Abstract
Protein ubiquitination plays an important role in activating the DNA damage response and maintaining genomic stability. In response to DNA double-strand breaks (DSBs), a ubiquitination cascade occurs at DNA lesions. Here, we show that checkpoint with Forkhead-associated (FHA) and RING finger domain protein (CHFR), an E3 ubiquitin ligase, is recruited to DSBs by poly(ADP-ribose) (PAR). At DSBs, CHFR regulates the first wave of protein ubiquitination. Moreover, CHFR ubiquitinates PAR polymerase 1 (PARP1) and regulates chromatin-associated PARP1 in vivo. Thus, these results demonstrate that CHFR is an important E3 ligase in the early stage of the DNA damage response, which mediates the crosstalk between ubiquitination and poly-ADP-ribosylation.
INTRODUCTION
Cells encounter numerous environmental and internal hazards that cause DNA lesions, such as DNA double-strand breaks (DSBs). Through evolution, cells have developed sophisticated cell cycle checkpoint system and DNA damage repair machineries to repair lesions and to maintain genomic stability (1,2). During these cellular events, protein post-translational modifications, including ubiquitination, phosphorylation, sumoylation, methylation, acetylation and ADP-ribosylation, play important roles in mediating DNA damage repair (2–17).
Among these modifications, a ubiquitination cascade mediated by a group of E2 and E3 enzymes occurs at DSBs. One major E3 ligase that controls this ubiquitination cascade is RNF8. In response to DSBs, RNF8 recognizes phospho-MDC1, a functional partner of γH2AX, and works together with Ubc13 to catalyze histone ubiquitination at DSBs (18–26). It is also reported that RNF8 associates with other E2 conjugases, such as UbcH5, which may be responsible for the displacement of KU80 at DNA damage sites (27). The RNF8-dependent ubiquitin signals recruit other E3 ligases including RNF168, RAD18 and HERC2, to DNA damage sites for the amplification of the ubiquitin cascade (4,28–38). The amplified ubiquitination at DSBs is important for recruiting DNA damage repair proteins such as BRCA1 and 53BP1 to DNA damage sites and for fulfilling their repair functions (19–22,26,31).
In addition to RNF8, another E3 ligase CHFR also participates in the DNA damage response initiation (39). Like RNF8, CHFR contains Forkhead-associated (FHA) domain and RING domain (40). Particularly, the RING domain of CHFR is interchangeable with the RING domain of RNF8 (41), suggesting that CHFR and RNF8 share the same E2 partners and have overlapping functions during the DNA damage response. Like the RING domain of RNF8, the RING domain of CHFR works together either with Ubc13 to catalyze K63-linked poly-ubiquitin chains or with UbcH5C to catalyze K48-linked poly-ubiquitin chains (42). Using genetic approach, we have shown that loss of CHFR and RNF8 additively induces genomic instability and suppresses the DNA damage response (39), which is in agreement with many previous reports that CHFR plays an important role in tumour suppression (40,43–47). In contrast with RNF8, CHFR contains a Cys-rich domain and a poly(ADP-ribose) (PAR)-binding zinc finger (PBZ) motif at the C-terminus that recognizes PAR (48,49). ADP-ribose is covalently conjugated at DNA damage sites as a branched polymer for DNA damage repair (48–50). It suggests that CHFR has a unique function during the DNA damage response distinct from that of RNF8. Here, we report that CHFR is recruited to DNA damage sites by PAR. CHFR ubiquitinates PAR polymerase 1 (PARP1), the major PAR polymerase and regulates chromatin-associated PARP1 in vivo. CHFR-dependent protein ubiquitination accounts for the first wave of protein ubiquitination at DNA damage sites.
MATERIALS AND METHODS
Generation and culture of mouse embryonic fibroblasts
The generation of wild-type, Rnf8−/−, Chfr−/− and double knockout (DKO) mouse embryonic fibroblasts (MEFs) was described (39,51). All the MEFs were maintained in the Dulbecco’s modified Eagle medium with 10% fetal bovine serum. For the ionizing radiation (IR) treatment, cells were irradiated with a JL Spepherd 137Cs radiation source with indicated doses. Following IR treatment, cells were maintained in the culture conditions for indicated time points. For the PARP1 inhibitor treatment, the cells were cultured in the Dulbecco’s modified Eagle medium with 10 µM PJ34 (EMD4Bioscience) for 1 h, then subjected to following experiments.
Plasmids and antibodies
CHFR, RNF8 and PARP1 cDNAs were subcloned into pEGFP-N1 vector. The deletion mutants of CHFR were generated by using the QuikChange site-directed mutagenesis kit (Stratagene). The primers were as follows: Δ-FHA-s: 5′-CGTCCTCCTGAGGAAGCGGGTTAAGAAGCAGACATGCC-3′, Δ-FHA-a: 5′-GGCATGTCTGCTTCTTAACCCGCTTCCTCAGGAGGACG-3′, Δ-RING-s: 5′-GACAAGATGGAGGAG-ACGGTGGAGCGGATCTGTAAA-3′, Δ-RING-a: 5′-TTTACAGATCCGCTCCACCGTCTCCTCCATCT-TGTC-3′, Δ-CRD-s: 5′-AGGCAGGCGGCGCAGCCTTTGCCAGTGGCCGTAACA-3′, Δ-CRD-a: 5′-TGTT-ACGGCCACTGGCAAAGGCTGCGCCGCCTGCCT-3′, Δ-PBZ-s: 5′-TGCCAGTGGCCGTAACATCCTGT-GAACAGACAAGGTTCAA-3′ and Δ-PBZ-a: 5′-TTGAACCTTGTCTGTTCACAGGATGTTACGGCCACTGG-CA-3′. siRNA for mouse Parp1: 5′-AAGCCCCCACUCCUGAACAACUU-3′. siRNA for human Parp1: 5′-AACCCCAAAGGAATTCCGAGAUU-3′.
Rabbit anti-CHFR antibody was raised against the RING domain of CHFR (residue 259–488) as described before (51). Monoclonal and polyclonal anti-mouse γH2AX antibodies, monoclonal anti-ubiquitin (FK2) antibody, polyclonal anti-histone H4, monoclonal and polyclonal anti-myc and anti-glutathione s-transferase (GST) antibodies, monoclonal anti-GAPDH antibodies were purchased from Upstate. Monoclonal anti-PAR antibody was purchased from Genetex. Rabbit monoclonal anti-PARP1 (46D11) antibody was purchased from Cell Signaling Technology. Human anti-K48 and anti-K63 poly-ubiquitin antibodies were purchased from Genentech. We performed cell transfection and immunoblotting using standard protocols.
Laser microirradiation, immunofluorescence staining and microscope image acquisition
For laser microirradiation, cells were grown on 35-mm glass bottom dishes (MatTek Corporation). Laser microirradiation was performed on OLYMPUS IX71 inverted fluorescence microscope with a Micropoint® Laser Illumination and Ablation System (Photonic Instruments). The laser output was set to 40%, which can reproducibly give a focused γH2AX stripe. For time-lapse microscopic analysis, cells were first transfected with corresponding plasmids. Then, green fluorescent protein (GFP) positive cells were subjected to microirradiation. The GPF strips were recorded at indicated time points and then analysed with Image J software. For the time course analysis of laser microirradiation, samples were subjected to continuous microirradiation along certain paths within the indicated time interval. Then, the samples were subjected to immunofluorescence staining with indicated antibodies. For immunofluorescence staining, cells were fixed in 3% paraformaldehyde for 10 min and permeabilized with 0.5% Triton X-100 in phosphate-buffered saline (PBS) for 5 min at room temperature. Samples were blocked with 8% goat serum and then incubated with the primary antibody for 1 h. Samples were washed for three times and incubated with the secondary antibody for 30 min. The coverslips were mounted onto glass slides and visualized with OLYMPUS IX71 inverted fluorescence microscope. All the images were acquired with cellSens standard (Version 1.3) software under OLYMPUS IX71 inverted fluorescence microscope equipped with a UPlanSApo 60×/1.35 oil immersion objective at room temperature. Identical contrast and brightness adjustments were used on images for all given experiments.
PARP1 auto-PARylation and in vitro and in vivo ubiquitination assay
To auto-PARylate His-PARP1, 100 µg purified His-PARP1 protein binding on the Ni Sepharose (GE healthcare) beads was incubated for 30 min at 30°C in the PARylation buffer (100 mM Tris–HCl (pH 7.6), 10 mM MgCl2, 50 µg DNA octamer (5′-GGAATTCC-3′) and 10 mM DTT), with or without 4 mM NAD+ (CALBIOCHEM). Then, the beads were washed for three times with PBS.
For in vitro ubiquitination assay, 1 µg HA-Ub, 200 ng E1, 300 ng UbcH5C or Ubc13/Uev1a (all from Boston Biochem), 500 ng GST-CHFR or other indicated mutant proteins purified from sf9 cells, 1 µg His-PARP1 or PARylated His-PARP1 binding on Ni Sepharose beads were incubated in the reaction buffer (50 mM Tris–HCl pH 7.5, 5 mM MgCl2, 100 mM NaCl and 0.5 mM DTT) at 30°C for 30 min. Then, the beads were thoroughly washed with ice-cold PBS and boiled with sodium dodecyl sulphate (SDS) sample buffer. Ubiquitinated proteins were resolved on 4–15% SDS–polyacrylamide gels (TGX™, BioRad).
For in vivo ubiquitination assay, 5 µg of myc-CHFR or other indicated mutant plasmids were transfected into HCT116 cells with Lipo2000 (Invitrogen). Twenty-four hours after transfection, the cells were treated with 10 Gy of IR and replaced with fresh media in the presence of dimethyl sulfoxide (DMSO) or 10 µM MG132 for 30 min. Then, the cells were lysed with NETN300 (20 mM Tris–HCl, pH 8.0, 300 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA) and 0.5% NP-40) on ice for 10 min. Equal amount of proteins from the cell lysates were incubated with protein A beads and anti-PARP1 antibody for 2 h at 4°C. Then, the beads were thoroughly washed with ice-cold PBS and boiled with SDS sample buffer. Proteins were resolved on 4–15% SDS-polyacrylamide gels (TGX™, BioRad) and analysed by immunoblotting with indicated antibodies.
Chromatin fraction
Cells were harvested at indicated time points after 10 Gy of IR treatment and washed twice with PBS. Cell pellets were subsequently resuspended in the NETN buffer (20 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA and 0.5% NP-40) and incubated on ice for 10 min. Thereafter, insoluble fraction was recovered and resuspended in 0.2 M HCl. The soluble fraction was neutralized with 1 M Tris–HCl pH 8.0 for further analysis.
Alkali comet assays
Single-cell gel electrophoretic comet assays were performed under alkaline conditions. Briefly, 24 h after electroporation of indicated plasmids or transfection with indicated siRNA, MEFs were irradiated with or without 5 Gy of IR and recovered in normal culture medium for indicated time at 37°C. Cells were collected and rinsed twice with ice-cold PBS; 2 × 104/ml cells were combined with 1% LMAgarose at 40°C at the ratio of 1:3 (v/v) and immediately pipetted onto slides. For cellular lysis, the slides were immersed in the alkali lysis solution (1.2 M NaCl, 100 mM EDTA, 0.1% SDS and 0.26 M NaOH, pH > 13) overnight at 4°C. Then, the slides were subjected to electrophoresis at 15 V for 25 min (0.6 V/cm) and stained in 10 µg/ml propidium iodide for 20 min. All images were taken with a fluorescence microscope and analysed by Comet Assay IV software.
Colony formation assay
One thousand cells were plated in the wells of a 6-well plate immediately after radiation. After incubation for 10 days, the surviving cell fractions were calculated by comparing the numbers of colonies formed in the irradiated cultures with those in untreated control.
GST pulldown assay
Two micrograms of GST or GST-CHFR proteins expressed and purified from Escherichia coli were incubated with 10 µg His-PARP1 or auto-PARylated His-PARP1 protein with Glutathione Sepharose 4B beads (GE Healthcare) at 4°C for 2 h with rotation. Then, the beads were thoroughly washed in ice-cold PBS for five times and then boiled in the SDS sample buffer for further analysis.
RESULTS
CHFR is rapidly recruited to laser-induced DNA damage sites
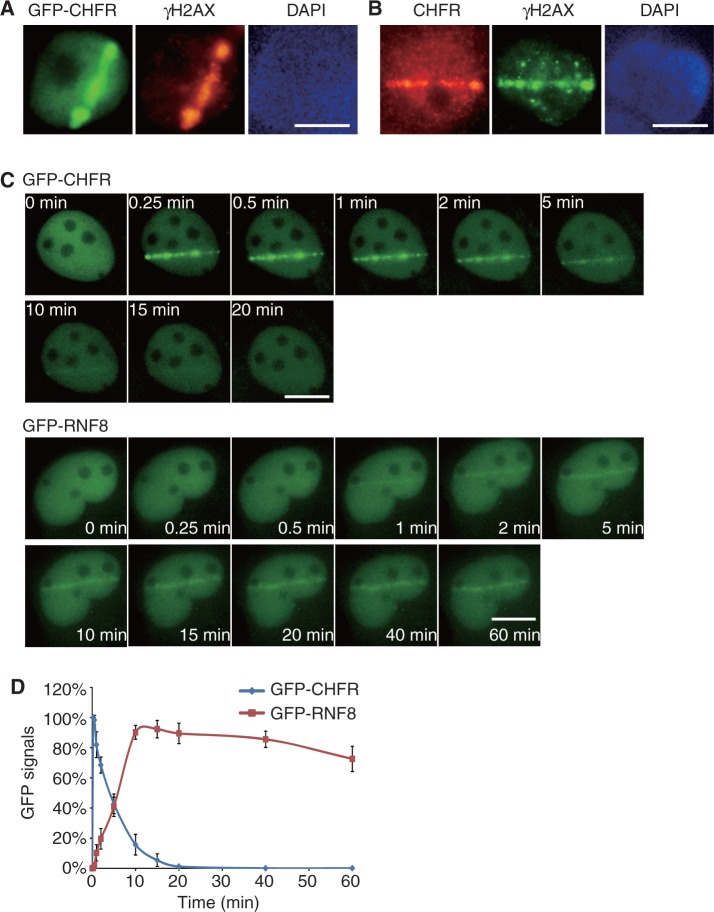
To search for the role of CHFR in the DNA damage response, we examined whether CHFR could be recruited to DNA damage sites. We engineered an EGFP tag at the C-terminus of CHFR and monitored CHFR’s localization in response to laser-induced DNA damage. Interestingly, CHFR is instantly recruited to DNA damage sites and colocalized with γH2AX, a surrogate marker of DNA damage sites (Figure 1A). Moreover, endogenous CHFR also relocated to DNA damage sites, suggesting that CHFR directly participates in the DNA damage response (Figure 1B and Supplementary Figure S1). We found that the recruitment of CHFR to DNA damage sites was very unique and different from other reported E3 ubiquitin ligases (21,31). We carefully measured the kinetics of the relocation of CHFR. As shown in Figure 1C, CHFR was recruited to lesions within a few seconds following DNA damage and was dropped off from DNA damage sites in 10 min. We checked multiple cells with different GFP-CHFR expression levels and found similar kinetics of recruitment to DNA damage sites (Supplementary Figure S2), which excludes the possibility that protein expression level affects the kinetics of recruitment. We also checked the endogenous CHFR kinetics after laser microirradiation and found similar kinetics to the exogenous over-expressed protein (Supplementary Figure S1B), suggesting that CHFR can be quickly recruited to DNA damage sites to participate in the DNA damage response. In contrast to the fast recruitment of CHFR to DNA damage sites, consistent with other reports, RNF8 started to be recruited to lesions 1 min after DNA damage, gradually accumulated at DNA damage sites in the first 15 min and was kept at DNA damage sites in a steady state at least for 1 h (Figure 1C and D) (21,31).
Figure 1.
CHFR is recruited to DNA damage sites. (A) Laser microirradiation induces recruitment of GFP-CHFR to DNA damage sites in U2OS cells. (B) Endogenous CHFR is recruited to DNA damage sites. U2OS cells were fixed immediately following laser microirradiation and subjected to immunofluorescence staining with polyclonal anti-CHFR and monoclonal anti-γH2AX antibodies. (C) Dynamic recruitment of GFP-CHFR and GFP-RNF8 to DNA damage sites in U2OS cells following laser microirradiation. (D) Different kinetics of the recruitment of CHFR and RNF8 to DNA damage sites are summarized. The highest GFP intensity was calculated as 100% in each cell, and kinetics of the recruitment were plotted. Data were analysed from 20 cells in each experiment. Data were presented as mean ± SD. Bars, 10 µm.
CHFR regulates the first wave of ubiquitination events at DNA damage sites
The difference in kinetics between CHFR and RNF8 at DNA damage sites indicates that these two E3 ligases may regulate ubiquitination events at different stages of DNA damage response. Using genetic tools that we have generated (39), we next examined the kinetics of ubiquitin conjugation at DNA damage sites in wild-type, Chfr−/−, Rnf8−/− and DKO MEFs. In wild-type MEFs, DNA damage-induced ubiquitination could be clearly visualized within a couple of minutes following DNA damage (Figure 2). However, this cellular process was significantly delayed in Chfr−/− MEFs; whereas γH2AX at DNA damage sites was not affected in the absence of CHFR. In contrast, in Rnf8−/− MEFs, although ubiquitin was still quickly conjugated at DNA damage sites, the intensity of ubiquitin signals was quickly reduced to undetectable levels. In the absence of CHFR and RNF8, we could not detect protein ubiquitination at DNA damage sites. Thus, these results indicate that CHFR mainly regulates early ubiquitination events during the DNA damage response, which accounts for the first wave of protein ubiquitination at DNA damage sites.
Figure 2.
CHFR regulates the first wave of protein ubiquitination at DNA damage sites. Wild-type (WT), Chfr−/−, Rnf8−/− or DKO MEFs were treated with laser microirradiation and then fixed at the indicated time points. The ubiquitin (detected by FK2 antibody) and γH2AX at DNA damage sites were then examined by immunofluorescence staining. (A) Representative cells at different time point in each MEF were shown. (B) Cells with colocalized ubiquitin and γH2AX signals were counted as Ub positive cells. The percentage (mean ± SD) of Ub positive cells at different time point was summarized from 100 cells. Bar, 10 µm.
PAR mediates the recruitment of CHFR to DNA damage sites
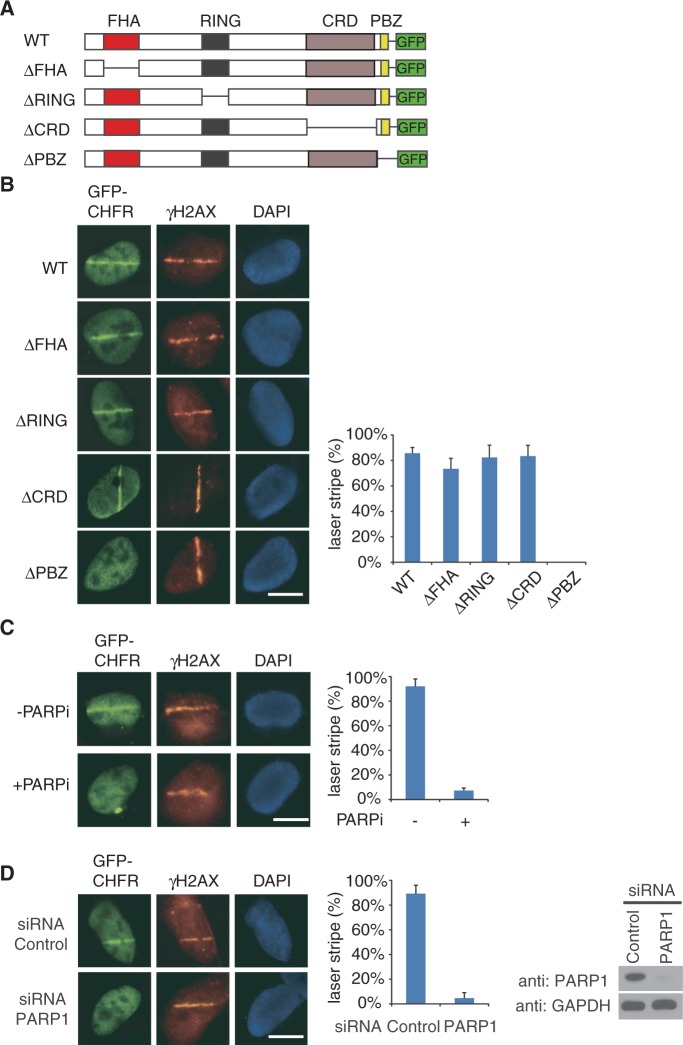
Next, we examined the mechanism by which CHFR is targeted to DNA damage sites. Interestingly, CHFR can be recruited to DNA damage sites in the absence of H2AX and MDC1 (Supplementary Figure S3), suggesting that other mechanisms instead of γH2AX target CHFR to DNA damage sites. As CHFR contains four different domains, namely the N-terminal FHA domain, the RING domain, the Cys-rich domain and the C-terminal PBZ motif (Figure 3A), we deleted each domain of CHFR and found that loss of PBZ motif but not other domains clearly abolished the relocation of CHFR to DNA damage sites (Figure 3B). As the PBZ motif of CHFR recognizes PAR (48), it is likely that PAR mediates the relocation of CHFR to DNA damage sites. PAR is mainly synthesized by PARP1 and is covalently conjugated to PARP1 itself at DNA lesions immediately following DNA damage (10,52). PARP inhibitor PJ34 can efficiently suppress PAR synthesis at DNA damage sites (53–55). Thus, with PJ34 treatment, CHFR failed to be recruited to DNA damage sites (Figure 3C). Moreover, we depleted PARP1 in U2OS cells by siRNA knockdown, the recruitment of CHFR to DNA damage sites was significantly suppressed (Figure 3D). Collectively, these results demonstrate that PAR mediates the relocation of CHFR to DNA damage sites.
Figure 3.
PAR mediates the recruitment of CHFR to DNA damage sites. (A) Domain architecture of GFP-tagged wild-type CHFR (WT), FHA domain deletion mutant (ΔFHA), RING domain deletion mutant (ΔRING), Cys-rich domain deletion mutant (ΔCRD) and PBZ motif deletion mutant (ΔPBZ). (B) The PBZ motif is required for the relocation of CHFR to DNA damage sites. U2OS cells expressing indicated plasmids were treated with laser microirradiation and immunostaining with anti-γH2AX antibody. (C) PARP1 inhibitor (PJ34) suppresses the relocation of CHFR to DNA damage sites. U2OS cells expressing GFP-CHFR were treated with laser microirradiation in the absence (−PARPi) or presence (+PARPi) of 10 µM PJ34, following immunostaining with anti-γH2AX antibody. (D) Knockdown of PARP1 significantly suppresses the recruitment of CHFR to DNA damage site. U2OS cells expressing GFP-CHFR were transfected with control or PARP1 siRNA. Then, cells were treated with laser microirradiation and immunostaining with anti-γH2AX antibody. PARP1 expression following siRNA treatment is shown in the right. Histograms in B, C and D summarize the percentage (mean ± SD) of cells with GFP laser stripes colocalized with γH2AX following laser-induced DNA damage. Data were analysed from 100 cells in each experiment from three independent experiments. Bar, 10 µm.
CHFR ubiquitinates PARP1 and regulates the chromatin-associated PARP1 following DNA damage
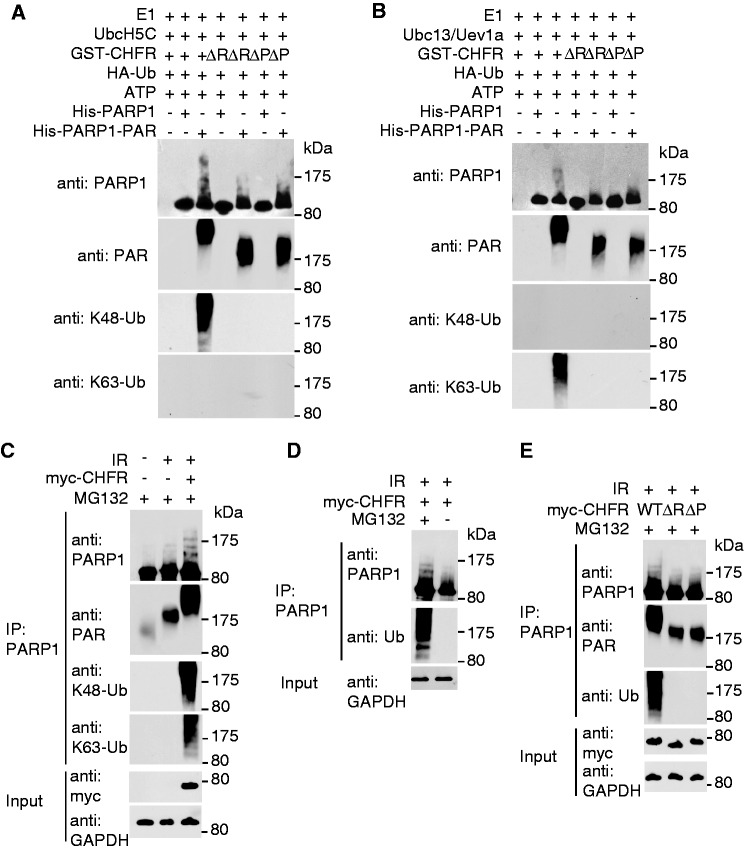
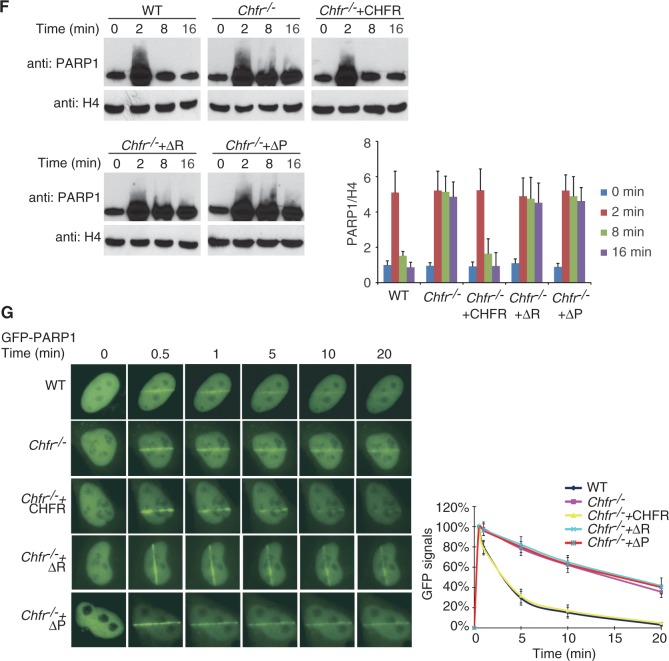
As we reported previously, histones are substrates of CHFR and RNF8 during DNA damage (39). We wondered whether CHFR has other substrates during its recruitment to DNA damage sites. As PARP1 itself is a major target of PARylation after DNA damage (52), we hypothesized that CHFR may ubiquitinate PARP1 through recognizing PAR on PARP1. We first purified His-tagged PARP1 or in vitro PARylated His-tagged PARP1 and performed in vitro ubiquitination assays. As shown in Figure 4A and B, CHFR can ubiquitinate PARylated PARP1 but not the unmodified PARP1 with either UbcH5C or Ubc13/Uev1a as the E2 enzyme. These two different E2 enzymes catalyze K48-linked and K63-linked poly-ubiquitin chain on PARylated PARP1, respectively. Moreover, the RING domain deletion (ΔR) or PBZ motif deletion (ΔP) mutants cannot ubiquitinate PARP1 or PARylated PARP1 in vitro, suggesting that both the E3 ligase of CHFR and the interaction between CHFR and PAR are important for the ubiquitination of PARP1. Consistently, recombinant CHFR directly bound PARylated PARP1 but not unmodified PARP1 (Supplementary Figure S4). To examine whether CHFR regulates PARP1 ubiquitination in vivo, we expressed CHFR, the RING domain deletion or the PBZ motif deletion mutants in HCT116 cells, which do not express endogenous CHFR (47). PARP1 was significantly PARylated after IR and large amounts of ubiquitinated PARP1 accumulated in cells expressing wild-type CHFR in the presence of MG132 (Figure 4C and D). Neither the RING domain deletion mutant nor the PBZ motif deletion mutant induced PARP1 ubiquitination under the same condition, indicating that both the E3 ligase of CHFR and the interaction between CHFR and PAR are important for the ubiquitination of PARP1 in vivo (Figure 4E). Moreover, the poly-ubiquitin chain on PARP1 could be recognized by both anti-K48 and K63-linked poly-ubiquitin chain antibodies, suggesting that CHFR mediates a mixed poly-ubiquitin chain linkage on PARP1. With MG132 treatment, ubiquitinated PARP1 was significantly accumulated (Figure 4D), suggesting that the ubiquitination of PARP1 is likely involved in protein degradation. Consistently, we found that following DNA damage, PARP1 quickly dissociated from the chromatin in the wild-type cells (Figure 4F). However, in the Chfr−/− cells, the dissociation of PARP1 from the chromatin was significantly delayed. Moreover, when the Chfr−/− cells were reconstituted with wild-type or mutant CHFR, only wild-type CHFR but neither the RING domain deletion mutant nor the PBZ motif deletion mutant facilitated the fast displacement of PARP1 from chromatin in response to DNA damage. We also examined the kinetics of the recruitment of PARP1 to DNA damage sites in wild-type and in Chfr−/− MEFs. As shown in Figure 4G, PARP1 in wild-type MEFs was quickly displaced from DNA damage sites. However, in Chfr−/− MEFs, the retention of PARP1 at DNA damage sites was significantly prolonged. Moreover, when Chfr−/− cells were reconstituted with wild-type or mutant CHFR, only wily type CHFR but neither the RING domain deletion mutant nor the PBZ motif deletion mutant restored the quick PARP1 displacement from DNA damage sites. To confirm the results, the kinetics of PARP1 at DNA damage sites was examined in HCT116 cells. Again, the displacement of PARP1 from DNA damage sites was significantly faster in HCT116 cells reconstituted with wild-type CHFR than that in mock-transfected cells or cells reconstituted with the RING domain or PBZ motif deletion mutants (Supplementary Figure S5). Taken together, our results demonstrate that both the E3 ligase activity and PAR-binding ability of CHFR are important for the removal of PARP1 at DNA damage sites.
Figure 4.
CHFR ubiquitinates PARylated PARP1 and regulates the chromatin-associated PARP1 following DNA damage. (A and B) CHFR ubiquitinates PARylated PARP1 in vitro. In vitro ubquitination assay was performed using His-PARP1 or PARylated His-PARP1 as the substrates. Ubiquitinated proteins were examined by SDS–PAGE and western blot by using anti-PARP1, anti-PAR, anti-K48 and anti-K63 poly-ubiquitin chain antibodies. (A) UbcH5c was used as E2 conjugase. (B) Ubc13/Uev1a was used as E2 conjugase. (C) CHFR regulates PARP1 ubiquitination in vivo. HCT116 cells expressing myc-CHFR or mock plasmids were treated with 10 Gy of IR in the presence of 10 µM MG132. PARP1 was immunoprecipitated from the cell lysates and subjected to SDS–PAGE and immunoblotting with anti-PARP1, anti-PAR, anti-K48 and anti-K63 poly-ubiquitin chain antibodies. (D) HCT116 cells expressing myc-CHFR were treated with 10 Gy of IR in the absence or presence of MG132. PARP1 was immunoprecipitated from the cell lysates and analysed by SDS–PAGE and immunoblotting with anti-PARP1 and anti-Ub (FK2) antibodies. (E) HCT116 cells expressing myc-tagged wild-type CHFR, the RING domain or PBZ motif deletion mutants were treated with 10 Gy of IR in the presence of MG132. PARP1 status was examined by indicated antibodies. (F) The chromatin retention of PARP1 is regulated by the E3 ligase activity and PAR-binding ability of CHFR. The chromatin-associated PARP1 was examined at the indicated time points following 10 Gy of IR treatments. The displacement of PARP1 from the chromatin was restored in Chfr−/− MEFs reconstituted with wild-type CHFR but not the RING domain or PBZ motif deletion mutants. Histone H4 was blotted as input control for chromatin-associated proteins. The relative amount of PARP1 in the chromatin fraction was quantitatively analysed. The data were obtained from three independent experiments and bar stands for SD. (G) The retention of GFP-PARP1 at DNA damage sites in wild-type (WT), Chfr−/− or Chfr−/− MEF reconstituted with wild-type CHFR, the RING domain deletion mutant or the PBZ motif deletion mutant was examined. The highest GFP intensity was calculated as 100% in each cell, and kinetics of the recruitment were plotted. Data were analysed from 20 cells in each experiment. Data were presented as mean ± SD. ΔR, RING domain deletion mutant of CHFR; ΔP, PBZ motif deletion mutant of CHFR. Bar, 10 µm.
CHFR participates in DNA damage repair
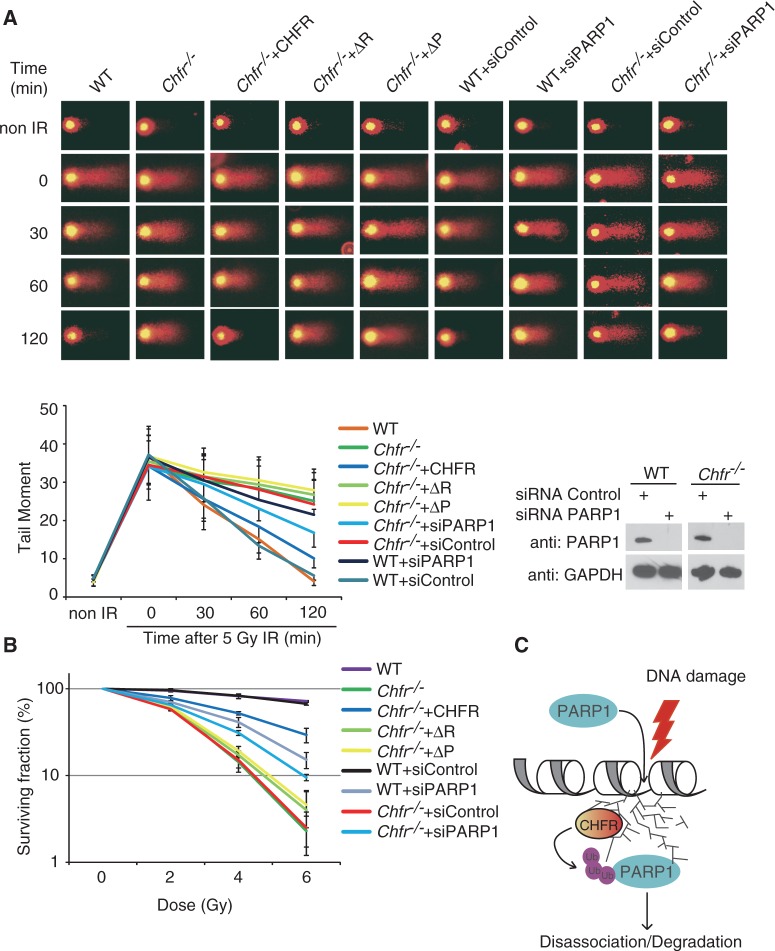
Removal of PAR from DNA damage sites is important for the next step of the DNA damage repair process (56–60). Since PARP1 is the major enzyme to synthesize PAR at DNA damage sites, the eviction of PARP1 is likely to be critical for the next step of DNA damage repair. To study the role of CHFR-dependent PARP1 eviction in the DNA damage response, we measured DNA damage repair kinetics in the Chfr−/− and wild-type MEFs by comet assay under alkaline condition. As shown in Figure 5A, within 2 h following low dose IR treatment (5 Gy), most DNA damage sites, including single- and double-strand breaks, as well as alkali-labile sites, were repaired in wild-type MEFs but not in Chfr−/− MEFs. Moreover, Chfr−/− MEFs reconstituted with wild-type CHFR, but not the RING domain deletion mutant nor the PBZ motif deletion mutant, rescued the DNA damage repair defects (Figure 5A), suggesting that both the E3 ligase activity of CHFR and the PAR-binding of CHFR are important for the DNA damage repair. We also depleted PARP1 expression with siRNA in wild-type and Chfr−/− MEFs. In wild-type MEFs, PARP1 depletion caused mild DNA damage repair defects in control MEFs, suggesting that PARP1 itself is important for DNA damage repair. However, in Chfr−/− MEFs, depletion of PARP1 partially rescued the DNA damage repair defect (Figure 5A), suggesting that the eviction of PARP1 from the chromatin is important for the DNA repair process, and CHFR-dependent PARP1 ubiquitination facilitates the dissociation of PARP1 from the chromatin following DNA damage. We confirmed these results using a long-term cell viability assay. Again, we found that both the E3 ligase activity of CHFR and the PAR-binding of CHFR are important for DNA damage repair. Moreover, depletion of PARP1 partially rescued the DNA damage repair defect in Chfr−/− MEFs (Figure 5B).
Figure 5.
CHFR protects against DNA damage. (A) Representative microphotographs of comet assay at indicated time points following 5 Gy of IR treatment. Cells with indicated genotypes were subjected to alkali comet assays. Tail moments were summarized from three independent experiments with at least 50 cells in single time point per sample. Data were presented as mean ± SD. PARP1’s expression in WT and Chfr−/− MEFs after siRNA treatment was shown in the lower right panel. (B) CHFR-deficient MEFs are sensitive to IR. Cells with indicated genotypes were subjected to cell survival assay. Data were presented as mean ± SD from three independent experiments. (C) A model of CHFR recruited by PAR and ubiquitinating PARP1 at DNA damage sites. ΔR, RING domain deletion mutant of CHFR; ΔP, PBZ motif deletion mutant of CHFR.
DISCUSSION
In this study, we demonstrated that CHFR is one of the earliest E3 ligases recruited to DNA damage sites. This cellular process is mediated by the interaction between the PBZ motif of CHFR and PAR at DNA damage sites. The CHFR-dependent protein ubiquitination represents the first wave of protein ubiquitination at DNA damage sites. Here, we show evidence that CHFR can ubiquitinate PARylated PARP1, which might be important for its displacement from DNA damage sites (Figure 5C). Following DNA damage, massive protein PARylation occurs at DNA damage sites catalyzed mainly by PARP1 and the major substrate of protein PARylation is PARP1 itself (52,61–63), which is important for chromatin relaxation (64). Recent studies suggest that PAR at DNA damage sites recruit DNA damage repair proteins to DNA lesions to fulfill their repair function (65–72). Meanwhile, the hyper-activated PARP1 may deplete intracellular pools of NAD+, resulting in impaired ATP production and genomic instability (73–76). Thus, the activity of PARP1 during the DNA damage response needs to be tightly controlled. Poly(ADP-ribose) glycohydrolase (PARG) is recruited to DNA damage sites and plays a critical role in hydrolyzing PAR and recycling PARP1 during DNA damage response (67,77,78). Thus, PARP1 has to be removed from DNA damage sites, or degraded to prevent the recycling of PARP1 through PARG. This process is to avoid the continuous activation of PARP1 during DNA damage response. Here, our results suggest that CHFR-dependent ubiquitination is important for the eviction of PARP1 from DNA damage sites for proteasomal degradation. This is one of the mechanisms by which cells control the activated PARP1 in response to DNA damage.
In addition to PARP1, CHFR may also ubiquitinate other substrates, such as nucleosomal histones (39). Interestingly, histones can be also PARylated by PARP1 in response to DNA damage (61,62,64,79). Thus, it is likely that CHFR ubiquitinates PARylated histones at DNA damage sites, which facilitates histone eviction at DNA damage sites. It would allow other DNA damage repair proteins to access DNA lesions for the next step of DNA damage repair. Removal of PARylated histones might also promote the displacement of CHFR from DNA damage sites.
Using mouse genetic approaches, we have demonstrated that CHFR and RNF8 additively regulate the DNA damage response and maintain genomic stability (39). The RING domains of CHFR and RNF8 are interchangeable (41). Both CHFR and RNF8 ubiquitinate histones and may share other substrates. However, the molecular mechanism of recruitment of these two E3 ligases are different, which determines that CHFR reaches DNA damage sites earlier than RNF8. Thus, CHFR-dependent protein ubiquitination represent the first wave of protein ubiquitination at DNA damage sites. Loss of CHFR only delays protein ubiquitination but not abolish protein ubiquitination, suggesting that RNF8- and the RNF8-dependent ubiquitin cascade have a redundant role of CHFR at DNA damage sites. Thus, it is possible that PARP1 eviction from DNA damage sites may also be regulated by RNF8- and the RNF8-dependent ubiquitin cascade in the absence of CHFR, albeit in a delayed manner. However, following loss of both CHFR and RNF8, protein ubiquitination is completely abolished at DNA damage sites, which significantly suppresses the DNA damage response and induces genomic instability (39).
Interestingly, in human cancer cells, it is CHFR but not RNF8 that is often silenced (40,43,47). Although this selection mechanism is not clear, long-term loss of CHFR prolongs the retention of PARP1 at DNA damage sites, which may induce the accumulation of DNA lesions and facilitate tumourigenesis. PARP1 inhibitor treatment could antagonize the defects generated by the prolonged PARP1 at DNA damage sites. Thus, it is possible that PARP inhibitors could be used in chemoprevention to suppress CHFR-deficiency-induced tumourigenesis. During the preparation of this article, Kashima et al. (80) reported that CHFR interacts with and ubiquitinates unmodified PARP1 during mitosis, although the function of PARP1 in mitosis remains elusive. In our study, we found that CHFR only recognizes PAR instead of unmodified PARP1. Since PAR is massively synthesized at DNA damage sites, this interaction induces the relocation of CHFR to DNA damage sites and facilitates the removal of PARP1 from the chromatin through ubiquitination. This process is important for DNA damage repair. Moreover, distinct from the previous report, CHFR only ubiquitinates PARylated PARP1 but not unmodified PARP1. Since PARP1 is heavily PARylated in response to DNA damage, we found that CHFR induced ubiquitination of PARylated PARP1 following DNA damage. But lacking CHFR did not alter the expression of PARP1 under normal conditions (Supplementary Figure S6). Consistently, we found that recombinant CHFR only interacts with PARylated PARP1 but does not recognize unmodified PARP1 (Supplementary Figure S4).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplemental Figures 1–6.
FUNDING
The American Cancer Society [IRG-58-010-52 to Z.Y.]; the National Institute of Health (NIH) [GM098535 to Z.Y., CA132755 and CA130899 to X.Y.] and a Siteman Career Award in Breast Cancer Research (to Z.Y.). Recipient of the Era of Hope Scholar Award from the Department of Defense (to X.Y.). Funding for open access charge: NIH [R01CA130899].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Henry Kuang and Zhonghao Wang for proof reading.
REFERENCES
- 1.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 2.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 3.Bekker-Jensen S, Mailand N. The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett. 2011;585:2914–2919. doi: 10.1016/j.febslet.2011.05.056. [DOI] [PubMed] [Google Scholar]
- 4.Panier S, Durocher D. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair. 2009;8:436–443. doi: 10.1016/j.dnarep.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Al-Hakim A, Escribano-Diaz C, Landry MC, O’Donnell L, Panier S, Szilard RK, Durocher D. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair. 2010;9:1229–1240. doi: 10.1016/j.dnarep.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Messick TE, Greenberg RA. The ubiquitin landscape at DNA double-strand breaks. J. Cell Biol. 2009;187:319–326. doi: 10.1083/jcb.200908074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma T, Keller JA, Yu X. RNF8-dependent histone ubiquitination during DNA damage response and spermatogenesis. Acta Biochim. Biophys. Sin. 2011;43:339–345. doi: 10.1093/abbs/gmr016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiriet C, Hayes JJ. Chromatin in need of a fix: phosphorylation of H2AX connects chromatin to DNA repair. Mol. Cell. 2005;18:617–622. doi: 10.1016/j.molcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Müller S, Ledl A, Schmidt D. SUMO: a regulator of gene expression and genome integrity. Oncogene. 2004;23:1998–2008. doi: 10.1038/sj.onc.1207415. [DOI] [PubMed] [Google Scholar]
- 10.Lindahl T, Satoh MS, Poirier GG, Klungland A. Post-translational modification of poly (ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem. Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 11.Dou H, Huang C, Van Nguyen T, Lu LS, Yeh ETH. SUMOylation and de-SUMOylation in response to DNA damage. FEBS Lett. 2011;585:2891–2896. doi: 10.1016/j.febslet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Kim KI, Baek SH. SUMOylation code in cancer development and metastasis. Mol. Cells. 2006;22:247–253. [PubMed] [Google Scholar]
- 13.Lake AN, Bedford MT. Protein methylation and DNA repair. Mutat. Res. 2007;618:91–101. doi: 10.1016/j.mrfmmm.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Van Attikum H, Gasser SM. The histone code at DNA breaks: a guide to repair? Nat. Rev. Mol. Cell Biol. 2005;6:757–765. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- 15.Wurtele H, Verreault A. Histone post-translational modifications and the response to DNA double-strand breaks. Curr. Opin. Cell Biol. 2006;18:137–144. doi: 10.1016/j.ceb.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 16.Morris JR. More modifiers move on DNA damage. Cancer Res. 2010;70:3861–3863. doi: 10.1158/0008-5472.CAN-10-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lisby M, Rothstein R. DNA damage checkpoint and repair centers. Curr. Opin. Cell Biol. 2004;16:328–334. doi: 10.1016/j.ceb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Plans V, Scheper J, Soler M, Loukili N, Okano Y, Thomson TM. The RING finger protein RNF8 recruits UBC13 for lysine 63-based self polyubiquitylation. J. Cell. Biochem. 2006;97:572–582. doi: 10.1002/jcb.20587. [DOI] [PubMed] [Google Scholar]
- 19.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 22.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl Acad. Sci. USA. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marteijn JA, Bekker-Jensen S, Mailand N, Lans H, Schwertman P, Gourdin AM, Dantuma NP, Lukas J, Vermeulen W. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol. 2009;186:835–847. doi: 10.1083/jcb.200902150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J, Huen MS, Lu LY, Ye L, Dou Y, Ljungman M, Chen J, Yu X. Histone ubiquitination associates with BRCA1-dependent DNA damage response. Mol. Cell. Biol. 2009;29:849–860. doi: 10.1128/MCB.01302-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol. Cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 26.Sakasai R, Tibbetts R. RNF8-dependent and RNF8-independent regulation of 53BP1 in response to DNA damage. J. Biol. Chem. 2008;283:13549–13555. doi: 10.1074/jbc.M710197200. [DOI] [PubMed] [Google Scholar]
- 27.Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat. Struct. Mol. Biol. 2012;19:201–206. doi: 10.1038/nsmb.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C, Bartek J, Lukas J, Mailand N. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell Biol. 2010;12:80–86. doi: 10.1038/ncb2008. [DOI] [PubMed] [Google Scholar]
- 29.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 30.Stewart GS. Solving the RIDDLE of 53BP1 recruitment to sites of damage. Cell Cycle. 2009;8:1532–1538. doi: 10.4161/cc.8.10.8351. [DOI] [PubMed] [Google Scholar]
- 31.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 32.Kang TH, Lindsey-Boltz LA, Reardon JT, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc. Natl Acad. Sci. USA. 2010;107:4890–4895. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu W, Sato K, Koike A, Nishikawa H, Koizumi H, Venkitaraman AR, Ohta T. HERC2 is an E3 ligase that targets BRCA1 for degradation. Cancer Res. 2010;70:6384–6392. doi: 10.1158/0008-5472.CAN-10-1304. [DOI] [PubMed] [Google Scholar]
- 34.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 36.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 37.Zhang S, Chea J, Meng X, Zhou Y, Lee EY, Lee MY. PCNA is ubiquitinated by RNF8. Cell Cycle. 2008;7:3399–3404. doi: 10.4161/cc.7.21.6949. [DOI] [PubMed] [Google Scholar]
- 38.Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat. Cell Biol. 2009;11:592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu J, Chen Y, Lu LY, Wu Y, Paulsen MT, Ljungman M, Ferguson DO, Yu X. Chfr and RNF8 synergistically regulate ATM activation. Nat. Struct. Mol. Biol. 2011;18:761–768. doi: 10.1038/nsmb.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scolnick DM, Halazonetis TD. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature. 2000;406:430–435. doi: 10.1038/35019108. [DOI] [PubMed] [Google Scholar]
- 41.Huen MS, Huang J, Yuan J, Yamamoto M, Akira S, Ashley C, Xiao W, Chen J. Noncanonical E2 variant-independent function of UBC13 in promoting checkpoint protein assembly. Mol. Cell. Biol. 2008;28:6104–6112. doi: 10.1128/MCB.00987-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bothos J, Summers MK, Venere M, Scolnick DM, Halazonetis TD. The Chfr mitotic checkpoint protein functions with Ubc13-Mms2 to form Lys63-linked polyubiquitin chains. Oncogene. 2003;22:7101–7107. doi: 10.1038/sj.onc.1206831. [DOI] [PubMed] [Google Scholar]
- 43.Mizuno K, Osada H, Konishi H, Tatematsu Y, Yatabe Y, Mitsudomi T, Fujii Y, Takahashi T. Aberrant hypermethylation of the CHFR prophase checkpoint gene in human lung cancers. Oncogene. 2002;21:2328–2333. doi: 10.1038/sj.onc.1205402. [DOI] [PubMed] [Google Scholar]
- 44.Shibata Y, Haruki N, Kuwabara Y, Ishiguro H, Shinoda N, Sato A, Kimura M, Koyama H, Toyama T, Nishiwaki T, et al. Chfr expression is downregulated by CpG island hypermethylation in esophageal cancer. Carcinogenesis. 2002;23:1695–1699. doi: 10.1093/carcin/23.10.1695. [DOI] [PubMed] [Google Scholar]
- 45.Corn PG, Summers MK, Fogt F, Virmani AK, Gazdar AF, Halazonetis TD, El-Deiry WS. Frequent hypermethylation of the 5′ CpG island of the mitotic stress checkpoint gene Chfr in colorectal and non-small cell lung cancer. Carcinogenesis. 2003;24:47–51. doi: 10.1093/carcin/24.1.47. [DOI] [PubMed] [Google Scholar]
- 46.Mariatos G, Bothos J, Zacharatos P, Summers MK, Scolnick DM, Kittas C, Halazonetis TD, Gorgoulis VG. Inactivating mutations targeting the chfr mitotic checkpoint gene in human lung cancer. Cancer Res. 2003;63:7185–7189. [PubMed] [Google Scholar]
- 47.Toyota M, Sasaki Y, Satoh A, Ogi K, Kikuchi T, Suzuki H, Mita H, Tanaka N, Itoh F, Issa JPJ. Epigenetic inactivation of CHFR in human tumors. Proc. Natl Acad. Sci. USA. 2003;100:7818–7823. doi: 10.1073/pnas.1337066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 49.Oberoi J, Richards MW, Crumpler S, Brown N, Blagg J, Bayliss R. Structural basis of poly(ADP-ribose) recognition by the multizinc binding domain of checkpoint with forkhead-associated and RING Domains (CHFR) J. Biol. Chem. 2010;285:39348–39358. doi: 10.1074/jbc.M110.159855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Isogai S, Kanno S, Ariyoshi M, Tochio H, Ito Y, Yasui A, Shirakawa M. Solution structure of a zinc-finger domain that binds to poly-ADP-ribose. Genes Cells. 2010;15:101–110. doi: 10.1111/j.1365-2443.2009.01369.x. [DOI] [PubMed] [Google Scholar]
- 51.Yu X, Minter-Dykhouse K, Malureanu L, Zhao WM, Zhang D, Merkle CJ, Ward IM, Saya H, Fang G, Van Deursen J. Chfr is required for tumor suppression and Aurora A regulation. Nat. Genet. 2005;37:401–406. doi: 10.1038/ng1538. [DOI] [PubMed] [Google Scholar]
- 52.Ogata N, Ueda K, Kawaichi M, Hayaishi O. Poly (ADP-ribose) synthetase, a main acceptor of poly (ADP-ribose) in isolated nuclei. J. Biol. Chem. 1981;256:4135–4137. [PubMed] [Google Scholar]
- 53.Haince JF, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, Poirier GG. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly (ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J. Biol. Chem. 2007;282:16441–16453. doi: 10.1074/jbc.M608406200. [DOI] [PubMed] [Google Scholar]
- 54.Madison DL, Stauffer D, Lundblad JR. The PARP inhibitor PJ34 causes a PARP1-independent, p21 dependent mitotic arrest. DNA Repair. 2011;10:1003–1013. doi: 10.1016/j.dnarep.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amé JC, Fouquerel E, Gauthier LR, Biard D, Boussin FD, Dantzer F, de Murcia G, Schreiber V. Radiation-induced mitotic catastrophe in PARG-deficient cells. J. Cell Sci. 2009;122:1990–2002. doi: 10.1242/jcs.039115. [DOI] [PubMed] [Google Scholar]
- 57.Cortes U, Tong WM, Coyle DL, Meyer-Ficca ML, Meyer RG, Petrilli V, Herceg Z, Jacobson EL, Jacobson MK, Wang ZQ. Depletion of the 110-kilodalton isoform of poly (ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol. Cell. Biol. 2004;24:7163–7178. doi: 10.1128/MCB.24.16.7163-7178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, Stöger T, Poirier GG, Dawson VL, Dawson TM. Failure to degrade poly (ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl Acad. Sci. USA. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Y, Feng X, Koh DW. Enhanced DNA accessibility and increased DNA damage induced by the absence of poly (ADP-ribose) hydrolysis. Biochemistry. 2010;49:7360–7366. doi: 10.1021/bi100979j. [DOI] [PubMed] [Google Scholar]
- 60.Zhou Y, Feng X, Koh DW. Activation of cell death mediated by apoptosis-inducing factor due to the absence of poly (ADP-ribose) glycohydrolase. Biochemistry. 2011;50:2850–2859. doi: 10.1021/bi101829r. [DOI] [PubMed] [Google Scholar]
- 61.Huletsky A, De Murcia G, Muller S, Hengartner M, Menard L, Lamarre D, Poirier G. The effect of poly (ADP-ribosyl) ation on native and H1-depleted chromatin. A role of poly (ADP-ribosyl) ation on core nucleosome structure. J. Biol. Chem. 1989;264:8878–8886. [PubMed] [Google Scholar]
- 62.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly (ADP-ribosyl) ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 63.Monaco L, Kolthur-Seetharam U, Loury R, Murcia JM, De Murcia G, Sassone-Corsi P. Inhibition of Aurora-B kinase activity by poly (ADP-ribosyl) ation in response to DNA damage. Proc. Natl Acad. Sci. USA. 2005;102:14244–14248. doi: 10.1073/pnas.0506252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poirier GG, De Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly (ADP-ribosyl) ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl Acad. Sci. USA. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly (ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruscetti T, Lehnert BE, Halbrook J, Le Trong H, Hoekstra MF, Chen DJ, Peterson SR. Stimulation of the DNA-dependent protein kinase by poly (ADP-ribose) polymerase. J. Biol. Chem. 1998;273:14461–14467. doi: 10.1074/jbc.273.23.14461. [DOI] [PubMed] [Google Scholar]
- 67.Mortusewicz O, Amé JC, Schreiber V, Leonhardt H. Feedback-regulated poly (ADP-ribosyl) ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haince JF, McDonald D, Rodrigue A, Déry U, Masson JY, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 69.Ahel D, Hořejší Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP. Poly (ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325: 1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EHK, Scheffzek K. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 71.Lukas J, Lukas C, Bartek J. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 72.Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl Acad. Sci. USA. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leist M, Single B, Castoldi AF, Kühnle S, Nicotera P. Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis. J. Exp. Med. 1997;185:1481–1486. doi: 10.1084/jem.185.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eguchi Y, Shimizu S, Tsujimoto Y. Intracellular ATP levels determine cell death fate by apoptosis or necrosis. Cancer Res. 1997;57:1835–1840. [PubMed] [Google Scholar]
- 75.Ha HC, Snyder SH. Poly (ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc. Natl Acad. Sci. USA. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Herceg Z, Wang ZQ. Functions of poly (ADP-ribose) polymerase (PARP) in DNA repair, genomic integrity and cell death. Mutat. Res. 2001;477:97–110. doi: 10.1016/s0027-5107(01)00111-7. [DOI] [PubMed] [Google Scholar]
- 77.Ueda K, Oka J, Narumiya S, Miyakawa N, Hayaishi O. Poly ADP-ribose glycohydrolase from rat liver nuclei, a novel enzyme degrading the polymer. Biochem. Biophys. Res. Commun. 1972;46:516–523. doi: 10.1016/s0006-291x(72)80169-4. [DOI] [PubMed] [Google Scholar]
- 78.Woodhouse BC, Dianov GL. Poly ADP-ribose polymerase-1: an international molecule of mystery. DNA Repair. 2008;7:1077–1086. doi: 10.1016/j.dnarep.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 79.De Murcia G, Huletsky A, Lamarre D, Gaudreau A, Pouyet J, Daune M, Poirier G. Modulation of chromatin superstructure induced by poly (ADP-ribose) synthesis and degradation. J. Biol. Chem. 1986;261:7011–7017. [PubMed] [Google Scholar]
- 80.Kashima L, Idogawa M, Mita H, Shitashige M, Yamada T, Ogi K, Suzuki H, Toyota M, Ariga H, Sasaki Y. CHFR regulates the mitotic checkpoint by targeting PARP-1 for ubiquitination and degradation. J. Biol. Chem. 2012;287:12975–12984. doi: 10.1074/jbc.M111.321828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.