Abstract
The yeast two-hybrid (Y2H) system is the most widely applied methodology for systematic protein–protein interaction (PPI) screening and the generation of comprehensive interaction networks. We developed a novel Y2H interaction screening procedure using DNA microarrays for high-throughput quantitative PPI detection. Applying a global pooling and selection scheme to a large collection of human open reading frames, proof-of-principle Y2H interaction screens were performed for the human neurodegenerative disease proteins huntingtin and ataxin-1. Using systematic controls for unspecific Y2H results and quantitative benchmarking, we identified and scored a large number of known and novel partner proteins for both huntingtin and ataxin-1. Moreover, we show that this parallelized screening procedure and the global inspection of Y2H interaction data are uniquely suited to define specific PPI patterns and their alteration by disease-causing mutations in huntingtin and ataxin-1. This approach takes advantage of the specificity and flexibility of DNA microarrays and of the existence of solid-related statistical methods for the analysis of DNA microarray data, and allows a quantitative approach toward interaction screens in human and in model organisms.
INTRODUCTION
Networks of protein–protein interactions (PPIs) underlie all cellular processes and are highly predictive for functional relationships among gene products. Consequently, one of the principal goals in modern systems biology is the generation of comprehensive maps for PPIs in human and model organisms (1). The most important tool for systematic mapping of binary PPIs is the well-established yeast two-hybrid (Y2H) methodology (2). In the classical implementation of the Y2H system, a split transcription factor, consisting of activation and DNA-binding domains, is functionally reconstituted via the physical interaction of bait and prey proteins (3). The reconstituted hybrid transcription factor drives the expression of reporter genes that are scored by growth and color phenotypes (typically HIS3 and lacZ). Traditionally, a specific bait protein is combined with a cDNA library encoding prey fusion proteins, and interacting bait–prey combinations are identified from yeast colonies that are grown on selective agar plates. Crucial for the generation of entire protein interactome networks have been matrix-based Y2H screening procedures using libraries of annotated open reading frames (ORFs). These have been applied for the exploration of PPI networks in eukaryotic model organisms, such as Saccharomyces cerevisiae (4,5), Drosophila melanogaster (6), Caenorhabditis elegans (7), and also for a first overview of the human interactome (8,9). Moreover, a number of other screens focused on specific disease-causing proteins, and signaling pathways were performed to obtain increased depth and coverage of relevant PPI networks (10–13).
So far, Y2H data have been reported as reproducible outcomes from repeated interaction screens and are not based on quantitative measurements, which contrasts with gene expression and protein–DNA interaction data that have been extensively addressed with DNA microarrays (14). The DNA microarray technology has also been instrumental in other applications, such as the high-throughput screening and quantitative measuring of drug sensitivity and resistance of yeast deletion strains (15,16). For these experiments, large populations of yeast strains comprising thousands of barcoded deletions are grown in the presence of diverse chemical compounds. The barcodes from compound-treated pools and untreated control pools are amplified by polymerase chain reaction (PCR) and hybridized to DNA microarrays to score deletion strains that are under- or overrepresented after selection. The same strategy is followed with pools of yeast cells that overexpress large collections of ORFs (17). A large number of template ORFs of different sizes can be PCR amplified in one pooled reaction when using a primer set that anneals to adjacent vector sequences.
Here, we apply a novel Y2H screening scheme that is based on pooling and competitive growth on selective plates. For proof-of-principle experiments, we explored PPI networks for the neurodegenerative disease proteins huntingtin (HTT) and ataxin-1 (ATXN1), which, as mutant variants, cause Huntington’s disease (HD) and spinocerebellar ataxia type 1 (SCA1), respectively (18,19). Both proteins contain polyglutamine tracts that, on expansion to a pathological length, cause protein misfolding and aggregation in neuronal cells. PPI networks for HTT and ATXN1 have already been generated previously with high-throughput Y2H screens (11–13). It was suggested previously that the underlying function of polyQ tracts in proteins is to mediate PPIs and that alterations in PPI patterns due to polyQ expansions are important for disease pathogenesis (20,21).
By screening the bait proteins HTT and ATXN1 against a large preassembled library of ORFs, we achieved an unprecedented throughput and parallelization of the Y2H procedure. Quantitative benchmarking and receiver operation characteristics (ROC) via repeated sampling revealed the distribution of known PPIs among the microarray scores and determined the empirical cutoffs for high-confidence PPIs. For HTT, a larger number of PPIs identified by microarray Y2H screening were further confirmed by LUminescence-based Mammalian intERactome mapping (LUMIER) co-immunoprecipitation assays. Importantly, the interpretation of Y2H interaction results as large sets of numerical scores not only allowed a systematic sampling for true positive results but also the exclusion of false positives. In addition, gene ontology (GO) term enrichment analysis predicted the functional involvement of HTT and ATXN1 in different cellular compartments and molecular functions, such as the involvement in cellular signaling pathways and protein binding. The screening approach presented here could be applied more broadly for the systematic mapping of human PPIs and to examine the effects of disease-specific mutations on PPI networks.
MATERIALS AND METHODS
Yeast strains and Y2H matrix
Individual bait strains were constructed by cloning DNA sequences encoding the huntingtin fragments HD506-Q23, ATXN1-Q32 and ATXN1-Q79 into Y2H plasmid pBTM116-D9, derived from pBTM116 (Clontech). Baits selected to perform the Y2H screens are the N-terminal fragment of the HD protein with a short polyglutamine tract (HD506-Q23), wild-type ataxin-1 (ATXN1-Q32), and mutant ataxin-1 that has an elongated polyglutamine tract (ATXN1-Q79). The bait constructs were transformed into yeast strain L40ccua (Mata). Identity of the individual bait clones was confirmed by PCR. The prey ORFs are in vector pACT4-DM (derived from pACT2, Clontech) and grown in strain L40ccα (MATα). Plasmid constructs and shuttling procedures are described elsewhere (22).
Composition of the ActMat v3 matrix
A large matrix of prey strains (ActMat collection, version 3) containing full-length ORFs was constructed by recombinational cloning using the Gateway system (Invitrogen). The ActMat v.3 is an expanded version of a prey matrix described earlier (8), containing a total of 14 119 full-length ORF clones from four different resources. The full MGC3 collection (23) comprises ca. 80% of all clones, and additional clones are from Harvard, SMP and RZPD clone repositories, as well as a collection of ORFs that were assembled in our lab (1–22 collection) (Supplementary Table S1). From the assembled clones, 13 405 ORFs are Entry clones that were transferred into pACT4-DM via Gateway LR-reactions. These entry clones in turn correspond to 11 083 unique Entrez GeneIDs according to NCBI annotations, and 11 685 unique gene annotations in the Ensembl v.58 release. Comparing the annotations in the ActMat v.3 collection with the probesets on the ST1.0 array that were mapped with the Ensembl v.58 release, we identified 10 929 corresponding Ensembl gene IDs (10 500 Entrez GeneIDs).
Pooling and selective growth
The ActMat v.3 strains were arrayed in 384-well microtiter plates and stamped out onto seven minimal medium Omnitrays. Arrayed strains were grown in 45 µl SD-Leu (384-well plates) until saturation and stamped out on Omnitrays containing SD-Leu agar medium using a KBio Systems K4 robot and grown for 2 days at 30°C. Freshly grown yeast strains were then washed off with SD-Trp medium (containing 10% glycerol), pooled and concentrated to 50 or 100 optical densities at 600 nm (OD600) per ml to generate pool aliquots. For each mating reaction, 20 µl of the concentrated stock, roughly corresponding to one OD600 (1−2 × 107 cells), were used. The amount of 1−2 × 107 cells per OD600 covers the complexity of the library (ca. 14 000 ORFs in the pools) several 100-fold.
For the bait screening procedure, freshly grown cultures (5 ml) of the bait strain were concentrated in a small volume to a maximum density of 50 ODs/ml. The concentrated bait strain was then combined with a prey pool aliquot in a 1:1 to 2:1 ratio and thoroughly mixed. Then, 10–20 µl of the mix was spotted on yeast extract peptone glucose (YPD) agar medium and incubated for 24 h at 30°C to allow for sufficient mating. To control mating efficiency, a small amount of cell material was diluted 1:20 000 in liquid SDIV (SD-His-Leu-Trp-Ura), and plated on SDII (SD-Leu-Trp), SD-Leu and SD-Trp plates for mating control. Because mating efficiency of the prey strains is generally diminished with every freeze-thaw cycle, prey pools were stored in 1 ml aliquots for single use. For the preselection of diploids, the mated cells were transferred onto SDII-agar medium using an incubation loop and incubated for 24 h at 30°C. Pools enriched for diploid cells were diluted to an OD600 of 0.05 in 5 ml SDIV medium and grown at 30°C with 250 rpm. For four of the screens (with HTT and pBTM), inoculation and culture volumes were 5 times the inoculation (OD600 = 0.25) and/or 10 times the volume (50 ml). After reaching an OD600 of 2–3 (ca. 48 h), cells were diluted back into a fresh culture to OD600 = 0.05 for a second round of selection (ca. 24 h). Considering the total incubation in SDIV medium and an average generation time of ∼3.5 h, the selection of the HIS3 reporter activation was expected to last for ca. 20–24 generations.
Preparation of DNA microarrays
DNA was extracted from 1 ml of each final SDIV culture using the Zymoprep II Yeast Plasmid Miniprep protocol (Zymo Research). To measure the full representation of all ORFs in ActMat v.3, an equivalent amount (∼2 × 107 cells) was extracted from the original pooled cells. To increase the yield of plasmid DNA elution from the column, 20 µl bdH2O was incubated in the column for 2 min before spinning into a new Eppendorf tube. The population of cloned ORFs in prey plasmid pACT4 was selectively amplified with primers pACT4-5-P3 (5′-TGC GGG GTT TTT CAG TAT CTA-3′) and pACT4-3-P4 (5′-ATG ATG AAG ATA CCC CAC CAA A-3′) using the Expand High Fidelity PCR kit (Roche). The PCR reaction (50 µl) contained one-tenth of the eluted DNA (2 µl), 300 nM of each primer, 200 µM of dNTP mix and 2.6 U/reaction Expand High Fidelity enzyme. Routine PCR reaction was 10 min initial denaturation, followed by 35 cycles of amplification (30 s 95°C, 45 s 55°C and 5 min 68°C), and 10 min final elongation at 68°C. The amplified DNA was measured with a NanoDrop spectrophotometer and checked on 1% agarose gels (Supplementary Figure S1).
PCR products were then purified with the PCR purification kit (Invitrogen) according the instructions of the manufacturer. Elution was done in 50 µl water after 1 min incubation. Half of the amplified PCR product (25 µl) was biotin labeled with the BioPrime DNA labeling system. Labeling was done according to the specifications of the manufacturer (Invitrogen). The total volume of 50 µl from the labeling reaction was used for microarray hybridization.
Affymetrix microarray processing
Hybridization to Affymetrix human gene ST1.0 arrays was done according to the WT Sense Target Labeling Assay Manual with minor modifications. Prehybridization was done for 10 min at 45°C and 60 rpm in the hybridization oven. The total hybridization mix (150 µl) contains ∼50 µl total labeled DNA, 3 nM B2-Oligo, 1× control RNAs (bioB, bioC, bioD, cre), 1× hybridization buffer and 7.5% dimethyl sulfoxide. The mix (150 µl) was denatured for 2 min at 95°C and stored for 2 min on ice to fill the array chamber completely. Washing and staining was done according to the Affymetrix Eukaryotic Antibody Staining protocol (protocol FS450-0007), but without the signal amplification using the biotinylated antibody. In the modified protocol, stain 1 contained streptavidin phycoerythrin (SAPE) solution [1× 2-(N-morpholino)ethanesulfonic acid (MES) stain buffer, 2 mg/ml acetylated bovine serum albumin and 10 µg/ml streptavidin-phycoerythrin], stain 2 consisted only of 1× MES stain and 2 mg/ml bovine serum albumin and stain 3 was 800 µl array holding buffer. See Supplementary information for microarray analysis.
LUMIER assay
LUMIER was developed as a comprehensive mammalian interactome screening strategy (24). Here we apply a modified version as a validation assay for PPI results. Protein A (PA)-Renilla luciferase (RL)-tagged fusion proteins were co-expressed with firefly luciferase (FL)- V5-tagged interactor proteins in HEK293 cells. After 48 h, protein complexes were co-immunoprecipitated from 70 µl cell extracts in IgG coated beads and subsequently washed with 100 µl phosphate buffered saline; interactions between bait (PA-RL) and prey proteins (FL fusions) were monitored by quantification of FL activities. Quantification of RL activity was used to confirm that PA-RL-tagged bait protein is successfully immunoprecipitated from cell extracts. To detect RL- and FL-based luminescence, Dual-Glo Luciferase Kit (Promega) was used. Bioluminescence was quantified in a luminescence plate reader (TECAN Infinite M1000).
For each protein pair, three co-immunoprecipitation experiments (Co-IPs) were performed in parallel (see Supplementary Figure S4). PA-RL and FL without a fusion protein were used as controls to examine background protein binding. After 48 h, protein complexes were co-immunoprecipitated in IgG-coated beads. By comparing the firefly luminescence activity measured in the Co-IP with the two fusion proteins with the controls, the R-op and R-ob binding ratios were obtained, which are a measure for the protein interaction specificity. Based on well-characterized interaction test pairs, an interaction was defined as positive when the calculated R-op and R-ob ratios were >1.25 and >2, respectively.
Bioinformatics
Quantitative scoring of the microarray data for known positive sets (literature) was performed using the QiSampler application (25). Literature interactions for binary classification in the QiSampler procedure were derived from the v1.2 release of the Human Integrated Protein–Protein Interaction rEference (HIPPIE) database (26), available at http://cbdm.mdc-berlin.de/tools/hippie. Sets of geneIDs, associated with specific GO-terms were downloaded from the Gene Ontology browser AmiGo (http://amigo.geneontology.org). Network graphs are drawn with Cytoscape 2.7.0. Venn diagrams were constructed with BioVenn online tool (http://www.cmbi.ru.nl/cdd/biovenn). GO-term enrichment was determined using the human Consensus Path Database (CPDB; http://cpdb.molgen.mpg.de) (27,28). For CPDB analysis, P < 0.01 were considered significant for the enrichment for a pathway or a functional group defined by GO. The analysis of the microarray data is described in the Supplementary information.
RESULTS
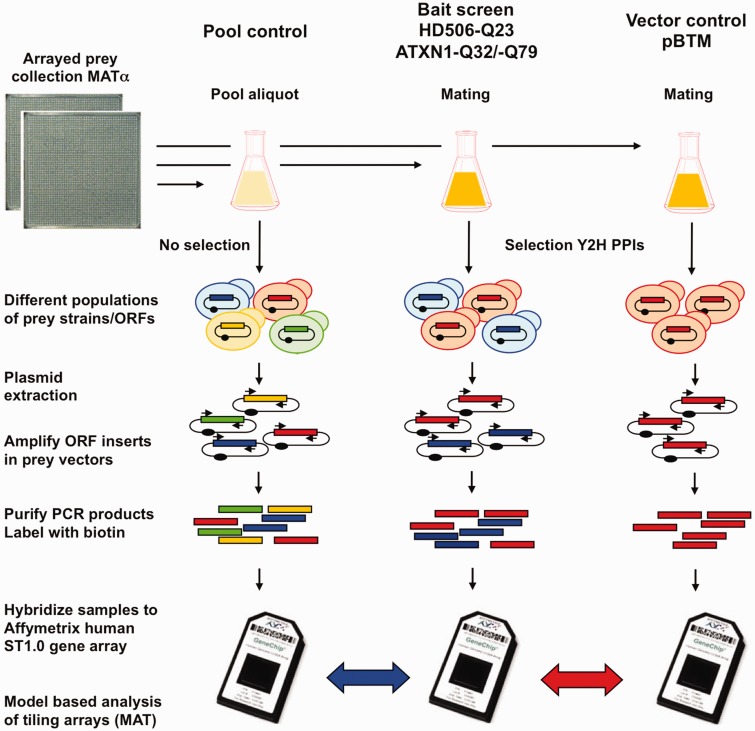
To apply DNA microarrays as a quantitative readout for PPI detection, we implemented a global mating and selection scheme for Y2H interaction screens (Figure 1). An arrayed collection of ∼14 000 ORFs in Y2H prey vectors (ActMat v.3; Supplementary Table S1) was pooled and small aliquots were used for mating reactions with bait constructs or the empty vector control (pBTM). As baits for interaction screening, a wild-type N-terminal HTT fragment (HD506-Q23) as well as both wild-type and mutant elongated ATXN1 (ATXN1-Q32 and ATXN1-Q79) were used. Mated diploid yeast cells were grown under selective conditions for HIS3 reporter gene activation, and plasmid DNA was isolated from selected and non-selected samples. Prey ORFs were amplified with a primer pair in the prey vector that flanks the recombination sites, yielding PCR products over the full range of expected sizes (Supplementary Figure S1). PCR products were then biotin labeled and hybridized to Affymetrix ST1.0 DNA microarrays. The hybridization signals were characterized with a number of parameters that measure the enrichment of bait–prey combinations (ratios) and different statistical tests (e.g. P-value and q-value Wilcoxon) to determine the significance of results and the screen-to-screen variations (Supplementary Tables S2 and S3). For any represented geneID, the hybridization signals in the ‘bait’ screens, representing the preys selected in combination with a defined bait, were compared with two sets of ‘control’ samples: the original unselected pooled prey collection (Pool), which represents the background signal for a given library, and the empty bait plasmid (pBTM) control, which reports reporter activation in the absence of a functional bait. Hence, whereas the ratiopool quantifies the (initial) Y2H interaction selection, the ratiopBTM displays the difference of the specific ‘bait’ selection to the unspecific self-activation of prey ORFs that interact with the DNA binding domain of the vector in the absence of a bait protein.
Figure 1.
Pooling and selective growth strategy for microarray Y2H screening. All strains of the Y2H prey collection (ActMat v.3 in Matα) with ∼14 000 full length ORF clones are combined into one large pool and aliquots are mated with Mata Y2H strains containing selected bait constructs (HD506-Q23, ATXN-Q32/-Q79) or empty vector control (pBTM116-D9) in 1:1 ratio. Because only a few prey ORFs are enriched after the Y2H procedure by HIS3 activation, the resulting pools differ in their composition from the original pool. Selection of preys is either bait-dependent (blue) or unspecific (red). After plasmid extraction and PCR amplification of the prey inserts, biotin-labeled PCR products are hybridized to Affymetrix ST1.0 microarrays. Using model-based analysis of tiling arrays (MAT), the hybridization results from the bait screens are compared with the pool control (pooled preys without selection or bait; Pool) and with the screening control (a selection screen with empty vector; pBTM). The first comparison shows the enrichment of interacting preys. The second comparison shows enrichment that is specific to the use of the bait.
Quantitative benchmarking of Y2H screening results
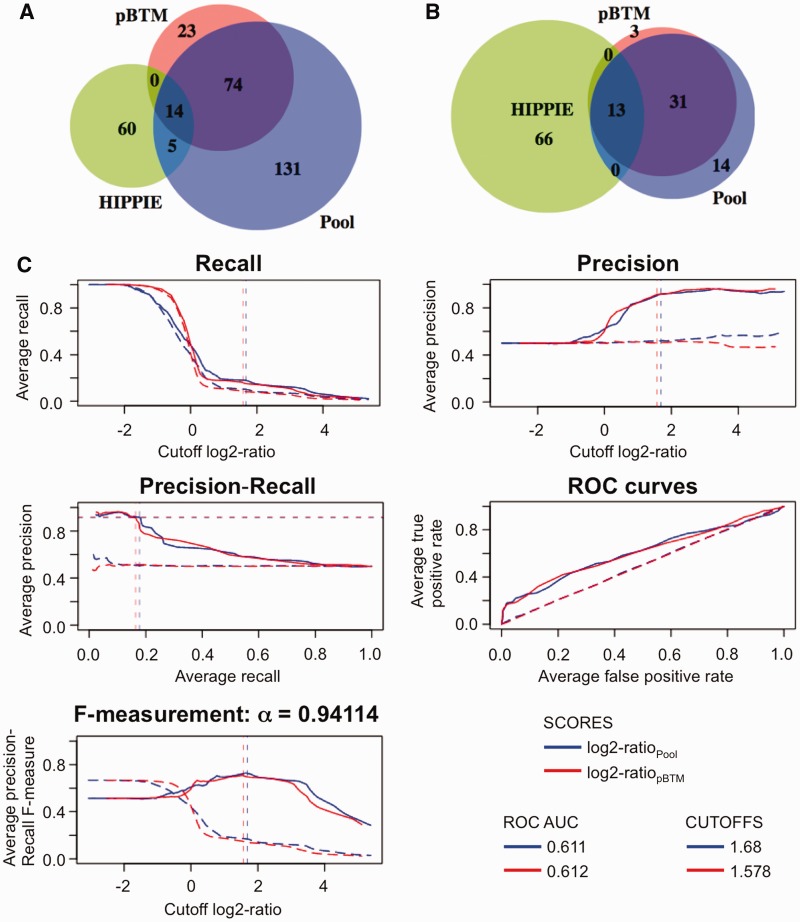
Screening of the ActMat v.3 library with HD506-Q23 in nine replicates revealed 9888 ORFs as ‘present’ on the microarray via detection calls or background tags (Supplementary Table S4). Moreover, the application of background tags and median probeset signals displayed a rather efficient amplification of ca. 90% of all ORFs and the absence of major biases and PCR artefacts (Supplementary Figure S2). In a second step, we found that differential enrichment of 2638 ORFs in the pool and pBTM comparisons was significant after multiple testing (q-value Wilcoxon ≤0.05). Through the application of this threshold, screen-to-screen variability is taken into account to determine bait-specific enrichment of ORFs compared with pool and pBTM controls (Supplementary Figure S3). Third, for a primary network analysis, low arbitrary cutoffs for bait-specific activation were set at log2-ratio ≥0.6 (ratio ≥1.516), which identified in total 224 preys in the pool and 111 preys in the pBTM comparison (Figure 2A). The restriction to 88 ORFs in the overlap between ratiopool and ratiopBTM scores excludes potential false positives, especially those from bait-independent reporter gene activation (see Materials and Methods section and Supplementary information for all technical descriptions).
Figure 2.
Quantitative benchmarking and cutoffs for HD506-Q23 screens. (A) Venn diagram for overlap of log2-ratio scores at arbitrary cutoff (log2-ratio ≥0.6), ratiopool (blue), ratiopBTM (red) and literature data (HIPPIE, green). (B) Venn diagram for overlap of log2-ratio scores at automated cutoffs based on repeat sampling with HIPPIE classifiers, log2-ratiopool (1.68) and ratiopBTM (1.578). Evaluations are based on 2638 high-confidence results (GeneIDs) after multiple testing (q-value Wilcoxon ≤0.05), including 79 known positives from the HIPPIE database. (C) Quantitative benchmarking and determination of automated cutoffs using QiSampler. Log2-ratios are the numerical scores and HIPPIE positives as binary classifiers. Precision, recall, F-measurements and ROC are displayed as repeat sampling curves. F-measurement is adjusted (β = 0.25, α = 0.94114). For ROC, area under the ROC curve (ROC AUC) is calculated. Sampling with HIPPIE positive set: log2-ratiopool (closed blue line), log2-ratiopBTM (closed red line). Control sampling with random set (no discrimination): log2-ratiopool (dashed blue line), log2-ratiopBTM (dashed red line). Positives correspond to HIPPIE and negatives to non-HIPPIE PPIs. Sampling is done in 1000 repetitions, with a rate of 0.5 (50% of classifiers sampled per run).
With the presumption that the occurrence of known positives increases the confidence in the overall screening results, Y2H interaction results are commonly benchmarked against a dataset of known literature interactions. We recently developed QiSampler, a statistical tool that allows the comparison of numerical scores (such as the ratios from Y2H microarrays obtained here) with binary classifiers using a repetitive random and balanced sampling strategy (25). The primary source of binary classifiers for the sampling analysis of Y2H interaction microarray data were known literature interactions in the HIPPIE database, a comprehensive collection of human PPIs with experiment-based quality scores (26). Control samplings were done with random sets from all other preys that are not contained in the HIPPIE dataset. Because PPIs for HTT were previously explored, notably also with Y2H and co-precipitation assays in high-throughput experiments (11,12), a rather large collection of 289 HTT interactions is available in the HIPPIE database, with 79 PPIs among the subset of filtered ORFs (2638) that were significant after multiple testing. Using the known PPIs as binary classifiers for the filtered HTT dataset in the QiSampler procedure, a high precision and a relatively low recall were found at increasing ratio cutoffs (Figure 2C). Moreover, the ROC curves displayed a clear discrimination for both ratiopool and ratiopBTM with respect to the diagonal representing randomness (area under the ROC curve equal 0.611 for ratiopool and 0.612 for ratiopBTM). The distinct quantitative effect with both ratios reflected, therefore, a specific enrichment of known HTT interactors with the HD506-Q23 bait protein.
Besides an overall characterization of the screening outcome, we sought to use the quantitative benchmarking procedures to estimate the ideal cutoff for high-confidence PPIs, based on the distribution of known positives across the entire range of ratiopool and ratiopBTM scores. Estimations for an ideal cutoff aim at the inclusion of the most possible true positives, while avoiding the inclusion of false positives (see Supplementary information for cutoff determination). Confronted with a high precision and low recall in sampling of Y2H interaction screening data, we emphasized on precision as the major determinant for the cutoff selection. A modified version of QiSampler allowed automated cutoff computation based on F-measurement (harmonic mean of the precision and the recall), with adjustment through the alpha (α) coefficient. After testing various α coefficients, we settled for a 4-fold emphasis of precision over recall, corresponding to α = 0.94114. This resulted in cutoffs at log2-ratiopool = 1.68 and ratiopBTM = 1.578, roughly corresponding to the log2-ratios at which 90% precision is reached (Figure 2B). Using this approach, 44 prey ORFs were found as interaction partners for HD506-Q23 in the overlap between pool and pBTM comparisons, which is a significant result (P = 1.1 × 10−42 for one-sided Fisher’s exact test). Compared with the low arbitrary cutoff (see Figure 2A), 14 out of the 15 known positive PPIs were retained, while the total overlap was narrowed by 50%. Sampling also further eliminated 90% of potential false-positive interactions (ratiopool only). Hence, repeat sampling defined a set of 44 high-confidence PPIs for HD506-Q23 that result from bait-specific Y2H interaction selection.
A high-confidence microarray-Y2H interaction network for HD506-Q23
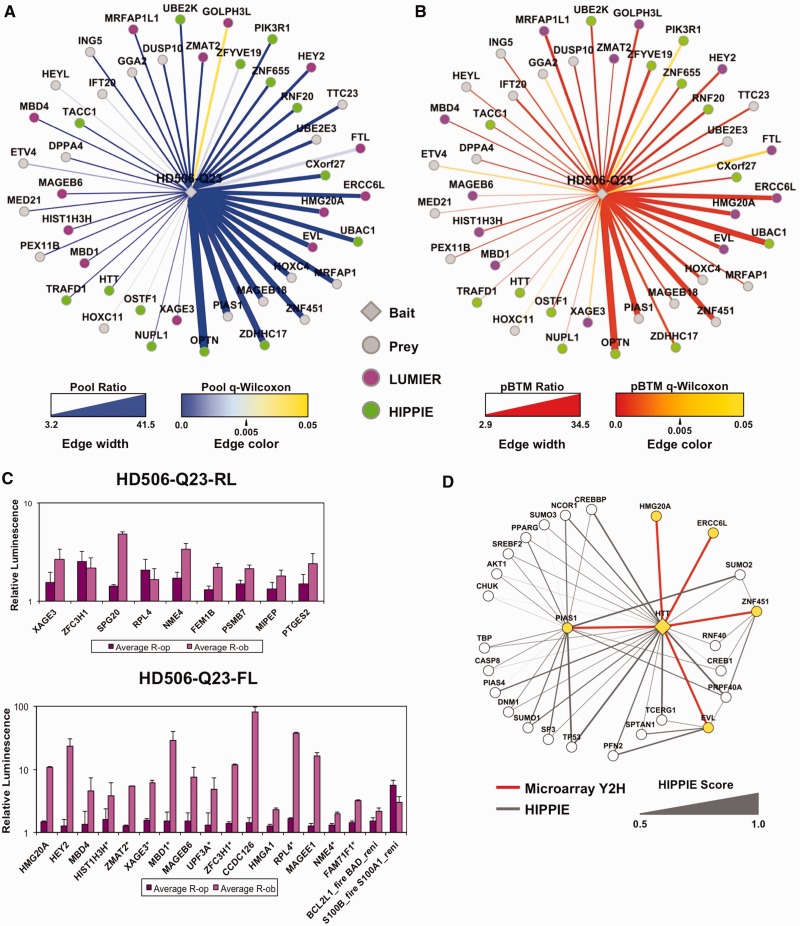
For network representation, the PPIs with HTT were displayed according to ratio and q-value Wilcoxon parameters (Figure 3A and B). High ratios and low q-values in the pool comparison reflected strong and reproducible reporter gene activation, whereas the pBTM comparison measured the specificity of the reporter activation with respect to the empty vector control. Importantly, about one-third of the high-confidence HTT interactions were known HIPPIE positives (13 and HTT self-interaction), which was also reflected in the high precision for the QiSampler (see Figure 2C). Known positives among the highest-scoring HD506-Q23-interacting proteins included optineurin (OPTN), palmitoyltransfease ZDHHC17 (HIP14) and, importantly, also enzymes with roles in the ubiquitin cycle, with activation (UBAC1), conjugation (UBE2K) and ligation (RNF20) (11,12,29).
Figure 3.
Analysis of high-confidence Y2H interactions for HD506-Q23. (A) Display of the ‘pool’ comparison, and (B) display of the ‘pBTM’ comparison above sampling cutoffs: log2-ratiopool ≥1.68 and log2-ratiopBTM ≥1.578 (34 overlapping scores). Edge line width: ratiopool and ratiopBTM (linear scale). Edge color: q-value Wilcoxon, with high confidence q-values <0.05 (blue, red). Nodes: baits in diamonds, preys in circles, HIPPIE (green), LUMIER confirmed novel interactions (purple). (C) Validation of HTT Y2H data by LUMIER (description in Supplementary Figure S5). LUMIER results are shown from selected positives with HD506-Q23 in the prey and bait orientation. Binding ratios R-op (deep purple) and R-ob (light purple) measure firefly (FL) signal with bait and prey combinations compared with bait- and prey-only controls. Asterisks (*) show FL-baits with positive LUMIER in two repeat screen sets. Positive controls are BCL2L1-firefly with BAD-renilla, and S100B-firefly with S100A1. (D) Indirect evidence for novel Y2H PPIs by HIPPIE comparison. Representation of shared PPIs between HTT and EVL, ERCC6L, HMG20A, PIAS1, ZNF451 according to HIPPIE. Nodes: microarray Y2H (yellow), HIPPIE (white), Edges: microarray Y2H (red lines), HIPPIE (grey, closed for HTT interactions), Edge line width: HIPPE evidence score (0.5–1).
We applied a modified version of the LUMIER method as an orthogonal PPI confirmation assay (Supplementary Figure S4). Baits tagged with protein-A and RL were co-expressed with FL-tagged prey proteins (30). To test for interactions, 73 candidate proteins were chosen based on the low arbitrary cutoff (not including 15 known positives) and co-expressed as bait and/or preys in HEK293 cells. Interactions were detected by quantification of FL luminescence from co-immunoprecipitated protein complexes. In total, with HD506-Q23 either in bait or prey orientation, 31 of the tested Y2H interactions (42%) were confirmed by LUMIER assays (Supplementary Table S5, Figure 3C). Among the high-confidence interactions, 27 out of the 44 PPIs (61%) were either confirmed with LUMIER or were HIPPIE positives (Figure 3A and B). If the cutoffs were further raised (log2-ratios ≥3), the overall precision is even higher with 14 out of 18 Y2H PPIs (78%) confirmed by LUMIER or HIPPIE. Hence, in general, the significance and enrichment of the microarray signals correlate well with confidence for genuine PPIs.
We further inspected five intriguing novel HTT interactions among the highest ratio scores (all ratios ≥15) in more detail: ERCC6L, EVL, HMG20A, PIAS1 and ZNF451. HMG20A is part of the high-mobility group proteins, EVL is an Ena/VASP family protein that links cell signaling to remodeling of the actin cytoskeleton (31) and ERCC6L is a member of the SNF2/RAD54 helicase family with a role in DNA repair (32). The other two proteins have roles in protein sumoylation; PIAS1 (Protein inhibitor of STAT) is a Sumo ligase and ZNF451 a transcriptional co-regulator associated with PML bodies and Sumo (33). The associations of ERCC6L, EVL and HMG20A with HTT were confirmed in LUMIER assays, whereas those with PIAS1 and ZNF451 were not. We used the HIPPIE database for further evaluation of these five proteins, looking for co-complex formation with previously identified HTT interactors as an indirect evidence for association (Figure 3D). We found that EVL shares four partners out of 22 with HTT, including the actin monomer-binding protein profilin-2 (PFN2), and a spectrin protein involved in actin crosslinking (SPTAN1), which is consistent with a functional involvement of HTT in actin remodeling (34). ZNF451 shares 3 out of its 13 known partners with HTT. These include also a shared interactor with EVL, the pre-mRNA processing factor 40 (PRPF40A), which was originally discovered as an HTT interacting protein (HIP10) (35). In addition, ZNF451 is linked to PIAS1 (33), which also shares 20% of its known partners with HTT, further corroborating the association of HTT with the sumoylation machinery. This is consistent with the observed regulation of HTT stability by sumoylation and ubiquitination (36).
For a functional analysis of PPI networks, we relied on a dual strategy: hypergeometric testing for overrepresentation of pathways and GO above determined cutoffs, and deep sampling of selected gene associations for true enrichment over the entire range of scores. For a global overview and functional enrichment analysis, it is preferable to increase the sensitivity using less stringent cutoffs, expanding also to less significant results (q-value Wilcoxon >0.05). When doing cutoff sampling and gene overrepresentation analysis for the total HD506-Q23 screening data (Supplementary Figure S5 and Table S6), the results are consistent with the multiple roles for HTT as a hub for PPIs and diverse functions such DNA binding, signaling and binding to ubiquitin-proteasome components (11,12,21).
Functional analysis of wild-type and mutant ATXN1 PPI networks
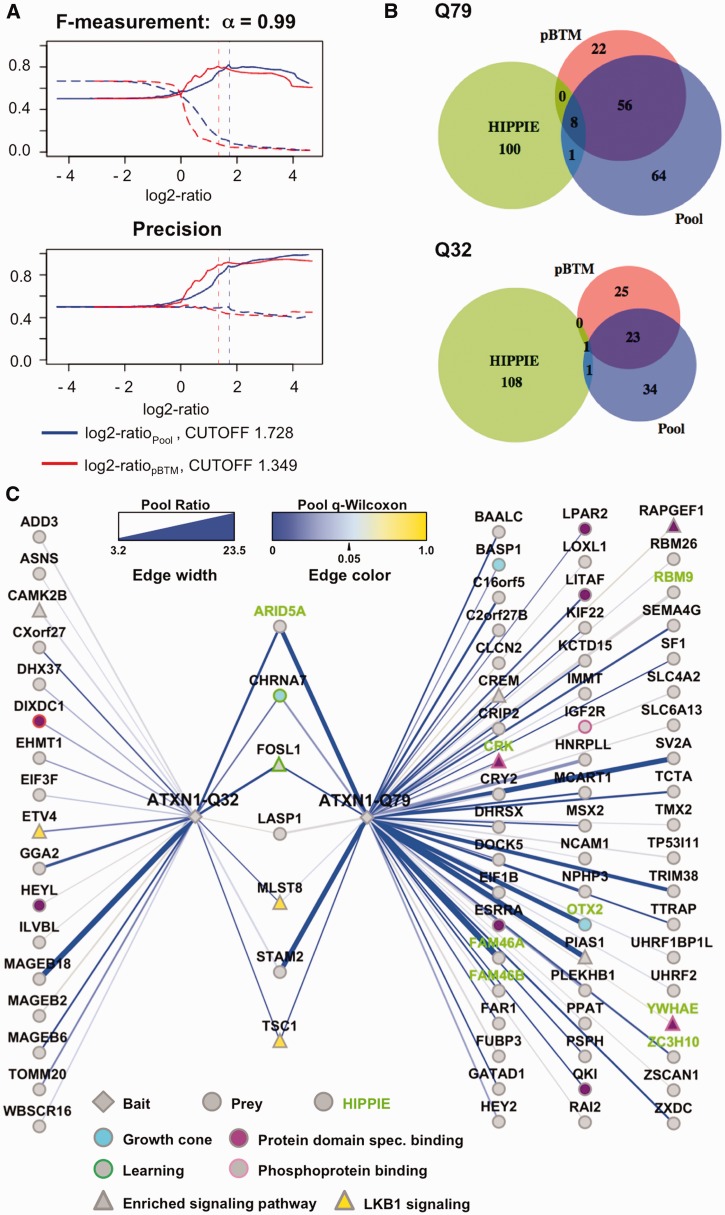
A major strength of the microarray-based Y2H method is the comprehensive readout of the total screening results, which allows the side-by-side comparison of PPI profiles for mutant and wild-type bait proteins. We compared the PPI patterns for the bait proteins ATXN1-Q32 and ATXN1-Q79 containing a non-pathogenic and a pathogenic polyQ tract, respectively (Figure 4). Inspecting the PPI data obtained for the two bait proteins revealed that ATXN1-Q79 interacts with two to three times more prey proteins than ATXN1-Q32 (Supplementary Table S7). Applying quantitative benchmarking for all 9941 ATXN1-Q79 scores with 109 known HIPPIE positives, we found a relatively stronger performance for the expanded ATXN1-Q79 protein (ROC-values: 0.596 and 0.585), whereas the performance for the short ATXN1-Q32 form was closer to random (ROC-values: 0.554 and 0.520) (Supplementary Figure S6). Considering the bias of known positives, automated cutoffs were generated only from the ATXN1-Q79 PPI data (log2-ratiopool = 1.728, log2-ratiopBTM = 1.329; α = 0.99) (Figure 4A), but then were also applied to the ATXN1-Q32 screen set (Figure 4B). We found that eight known positive interactions were among the ATXN1-Q79 PPIs, whereas for ATXN1-Q32, only one known prey protein (ARID5A) was selected. In total, 64 PPIs were found for the expanded ATXN1-Q79 and 24 for the short -Q32 form with a significant overlap of 7 interactions (P-value Fisher exact test: 1.5 × 10−9) (Figures 4B and C). Hence, the results for ATXN1-Q32 and -Q79 differ in respect with the overall yield of scores and the enrichment of known literature positives. A possible explanation for the increased number of interaction partners observed with mutant ATXN1 is provided by the notion that the expanded glutamine tract alters the conformation of ATXN1 and may promote the formation of abnormal PPIs with multiple cellular proteins (20,37). But it also might enhance the strength of interaction with partners of the wild-type form, leading to an increased detection of true biological positives.
Figure 4.
Benchmarking and network display for ATXN1 interaction partners. Results are from ATXN1-Q32 and ATXN1-Q79 Y2H screens. (A) Quantitative benchmarking and determination of automated cutoffs for the ATXN1-Q79 screen set. Evaluations are based on 9941 comparisons including 109 and 110 known positives from the HIPPIE database. Log2-ratios represented as numerical scores and HIPPIE positives as binary classifiers. F-measurements (β = 0.1, α = 0.99) and precision are displayed as repeat sampling curves: sampling with HIPPIE positives: log2-ratiopool (closed blue line), log2-ratiopBTM (closed red line); control sampling with random set: log2-ratiopool (dashed blue line), log2-ratiopBTM (dashed red line). Positives correspond to HIPPIE and negatives to non-HIPPIE PPIs. Sampling is done in 100 repetitions, with a rate of 0.5 (50% of classifiers sampled per run). (B) Venn diagram for overlap of log2-ratio scores for ATXN1-Q79 and ATXN1-Q32. Cutoff is based on repeat sampling of the ATXN1-Q79 screen set, log2-ratiopool (1.728) and ratiopBTM (1.349). (C) Network display of the ‘pool’ comparison above sampling cutoffs: edge line width: ratiopool (linear scale). Edge color: q-value Wilcoxon, with high confidence q-values <0.05 (blue). Node and node border colors for pathway and GO-term associations: baits (diamonds), preys (circles), HIPPIE (green labels), enriched CPDB pathway components (triangles), Lkb1 pathway (yellow), growth cone (light blue), protein domain-specific binding (purple), phosphoprotein binding (light purple border), learning (green border).
In the functional enrichment analysis for ATXN1-Q32 and ATXN1-Q79, we found overrepresentations of different signaling pathways and several interesting targets (Supplementary Table S6). Indeed, at least 10 out of 81 proteins detected in the ATXN1 screens take part in one or several signaling pathways, such as Lkb1, IFNy, IGF1, mTOR and more others (enrichment for Lkb1 pathway: P = 4.6 × 104 for –Q79 and P = 2.6 × 105 for –Q32). Examples for signaling proteins that were found with both isoforms include the signal transducing adaptor molecule 2 (STAM2), the mTOR associated protein LST8 homolog (MLST8) and the hamartin protein TSC1 (Figure 4C). We also found association with 14–3–3 proteins (YWHAE, YWHAZ and YWHAQ above or slightly below the chosen cutoffs), which are known modulators of ATXN1-mediated neurodegeneration (38), confirming previously published results (see Supplementary Table S7). For both wild-type and mutant ATXN1 isoforms, we found GO-terms enriched that are related to neuronal cell growth and brain development, such as ‘growth cone’, ‘pallium development’, and ‘neuron projection’, suggesting that ATXN1 function is critical for these processes. For example, genes among the high confidence scores associated with ‘growth cone’ included TSC1, orthodenticle homeobox 2 (OTX2), brain acid soluble protein 1 (BASP1) and the neuronal acetylcholine receptor subunit alpha-7 (CHRNA7) (Figure 4C). Overall, the GO analysis suggests a role for ATXN-1 in cell signaling and neuronal functions.
Sampling the distribution of gene sets with QiSampler using GO-annotated genes instead of literature-positive interactors allows a global comparison of quantitative enrichments in PPI patterns for both ATXN1 isoforms (Supplementary Figure S6). When sampling for the Lkb1 signaling pathway and the GO-terms ‘growth cone’ and ‘learning’, we found similar ROC performances for both mutant and wild-type ATXN1 baits. For Lkb1 gene associations, for example, ratiopool ROC AUC values were in the same range for ATXN1-Q32 and ATXN1-Q79 (0.626 and 0.64). Likewise, for most other GO-terms investigated (not shown), sampling reflects a similar distribution of classifiers among the scores. In a further attempt to quantify the enrichments, we sampled the Y2H scores with two ‘molecular function’ GO-associations, ‘protein domain specific binding’ and ‘phosphoprotein binding’ (Supplementary Table S6). Here, ROC performances show a selective association of ‘phosphoprotein binding’ with the expanded ATXN1-Q79 form, while for ‘protein domain specific binding’, a similar result for wild-type and mutant ATXN1 proteins was obtained. In conclusion, GO term enrichments and individual samplings revealed that the overall PPI pattern of ATXN1 is similar for the Q32 and Q79 forms. This indicates that enhancement of wild-type protein binding determines pathogenesis of ATXN1 on polyglutamine expansion, as opposed to pathogenesis being due to binding the wrong partners.
DISCUSSION
We describe a novel approach for the detection of high quality Y2H PPIs using DNA microarrays and quantitative statistics. The concept study presented here takes full advantage of the established tools for the analysis of DNA microarray data and could have important implications on how future research on protein interactomes is being conducted.
We concentrated our proof-of-principle experiments on the HTT and ATXN1 proteins, which are both neurotoxic on polyglutamine repeat expansion (18). The approach was validated by the generation of a set of high-confidence PPIs for the HTT protein, which were based on microarray data after multiple testing for significance. These results were benchmarked against sets of known positive PPIs using a quantitative sampling strategy. F-statistics based on precision-recall distributions was used to determine automated cutoffs for high-confidence interactions. PPIs were further restricted by applying two distinct background controls (pool and vector), which allows the simultaneous selection of Y2H positives and the filtering of unspecific autoactivators. Notably, almost two-thirds of the final high-confidence PPIs for a HTT bait protein were known positives or validated by a modified LUMIER assay. Hence, by using quantitative benchmarking and F-statistics, we established a microarray-based Y2H screening method for the high-confidence mapping of PPI networks. However, we also advocate that results may be interpreted with different procedures, depending on the overall screening performance, the availability of sets of known positives and also on the specific aims intended by individual researchers (see Supplementary information).
Besides the mapping of individual high-confidence PPIs, microarray Y2H screening data can be more broadly interpreted for enrichments of pathways and functional associations. This may be important when addressing biological consequences of mutations that alter structural properties in proteins and thus underlie global perturbations in PPI networks and potentially influence the outcome of disease (39). Specifically, we addressed here potential differences in PPI patterns between protein isoforms (ATXN1-Q32 and ATXN1-Q79, containing short and expanded polyQ tracts). In this assay, ATXN1-Q79 exhibits more and stronger Y2H interactions than ATXN1-Q32. On the other hand, our data analysis also shows that the overall PPI patterns of wild-type and mutant ATXN1 are not radically different, suggesting that ATXN1 pathology results from abnormally strong interactions with its biological partners. Although resulting from a screening effort in a heterologous system (yeast), this finding is consistent with previously observed effects of expanded polyQ tracts in ATXN1 and other polyQ disease proteins (20,21,37). This example demonstrates how microarray-based Y2H procedures can be used in conjunction with extensive data-mining strategies to predict the biological consequences of altered proteins.
While DNA microarrays were used to address Y2H results in an earlier study (40), a quantitative procedure, such as the one presented here with large-scale pooling of a prey library, unbiased selection by competitive growth and systematic control measurements, was not attempted before. This approach has two major advantages over matrix-based Y2H screenings. First, PPIs are characterized as scores with different parameters (ratios, P-values, etc.) over a wide dynamic range, instead of being simple counts from identifications in replicate screens. Repetitive sampling strategies and the application of two background controls (pool and pBTM comparisons) have the important consequence that potential false-positive interactions can be addressed and eliminated (see Supplementary information for discussion of false positive interactions). Because false positives are sometimes estimated up to 50% of all reported interactions (41,42), their minimization would constitute a major advantage for mapping of high-confidence PPIs, reducing also the need for confirmation with orthogonal assays. Second, smaller volumes of medium for yeast mating and selection as well as the efficient readout provided by DNA microarrays greatly reduce labor and material costs. Simplifying the screening procedure increases potential throughput, and therefore larger numbers of Y2H screens can be performed in parallel. However, while our system is superior over the ‘classical’ Y2H method with respect of quantitative measurements, it has also some limitations. First, ‘color’-based scoring of interactions via lacZ activation is not possible for the pool-based screening scheme. Second, some ORFs may not undergo proper PCR amplification, which could lead to a fraction of putative PPIs that are undetectable in microarray-Y2H assays. Indeed, a bias against longer DNA sequences is evidenced by the lesser representation of ORFs >2 kb in sizes on the microarray (Supplementary Figure S2). Third, prey proteins in the complex pool that occur as different isoforms or with individual mutations may be indistinguishable on the DNA microarray. Hence, for optimal coverage of potential PPIs, DNA microarray and matrix-based robotic Y2H procedures should be envisioned as complementary approaches.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Information and Methods, Supplementary Tables 1–7, Supplementary Figures 1–6 and Supplementary References [43–50].
FUNDING
German Ministry of Education and Science (BMBF) MedSys project “PREDICT” [0315428 to R.H.] and other grants [01GS08170, MooDS, Mutanom, GoBio to E.E.W.]; the Max Planck Society (to R.H.) (to E.E.W.): Deutsche Forschungsgemeinschaft (DFG) [SFB740], Huntington’s Disease Society of America (HDSA) Coalition for the Cure, EU [EuroSpin, SynSys], the Helmholtz Association [HelMA], CHDI Foundation. (to E.E.W. and M.A.A.-N.): German Ministry of Education and Science (BMBF) NGFN-Plus, NeuroNet; Deutsche Forschungsgemeinschaft (DFG) [SFB618]; the Helmholtz Association [MSBN]. Funding for open access charge: NGFN-Plus grant Neuronet [01GS08170].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Gabriele Born for processing the Affymetrix arrays and the other members of the Andrade, Wanker and Huebner groups at the Max Delbrück Center for assistance and support.
The interactions found in this study were submitted to the IntAct database with the IMEx registry number IM-17394. Microarray data were submitted to GEO according to MIAME standards.
REFERENCES
- 1.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stelzl U, Wanker EE. The value of high quality protein-protein interaction networks for systems biology. Curr. Opin. Chem. Biol. 2006;10:551–558. doi: 10.1016/j.cbpa.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 4.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 5.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 7.Simonis N, Rual JF, Carvunis AR, Tasan M, Lemmens I, Hirozane-Kishikawa T, Hao T, Sahalie JM, Venkatesan K, Gebreab F, et al. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nature Methods. 2009;6:47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 9.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 10.Bandyopadhyay S, Chiang CY, Srivastava J, Gersten M, White S, Bell R, Kurschner C, Martin CH, Smoot M, Sahasrabudhe S, et al. A human MAP kinase interactome. Nature Methods. 2008;7:801–805. doi: 10.1038/nmeth.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goehler H, Lalowski M, Stelzl U, Waelter S, Stroedicke M, Worm U, Droege A, Lindenberg KS, Knoblich M, Haenig C, et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol. Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 12.Kaltenbach LS, Romero E, Becklin RR, Chettier R, Bell R, Phansalkar A, Strand A, Torcassi C, Savage J, Hurlburt A, et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 14.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nature Rev. Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 15.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae. genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 17.Hoon S, Smith AM, Wallace IM, Suresh S, Miranda M, Fung E, Proctor M, Shokat KM, Zhang C, Davis RW, et al. An integrated platform of genomic assays reveals small-molecule bioactivities. Nat. Chem. Biol. 2008;4:498–506. doi: 10.1038/nchembio.100. [DOI] [PubMed] [Google Scholar]
- 18.Shao J, Diamond MI. Polyglutamine diseases: emerging concepts in pathogenesis and therapy. Hum. Mol. Genet. 2007;16(Spec No. 2):R115–R123. doi: 10.1093/hmg/ddm213. [DOI] [PubMed] [Google Scholar]
- 19.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 20.Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaefer MH, Wanker EE, Andrade-Navarro MA. Evolution and function of CAG/polyglutamine repeats in protein-protein interaction networks. Nucleic Acids Res. 2012;40:4273–4287. doi: 10.1093/nar/gks011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Worseck J, Grossmann A, Weimann M, Hegele A, Stelzl U. In: Two-Hybrid Technologies. Suter B, editor. New York, NY: Humana Press; 2011. [Google Scholar]
- 23.Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P, et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 25.Fontaine JF, Suter B, Andrade-Navarro MA. QiSampler: evaluation of scoring schemes for high-throughput datasets using a repetitive sampling strategy on gold standards. BMC Res. Notes. 2011;4:57. doi: 10.1186/1756-0500-4-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaefer MH, Fontaine JF, Vinayagam A, Porras P, Wanker EE, Andrade-Navarro MA. HIPPIE: Integrating protein interaction networks with experiment based quality scores. PloS One. 2012;7:e31826. doi: 10.1371/journal.pone.0031826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2010;39:D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pentchev K, Ono K, Herwig R, Ideker T, Kamburov A. Evidence mining and novelty assessment of protein-protein interactions with the ConsensusPathDB plugin for Cytoscape. Bioinformatics. 2010;26:2796–2797. doi: 10.1093/bioinformatics/btq522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanai A, Huang K, Kang R, Singaraja RR, Arstikaitis P, Gan L, Orban PC, Mullard A, Cowan CM, Raymond LA, et al. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat. Neurosci. 2006;9:824–831. doi: 10.1038/nn1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palidwor GA, Shcherbinin S, Huska MR, Rasko T, Stelzl U, Arumughan A, Foulle R, Porras P, Sanchez-Pulido L, Wanker EE, et al. Detection of alpha-rod protein repeats using a neural network and application to huntingtin. PLoS Comput. Biol. 2009;5:e1000304. doi: 10.1371/journal.pcbi.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwiatkowski AV, Gertler FB, Loureiro JJ. Function and regulation of Ena/VASP proteins. Trends Cell Biol. 2003;13:386–392. doi: 10.1016/s0962-8924(03)00130-2. [DOI] [PubMed] [Google Scholar]
- 32.Xu Y, Chen X, Li Y. Ercc6l, a gene of SNF2 family, may play a role in the teratogenic action of alcohol. Toxicol. Lett. 2005;157:233–239. doi: 10.1016/j.toxlet.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Karvonen U, Jaaskelainen T, Rytinki M, Kaikkonen S, Palvimo JJ. ZNF451 is a novel PML body- and SUMO-associated transcriptional coregulator. J. Mol. Biol. 2008;382:585–600. doi: 10.1016/j.jmb.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 34.Munsie L, Caron N, Atwal RS, Marsden I, Wild EJ, Bamburg JR, Tabrizi SJ, Truant R. Mutant huntingtin causes defective actin remodeling during stress: defining a new role for transglutaminase 2 in neurodegenerative disease. Hum. Mol. Genet. 2011;20:1937–1951. doi: 10.1093/hmg/ddr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME. Huntingtin interacts with a family of WW domain proteins. Hum. Mol. Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 36.Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, Illes K, Lukacsovich T, Zhu YZ, Cattaneo E, et al. SUMO modification of Huntingtin and Huntington's disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 37.Tsuda H, Jafar-Nejad H, Patel AJ, Sun Y, Chen HK, Rose MF, Venken KJ, Botas J, Orr HT, Bellen HJ, et al. The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/Senseless proteins. Cell. 2005;122:633–644. doi: 10.1016/j.cell.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 38.Chen HK, Fernandez-Funez P, Acevedo SF, Lam YC, Kaytor MD, Fernandez MH, Aitken A, Skoulakis EM, Orr HT, Botas J, et al. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113:457–468. doi: 10.1016/s0092-8674(03)00349-0. [DOI] [PubMed] [Google Scholar]
- 39.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho RJ, Fromont-Racine M, Wodicka L, Feierbach B, Stearns T, Legrain P, Lockhart DJ, Davis RW. Parallel analysis of genetic selections using whole genome oligonucleotide arrays. Proc. Natl Acad. Sci. USA. 1998;95:3752–3757. doi: 10.1073/pnas.95.7.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 42.Huang H, Jedynak BM, Bader JS. Where have all the interactions gone? Estimating the coverage of two-hybrid protein interaction maps. PLoS Comput. Biol. 2007;3:e214. doi: 10.1371/journal.pcbi.0030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart–biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rasche A, Herwig R. ARH: predicting splice variants from genome-wide data with modified entropy. Bioinformatics. 26:84–90. doi: 10.1093/bioinformatics/btp626. [DOI] [PubMed] [Google Scholar]
- 46.Dabney A, Storey J, Warnes G. q-value: Q-value estimation for false discovery rate control. 2006 R package version 1. [Google Scholar]
- 47.Storey J, Taylor J, Siegmund D. Strong control, conservative point estimation and simultaneous conservative consistency of false discovery rates: a unified approach. J. R. Stat. Soc. Series B. 2004:187–205. [Google Scholar]
- 48.Rajagopala SV, Hughes KT, Uetz P. Benchmarking yeast two-hybrid systems using the interactions of bacterial motility proteins. Proteomics. 2009;9:5296–5302. doi: 10.1002/pmic.200900282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang H, Bader JS. Precision and recall estimates for two-hybrid screens. Bioinformatics. 2009;25:372–378. doi: 10.1093/bioinformatics/btn640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vidalain PO, Boxem M, Ge H, Li S, Vidal M. Increasing specificity in high-throughput yeast two-hybrid experiments. Methods. 2004;32:363–370. doi: 10.1016/j.ymeth.2003.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.