The optimal temperature for threonine incorporation was 50°C with 65% of activity still retained at 73°C (Supplementary Figure S3C). Because the optimal growth temperature of P. abyssi GE5 is 96°C, it is likely that the reaction is limited by thermal denaturation of tRNA in non-physiological in vitro conditions.
Importantly, the threonine incorporation test was negative for non-ANN decoding tRNAVal from E. coli and P. abyssi, thus confirming that only ANN-decoding tRNA molecules are valid substrates. Moreover, Ec_tRNAAsn failed to act as a substrate when adenosine at position 37 was replaced by cytosine, showing that t6A is specifically added on the adenosine 37.
The incorporation of threonine into tRNAs was confirmed by purification of tRNAs after t6A reaction. As expected, only tRNAs from the reaction containing PaSua5 and PaKEOPS were radioactive (Supplementary Figure S3B).
Finally, we performed 2D TLC to demonstrate that the incorporation of threonine into tRNA corresponded to t6A formation. For this, tRNAs were isolated from the reaction mixture, hydrolysed to individual nucleotides and separated by 2D TLC. The position of the radioactive spot on the plate was reproducible and consistent with the position of the t6A-modified nucleotide according to previously published reference maps (33) (Figure 2B).
Next, it was important to verify that the in vitro assay we established can generate a significant amount of t6A-modified tRNA. Standard assay conditions yielded 25–32% of modified Pa_tRNALys (i.e. mole t6A37 per mole of tRNA present in the incubation mixture). This result indicated that the in vitro reaction could still be optimized or that the enzymatic machinery may work better in vivo under the physiological conditions of the cell. To optimize the reaction, we added manganese divalent ions, which were reported to be required for optimal activity of Bud32 (34). Indeed, manganese had a stimulating effect because we could increase the yield of the reaction to 50% for archaeal system and to 38% for the S. cerevisiae system (as explained later) (Supplementary Figure S5).
However, the yield reported is the lowest estimate for our assay conditions because, as noted previously, it is possible that a subpopulation of the in vivo produced Pa_tRNAsLys already carried the t6A modification. The yield could not be improved by adding a fresh aliquot of PaSua5 and PaKEOPS or ATP into the reaction mixture (not shown), which indicated that the amount of substrate tRNA was the limiting factor.
To further characterize the range of substrate tRNAs used by PaKEOPS and PaSua5, we also tested overexpressed E. coli tRNAs, total tRNAs isolated from a S. cerevisiae Sua5 deletion mutant (19) and in vitro transcribed tRNAs. All of these acted as equivalent substrates (Figure 2C), demonstrating that (i) universally conserved structural and/or sequence features of tRNAs decoding ANN codons are recognized by PaKEOPS and PaSua5; and (ii) presence of modified nucleotides on ANN-decoding tRNA is not required for the t6A37 modification to take place.
Two distinct ATP-dependent reactions occur in the course of t6A synthesis by PaSua5 and PaKEOPS
To investigate the ATP-dependent activation steps undertaken by PaKEOPS and PaSua5, we assayed the ATPase activity using α-32P-ATP and TLC to visualize the resulting products (Figure 3A).
Figure 3.
ATPase activity of Sua5 and KEOPS. (A) P. abyssi proteins and (B) S. cerevisiae proteins. Produced radiolabelled (α-32P) ADP and AMP were separated by TLC and visualized by phosphorimaging. Presence (+) or absence (−) of different reactants in reaction mixtures is indicated. Pa_tRNALys produced in overexpressing E. coli strain was used as a substrate.
When all the proteins and substrates were put together in a reaction that leads to t6A synthesis and under saturating concentration of ATP, we observed that both AMP and ADP were produced. The production of AMP occurred when PaSua5 was present in the reaction mixture alone. This activity was significantly increased when threonine and carbonate were added but did not require tRNA (Figure 3A and Supplementary Figure S7A and S7B). In contrast, PaSua5 activity was not stimulated by glycine, phenylalanine and homoserine, an isomer of threonine (Supplementary Figure S4B).
In contrast to PaSua5, incubation of PaKEOPS alone with ATP generated ADP (Figure 3A). This basal activity is detected in the absence of tRNA; however, the addition of substrate tRNA or non-substrate Ec_tRNAAsn (C37) and Pa_tRNAVal significantly increased this activity (Figure 3A and Supplementary Figure S4A). Notably, such activation was not observed when tRNA was replaced by random sequence 65mer DNA or RNA, although this RNA was bound by KEOPS with an affinity similar to that observed for tRNA (see Supplementary Figures S2C and S4A).
The KEOPS-dependent ATP hydrolysis into ADP seems to be required for t6A synthesis because the t6A reaction was completely inhibited when ATP was replaced by AMP-PNP, an ATP analogue that can be hydrolysed into AMP (data not shown).
Taken together, our data indicate that two distinct ATP-dependent reactions, one generating AMP and another generating ADP, are catalysed by PaSua5 and PaKEOPS, respectively, in the course of t6A synthesis.
In vitro synthesis of t6A by Sua5 and KEOPS from S. cerevisiae
The S. cerevisiae KEOPS (ScKEOPS) complex was heterologously expressed and purified as previously described (23). In contrast to PaKEOPS, ScKEOPS was not associated with tRNAs when purified from E. coli cells (not shown). Consistent with observations made for PaSua5, the purified recombinant yeast homologue ScSua5 did not co-purify with tRNA either.
To test whether ScSua5 and ScKEOPS could also catalyse the biosynthesis of the t6A modification, we used the same experimental approach as for P. abyssi proteins except that the assay was performed at 30°C. As for P. abyssi proteins, we observed the incorporation of 14C-threonine into TCA-precipitated material, and 2D TLC analysis confirmed that t6A was formed (Figure 4B). This reaction was ATP and tRNA dependent and required the presence of ScSua5 and ScKEOPS. Similar as for the P. abyssi system, the omission of carbonate did not have an effect on the amount of incorporated threonine (Supplementary Figure S7C). The amount of threonine incorporation was similar for endogenous tRNAs and E. coli tRNAs, indicating that, like P. abyssi proteins, the S. cerevisiae proteins recognize features common to all ANN-decoding tRNA molecules.
Figure 4.
In vitro synthesis of t6A modification using S. cerevisiae KEOPS and Sua5 proteins. (A) Incorporation of radiolabelled threonine in tRNAs: the amount of incorporated 14C-threonine was measured as in Figure 2A. Total tRNAs were purified from a ΔSua5 strain; for comparison, in vitro transcribed E. coli tRNALys was used when indicated. Error bars indicate standard deviation from the mean obtained from three independent experiments. (B) Detection of t6A-modified nucleotide using 2D TLC. The chromatography was done as described in Figure 2B.
We next examined the ATPase activity of ScSua5 and ScKEOPS using TLC (Figure 3B). We observed weak ATPase activity of ScSua5 of which the product was AMP. This activity was threonine dependent and carbonate dependent (Supplementary Figure S7A and S7B) but did not require tRNA, and addition of tRNA did not have a stimulating effect. Thus, the ScSua5 behaved in the same manner as the PaSua5. In contrast, the behaviour of ScKEOPS was different from that of PaKEOPS because we observed the production of ADP and AMP in a tRNA-independent manner. Note, however, that we cannot exclude a contaminating ATPase activity from the E. coli expression strain.
Interdomain exchange of t6A machinery components
The similar behaviour of archaeal, yeast and bacterial t6A machineries suggested that the function of KEOPS/YgjD-YjeE-YeaZ (hereafter called DEZ) and Sua5/YrdC has been conserved throughout evolution. To test this hypothesis, we purified bacterial proteins and set up a functional in vitro system to perform complementation assays. We combined Sua5/YrdC proteins and KEOPS/DEZ complexes. TCL analysis confirmed that the mixed systems produce t6A-modified tRNA (Supplementary Figure S6). Remarkably, despite the non-optimal temperatures used in the assays, the amount of t6A produced by heterologous combinations was comparable with the amount produced by the homologous systems at the respective reaction temperatures (43°C for Pa and Ec proteins, and 35°C for Sc proteins) (Figure 5). Lower values were obtained in complementation with PaKEOPS, owing to suboptimal temperatures because at 43°C and 35°C, PaKEOPS retains 40% and 25% of its activity (Supplementary Figure S3C). Nevertheless, those values were reproducible and are still fairly above the background signal. These results indicate that Sua5/YrdC and KEOPS/DEZ components can be exchanged, suggesting that t6A-synthesizing machineries from the three domains of life share a common catalytic mechanism.
Figure 5.
Functional complementations assays between bacterial, archaeal and eukaryotic proteins. Complementations were tested using PaKEOPS, ScKEOPS or EcDEZ combined with PaSua5, ScSua5 or EcYgjD. Incorporation of radiolabelled threonine was measured as in Figure 2A. Activities of archaeal (A), bacterial (B) and eukaryotic (E) complexes were measured at 43°C (A, B and A/B) and 35°C (E, B/E and A/E). The dotted line indicates the background level, obtained without ATP.
DISCUSSION
Archaeal and eukaryal Sua5 and KEOPS together catalyse the synthesis of t6A-modified tRNA
In the present study, we established an in vitro system for enzymatic synthesis of a universal and essential t6A tRNA modification by archaeal and eukaryotic KEOPS and Sua5 proteins. This work thus provides for the first time direct evidence for the biological function of KEOPS and Sua5 in archaeal and eukaryotic cells and confirms the results of previous genetic and transcriptome studies in S. cerevisiae and H. volcanii (13,14,19,20,21). In the light of these findings, it is possible that the pleiotropic phenotypes described for yeast KEOPS and Sua5 deletion mutants (17,18,35) are indirect effects owing to the translational defects occurring in cells lacking t6A modification. This is well illustrated by a recent study that showed that the diverse cellular dysfunctions linked to the development of type 2 diabetes in mice can be attributed to abnormal protein synthesis caused by the lack of 2-methylthio-N6-threonylcarbamoyladenosine (ms2t6A)-modified tRNALys (36).
Purified proteins and a reconstituted in vitro system allowed us to get the first insights onto the structural and mechanistic features of the t6A modification process. We made two important observations: firstly, PaKEOPS, but not PaSua5, binds tRNA tightly; and secondly, the reaction requires the hydrolysis of ATP into AMP by Sua5 and the hydrolysis of ATP into ADP by KEOPS.
The KEOPS complex is a tRNA binding platform
PaKEOPS binds tRNA of E. coli in vivo and in vitro, and this binding is strong because it is preserved throughout the purification process. It therefore seems plausible, suggesting that the KEOPS complex is responsible for tRNA binding in the course of the t6A synthesis. Interestingly, PaKEOPS binds tRNA in a positive cooperative manner (Supplementary Figure S2). It is unlikely that PaKEOPS binds more than one molecule of tRNA simultaneously because of the large size of tRNA. An alternative explanation is that one tRNA molecule is bound by each subunit of a PaKEOPS homodimer. We have previously shown that Kae1 from P. abyssi binds DNA in vitro (24). This suggested that Kae1 could have a general affinity for nucleic acids. Indeed, we could show using electrophoretic mobility shift assays that purified P. abyssi Kae1 can bind ssRNA and tRNA, albeit with lower affinity as compared with KEOPS (unpublished data). This suggests that tRNA interacts with Kae1 in the KEOPS/tRNA complex. However, because purified Kae1 is not bound to E. coli tRNA, it is likely that other proteins of the complex are also involved in tRNA binding. Interestingly, the archaeal/eukaryal KEOPS and bacterial t6A machinery can use as substrate tRNA from the three domains of life. This again suggests that Kae1 is directly involved in tRNA binding because it is the only KEOPS protein with a bacterial homologue (YgjD). This also indicates that universal structural and/or sequence features of tRNA are recognized by all systems, suggesting a similar mode of interaction between tRNA and the proteins.
The catalytic mechanism for t6A synthesis seems to be conserved in the three domains of life
It was suggested that t6A synthesis, which is a complex modification, required at least two ATP-dependent activation steps (37). In agreement with this proposal, Deutsch et al. reported that both AMP and ADP were produced when all four proteins of the bacterial system were mixed (15). They observed that YrdC alone hydrolysed ATP into AMP in a threonine-dependent reaction, and that YgjD, YjeE and YeaZ (DEZ) generated ADP. These results were confirmed by a recent study that reconstituted in vitro the t6A biosynthesis by B. subtilis proteins YwlC (YrdC), YdiB (YjeE), YdiC (YeaZ) and YdiE (YgjD) (16). One common point between the bacterial system and our archaeal/eukaryal systems is the production of both AMP and ADP in the reaction that generates t6A-modified tRNA. The conservation of this feature across the three domains of life corroborates the hypothesis of at least two distinct ATP-dependent steps in the course of t6A synthesis, suggesting a conserved catalytic mechanism for the biosynthesis of t6A in nature, despite the fact that some of the proteins are unrelated. The second common point of all three systems is the carbonate and threonine dependence of AMP generation by YrdC/Sua5. This observation is in line with the universal character of these proteins and indicates that they perform the same catalytic role in the three domains of life.
The three systems diverge significantly in terms of complexes involved in the ADP-generating reaction step. In Bacteria, this reaction is catalysed by DEZ, whereas it is catalysed by KEOPS in Archaea and Eukarya. Kae1/YgjD is the only common protein between these different systems, suggesting that it might be responsible for this activity.
Finally, using in vitro complementation experiments, we have demonstrated that Sua5 can replace YrdC and vice versa and that KEOPS is interchangeable with DEZ. These data corroborate the notion of a common catalytic mechanism for t6A synthesis catalysed by universal proteins Sua5/YrdC and Kae1/YgjD.
A putative mechanism for t6A synthesis
The threonine-dependent hydrolysis of ATP into AMP by Sua5 and YrdC suggests the formation of an adenylate reaction intermediate, such as threonylcarbamoyladenylate. The fact that we do not directly observe threonylcarbamoyladenylate is probably because of its unstable nature under acidic conditions used for TLC analysis. This intermediate was isolated using HPLC under neutral pH conditions with B. subtilis YwlC (YrdC), which catalysed the conversion of l-threonine, bicarbonate/CO2 and ATP to give l-threonylcarbamoyl-AMP intermediate and pyrophosphate as products (16). Moreover, l-threonylcarbamoyl-AMP was found in the 3D crystal structure of Sua5 from Sulfolobus tokodaii, and a recent study of TobZ protein also suggested a putative model for t6A synthesis involving a threonylcarbamoyladenylate intermediate (38,39). This protein, which synthesizes the antibiotic nebramycin, consists of two domains homologous to YrdC and Kae1 fused into a single polypeptide. In the proposed model for the reaction catalysed by TobZ, the YrdC-like domain catalyses the ATP-dependent formation of the carbamoyladenylate intermediate, followed by the transfer of the carbamoyl moiety to tobramycin to form nebramycin with concomitant release of AMP. This final step is performed by the Kae1-like domain of TobZ, which positions the adenylate intermediate and the sugar substrate in the correct orientation. By analogy, Parthier et al. (39) suggested that Sua5 catalyses the formation of threonylcarbamoyladenylate, which is then accommodated in the active site of Kae1, together with the anticodon loop of tRNA, to form t6A-modified tRNA. Our results are compatible with this model. However, Parthier et al. further suggested that N-carbamoylthreonine, the precursor for the synthesis of the threonylcarbamoyladenylate intermediate, is generated from bicarbonate and threonine by an unknown enzymatic activity. Our data prove that it is not the case because Sua5 and KEOPS are sufficient to synthesize t6A. Moreover, the observation that ATPase activity of PaSua5 [and of YrdC and YwlC in bacterial systems (15,16)] is carbonate and threonine dependent strongly suggests the involvement of Sua5/YrdC in the early steps of the catalysis, leading to the formation of threonylcarbamoyladenylate. In addition, our data imply that KEOPS is involved in the last step of the catalysis, which requires the presence of tRNA. This is supported by the finding that the reaction of tRNA with l-threonylcarbamoyl-AMP can be catalysed by the B. subtilis YdiB (YjeE), YdiC (YeaZ) and YdiE (YgjD) complex in the absence of YwlC (YrdC) (16). Based on these data and previous studies, we propose a novel mechanism for the synthesis of t6A (Figure 6). In the first step, bicarbonate is activated by Sua5/YrdC to carboxy-AMP [1], which can then condense with the threonine to form carbamoylthreonine [2]. Sua5/YrdC uses a second ATP to activate this product and generates threonylcarbamoyladenylate [3]. Finally, this intermediate is transferred to Kae1/YgjD active site where second condensation reaction occurs to form t6A-modified tRNA. This proposal is compatible with the data reported by Lauhon (16), but it is in disagreement with the work of El Yacoubi et al. (14), who proposed the activation of bicarbonate to carboxyphosphate catalysed by Kae1. We do observe generation of ADP by KEOPS, and this activity is mandatory. However, KEOPS activity is sensible to neither carbonate nor threonine, suggesting strongly that the complex, and thus Kae1, is not involved in early steps of catalysis. In favour of our proposal, YrdC-homologous domain of TobZ protein was shown to catalyse formation of carbamoyl-AMP, a compound that is structurally reminiscent of carboxy-AMP. Our model is compatible with the data obtained for the bacterial t6A systems, and carboxy-AMP was included in the proposed mechanism scheme as an alternative to the formation of carboxyphosphate (15). The present data do not allow assigning convincingly the ADP-generating ATPase activity of KEOPS/DEZ to a particular process. However, in archaeal system, we observed that the active site responsible for ATP hydrolysis is not in a catalytically competent conformation until tRNA is bound, indicating that a set of mutually induced conformational changes occur between PaKEOPS and tRNA.
Figure 6.
Proposed reaction scheme for t6A synthesis. Different catalytic steps are catalysed by proteins indicated above arrows. Activation of bicarbonate to carboxy-AMP [1]; condensation of carboxy-AMP with threonine to form carbamoylthreonine [2]; activation of carbamoylthreonine to threonylcarbamoyladenylate [3]; transfer of threonylcarbamoyladenylate to Kae1/YgjD active site where second condensation reaction occurs to form t6A-modified tRNA [4]. The participation of Sua5 in the last catalytic step is currently unclear. Further details are in the text.
Therefore, we propose that KEOPS and DEZ must undergo conformational changes to attain a catalytically relevant conformation and that ATP hydrolysis is needed to compensate for the free energy cost of rearrangements. Further experimental work is needed to verify this hypothesis.
Possible role of archaeal Sua5 and KEOPS in the synthesis of N6-hydroxynorvalylcarbamoyladenosine (hn6A)
Two other modified adenosines occur at position 37 and contain carbamoyl and amino acid moieties: N6-glycylcarbamoyladenosine (g6A) and N6-hydroxynorvalylcarbamoyladenosine (hn6A) (3,40), but the enzymes catalysing the synthesis of these modified nucleosides have not been identified yet. g6A is present in certain cytoplasmic tRNAs of eukaryotes, whereas hn6A is found in tRNAs of Bacteria and Archaea (41). In agreement with this phylogenetic distribution, we found that PaSua5 hydrolyses ATP to AMP in presence of hydroxynorvaline, but not in presence of glycine (Supplementary Figure S4B). This suggested that Sua5 proteins could be responsible for the synthesis of the hydroxynorvalylcarbamoyladenylate and that, together with KEOPS, they may be responsible for the formation of hn6A in Archaea. Further experimental work is needed to confirm this hypothesis, which if true, would extend the role of archaeal KEOPS and Sua5 proteins to the synthesis of another tRNA modification.
Evolutionary implications
The present work and the previous work on bacterial system (15) revealed that the universal proteins Sua5/YrdC and Kae1/YgjD alone are not sufficient to perform t6A modification but require two sets of non-universal proteins, one specific to Bacteria (YeaZ, YjeE) and the other common to Archaea and Eukarya (Bud32, Cgi121 and Pcc1). A similar situation is found in many other fundamental molecular systems, e.g. translation initiation and transcription, in which a core of universally conserved proteins is supplemented by factors specific for Bacteria on one side and for Archaea/Eukarya on the other. It is often argued that such a situation suggests a simple LUCA [a progenote sensu (42) and subsequent independent refinement of molecular processes (i.e. translation)] in Bacteria and in Archaea/Eukarya. In case of t6A, it seems unlikely that LUCA lacked this modification. Indeed, several studies identified t6A as a part of a minimal tRNA modifications set required for life (43–45). Moreover, the genes responsible for the synthesis of t6A are resistant to loss in insect symbionts and mollicutes that have reduced genomes, suggesting that these genes emerged early during cellular evolution, and once acquired became essential for the cell (45,46). Finally, it has been argued convincingly that t6A modification is necessary for the accurate reading of the three letters genetic code (7,8,19). Because this code is universal, it was likely present in LUCA, and its accuracy was probably already optimal (47).
If t6A was already present in LUCA, one should imagine evolutionary scenarios to explain how an ancestral tRNA modification mechanism evolved into two distinct modern versions. The idea that these two versions evolved from a common mechanism is supported by the existence of two universal proteins in both versions, by the universality of the two distinct ATP-dependent steps and by the interdomain functional complementations. One possible scenario is that the core proteins (Sua5/YrdC and Kae1/YgjD) are the remnants of an ancient synthetase operating in LUCA, which could have performed the complete reaction without accessory proteins. After the separation of the three domains, additional proteins were added independently in Bacteria and in ancestors of Archaea and Eukarya to improve speed and accuracy of protein translation in increasingly complex cells. In this scenario, we propose that these proteins were selected to provide additional surfaces for tRNA binding. This suggestion fits well with the observation that Kae1 alone binds tRNA less tightly as compared with KEOPS (as explained previously). Alternatively, LUCA might have possessed either DEZ or KEOPS to work with the ancestor of Sua5/YrdC; later on, YeaZ and YjeE were replaced by Bud32, Pcc1 and Cgi121 in Archaea and Eukarya or vice versa, and KEOPS was replaced by DEZ in Bacteria. Our complementation experiments render this hypothesis plausible because we show that both KEOPS and DEZ can complement in vitro the Sua5/YrdC protein of the three different domains, suggesting that such situation could have happened in vivo in ancestors of modern organisms.
The study of t6A synthesis in mitochondria could contribute to the comprehension of t6A machinery evolution. Eukaryotic genomes encode mitochondrial homologue of Kae1 called Qri7 in S. cerevisiae. Genetic data from two independent studies indicated that S. cerevisiae Qri7 functions in mitochondrial t6A synthesis. Interestingly, Sua5 is required for both cytoplasmic and mitochondrial t6A synthesis, indicating that it is imported into mitochondria despite the lack of a recognizable import signal (14,20). It remains to be investigated if Qri7 and Sua5 alone could catalyse the formation of t6A-modified tRNA or if other proteins are required. In the case of the second hypothesis, the proteins of KEOPS are the obvious candidates. The first hypothesis, if true, would substantially argue in favour of the scenario in which the two universal proteins formed the ancient t6A synthetic machinery.
In conclusion, this study established an in vitro system for the synthesis of essential t6A tRNA modification by eukaryotic and archaeal t6A synthetases. Characterization of these systems allowed us to propose a novel reaction scheme for the synthesis of t6A. Our data suggest that the catalytic mechanism for t6A synthesis is conserved in nature. Our work paves the way for further mechanistic and structural studies of these enzymatic complexes. The full comprehension of how this ancient modification is synthesized should result in better understanding of evolutionary mechanisms that contributed to increased translation efficiency and fidelity in present day cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–7, Supplementary Methods.
FUNDING
Agence Nationale de Recherche [ANR-09-BLAN-0349 to P.F. and H.vT.]; Marie Curie postdoctoral fellowship (to E.C.); ENS Lyon (PhD fellowship to L.P.). Funding for open access charge: Agence Nationale de Recherche [ANR-09-BLAN-0349].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are indebted to Henri Grosjean for his encouragement, stimulating discussions and critical reading of the manuscript. The authors thank Domenico Libri and Marie-Claire Daugeron for helpful discussions and for providing the Sua5 yeast deletion strain; Dorian Guetta, Philippe Bouloc, Chantal Bohn and Emmanuelle Schmitt for sharing protocols and for helpful suggestions. B.C. wishes to warmly thank Magali Blaud for her helpful advices and protocols and François Mallet for his generous gift of pMRT-7 wt.
REFERENCES
- 1.Grosjean H, Sprinzl M, Steinberg S. Posttranscriptionally modified nucleosides in transfer RNA: their locations and frequencies. Biochimie. 1995;77:139–141. doi: 10.1016/0300-9084(96)88117-x. [DOI] [PubMed] [Google Scholar]
- 2.Gustilo EM, Vendeix FA, Agris PF. tRNA's modifications bring order to gene expression. Curr. Opin. Microbiol. 2008;11:134–140. doi: 10.1016/j.mib.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grosjean H. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin, Texas, USA: Landes Biosciences; Nucleic acids are not boring long polymers of only four types of nucleotides; pp. 1–18. [Google Scholar]
- 4.Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giegé R, Jühling F, Pütz J, Stadler P, Sauter C, Florentz C. Structure of transfer RNAs: similarity and variability. Wiley Interdiscipl. Rev.RNA. 2012;3:37–61. doi: 10.1002/wrna.103. [DOI] [PubMed] [Google Scholar]
- 6.Auffinger P, Westhof E. Location and distribution of modified nucleotides in tRNA. In: Grosjean H, Benne R, editors. Modification and editing of RNA. Washington DC, USA: ASM press; pp. 103–112. [Google Scholar]
- 7.Stuart JW, Gdaniec Z, Guenther R, Marszalek M, Sochacka E, Malkiewicz A, Agris PF. Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry. 2000;39:13396–13404. doi: 10.1021/bi0013039. [DOI] [PubMed] [Google Scholar]
- 8.Murphy FV, Ramakrishnan V, Malkiewicz A, Agris PF. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. StructMol. Biol. 2004;11:1186–1191. doi: 10.1038/nsmb861. [DOI] [PubMed] [Google Scholar]
- 9.Chheda GB, Hall RH, Mozejko J, Magrath DI, Schweizer MP, Stasiuk L, Taylor PR. Aminoacyl nucleosides. VI. Isolation and preliminary characterization of threonyladenine derivatives from transfer ribonucleic acid. Biochemistry. 1969;8:3278–3282. doi: 10.1021/bi00836a022. [DOI] [PubMed] [Google Scholar]
- 10.Chheda G, Hong C, Piskorz C. Biosynthesis of N-(purin-6-ylcarbamoyl)-l-threonine riboside. Incorporation of l-threonine in vivo into modified nucleoside of transfer ribonucleic acid. Biochem. J. 1972;127:515–519. doi: 10.1042/bj1270515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Körner A, Söll D. N-(purin-6-ylcarbamoyl)threonine: biosynthesis in vitro in transfer RNA by an enzyme purified from Escherichia coli. FEBS Lett. 1974;39:301–306. doi: 10.1016/0014-5793(74)80135-3. [DOI] [PubMed] [Google Scholar]
- 12.Galperin MY. Conserved hypothetical proteins: prioritization of targets for experimental study. Nucleic Acids Res. 2004;32:5452–5463. doi: 10.1093/nar/gkh885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, Williamson JR, Schimmel P, Swairjo MA, de Crecy-Lagard V. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 2009;37:2894–2909. doi: 10.1093/nar/gkp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset J, Iwata-Reuyl D, Murzin AG, de Crecy-Lagard V. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J. 2011;30:882–893. doi: 10.1038/emboj.2010.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deutsch C, El Yacoubi B, de Crecy-Lagard V, Iwata-Reuyl D. Biosynthesis of Threonylcarbamoyl Adenosine (t6A), a Universal tRNA Nucleoside. J Biol. Chem. 2012;287:13666–13673. doi: 10.1074/jbc.M112.344028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauhon C. Mechanism of N6-threonylcarbamoyladenonsine (t6a) biosynthesis: isolation and characterization of the intermediate threonylcarbamoyl-AMP. Biochemistry. 2012;51:8950–8963. doi: 10.1021/bi301233d. [DOI] [PubMed] [Google Scholar]
- 17.Downey M, Houlsworth R, Maringele L, Rollie A, Brehme M, Galicia S, Guillard S, Partington M, Zubko MK, Krogan NJ, et al. A Genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell. 2006;124:1155–1168. doi: 10.1016/j.cell.2005.12.044. [DOI] [PubMed] [Google Scholar]
- 18.Kisseleva-Romanova E, Lopreiato R, Baudin-Baillieu A, Rousselle J, Ilan L, Hofmann K, Namane A, Mann C, Libri D. Yeast homolog of a cancer-testis antigen defines a new transcription complex. EMBO J. 2006;25:3576–3585. doi: 10.1038/sj.emboj.7601235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daugeron M, Lenstra TL, Frizzarin M, El Yacoubi B, Liu X, Baudin-Baillieu A, Lijnzaad P, Decourty L, Saveanu C, Jacquier A, et al. Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t(6)A modification of tRNAs. Nucleic Acids Res. 2011;39:1–13. doi: 10.1093/nar/gkr178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin VE, Karzai AW, Sternglanz R. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J. 2011;30:873–881. doi: 10.1038/emboj.2010.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naor A, Thiaville PC, Altman-Price N, Cohen-Or I, Allers T, de Crécy-Lagard V, Gophna U. A genetic investigation of the KEOPS complex in halophilic Archaea. PLoS One. 2012;7:e43013. doi: 10.1371/journal.pone.0043013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdullah KM, Lo RY, Mellors A. Cloning, nucleotide sequence, and expression of the Pasteurella haemolytica A1 glycoprotease gene. J. Bacteriol. 1991;173:5597–5603. doi: 10.1128/jb.173.18.5597-5603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collinet B, Friberg A, Brooks MA, Elzen T, Henriot V, Dziembowski A, Graille M, Dur D, Leulliot N, Saint André CS, et al. Strategies for the structural analysis of multi-protein complexes: lessons from the 3D-Repertoire project. J. Struct. Biol. 2011;175:147–158. doi: 10.1016/j.jsb.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 24.Hecker A, Leulliot N, Gadelle D, Graille M, Justome A, Dorlet P, Brochier C, Quevillon-Cheruel S, Le Le Cam E, Le Cam E, et al. An archaeal orthologue of the universal protein Kae1 is an iron metalloprotein which exhibits atypical DNA-binding properties and apurinic-endonuclease activity in vitro. Nucleic Acids Res. 2007;35:6042–6051. doi: 10.1093/nar/gkm554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mougin A, Grégoire A, Banroques J, Ségault V, Fournier R, Brulé F, Chevrier-Miller M, Branlant C. Secondary structure of the yeast Saccharomyces cerevisiae pre-U3A snoRNA and its implication for splicing efficiency. RNA. 1996;2:1079–1093. [PMC free article] [PubMed] [Google Scholar]
- 27.Arnaud N, Cheynet V, Oriol G, Mandrand B, Mallet F. Construction and expression of a modular gene encoding bacteriophage T7 RNA polymerase. Gene. 1997;199:149–156. doi: 10.1016/s0378-1119(97)00362-4. [DOI] [PubMed] [Google Scholar]
- 28.Grosjean H, Droogmans L, Roovers M, Keith G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 2007;425:55–101. doi: 10.1016/S0076-6879(07)25003-7. [DOI] [PubMed] [Google Scholar]
- 29.Mao DYL, Neculai D, Downey M, Orlicky S, Haffani YZ, Ceccarelli DF, Ho JSL, Szilard RK, Zhang W, Ho CS, et al. Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine. Mol. Cell. 2008;32:259–275. doi: 10.1016/j.molcel.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morin A, Auxilien S, Senger B, Tewari R, Grosjean H. Structural requirements for enzymatic formation of threonylcarbamoyladenosine (t6A) in tRNA: an in vivo study with Xenopus laevis oocytes. RNA. 1998;4:24–37. [PMC free article] [PubMed] [Google Scholar]
- 31.Madore E, Florentz C, Giegé R, Sekine S, Yokoyama S, Lapointe J. Effect of modified nucleotides on Escherichia coli tRNAGlu structure and on its aminoacylation by glutamyl-tRNA synthetase. Predominant and distinct roles of the mnm5 and s2 modifications of U34. Eur. J. Biochem. 1999;266:1128–1135. doi: 10.1046/j.1432-1327.1999.00965.x. [DOI] [PubMed] [Google Scholar]
- 32.Tisné C, Rigourd M, Marquet R, Ehresmann C. NMR and biochemical characterization of recombinant human tRNA (Lys) 3 expressed in Escherichia coli: identification of posttranscriptional nucleotide modifications. RNA. 2000;6:1403–1412. doi: 10.1017/s1355838200000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grosjean H, Keith G, Droogmans L. Detection and quantification of modified nucleotides in RNA using thin-layer chromatography. Methods Mol. Biol. 2004;265:357–391. doi: 10.1385/1-59259-775-0:357. [DOI] [PubMed] [Google Scholar]
- 34.Stocchetto S, Marin O, Carignani G, Pinna LA. Biochemical evidence that Saccharomyces cerevisiae YGR262c gene, required for normal growth, encodes a novel Ser/Thr-specific protein kinase. FEBS Lett. 1997;414:171–175. doi: 10.1016/s0014-5793(97)00980-0. [DOI] [PubMed] [Google Scholar]
- 35.Meng F, Chen X, Hu Y, Tang H, Dang W, Zhou J. Sua5p is required for telomere recombination in Saccharomyces cerevisiae. Cell Res. 2010;20:495–498. doi: 10.1038/cr.2010.34. [DOI] [PubMed] [Google Scholar]
- 36.Wei F, Suzuki T, Watanabe S, Kimura S. Deficit of tRNALys modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Invest. 2011;121:3598–3608. doi: 10.1172/JCI58056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia GA, Goodenough-Lashua DM. Mechanisms of RNA modifying and editing enzymes. In: Dans Grosjean H, Benne R, editors. Modification and Editing of RNA. Washington DC: ASM Press; pp. 135–168. [Google Scholar]
- 38.Kuratani M, Kasai T, Akasaka R, Higashijima K, Terada T, Kigawa T, Shinkai A, Bessho Y, Yokoyama S. Crystal structure of sulfolobus tokodaii sua5 complexed with l-threonine and AMPPNP. Proteins: Structure, Function, and Bioinformatics. 2011;79:2065–2075. doi: 10.1002/prot.23026. [DOI] [PubMed] [Google Scholar]
- 39.Parthier C, Görlich S, Jaenecke F, Breithaupt C, Bräuer U, Fandrich U, Clausnitzer D, Wehmeier UF, Böttcher C, Scheel D, et al. The O-Carbamoyltransferase TobZ catalyzes an ancient enzymatic reaction. Angew. Chem. Int. Ed. Engl. 2012;51:4046–4052. doi: 10.1002/anie.201108896. [DOI] [PubMed] [Google Scholar]
- 40.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37:D118–D121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl Acad. Sci. USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Björk GR, Ericson JU, Gustafsson CE, Hagervall TG, Jönsson YH, Wikström PM. Transfer RNA modification. Annu. Rev. Biochem. 1987;56:263–287. doi: 10.1146/annurev.bi.56.070187.001403. [DOI] [PubMed] [Google Scholar]
- 44.Forster AC, Church GM. Towards synthesis of a minimal cell. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100090. doi: 10.1038/msb4100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Crecy-Lagarde V, Marck C, Grosjean H. Decoding in Candidatus Riesia pediculicola, close to minimal tRNA modification set? Trends Cell Mol. Biol. 2012; 7:11–34. [PMC free article] [PubMed] [Google Scholar]
- 46.de de Crécy-Lagard V, Marck C, Brochier-Armanet C, Grosjean H. Comparative RNomics and modomics in mollicutes: prediction of gene function and evolutionary implications. IUBMB Life. 2007;59:634–658. doi: 10.1080/15216540701604632. [DOI] [PubMed] [Google Scholar]
- 47.Vetsigian K, Woese C, Goldenfeld N. Collective evolution and the genetic code. Proc. Natl Acad. Sci. USA. 2006;103:10696–10701. doi: 10.1073/pnas.0603780103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.